NobZIP1 governs the expression of various key genes, leading to lipid overproduction and secretion in Nannochloropsis oceanica.
Abstract
Commercialization of algal lipids and biofuels is still impractical owing to the unavailability of lipogenic strains and lack of economically viable oil extraction strategies. Because lipogenesis is governed by multiple factors, success in generating industrial-suitable algal strains using conventional strategies has been limited. We report the discovery of a novel bZIP1 transcription factor, NobZIP1, whose overexpression results in a remarkable elevation of lipid accumulation and lipid secretion in a model microalga Nannochloropsis oceanica, without impairing other physiological properties. Chromatin immunoprecipitation–quantitative PCR analysis revealed that the key genes up- and down-regulated by NobZIP1 are involved in lipogenesis and cell wall polymer synthesis, respectively, which, in turn, induce lipid overproduction and secretion. Among these regulated genes, UDP-glucose dehydrogenase was shown to alter cell wall composition, thus also boosting lipid secretion. In summary, these results offer a comprehensive strategy for concurrent lipid overproduction and secretion, strongly increasing the commercial potential of microalgae.
INTRODUCTION
Microalgae have emerged as potential biofuel feedstock owing to depleting fossil fuel reserves, burgeoning environmental concerns, and their inherent beneficial characteristics such as higher growth rate, oleaginicity, non-encroachment of arable land, and capability to grow in a wide range of waters (1, 2). Despite these potential characteristics, the commercialization of microalgal lipids and biofuels is constrained by lack of strains with high lipid content and economically feasible methodology for intracellular oil extraction from the cell wall–protected microalgal biomass (3). Disruption of the algal cell wall is essential for efficient oil extraction that has been considered as high energy consuming and expensive process (4). These biological and economic obstacles have to be tackled to achieve economically sustainable microalgal oil production. Consequently, considerable efforts have been devoted to engineer microalgae by targeting various key metabolic nodes such as glycerol-3-phosphate acyltransferase (5), 1-acyl-sn-glycerol-3-phosphate acyltransferase [AGPAT/lysophosphatidic acid acyltransferase (LPAAT)] (6), and diacylglycerol acyltransferase for lipid overproduction (7, 8). Despite copious literature on lipid overproduction by metabolic engineering strategy, the promising candidate has not been exemplified to obviate the major bottlenecks exist in the commercial production of microalgal fuels. Because of the existing intricacies in lipogenesis, feasibility of the potential metabolic target is often overlooked. Another potential strategy for alleviating the expensive algal harvesting and oil extraction is to secrete the desired products directly into the culture medium, thereby obviating the necessity for cell disruption and keeping the continuous production from the cells. Although several nondestructive extraction strategies have been reported (3, 9, 10), these methods may likely not to be applicable for the extraction of desired products that are not secreted. Hence, there exists a pressing need to engineer a strain with the potential to secrete overproduced lipids. So far, algal strain with such characteristics has not been reported.
Availability of completely sequenced genome and in silico identification of transcription factors (TFs) and their binding sites (11) have paved the way to identify potential metabolic targets to enhance the industrial potential in a few microalgal species. Recently, transcriptional engineering has been recruited to regulate transcription of various key genes (12) and, nevertheless, substantial progress has not yet been attained in microalgae. Among other reported TFs, basic-region/leucine zipper (bZIP) TFs are evolutionary conserved family of eukaryotic TFs, and various studies have reported its crucial role in regulating signaling, lipid, and carbohydrate metabolic pathways (13). Previous reports demonstrated the crucial role of shoot apex–expressed bZIP TF in regulating the expression of floral meristem identity gene, AP1, thereby inducing floral transition and flowering in Arabidopsis (14, 15). TF bZIP11 has been reported to govern the transcriptional activation of auxin-responsive genes by recruiting the Spt/Ada/GCN5/acetyltransferase components in Arabidopsis (16). However, there are very few experimental reports on the characterization of bZIP TFs and their functional significance in microalgae.
On the basis of our previous transcriptomic analysis, we identified a bZIP1 TF (designated NobZIP1) in the model microalga Nannochloropsis oceanica and characterized its role by using overexpression and RNA interference (RNAi)–mediated silencing. Mechanistically, NobZIP1 up- and down-regulates several key genes involved in lipid and cell wall carbohydrate metabolism, respectively, which, in turn, lead to concomitant lipid overproduction and oil secretion across the weakened cell wall in the NobZIP1-overexpressing cells. Besides, we here uncover that NobZIP1-targeted UDP-glucose dehydrogenase (UGDH) is the predominant protein responsible for cell wall structural changes and lipid secretion in N. oceanica. Collectively, our successful proof of concept exemplifies the discovery of concurrent lipid overproduction and secretion phenomenon in microalgae and an important biotechnological breakthrough in economical feasible microalgal biofuel production.
RESULTS
Sequence analysis of NobZIP1 and expression vector construction
We annotated NobZIP1 in the genome of N. oceanica and predicted the conserved domains of NobZIP1 by using bioinformatic approaches. As shown in fig. S1A, NobZIP1 has fructose-1,6-bisphosphatase and bZIP domains. Phylogenetic analysis revealed that NobZIP1 from N. oceanica and that from N. gaditana were clustered into the same group while distinctly separated from other organisms (fig. S1B). In an effort to characterize the role of NobZIP1 in governing the expression of key metabolic nodes in microalgae, we overexpressed (Fig. 1A) and silenced NobZIP1 by using RNAi strategy (Fig. 1B) in N. oceanica. First, we cloned NobZIP1 fused with a 2×-FLAG tag into the expression vector pNa03 under the control of Hsp20 promoter, which has higher activity in the stationary phase, and the resultant recombinant expression vector (pNa03-NobZIP1) was electroporated into N. oceanica. Putative algal transformants were initially screened by antibiotic resistance and genomic polymerase chain reaction (PCR) (fig. S1C) and further evaluated by molecular approaches such as quantitative PCR (qPCR) and Western blotting. qPCR analysis demonstrated that relative transcript level of NobZIP1 was increased by 2.7- and 2.1-fold in overexpressing cells NobZIP1-1 and NobZIP1-2, respectively, than that of wild type (WT) (fig. S1D). Western blot analysis was performed by using anti-FLAG antibody, which showed that a cross-reacting band of ~95.74 kDa was present in the transformants, which is in accordance with the expected molecular weight of NobZIP1 (fig. S1E). However, WT did not exhibit such bands. These results suggested that NobZIP1 was successfully transcribed and expressed in overexpressing cells.
Fig. 1. Physiological and biochemical analyses of NobZIP1-engineered cells.
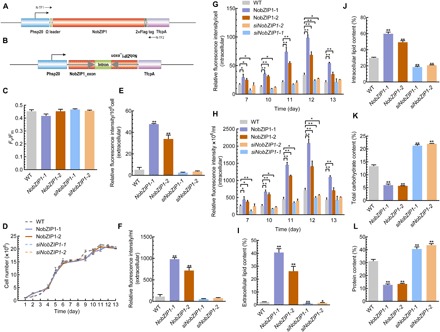
(A) Schematic map of the NobZIP1 overexpression cassette. The NobZIP1-coding region was cloned under the control of an Hsp20 promoter and a TfcpA terminator. An omega leader motif was inserted in between the promoter and NobZIP1 gene to enhance the translation. (B) Schematic map of the hpNobZIP1 expression cassette to silence NobZIP1. (C) Maximum quantum yield of photosystem II (PSII) as measured by Fv/Fm. (D) Growth curve analysis. Relative fluorescence intensity of extracellular lipids (E) per 106 cell and (F) per milliliter, as determined by Nile red fluorescence analysis. Relative fluorescence intensity of intracellular lipids as determined by Nile red fluorescence analysis (G) per cell and (H) fluorescence intensity ×106 per milliliter. Total extracellular (percentage of total cell dry weight) (I) and intracellular (J) lipid content (percentage of total cell dry weight). (K) Total carbohydrate content, as determined by the phenol-sulfuric acid method. (L) Total protein content, as measured by bicinchoninic acid (BCA) assay. Error bars represent mean values ± SD for three separate experiments. Significant difference is indicated at P < 0.05 (*) or P < 0.01 (**) level.
NobZIP1 overexpression altered lipid and carbohydrate content without impairing other physiological properties
Enhancing the desired metabolites without compromising cellular biomass has been considered a crucial factor in large-scale production of microalgal biofuels (17); therefore, we further evaluated growth and photosynthetic efficiency of overexpressing strains. As shown in Fig. 1 (C and D), there was no change observed in terms of photosynthesis and growth, respectively, between overexpressing and WT strains. Maximum quantum yield of photosystem II (PSII) (Fv/Fm), the sensitive method to assess the photosynthetic efficiency of PSII was measured, which revealed that there was no significant change observed in terms of Fv/Fm between WT and overexpressing cells (Fig. 1C). It has been reported that heterologous expression of TF GmDof4 from Glycine max did not affect the growth rate of engineered alga Chlorella ellipsoidea (18). Consistently, TFs have been reported to play a crucial role during adverse conditions in regulating defensive mechanisms and cellular growth and development (19). Despite the previous reports on the role of bZIP TFs on biochemical content, relatively little is explored about its multifaceted role in regulating key metabolic nodes and biochemical composition in microalgae. Hence, we sought to investigate the impact of NobZIP1 overexpression on lipid, carbohydrate, and protein content. In this regard, we first determined the relative lipid content by using Nile red fluorometric analysis and found that lipid content was significantly increased in overexpressing cells than that of WT (Fig. 1, E to H). Further, we qualitatively examined the lipid increment by observing the Nile red–stained cells under laser scanning confocal microscopy. Confocal microscopic analysis of Nile red–stained overexpressing cells revealed that number and volume of oil bodies were concomitantly increased in overexpressing cells and, intriguingly, oil bodies were secreted from cells and numerous oil bodies were observed in the culture medium (fig. S2 and movies S1 and S2), which notably provide additional insights into the previously reported role of bZIP in lipid enhancement (20). Besides, confocal microscopic analysis revealed that there was no physical aberration observed between WT and overexpressing cells, which implied that cells were not damaged and NobZIP1-mediated gene regulation possibly facilitates lipid secretion, which is also consistent with unaltered growth rate of the cells (Fig. 1D). The secretion of lipid droplets in NobZIP1-overexpressing cells through weakened cell wall can be seen in movie S1, which visualized the mobilization of lipid droplets across the cell wall (red fluorescence) and consequent secretion across the cell wall, which was apparently missing in WT. Intrigued by this observation, we further determined the intra- and extracellular lipids by conventional gravimetric analysis. The gravimetric lipidomic analysis confirmed that both intracellular and secreted lipids of overexpressing strains were increased significantly by 1- and 16.2-fold, respectively, than WT (Fig. 1, I and J). Notably, the extracellular lipid content was found to be negligible in WT cells when compared to overexpressing cells (Fig. 1I, fig. S2, and movies S1 and S2), which is in line with our postulation that the lipid secretion is due to the functional consequences of NobZIP1 overexpression. Besides, we also quantified total carbohydrate and protein content in overexpressing cells. Total carbohydrate content was significantly decreased by 1.24-fold (Fig. 1K), and congruently, total protein content was reduced by 1.37-fold (Fig. 1L) in overexpressing cells than WT. Consistent with the previous reports, these results demonstrated that NobZIP1 overexpression redirected energy and carbon precursors toward lipogenesis from protein and carbohydrate synthesis (6, 17, 21). Thereafter, we sought to gain further insight into structural changes of overexpressing cells, and hence, we examined the cell morphology and subcellular changes by transmission electron microscopy (TEM), which revealed significant ultrastructural changes in overexpressing strains (Fig. 2). TEM images of WT showed that oil bodies and other granules were present, and the cell was observed to be surrounded by an organized cell wall (Fig. 2). In contrast, the volume and number of oil bodies were increased remarkably, and few other granules were observed in a typical overexpressing cell compared to WT. The cell walls appeared thinner in overexpressing strains than WT (Fig. 2), and the thickness of the cell wall of WT was observed to be ~81 nm, whereas the thickness of NobZIP1-overexpressing cells was found to be ~8 nm, which is consistent with the confocal analysis of overproduced oil secretion and also compatible with the speculation that the dissolution of cell wall polymer influenced the penetration of alkaline lignin in Eucalyptus (22).
Fig. 2. Ultrastructural analyses of NobZIP1-engineered cells.
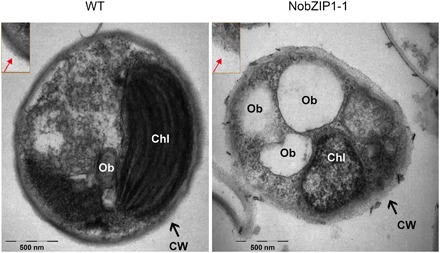
WT cells were encapsulated by rigid cell wall (left), and the enlarged image of cell wall was given in the box; NobZIP1-overexpressing cells exhibited loosen cell wall and highly enriched in oil bodies (middle). Enlarged image of cell wall was given in the box and the alteration in the cell wall structure was indicated by arrow marks. CW, cell wall; Chl, chloroplast; Ob, oil bodies. Scale bars, 500 nm.
To further confirm the regulatory role of NobZIP1 on cellular physiological and biochemical properties of the cell, and also to describe its mechanistic role in lipid production and secretion, we silenced NobZIP1 by using RNAi. The silencing efficiency of the constructed cassette in mutants siNobZIP1-1 and siNobZIP1-2 was evaluated by determining the relative transcript level of NobZIP1 by qPCR, which showed that relative transcript abundance of NobZIP1 was significantly reduced by 2.17- and 3.82-fold in siNobZIP1-1 and siNobZIP1-2, respectively, than WT (fig. S1D). Physiological analysis of NobZIP1-silencing lines revealed no significant difference in terms of photosynthetic and growth parameters when compared to overexpressing and WT cells (Fig. 1, C and D), which is in accordance with the previous report that knockdown of rice bZIP TF did not impair physiological properties under optimal conditions (23).
ChIP analysis demonstrated the unprecedented role of NobZIP1 in governing the expression of key genes in lipid and carbohydrate metabolism
We next sought to elucidate the mechanism underlying concurrent lipid overproduction and secretion in NobZIP1-overexpressing strains. It is well known that TFs play a potential role in regulating the expression of various genes (20). In silico analysis predicted the putative target of bZIP1 TFs in Nannochloropsis (11). However, there is no experimental evidence to determine the impact of NobZIP1 on the regulation of its key target genes, and hence, we carried out chromatin immunoprecipitation (ChIP) to investigate the regulatory role of NobZIP1 on the expression of other genes. ChIP analysis revealed that NobZIP1 regulates the expression of key genes involved in fatty acid, lipid, and carbohydrate metabolism, such as ACBP [acyl–coenzyme A (CoA)–binding protein] (24), KAS [3-ketoacyl-acyl carrier protein synthase] (25), LC-FACS (long-chain acyl-CoA synthetase) (26), LPAAT (26), and CPS (putative capsular polysaccharide synthesis gene) (11), and UGDH (27), the key genes involved in cell wall carbohydrate polymer metabolism (Fig. 3, A to F). Thus, it would be of great interest to interrogate the impact of NobZIP1 on transcriptional regulation of those detected target genes by qPCR analysis. qPCR data demonstrated that the relative transcript abundance of key lipid and fatty acid biosynthetic genes (ACBP, KAS, LC-FACS, and LPAAT) was significantly increased, while the expression of CPS and UGDH was reduced (Fig. 3, G to L). We also determined the relative expression of key lipogenic and carbohydrate genes in siNobZIP1 cells, which revealed that NobZIP1 silencing did not induce the expression of those lipogenic genes; meanwhile, expression of carbohydrate genes such as CPS and UGDH was increased (Fig. 3, G to L), thereby providing substantial proof for the novel mechanistic role for NobZIP1 in simultaneously regulating intricate lipid and carbohydrate metabolic nodes.
Fig. 3. ChIP-qPCR analysis.
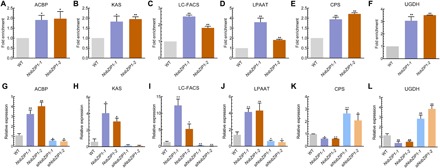
ChIP analysis of (A) ACBP, (B) KAS, (C) LC-FACS, (D) LPAAT, (E) CPS, and (F) UGDH. Each value represents mean ± SD (n = 3). qPCR was carried out to elucidate the regulatory role of NobZIP1 in regulating transcription of predicted key target genes. (G) ACBP, (H) KAS, (I) LC-FACS, (J) LPAAT, (K) CPS, and (L) UGDH. β-Actin was used as an internal reference gene. Significant difference is indicated at P < 0.05 (*) or P < 0.01 (**) level. Each value represents mean ± SD (n = 3).
The NobZIP1-mediated differential expression of lipogenic and carbohydrate genes is in line with the increased and decreased lipid and carbohydrate content, respectively, in overexpressing cells. On the other hand, NobZIP1 silencing resulted in increased lipid content, whereas protein and carbohydrate were found to be increased in silencing strains than that of overexpressing cells (Fig. 1, J to L), which further corroborated that the lipogenic and cell wall carbohydrate metabolic genes could be the target of NobZIP1. Among the differentially expressed genes, LPAAT has been considered as a key enzyme in de novo TAG biosynthetic pathway (6). ACBP, the protein responsible for the formation of acyl-CoA pool and regulation of long-chain fatty acid (LC-FA) metabolism (24), was found to be up-regulated in the engineered cells, which is in accordance with the increment of LC-FAs as shown in fig. S1F. These data uncovered the potential role of NobZIP1 in regulating the differential expression of key genes of lipid and carbohydrate metabolism, thereby ameliorating cells to simultaneously overproduce and secrete lipids without impairing cellular and physiological characteristics.
The distinct role of cell wall carbohydrate metabolic genes in mitigating simultaneous lipid overproduction and secretion
To substantiate the NobZIP1-mediated lipid overproduction and secretion and also to examine whether the reduced carbohydrate metabolic enzymes are responsible for lipid secretion, we next characterized the down-regulated cell wall carbohydrate metabolic genes. To this end, we silenced CPS and UGDH genes individually by using RNAi strategy. The recombinant cassette was cloned into the expression vector pNa03, yielding pNa03-siUGDH and pNa03-SiCPS, which harbor the self-complimentary partial UGDH and CPS genes, thereby generating double-stranded hairpin RNA structure. As determined by qPCR, relative transcript abundance of CPS and UGDH was significantly reduced in siCPS and siUGDH mutants, respectively, than that of WT (fig. S3, A and B). Besides, enzymatic assay of UGDH was found to be significantly reduced by 2.27-fold in silencing cells than WT (fig. S3C). Thereafter, we preliminarily evaluated the lipid secretion capability of these cells by Nile red staining. Unexpectedly, we found that UGDH silencing concomitantly increased the volume and number of lipid droplets and the lipid droplets were found to be secreted extracellularly (fig. S4), whereas silencing of CPS did not result in lipid secretion despite the increased lipid droplets in CPS-silencing cells (fig. S4). These data uncovered that UGDH plays a potential role in lipid secretion rather than CPS and exemplified a potential candidate for lipid metabolic engineering. Hence, we examined the impact of UGDH on microalgal cellular physiology and carbohydrate and lipid content by individually silencing (Fig. 4A) and overexpressing (Fig. 4B) the UGDH gene in N. oceanica. In this regard, to silence UGDH gene, the recombinant vector harbors UGDH gene corresponding to sense and antisense fragment UGDH under the control of promoter Phsp20 (Fig. 4A). Congruently, we cloned UGDH fused with 2×-FLAG tag into the expression vector pNa03 under the control of Hsp20 promoter to overexpress in N. oceanica (Fig. 4B).
Fig. 4. Physiological and biochemical analyses of UGDH-overexpressing and UGDH-silencing cells.
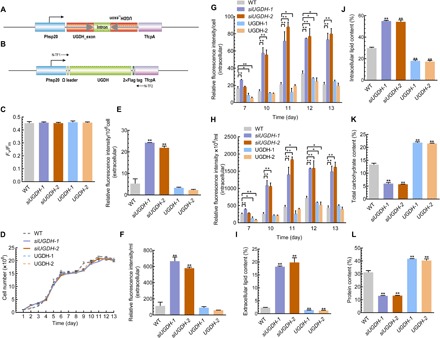
(A) Schematic map of the siUGDH expression cassette to silence NobZIP1. (B) Schematic representation of the UGDH overexpression cassette. The UGDH-coding region was under the control of an Hsp20 promoter and a TfcpA terminator. An omega leader motif was inserted in between the promoter and the UGDH gene to enhance the translation. (C) Maximum photochemical efficiency of PSII, as measured by Fv/Fm. (D) Growth curve analysis. Relative fluorescence intensity of extracellular lipids (E) per 106 cell and (F) per milliliter, as determined by Nile red fluorescence analysis. Relative fluorescence intensity of intracellular lipids, as determined by Nile red fluorescence analysis (G) per cell and (H) fluorescence intensity ×106 per milliliter. Total extracellular (percentage of total cell dry weight) (I) and intracellular (percentage of total cell dry weight) (J) lipid content (%). (K) Total carbohydrate content as determined by the phenol-sulfuric acid method. (L) Total protein content as determined by the BCA method. Error bars represent mean values ± SD for three separate experiments. Significant difference is indicated at P < 0.05 (*) or P < 0.01 (**) level.
UGDH silencing did not affect growth and photosynthetic performance but redirected the carbon flux toward lipid biosynthesis
Given the significance of UGDH on lipid enhancement and secretion, we analyzed the protein sequence by using bioinformatic methods. Domain analysis of UGDH protein reveals that UGDH harbors three domains [amino acids 6 to 195 (dh-N), 214 to 309 (dh), and 332 to 540 (dh-C)], and the region between the first and second domain (amino acids 78 to 231) is responsible for the interaction with malic enzyme and NAD (nicotinamide adenine dinucleotide) (fig. S3D). A phylogenetic tree was constructed by MEGA6 for the deduced amino acid sequences of UGDH from various species (fig. S3E), which revealed significant homology to UGDH from N. gaditana. Perturbing carbohydrate metabolism has been reported to affect growth and photosynthesis. To test whether overexpression and silencing of UGDH affect these parameters in microalgae, we measured growth and photosynthetic efficiency in both overexpressing and silencing cells. Chlorophyll fluorescence parameter (Fv/Fm) was measured to indicate the maximum photochemical efficiency of PSII, which showed no significant changes (Fig. 4C). As shown in Fig. 4D, all strains exhibited similar growth rate, which is consistent with previous reports that document the similar effects of silencing the carbohydrate metabolic pathway genes in microalgae under optimal conditions (28, 29). As carbohydrate and lipid compete for carbon metabolic precursors (28, 30), we determined the lipid content to gain further insights into the role of UGDH silencing on lipid biosynthesis. Nile red fluorometric analysis showed that both intra- and extracellular lipid content in silencing strains were markedly increased, especially extracellular lipid content per milliliter and per 106 cells (Fig. 4, E to H). On the other hand, lipid content in UGDH-overexpressing cells was significantly reduced compared to silencing cells, and notably, extracellular lipid content was negligible in both UGDH-overexpressing and WT cells (Fig. 4, E to H). Consistently, lipid content was found to be gradually increased in UGDH-silencing cells over the cultivation period (Fig. 4, G and H). The extra- and intracellular lipid content in UGDH-silencing cells were further quantified gravimetrically, which showed that both extra- and intracellular lipid content were significantly increased (Fig. 4, I and J). On the other hand, lipid content in UGDH-overexpressing cells was significantly reduced compared to silencing cells (fig. S4), and notably, extracellular lipid content was negligible in both UGDH overexpressing and WT cells (Fig. 4, E to H). In addition, confocal microscopy of UGDH overexpression implied that lipid droplets were significantly reduced than that of UGDH-silencing cells (fig. S4). We further profiled the intracellular fatty acid composition of UGDH-silencing cells by gas chromatography–mass spectrometry (GC-MS), which revealed a significant alteration in fatty acid composition between WT and silencing cells. A significant decrease was found in intracellular total polyunsaturated fatty acids, whereas total saturated fatty acid was found to be increased in silencing cells (fig. S3F). Given the significant role of UGDH in carbohydrate biosynthesis, we determined the total carbohydrate content. UGDH silencing significantly reduced total carbohydrate content by 1.23- and 1.34-fold in siUGDH-1 and siUGDH-2, respectively (Fig. 4K). Meanwhile, UGDH overexpression significantly enhanced carbohydrate content (Fig. 4K). We determined the protein content in both UGDH overexpression and silencing in the stationary phase, which exhibited a similar trend as carbohydrate content. Protein content was significantly decreased and increased in UGDH-silencing and UGDH-overexpressing cells, respectively, compared to WT (Fig. 4L). Consistent with the previous reports, these results suggested the crucial role of UGDH in critically reallocating carbon flux toward lipid and carbohydrate metabolism. Collectively, these data indicate that UGDH silencing did not alter the microalgal growth rate and photosynthetic efficiency significantly, whereas significantly altered carbohydrate and lipid content, which was corroborated by UGDH overexpression.
Ultrastructural analysis demonstrated that UGDH silencing altered the cell wall structure
We next attempted to interrogate the impact of UGDH silencing on cellular morphology and ultrastructure by TEM. TEM showed a notable change in cell wall thickness besides the apparently increased amount of oil bodies. Cell wall of the siUGDH strains was apparently thinner, and the cell wall thickness was observed to be ~27 nm, whereas it was ~81 nm in WT (Fig. 5). Collectively, these analyses demonstrated that UGDH silencing enhanced lipid content while, at the same time, significantly constrained the cell wall formation, corroborating the functional consequence of NobZIP1 in regulating cell wall alteration and consequent lipid secretion.
Fig. 5. Ultrastructural analyses of UGDH-engineered cells by TEM.
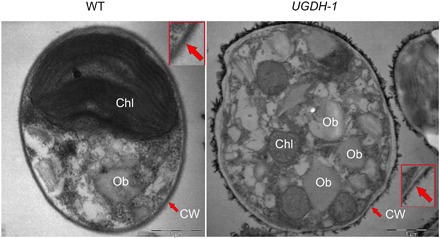
WT cells were encapsulated by rigid cell wall (left), and the enlarged image of cell wall was given in the box; UGDH-silencing cells exhibited thin cell wall and were observed to have large oil bodies than that of WT (right). Enlarged view of cell wall was given in the box, and the alteration in the cell wall structure was indicated by arrow marks. Scale bars, 1 μm.
DISCUSSION
In response to longstanding interest in commercialization of microalgal lipids and biofuels, previous studies have been focused on metabolic engineering to enhance lipid accumulation (17), enzymatic cell wall digestion (31), and on nondestructive lipid extraction strategies to extract algal lipids (3); yet increased lipid yield and economically feasible extraction of unsecreted products still have only limited success. Hence, economically viable lipid production from microalgae remains a major challenge. Here, we demonstrate the novel unprecedented role of TF in regulating a series of key genes involved in lipid and cell wall carbohydrate polymer metabolism, thereby significantly elevating and secreting lipids.
Although NobZIP1 overexpression significantly elevated lipid content, neither overexpression nor silencing of NobZIP1 impaired growth and photosynthetic rate (Fig. 1 C, D, I, and J). TFs have been reported to play a crucial role during adverse conditions in regulating defensive mechanisms and cellular growth and development (19). Consistently, overexpression of NobZIP1 did not impair growth rate, which is pivotal for the large-scale production of economic products from microalgae. Previous studies have demonstrated the functional role of TFs in regulating the transcription of key lipid metabolic genes in oleaginous microalgae; however, the target genes and their expression pattern regulated by the corresponding TFs remained largely unknown. The present study reveals that NobZIP1 acts as the regulator of the key genes involved in lipid and carbohydrate metabolism. This study extends the findings of previous research by demonstrating the ability of NobZIP1 to differentially regulate the expression of target genes. Our data on the effects of RNAi-mediated silencing of NobZIP1 in mutants confirm the role of NobZIP1 in regulating lipid accumulation and secretion.
ChIP analysis revealed that NobZIP1 regulates the key genes involved in fatty acid and lipid metabolism such as ACBP (24), KAS (25), LC-FACS (26), LPAAT (6), UGDH (27), and CPS (11) involved in cell wall carbohydrate polymer metabolism. We found that LC-FAs were significantly increased in the secreted fatty acids in overexpressing cells, which is consistent with the previous findings, and our engineered strains with increased ACBP and LC-FACS expression showed increased content of LC-FAs and lipids (24, 26, 32, 33). NobZIP1 negatively regulates the expression of CPS and UGDH, the genes encoding the enzymes for cell wall polymer metabolism, and provides valuable insight into the previously predicted role of NobZIP1 in regulating the lipogenic genes (11). The result is in accordance with the laser scanning confocal microscopy (fig. S4) and ultrastructural analysis (Fig. 5), which revealed the secretion of oil bodies and alteration in the cell wall structure, respectively.
Given the interesting role of NobZIP1 in negatively regulating the expression of key cell wall carbohydrate–synthesizing genes such as CPS and UGDH, we investigated whether this down-regulation resulted in lipid secretion by overexpressing and silencing these genes. First, we silenced these genes individually in the mutant strains by using RNAi and assessed the lipid secretion capability of these cells under laser scanning confocal microscopy, which notably demonstrated the potential role of UGDH in lipid secretion. As UGDH is a key enzyme in the biosynthesis of cell wall polysaccharides (27), UGDH silencing impaired the cell wall polysaccharide formation, thereby performing dual roles in redirecting the carbon flux toward lipogenesis and weakened the cell wall. Relationship between enzymatic degradation of the cell wall polysaccharide and extracellular secretion has been observed previously in plant cells (34–38) but has not been reported in lipogenic microalgae with reference to lipid secretion. These analyses corroborate the fact that NobZIP1-mediated down-regulation of UGDH has structural and functional consequences, which, in turn, facilitate concurrent lipid overproduction and secretion (Fig. 6).
Fig. 6. Schematic representation of the mechanistic role of NobZIP1 in N. oceanica.
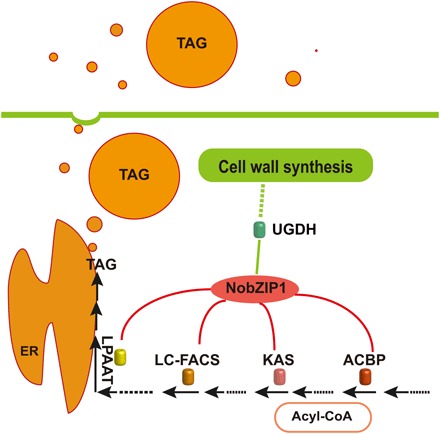
Expression of NobZIP1 up-regulates the expression of KAS, ACBP, LC-FACS, LPAAT (red) and down-regulates the expression of UGDH (green). The targets of NobZIP1 were represented by blue arrow marks, as revealed by ChIP analysis. The role of UGDH on cell wall formation and carbon flux direction toward lipogenesis was revealed by genetic engineering of UGDH in transgenic cells, which substantiate the NobZIP1-mediated concurrent lipid overproduction and secretion in transgenic cells. NobZIP that remarkably up- and down-regulated the target genes are indicated by red and green lines, respectively. TAG, triacylglyceride; ER, endoplasmic reticulum.
In summary, we characterize NobZIP1, a novel regulator of concurrent lipid overproduction and secretion in model microalga N. oceanica and demonstrate its multifaceted role in governing several key genes associated with lipid and carbohydrate metabolism (Fig. 6), thereby up- and down-regulating the expression of key lipogenic and carbohydrate genes, respectively. Ultrastructural analyses reveal that NobZIP1-associated gene regulation alters and weakens the cell wall structure, which, in turn, facilitates lipid secretion from cells. These findings are corroborated by UGDH silencing, the gene which is significantly down-regulated by NobZIP1 that uncovers the potential role of NobZIP1 in governing the intricate lipogenic metabolic circuit in microalgae. Significantly, our data reveal the molecular mechanisms underpinning the secretion of metabolites across the weakened cell wall. Further investigations of this phenomenon would pave the way to explore its feasibility for secreting other valuable products from microalgae, which could even facilitate the use of microalgae for production of various commercial commodities. Together, these findings not only elucidate the unprecedented role of NobZIP1 in regulating key multiple metabolic pathways but also reveal the molecular mechanisms underlying this intricate phenomenon.
MATERIALS AND METHODS
Microalgal culture conditions
N. oceanica CCAP 849/10 (formerly CCMP1779) was obtained from National Center for Marine Algae and Microbiota, USA. Microalgae were cultured in f/2 liquid medium (without Na2SiO3 •9H2O) at 25° ± 1°C in an artificial climate incubator (Ningbo, China), provided with cool white fluorescent illumination of 200 μmol photons m−2 s−1 under a photoperiod of 12/12-hour light/dark cycle. Cell concentration was determined by direct count method using a Brightline hemocytometer.
Cloning and construction of NobZIP1 expression vector
The full-length coding region of NobZIP1 (GenBank: MH118545) and UGDH (GenBank: KU745463) were PCR-amplified from genomic DNA of N. oceanica with primers Na07F and Na08R and UG-OEF and UG-OER, respectively. The amplified PCR products were cloned separately into vector pNa03 downstream of Hsp20 promoter by using the ClonExpress II One Step Cloning Kit (Vazyme, China). FLAG tag was fused at the C terminus of the target genes for the detection of protein expression, which yielded recombinant overexpression vectors pNa03-NobZIP1 and pNa03-UGDH that were electroporated into microalgae using a Gene Pulser Xcell electroporation system (Bio-Rad) as previously described (8). RNAi vector was constructed as described previously (39). Briefly, primer sets were designed to amplify the sense and antisense fragments from the full-length target genes, and subsequently, these fragments were cloned into the pNa03 vector in sense and antisense orientations, respectively, to silence the target gene by using the ClonExpress MultiS One Step Cloning Kit (Vazyme, China) following the supplier’s instruction. The primer sequences for silencing NobZIP1 are as follows: siNobZIPF1 and siNobZIPR1 primers were used to amplify NobZIP1 gene sequence from 2597 to 3017 bp and the primers siNobZIPF2 and siNobZIPR2 were used to amplify the inverted repeats of NobZIP1 from 2800 to 2597 bp, respectively. The primer sequences for silencing UGDH are as follows: siUGDHF1 and siUGDHR1 primers were used to amplify the UGDH sequence from 163 to 662 bp, and the primers siUGDHF2 and siUGDHR2 were used to amplify the inverted repeats of UGDH from 386 to 163 bp, respectively. The primer sequences for silencing CPS are as follows: siCPSF1 and siCPSR1 primers were used to amplify the CPS sequence from 1 to 240 bp, and the primers siCPSF2 and siCPSR2 were used to amplify the inverted repeats of CPS from 240 to 1 bp, respectively. As the CPS gene lacks the intron, we used the first intron of NobZIP1 to generate hairpin loop formation as described (39). Thereafter, these fragments were cloned into the vector pNa03 separately in sense and antisense orientations to silence the respective genes, and the resultant recombinant vectors pNa03-siNobZIP1, pNa03-siUGDH, and pNa03-siCPS were electroporated into microalgae cells using a Gene Pulser Xcell electroporation system (Bio-Rad) as described previously (8).
Evaluation of transformants by molecular approaches
The integration of transgene into the host genome was confirmed by genomic PCR to amplify the backbone of the expression cassette that integrated into the host genome by using the primers in the overexpressing cells (N-TF1 and N-TF2). Besides, genome integration of the expression cassettes to silence the NobZIP1, UGDH, and CPS was confirmed in the silencing cells by genomic PCR amplification to amplify the vector backbone and introns of the respective genes by using the primers siNobZIPF3-siNobZIPR3, siUGDHF3-siUGDHR3, and siCPSF3-siCPSR3, respectively. The efficacy of overexpression and silencing of the target genes in the host cells were initially assessed by real-time qPCR using a SYBR green qPCR SuperMix (Invitrogen, USA) on ABI PRISM 7500 Sequence Detection System (Applied Biosystems, USA). Total RNA was extracted from overexpressing strains (NobZIP1 and UGDH), silencing cells (siNobZIP1, siUGDH, and siCPS), and WT by using RNAiso Plus (Takara, Japan). The prepared RNA was served as a template for first-strand complementary DNA synthesis using the PrimeScript RT Reagent Kit (Takara, Japan) according to the manufacturer’s instruction. The qPCR was performed in a 96-well reaction plate in a total reaction volume of 20 μl, according to the manufacturer’s specification (Applied Biosystems, USA) by using the primers Q-TF1 and Q-TF2, Q-Cs1 and Q-Cs2, and Q-UG1 and Q-UG2 to determine the transcripts of NobZIP1, CPS, and UGDH, respectively. Tubulin gene was used as the internal control with a forward (tublinF) and reverse primer (tublinR). The threshold cycle (Ct) value in each sample was normalized by using the corresponding internal reference gene. The list of primers and their sequences used in this study are given in table S1. To examine the expression of target proteins in the overexpressing cells, Western blotting was performed using anti-Flag antibody (1:3000; Sigma-Aldrich, USA) against the recombinant Flag-tagged NobZIP1, as described previously (17). The anti-actin antibody was used as a reference (1:3000; Sigma-Aldrich, USA). Horseradish peroxidase–conjugated goat anti-rabbit antibody (CST, USA) at a dilution of 1:5000 was used as a secondary antibody. β-Actin protein was used as an internal control. Membrane development was performed using chemiluminescent system. Further, enzymatic activity of UGDH was determined by using the sandwich enzyme-linked immunosorbent assay kit for UGDH following the manufacturer’s instruction (Jialai, China).
Analysis of growth rate and photosynthetic efficiency
The engineered microalgae were cultivated in fresh medium for several times to avoid the influence of antibiotic zeocin. Thereafter, engineered and WT cells were harvested at 3000g for 10 min and resuspended in fresh f/2 medium without antibiotic, and algal cell concentration was made up to 1 × 106 cells/ml. Growth curve was determined by counting cells with a Brightline hemocytometer under a light microscope every day. Both overexpressed and WT cells on 11th day were incubated in dark for at least 40 min, and thereafter, the chlorophyll fluorescence (Fv/Fm; the maximum photochemical efficiency of PSII) was measured with a TD-700 fluorometer (Turner Design, USA) (17).
Analysis of neutral lipids and fatty acid composition
Lipid content of the microalgae was analyzed by both fluorometric and gravimetric analyses. Extracellular and intracellular relative neutral lipid content was determined by fluorescent dye Nile red staining as described (17) with slight modifications. Briefly, microalgae were harvested, and both the pellet and the supernatant were separated. The supernatant was considered as the cell-free fraction, and the resultant pellet was washed thrice with f/2 medium and resuspended in the fresh f/2 media. For intracellular relative neutral lipid measurement, 300 μl of resuspended pellet was added with 75 μl of glycerol (0.5 g/ml) and 3.75 μl of Nile red (0.3 μg/ml, in acetone; Sigma-Aldrich, USA), mixed by rapid inversion and lucifugally incubated for 5 min at room temperature. The fluorescence intensities of stained cells were detected by flow cytometry (Beckman Gallios), with excitation and emission wavelengths of 488 and 575 nm, respectively. Extracellular lipid content of the supernatant was measured as mentioned above using a microplate reader (Biotek H1, Synergy), with excitation and emission wavelengths of 488 and 575 nm, respectively. The intracellular and extracellular lipids from WT and engineered cells (overexpressing and silencing cells) were extracted, as described previously (6), and subsequently determined gravimetrically. Briefly, for the intracellular lipid content measurement, microalgae were lyophilized, ground into powder, and extracted with 3.6 ml of methanol/chloroform/water (2:1:0.6, v/v) using sonication, and 1 ml of water/chloroform (1:1, v/v) was added to the content, mixed, and vortexed. The upper phase (aqueous phase) was discarded after centrifugation at 1400g for 5 min, and the lower chloroform phase was collected and concentrated under nitrogen. For the extracellular lipid content measurement, the supernatant was freeze-dried and extracted with 3.6 ml of methanol/chloroform/water (2:1:0.6, v/v), and then, the measurement was performed as mentioned above. Lyophilized biomass was used for the cell dry weight determination, the extracted lipids were further dried in oven at 60°C, and the lipid content was determined as a percentage of cell dry weight (17). The fatty acid composition was analyzed as fatty acid methyl esters by GC-MS (17). Fatty acids were identified according to the equipped National Bureau of Standards spectrum library. The integrated peak areas were determined and calculated by normalization to obtain the contents of fatty acid composition (%).
Measurement of total carbohydrate and protein content
Total carbohydrate content was quantified using the phenol-sulphuric acid method (40). Briefly, 5 ml of microalgal culture in the stationary phase was harvested at 13,000 rpm for 10 min and resuspended in 1 ml of deionized water. This algal suspension was added with 1 ml of 5% (w/v) phenol solution. Five milliliters of concentrated sulphuric acid solution (95 to 98%, v/v) was rapidly added directly to the mixture without any drops on the wall, and the mixture was incubated for 10 min at room temperature. The content was mixed thoroughly, followed by incubation in a water bath at 30°C for 20 min. As phenol reacted with carbohydrates, it turned orange and was detectable at 483 nm. Glucose was used as standard at gradients (up to 1 g liter−1). Total protein was also extracted from the NobZIP1-overexpressing cells in the stationary phase by using a protein extraction kit (KeyGEN, Nanjing, China) following the manufacturer’s instructions, and the protein concentration was determined by the bicinchoninic acid (BCA) assay.
Laser scanning confocal microscopic observation of microalgae
Morphological changes of N. oceanica cells and oil bodies were observed under a laser confocal microscope, as described previously with slight modifications (17). Briefly, 300 μl of culture was added with 75 μl of glycerol (0.5 g/ml) and 3.75 μl of Nile red (0.3 μg/ml, in acetone; Sigma-Aldrich, USA) and lucifugally incubated for 5 min at room temperature. Stained cells were observed under a laser scanning microscope LSM 510 META (Zeiss, Germany), with excitation and emission wavelength of 525 and 550 to 575 nm, respectively.
Ultrastructural analysis by TEM
TEM analysis was carried out as described previously (17). Briefly, cells were fixed with 2% (v/v) paraformaldehyde and 2% (v/v) glutaraldehyde in 100 mM sodium cacodylate buffer (pH 7.4) for 3 hours at 4°C. The supernatant was gently removed, and the cells were rinsed thrice with 130 mM sucrose, 10 mM 2-mercaptoethanol, and 100 mM sodium cacodylate buffer (pH 7.4) for 20 min. The samples were then fixed with 1% (w/v) osmium tetroxide in 100 mM sodium cacodylate buffer (pH 7.4) and rinsed thrice with ultrapure water (Milli-Q; Millipore, Billerica, MA, USA). The samples were then dehydrated in a graded series of acetone [20, 50, 70, 90, and 100% (v/v)] for 10 min at each concentration of acetone. Thereafter, samples were infiltrated through a series of graded acetone/Epon/Spurr’s epoxy resin, followed by embedding in 100% (w/v) Spurr’s epoxy resin and polymerization at 60°C for 24 hours. Ultrathin sections were cut on an ultramicrotome LKB 8800 (LKB Instruments, USA) using a diamond knife (Diatome, Hatfield, USA). Subsequently, sections were stained with uranyl acetate and lead citrate and observed under a JEM-1200EX transmission electron microscope (JEOL, Japan), and images were recorded on EM (electron microscopy) film 4489 (Eastman-Kodak, NY, USA).
ChIP-qPCR analysis
We performed the ChIP analysis, which has been considered as a robust tool to analyze the interactions between the TFs and their target genes of the DNA in vivo. ChIP and subsequent qPCR analyses were performed (41). Briefly, 100 ml of algal cells was ground into fine powder with liquid nitrogen, and thereafter, chromatin was extracted and sheared into 200- to 800-bp fragments by sonication. Subsequently, sheared chromatin was immunoprecipitated with antibodies (anti-Flag). The precipitated DNA was then purified by using an Omega purification kit, as per the manufacturer’s specification. Relative enrichment of the target region was normalized against β-actin. The following primers were used: Na-Ks1 and Na-Ks2 for KAS, Na-Ap1 and Na-Ap2 for ACBP, Na-Lt1 and Na-Lt2 for LPAAT, Na-Ls1 and Na-Ls2 for LC-FACS, Na-Cs1 and Na-Cs2 for putative CPS, and Na-UG1 and Na-UG2 for UGDH. The relative transcript abundance of the predicted key target genes such as KAS, ACBP, LPAAT, LC-FACS, CPS, and UGDH was determined by qPCR, as mentioned above by using the primers. The following primers were used: Q-Ks1 and Q-Ks2 for KAS, Q-Ap1 and Q-Ap2 for ACBP, Q-Lt1 and Q-Lt2 for LPAAT, Q-Ls1 and Q-Ls2 for LC-FACS, Q-Cs1 and Q-Cs2 for putative CPS, and Q-UG1 and Q-UG2 for UGDH. The sequences of the oligonucleotide primers used are given in table S1. β-Actin was used as the internal control. All the primers used in this study are given in table S1.
Data analysis
Several overexpressing and silencing cells with uniform colonial phenotypes were randomly selected and subjected for further analysis. For clarity of presentation, results from two representative lines of both overexpressing and silencing algae are shown. Data from at least three independent experiments are given. Data were expressed as means ± SD (SD of the mean). Data were statistically analyzed by two-tailed unpaired Student’s t test. Significant difference is indicated at P < 0.05 (*) or P < 0.01 (**) level.
Supplementary Material
Acknowledgments
Funding: This work was supported by the Natural Science Foundation of China (31870027 and 41576132), the Guangdong Natural Science Foundation (2014A030308010), and the China Postdoctoral Science Foundation (2017M612838). Author contributions: D.-W.L. and H.-Y.L. conceived of and designed experiments; D.-W.L., S.B., J.-W.Z., D.H., L.-G.Z., and Y.-F.Y. performed experiments; Y.G. contributed to the bioinformatics analysis; D.-W.L., S.B., D.H., and W.-D.Y. analyzed data; D.-W.L., J.-S.L., Y.G., and H.-Y.L. wrote the manuscript. Competing interests: H.-Y.L., D.-W.L., W.-D.Y., and J.-S.L. are inventors on patents covering the TF held by the Jinan University (including no. CN106834309A, priority date: 13 June 2017). All other authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Sequencing data are deposited in the NCBI GenBank (accession numbers MH118545 and KU745463). Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/5/1/eaau3795/DC1
Fig. S1. Sequence analysis and characterization of NobZIP1 engineered cells.
Fig. S2. Laser scanning confocal microscopic analysis of Nile red–stained NobZIP1-engineered cells.
Fig. S3. Sequence analysis and characterization of UGDH overexpressing and silencing cells.
Fig. S4. Confocal microscopic analysis of Nile red–stained UGDH-engineered and CPS-silencing cells.
Table S1. List of oligonucleotide primers and their sequences used in this study.
Movie S1. Laser scanning confocal fluorescence of Nile red–stained cells identifying the lipid droplets mobilization across the cell wall and revealing the lipid secretion phenomenon across the weakened cell wall in overexpressing cells NobIP1-1 (movie S1), whereas no such mechanism was observed in WT (movie S2).
Movie S2. Laser scanning confocal fluorescence of Nile red–stained cells identifying the lipid droplets mobilization across the cell wall and revealing the lipid secretion phenomenon across the weakened cell wall in overexpressing cells NobIP1-1 (movie S1), whereas no such mechanism was observed in WT (movie S2).
REFERENCES AND NOTES
- 1.Jasny B. R., Bulking up algae for biofuels. Science 357, 160–161 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Duffy J. E., Canuel E. A., Adey W., Swaddle J. P., Biofuels: Algae. Science 326, 1345 (2009). [DOI] [PubMed] [Google Scholar]
- 3.Moheimani N. R., Matsuura H., Watanabe M. M., Borowitzka M. A., Non-destructive hydrocarbon extraction from Botryococcus braunii BOT-22 (race B). J. Appl. Phycol. 26, 1453–1463 (2014). [Google Scholar]
- 4.Grima E. M., Belarbi E.-H., Fernández F. A., Medina A. R., Chisti Y., Recovery of microalgal biomass and metabolites: Process options and economics. Biotechnol. Adv. 20, 491–515 (2003). [DOI] [PubMed] [Google Scholar]
- 5.Niu Y.-F., Wang X., Hu X., Balamurugan S., Li D.-W., Yang W.-D., Liu J.-S., Li H.-Y., Molecular characterization of a glycerol-3-phosphate acyltransferase reveals key features essential for triacylglycerol production in Phaeodactylum tricornutum. Biotechnol. Biofuels 9, 60 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Balamurugan S., Wang X., Wang H.-L., An C.-J., Li H., Li D.-W., Yang W.-D., Liu J.-S., Li H.-Y., Occurrence of plastidial triacylglycerol synthesis and the potential regulatory role of AGPAT in the model diatom Phaeodactylum tricornutum. Biotechnol. Biofuels 10, 97 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deng X.-D., Gu B., Li Y.-J., Hu X.-W., Guo J.-C., Fei X.-W., The roles of acyl-CoA: Diacylglycerol acyltransferase 2 genes in the biosynthesis of triacylglycerols by the green algae Chlamydomonas reinhardtii. Mol. Plant 5, 945–947 (2012). [DOI] [PubMed] [Google Scholar]
- 8.Li D.-W., Cen S.-Y., Liu Y.-H., Balamurugan S., Zheng X.-Y., Alimujiang A., Yang W.-D., Liu J.-S., Li H.-Y., A type 2 diacylglycerol acyltransferase accelerates the triacylglycerol biosynthesis in heterokont oleaginous microalga Nannochloropsis oceanica. J. Biotechnol. 229, 65–71 (2016). [DOI] [PubMed] [Google Scholar]
- 9.Eroglu E., Melis A., Extracellular terpenoid hydrocarbon extraction and quantitation from the green microalgae Botryococcus braunii var. Showa. Bioresour. Technol. 101, 2359–2366 (2010). [DOI] [PubMed] [Google Scholar]
- 10.Hejazi M., Holwerda E., Wijffels R., Milking microalga Dunaliella salina for β-carotene production in two-phase bioreactors. Biotechnol. Bioeng. 85, 475–481 (2004). [DOI] [PubMed] [Google Scholar]
- 11.Hu J., Wang D., Li J., Jing G., Ning K., Xu J., Genome-wide identification of transcription factors and transcription-factor binding sites in oleaginous microalgae Nannochloropsis. Sci. Rep. 4, 5454 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bajhaiya A. K., Ziehe Moreira J., Pittman J. K., Transcriptional engineering of microalgae: Prospects for high-value chemicals. Trends Biotechnol. 35, 95–99 (2017). [DOI] [PubMed] [Google Scholar]
- 13.Dröge-Laser W., Weiste C., The C/S1 bZIP network: A regulatory hub orchestrating plant energy homeostasis. Trends Plant Sci. 23, 422–433 (2018). [DOI] [PubMed] [Google Scholar]
- 14.Wigge P. A., Kim M. C., Jaeger K. E., Busch W., Schmid M., Lohmann J. U., Weigel D., Integration of spatial and temporal information during floral induction in Arabidopsis. Science 309, 1056–1059 (2005). [DOI] [PubMed] [Google Scholar]
- 15.Abe M., Kobayashi Y., Yamamoto S., Daimon Y., Yamaguchi A., Ikeda Y., Ichinoki H., Notaguchi M., Goto K., Araki T., FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 309, 1052–1056 (2005). [DOI] [PubMed] [Google Scholar]
- 16.Weiste C., Dröge-Laser W., The Arabidopsis transcription factor bZIP11 activates auxin-mediated transcription by recruiting the histone acetylation machinery. Nat. Commun. 5, 3883 (2014). [DOI] [PubMed] [Google Scholar]
- 17.Xue J., Balamurugan S., Li D.-W., Liu Y.-H., Zeng H., Wang L., Yang W.-D., Liu J.-S., Li H.-Y., Glucose-6-phosphate dehydrogenase as a target for highly efficient fatty acid biosynthesis in microalgae by enhancing NADPH supply. Metab. Eng. 41, 212–221 (2017). [DOI] [PubMed] [Google Scholar]
- 18.Zhang J., Hao Q., Bai L., Xu J., Yin W., Song L., Xu L., Guo X., Fan C., Chen Y., Overexpression of the soybean transcription factor GmDof4 significantly enhances the lipid content of Chlorella ellipsoidea. Biotechnol. Biofuels 7, 128 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kang N. K., Jeon S., Kwon S., Koh H. G., Shin S.-E., Lee B., Choi G.-G., Yang J.-W., Jeong B.-r., Chang Y. K., Effects of overexpression of a bHLH transcription factor on biomass and lipid production in Nannochloropsis salina. Biotechnol. Biofuels 8, 200 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kwon S., Kang N. K., Koh H. G., Shin S.-E., Lee B., Jeong B.-r., Chang Y. K., Enhancement of biomass and lipid productivity by overexpression of a bZIP transcription factor in Nannochloropsis salina. Biotechnol. Bioeng. 115, 331–340 (2018). [DOI] [PubMed] [Google Scholar]
- 21.Li H.-Y., Wang C.-Z., Chen X., Cao X.-F., Sun S.-N., Sun R.-C., Structural elucidation of Eucalyptus lignin and its dynamic changes in the cell walls during an integrated process of ionic liquids and successive alkali treatments. Bioresour. Technol. 222, 175–181 (2016). [DOI] [PubMed] [Google Scholar]
- 22.Lv J.-M., Cheng L.-H., Xu H., Zhang L., Chen H.-L., Enhanced lipid production of Chlorella vulgaris by adjustment of cultivation conditions. Bioresour. Technol. 101, 6797–6804 (2010). [DOI] [PubMed] [Google Scholar]
- 23.Liu C., Mao B., Ou S., Wang W., Liu L., Wu Y., Chu C., Wang X., OsbZIP71, a bZIP transcription factor, confers salinity and drought tolerance in rice. Plant Mol. Biol. 84, 19–36 (2014). [DOI] [PubMed] [Google Scholar]
- 24.Xie L. J., Yu L. J., Chen Q. F., Wang F. Z., Huang L., Xia F. N., Zhu T. R., Wu J. X., Yin J., Liao B., Arabidopsis acyl-CoA-binding protein ACBP3 participates in plant response to hypoxia by modulating very-long-chain fatty acid metabolism. Plant J. 81, 53–67 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maier T., Jenni S., Ban N., Architecture of mammalian fatty acid synthase at 4.5 A resolution. Science 311, 1258–1262 (2006). [DOI] [PubMed] [Google Scholar]
- 26.Jia B., Song Y., Wu M., Lin B., Xiao K., Hu Z., Huang Y., Characterization of long-chain acyl-CoA synthetases which stimulate secretion of fatty acids in green algae Chlamydomonas reinhardtii. Biotechnol. Biofuels 9, 184 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oka T., Jigami Y., Reconstruction of de novo pathway for synthesis of UDP-glucuronic acid and UDP-xylose from intrinsic UDP-glucose in Saccharomyces cerevisiae. FEBS J. 273, 2645–2657 (2006). [DOI] [PubMed] [Google Scholar]
- 28.Krishnan A., Kumaraswamy G. K., Vinyard D. J., Gu H., Ananyev G., Posewitz M. C., Dismukes G. C., Metabolic and photosynthetic consequences of blocking starch biosynthesis in the green alga Chlamydomonas reinhardtii sta6 mutant. Plant J. 81, 947–960 (2015). [DOI] [PubMed] [Google Scholar]
- 29.Zhu B.-H., Shi H.-P., Yang G.-P., Lv N.-N., Yang M., Pan K.-H., Silencing UDP-glucose pyrophosphorylase gene in Phaeodactylum tricornutum affects carbon allocation. N. Biotechnol. 33, 237–244 (2016). [DOI] [PubMed] [Google Scholar]
- 30.Eastmond P. J., Rawsthorne S., Coordinate changes in carbon partitioning and plastidial metabolism during the development of oilseed rape embryos. Plant Physiol. 122, 767–774 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim D.-Y., Vijayan D., Praveenkumar R., Han J.-I., Lee K., Park J.-Y., Chang W.-S., Lee J.-S., Oh K., Cell-wall disruption and lipid/astaxanthin extraction from microalgae: Chlorella and Haematococcus. Bioresour. Technol. 199, 300–310 (2016). [DOI] [PubMed] [Google Scholar]
- 32.Bouyakdan K., Taïb B., Budry L., Zhao S., Rodaros D., Neess D., Mandrup S., Faergeman N. J., Alquier T., A novel role for central ACBP/DBI as a regulator of long-chain fatty acid metabolism in astrocytes. J. Neurochem. 133, 253–265 (2015). [DOI] [PubMed] [Google Scholar]
- 33.Yagita Y., Shinohara K., Abe Y., Nakagawa K., Al-Owain M., Alkuraya F. S., Fujiki Y., Deficiency of a retinal dystrophy protein, acyl-CoA binding domain-containing 5 (ACBD5), impairs peroxisomal β-oxidation of very-long-chain fatty acids. J. Biol. Chem. 292, 691–705 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dixon R. A., Microbiology: Break down the walls. Nature 493, 36–37 (2013). [DOI] [PubMed] [Google Scholar]
- 35.Gilbert H. J., Knox J. P., Boraston A. B., Advances in understanding the molecular basis of plant cell wall polysaccharide recognition by carbohydrate-binding modules. Curr. Opin. Struct. Biol. 23, 669–677 (2013). [DOI] [PubMed] [Google Scholar]
- 36.Hansen M. A., Ahl L. I., Pedersen H. L., Westereng B., Willats W. G., Jørgensen H., Felby C., Extractability and digestibility of plant cell wall polysaccharides during hydrothermal and enzymatic degradation of wheat straw (Triticum aestivum L.). Ind. Crops Prod. 55, 63–69 (2014). [Google Scholar]
- 37.Wang L., Jin P., Wang J., Jiang L., Shan T., Zheng Y., Effect of β-aminobutyric acid on cell wall modification and senescence in sweet cherry during storage at 20 °C. Food Chem. 175, 471–477 (2015). [DOI] [PubMed] [Google Scholar]
- 38.Zhou R., Li Y., Yan L., Xie J., Effect of edible coatings on enzymes, cell-membrane integrity, and cell-wall constituents in relation to brittleness and firmness of Huanghua pears (Pyrus pyrifolia Nakai, cv. Huanghua) during storage. Food Chem. 124, 569–575 (2011). [Google Scholar]
- 39.Xu G., Sui N., Tang Y., Xie K., Lai Y., Liu Y., One-step, zero-background ligation-independent cloning intron-containing hairpin RNA constructs for RNAi in plants. New Phytol. 187, 240–250 (2010). [DOI] [PubMed] [Google Scholar]
- 40.Dubois M., Gilles K. A., Hamilton J. K., Rebers P. A., Smith F., Colorimetric method for determination of sugars and related substances. Anal. Chem. 28, 350–356 (1956). [Google Scholar]
- 41.C. L. Ricupero, M. R. Swerdel, R. P. Hart, Pluripotent Stem Cells (Springer, 2013), pp. 203–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/5/1/eaau3795/DC1
Fig. S1. Sequence analysis and characterization of NobZIP1 engineered cells.
Fig. S2. Laser scanning confocal microscopic analysis of Nile red–stained NobZIP1-engineered cells.
Fig. S3. Sequence analysis and characterization of UGDH overexpressing and silencing cells.
Fig. S4. Confocal microscopic analysis of Nile red–stained UGDH-engineered and CPS-silencing cells.
Table S1. List of oligonucleotide primers and their sequences used in this study.
Movie S1. Laser scanning confocal fluorescence of Nile red–stained cells identifying the lipid droplets mobilization across the cell wall and revealing the lipid secretion phenomenon across the weakened cell wall in overexpressing cells NobIP1-1 (movie S1), whereas no such mechanism was observed in WT (movie S2).
Movie S2. Laser scanning confocal fluorescence of Nile red–stained cells identifying the lipid droplets mobilization across the cell wall and revealing the lipid secretion phenomenon across the weakened cell wall in overexpressing cells NobIP1-1 (movie S1), whereas no such mechanism was observed in WT (movie S2).


