Abstract
Crop productivity is highly dependent on the application of N fertilizers, but ever-increasing N application is causing serious environmental impacts. To facilitate the development of new wheat cultivars that can thrive in low N growth conditions, key loci and genes associated with wheat responses to low N must be identified. In this GWAS and t-test study of 190 M6 mutant wheat lines (Jing 411-derived) based on genotype data from the wheat 660k SNP array, we identified a total of 221 significant SNPs associated four seedling phenotypic traits that have been implicated in resistance to low N: relative root length, relative shoot length, relative root weight, and relative shoot weight. Notably, we detected large numbers of significantly associated SNP in what appear to be genomic ‘hotspots’ for resistance to low N on chromosomes 2A and 6B, strongly suggesting that these regions are functionally related to the resistance phenotypes that we observed in some of the mutant lines. Moreover, the candidate genes, including genes encoding high-affinity nitrate transporter 2.1, gibberellin responsive protein, were identified for resistance to low N. This study raises plausible mechanistic hypotheses that can be evaluated in future applied or basic efforts by breeders or plant biologists seeking to develop new high-NUE wheat cultivars.
Introduction
Nitrogen (N) is one of the essential elements for plant growth, and the application of N fertilizer to crops results in a dramatically increased yield [1]. However, 50% to 70% of the N fertilizer applied to production fields is not actually utilized by crops; this results in negative impacts to the environment such as the eutrophication of water supplies [2]. The improvement of nitrogen use efficiency (NUE) in crops is therefore of enormous importance [3]. Viewed in this context, the identification of any genetic loci or significant molecular markers associated with resistance to low N will be useful for the improvement of NUE in crops [4].
Wheat (Triticum aestivum L.) is one of the most important food crops worldwide. Genetic mapping of quantitative trait loci (QTLs) for nitrogen-related traits in wheat has been reported in previous studies [5–10]. These studies have identified QTLs for yield under a variety of different nitrogen conditions, including 12 QTLs distributed on chromosomes 1A, 2A, 2B, 2D, 3D, 4B, 5B, 6A, and 7A [8]. Another study of QTLs related to differentially responsive grain yields in response to N application identified QTLs on chromosomes 1A, 1B, 2A, 2B, 3B, 3D, 5A, 6A, 6B, 7A, and 7B, with LOD scores ranging from 3–11.8 [5]. There have also been 25 QTLs identified for differences in the value of kernel-related traits between high-N and low-N treatments on chromosomes 1B, 2A, 2B, 3B, 4B, 5D, 6B, and 7A (including 6 QTLs on chromosome 4B and 5 QTLs on chromosome 2A) [6]. It is well-established that the index of biomass and nutrient content at the seedling stage in hydroponic treatments is highly correlated with yield-related traits at the mature stage in wheat [9]. Another study reported 15 QTLs related to wheat seedling traits in response to low-N treatments on chromosomes 1B, 1D, 2B, 4A, 4B, 5D, 6A, 6B, 7A, and 7B [10].
Genome-wide association studies (GWAS) are widely used for mapping significant genetic variability from large germplasm diversity panels; such studies provide important genetic information about complex traits (i.e., traits controlled by more than one gene) in crops [11, 12]. Previous GWAS studies have implicated numerous loci with yield-related traits [13–19], disease resistance [20–23], drought resistance [24], heat tolerance [25], and so on. Several GWAS studies about nitrogen-related traits in crops have also been reported [26–28]. An association mapping study of NUE-related traits in rice identified 8 statistically significant marker loci; and two of these loci harbored known NUE-related genes [26]. A study in wheat that used 214 European winter wheat varieties identified 333 genomic regions associated with 28 NUE-related traits [27]. Additionally, a study of a global wheat core collection comprised of both land races and modern cultivars identified 23 genetic regions distributed across 16 wheat chromosomes that were putatively involved in the plant responses to different N levels [28]. Hydroponic treatment is a useful experimental approach for nutrient-responsive studies, and it is accepted that hydroponic studies can reflect strong performance for high nutrient efficiency in mature plants that are grown in field conditions [29]. However, there have been very few GWAS studies of wheat seedling traits in response to differential N treatments.
Plant genes involved in N nutrition have been studied extensively [2, 30]. Major genes for N acquisition and translocation include genes of the ammonium transporter (AMT) [31] and nitrate transporter (NRT) families [32]. It is also known that complex signaling pathways are important component for the regulation of N nutrition in plants; known signaling components include a MADS-box transcription factor [33], the Nitrate Regulatory Gene2 (NRG2) [34], NIN-Like Protein 6 (NLP6), NLP7 [35], and the E3 SUMO ligase (AtSIZ1) gene [36]. Molecular genetics studies of wheat responses to N starvation have noted differential expression patterns for TaNRT1, TaNRT2 [37], and 23 different TaAMTs [38]. In addition, the TaNAC [39] and TabHLH1 [40] transcription factors in wheat were shown to be important regulators of N signaling.
Mutagenesis has long been a centrally important experimental tool in crop improvement research programs [41]. Indeed, to date, more than 3,200 crop cultivars have been developed based on induced mutations [42]. Here, using a diversity panel comprised of 190 stable wheat mutant lines (M6 generation) in the Jing411 background, we identified significant SNPs associated with resistance to low N of wheat seedlings by combining GWAS and t-test method. This study revealed novel allelic variation and key genes affecting resistance to N limitation, particularly on chromosomes 2A and 6B, and thus provides important insights for breeders using molecular methods to improve NUE performance in wheat.
Materials and methods
Plant materials
The seeds of Chinese winter wheat (Triticum aestivum L.) cultivar Jing411 were used for EMS and γ-rays mutagenesis, and the methods for EMS and γ-rays mutagenesis were the same as previously described [43]. After phenotypic selection for several generations, 190 individual M6 mutant lines showing observable phenotypic changes such as plant height, flowering time, were used for genotyping and N treatment.
Experimental design for N treatment, phenotyping, and data analysis
After germination in water for three days, the WT and mutant lines were grown in nutrient solution with either normal (4 mM) or low N (1/50 of the normal amount N) for 13 days; the composition of the nutrient solution was according to a previous study in wheat [10]. This experiment was conducted in a greenhouse (temperature 20–26°C and humidity 60%) with sunlight and two additional hours of 200–300 μmol m-2 s-1 light. For each genotype, 15 plants were treated, and finally a total of 8 plants with similar growth status from each genotype and each experimental group were sampled as replicates for data measurement. The following phenotypic values were measured for each genotype under low and normal N conditions: root length, shoot length, root weight, and shoot weight. The experiment was independently conducted twice. Based on the measured data, the relative root length (RRL), relative shoot length (RSL), relative root weight (RRW), and relative shoot weight (RSW) values were calculated according to the following formulas, respectively: RRL = Root length in low N / Root length in normal N; RSL = Shoot length in low N / Shoot length in normal N; RRW = Root weight in low N / Root weight in normal N; RSW = Shoot weight in low N / Shoot weight in normal N. The best linear unbiased estimates (BLUE) values for RRL, RSL, RRW, and RSW from 8 replicates and 2 independent experiments were used for marker-trait association.
Analyses of BLUE, variance, correlation coefficients, and broad sense heritability were performed by using the ANOVA analysis tools of the QTL IciMapping v4.1 program (http://www.isbreeding.net/).
Genotyping and filtering
The Axiom® Wheat 660K Genotyping Array (Thermo) was used to genotype the WT and 190 mutant lines; this wheat SNP genotyping array was described in a previous study [44], and the genotyping was performed by China Golden Marker (Beijing) Biotech Co. Ltd. (CGMB, http://www.cgmb.com.cn/). Quality filtering of the genotyping data (‘pruning’) was performed using R 3.4.1 (http://www.r-project.org/). This filtration identified 463,826 SNPs that failed a ‘frequency test’ (minor allele frequency, MAF < 0.05) and 66,381 SNPs that failed a ‘missing test’ (Call-Rate < 0.97); these putative SNPs were thus excluded from further analysis. Finally, 67,402 SNP markers were used for GWAS.
Genome-wide association study
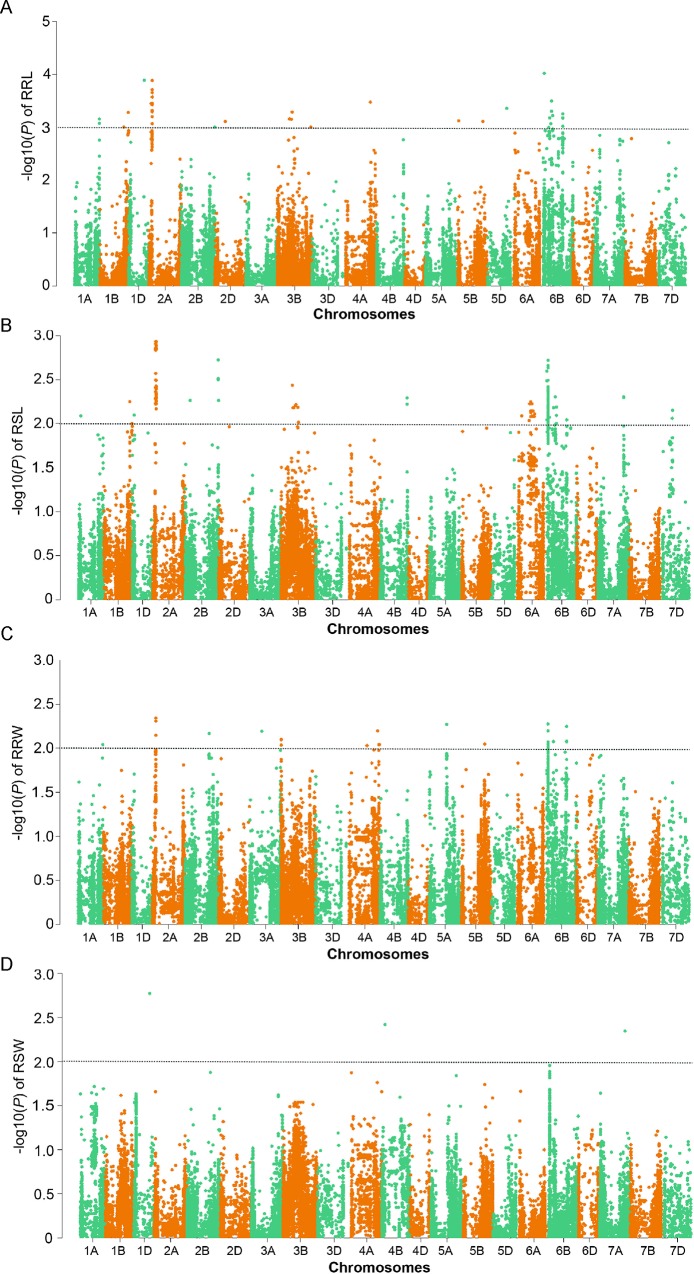
GWAS was performed using the General Linear Model (GLM) and the Mixed Linear Model (MLM) in TASSEL 5.0 [44–46]; based on the deviation of the observed statistic values from the expected statistic values in Q-Q plots, we selected the best model MLM from the GWAS analysis of the RRL, RSL, RRW, and RSW traits. Marker-trait association examined relationships between 67,402 SNP markers and the BLUE values of RRL, RSL, RRW, and RSW trait data calculated from 8 replications and 2 independent experiments. According to the general distribution of all p values of the SNPs for each trait, we selected a suggestive significance threshold of p values ≤ 0.001 for RRL and p values ≤ 0.01 for RSL, RRW, and RSW. The p value distributions of SNPs across the chromosomes were visualized using Manhattan plots that were constructed using R. Finally, 364 SNPs were identified.
Identification of significant SNPs by t-test
The identified 364 SNPs were further screened by statistically analyses of phenotypic data. The RRL, RSL, RRW and RSW from WT and mutant allele groups were compared. The phenotypic data in the two allele groups with significant difference of p ≤ 0.05 by t-test were detected as significant SNPs.
Identification of candidate genes
By using the flanking sequencing of the significant SNP markers, a BLAST search was performed against the reference genome Chinese Spring wheat v1.0 (http://www.wheatgenome.org). The genes containing the significant SNP markers were identified as the ‘candidate genes,’ and the gene annotations were based on the BLAST searching against gene sequence from other cereal plants in NCBI (https://www.ncbi.nlm.nih.gov/).
Results
Assessment and correlations among wheat seedling traits related to plant resistance to low nitrogen
Plants of the M6 generation of 190 wheat mutant lines from the mutant library of Jing411 background, which showed observable phenotypic changes (eg. plant height, flowering time), were grown hydroponically with either normal or low N concentrations. Unsurprisingly, wheat seedlings grown in the low-N treatment were much smaller than the seedlings grown with normal N treatment. Some of the mutants showed resistance to the low-N treatment (as evidence by taller growth stature and/or increased fresh weight) (Fig 1). The relative root length (RRL), relative shoot length (RSL), relative root weight (RRW), and relative shoot weight (RSW) were calculated and used as indices for resistance to the low N treatment. The mean values of RRL, RSL, RRW, and RSW in WT and mutants of two independent experiments were shown in S1 Table. Among the 190 mutant lines, 5 lines showed higher relative shoot length and shoot weight (more than 26% higher than that of WT in both experiments), indicating the effects of low N treatment on the shoot growth of these lines were lower. Therefore, these mutants were considered as resistance to the low N treatment. ANOVA analysis indicated that the variance among the WT and the 190 mutant genotypes for all four investigated traits (in two independent experiments) were significant at the p ≤ 0.05 level (Table 1). Further, Pearson correlation coefficient analysis between analytical pairings of each of the traits ranged from 0.52 to 0.77 (RRL, RSL, RRW, and RSW) (Table 2), indicating positive correlations among these phenotypic traits. Best linear unbiased estimates (BLUE) value analysis showed that variation ranged from 0.93 to 2.61 for RRL, from 0.55 to 1.18 for RSL, from 0.68 to 2.28 for RRW, and from 0.12 to 1.06 for RSW; the coefficient of variation for the four traits ranged from 12.75–20.22% (Table 3), highlighting wide variation for each trait among the different wheat mutant lines. Further, each of these four phenotypic traits related to resistance to low N exhibited high broad sense heritability, with H2 values ranging from 0.48–0.71.
Fig 1. Phenotypes of wheat plants grown in low or normal N conditions.
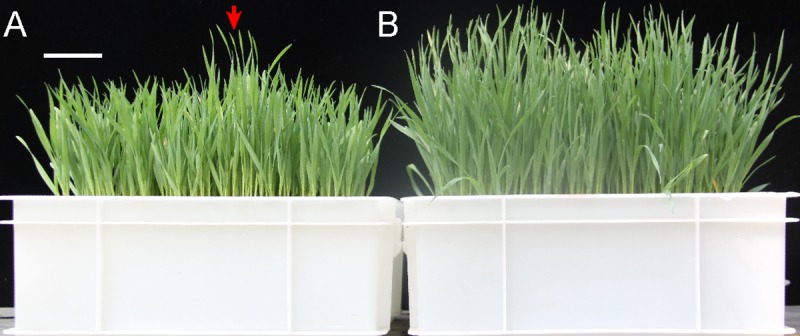
(A) WT and mutant wheat plants were grown in low-N nutrient solutions for 13 days. The red arrow indicates the wheat mutant lines that exhibited resistance to low N (i.e., taller than average lines). (B) The control WT and mutant plants grown under normal N condition. Bar = 5 cm.
Table 1. Analysis of variance for the investigated traits.
| Trait | Genotype | Experiment |
|---|---|---|
| RRL | 10.9368*** | 3.8036* |
| RSL | 15.1476*** | 286.9342*** |
| RRW | 3.381*** | 9.4928** |
| RSW | 4.1495*** | 22.068*** |
The following traits were investigated in this study: the relative root length (RRL), relative shoot length (RSL), relative root weight (RRW), and relative shoot weight (RSW).
* indicates statistical significance at p ≤ 0.05
** indicates statistical significance at p ≤ 0.01
*** indicates statistical significance at p ≤ 0.001.
Table 2. Correlation coefficients among the four phenotypic traits.
*** Statistical significance at p ≤ 0.001.
Table 3. Summary statistics for the four phenotypic traits among the WT and mutant lines.
| Trait | RRL | RSL | RRW | RSW |
|---|---|---|---|---|
| Mean | 1.31 | 0.73 | 1.21 | 0.63 |
| Minimum | 0.93 | 0.55 | 0.68 | 0.12 |
| Maximum | 2.61 | 1.18 | 2.28 | 1.06 |
| SD | 0.23 | 0.09 | 0.25 | 0.12 |
| CV% | 17.28 | 12.75 | 20.22 | 19.41 |
| H2 | 0.69 | 0.71 | 0.48 | 0.50 |
SD, standard deviation; CV, coefficients of variation; H2, broad sense heritability.
The SNPs loci associated with seedling resistance to low N by MLM analysis
To obtain reliable marker-trait associations, we based the analysis on BLUE values for the four traits from two independent experiments. Based on the deviation of the observed statistic values from the expected statistic values in the Q-Q plots, we determined that a MLM model was superior to a GLM model for GWAS of RRL, RSL, RRW, and RSW (S1 Fig). Finally, a total of 364 SNPs were detected for association with RRL, RSL, RRW, and RSW.
For RRL, a total of 95 SNPs, distributed on chromosomes 1A, 1B, 1D, 2A, 2B, 2D, 3B, 4A, 5B, 5D, and 6B, were identified at the p ≤ 0.001 level (S2 Table). Among these, the highest number of SNPs was detected on chromosome 2A (Table 4 and Fig 2A). Strikingly, fully 10% of the total phenotypic variation for RRL could be explained by marker AX-111453204 on chromosome 2A (Table 4). Additionally, on chromosome 6B, 19 significant SNPs were detected, and the marker AX-110666921 also account for 10% of the phenotypic variation for RRL (Table 4).
Table 4. List of markers associated with RRL, RSL, RRW, and RSW by GWAS.
| Trait | Marker | SNP | Chr. | Position | P-value | R2 |
|---|---|---|---|---|---|---|
| RRL | AX-110941028 | 2 | 1A | 587818235–588491731 | 6.95E-04 | 0.08 |
| AX-108868526 | 2 | 1B | 574569218–675535984 | 5.24E-04 | 0.08 | |
| AX-108811005 | 1 | 1D | 372812518 | 1.29E-04 | 0.10 | |
| AX-111453204 | 60 | 2A | 58538037–77335120 | 1.31E-04 | 0.10 | |
| AX-111611740 | 1 | 2B | 793179533 | 9.94E-04 | 0.08 | |
| AX-110467352 | 1 | 2D | 258384771 | 7.76E-04 | 0.08 | |
| AX-109951689 | 5 | 3B | 283539431–800088745 | 5.19E-04 | 0.08 | |
| AX-89472629 | 1 | 4A | 595265943 | 3.38E-04 | 0.07 | |
| AX-109031151 | 2 | 5B | 29569824–606299220 | 7.59E-04 | 0.08 | |
| AX-111358076 | 1 | 5D | 458076450 | 4.41E-04 | 0.09 | |
| AX-110666921 | 19 | 6B | 25806926–476960396 | 9.61E-05 | 0.10 | |
| RSL | AX-110618032 | 1 | 1A | 53447083 | 8.20E-03 | 0.05 |
| AX-110376696 | 2 | 1B | 629148059–629151382 | 5.62E-03 | 0.04 | |
| AX-94723578 | 1 | 1D | 46631942 | 7.98E-03 | 0.05 | |
| AX-108805657 | 103 | 2A | 75143409–77335120 | 1.18E-03 | 0.06 | |
| AX-110431432 | 5 | 2B | 111285723–793179533 | 1.90E-03 | 0.07 | |
| AX-111638430 | 7 | 3B | 261150728–413095187 | 3.66E-03 | 0.06 | |
| AX-111039474 | 2 | 4B | 643481153–643501106 | 5.12E-03 | 0.06 | |
| AX-111490103 | 18 | 6A | 104119755–405861085 | 5.68E-03 | 0.06 | |
| AX-111450720 | 85 | 6B | 2447095–476960396 | 1.91E-03 | 0.07 | |
| AX-110475574 | 2 | 7A | 610616256–610872015 | 4.97E-03 | 0.06 | |
| AX-110829820 | 3 | 7D | 231185616–239849103 | 7.06E-03 | 0.04 | |
| RRW | AX-110941028 | 1 | 1A | 587818235 | 9.06E-03 | 0.05 |
| AX-111022688 | 6 | 2A | 77267434–77335120 | 4.58E-03 | 0.04 | |
| AX-109979330 | 1 | 2B | 574720198 | 6.78E-03 | 0.04 | |
| AX-109915920 | 1 | 3A | 287684775 | 6.41E-03 | 0.05 | |
| AX-110125256 | 3 | 3B | 667972–2235874 | 7.95E-03 | 0.05 | |
| AX-108888540 | 4 | 4A | 427992360–723204067 | 6.34E-03 | 0.05 | |
| AX-109984948 | 1 | 5A | 409357352 | 5.34E-03 | 0.04 | |
| AX-110913918 | 1 | 5B | 559445291 | 8.97E-03 | 0.04 | |
| AX-110666921 | 19 | 6B | 25806926–476042649 | 5.31E-03 | 0.06 | |
| RSW | AX-108811005 | 1 | 1D | 372812518 | 1.69E-03 | 0.07 |
| AX-111456285 | 1 | 4B | 68900101 | 3.81E-03 | 0.06 | |
| AX-111539752 | 1 | 7A | 603542835 | 4.52E-03 | 0.06 |
For each phenotypic trait, the total numbers and the range of positions of the SNPs distributed across the chromosomes are given. The SNPs showing the lowest p value for each chromosome are listed with marker names, p-value, and R2.
Fig 2. Manhattan plots of the four phenotypic traits.
(A) RRL; (B) RSL; (C) RRW; (D) RSW. The black dashed lines indicate–log10 transforms of the suggestive p values.
For RSL, a total of 229 markers were detected at the p ≤ 0.01 level, and these were distributed on chromosomes 1A, 1B, 1D, 2A, 2B, 3B, 4B, 6A, 6B, 7A, and 7D (S3 Table). As with RRL, many of these SNPs (103) were located on chromosome 2A and many (85) were on chromosome 6B (Table 4 and Fig 2B). The SNP marker AX-108805657 on chromosome 2A and the marker AX-111450720 on chromosome 6B explained, respectively, 6% and 7% of the phenotypic variation for RSL (Table 4).
There were relatively fewer SNPs at the p ≤ 0.01 level for RRW: 37 SNPs (S4 Table), distributed on chromosomes 1A, 2A, 2B, 3A, 3B, 4A, 5A, 5B, and 6B, were associated with RRW at the p ≤ 0.01 level (Table 4 and Fig 2C). Fully half of these SNPs for RRW were located on chromosome 6B, and the AX-110666921 marker explained 6% of the phenotypic variation for RRW (Table 4). Finally, there were only 3 SNPs for RSW at the p ≤ 0.01 level (S5 Table) distributed on chromosomes 1D, 4B, and 7A, explained 7%, 6%, and 6% of the phenotypic variation, respectively (Table 4 and Fig 2D).
The significant SNP loci resulted in phenotypic variations between WT and mutants by t-test
We further investigated the significant SNPs among the detected 364 SNPs, which statistically resulted in phenotypic variations by t-test. Finally, a total of 221 SNPs significantly increased the RRL, RSL, and RRW, respectively, in the allele of mutant group compared to that of WT group (S6 Table). Generally, the number of lines with mutant allele ranging from 7 to 88 was observed among the significant SNP loci. For RRL, two SNPs were distributed on chromosomes 1A and 6B, respectively. For RSL, the significant SNPs were detected on chromosomes 2A, 2B, 4B, 6A, 6B, and 7A, including 103 and 81 SNPs on chromosome 2A and 6B, respectively. For RRW, the significant SNPs were distributed on chromosomes 2A, 2B, 4A, 5A, and 6B. Additionally, 6 significant SNPs (marker AX-110666921, AX-111453204, AX-110574568, AX-111023022, AX-110530579, AX-95011058) were associated with two of the three traits.
Candidate genes associated with resistance to low N
Genes containing the significant SNPs that resulted in statistically variation of RRL, RSL, RRW, and RSW in the mutant allele would be important for resistance to low N and were further examined in this study. A total of 41 SNPs occurred in genic sequences, including 19 on chromosome 6B and 22 on chromosome 2A (Table 5). BLAST-based annotation of these candidate genes suggested that 1 significant SNP (AX-94852973, mutation in 71 lines) resulted in amino acid change of a gene encoding high-affinity nitrate transporter 2.1, and another significant SNP (AX-95011058, mutation in 88 lines) occurred in a gene encoding gibberellin responsive protein; 11 significant SNPs occurred in three genes encoding disease resistance protein RPP13-like; 2 SNPs occurred in a gene encoding UDP-N-acetylglucosamine—dolichyl-phosphate N-acetylglucosaminephosphotransferase-like, RNA pseudouridine synthase 6, DEAD-box ATP-dependent RNA helicase 10, respectively; and 3 SNPs occurred in two genes encoding L-type lectin-domain containing receptor kinase. Additionally, the significant SNPs were also observed in a gene involving in bifunctional protein-serine/threonine kinase/phosphatase, transcription termination factor MTERF15, pre-mRNA-processing factor 39-like, protein STRUBBELIG-RECEPTOR FAMILY 5-like, UPF0481 protein At3g47200-like, G-type lectin S-receptor-like serine/threonine-protein kinase, ABC transporter C family member 10-like, and cis-zeatin O-glucosyltransferase 1-like (Table 5).
Table 5. The candidate genes associated with resistance to low N.
| Marker | Gene ID | Gene annotation |
|---|---|---|
| AX-94852973 | TraesCS6B01G044000 | high-affinity nitrate transporter 2.1-like |
| AX-95011058 | TraesCS6B01G050700 | gibberellin responsive protein gene |
| AX-109488831 | Traes_6BS_6C812D341 | disease resistance protein RPP13-like |
| AX-108831423 | Traes_6BS_6C812D341 | disease resistance protein RPP13-like |
| AX-111068223 | Traes_6BS_6C812D341 | disease resistance protein RPP13-like |
| AX-110666921 | TraesCS6B01G041800 | disease resistance RPP13-like protein 3 |
| AX-111581633 | TraesCS6B01G041800 | disease resistance RPP13-like protein 3 |
| AX-108891336 | TraesCS6B01G041800 | disease resistance RPP13-like protein 3 |
| AX-109404235 | TraesCS6B01G041800 | disease resistance RPP13-like protein 3 |
| AX-111078323 | TraesCS6B01G041800 | disease resistance RPP13-like protein 3 |
| AX-89334353 | TraesCS6B01G041800 | disease resistance RPP13-like protein 3 |
| AX-110475152 | TraesCS6B01G041800 | disease resistance RPP13-like protein 3 |
| AX-109464253 | TraesCS6B01G043500 | disease resistance protein RPP13-like |
| AX-111518118 | Traes_6BS_143FEF476 | Bifunctional protein-serine/threonine kinase/phosphatase |
| AX-111175044 | Traes_6BS_A5FB0FF9D | transcription termination factor MTERF15 |
| AX-110071044 | Traes_6BS_FF3A794F8 | pre-mRNA-processing factor 39-like |
| AX-108918819 | TraesCS6B01G051000 | protein STRUBBELIG-RECEPTOR FAMILY 5-like |
| AX-111450720 | Traes_6BS_26326DD6C | UPF0481 protein At3g47200-like |
| AX-110544749 | Traes_2AS_8DF7D907F | G-type lectin S-receptor-like serine/threonine-protein kinase |
| AX-109010730 | Traes_2AS_1CBBCBAA2 | ABC transporter C family member 10-like |
| AX-108805657 | Traes_2AS_FB8C94F8E | UDP-N-acetylglucosamine—dolichyl-phosphate N-acetylglucosaminephosphotransferase-like |
| AX-111519665 | Traes_2AS_FB8C94F8E | UDP-N-acetylglucosamine—dolichyl-phosphate N-acetylglucosaminephosphotransferase-like |
| AX-110360537 | Traes_2AS_C3CF1C55F | RNA pseudouridine synthase 6, chloroplastic |
| AX-110428187 | Traes_2AS_C3CF1C55F | RNA pseudouridine synthase 6, chloroplastic |
| AX-111040684 | Traes_2AS_617EE2FA7 | DEAD-box ATP-dependent RNA helicase 10 |
| AX-110396519 | Traes_2AS_617EE2FA7 | DEAD-box ATP-dependent RNA helicase 10 |
| AX-111749124 | Traes_2AS_1FA413046 | L-type lectin-domain containing receptor kinase IV.1-like |
| AX-111465844 | Traes_2AS_1FA413046 | L-type lectin-domain containing receptor kinase IV.1-like |
| AX-109345931 | Traes_2AS_B834BD325 | L-type lectin-domain containing receptor kinase IV.1-like |
| AX-110365502 | TraesCS2A01G128200 | cis-zeatin O-glucosyltransferase 1-like |
| AX-109973857 | TraesCS2A01G127800 | uncharacterized protein |
| AX-110609989 | TraesCS2A01G127800 | uncharacterized protein |
| AX-110652048 | TraesCS2A01G128400 | uncharacterized protein |
| AX-110907563 | TraesCS2A01G127800 | uncharacterized protein |
| AX-109833412 | Traes_2AS_4ACFB257F | uncharacterized protein |
| AX-110383858 | Traes_2AS_4ACFB257F | uncharacterized protein |
| AX-108939488 | TraesCS2A01G130100LC | uncharacterized protein |
| AX-109419490 | TraesCS6B01G194500 | uncharacterized protein |
| AX-110504653 | Traes_2AS_42C13EE1C | uncharacterized protein |
| AX-111715384 | Traes_2AS_42C13EE1C | uncharacterized protein |
| AX-108770012 | Traes_2AS_42C13EE1C | uncharacterized protein |
Genes containing the significant markers are identified as the candidate gene and listed.
Discussion
Understanding the genetic basis of resistance to low N in crops is an important building block for NUE improvement strategies [3]. In this study, using a population derived from induced mutagenesis in wheat, we characterized allelic variation that affects seedling resistance to low N. Induced mutagenesis methods reliably produce large numbers of genetic and thus phenotypic variations [47, 48]. Compared to the diploid species Arabidopsis, treatment of hexaploid wheat with common mutagens results in considerably higher mutation frequencies (~one mutation per 30 kb) [42]. The combining of the MLM analysis and t-test of phenotypic traits in the wheat mutant population for identification of the significant SNPs provides an effective route for investigation the novel SNP loci and/or genes in resistance to low N.
N use efficiency is tightly connected with the agronomic traits such as plant height and flowering time [49]. In this study, we used 190 mutant lines showing observable phenotypic changes (eg. plant height, flowering time) for genotyping and N treatment. It is reasonable to speculate that more genomic variations exist in these mutants. Interestingly, we found that the phenotypic data for four traits for resistance to low N were highly variable among the 190 mutant lines (Tables 1 and 3). Hydroponic methods are often used in studies of nutrient metabolism and signaling in plants, because they are relatively easy to use and enable very precise control of nutrient delivery. Importantly, it has been reported that the nutrient-related traits observed in hydroponic system in seedling-stage plants are significantly positively correlated to N and P uptake efficiency traits monitored for mature plants grown in field conditions [9, 29]. The four traits measured in this study (RRL, RSL, RRW, and RSW) reflect the NUE levels. We observed mutant wheat lines with relatively higher seedling length and/or fresh weights under low-N treatment (Fig 1 and S1 Table), indicating the potential of identifying high-NUE performers from induced mutagenesis populations. Obviously, the detailed mechanisms underlying the observed resistance to low N, and any practical application of the mutant lines of interest under the field condition will require further characterization in future studies.
Although there have been few GWAS of NUE in wheat, the limited information available suggests that the almost all chromosomes have at least some regions that affect NUE [27, 28]. In our study, the loci associated with the four resistance to low N traits were located on 17 chromosomes; that is, all chromosomes excepting 3D, 4D, 6D, and 7B (Table 4 and Fig 2). By using mutant population in this study, the allele could be easily classified into two groups (WT and mutant allele groups). Therefore, the changes of phenotypic data resulted from allele variation would be statistically detected by t-test. The combining of GWAS and t-test restricted the significant markers associated with resistance in low N to chromosome 1A, 2A, 2B, 4A, 4B, 5A, 6A, 6B, 7A, and most of the significant SNPs were located on chromosome 2A and 6B (S6 Table). Previous QTL mapping studies of wheat grain yield in response to varying N application indicated that a QTL on chromosome 2A explained a high proportion of phenotypic variance as evaluated across three field test sites [5]. This is consistent with our finding that the highest number of significant SNPs associated with RSL was observed on chromosome 2A (S6 Table). Additionally, a QTL study of kernel-related traits in plants grown under different N conditions also identified multiple QTLs on chromosome 2A [6]. The chromosome 6B also exhibited higher amounts of significant SNPs associated with RSL and RRW (S6 Table). Meanwhile, the genomic regions that were associated with more than one of the four traits suggested that loci on chromosomes 2A and 6B were significantly associated with RRL, RSL, and RRW (Fig 2 and Table 4). These results clearly suggested that these regions appear to somehow confer resistance to low N. Previous QTL mapping studies of seedling traits related to N nutrition also identified significant QTLs on chromosome 6B, but did not report QTLs for chromosome 2A [9, 10].
The mutated genes resulting in relative phenotypic data variations under low N to normal N condition would be important for resistance to low N. Interestingly, 1 significant SNP occurred in a gene encoding high-affinity nitrate transporter 2.1 (NRT2.1) and the mutation was found in 71 lines (S6 Table and Table 5). It is well documented that NRT1 and NRT2 family transporters mediate nitrate uptake from soil [32] and the Arabidopsis NRT2.1 play a central role in coordinating root response to N limitation [50]. Moreover, it has been suggested that the transcript level of wheat NRT2.1 was significantly induced by N starvation [37]. In this study, the mutation of NRT2.1 in the 71 lines leaded to the encoded amino acid changes at the site of 402, which probably resulted in the phenotypic variation in response to low N. Gibberellins are essential regulators for plant development and closely related to N acquisition in plant [51]. It has been suggested that GA signaling pathway participated in regulation of N deficiency-induced anthocyanin accumulation [52]. Conversely, N availability modulates the activity of GA transporter NPF3.1 in Arabidopsis [53]. Therefore, it is reasonable to observe that the mutation of gibberellin responsive protein gene resulted in resistance to low N compared to that of WT- allele group (S6 Table and Table 5). Additionally, 11 significant SNPs occurred in three genes encoding disease resistance protein RPP13-like, suggesting the important roles of RPP13 genes for resistance in low N. Disease resistance proteins are a well-known large family of proteins that are important in the regulation of plant disease resistance responses [54, 55]. A previous study showed that expression of the gene encoding a disease resistance protein was differentially regulated by different forms of N [56]. Clearly, the possible low N resistance function of the candidate disease resistance protein RPP13-like identified in the present study will require further investigation.
Conclusions
We here identified SNPs that were significantly association with the low N resistance traits RRL, RSL, RRW, and RSW in wheat by combining GWAS and t-test methods. Of particular note, loci on chromosomes 2A and 6B were found to be especially impactful for resistance to low N. Several candidate genes, including genes encoding a high-affinity nitrate transporter 2.1 and a gibberellin responsive protein, were implicated as having possible functions associated with resistance to low N. Future work can validate the significant markers we identified here and can determine whether or not any of these markers will be effective in NUE-improvement efforts in wheat. Finally, this study deepens our knowledge about the genetic basis of a core metabolic nexus in arguably the world’s most important food crop.
Supporting information
Comparison of Q-Q plots for RRL using GLM (A) and MLM (B). The black line indicates the expected values.
(TIF)
The five mutant lines with bold font showing higher RSL and RSW in both experiments were considered as resistance to the low N treatment. The 1st or 2nd in the brackets indicate the values in the column are from the first or the second experiment.
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
a, b indicate the statistically significant difference of the traits in two allele groups by t-test; c, e represent the allele in WT and mutant, respectively; d, f show the mean values and SD of RRL, RSL and RRW in the WT and mutant allele groups, respectively.
(XLSX)
Acknowledgments
We thank Dr. Luyan Zhang, Prof. Jiankang Wang, and Dr. Junjie Fu (Institute of Crop Sciences, Chinese Academy of Agricultural Sciences) for advice for our data analysis.
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
This work was financially supported by The National Key Research and Development Program of China (Grant No. 2016YFD0101802 to LL) and the China Agriculture Research System (Grant No. CARS-03 to LL). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Hirel B, Tétu T, Lea PJ, Dubois F. Improving nitrogen use efficiency in crops for sustainable agriculture. Sustainability. 2011; 3:1452. [Google Scholar]
- 2.Gutierrez RA. Systems biology for enhanced plant nitrogen nutrition. Science. 2012; 336: 1673–1675. 10.1126/science.1217620 [DOI] [PubMed] [Google Scholar]
- 3.Han M, Okamoto M, Beatty PH, Rothstein SJ, Good AG. The genetics of nitrogen use efficiency in crop plants. Annu Rev Genet. 2015; 49: 269–289. 10.1146/annurev-genet-112414-055037 [DOI] [PubMed] [Google Scholar]
- 4.Garnett T, Plett D, Heuer S, Okamoto M. Genetic approaches to enhancing nitrogen-use efficiency (NUE) in cereals: challenges and future directions. Funct Plant Biol. 2015; 42: 921–941. [DOI] [PubMed] [Google Scholar]
- 5.Mahjourimajd S, Taylor J, Sznajder B, Timmins A, Shahinnia F, Rengel Z, et al. Genetic basis for variation in wheat grain yield in response to varying nitrogen application. PLoS ONE. 2016; 11: e0159374 10.1371/journal.pone.0159374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cui F, Fan X, Chen M, Zhang N, Zhao C, Zhang W, et al. QTL detection for wheat kernel size and quality and the responses of these traits to low nitrogen stress. Theor Appl Genet. 2016; 129: 469–484. 10.1007/s00122-015-2641-7 [DOI] [PubMed] [Google Scholar]
- 7.Xu Y, Wang R, Tong Y, Zhao H, Xie Q, Liu D, et al. Mapping QTLs for yield and nitrogen-related traits in wheat: influence of nitrogen and phosphorus fertilization on QTL expression. Theor Appl Genet. 2014; 127: 59–72. 10.1007/s00122-013-2201-y [DOI] [PubMed] [Google Scholar]
- 8.Cui F, Fan XL, Zhao CH, Zhang W, Chen M, Ji J, et al. A novel genetic map of wheat: utility for mapping QTL for yield under different nitrogen treatments. BMC Genet. 2014; 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sun JJ, Guo Y, Zhang GZ, Gao MG, Zhang GH, Kong FM, et al. QTL mapping for seedling traits under different nitrogen forms in wheat. Euphytica. 2013; 191: 317–331. [Google Scholar]
- 10.Guo Y, Kong FM, Xu YF, Zhao Y, Liang X, Wang YY, et al. QTL mapping for seedling traits in wheat grown under varying concentrations of N, P and K nutrients. Theor Appl Genet. 2012; 124: 851–865. 10.1007/s00122-011-1749-7 [DOI] [PubMed] [Google Scholar]
- 11.Zhu C, Gore M, Buckler ES, Yu J. Status and prospects of association mapping in plants. Plant Genome. 2008; 1: 5–20. [Google Scholar]
- 12.Nordborg M, Weigel D. Next-generation genetics in plants. Nature 2008; 456: 720–723. 10.1038/nature07629 [DOI] [PubMed] [Google Scholar]
- 13.Sun CW, Zhang FY, Yan XF, Zhang XF, Dong ZD, Cui DQ, et al. Genome-wide association study for 13 agronomic traits reveals distribution of superior alleles in bread wheat from the Yellow and Huai Valley of China. Plant Biotechnol J. 2017; 15: 953–969. 10.1111/pbi.12690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu YX, Lin Y, Gao S, Li ZY, Ma J, Deng M, et al. A genome-wide association study of 23 agronomic traits in Chinese wheat landraces. Plant J. 2017; 91: 861–873. 10.1111/tpj.13614 [DOI] [PubMed] [Google Scholar]
- 15.Chen GF, Zhang H, Deng ZY, Wu RG, Li DM, Wang MY, et al. Genome-wide association study for kernel weight-related traits using SNPs in a Chinese winter wheat population. Euphytica. 2016; 212: 173–185. [Google Scholar]
- 16.Tadesse W, Ogbonnaya FC, Jighly A, Sanchez-Garcia M, Sohail Q, Rajaram S, et al. Genome-wide association mapping of yield and grain quality traits in winter wheat genotypes. PLoS ONE. 2015; 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sukumaran S, Dreisigacker S, Lopes M, Chavez P, Reynolds MP. Genome-wide association study for grain yield and related traits in an elite spring wheat population grown in temperate irrigated environments. Theor Appl Genet. 2015; 128: 353–363. 10.1007/s00122-014-2435-3 [DOI] [PubMed] [Google Scholar]
- 18.Sajjad M, Khan SH, Ahmad MQ, Rasheed A, Mujeeb-Kazi A, Khan IA. Association mapping identifies QTLs on wheat chromosome 3A for yield related traits. Cereal Res Commun. 2014; 42: 177–188. [Google Scholar]
- 19.Edae EA, Byrne PF, Haley SD, Lopes MS, Reynolds MP. Genome-wide association mapping of yield and yield components of spring wheat under contrasting moisture regimes. Theor Appl Genet. 2014; 127: 791–807. 10.1007/s00122-013-2257-8 [DOI] [PubMed] [Google Scholar]
- 20.Pasam RK, Bansal U, Daetwyler HD, Forrest KL, Wong D, Petkowski J, et al. Detection and validation of genomic regions associated with resistance to rust diseases in a worldwide hexaploid wheat landrace collection using BayesR and mixed linear model approaches. Theor Appl Genet. 2017; 130: 777–793. 10.1007/s00122-016-2851-7 [DOI] [PubMed] [Google Scholar]
- 21.Gao LL, Rouse MN, Mihalyov PD, Bulli P, Pumphrey MO, Anderson JA. Genetic characterization of stem rust resistance in a global spring wheat germplasm collection. Crop Sci. 2017; 57: 2575–2589. [Google Scholar]
- 22.Li GQ, Xu XY, Bai GH, Carver BF, Hunger R, Bonman JM, et al. Genome-wide association mapping reveals novel QTL for seedling leaf rust resistance in a worldwide collection of winter wheat. Plant Genome. 2016; 9. [DOI] [PubMed] [Google Scholar]
- 23.Maccaferri M, Zhang JL, Bulli P, Abate Z, Chao SM, Cantu D, et al. A Genome-wide association study of resistance to stripe rust (Puccinia striiformis f. sp tritici) in a worldwide collection of hexaploid spring wheat (Triticum aestivum L.). G3-Genes Genom Genet. 2015; 5: 449–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mwadzingeni L, Shimelis H, Rees DJG, Tsilo TJ. Genome-wide association analysis of agronomic traits in wheat under drought-stressed and non-stressed conditions. PLoS ONE. 2017; 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pandey GC, Sareen S, Siwach P, Tiwari R. Molecular characterization of heat tolerance in bread wheat (Triticum aestivum L.) using differences in thousand-grain weights (dTGW) as a potential indirect selection criterion. Cereal Res Commun. 2014; 42: 38–47. [Google Scholar]
- 26.Liu ZY, Zhu CS, Jiang Y, Tian YL, Yu J, An HZ, et al. Association mapping and genetic dissection of nitrogen use efficiency-related traits in rice (Oryza sativa L.). Funct Integr Genomic. 2016; 16: 323–333. [DOI] [PubMed] [Google Scholar]
- 27.Cormier F, Le Gouis J, Dubreuil P, Lafarge S, Praud S. A genome-wide identification of chromosomal regions determining nitrogen use efficiency components in wheat (Triticum aestivum L.). Theor Appl Genet. 2014; 127: 2679–2693. 10.1007/s00122-014-2407-7 [DOI] [PubMed] [Google Scholar]
- 28.Bordes J, Ravel C, Jaubertie JP, Duperrier B, Gardet O, Heumez E, et al. Genomic regions associated with the nitrogen limitation response revealed in a global wheat core collection. Theor Appl Genet. 2013; 126: 805–822. 10.1007/s00122-012-2019-z [DOI] [PubMed] [Google Scholar]
- 29.Liu ZG, Gao K, Shan S, Gu RC, Wang ZK, Craft EJ, et al. Comparative analysis of root traits and the associated QTLs for maize seedlings grown in paper roll, hydroponics and vermiculite culture system. Front Plant Sci. 2017; 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xuan W, Beeckman T, Xu G. Plant nitrogen nutrition: sensing and signaling. Curr Opin Plant Biol. 2017; 39: 57–65. 10.1016/j.pbi.2017.05.010 [DOI] [PubMed] [Google Scholar]
- 31.Ninnemann O, Jauniaux JC, Frommer WB. Identification of a high affinity NH4+ transporter from plants. EMBO J. 1994; 13: 3464–3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miller AJ, Fan X, Orsel M, Smith SJ, Wells DM. Nitrate transport and signalling. J Exp Bot. 2007; 58: 2297–2306. 10.1093/jxb/erm066 [DOI] [PubMed] [Google Scholar]
- 33.Zhang H, Forde BG. An Arabidopsis MADS box gene that controls nutrient-induced changes in root architecture. Science. 1998; 279: 407 [DOI] [PubMed] [Google Scholar]
- 34.Xu N, Wang R, Zhao L, Zhang C, Li Z, Lei Z, et al. The Arabidopsis NRG2 protein mediates nitrate signaling and interacts with and regulates key nitrate regulators. Plant Cell. 2016; 28: 485 10.1105/tpc.15.00567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bellegarde F, Gojon A, Martin A. Signals and players in the transcriptional regulation of root responses by local and systemic N signaling in Arabidopsis thaliana. J Exp Bot. 2017; 68: 2553–2565. 10.1093/jxb/erx062 [DOI] [PubMed] [Google Scholar]
- 36.Park BS, Song JT, Seo HS. Arabidopsis nitrate reductase activity is stimulated by the E3 SUMO ligase AtSIZ1. Nat Commun. 2011; 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guo TC, Xuan HM, Yang YY, Wang LN, Wei LT, Wang YH, et al. Transcription analysis of genes encoding the wheat root transporter NRT1 and NRT2 families during nitrogen starvation. J Plant Growth Regul. 2014; 33: 837–848. [Google Scholar]
- 38.Li T, Liao K, Xu X, Gao Y, Wang Z, Zhu X, et al. Wheat ammonium transporter (AMT) gene family: diversity and possible role in host-pathogen interaction with stem rust. Front Plant Sci. 2017; 8: 1637 10.3389/fpls.2017.01637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.He X, Qu B, Li W, Zhao X, Teng W, Ma W, et al. The nitrate-inducible NAC transcription factor TaNAC2-5A controls nitrate response and increases wheat yield. Plant Physiol 2015; 169: 1991–2005. 10.1104/pp.15.00568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang T, Hao L, Yao S, Zhao Y, Lu W, Xiao K. TabHLH1, a bHLH-type transcription factor gene in wheat, improves plant tolerance to Pi and N deprivation via regulation of nutrient transporter gene transcription and ROS homeostasis. Plant Physiol Biochem. 2016; 104: 99–113. 10.1016/j.plaphy.2016.03.023 [DOI] [PubMed] [Google Scholar]
- 41.Parry MAJ, Madgwick PJ, Bayon C, Tearall K, Hernandez-Lopez A, Baudo M, et al. Mutation discovery for crop improvement. J Exp Bot. 2009; 60: 2817–2825. 10.1093/jxb/erp189 [DOI] [PubMed] [Google Scholar]
- 42.Nawaz Z, Shu Q. Molecular nature of chemically and physically induced mutants in plants: a review. Plant Genetic Resources. 2014; 12: S74–S78. [Google Scholar]
- 43.Xiong H, Guo H, Xie Y, Zhao L, Gu J, Zhao S, et al. Enhancement of dwarf wheat germplasm with high-yield potential derived from induced mutagenesis. Plant Genet Resour-C. 2018;16: 74–81. [Google Scholar]
- 44.Zhou S, Zhang J, Che Y, Liu W, Lu Y, Yang X, et al. Construction of Agropyron Gaertn. genetic linkage maps using a wheat 660K SNP array reveals a homoeologous relationship with the wheat genome. Plant Biotechnol J. 2018; 16: 818–827. 10.1111/pbi.12831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang Z, Ersoz E, Lai CQ, Todhunter RJ, Tiwari HK, Gore MA, et al. Mixed linear model approach adapted for genome-wide association studies. Nat Genet. 2010; 42: 355–360. 10.1038/ng.546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yu J, Pressoir G, Briggs WH, Vroh Bi I, Yamasaki M, Doebley JF, et al. A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nat Genet. 2006; 38: 203–208. 10.1038/ng1702 [DOI] [PubMed] [Google Scholar]
- 47.Guo H, Yan Z, Li X, Xie Y, Xiong H, Liu Y, et al. Development of a high-efficient mutation resource with phenotypic variation in hexaploid winter wheat and identification of novel alleles in the TaAGP.L-B1 gene. Front Plant Sci. 2017; 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li GT, Jain R, Chern M, Pham NT, Martin JA, Wei T, et al. The sequences of 1504 mutants in the model rice variety kitaake facilitate rapid functional genomic studies. Plant Cell. 2017; 29: 1218–1231. 10.1105/tpc.17.00154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gooding MJ, Addisu M, Uppal RK, Snape JW, Jones HE. Effect of wheat dwarfing genes on nitrogen-use efficiency. J Agric Sci. 2012; 150: 3–22. [Google Scholar]
- 50.Remans T, Nacry P, Pervent M, Girin T, Tillard P, Lepetit M, et al. A central role for the nitrate transporter NRT2.1 in the integrated morphological and physiological responses of the root system to nitrogen limitation in Arabidopsis. Plant Physiol. 2006; 140: 909–921. 10.1104/pp.105.075721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bai L, Deng H, Zhang X, Yu X, Li Y. Gibberellin is involved in inhibition of cucumber growth and nitrogen uptake at suboptimal root-zone temperatures. PLoS ONE 2016; 11: e0156188 10.1371/journal.pone.0156188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang Y, Liu Z, Liu J, Lin S, Wang J, Lin W, et al. GA-DELLA pathway is involved in regulation of nitrogen deficiency-induced anthocyanin accumulation. Plant Cell Rep. 2017; 36: 557–569. 10.1007/s00299-017-2102-7 [DOI] [PubMed] [Google Scholar]
- 53.David LC, Berquin P, Kanno Y, Seo M, Daniel-Vedele F, Ferrario-Mery S. N availability modulates the role of NPF3.1, a gibberellin transporter, in GA-mediated phenotypes in Arabidopsis. Planta. 2016; 244: 1315–1328. 10.1007/s00425-016-2588-1 [DOI] [PubMed] [Google Scholar]
- 54.McHale L, Tan X, Koehl P, Michelmore RW. Plant NBS-LRR proteins: adaptable guards. Genome Biol. 2006; 7: 212 10.1186/gb-2006-7-4-212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Belkhadir Y, Subramaniam R, Dangl JL. Plant disease resistance protein signaling: NBS-LRR proteins and their partners. Curr Opin Plant Biol. 2004; 7: 391–399. 10.1016/j.pbi.2004.05.009 [DOI] [PubMed] [Google Scholar]
- 56.Wang YQ, Zhang JJ, Zhu GH, Peng XX. Differential expression of proteins in rice leaves cultivated with different forms of nitrogen nutrients. Journal of plant physiology and molecular biology. 2006; 32: 403–410. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Comparison of Q-Q plots for RRL using GLM (A) and MLM (B). The black line indicates the expected values.
(TIF)
The five mutant lines with bold font showing higher RSL and RSW in both experiments were considered as resistance to the low N treatment. The 1st or 2nd in the brackets indicate the values in the column are from the first or the second experiment.
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
a, b indicate the statistically significant difference of the traits in two allele groups by t-test; c, e represent the allele in WT and mutant, respectively; d, f show the mean values and SD of RRL, RSL and RRW in the WT and mutant allele groups, respectively.
(XLSX)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.