Abstract
Metformin is one of the first-line and most widely prescribed drugs to treat type 2 diabetes (T2D). Its clearance from circulation is mostly facilitated by SLC22A2 (OCT2) in the renal cells. SLC22A2 is a polyspecific organic cation transporter and mediate transport of structurally unrelated endogenous and exogenous compounds including many drugs. rs316019 (p.270A > S) is the most common variant of SLC22A2 with a frequency as high as 15% or more in many populations. The 270S form of SLC22A2 clears metformin from circulation at much reduced level compared to the 270A form. If accumulated, metformin increases plasma lactate level in a concentration-dependent manner which can lead to a condition known as metformin-associated lactic acidosis (MALA). MALA is a potentially life-threatening complication with a mortality rate of 30–50%. Pre-existing clinical conditions, such as renal impairment, sepsis, anoxia, etc may make individuals more prone to MALA. In this study, we used computational approaches to investigate the effect of 270A > S change in SLC22A2 on interaction with metformin and other drugs. Based on the structural models, all substrates bind to the same pocket of SLC22A2. The substrates fit better to the binding site of 270A form of SLC22A2. The binding site has a few core interacting residues, among which SER358 appears to be the most important. It is an in silico prediction that the T2D patients, who are under metformin regimen, should be cautious in taking ranitidine (an over-the-counter sold drug) on a regular basis as it may lead to metformin associated lactate accumulation in blood.
Keywords: SLC22A2, Rs316019, Metformin, Metformin associated lactic acidosis, Type 2 diabetes
1. Introduction
Metformin is one of the first-line and most widely prescribed drugs to treat type 2 diabetes (T2D) [1], [2], [3]. It is an organic cation and belongs to the biguanide family [4], [5]. It lowers both basal and postprandial plasma glucose levels by inhibiting the production of hepatic glucose, reducing intestinal glucose absorption, and improving glucose uptake and utilization in the peripheral tissues particularly in muscle [1], [4]. Although it is in use since 1957, the direct molecular target of metformin remains unknown [6].
Metformin circulates unbound in the plasma and is not metabolized by the enzymes in liver [1]. Its clearance from circulation is mostly facilitated by SLC22A2 (OCT2) in the renal cells [6], [7], [8]. SLC22A2 is a member of the solute carrier (SLC) super-family of proteins which comprises over 300 members in human [9], [10]. The human SLC22A2 gene is localized on chromosome 6 and consists of 11 exons [11]. It is primarily expressed in the kidney with some level of expression in the brain, placenta, lungs, spleen, small intestine and skin [11], [12], [13], [14]. In the kidney, SLC22A2 protein is localized in the basolateral membrane of proximal tubular cells [15]. SLC22A2 is a polyspecific transporter and mediate passive facilitated bi-directional transport of structurally unrelated small organic cations down their electrochemical gradients [11], [12], [15]. It transports both endogenous and exogenous compounds including many drugs which have at least one positively charged moiety at physiological pH [3], [11], [12], [13], [16], [17]. Considering the fact that 40% or more of the prescribed drugs are positively charged and belong to the group of organic cations, it is obvious that the function of this transporter in the kidney has important pharmacological implications [15], [18].
Considerable inter-individual variability in clinical efficacy exists in the treatment of T2D patients with metformin [1], [6], [19], [20]. rs316019 (c.808 G > T) is the most common variant of SLC22A2 and it significantly influences the pharmacokinetics of orally administered metformin [6], [21]. According to the sequence data in the 1000 genomes browser, the 270S variant of SLC22A2 is present with a frequency as high as 15% or more in at least ten different populations [22]. The c.808G > T polymorphism in the SLC22A2 results in a protein (p.270A > S), which clears metformin from circulation much slower than the 270A form [6], [19], [21]. If accumulated, metformin and other drugs of the biguanide class increase plasma lactate level in a concentration-dependent manner by inhibiting mitochondrial respiration [1], [23]. The 270A > S change in SLC22A2 can lead to hyperlactacidemia in patients undergoing metformin therapy, particularly in the females carrying the TT genotype [19]. In metformin-associated lactic acidosis (MALA) arterial pH and blood lactate concentration may vary from ≤7.34 to 6.4 and from >5 mmol/L to 35.5 mmol/L, respectively [24]. MALA is a potentially life-threatening complication with a mortality rate of 30–50% [25]. Although prevalence of MALA has been reported to be approximately 1–10 cases per 100,000 patients [26], the actual number is speculated to be several times higher [27]. The incidence of MALA is caused within the clinical doses of metformin [23], [26]. Pre-existing clinical conditions, such as renal impairment, sepsis, anoxia, etc may make individuals under metformin regimen more prone to MALA [1], [2], [21], [28]. Therefore, dose adjustments based on the SLC22A2 rs316019 variant may be beneficial to maximize the efficacy and minimize the toxicity of metformin [6], [20].
In this study, we used computational approaches to investigate the effect of p.270A > S change in SLC22A2 on interaction with metformin and other drugs.
2. Materials and methods
2.1. Sequence retrieval and protein secondary structure prediction
SLC22A2 protein and rs316019 SNP sequence information were retrieved from the database resources of the National Center for Biotechnology Information [29]. Secondary structure of the SLC22A2 rs316019 protein variants were predicted using PSIPRED [30], PredictProtein [31] and RaptorX Structure Prediction [32] tools. Transmembrane segments of SLC22A2 variants were predicted using TMHMM [33] and SPOCTOPUS [34]. Sequences were aligned using BLAST [35] and MUSCLE [36] and analyzed with Jalview [37].
2.2. Protein tertiary structure prediction and analysis
Since no 3D-structural data were available in literature and databases, conformation of SLC22A2 rs316019 protein variants were predicted using I-TASSER [38] and RaptorX Structure Prediction [32] tools. Zeus PDB Viewer was used to derive the Ramachandran plots of the protein variants. Protein structures were analyzed using CCP4mg [39]. 3D structures of SLC22A2 rs316019 protein variants were aligned using RaptorX Structure Alignment tool [32].
2.3. Protein-drug interaction prediction
Drug structures were retrieved from either Pubchem [40] or Drugbank [41]. Drug bound models of SLC22A2 rs316019 protein variants were predicted using BSP-SLIM [42] and analyzed with LigPlot+ [43] and CCP4mg [39]. BSP-SLIM uses a blind docking method and tries to fit small molecules into the structure of proteins and evaluate their binding affinity using a scoring system [42]. Drug (ligand) binding sites were searched within the entire protein structure.
2.4. Oligomeric structure prediction
GalaxyHomomer [48] was used to predict whether the SLC22A2 protein oligomerizes. Protein homo-oligomer structures were predicted without using template information (ab initio) and providing only the monomer structure as input rather than a sequence. The oligomeric state was determined by the tool itself. Predicted oligomeric structures were analyzed using CCP4mg [39].
3. Results and discussion
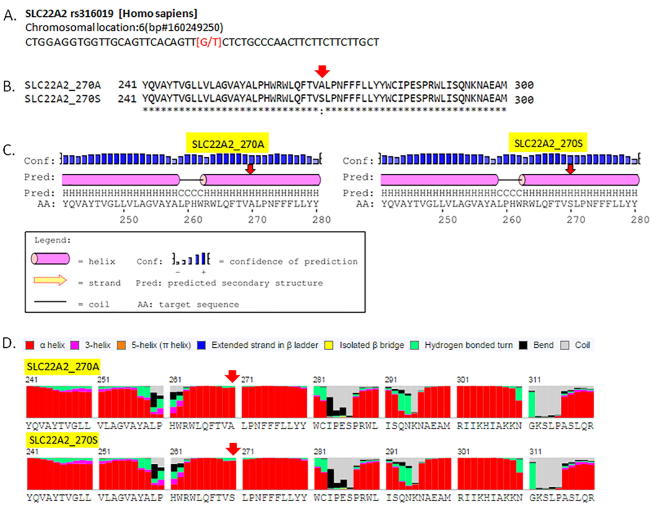
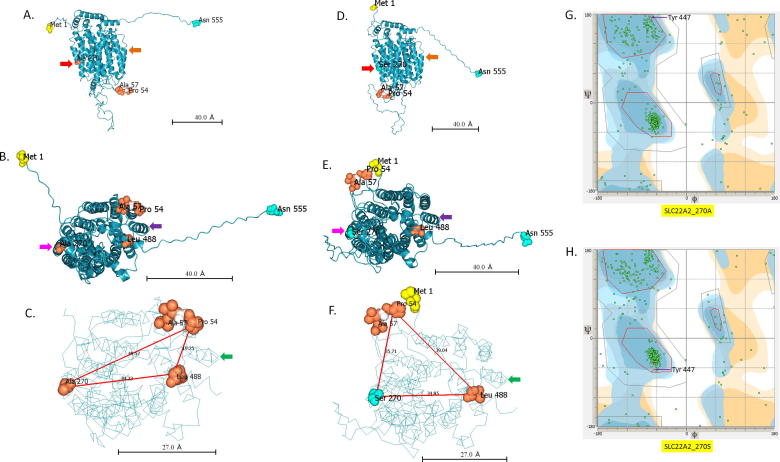
rs316019 results in single amino acid change in SLC22A2, but it does not affect the local secondary structure (Fig. 1). rs316019, however, affects the global tertiary structure of SLC22A2 protein (Fig. 2). As shown in Fig. 2, position of Leu488 changes relative to Pro54 and Ala/Ser270 in the protein variants. Ramachandran plots also suggest a global change in conformation due to a single amino acid change (A > S) at position 270.
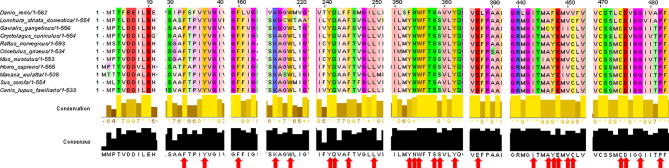
Fig. 1.
Secondary structure of SLC22A2 rs316019 variants. A. rs316019 at the genomic DNA level. B. Pair-wise protein sequence alignment of rs316019 variants. C and D. Secondary sequence prediction of rs316019 variants by PSIPRED and RaptorX Structure prediction tools. Secondary structures predicted with PredictProtein are not shown here.
Fig. 2.
Tertiary structural models of SLC22A2 rs316019 protein variants. A, B and C. Models of SLC22A2 with Alanine at position 270. D, E and F. Models of SLC22A2 with Serine at position 270. A single amino acid alteration at position 270 causes a global conformational change in SLC22A2. As an example, distances of Leu 488 from Pro 54 and Ala/Ser 270 are shown in figure C and F. Red lines represent the distance between selected amino acid residues in Angstrom (Å). Solid arrows (matched colour) denote reference points to compare structures. G and H. Ramachandran plots of SLC22A2 rs316019 protein variants.
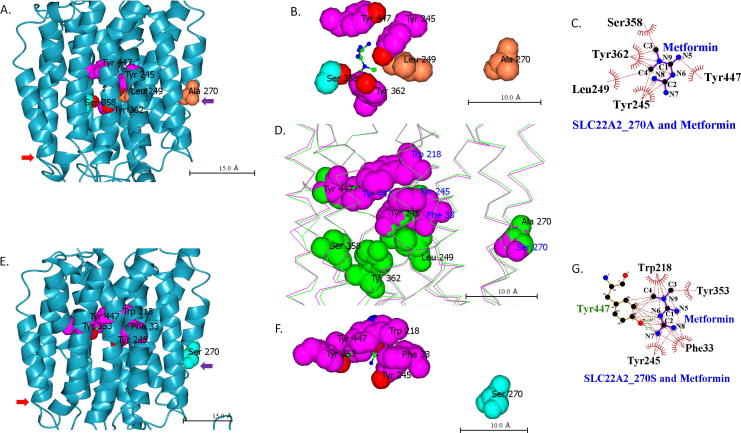
Analysis of BSP-SLIM generated 3D-model of metformin bound SLC22A2 270A variant demonstrates that Tyr245, Leu249, Ser358, Tyr362 and Tyr447 interact with the drug (Fig. 3). A different set of residues interact with metformin in SLC22A2 variant with serine at position 270 (Fig. 3). 270A variant of SLC22A2 has more open and wider space for metformin binding compared to the 270S variant. Table 1 shows the predicted docking scores of metformin with SLC22A2 variants. While 270A variant has a docking score of 5.18 for metformin, no score could be calculated by BSP-SLIM for the 270S variant. Similar interaction takes place between the rs316019 protein variants and creatinine (Fig. 4). Creatinine is a known substrate of SLC22A2 [44]. While the SLC22A2 270A variant has a docking score of 3.911 for creatinine, the score is 0.619 for the 270S variant (Table 1).
Fig. 3.
Metformin binding site in SLC22A2 protein. A, B and C. Metformin binding site in SLC22A2 270A variant. E, F and G. Metformin binding site in SLC22A2 270S variant. Solid arrows (matched colour) denote reference points to compare structures. Colour of amino acids is based on residue type. D. Structural alignment of SLC22A2 rs316019 protein variants with binding site amino acid residues shown as spheres. Green and purple indicate amino acids of SLC22A2 variants with 270A and 270S, respectively. In C and G, red and green dotted lines denote hydrophobic interactions and hydrogen bonds, respectively.
Table 1.
Interacting amino acid residues at the substrate/inhibitor binding site of SLC22A2.
| SLC22A2 variant | Substrate/inhibitor | Phe 33 | Tyr 37 | Phe 160 | Lys 215 | Trp 218 | Tyr 241 | Gln 242 | Tyr 245 | Leu 249 | Tyr 353 | Asn 354 | Trp 355 | Ser 358 | Ser 359 | Tyr 362 | Glu 387 | Ile 443 | Ala 446 | Tyr 447 | Glu 448 | Val 450 | Cys 451 | Cys 474 | Asp 475 | Gly 478 | Pro 482 | Docking score |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 270A | Metformin | + | + | + | + | + | 5.18 | |||||||||||||||||||||
| 270A | Agmatine | + | + | + | + | + | + | + | + | + | 3.818 | |||||||||||||||||
| 270A | Ifosfamide | + | + | + | + | + | + | + | + | 5.79 | ||||||||||||||||||
| 270A | Paraquat | + | + | + | + | + | + | + | + | + | 4.007 | |||||||||||||||||
| 270A | Trimethoprim | + | + | + | + | + | + | + | + | + | + | + | 7.581 | |||||||||||||||
| 270A | Cimetidine | + | + | + | + | + | + | + | + | + | 5.733 | |||||||||||||||||
| 270A | Creatinine | + | + | + | 3.911 | |||||||||||||||||||||||
| 270A | Ranitidine | + | + | + | + | + | + | + | + | + | + | + | + | + | 5.591 | |||||||||||||
| 270S | Metformin | + | + | + | + | + | nan* | |||||||||||||||||||||
| 270S | Cimetidine | + | + | + | + | + | + | + | + | + | + | 4.385 | ||||||||||||||||
| 270S | Creatinine | + | + | + | + | + | 0.619 | |||||||||||||||||||||
| 270S | Ranitidine | + | + | + | + | + | + | + | + | 4.588 |
nan = not-a-number.
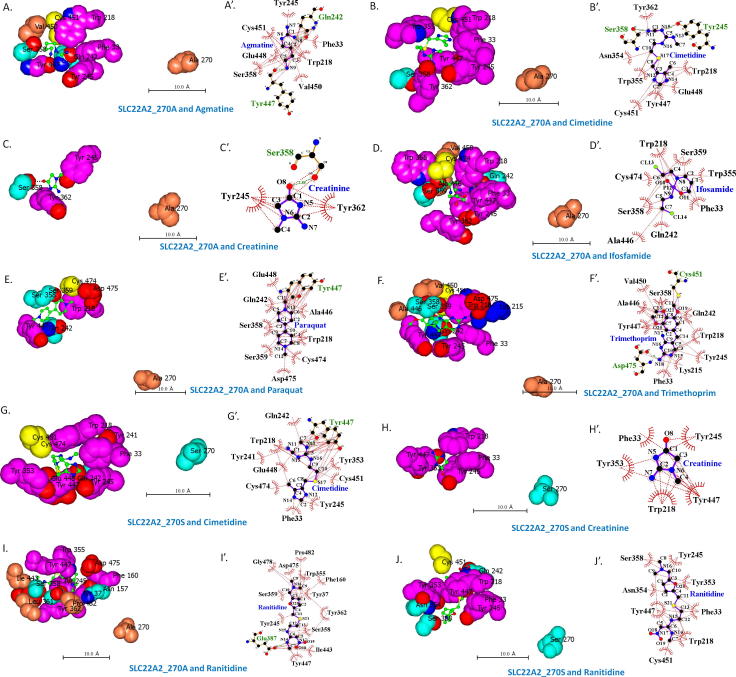
Fig. 4.
Interacting residues at the substrate binding site of SLC22A2 protein. A–J and A′–J′. Interacting amino acid residues with different compounds at the binding sites of SLC22A2 270A and 270S variants. All models were generated using BSP-SLIM. A–J and A′–J′ were analyzed with CCP4mg and LigPlot+, respectively. All scale bars are shown as 10 Å. In A′–J′, red and green dotted lines denote hydrophobic interactions and hydrogen bonds, respectively.
BSP-SLIM calculated docking score is based on structural complementarities and chemical feature similarities between the ligand and the binding pocket [42]. The ligand bound protein model with the highest docking score is the best fit model. BSP-SLIM generates reliable docking results using low-resolution predicted protein structural models [42]. Since the ligand poses are determined by global topology similarity of protein structures and low-resolution docking method, the performance of BSP-SLIM is much less sensitive to the local structural errors in the predicted model structure and its ability to predict binding site outperforms geometry-based method for both experimentally solved and theoretically predicted protein structures [42]. BSP-SLIM even demonstrated remarkable performance with docking on low-resolution structures over the widely-used blind docking tool, AutoDock [42].
In this study, ligand bound structural models of SLC22A2 were generated with other known substrates and inhibitors including agmatine, cimetidine, ifosfamide, paraquat, ranitidine and trimethoprim [11], [14], [16], [45] (Fig. 4). Amino acid residues that interact with these compounds are listed in Table 1. Based on these models Ser358 appears to be the most important residue at the binding site of 270A form of SLC22A2 followed by Tyr245, Tyr447 and Trp218 (Table 1). Larger compounds interact with more residues at the same binding pocket.
SLC22A2 protein sequences from different species were aligned (Fig. 5). All binding site residues shown in Table 1, except Tyr 245, are highly conserved among different species. SLC22A2 has 12 α-helical transmembrane domains (TMDs) [11]. The interacting residues at the ligand binding site of SLC22A2 come from multiple TMDs (Fig. 3, Fig. 4 and Table 1). Similar observation was reported for the SLC22A1 homolog in rat [11]. Amino acid residue at position 270 is distantly localized from the binding pocket residues (Fig. 3). Change in amino acid residues distantly localized from the protein-protein interfaces and outside of ligand-binding pockets can change protein conformation and affect functionality [46].
Fig. 5.
Conservation of predicted binding site residues. SLC22A2 protein sequences from different species were aligned using the multiple sequence alignment tool MUSCLE and analyzed with Jalview. Scale above the sequences represents the actual amino acid position in human SLC22A2 protein. All binding site residues (red arrow), except Tyr 245, are highly conserved among the species.
Trimethoprim has been shown to significantly reduce systemic clearance of metformin and creatinine and increase plasma lactate concentration [45]. Trimethoprim bound SLC22A2 has the largest docking score among the compounds analyzed in this study (Table 1). A larger docking score suggest that trimethoprim fits better to the binding pocket and, therefore, should preferentially bind to the ligand binding site of SLC22A2 in presence of metformin and creatinine.
Cimetidine and ranitidine are also known inhibitors of SLC22A2 [7], [14], [47]. Compared to trimethoprim, lower or absent effects on metformin clearance from circulation was observed in healthy Asian volunteers treated with cimetidine [45]. Our analyses show that cimetidine and ranitidine bound SLC22A2 has larger docking scores than metformin, but much lower than trimethoprim (Table 1). This may explain the weaker inhibition of metformin clearance by cimetidine. Ranitidine is sold as an over-the-counter drug. It is an in silico prediction that the T2D patients, who are under metformin regimen, should be cautious in taking ranitidine on a regular basis as it may lead to metformin accumulation in blood and subsequently increase lactate concentration. However, this hypothesis needs to be tested in vivo.
It was suggested that SLC22A2 has a size dependent ‘selectivity filter’ and cannot transport compounds larger than 4°A for sterical hindrances [11], [16]. Attachment to the substrate binding site, however, is also possible for large molecules [16]. Stronger binding of such large molecules along with size dependent selectivity may occlude transport through SLC22A2. This may explain why large compounds like trimethoprim, cimetidine and ranitidine with a larger docking score inhibit metformin transport through SLC22A2 in a competitive fashion.
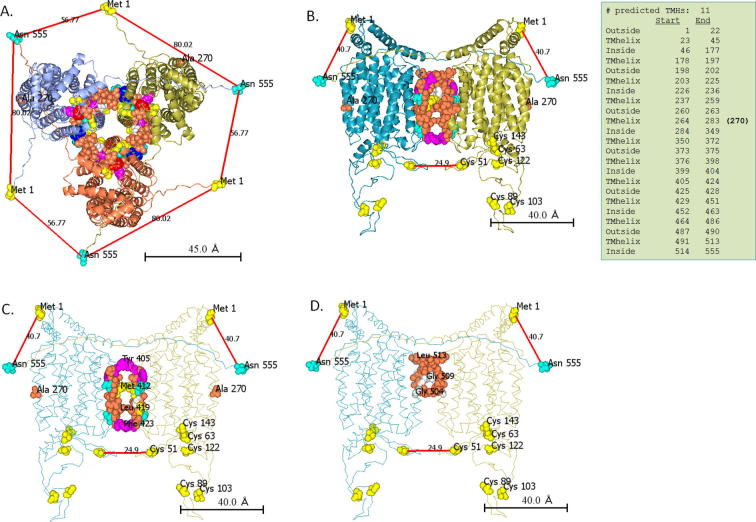
Individuals, who are heterozygous for the SLC22A2 rs316019 genotype (808GT), show slower metformin clearance rate compared to those with the 808GG genotype [21]. This raises a possibility among others that SLC22A2 forms oligomer. An earlier study also suggested that SLC22A2 forms oligomers [15]. Homo-oligomerization of proteins is quite common in nature and is often a prerequisite to physiological functions of proteins [48]. Members of SLC family are known to form homodimer or homotetramer [15], [49]. In this study, we computationally predicted the homo-oligomeric structural model of SLC22A2. The monomeric structure rather than the sequence of SLC22A2 was used as input in GalaxyHomomer to predict the oligomeric state. Although the structure based predictions are more restrictive, oligomer structures predicted by template-based methods may have errors due to sequence differences between the target and template proteins [48].
The predicted SLC22A2 oligomeric models include both dimer and trimers (Fig. 6). Since the protein-protein interface strength is proportional to the interface area [50], we selected the dimer and trimer models of SLC22A2 with the largest interacting interface for further analysis. Based on fluorescence resonance energy transfer (FRET) experiment, it was suggested that in the quaternary conformation the N and C termini of the SLC22A2 oligomers are in close proximity [15]. Energy transfer in FRET occurs only if the distance is <50 Å [15]. Only in the dimeric model, the N and C termini of the oligomer are closer than 50 Å (Fig. 6). In this model, the adjacent N and C termini come from different monomeric subunits. Interacting residues at the interface of this dimeric model are localized at position 405–423 and 504–513. As shown in Fig. 6, these amino acid residues appear to make an intertwined structure. Most homodimeric proteins have symmetric structure [51]. The predicted dimeric model of SLC22A2 appears symmetric as well.
Fig. 6.
Oligomeic models of SLC22A2. GalaxyHomomer predicted two types of homo-oligomers (3-mer and 2-mer) of SLC22A2. Only models with maximum interacting surfaces are shown here. A. In the trimer model, transmembrane (TM) helices with residues from 429-451 and 491–513 interact at the interface (shown as spheres). B. In the dimer model, transmembrane (TM) helices with residues from 405-423 and 504–513 interact at the interface (shown as spheres). Red lines show the distance between selected amino acid residues in Angstrom (Å). C and D show the dimeric model with residues from 405-423 and 504–513 (shown as spheres), respectively.
4. Conclusion
Molecular docking is one of the most commonly used computational approaches to study protein-drug interactions. It is based on the theoretical prediction of the binding mode as well as the binding affinity of small molecules for given target proteins. In this study, we used computational approaches to investigate the effect of 270A > S change in SLC22A2 with interaction with Metformin. Our analyses suggest that all substrates and inhibitors bind to the same pocket and these molecules fit better to the binding site of SLC22A2 with alanine at position 270 than serine. The binding site has a few core interacting residues, among which serine 358 is the most important. But the number and position of the interacting residues is also dependent on the size and structure of the compound. This may be true for other polyspecific transporters as well. Based on the docking scores, it is a suggestion that the T2D patients, who are under metformin regimen, should be cautious in taking ranitidine (an over-the-counter drug) on a regular basis as it may lead to metformin accumulation in blood and subsequently increase lactate concentration.
Footnotes
Peer review under responsibility of National Research Center, Egypt.
References
- 1.DeFronzo R., Fleming G.A., Chen K., Bicsak T.A. Metab Clin Exp. 2016;65:20–29. doi: 10.1016/j.metabol.2015.10.014. [DOI] [PubMed] [Google Scholar]
- 2.Protti A., Russo R., Tagliabue P., Vecchi S., Singer M., Rudiger A. Crit Care. 2010;14:R22. doi: 10.1186/cc8885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shikata E., Yamamoto R., Takane H., Shigemasa C., Ikeda T., Otsubo K. J Hum Genet. 2007;52:117–122. doi: 10.1007/s10038-006-0087-0. [DOI] [PubMed] [Google Scholar]
- 4.Rojas L.B., Gomes M.B. Diabetol Metab Syndr. 2013;5:6. doi: 10.1186/1758-5996-5-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dykens J.A., Jamieson J., Marroquin L., Nadanaciva S., Billis P.A., Will Y. Toxicol Appl Pharmacol. 2008;233:203–210. doi: 10.1016/j.taap.2008.08.013. [DOI] [PubMed] [Google Scholar]
- 6.Zolk O. Ann Med. 2012;44:119–129. doi: 10.3109/07853890.2010.549144. [DOI] [PubMed] [Google Scholar]
- 7.Zu Schwabedissen H.E.M., Verstuyft C., Kroemer H.K., Becquemont L., Kim R.B. Am J Physiol Renal Physiol. 2010;298:F997–F1005. doi: 10.1152/ajprenal.00431.2009. [DOI] [PubMed] [Google Scholar]
- 8.Pernicova I., Korbonits M. Nat Rev Endocrinol. 2014;10:143–156. doi: 10.1038/nrendo.2013.256. [DOI] [PubMed] [Google Scholar]
- 9.Schlessinger A., Khuri N., Giacomini K.M., Sali A. Curr Top Med Chem. 2013;13:843–856. doi: 10.2174/1568026611313070007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lin L., Yee S.W., Kim R.B., Giacomini K.M. Nat Rev Drug Discov. 2015;14:543–560. doi: 10.1038/nrd4626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koepsell H., Lips K., Volk C. Pharm Res. 2007;24:1227–1251. doi: 10.1007/s11095-007-9254-z. [DOI] [PubMed] [Google Scholar]
- 12.Roth M., Obaidat A., Hagenbuch B. Br J Pharmacol. 2012;165:1260–1287. doi: 10.1111/j.1476-5381.2011.01724.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jonker J.W., Schinkel A.H. J Pharmacol Exper Therap. 2004;308:2–9. doi: 10.1124/jpet.103.053298. [DOI] [PubMed] [Google Scholar]
- 14.Bergen A.W., Javitz H.S., Krasnow R., Michel M., Nishita D., Conti D.V., Edlund C.K. Nicotine Tobacco Res: Off J Soc Res Nicotine Tobacco. 2014;16:1638–1646. doi: 10.1093/ntr/ntu161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brast S., Grabner A., Sucic S., Sitte H.H., Hermann E., Pavenstädt H. FASEB J. 2012;26:976–986. doi: 10.1096/fj.11-180679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Volk C. Wiley Interdiscip Rev: Membr Transp Signal. 2014;3:1–13. [Google Scholar]
- 17.Ingoglia F., Visigalli R., Rotoli B.M., Barilli A., Riccardi B., Puccini P. Biochimica et Biophysica Acta. 2012;1848:1563–1572. doi: 10.1016/j.bbamem.2015.04.001. [DOI] [PubMed] [Google Scholar]
- 18.Hacker K., Maas R., Kornhuber J., Fromm M.F., Zolk O. PLoS ONE. 2015;10:e0136451. doi: 10.1371/journal.pone.0136451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li Q., Liu F., Zheng T.S., Tang J.L., Lu H.J., Jia W.P. Acta Pharmacol Sin. 2010;31:184–190. doi: 10.1038/aps.2009.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Avery P., Mousa S.S., Mousa S.A. Pharmgenomics Pers Med. 2009;2:79–91. doi: 10.2147/pgpm.s5806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yoon H., Cho H.Y., Yoo H.D., Kim S.M., Lee Y.B. AAPS J. 2013;15:571–580. doi: 10.1208/s12248-013-9460-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.1000 Genomes Project Consortium. Auton A., Brooks L.D., Durbin R.M., Garrison E.P., Kang H.M., Korbel J.O. Nature. 2015;526:68–74. [Google Scholar]
- 23.Piel S., Ehinger J.K., Elmer E., Hansson M.J. Acta Physiol. 2015;213:171–180. doi: 10.1111/apha.12311. [DOI] [PubMed] [Google Scholar]
- 24.Kajbaf F., Lalau J.D. BMC Pharmacol Toxicol. 2013;14 doi: 10.1186/2050-6511-14-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Friesecke S., Abel P., Roser M., Felix S.B., Runge S. Crit Care. 2010;14:R226. doi: 10.1186/cc9376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wills B.K., Bryant S.M., Buckley P., Seo B. Am J Emerg Med. 2010;28:857–861. doi: 10.1016/j.ajem.2009.04.012. [DOI] [PubMed] [Google Scholar]
- 27.van Berlo-van de Laar I.R., Vermeij C.G., Doorenbos C.J. J Clin Pharm Ther. 2011;36:376–382. doi: 10.1111/j.1365-2710.2010.01192.x. [DOI] [PubMed] [Google Scholar]
- 28.Huang W., Castelino R.L., Peterson G.M. J Diabetes Complications. 2015;29:1261–1265. doi: 10.1016/j.jdiacomp.2015.06.001. [DOI] [PubMed] [Google Scholar]
- 29.NCBI Resource Coordinators Nucl Acids Res. 2017;45:D12–D17. doi: 10.1093/nar/gkw1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Buchan D.W., Minneci F., Nugent T.C., Bryson K., Jones D.T. Nucl Acids Res. 2013;41:W349–W357. doi: 10.1093/nar/gkt381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yachdav G., Kloppmann E., Kajan L., Hecht M., Goldberg T., Hamp T. Nucl Acids Res. 2014;42:W337–W343. doi: 10.1093/nar/gku366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Källberg M., Wang H., Wang S., Peng J., Wang Z., Lu H. Nat Protoc. 2012;7:1511–1522. doi: 10.1038/nprot.2012.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krogh A., Larsson B., von Heijne G., Sonnhammer E.L. J Mol Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 34.Viklund H., Bernsel A., Skwark M., Elofsson A. Bioinformatics. 2008;24:2928–2929. doi: 10.1093/bioinformatics/btn550. [DOI] [PubMed] [Google Scholar]
- 35.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 36.Edgar R.C. Nucl Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Waterhouse A.M., Procter J.B., Martin D.M.A., Clamp M., Barton G.J. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang J., Yan R., Roy A., Xu D., Poisson J., Zhang Y. Nat Methods. 2015;12:7–8. doi: 10.1038/nmeth.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McNicholas S., Potterton E., Wilson K.S., Noble M.E.M. Acta Cryst D. 2011;67:386–394. doi: 10.1107/S0907444911007281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim S., Thiessen P.A., Bolton E.E., Chen J., Fu G., Gindulyte A. Nucl Acids Res. 2016;44:D1202–D1213. doi: 10.1093/nar/gkv951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Law V., Knox C., Djoumbou Y., Jewison T., Guo A.C., Liu Y. Nucl Acids Res. 2014;42:D1091–D1097. doi: 10.1093/nar/gkt1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee H.S., Zhang Y. Proteins. 2012;80:93–110. doi: 10.1002/prot.23165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Laskowski R.A., Swindells M.B. J Chem Inf Model. 2011;51:2778–2786. doi: 10.1021/ci200227u. [DOI] [PubMed] [Google Scholar]
- 44.Reznichenko A., Sinkeler S.J., Snieder H., van den Born J., de Borst M.H., Damman J. Physiol Genomics. 2013;45:201–209. doi: 10.1152/physiolgenomics.00087.2012. [DOI] [PubMed] [Google Scholar]
- 45.Grun B., Kiessling M.K., Burhenne J., Riedel R.D., Weiss J., Rauch G., Haefeli W.E. Br J Clin Pharmacol. 2013;76:787–796. doi: 10.1111/bcp.12079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Perica T., Kondo Y., Tiwari S.P., McLaughlin S.H., Kemplen K.R., Zhang X. Science. 2014;346:12543–12546. doi: 10.1126/science.1254346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li Q., Shu Y. Mol Cell Ther. 2014;2 doi: 10.1186/2052-8426-2-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Baek M., Park T., Heo L., Park C., Seok C. Nucl Acids Res. 2017;45:W320–W324. doi: 10.1093/nar/gkx246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Alexander S.P., Kelly E., Marrion N., Peters J.A., Benson H.E., Faccenda E. Br J Pharmacol. 2015;172:6110–6202. doi: 10.1111/bph.13355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Quintyn R.S., Yan J., Wysocki V.H. Chem Biol. 2015;22:583–592. doi: 10.1016/j.chembiol.2015.03.019. [DOI] [PubMed] [Google Scholar]
- 51.Swapna L.S., Srikeerthana K., Srinivasan N. PLoS ONE. 2012;7:e36688. doi: 10.1371/journal.pone.0036688. [DOI] [PMC free article] [PubMed] [Google Scholar]