Abstract
The circadian clock regulates numerous biological processes in plants, especially development and stress responses. CIRCADIAN CLOCK-ASSOCIATED 1 (CCA1) is one of the core components of the day–night rhythm response and is reportedly associated with ambient temperature in Arabidopsis thaliana. However, it remains unknown if alternative splicing of ZmCCA1 is modulated by external stress in maize, such as drought stress and photoperiod. Here, we identified three ZmCCA1 splice variants in the tropical maize line CML288, which are predicted to encode three different protein isoforms, i.e., ZmCCA1.1, ZmCCA1.2, and ZmCCA1.3, which all retain the MYB domain. In maize, the expression levels of ZmCCA1 splice variants were influenced by photoperiod, tissue type, and drought stress. In transgenic A. thaliana, ZmCCA1.1 may be more effective than ZmCCA1.3 in increasing drought tolerance while ZmCCA1.2 may have only a small effect on tolerance to drought stress. Additionally, although CCA1 genes have been found in many plant species, alternative CCA1 splicing events are known to occur in species-specific ways. Our study provides new sight to explore the function of ZmCCA1 splice variants’ response to abiotic stress, and clarify the linkage between circadian clock and environmental stress in maize.
Introduction
Subtropical and tropical maize lines have been used as germplasm to improve maize quality and yield compared with temperate maize lines due to their abundant genetic variations. However, photoperiod sensitivity in maize restricts the utilization of subtropical and tropical germplasms under long day (LD) conditions because of a delayed floral transition [1, 2]. The circadian clock is involved in photoperiod-mediated flowering and accurately perceives external input signals to generate endogenous rhythmic outputs during an approximate 24 h cycle, which can be synchronized with the environment by regulating key basic metabolic processes including photosynthesis, hypocotyl elongation, and floral transition [3]. Moreover, the clock can also regulate the stomatal aperture, rhythmic leaf movement, and roots and stem circumnutating in plants [4, 5].
In recent years, the transcriptional regulation and molecular functions of circadian clock genes have been studied in Arabidopsis thaliana, including CCA1 [6], LATE ELONGATED HYPOCOTYL (LHY) [7], TIMING OF CAB EXPRESSION 1 (TOC1), and GIGANTEA (GI). Feedback loops mediated by CCA1, LHY, and TOC1 constitute one of the circadian clock regulatory models whereby CCA1 and LHY can bind directly to the TOC1 promoter as the transcriptional repressors that suppress TOC1 accumulation. Conversely, TOC1 represses the expression of CCA1 and LHY through directly binding their promoters [8]. However, the molecular mechanisms of the circadian clock gene in maize are yet to be reported; therefore, cloning of these associated genes will provide a theoretical foundation for understanding the maize circadian clock.
Alternative splicing (AS) events are always associated with growth, signal transduction, circadian rhythms, and abiotic stresses in plants [9]. In moss, 1779 AS events identified by genome-wide analysis and accounting for nearly half of the expressed genes showed significant responses to high-temperature stress [10]. Additionally, approximately 7500 genes identified in tomato pollen demonstrated heat-dependent accumulation of intron retention and exon skipping, including six heat-shock factors and 29 heat-shock proteins that play important roles in plant responses to heat stress [11]. Furthermore, AS phenomena of some genes related to the A. thaliana circadian clock are reportedly involved in defense responses [12]. For example, the SNW/Ski-interacting protein gene, coding a SNW/Ski-interacting protein component of the spliceosome, is regulated by salt, mannitol, and abscisic acid (ABA) treatment. It serves as a major link between the circadian clock and AS under biotic stress conditions [13]. Moreover, two splice variants of CCA1, CCA1α and CCA1β, were shown to be modulated by low temperature in A. thaliana [14]. However, only limited studies are available regarding the function of maize ZmCCA1 splice variants under other environmental stresses, such as drought.
In this study, we cloned ZmCCA1 from the tropical maize line CML288 using homologous cloning and the 3′RACE technique and identified three splice variants encoding three protein isoforms. We systematically investigated the expression of ZmCCA1 splice variants in leaves and leaf sheaths of CML288 plants at five fully expanded-leaf stage under various environmental conditions. Compared with wild-type (WT) plants, transgenic A. thaliana plants overexpressing ZmCCA1.1 and ZmCCA1.3 obviously showed higher tolerance to drought stress while transgenic A. thaliana plants overexpressing ZmCCA1.2 exhibited slightly enhanced drought tolerance. Cross-species examination suggests that AS events of CCA1 are present in numerous species, but occur with species specificity. Our data will help clarify the linkage between circadian clock and environmental stress in maize. Moreover, the AS of ZmCCA1 should play more complex roles than previously expected.
Materials and methods
The maize inbred line CML288 used in this study was collected from the International Maize and Wheat Improvement Center (CIMMYT) in Mexico. All A. thaliana were Columbia-0 (Col-0) unless otherwise specified.
Total RNA of leaves was isolated from CML288 five-leaf seedlings in a controlled culture room under LD conditions (16-h light/8-h dark) at 28°C with the TRIzol reagent (Invitrogen, Carlsbad, CA, USA). For 3′RACE, the first-strand cDNA was synthesized via the 3′-Full RACE Core Set kit (v. 2.0; Takara Bio, Dalian, China). For homologous cloning, first-strand cDNA was synthesized via the PrimeScript 1st Strand cDNA Synthesis Kit (Takara Bio). Based on the cDNA sequence of ZmCCA1 (GenBank number: EU954568.1), 3′RACE sequencing results, and gDNA sequence (GenBank number: AC215881.5), the primers were designed using Primer Premier 6 software (S1 Table).
The sequence analysis and multiple sequence alignments were analyzed by DNAMAN (v. 9.0; Lynnon Biosoft, San Ramon, CA, USA). The open reading frame (ORF) was acquired by the ORF Finder tool (https://www.ncbi.nlm.nih.gov/orffinder/). Molecular weight and isoelectric point (pI) were predicted by ExPASy (http://www.expasy.org/). Domain prediction was performed by Conserved Domain Search Service (CD Search) and SMART software (http://smart.embl-heidelberg.de/). The phylogenetic tree was constructed by MEGA 6.0 software (http://www.megasoftware.net/) using the maximum-likelihood method.
To produce transgenic A. thaliana lines with overexpression of ZmCCA1.1, ZmCCA1.2, and ZmCCA1.3, the entire ORF of these splice variants were subcloned into the pCAMBIA1304 vector under control of the CaMV 35S promoter, respectively. Agrobacterium tumefaciens-mediated A. thaliana transformation was performed according to a modified floral dip method [15]. Homozygotic lines were obtained by hygromycin selection and analysis of segregation ratios, and T4 homozygotic seeds were used for the stress treatment.
CML288 seeds were grown in a controlled culture room at 28°C under short day (SD) conditions (8-h light/6-h dark) or LD conditions (16-h light/8-h dark). At the five fully expanded leaf stage, the leaves and leaf sheaths of the fifth leaf obtained from the same seedling were harvested over a 24-h period (ZT0–ZT8), starting at Zeitgeber time 0 (ZT0) with 3 h intervals between harvests, then immediately stored at −80°C until use. Three biological replicates were performed in this experiment, and three seedlings were mixed per biological replicate.
CML288 seeds were grown in vermiculite in a controlled culture room under SD conditions (28°C 8-h light/22°C 16-h dark). Then the grown uniform seedlings at two fully expanded leaf stage were then transferred to the pots with 2-L full-strength Hoagland’s nutrient solution. At the five fully expanded leaf stage, seedlings were treated with 20% (w/v) polyethylene glycol (PEG) 6000 before light irradiation under 24-h light conditions [16, 17]. Leaves (the fifth leaf) and their leaf sheaths were harvested every 3 h after 20% PEG-6000 treatment over a 24-h period (ZT0–ZT8), starting at ZT0, and immediately stored at −80°C until use. Three biological replicates were collected at each time point. Each tissue sample contained three different randomly uniform selected plants.
Relative water contents (RWCs) were measured under drought stress conditions compared to that under normal conditions according to our previous report [17]. Two days after drought stress treatment, the RWC was calculated using the following formula: RWC (%) = [(FM − DM)/(TM − DM)] × 100, where FM, DM, and TM refer to the fresh, dry, and turgid masses of the tissue, respectively [17].
As described in previous reports with minor modification [18, 19], the seeds of WT A. thaliana and transgenic T4 lines were surface sterilized with 5% NaClO for 5 min and washed five times with sterile distilled water. Approximately 50 seeds were placed on 1/2 MS medium with 0, 50, 150, 250 mmol/L mannitol with three replicates and incubated at 4°C in the dark for 2 days. Then the medium plates were then vertically cultured at 20°C for 2 weeks under LD conditions (16-h light/8-h dark). Germination (emergence of radicle) was calculated daily for 7 days after transferring to LDs. The length of the primary root was investigated after 2 weeks.
Total RNAs were extracted using TRIzol reagent according to the manufacturer’s instructions (Invitrogen), and first-strand cDNA was prepared by using HiScript Q RT SuperMix for qPCR with gDNA Eraser as per the manufacturer’s instructions (Vazyme, Nanjing, China). Quantitative real-time RT-PCR (qRT-PCR) was conducted on a Bio-Rad iQ5 Real-Time PCR System (Bio Rad Laboratories, Richmond, CA, USA). Each PCR reaction mix consisted of 1.0 μL diluted cDNA, 0.5 μL forward/reverse primers (10 μM), 12.5 μL 2 × Taq Plus Master Mix (Vazyme), 1.5 μL 10 × SYBR Green I (Invitrogen), and 9.0 μL RNase-free water in a total volume of 25 μL. Relative expression levels of the target genes were analyzed using the 2−ΔΔCt method [20]. The 18s rRNA gene was used as an endogenous reference, and all experiments were performed using three biological replicates.
Results
Cloning and sequence analysis of ZmCCA1
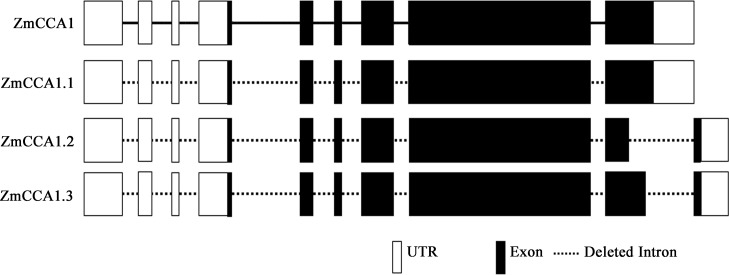
In our study, we amplified three PCR products by homologous cloning and the 3′RACE technique. Sequence analysis revealed that ZmCCA1 was located on chromosome 10 and had three splice variants, i.e., named ZmCCA1.1, ZmCCA1.2, and ZmCCA1.3. ZmCCA1.1 consisted of a 2157 bp ORF encoding a protein of 718 amino acids with a molecular weight of 78.2 kDa, a pI of 6.08, more than 147 bp 5′untranslated region (UTR), and 282 bp 3′UTR, whereas ZmCCA1.2 and ZmCCA1.3 were found to encode proteins of 676 and 709 amino acids with 249 and 87 bp 3′UTR, respectively. Notably, the latter two splice variants had the same length of 5′UTR (>629 bp) (Fig 1, S1–S3 Figs). These results indicated that three splice variants had different structural characteristics.
Fig 1. Characterization of three ZmCCA1 splice variants.
Diagrams showing the predicated genomic structures of ZmCCA1 and three ZmCCA1 splice variants. White boxes are 3′ and 5′ UTRs. Black boxes depict exons. The deleted introns in three transcripts were represented by dash lines. The introns in ZmCCA1 genome sequence were indicated using solid lines.
ZmCCA1 splice variants are generated by alternative 5′ splice sites, alternative poly (A), and a combination of the above two or more AS types (Fig 1, S2 Fig). We found ZmCCA1.1 encoded authentic proteins, while ZmCCA1.2 and ZmCCA1.3 both encoded truncated proteins. ZmCCA1.1, ZmCCA1.2, and ZmCCA1.3 shared 99.7%, 92.3%, and 97.8% identity with ZmCCA1, respectively. Moreover, we also showed that the N-terminus of the three ZmCCA1 protein isoforms was highly conserved while the C-terminus differed, indicating that individual splice variant potentially possess distinctive functions (S3 Fig).
Expression of the three ZmCCA1 splice variants was affected by photoperiod and tissue type
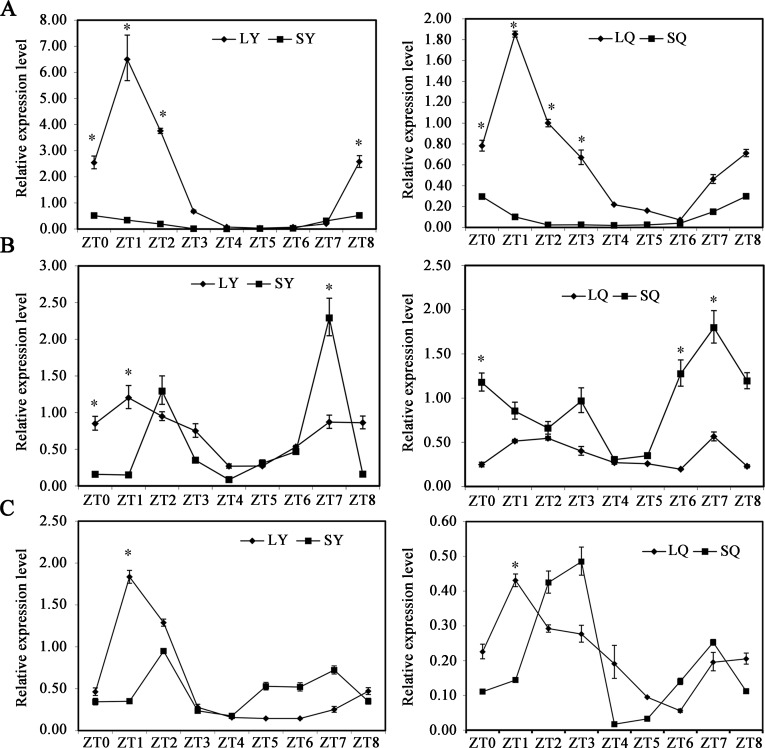
The expression level of ZmCCA1 is affected by photoperiod [21]. Moreover, CML288 is much sensitive to night break (NB) treatment under SDs at the five-leaf stage [22, 23]. In this study, we quantified the expression levels of three ZmCCA1 splice variants in leaves and leaf sheaths at the five fully expanded leaf stage under SD and LD conditions. ZmCCA1.1 transcript levels in leaves and leaf sheaths showed a rhythmic pattern under SDs and LDs and peaked at ZT0 under SDs and at ZT1 under LDs (Fig 2A). ZmCCA1.2 was also expressed rhythmically in leaves under LDs and in leaf sheaths under SDs, respectively peaking at ZT1 and ZT7; transcript levels of ZmCCA1.2 in leaves under SDs or in leaf sheaths under LDs, however, peaked at both ZT2 and ZT7 (Fig 2B). ZmCCA1.3 was expressed rhythmically in leaves and leaf sheaths under LDs, peaking at ZT1. Under SDs, however, transcript levels of ZmCCA1.3 in leaves and leaf sheaths presented two peaks: the former peaking at ZT2 and ZT7 and the latter at ZT3 and ZT7 (Fig 2C). Thus, these results indicate that AS of ZmCCA1 is affected by photoperiod and tissue type.
Fig 2. Effects of photoperiod and tissue types on the alternative splicing of ZmCCA1 in maize.
The plant seedlings at five fully-expanded leaf stage on either LDs or SDs were harvested at the indicated ZT points of the extraction of total RNA samples. Relative expression level of ZmCCA1.1 (A), ZmCCA1.2 (B), and ZmCCA1.3 (C) in leaves and leaf sheaths were determined by qRT-PCR. At ZT2, the expression of plant seedlings in the leaf sheaths under LD condition, the leaves under LD condition, and the leaves under SD condition were set to 1 in A, B, and C, respectively. LD, long day conditions; SD, short day conditions; Q, leaf sheaths; Y, leave; ZT, Zeitgeber time. Biological triplicates were averaged. Bars indicate the standard error of the mean. Significant differences between normal and drought treatment condition were assessed using Student’s t-test; * P < 0.05.
ZmCCA1 transcript levels in leaves and leaf sheaths showed a rhythmic pattern under SDs and LDs and peaked at ZT0 under SDs and at ZT1 under LDs (S4A Fig). At the corresponding peak expression of ZmCCA1 in leaves and leaf sheaths under SDs or LDs, we compared the expression levels of three ZmCCA1 splice variants. The results showed that expression levels of ZmCCA1.1 in leaves and leaf sheaths were significantly higher under SDs or LDs, followed by ZmCCA1.2 and ZmCCA1.3, respectively. Moreover, expression levels of the three ZmCCA1 splice variants in leaves were higher than those in leaf sheaths under SDs or LDs except for ZmCCA1.2 under SDs (S4B and S4C Fig).
Expression of three ZmCCA1 splice variants under drought stress in maize
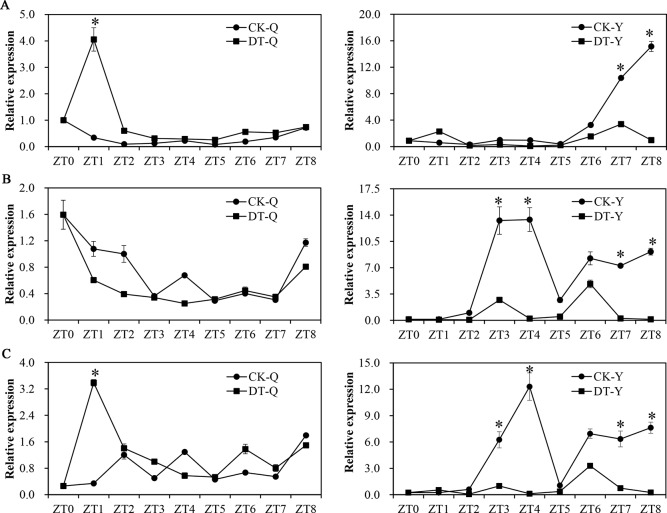
To verify the linkage between AS of ZmCCA1 and environmental stress responses in maize, CML288 seedlings were treated with 20% PEG-6000 for 1 day at the five fully expanded leaf stage. The RWC of maize leaves grown under normal condition (85.7%) was significantly higher than that under drought stress (57.3%) leaves after 1 day of drought stress (P < 0.01) (S5 Fig). After 20% PEG-6000 treatment, compared with the untreated control, expression levels of ZmCCA1.1 first increased in the leaves and leaf sheaths at ZT1, and then the expression in leaf sheath remained continually higher under drought stress; however, in leaves, ZmCCA1.1 showed relatively higher expression level under normal conditions. The expression of ZmCCA1.3 in leaves first increased, then maintained lower levels; however, the expression of ZmCCA1.3 in leaf sheaths first increased, then decreased slowly. Interestingly, transcript levels of ZmCCA1.2 in leaves and leaf sheaths were always lower under drought than that in untreated seedlings (Fig 3). Together, these results indicate that AS of ZmCCA1 is suppressed under drought stress and that ZmCCA1.1 and ZmCCA1.3 are both involved in drought stress regulation of maize, with ZmCCA1.1 potentially playing a major role. ZmCCA1.2 does not appear to be involved in the drought stress response of maize.
Fig 3. Effects of drought stress on the alternative splicing of ZmCCA1 in maize.
Expression level of ZmCCA1.1 (A), ZmCCA1.2 (B), and ZmCCA1.3 (C) in leaves and leaf sheaths at the five-fully expanded leaf stage after 20% PEG-6000 treatment under continuous light conditions were determined by qRT-PCR. The leaf sheath of seedlings under normal growth condition at ZT0 was considered as 1. Samples were harvested, starting at ZT0 with 3 h intervals for 1 day. CK, seedling grown under normal condition; DT, seedlings grown under 20% PEG-6000 treatment; Q, leaf sheaths; Y, leaves; ZT, Zeitgeber time. Biological triplicates were averaged. Bars indicate the standard error of the mean. Significant differences between normal and drought treatment condition were assessed using Student’s t-test; * P < 0.05.
Overexpression of ZmCCA1.1 and ZmCCA1.3 enhanced tolerance towards drought stress in transgenic A. thaliana
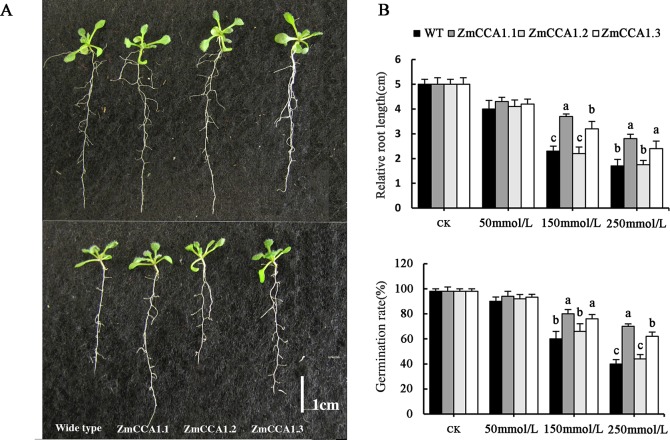
Numerous MYB transcription factors (TFs) have been found to function in cell development and cycling, hormone synthesis, primary and secondary metabolism, and various biotic and abiotic stresses [24–27]. To further determine whether overexpression of ZmCCA1 splice variants affected tolerance to drought stress in transgenic Arabidopsis, we first generated transgenic A. thaliana plants with overexpression of ZmCCA1.1, ZmCCA1.2, and ZmCCA1.3, which were expressed 15 times higher than in WTs (S6 Fig). Under 50 mmol/L mannitol, the relative primary root lengths of 35S:ZmCCA1.1, 35S:ZmCCA1.2, and 35S:ZmCCA1.3 were 12.5%, 2.5%, and 7.5% longer than that of WT plants, respectively, and relative germination rates increased by 3%, 1.6%, and 1.9%, respectively. Under 150 mmol/L mannitol, compared with WT, relative root lengths of transgenic plants were elevated by 39.2%, 7.1%, and 32.1%, respectively, and relative germination rates were 20%, 5%, and 16% higher, respectively. We also found significantly more fibrous roots in ZmCCA1.1 and ZmCCA1.3 transgenic plants than in WT plants under 150 mmol/L mannitol (Fig 4A). Finally, under 250 mmol/L mannitol, relative root lengths were 52.3%, 9.5%, and 33.3% longer than WT, respectively, and relative germination rates were 31%, 5%, and 23% higher, respectively. However, under normal growth conditions, root lengths and germination rates of transgenic A. thaliana were all consistent with WT (Fig 4B) while the growth of WT A. thaliana was severely affected under 250 mmol/L mannitol. These results suggest that ZmCCA1.1 may be more effective than ZmCCA1.3 in improving the tolerance of transgenic A. thaliana plants to drought stress, especially under severe drought conditions, while ZmCCA1.2 may have only a limited effect on increasing tolerance to drought stress in transgenic A. thaliana.
Fig 4. Phenotype analysis of transgenic A. thaliana lines under drought stress for 2 weeks under LDs.
(A) Representative images of 14-day-old transgenic A. thaliana overexpression lines and wild-type (WT) seedlings under normal (upper panel) and 150 mmol/L mannitol treatment condition (lower panel). (B) Relative root lengths and germination rates of WT and transgenic A. thaliana overexpression lines under 0–250 mmol/L mannitol. Vertical bars represent standard deviations from the mean. CK, seedlings grown under normal condition. Scale bars in (A) represent 0.5 cm. The different letters (a, b, and c) represent significant differences by ANOVA (P < 0.05).
Evolutionary analysis of CCA1 genes and their AS events
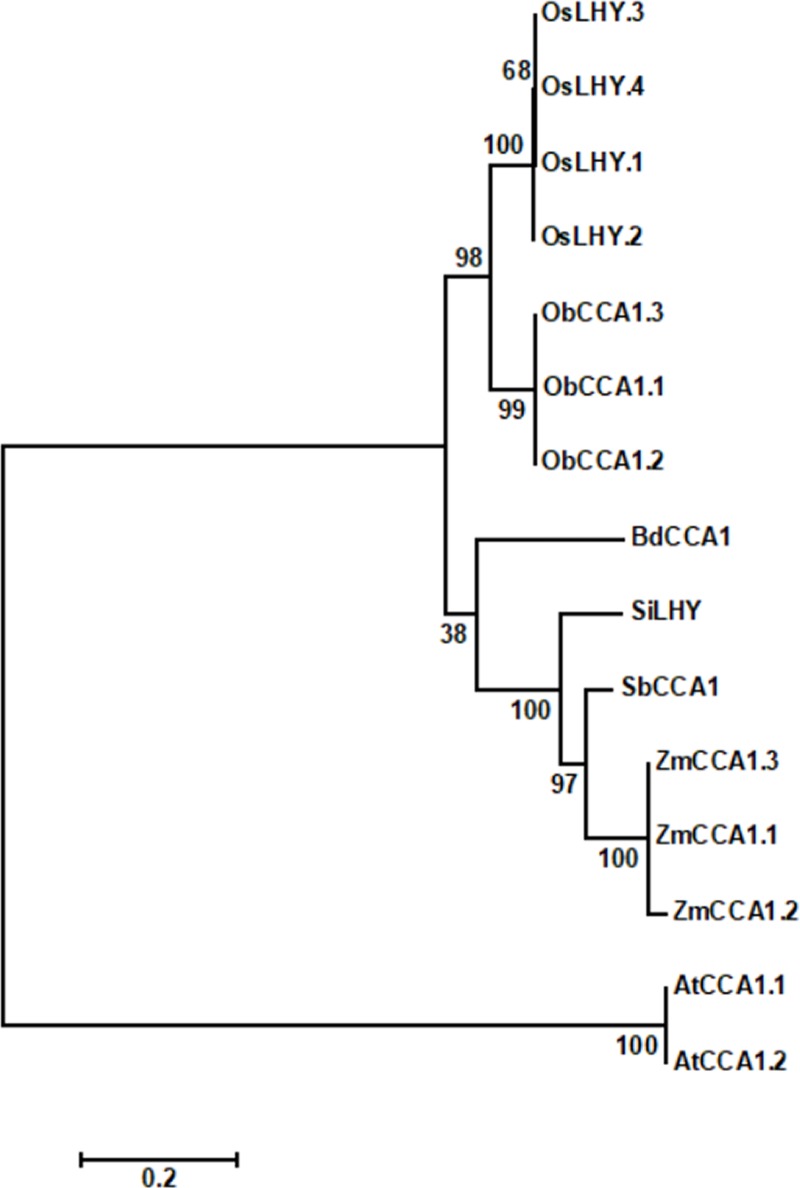
To determine the evolutionary relationship of ZmCCA1 and those from other species, a phylogenetic tree was constructed using overall protein sequences (Fig 5). ZmCCA1 splice variants and ZmCCA1 were located in the same subfamily. Compared with A. thaliana, a dicotyledonous species, CCA1 genes were more homologous among the monocotyledon species, especially in C4 plants.
Fig 5. Phylogenetic analysis of CML288 ZmCCA1 splice variants and related genes.
Protein sequences were aligned by DNAMAN version 9.0. The phylogenetic tree was constructed using the maximum-likelihood method with full-length protein sequences. Species names and accession numbers are indicated: Zea mays, ZmCCA1 (ACG26686); A. thaliana, AtCCA1.1 (NP_850460), AtCCA1.2 (NP_001318437); Sorghum bicolor, SbCCA1 (LOC8080836); Setaria italica, SiLHY (LOC101764638); Oryza sativa Japonica Group, OsLHY.1 (XP_015649835), OsLHY.2 (XP_015649844), OsLHY.3 (XP_015649846), OsLHY.4 (XP_015649847); Oryza brachyantha, ObCCA1.1 (XP_006659138), ObCCA1.2 (XP_006659145), ObCCA1.3 (XP_015695906); Brachypodium distachyon, BdCCA1 (LOC100838310). Scale bar indicates 0.2 of amino acid substitutions per site.
We provided experimental evidence to support the occurrence of three ZmCCA1 splice variants on chromosome 10 in maize (Fig 5). AtCCA1 and OsCCA1 have also been consistently found to undergo AS [28, 29]. To explore whether AS events of CCA1 genes are conserved across species, we identified CCA1 homolog genes in several well-annotated genomes (Table 1). We found that there is only one homologous gene in numerous species, producing three splice variants encoding three protein isoforms on chromosome 10 in Zea mays, 21 splice variants in Sorghum bicolor, 7 splice variants in Setaria italica, 12 splice variants in Oryza sativa Japonica Group, 9 splice variants in Oryza brachyantha, 4 splice variants in Brachypodium distachyon, and 3 splice variants in Arabidopsis, respectively encoding 1, 1, 4, 3, 1, and 2 protein isoforms. Collectively, these results suggest that AS events of CCA1 are present in numerous species, but occur with species specificity.
Table 1. List of CCA1 homologous genes and their predicted splice variants in plants.
| Organism | Annotated gene number in each species | Chromosome | Locus ID | Number of AS for each gene | Number of protein isoforms |
|---|---|---|---|---|---|
| Zea mays | 1 | 10 | GRMZM2G474769 | 3 | 3 |
| Sorghum bicolor | 1 | 7 | LOC8080836 | 21 | 1 |
| Setaria italica | 1 | 6 | LOC101764638 | 7 | 1 |
| Oryza sativa Japonica Group | 1 | 8 | LOC4344703 | 12 | 4 |
| Oryza brachyantha | 1 | 8 | LOC102717637 | 9 | 3 |
| Brachypodium distachyon | 1 | 3 | LOC100838310 | 4 | 1 |
| Arabidopsis thaliana | 1 | 2 | AT2G46830 | 3 | 2 |
Predicted splice variants were collected from the National Center for Biotechnology Information (NCBI; https://www.ncbi.nlm.nih.gov/; NCBI B73-v4 annotation release 101), the Gramene (http://www.gramene.org/), the Maize Genetics and Genomics Database (MaizeGDB; https://www.maizegdb.org/; Zm-B73-REFERENCE-GRAMENE-4.0), the National Center for Biotechnology Information (NCBI; https://www.ncbi.nlm.nih.gov/), and the A. thaliana Information Resource (TAIR, http://www.arabidopsis.org/).
Discussion
Alternative splicing (AS) is a widespread phenomenon in higher eukaryotes. In Arabidopsis, at least 61% of intron-containing genes are AS [30]. In O. sativa, 33% of all rice genes undergo AS [31]. Approximatively, 55.3% of maize genes may be subjected to AS [32]. In this study, we confirmed by homologous cloning and the 3′RACE technique that ZmCCA1 undergoes extensive AS, which occurred at alternative 5′ splice site, alternative poly (A), or by a combination of two or more AS types, encoding three protein isoforms, i.e., ZmCCA1.1, ZmCCA1.2, and ZmCCA1.3. AS isoforms were found to be conserved between one maize homolog and its sorghum ortholog in our study, but absent from the second maize homolog. These results may support the published predication that AS isoforms may have been lost after the maize whole genome duplication event [33].
There are two hypotheses about the function of splice variants: i.e., the “noise” hypothesis, and the “regulated unproductive splicing and translation (RUST)” hypothesis. Based on the former hypothesis, NMD-sensitive RNA variants are generated due to splicing error and are finally removed through the NMD pathway [34–36]. However, the later hypothesized that alternative mRNA splicing coupled with NMD is also known to dynamically monitor the abundance of full-size RNA splicing, particularly during various forms of cellular stress [37–39]. Interesting, recent reports have provided an overview that TF genes can generate protein isoforms without specific functional domains by AS, which competitively inhibit the authentic TFs by forming nonfunctional heterodimers [14, 40–42]. In this study, we confirmed that AS of ZmCCA1 is influenced by tissue type, photoperiod, and drought stress in maize. By constructing the overexpression vector of these three variants, we found that 35S:ZmCCA1.1 and 35S:ZmCCA1.3 (Col-0) transgenic plants all showed significantly higher drought tolerance whereas 35S:ZmCCA1.2 (Col-0) exhibited only slightly higher drought tolerance, indicating that these three splice variants were identified to perform different tolerance abilities in response to drought, and further support that the RUST hypothesis potentially fits well into the AS of ZmCCA1.
In addition, AS events of CCA1 were identified across various species but occurred with species specificity, which helps to further explore the AS function of circadian-associated genes in other plant species. Moreover, AS could be affected by tissue types, photoperiod, and abiotic stress [16, 33, 43–46]. Given that in maize hybrids, CCA1 proteins target thousands of output genes early in the morning, as if the hybrids wake up early to promote photosynthesis, starch metabolism, and biomass accumulation [37–39]; therefore, AS of ZmCCA1 should play more complex roles than previously expected.
In conclusion, three ZmCCA1 splice variants, i.e., ZmCCA1.1, ZmCCA1.2, and ZmCCA1.3, were cloned and obtained a highly conserved N-termini and different C-termini, which were influenced by photoperiod, tissue type, and drought stress in maize. Overexpression of these three ZmCCA1 variants resulted in increased tolerance to drought stress. ZmCCA1.1 may be more effective than ZmCCA1.3 with higher drought tolerance, whereas ZmCCA1.2 performed with a relatively limited effect on increasing tolerance to drought stress. Additionally, AS events of CCA1 genes may occur with species specificity. Thus, this study lays a basic foundation to understand the function of ZmCCA1 in drought resistance mediated by AS in maize, and further provides new evidence to link the circadian clock, AS, and abiotic stress.
Supporting information
(XLSX)
A large fragment insertion was indicated by “X”.
(TIF)
(TIF)
The MYB domain is marked by a black line.
(TIF)
(A) Relative expression of ZmCCA1 in leaves and leaf sheaths at the five fully expanded leaf stage under LDs and SDs. Significant differences between leaf sheaths and leaves were assessed using Student’s t-test; * P < 0.05. (B, C) Relative expression of three ZmCCA1 splice variants in leaves and leaf sheaths at the five fully expanded leaf stage at the corresponding peak expression of ZmCCA1 under LDs (B) and SDs (C). L, LDs; S, SDs; Q, leaf sheaths; Y, leaves. Vertical bars represent standard deviations from the mean.
(TIF)
RWC was measured with detached leaves from seedlings. Significant differences between normal and drought treatments condition were assessed using Student’s t-test; * P < 0.05.
(TIF)
Significant differences between wild type and three splice variants were assessed using Student’s t-test; ** P < 0.01. WT, wild type.
(TIF)
Acknowledgments
The authors thank everyone who provided the materials included in this study. Additionally, we thank Sarah Williams, PhD, from Liwen Bianji, Edanz Group China (www.liwenbianji.cn), for critically reviewing and revising the manuscript and Emma Tacken, PhD, for editing the English text of a draft of this manuscript.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was funded by the Major Science and Technology Projects of Henan Province (grant no. 161100110500 to Yanhui Chen). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Muchow RC, Carberry PS: Environmental control of phenology and leaf growth in a tropically adapted maize. Field Crops Research 1989, 20(3):221–236. [Google Scholar]
- 2.Wang CL, Chen YH, Lixia K, Wang TG, Sun ZH, Cheng FF, Wu LC: Mapping QTL associated with photoperiod sensitivity and assessing the importance of QTL × environment interaction for flowering time in maize. Plos One 2010, 5(11):e14068 10.1371/journal.pone.0014068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Niwa Y, Yamashino T, Mizuno T: The circadian clock regulates the photoperiodic response of hypocotyl elongation through a coincidence mechanism in Arabidopsis thaliana. Plant & Cell Physiology 2009, 50(4):838–854. [DOI] [PubMed] [Google Scholar]
- 4.Mcclung CR: Comes a time. Current Opinion in Plant Biology 2008, 11(5):514–520. 10.1016/j.pbi.2008.06.010 [DOI] [PubMed] [Google Scholar]
- 5.PrunedaPaz Jose L, Kay Steve A: An expanding universe of circadian networks in higher plants. Trends in plant science 2010, 15(5):259–265. 10.1016/j.tplants.2010.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang ZY, Tobin EM: Constitutive expression of the CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) gene disrupts circadian rhythms and suppresses its own expression. Cell 1998, 93(7):1207 [DOI] [PubMed] [Google Scholar]
- 7.Schaffer R, Ramsay N, Samach A, Corden S, Putterill J, Carré IA, Coupland G: The late elongated hypocotyl mutation of Arabidopsis disrupts circadian rhythms and the photoperiodic control of flowering. Cell 1998, 93(7):1219–1229. [DOI] [PubMed] [Google Scholar]
- 8.Huang W, Pérezgarcía P, Pokhilko A, Millar AJ, Antoshechkin I, Riechmann JL, Mas P: Mapping the Core of the Arabidopsis Circadian Clock Defines the Network Structure of the Oscillator. Science 2012, 336(6077):75–79. 10.1126/science.1219075 [DOI] [PubMed] [Google Scholar]
- 9.Li C, Zheng L, Zhang J, Lv Y, Liu J, Wang X, Palfalvi G, Wang G, Zhang Y: Characterization and functional analysis of four HYH splicing variants in Arabidopsis hypocotyl elongation. Gene 2017, 619:44–49. 10.1016/j.gene.2017.04.001 [DOI] [PubMed] [Google Scholar]
- 10.Chang CY, Lin WD, Tu SL: Genome-Wide Analysis of Heat-Sensitive Alternative Splicing in Physcomitrella patens. Plant Physiology 2014, 165(2):826 10.1104/pp.113.230540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Keller M, Hu Y, Mesihovic A, Fragkostefanakis S, Schleiff E, Simm S: Alternative splicing in tomato pollen in response to heat stress†. Dna Research 2017, 24(2):205–217. 10.1093/dnares/dsw051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kwon Y-J, Park M-J, Kim S-G, Baldwin IT, Park C-M: Alternative splicing and nonsense-mediated decay of circadian clock genes under environmental stress conditions in Arabidopsis. BMC Plant Biology 2014, 14(1):136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang X, Wu F, Xie Q, Wang H, Wang Y, Yue Y, Gahura O, Ma S, Liu L, Cao Y et al. : SKIP Is a Component of the Spliceosome Linking Alternative Splicing and the Circadian Clock in Arabidopsis. The Plant Cell 2012, 24(8):3278–3295. 10.1105/tpc.112.100081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Park MJ, Seo PJ, Park CM: CCA1 alternative splicing as a way of linking the circadian clock to temperature response in Arabidopsis. Plant Signaling & Behavior 2012, 7(9):1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clough S, Bent AF: Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thal-iana, vol. 16; 1999. [DOI] [PubMed] [Google Scholar]
- 16.Kwon YJ, Park MJ, Kim SG, Baldwin IT, Park CM: Alternative splicing and nonsense-mediated decay of circadian clock genes under environmental stress conditions in Arabidopsis. Bmc Plant Biology 2014, 14(1):136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ku L, Tian L, Su H, Wang C, Wang X, Wu L, Shi Y, Li G, Wang Z, Wang H: Dual functions of the ZmCCT-associated quantitative trait locus in flowering and stress responses under long-day conditions. Bmc Plant Biology 2016, 16(1):239 10.1186/s12870-016-0930-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ying S, Zhang DF, Fu J, Shi YS, Song YC, Wang TY, Li Y: Cloning and characterization of a maize bZIP transcription factor, ZmbZIP72, confers drought and salt tolerance in transgenic Arabidopsis. Planta 2012, 235(2):253–266. 10.1007/s00425-011-1496-7 [DOI] [PubMed] [Google Scholar]
- 19.Yu HQ, Han N, Zhang YY, Tao Y, Chen L, Liu YP, Zhou SF, Fu FL, Li WC: Cloning and characterization of vacuolar H + -pyrophosphatase gene (AnVP1) from Ammopiptanthus nanus and its heterologous expression enhances osmotic tolerance in yeast and Arabidopsis thaliana. Plant Growth Regulation 2017, 81(3):385–397. [Google Scholar]
- 20.Livak KJ, Schmittgen TD: Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods (San Diego, Calif) 2001, 25(4):402–408. [DOI] [PubMed] [Google Scholar]
- 21.Wang X, Wu L, Zhang S, Wu L, Ku L, Wei X, Xie L, Chen Y: Robust expression and association of ZmCCA1 with circadian rhythms in maize. Plant Cell Reports 2011, 30(7):1261–1272. 10.1007/s00299-011-1036-8 [DOI] [PubMed] [Google Scholar]
- 22.Wei X, Ku L, Li S, Zhang S, Wang X, Chen Y: Effects of night break on accumulation of HD6 mRNA in tropical photoperiod-sensitive maize. African Journal of Agricultural Research 2011, 6(21):4871–4878. [Google Scholar]
- 23.Zhao X, Liu H, Wei X, Wu L, Cheng F, Ku L, Zhang ZZ, Han Z, Cao L, Cui X: Promoter region characterization of ZmPhyB2 associated with the photoperiod-dependent floral transition in maize (Zea mays L.). Molecular Breeding 2014, 34(3):1413–1422. [Google Scholar]
- 24.Dubos C, Stracke R, Grotewold E, Weisshaar B, Martin C, Lepiniec L: MYB transcription factors in Arabidopsis. Trends in plant science 2010, 15(10):573–581. 10.1016/j.tplants.2010.06.005 [DOI] [PubMed] [Google Scholar]
- 25.Ambawat S, Sharma P, Yadav NR, Yadav RC: MYB transcription factor genes as regulators for plant responses: an overview. Physiology & Molecular Biology of Plants 2013, 19(3):307–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gao F, Zhou J, Deng RY, Zhao HX, Li CL, Chen H, Suzuki T, Park SU, Wu Q: Overexpression of a tartary buckwheat R2R3-MYB transcription factor gene, FtMYB9, enhances tolerance to drought and salt stresses in transgenic Arabidopsis. Journal of Plant Physiology 2017, 214:81 10.1016/j.jplph.2017.04.007 [DOI] [PubMed] [Google Scholar]
- 27.Chen YH, Cao YY, Wang LJ, Li LM, Yang J, Zou MX: Identification of MYB transcription factor genes and their expression during abiotic stresses in maize. Biologia Plantarum 2017:1–9. [Google Scholar]
- 28.Filichkin SA, Priest HD, Givan SA, Shen R, Bryant DW, Fox SE, Wong WK, Mockler TC: Genome-wide mapping of alternative splicing in Arabidopsis thaliana. Genome Research 2010, 20(1):45–58. 10.1101/gr.093302.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bartok O, Kyriacou CP, Levine J, Sehgal A, Kadener S: Adaptation of molecular circadian clockwork to environmental changes: a role for alternative splicing and miRNAs. Proceedings of the Royal Society B Biological Sciences 2013, 280(1765):20130011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marquez Y, Brown JW, Simpson C, Barta A, Kalyna M: Transcriptome survey reveals increased complexity of the alternative splicing landscape in Arabidopsis. Genome Research 2012, 22(6):1184–1195. 10.1101/gr.134106.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang G, Guo G, Hu X, Yong Z, Li Q, Li R, Zhuang R, Lu Z, He Z, Fang X: Deep RNA sequencing at single base-pair resolution reveals high complexity of the rice transcriptome. Genome Research 2010, 20(5):646–654. 10.1101/gr.100677.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Min XJ: Comprehensive Cataloging and Analysis of Alternative Splicing in Maize. 2017.
- 33.Mei W, Liu S, Schnable JC, Yeh CT, Springer NM, Schnable PS, Barbazuk WB: A Comprehensive Analysis of Alternative Splicing in Paleopolyploid Maize. Frontiers in Plant Science 2017, 8:694 10.3389/fpls.2017.00694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saltzman AL, Kim YK, Pan Q, Fagnani MM, Maquat LE, Blencowe BJ: Regulation of Multiple Core Spliceosomal Proteins by Alternative Splicing-Coupled Nonsense-Mediated mRNA Decay. Molecular & Cellular Biology 2008, 28(13):4320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sureshkumar S, Dent C, Seleznev A, Tasset C, Balasubramanian S: Nonsense-mediated mRNA decay modulates FLM-dependent thermosensory flowering response in Arabidopsis. Nature Plants 2016, 2(5):16055 10.1038/nplants.2016.55 [DOI] [PubMed] [Google Scholar]
- 36.Karousis ED, Nasif S, Mühlemann O: Nonsense-mediated mRNA decay: novel mechanistic insights and biological impact. Wiley Interdisciplinary Reviews Rna 2016, 7(5):661 10.1002/wrna.1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lareau LF, Brooks AN, Soergel DA, Meng Q, Brenner SE: The coupling of alternative splicing and nonsense-mediated mRNA decay. Oxygen Transport to Tissue XXXIII 2007, 623:190–211. [DOI] [PubMed] [Google Scholar]
- 38.Cheng Q, Zhou Y, Liu Z, Zhang L, Song G, Guo Z, Wang W, Qu X, Zhu Y, Yang D: An alternatively spliced heat shock transcription factor, OsHSFA2dI, functions in the heat stress-induced unfolded protein response in rice. Plant Biol 2015, 17(2):419–429. 10.1111/plb.12267 [DOI] [PubMed] [Google Scholar]
- 39.Qin Z, Wu J, Geng S, Nan F, Chen F, Kong X, Song G, Kai C, Li A, Long M: Regulation of FT splicing by an endogenous cue in temperate grasses. Nature Communications 2017, 8:14320 10.1038/ncomms14320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Seo PJ, Kim MJ, Ryu JY, Jeong EY, Park CM: Two splice variants of the IDD14 transcription factor competitively form nonfunctional heterodimers which may regulate starch metabolism. Nature Communications 2011, 2(303):303. [DOI] [PubMed] [Google Scholar]
- 41.Ryu JY, Kim JY, Park CM: Adaptive thermal control of stem gravitropism through alternative RNA splicing in Arabidopsis. Plant Signaling & Behavior 2015, 10(11):-. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Seo PJ, Park MJ, Lim MH, Kim SG, Lee M, Baldwin IT, Park CM: A self-regulatory circuit of CIRCADIAN CLOCK-ASSOCIATED1 underlies the circadian clock regulation of temperature responses in Arabidopsis. Plant Cell 2012, 24(6):2427–2442. 10.1105/tpc.112.098723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li J, Li X, Guo L, Lu F, Feng X, He K, Wei L, Chen Z, Qu LJ, Gu H: A subgroup of MYB transcription factor genes undergoes highly conserved alternative splicing in Arabidopsis and rice. Journal of Experimental Botany 2006, 57(6):1263–1273. 10.1093/jxb/erj094 [DOI] [PubMed] [Google Scholar]
- 44.Hradilová J, Brzobohatý B: Expression pattern of the AHP gene family from Arabidopsis thaliana and organ specific alternative splicing in the AHP5 gene. Biologia Plantarum 2007, 51(2):257–267. [Google Scholar]
- 45.Liu Z, Qin J, Tian X, Xu S, Wang Y, Li H, Wang X, Peng H, Yao Y, Hu Z: Global Profiling of Alternative Splicing Landscape Responsive to Drought, Heat and Their Combination in Wheat (Triticum asetivum L.). Plant Biotechnology Journal 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhu G, Li W, Zhang F, Guo W: RNA-seq analysis reveals alternative splicing under salt stress in cotton, Gossypium davidsonii. Bmc Genomics 2018, 19(1):73 10.1186/s12864-018-4449-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX)
A large fragment insertion was indicated by “X”.
(TIF)
(TIF)
The MYB domain is marked by a black line.
(TIF)
(A) Relative expression of ZmCCA1 in leaves and leaf sheaths at the five fully expanded leaf stage under LDs and SDs. Significant differences between leaf sheaths and leaves were assessed using Student’s t-test; * P < 0.05. (B, C) Relative expression of three ZmCCA1 splice variants in leaves and leaf sheaths at the five fully expanded leaf stage at the corresponding peak expression of ZmCCA1 under LDs (B) and SDs (C). L, LDs; S, SDs; Q, leaf sheaths; Y, leaves. Vertical bars represent standard deviations from the mean.
(TIF)
RWC was measured with detached leaves from seedlings. Significant differences between normal and drought treatments condition were assessed using Student’s t-test; * P < 0.05.
(TIF)
Significant differences between wild type and three splice variants were assessed using Student’s t-test; ** P < 0.01. WT, wild type.
(TIF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.