Abstract
MicroRNAs (miRNAs) are small noncoding RNAs of about 19-25 nt that regulate gene expression posttranscriptionally under various cellular conditions, including apoptosis. The miRNAs involved in modulation of apoptotic events in T cells are partially known. However, heterogeneity associated with cell lines makes it difficult to interpret gene expression signatures, especially in cancer-related cell lines. Treatment of the Jurkat T-cell leukemia cell line with the universal apoptotic drug, camptothecin, resulted in identification of two Jurkat subpopulations: one that is sensitive to camptothecin and another that is rather intrinsically resistant. We sorted apoptotic Jurkat cells from nonapoptotic ones prior to profiling miRNAs through deep sequencing. Our data showed that a total of 184 miRNAs were dysregulated. Interestingly, the apoptotic and nonapoptotic subpopulations exhibited distinct miRNA expression profiles. In particular, 6 miRNAs were inversely expressed in these two subpopulations. The pyrosequencing results were validated by real-time qPCR. Altogether, these results suggest that miRNAs modulate apoptotic events in T cells and that cellular heterogeneity requires careful interpretation of miRNA expression profiles obtained from drug-treated cell lines.
Keywords: Apoptosis, microRNAs, Jurkat, deep sequencing
1. Introduction
Apoptosis is programmed cell death triggered by various stimuli from outside or inside the cell, such as ligation of cell surface receptors, treatment with cytotoxic drugs, or irradiation and it results in transcriptionally regulated activation of a number of regulatory proteins (Blank and Shiloh, 2007) . Apoptosis is characterized by exposure of phosphatidylserine on the plasma membrane outer leaflet, membrane blebbing, cellular shrinkage, chromatin condensation, and fragmentation of nuclear DNA, leading to formation of apoptotic bodies (Blagosklonny, 2000; Baumann et al., 2002) .
T cells constitute a vital branch of cell-mediated immunity and homeostasis of the immune response is sustained through a balance between proliferation and apoptosis of T cells. A wealth of information has accumulated over the past two decades about the transcriptional regulation of genes mediating apoptosis in T cells. The recent discovery of small RNAs, however, suggests that posttranscriptional gene regulatory networks might have prominent effects on modulation of apoptotic pathways of T cells (Lodish et al., 2008; O’Connell et al., 2010) . MicroRNAs (miRNAs), a type of those small RNAs, are noncoding small RNAs of about 19–25 nucleotides in length, which are transcribed by RNA polymerase II (Bartel, 2004, 2009) . Followed by nuclear processing by Drosha and cytoplasmic processing by Dicer, the mature miRNA strand negatively regulates gene expression by translational inhibition or destabilization of mRNAs (Miska, 2005; Lawrie, 2007; Stefani and Slack, 2008) . MiR14 and bantam were the first members of miRNAs shown to modulate apoptotic functions in Drosophila (Brennecke et al., 2003; Xu et al., 2003) . Over the past few years, a clear link has been established between apoptosis and miRNAs (Su et al., 2015) , particularly in cancer development (Kumar et al., 2007; Marcucci et al., 2011) .
Besides the significance of miRNAs in T-cell functions, miRNA-mediated T-cell apoptosis has also been associated with oncogenic miRNAs in leukemogenesis (Calin et al., 2009; Pekarsky et al., 2009; Chen et al., 2010) . In fact, miRNA profiling studies particularly on cancerous tissues have clearly shown that each leukemia type (for instance, CLL vs. ALL) possesses a prominent miRNA expression signature (Zanette et al., 2007) . Additional studies showed that these miRNAs regulate the expression of apoptotic or antiapoptotic mRNAs (Mott et al., 2007; Xiao et al., 2008; Akao et al., 2009) .
Although the use of cell lines has led to the identification of a number of dysregulated miRNAs involved in apoptosis and/or leukemogenesis (Li et al., 2013; Yamada et al., 2014; Zhou et al., 2014; Fan et al., 2016) , cellular heterogeneity associated with cancerous tissues requires careful interpretation of the data acquired from human studies. Cellular heterogeneity is also an important issue that needs to be taken into account while interpreting the data collected from cell lines. It is well documented that cells use cellular heterogeneity to function properly and survive (Benchaouir, 2004; Stockholm et al., 2007; Walling et al., 2012) . Jurkat cells were also reported to be heterogeneous as sublines from the same clones displayed different cellular morphologies and growth patterns (Snow and Judd, 2009) . Although miRNA heterogeneity is known to exist across different cell lines that originate from the same cancer type (Lu et al., 2015) , the potential for differential miRNA expression across the cells in the same cell line has not been reported before. To identify miRNAs that regulate apoptosis in Jurkat cells and also to test the effect of cellular heterogeneity on drug response and miRNA expression profiles, we triggered apoptosis in Jurkat cells with camptothecin, an inhibitor of DNA topoisomerase I and a potent inducer of apoptosis (Li and Liu, 2001; Pommier et al., 2003) . Following the drug treatment, we sorted the apoptotic subpopulation (as defined by Annexin V positivity) from the nonapoptotic subpopulation by magnetic beads. Deep sequencing of small RNAs isolated from each subpopulation revealed that each subpopulation possessed a distinct miRNA expression signature that might be associated with a population-specific apoptotic response.
2. Materials and methods
2.1. Cell culture, drug treatment, and transfection
Jurkat human leukemic T cells (American Type Culture Collection clone E6.1) were maintained in RPMI 1640 (GIBCO) supplemented with 2 mM L-glutamine, 10% fetal bovine serum (GIBCO), and 100 U penicillin/100 µg streptomycin (Biochrom AG) in an atmosphere of 5% CO2 at 37 °C. Cells (n = 106) were treated with different concentrations of camptothecin (Sigma) and incubated at 37 °C and 5% CO2 to determine the dose kinetics. Cells in both the treatment and control groups were labeled with PE-conjugated Annexin V and 7-AAD (BD Pharmingen) and analyzed by flow cytometry (BD FACSArray) to identify stages of apoptosis. Cells that were Annexin V-positive but 7-AAD-negative were defined as early apoptotic ones. Annexin-V-positive cells were then separated by Annexin V Microbead Kit (Miltenyi Biotech) according to the manufacturer’s instructions. Four fractions were obtained as follows: untreated Annexin-negative (JNN), untreated Annexin-positive (JNP), treated Annexin-positive (JAP), and treated Annexin-negative (JAN) cells. Because suficient cells could not be obtained from the JNP cells, they were excluded from the study.
2.2. Total RNA isolation and deep sequencing
Total RNA was isolated with the mirVana miRNA Isolation Kit (Ambion) according to the manufacturer’s instructions. Total RNA samples were treated with the TurboDNase DNA-free Kit (Ambion) to remove traces of genomic DNA contamination. RNA integrity was determined by a bioanalyzer (Agilent 2100) using the RNA 6000 Nano Kit (average RIN: ~9–10).
Three replicates from JNN, JAP, and JAN cells were mixed in equal amounts and sequenced using the Illumina Genome Analyzer by Fasteris (Switzerland). The fragments missing either adaptor were excluded from further analyses. The adaptor sequences of 15–29 bp inserts were removed prior to the subsequent data analyses. The Nexalign program (http://genome.gsc.riken. jp/osc/english/dataresource/) was used to align reads to rRNA (NCBI, U133169) and hairpin and mature miRNAs (miRBase, R18) (Vaz et al., 2010) . The sequences were aligned first for exact matches and then the remaining sequences were used to identify matches with up to three mismatches as described previously (De Hoon et al., 2010) . The data were deposited in GEO under accession number GSE35442.
2.3. Real-time qPCR analyses
RNAs smaller than 200 nt were isolated directly from the cells using the miRVana miRNA Isolation Kit according to the manufacturer’s instruction (Ambion). cDNA was prepared from the small RNAs using the RT2 miRNA cDNA Kit (SA Biosciences). qPCR was performed in duplicates of three biological replicates (Roche, LightCycler 480). U6 ncRNA was used for normalization.
2.4. Statistical analyses
Student’s t-test was used to statistically analyze the biological replicates in flow cytometry and qPCR analyses. P ≤ 0.05 was accepted as statistically significant.
3. Results
3.1. Identification of two Jurkat subpopulations with different apoptotic properties
We first performed dose-response (0.5–1024 µM) kinetics to determine the optimal drug treatment conditions for capturing cells at the apoptotic stage (Annexin V-positive and 7-AAD-negative). When Jurkat cells were treated with camptothecin for 4 h, apoptosis was observed in direct proportion to the dose applied up to 8 µM (Figure 1A).
Figure 1.
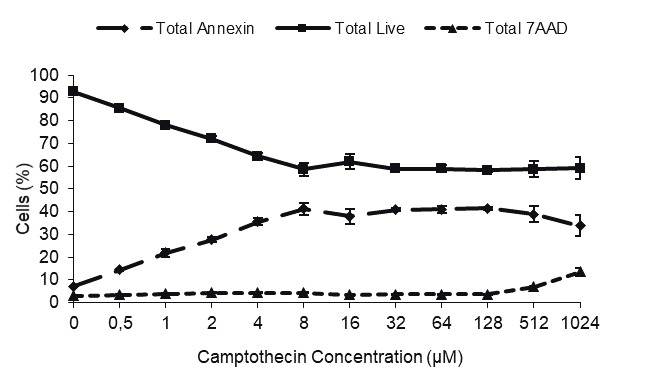
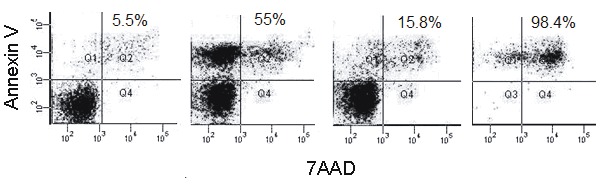
Dose response and identification of apoptotic cells by flow cytometry. A) Dose kinetics of camptothecin. Jurkat cells were treated with a range of camptothecin (0.5–1024 μM) for 4 h and the apoptotic cells were determined with Annexin V/7-AAD labeling. B–E) Enrichment of apoptotic cells with magnetic bead separation. Jurkat cells were treated with 8 μM camptothecin for 4 h and the apoptotic cells were sorted using an Annexin V magnetic bead separation kit. The apoptosis rate of the following samples was determined by flow cytometry: B) control, untreated cells (JNN); C) camptothecin-treated cells, D) camptothecin-treated and magnetic bead-sorted Annexin V-negative cells (JAN); E) camptothecin-treated, magnetic bead-sorted Annexin V-positive cells (JAP). It is important to note that the cells became Annexin V/7-AAD-double positive following the sorting.
Percentage of apoptotic cells did not significantly change and reached a plateau after 8 µM. Apoptosis was observed in 41% of the cells in camptothecin-treated group compared to 5.5% of the control cells (Figure 1A, P < 0.05).
The unresponsiveness of some cells to the drug led us to hypothesize that Jurkat cells may contain additional subpopulations, each of which might have a distinct camptothecin-mediated apoptotic property. uThs, we increased the concentration of the drug up to 1 mM (128fold in excess of the maximum 8 µM concentration). Interestingly, over 50% of the camptothecin-treated cells still remained relatively resistant to the drug treatment despite over 100-fold camptothecin concentrations, suggesting the presence of a drug-resistant second subpopulation (Figure 1A). The apparent difference in the apoptotic response could stem either from an uneven exposure of the cells to the drug or from intrinsic resistance of a subpopulation to the drug. To ensure that the differential apoptotic capacity of two subpopulations is not due to uneven drug treatment, we sorted the nonapoptotic cells and retreated them with the drug. To this end, Jurkat cells were first treated with 8 µM camptothecin for 4 h and the Annexin V-negative nonapoptotic cells (JAN) were separated from the apoptotic cells (JAP) using Annexin V-conjugated magnetic beads (Figures 1B–1E). Flow cytometry analysis of the sorted cells showed that the sorting eficiency was as high as 98.4% (purity > 85%, 95% on average) (Figure 1E). Retreatment of the nonapoptotic cells (JAN) with 8 µM camptothecin for 4 h showed that this fraction of the cells was indeed intrinsically resistant to induction of apoptosis by camptothecin. In the first round of treatment, 42.4% of the cells underwent apoptosis, while the second camptothecin treatment triggered apoptosis in only 7.4% of the JAN cells (Figure 2, P < 0.05). We concluded that there are intrinsic gene expression properties, e.g., miRNAs, associated with resistance to the camptothecininduced apoptosis in Jurkat T cells apparently composed of at least two subpopulations.
Figure 2.
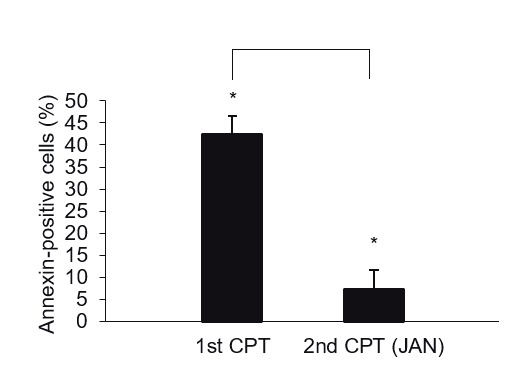
Two subpopulations of Jurkat cells with different camptothecin-mediated apoptotic capacity. Following the magnetic bead separation of camptothecin-treated and magnetic bead-sorted Annexin V-negative cells (Figure 1D, JAN cells), these cells were retreated with camptothecin for 4 h. Apoptosis was measured with Annexin V/7-AAD labeling. 1st CPT: Cells treated with camptothecin once; 2nd CPT (JAN): sorted cells that were retreated with camptothecin.
To investigate whether each Jurkat subpopulation possesses a distinct miRNA expression profile, we compared the miRNA expression patterns in 3 replicates of each subpopulation. The JNN sample contained the Annexin V-negative cells, which were not treated with the drug (negative control), whereas the JAN and JAP samples contained the Annexin V-negative (intrinsically camptothecin-resistant) and -positive (intrinsically camptothecin-susceptible) cells, respectively, which were treated with the drug. It should be noted that although the majority of the camptothecin-treated cells were Annexin V-positive and 7-AAD-negative prior to sorting, they shieftd to a Annexin V/7-AAD-double positive phenotype after sorting. We used the Illumina platform (Fasteris, Switzerland) to quantitatively measure the amounts of small RNAs in each sample. After the removal of the adapter sequences, 91.3% of all reads contained 15–29 bp inserts. Based on the size of the inserts, there appeared to be two major small RNA populations, one of 22–23 bp and the other 28 bp, each representing miRNAs and tRNA-derived small RNAs, respectively (Figure 3). The alignment of the reads to the known RNAs revealed two striking points with respect to the small RNA content of each sample: 1) The control JNN cells are rich in miRNA, which constitutes 60% of small RNAs. The miRNA content plummets to 26% and 16% in the camptothecin-treated JAN and JAP samples, respectively. 2) The drug treatment induces major tRNA fragmentation, constituting up to 45% of all small RNAs (Figure 3; nt 27–29 region). We did not notice a major difference in the proportion of other small RNA categories, although there may be differences in the expression of individual small RNAs.
Figure 3.
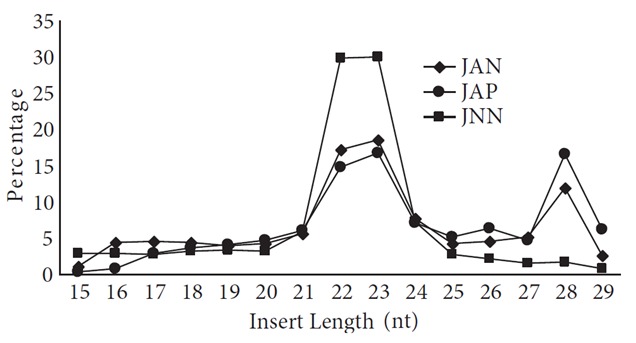
Small RNA profiles of camptothecin-treated Jurkat cells. The 15–29 bp fragments were sorted based on their size and plotted as percentage. JNN: Small RNA population in the drug-free control cells; JAP: camptothecin-treated and Annexin V-positive cells; JAN: camptothecin-treated and Annexin V-negative cells.
3.2. Apoptosis is regulated by differential miRNA expression
The alignment of reads to the known miRNAs in miRBase (R18) resulted in identification of 184 miRNAs differentially expressed among the three samples (Table 1). Our list includes the differentially expressed miRNAs, whose expression are greater than 10 reads per million (RPM) in all three samples. uThs, the number of differentially expressed genes could be greater. Camptothecin treatment usually suppresses miRNA expression compared to the control cells (Table 2: 38 induced miRNAs versus 144 downregulated miRNAs in the JAN or JAP samples). We identified a single miRNA, miR-1246, overexpressed in the camptothecin-treated JAN and JAP samples. However, a total of 79 miRNAs were downregulated in response to the camptothecin treatment. Interestingly, 16 and 30 miRNAs were down- and upregulated in the drug-resistant JAP sample, respectively, whereas they were equally expressed in the drug-sensitive JAP sample. More interestingly, a total of 6 miRNAs (let-7b-5p, miR-15a-5p, 324-5p, 128, 425, and 720) were reciprocally expressed in the drugsensitive and resistant cells. To validate our findings from deep sequencing, we randomly chose 7 miRNAs for validation by real-time qPCR, namely miR-7, 17, 18a, 25, 26a, 93, and 425. As shown in Figure 4, the qPCR results were quite consistent with the deep-sequencing data.
Table 1.
A list of miRNAs deregulated in camptothecin-treated Jurkat cells. The cloning frequency of miRNAs was calculated as reads per million (RPM) following the removal of the adapter sequences. The miRNAs with a cloning frequency of 10 RPM in all three samples were used in calculations. The fold of expression is presented in a log ratio. JNN: Drug-free control cells; JAN: Annexin V-negative fraction of drug-treated cells; JAP: Annexin V-positive fraction of drug-treated cells.
| Upregulated in JAN | Suppressed in JAN vs JNN | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| miRNA | JAN/JNN | JAP/JNN | JAP/JAN | miRNA | JAN/JNN | JAP/JNN | JAP/JAN | ||||
| hsa-miR-128 | 4.82 | –2.94 | –7.75 | hsa-miR-19b-3p | –1.63 | –4.34 | –2.71 | ||||
| hsa-miR-17-3p | 7.14 | 0.51 | –6.62 | hsa-miR-19a-3p | –1.73 | –4.31 | –2.59 | ||||
| hsa-miR-720 | 1.74 | –3.42 | –5.15 | hsa-miR-17-5p | –1.59 | –3.87 | –2.28 | ||||
| hsa-miR-7-5p | 3.53 | –0.52 | –4.05 | hsa-miR-20b-5p | –1.10 | –3.23 | –2.13 | ||||
| hsa-miR-1268b | 3.79 | –0.26 | –4.05 | hsa-miR-210 | –1.91 | –3.97 | –2.06 | ||||
| hsa-miR-1268a | 3.77 | –0.26 | –4.04 | hsa-miR-374a-5p | –1.09 | –2.93 | –1.84 | ||||
| hsa-miR-25-3p | 3.49 | –0.41 | –3.90 | hsa-miR-320a | –2.57 | –4.29 | –1.72 | ||||
| hsa-let-7b-5p | 1.60 | –2.19 | –3.79 | hsa-miR-29b-3p | –5.01 | –6.44 | –1.43 | ||||
| hsa-miR-324-5p | 1.87 | –1.85 | –3.72 | hsa-let-7d-5p | –1.95 | –3.29 | –1.34 | ||||
| hsa-miR-106b-3p | 2.76 | –0.86 | –3.62 | hsa-miR-185-5p | –1.27 | –2.34 | –1.08 | ||||
| hsa-miR-101-3p | 2.62 | –0.66 | –3.28 | hsa-miR-484 | –1.83 | –2.77 | –0.94 | ||||
| hsa-miR-221-3p | 3.17 | –0.07 | –3.23 | hsa-miR-150-3p | –1.02 | –1.86 | –0.83 | ||||
| hsa-miR-331-3p | 2.85 | –0.36 | –3.21 | hsa-miR-20b-3p | –1.34 | –2.14 | –0.80 | ||||
| hsa-miR-15a-5p | 1.88 | –1.09 | –2.97 | hsa-miR-106a-5p | –2.50 | –3.24 | –0.74 | ||||
| hsa-miR-28-3p | 1.75 | –0.95 | –2.70 | hsa-miR-153 | –1.56 | –2.22 | –0.66 | ||||
| hsa-let-7d-3p | 1.52 | –0.99 | –2.51 | hsa-miR-542-5p | –1.77 | –2.43 | –0.65 | ||||
| hsa-miR-16-2-3p | 1.62 | –0.68 | –2.30 | hsa-miR-1301 | –2.37 | –3.01 | –0.64 | ||||
| hsa-miR-21-5p | 1.63 | –0.63 | –2.26 | hsa-miR-148a-5p | –1.35 | –1.87 | –0.53 | ||||
| hsa-miR-3613-5p | 2.02 | –0.21 | –2.23 | hsa-miR-454-3p | –1.35 | –1.85 | –0.49 | ||||
| hsa-miR-181d | 1.87 | –0.33 | –2.20 | hsa-miR-26b-5p | –3.17 | –3.58 | –0.41 | ||||
| hsa-miR-362-5p | 2.13 | –0.07 | –2.20 | hsa-miR-181b-3p | –1.20 | –1.54 | –0.34 | ||||
| hsa-miR-548d-5p | 1.66 | –0.50 | –2.16 | hsa-miR-191-5p | –4.24 | –4.57 | –0.33 | ||||
| hsa-miR-301a-5p | 1.54 | –0.37 | –1.92 | hsa-miR-301b | –1.86 | –2.17 | –0.31 | ||||
| hsa-miR-186-3p | 1.68 | –0.17 | –1.84 | hsa-miR-20a-5p | –3.54 | –3.83 | –0.30 | ||||
| hsa-miR-1307-3p | 1.08 | –0.63 | –1.71 | hsa-miR-222-3p | –1.69 | –1.93 | –0.24 | ||||
| hsa-miR-671-3p | 1.07 | –0.54 | –1.61 | hsa-miR-30c-1-3p | –2.58 | –2.81 | –0.22 | ||||
| hsa-miR-32-3p | 1.30 | –0.27 | –1.57 | hsa-miR-590-3p | –1.19 | –1.41 | –0.21 | ||||
| hsa-miR-378d | 1.32 | –0.03 | –1.35 | hsa-miR-183-5p | –1.08 | –1.29 | –0.21 | ||||
| hsa-miR-589-5p | 1.01 | –0.30 | –1.31 | hsa-miR-342-5p | –1.13 | –1.30 | –0.17 | ||||
| hsa-miR-4677-3p | 1.13 | –0.16 | –1.28 | hsa-miR-545-5p | –1.03 | –1.19 | –0.17 | ||||
| hsa-miR-548w | 1.35 | 0.09 | –1.25 | hsa-miR-625-3p | –1.34 | –1.51 | –0.16 | ||||
| hsa-miR-378c | 1.14 | –0.09 | –1.23 | hsa-miR-30b-3p | –2.59 | –2.71 | –0.12 | ||||
| hsa-miR-29b-1-5p | 1.06 | 0.00 | –1.06 | hsa-miR-146b-5p | –1.84 | –1.92 | –0.07 | ||||
| hsa-miR-193a-5p | 1.75 | 0.69 | –1.06 | hsa-miR-4454 | –1.13 | –1.19 | –0.06 | ||||
| hsa-miR-424-3p | 1.20 | 0.21 | –0.99 | hsa-miR-503 | –1.61 | –1.65 | –0.04 | ||||
| hsa-miR-1246 | 1.86 | 2.73 | 0.87 | hsa-miR-99b-5p | –1.13 | –1.14 | –0.01 | ||||
| hsa-miR-106a-3p | –1.04 | –1.04 | 0.00 | ||||||||
| hsa-miR-1285-3p | –1.39 | –1.39 | 0.00 | ||||||||
| Equal in JAN vs. JNN | hsa-miR-331-5p | –1.78 | –1.78 | 0.00 | |||||||
| miRNA | JAN/JNN | JAP/JNN | JAP/JAN | hsa-miR-28-5p | –1.12 | –1.08 | 0.03 | ||||
| hsa-miR-92a-1-5p | –0.87 | –4.54 | –3.67 | hsa-miR-19b-1-5p | –2.85 | –2.80 | 0.05 | ||||
| hsa-miR-26a-5p | –0.35 | –3.69 | –3.35 | hsa-miR-1277-3p | –3.02 | –2.97 | 0.05 | ||||
| hsa-miR-18a-5p | –0.41 | –3.62 | –3.21 | hsa-miR-96-5p | –2.39 | –2.34 | 0.05 | ||||
| hsa-miR-301a-3p | 0.86 | –2.17 | –3.03 | hsa-miR-744-3p | –1.24 | –1.17 | 0.07 | ||||
| hsa-miR-125a-5p | 0.69 | –2.26 | –2.94 | hsa-miR-29a-3p | –3.01 | –2.94 | 0.07 | ||||
| hsa-miR-18b-5p | 0.59 | –2.15 | –2.74 | hsa-miR-532-5p | –1.20 | –1.12 | 0.08 | ||||
| hsa-let-7a-5p | 0.33 | –2.18 | –2.52 | hsa-miR-26b-3p | –1.28 | –1.19 | 0.10 | ||||
| hsa-miR-296-3p | –0.01 | –2.48 | –2.47 | hsa-let-7g-3p | –1.98 | –1.88 | 0.11 | ||||
| hsa-miR-27b-3p | 0.71 | –1.74 | –2.45 | hsa-miR-130b-3p | –4.70 | –4.58 | 0.12 | ||||
| hsa-miR-148a-3p | –0.59 | –2.86 | –2.27 | hsa-miR-140-3p | –1.46 | –1.33 | 0.13 | ||||
| hsa-miR-374b-5p | 0.44 | –1.81 | –2.26 | hsa-miR-16-1-3p | –2.84 | –2.69 | 0.15 | ||||
| hsa-miR-92a-3p | 0.53 | –1.68 | –2.21 | hsa-miR-151a-5p | –4.74 | –4.55 | 0.19 | ||||
| hsa-miR-744-5p | 0.46 | –1.68 | –2.14 | hsa-miR-424-5p | –4.79 | –4.59 | 0.20 | ||||
| hsa-miR-150-5p | 0.61 | –1.43 | –2.04 | hsa-miR-223-5p | –1.71 | –1.50 | 0.21 | ||||
| hsa-miR-106b-5p | –0.40 | –2.38 | –1.98 | hsa-miR-30b-5p | –2.58 | –2.34 | 0.24 | ||||
| hsa-miR-191-3p | –0.03 | –1.99 | –1.96 | hsa-let-7i-3p | –2.75 | –2.49 | 0.25 | ||||
| hsa-miR-223-3p | 0.50 | –1.42 | –1.92 | hsa-miR-197-3p | –2.08 | –1.80 | 0.28 | ||||
| hsa-miR-18a-3p | –0.35 | –2.11 | –1.76 | hsa-let-7i-5p | –1.92 | –1.62 | 0.30 | ||||
| hsa-miR-92b-3p | 0.20 | –1.54 | –1.75 | hsa-miR-30d-3p | –1.37 | –1.07 | 0.30 | ||||
| hsa-miR-29c-3p | –0.66 | –2.41 | –1.74 | hsa-miR-1277-5p | –2.22 | –1.88 | 0.34 | ||||
| hsa-miR-181a-3p | –0.36 | –1.89 | –1.52 | hsa-miR-194-5p | –2.13 | –1.76 | 0.38 | ||||
| hsa-let-7e-5p | –0.72 | –2.24 | –1.52 | hsa-miR-627 | –2.09 | –1.72 | 0.38 | ||||
| hsa-miR-219-1-3p | 0.53 | –0.93 | –1.46 | hsa-miR-200c-3p | –2.41 | –1.96 | 0.45 | ||||
| hsa-miR-27a-3p | –0.14 | –1.59 | –1.45 | hsa-miR-500a-5p | –1.05 | –0.57 | 0.48 | ||||
| hsa-let-7c | 0.22 | –1.21 | –1.43 | hsa-miR-93-3p | –1.42 | –0.90 | 0.52 | ||||
| hsa-miR-98 | –0.21 | –1.63 | –1.42 | hsa-miR-146a-5p | –3.25 | –2.72 | 0.53 | ||||
| hsa-miR-942 | 0.24 | –1.17 | –1.41 | hsa-miR-320b | –1.52 | –0.94 | 0.57 | ||||
| hsa-miR-7-1-3p | 0.12 | –1.25 | –1.38 | hsa-miR-9-3p | –1.39 | –0.73 | 0.66 | ||||
| hsa-let-7f-5p | 0.10 | –1.19 | –1.29 | hsa-miR-374a-3p | –2.08 | –1.42 | 0.66 | ||||
| hsa-miR-181a-2-3p | 0.31 | –0.98 | –1.28 | hsa-miR-320c | –1.59 | –0.88 | 0.71 | ||||
| hsa-miR-652-3p | 0.44 | –0.78 | –1.22 | hsa-miR-450b-5p | –1.78 | –1.04 | 0.74 | ||||
| hsa-miR-548b-5p | 0.90 | –0.31 | –1.21 | hsa-miR-320d | –1.04 | –0.24 | 0.79 | ||||
| hsa-miR-9-5p | 0.57 | –0.61 | –1.18 | hsa-miR-542-3p | –1.44 | –0.62 | 0.83 | ||||
| hsa-miR-93-5p | 0.09 | –1.08 | –1.17 | hsa-miR-629-3p | –1.24 | –0.38 | 0.86 | ||||
| hsa-miR-760 | 0.81 | –0.33 | –1.14 | hsa-miR-148b-3p | –3.28 | –2.36 | 0.92 | ||||
| hsa-miR-184 | 0.96 | –0.15 | –1.12 | hsa-miR-340-3p | –1.36 | –0.44 | 0.93 | ||||
| hsa-miR-151a-3p | –0.30 | –1.32 | –1.01 | hsa-miR-339-5p | –1.34 | –0.27 | 1.07 | ||||
| hsa-miR-95 | 0.01 | –1.00 | –1.01 | hsa-miR-345-5p | –1.41 | –0.30 | 1.11 | ||||
| hsa-miR-152 | 0.12 | –0.88 | –1.01 | hsa-miR-342-3p | –2.72 | –1.60 | 1.11 | ||||
| hsa-miR-501-3p | –0.40 | –1.26 | –0.86 | hsa-miR-126-3p | –1.96 | –0.78 | 1.18 | ||||
| hsa-miR-27b-5p | –0.29 | –1.07 | –0.78 | hsa-miR-193b-3p | –2.05 | –0.82 | 1.23 | ||||
| hsa-miR-15b-3p | –0.55 | –1.33 | –0.77 | hsa-miR-363-3p | –2.82 | –1.55 | 1.27 | ||||
| hsa-miR-30e-3p | –0.40 | –1.13 | –0.73 | hsa-miR-361-5p | –6.52 | –5.24 | 1.28 | ||||
| hsa-miR-203 | –0.36 | –1.04 | –0.69 | hsa-miR-126-5p | –2.77 | –1.42 | 1.36 | ||||
| hsa-miR-340-5p | –0.82 | –1.34 | –0.52 | hsa-miR-769-5p | –3.28 | –1.87 | 1.41 | ||||
| hsa-miR-196a-5p | –0.99 | –1.39 | –0.40 | hsa-miR-140-5p | –6.36 | –4.74 | 1.61 | ||||
| hsa-miR-625-5p | –0.88 | –1.19 | –0.31 | hsa-miR-30c-5p | –4.80 | –2.90 | 1.90 | ||||
| hsa-miR-580 | –0.85 | –1.13 | –0.29 | hsa-miR-455-3p | –1.37 | 0.66 | 2.03 | ||||
| hsa-miR-454-5p | –0.93 | –1.01 | –0.08 | hsa-let-7g-5p | –6.14 | –3.78 | 2.37 | ||||
| hsa-miR-1292 | –1.00 | –1.02 | –0.02 | hsa-miR-186-5p | –5.45 | –2.80 | 2.65 | ||||
| hsa-miR-1248 | 0.06 | 1.03 | 0.97 | hsa-miR-24-3p | –3.48 | –0.81 | 2.67 | ||||
| hsa-miR-181a-5p | –0.81 | 0.31 | 1.13 | hsa-miR-142-5p | –6.58 | –3.89 | 2.70 | ||||
| hsa-miR-32-5p | –3.12 | –0.11 | 3.01 | ||||||||
| hsa-miR-142-3p | –4.60 | –1.08 | 3.52 | ||||||||
| hsa-miR-766-3p | –4.79 | –1.05 | 3.74 | ||||||||
| hsa-miR-425-5p | –2.48 | 1.74 | 4.22 | ||||||||
Table 2.
A summary of differentially expressed miRNAs and qPCR validation. The dysregulated miRNAs from Table 1 are summarized. JNN: Control Jurkat cells not treated with camptothecin; JAN and JAP: Annexin V-negative and -positive populations of camptothecin-treated cells, respectively; +: induced miRNAs; -: suppressed miRNAs; =: equally expressed miRNAs.
| JAN/JNN | JAN/JNN | JAP/JAN | # |
|---|---|---|---|
| - | - | = | 57 |
| = | - | - | 30 |
| + | = | - | 29 |
| - | - | + | 12 |
| = | - | = | 11 |
| - | - | - | 10 |
| - | = | = | 9 |
| = | = | - | 9 |
| - | = | + | 7 |
| + | - | - | 5 |
| + | + | = | 1 |
| + | = | = | 1 |
| - | + | + | 1 |
| = | + | = | 1 |
| = | = | + | 1 |
Figure 4.
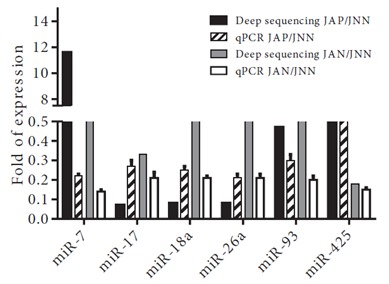
T-98G cells, treated or nontreated with 25 or 50 μM ceranib-2 for 24 (A) or 48 (B) h, then stained with FITC annexin V apoptosis assay kit with PI (Invitrogen). Lower left sections: annexin V/PI ( − ), living cells; lower right sections: annexin V ( + )/PI ( − ), early apoptotic cells; upper left sections: annexin ( − )/PI ( + ), necrotic cells; upper right sections: annexin ( + )/PI ( + ), late stages of apoptosis and secondary necrosis. Results of only one independent experiment out of 3 are depicted.
4. Discussion
The balance between proliferation and apoptosis is important for the overall cellular homeostasis. Apoptosis is particularly important in modulating the fate of immune cells, including T cells. Microarray and deepsequencing studies have been instrumental in identifying several miRNAs involved in cell proliferation or apoptosis (Subramanian and Steer, 2010) . However, these studies were mainly conducted with heterogeneous cancerous tissues in which the apoptotic states of the cells were not synchronized. uThs, we used the Jurkat cell line and the universal apoptosis inducer camptothecin to identify the miRNAs involved in apoptosis. The apoptotic cells were identified by marking the cells in which phosphatidylserine was exposed to the cell surface, which is readily detected by Annexin V labeling. Sorting cells based on their Annexin-V labeling allowed us to obtain the miRNA expression profile of a purely apoptotic cell population (Annexin V/7-AAD-double positive).
MiR-14 and bantam were the first miRNAs shown to have apoptotic function in Drosophila (Brennecke et al., 2003; Xu et al., 2003) . Studies on various cancer cells showed a prominent p53-mediated regulation of miRNAs, particularly the miR-34 family, miR-215 and 192, with proapoptotic capacity (He et al., 2007; Georges et al., 2008) . However, we did not detect any differential expression of these miRNAs in our study. Let-7 and miR15/16 were also reported to be associated with apoptosis (Ghodgaonkar et al., 2009) . MiR-16-1 and let-7d, -7g, and -7i were suppressed in the camptothecin-resistant JAN cells. MiR-16-2 and miR-15a, on the other hand, were slightly upregulated. These miRNAs were suppressed in camptothecin-treated Jurkat cells. The miR-17-92 cluster, which encodes miR-17-3p, -17-5p, -18a, -19a, -20a, -19b, and 92, is one of the well-established miRNA clusters with antiapoptotic activity (He et al., 2007; Georges et al., 2008) . We observed nearly 4- to 5-fold downregulation of these miRNAs in camptothecin-treated Jurkat cells, parallel to the published results. MiR-125 and -128, which are reported to be antiapoptotic miRNAs in Jurkat cells (Yamado et al., 2014; Zhou et al., 2014) , are downregulated in the camptothecintreated cells, as expected. MiR-143, which is expressed at an extremely low level in cancer cell lines compared to normal tissues, is involved in Fas-mediated apoptosis in Jurkat cells (Akao et al., 2009) . Accordingly, this particular miRNA was nearly undetectable in Jurkat cells. However, its expression did not change in response to the camptothecin treatment.
Our study revealed the involvement of additional miRNAs that may modulate apoptosis in human Jurkat T cells (Table 1). The most striking dysregulations were detected in the expression of miR-17*, -128, -140, -142, -161, -186, -766, and-1268. Although these miRNAs are dysregulated in camptothecin-treated Jurkat T cells, we cannot unequivocally state that they are directly involved in modulating apoptotic pathways. It is possible that they may be associated with response to drug treatment, such as drug euflx. Further studies are required to determine their involvement, if any, in apoptosis and related signaling pathways.
Although camptothecin is known to be a potent apoptosis inducer, a portion of the cells were unresponsive to the drug despite over 100-fold increase in the drug concentration (Figures 1A and 1C). This observation was consistent with previous reports that there may be phenotypic and genotypic heterogeneity in clonal cell populations (Stockholm et al., 2007; Walling and Shepard, 2012) . To validate the cellular heterogeneity in the Jurkat cell line, we sorted the Annexin-V-positive cells (JAP) from the Annexin-V-negative cells (JAN) following the initial drug treatment and retreated the Annexin-V-negative, apoptosis-resistant cells with camptothecin. Although the inclusion of the Annexin-V-positive fraction of the untreated control cells (JNP) would provide valuable insight into the potential role of miRNAs in natural or accidental death (i.e. drug-independent), the number of these cells was always insuficient for downstream experiments. uThs, we focused on the drug-treated samples. Interestingly, some of the drug-tested cells were intrinsically resistant to the drug (Figure 1C and Figure 2). To investigate whether the differential apoptotic response of each subpopulation is associated with the differential miRNA expression, we subjected the total RNAs from each subpopulation to deep sequencing using the untreated cells as control. There were 79 miRNAs commonly downregulated in both populations (Table 2). Quite interestingly, although 16 and 30 miRNAs were down- and upregulated in the drug-resistant JAN sample, respectively, they were equally expressed in the drug-sensitive JAP sample. More interestingly, a total of 6 miRNAs (let-7b-5p, miR-15a-5p, 324-5p, 128, 425, and 720) were reciprocally expressed in the drug-sensitive JAP and resistant JAN subpopulations (Table 1). Pending the direct demonstration of their role in cell-specific response to drugs, this observation suggests that the miRNA expression data obtained from drug treatments should be carefully interpreted if the cells of the desired phenotype are not sorted from the other cells. The presence of cells with undesired phenotypes may introduce noise that may interfere with the accurate assessment of the phenotypespecific miRNA expression profiles.
We then carried out gene ontology analyses using DIANA to examine which biological processes could be affected by the miRNAs inversely expressed in the JAN and JAP samples (Vlachos et al., 2015) . MiR-720 was shown to originate from a tRNA (Schopman et al., 2010) . Thus, we excluded this miRNA in our gene ontology analyses. Of the other vfie miRNAs, let-7b-5p targets modulate the cell cycle, chronic myeloid leukemia, p53 signaling, hippo signaling, and thyroid hormone signaling. MiR-15 targets are involved in the cell cycle, pathways in cancer, and several signaling pathways such as p53, TGF-beta, hippo, and thyroid hormone. MiR128 target genes are interesting in that they typically modulate extracellular matrix organization, focal adhesion, or proteoglycan in cancer. MiR-324 has binding sites on mRNAs whose biological functions include hippo signaling, adherence junction, and miRNAs in cancer while miR-425 target genes function in N-glycan biosynthesis, adherence junction, thyroid hormone, and hippo signaling. From these analyses, it is possible to state that the target genes of these vfie miRNAs appear to modulate common biological processes, such as the cell cycle, focal adhesion, and several key signaling pathways, such as p53, hippo, and thyroid hormones. Interestingly, recent studies demonstrate, for example, the importance of cell-to-cell variation in p53 expression in inducing apoptosis (Paek et al., 2016) . Further studies would be required, however, to directly show whether these biological processes modulate cellular heterogeneity through the differentially expressed miRNAs reported in our study.
Acknowledgments
The authors wish to thank İYTE BİYOMER for the instrumental support. This research was supported by TÜBİTAK grant 107T475 to BA.
References
- Akao Y , Nakagawa Y , Iio A , Naoe T ( 2009. ). Role of microRNA-143 in Fas-mediated apoptosis in human T-cell leukemia Jurkat cells . Leukemia Res 33 : 1530 - 1538 . [DOI] [PubMed] [Google Scholar]
- Bartel DP ( 2004. ). MicroRNAs: genomics, biogenesis, mechanism, and function . Cell 116 : 281 - 297 . [DOI] [PubMed] [Google Scholar]
- Bartel DP ( 2009. ). MicroRNAs: target recognition and regulatory functions . Cell 136 : 215 - 233 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann S , Krueger A , Kirchhof S , Krammer PH ( 2002. ). Regulation of T cell apoptosis during the immune response . Curr Mol Med 2 : 257 - 272 . [DOI] [PubMed] [Google Scholar]
- Benchaouir R ( 2004. ). Evidence for a resident subset of cells with SP phenotype in the C2C12 myogenic line: a tool to explore muscle stem cell biology . Exp Cell Res 294 : 254 - 268 . [DOI] [PubMed] [Google Scholar]
- Blagosklonny MV ( 2000. ). Cell death beyond apoptosis . Leukemia 14 : 1502 - 1508 . [DOI] [PubMed] [Google Scholar]
- Blank M , Shiloh Y ( 2007. ). Programs for cell death: apoptosis is only one way to go . Cell Cycle 6 : 686 - 695 . [DOI] [PubMed] [Google Scholar]
- Brennecke J , Hipfner DR , Stark A , Russell RB , Cohen SM ( 2003. ). Bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila . Cell 113 : 25 - 36 . [DOI] [PubMed] [Google Scholar]
- Calin GA , Croce CM ( 2009. ). Chronic lymphocytic leukemia: interplay between noncoding RNAs and protein-coding genes . Blood 114 : 4761 - 4770 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J , Odenike O , Rowley JD ( 2010. ). Leukaemogenesis: more than mutant genes . Nat Rev Cancer 10 : 23 - 36 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Hoon MJL , Taft, RJ , Hasimoto T , Kanamori-Katayama M , Kawaji H , Kawano M , Kishima M , Lassmann T , Faulkner GJ , Faulkner GJ et al. ( 2010. ). Cross-mapping and the identification of editing sites in mature microRNAs in high-throughput sequencing libraries . Genome Res 20 : 257 - 264 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan SJ , Li HB , Cui G , Kong XL , Sun LL , Zhao YQ , Li YH , Zhou J ( 2016. ). miRNA- 149 * promotes cell proliferation and suppresses apoptosis by mediating JunB in T-cell acute lymphoblastic leukemia . Leuk Res 41 : 62 - 70 . [DOI] [PubMed] [Google Scholar]
- Georges SA , Biery MC , Kim SY , Schelter JM , Guo J , Chang AN , Jackson AL , Carleton MO , Linsley PS , Cleary MA et al. ( 2008. ). Coordinated regulation of cell cycle transcripts by p53- inducible microRNA , miR-192 and miR-215. Cancer Res 68 : 10105 - 10112 . [DOI] [PubMed] [Google Scholar]
- Ghodgaonkar MM , Shah RG , Kandan-Kulangara F , Afar EB , Qi HH , Wiemer E , Shah GM ( 2009. ). Abrogation of DNA vector-based RNAi during apoptosis in mammalian cells due to caspasemediated cleavage and inactivation of Dicer-1 . Cell Death Diefr 16 : 858 - 868 . [DOI] [PubMed] [Google Scholar]
- He L , He X , Lim LP , de Stanchina E , Xuan Z , Liang Y , Xue W , Zender L , Magnus J , Ridzon D et al. ( 2007. ). A microRNA component of the p53 tumour suppressor network . Nature 447 : 1130 - 1134 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar MS , Lu J , Mercer KL , Golub TR , Jacks T ( 2007. ). Impaired microRNA processing enhances cellular transformation and tumorigenesis . Nat Genet 39 : 673 - 677 . [DOI] [PubMed] [Google Scholar]
- Lawrie CH ( 2007. ). MicroRNAs and hematology: small molecules, big function . Brit J Haematol 137 : 503 - 512 . [DOI] [PubMed] [Google Scholar]
- Li TK , Liu LF ( 2001. ). Tumor cell death induced by topoisomerasetargeting drugs . Annu Rev Pharmacol 41 : 53 - 77 . [DOI] [PubMed] [Google Scholar]
- Li XJ , Luo XQ , Han BW , Duan FT , Wei PP , Chen YQ ( 2013. ). MicroRNA- 100 /99a, dysregulated in acute lymphoblastic leukaemia, suppress proliferation and promote apoptosis by regulating the FKBP51 and IGF1R/mTOR signaling pathways . Brit J Cancer 109 : 2189 - 2198 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodish HF , Zhou B , Liu G , Chen CZ ( 2008. ). Micromanagement of the immune system by miRNAs . Nature Rev 8 : 120 - 130 . [DOI] [PubMed] [Google Scholar]
- Lu J , Zhang X , Zhang R , Ge Q ( 2015. ). MicroRNA heterogeneity in endometrial cancer cell lines revealed by deep sequencing . Oncol Letters 10 : 3457 - 3465 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcucci G , Mrozek K , Radmacher MD , Garzon R , Bloomfield CD ( 2011. ). The prognostic and functional role of microRNAs in acute myeloid leukemia . Blood 117 : 1121 - 1129 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miska EA ( 2005. ). How microRNAs control cell division, differentiation and death . Curr Opinion Genet Dev 15 : 563 - 568 . [DOI] [PubMed] [Google Scholar]
- Mott JL , Kobayashi S , Bronk SF , Gores GJ ( 2007. ). Mir-29 regulates Mcl-1 protein expression and apoptosis . Oncogene 26 : 6133 - 6140 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell R , Kahn D , Gibson WSJ , Round JL , Schotz RL , Chadhuri AA , Kahn ME , Rao DS , Baltimore D ( 2010. ). MicroRNA-155 promotes autoimmune inflammation by enhancing inflammatory T cell development . Immunity 33 : 607 - 619 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paek AL , Liu JC , Loewer A , Forrester WC , Lahav G ( 2016. ). Cell-tocell variation in p53 dynamics leads to fractional killing . Cell 165 : 631 - 642 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pekarsky Y , Santanam U , Cimmino A , Palamarchuk A , Efanov A , Maximov V , Volinia S , Alder H , Liu CG , Rassenti L et al. ( 2006. ). Tcl1 expression in chronic lymphocytic leukemia is regulated by miR- 29 and miR-181. Cancer Res 66 : 24 . [DOI] [PubMed] [Google Scholar]
- Pommier Y , Redon C , Rao VA , Seiler JA , Sordet O , Takemura H , Antony S , Meng L , Liao Z , Kohlhagen G et al. ( 2003. ). Repair of and checkpoint response to topoisomerase I-mediated DNA damage . Mutat Res 532 : 173 - 203 . [DOI] [PubMed] [Google Scholar]
- Schopman NC , Heynen S , Haasnoot J , Berkhout , B ( 2010. ). A miRNA-tRNA mix-up: tRNA origin of proposed miRNA . RNA Biol 7 : 573 - 576 . [DOI] [PubMed] [Google Scholar]
- Snow K , Judd W ( 2009. ). Heterogeneity of a human T-lymphoblastoid cell line . Exp Cell Res 171 : 389 - 403 . [DOI] [PubMed] [Google Scholar]
- Stefani G , Slack FJ ( 2008. ). Small non-coding RNAs in animal development . Nature Rev 9 : 219 - 230 . [DOI] [PubMed] [Google Scholar]
- Stockholm D , Benchaouir R , Picot J , Maneau P , Neildez T , Landini G , Laplace-Builhe C , Paldi A ( 2007. ). The origin of phenotypic heterogeneity in a clonal cell population in vitro . PLoS One 4 : e394 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Z , Yang Z , Xu Y , Chen Y , Yu Q ( 2015. ). MicroRNAs in apoptosis, autophagy and necroptosis . Oncotarget 6 : 8474 - 8490 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian S , Steer CF ( 2010. ). MicroRNAs as gatekeepers of apoptosis . J Cell Physiol 223 : 289 - 98 . [DOI] [PubMed] [Google Scholar]
- Vaz C , Ahmad HM , Sharma P , Gupta R , Kumar L , Kulshreshtha R , Bhattacharya A ( 2010. ). Analysis of microRNA transcriptome by deep sequencing of small RNA libraries of peripheral blood . BMC Genomics 11 : 288 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlachos IS , Zagganas K , Parashevopoulou MD , Georgakilas G , Karagkouni D , Vergoulis T , Dalamagas T , Hatzigeorgiou AG ( 2015. ). DIANA-miRPath v3. . 0: deciphering microRNA function with experimental support . Nucleic Acids Res 43 : W460 - W466 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walling MA , Shepard JRE ( 2012. ). Cellular heterogeneity and live cell arrays . Chem Soc Rev 40 : 4049 - 4076 . [DOI] [PubMed] [Google Scholar]
- Xiao C , Srinivasan L , Calado DP , Patterson HC , Zhang B , Wang J , Henderson JM , Kutok JL , Rajewsky K ( 2008. ). Lymphoproliferative disease and autoimmunity in mice with increased miR-17-92 expression in lymphocytes . Nat Immunol 9 : 405 - 414 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P , Vernooy SY , Guo M , Hay BA ( 2003. ). The Drosophila microRNA mir-14 suppresses cell death and is required for normal fat metabolism . Curr Biol 13 : 790 - 795 . [DOI] [PubMed] [Google Scholar]
- Yamada N , Noquichi S , Kumazaki M , Shinohara H , Miki K , Naoe T , Akao Y ( 2014. ). Epigenetic regulation of microRNA-128a expression contributes to the apoptosis-resistance of human T-cell Jurkat cells by modulating expression of fas-associated protein with death domain (FADD) . Biochim Biophys Acta 1843. : 590 - 602 . [DOI] [PubMed]
- Zanette DL , Rivadavia F , Molfetta GA , Barbuzano FG , Proto-Siqueira R , Falcão RP , Zago MA , Silva-Jr WA ( 2007. ). MiRNA expression profiles in chronic lymphocytic and acute lymphocytic leukemia . Brazilian J Med Biol Res 40 : 1435 - 1440 . [DOI] [PubMed] [Google Scholar]
- Zhou L , Bai H , Wang C , Wei D , Qin Y , Xu U ( 2014. ). MicroRNA125b promotes leukemia cell resistance to daunorubicin by inhibiting apoptosis . Mol Med Rep 9 : 1909 - 1916 . [DOI] [PubMed] [Google Scholar]


