Abstract
Document retrieval (DR) forms an important component in end-to-end question-answering (QA) systems where particular answers are sought for well-formed questions. DR in the QA scenario is also useful by itself even without a more involved natural language processing component to extract exact answers from the retrieved documents. This latter step may simply be done by humans like in traditional search engines granted the retrieved documents contain the answer. In this paper, we take advantage of datasets made available through the BioASQ end-to-end QA shared task series and build an effective biomedical DR system that relies on relevant answer snippets in the BioASQ training datasets. At the core of our approach is a question-answer sentence matching neural network that learns a measure of relevance of a sentence to an input question in the form of a matching score. In addition to this matching score feature, we also exploit two auxiliary features for scoring document relevance: the name of the journal in which a document is published and the presence/absence of semantic relations (subject-predicate-object triples) in a candidate answer sentence connecting entities mentioned in the question. We rerank our baseline sequential dependence model scores using these three additional features weighted via adaptive random research and other learning-to-rank methods. Our full system placed 2nd in the final batch of Phase A (DR) of task B (QA) in BioASQ 2018. Our ablation experiments highlight the significance of the neural matching network component in the full system.
I. INTRODUCTION
Question answering (QA) has emerged as an important field within natural language processing (NLP) and information retrieval (IR) communities to handle the explosion in curated textual and structured datasets. Modern search engines heavily use QA methods under the hood to deliver precise answers to different types of questions. In Google, simple factoid questions whose answers are usually fixed (e.g., “What is the capital of USA?”) directly result in a bold font phrase that captures the answer (e.g., Washington, D.C.) displayed just below the search box. More complex questions may result in small Web text snippets that often contain the answer. For the question “What causes constipation?”, Bing shows an HTML list from WebMD of various causes. In specialized fields such as biomedicine, questions can be much more complex where the answers may not be readily available on Web pages but may need to be gleaned from scientific literature indexed by NIH search engine PubMed. To address challenges in biomedical QA, the U.S. National Library of Medicine (NLM) has been sponsoring a series of community shared tasks under the name BioASQ (http://www.bioasq.org) since 2013 [1]. For a recent BioASQ example question, “Which currently known mitochondrial diseases have been attributed to POLG mutations?”, Google and Bing do not have any straightforward responses but instead point to some research articles. However, what is expected as an answer in BioASQ is a list of diseases.
In the BioASQ QA task, the question types include yes/no (Boolean response to a statement), factoid (answer is a single entity), list (response is a list of entities), and summary, which involves a detailed narrative response. Results are evaluated at various levels of granularity including the relevant documents (PubMed abstracts) retrieved, various snippets (small blurbs of text) retrieved from selected documents, specific biomedical concepts that may directly answer a question, and a so-called “ideal” answer to a question (which is usually a precise English description of the answer). That is, although the eventual goal is the ideal answer(s), documents that contain answers, smaller snippets within in them that contain the answer, and biomedical concepts relevant to the answer are also expected as output and evaluated separately. The corpus available for all retrieval tasks in BioASQ tasks is the set of all PubMed indexed biomedical article citations (title, abstract, and additional metadata such as authors, journal name, and indexing terms). Hence throughout this paper, by document, we mean the title+abstract and any other associated metadata.
In this paper, we specifically focus on the high level document retrieval (DR) component of the BioASQ shared task on QA (task B). This is a natural first step because most end-to-end QA systems first need to identify documents that potentially contain answers. Subsequently, more sophisticated NLP methods are used to identify smaller snippets and next spans of particular phrases representing the answers within them. Also, superior performance in the DR task will lead to overall better end-to-end system performance, all other factors being equal. Hence we focus on this task in our preliminary foray into the BioASQ series. Our approach to DR involves a traditional IR model to get a list of documents and then rerank this list using neural question-answer sentence matching and some auxiliary features involving journal names (of documents) and an external knowledge base of relations extracted from biomedical articles. Specifically, we make the following contributions.
We train a neural sentence matching network to learn a matching score of the question sentence with each sentence in a candidate relevant document. We do this by exploiting the training data that includes the relevant snippets from prior years in the BioASQ series.
We devise a feature that exploits the thematic overlap of a journal in which a candidate document is published and the question at hand, using medical subject headings (MeSH terms) as proxies for thematic content.
We also use an external knowledge base of relations called SemMedDB [2] extracted by applying rule-based relation extraction algorithms to the BioASQ corpus. The main intuition is that documents containing binary relations involving a pair of entities mentioned in the question may have a higher chance of being relevant.
With features discussed thus far in this list, by using adaptive random search and learning-to-rank algorithms, we rerank documents retrieved by a traditional sequential dependence model implemented as part of the open source Galago search engine [3].
Overall, we find that our reranking approach performs consistently better than the baseline retrieval system when tested on the 2016 and 2017 BioASQ test sets. We also participated in BioASQ 2018, and our system came in 2nd (among 26 different entries) in the final batch as shown in Table I (based on the mean average precision (MAP1) measure used by the task organizers).
TABLE I:
The official BioASQ results of the top 5 different systems (2018, task 6b phaseA batch 5)
| System | Mean Precision | Recall | F1 | MAP | GMAP |
|---|---|---|---|---|---|
| aueb-nlp-4 | 0.1145 | 0.3790 | 0.1590 | 0.0695 | 0.0012 |
| ours | 0.1085 | 0.3539 | 0.1513 | 0.0680 | 0.0009 |
| sys2 | 0.1055 | 0.3331 | 0.1458 | 0.0633 | 0.0008 |
| ustb_prir4 | 0.1105 | 0.3441 | 0.1532 | 0.0622 | 0.0009 |
| testtext | 0.1115 | 0.3540 | 0.1550 | 0.0618 | 0.0009 |
II. METHODOLOGICAL DETAILS
We use the BioASQ [1] QA datasets from years 2014 through 2017. When using a certain year’s dataset as test set, we use all preceding years’ datasets for training.
A. Baseline Document Retrieval Model
We use the sequential dependence model (SDM) [4] in the initial document retrieval process as implemented in the open source Galago search engine [3]. Unlike the traditional bag-of-words models, the order of terms in a query is also taken into account in the SDM model. SDM is based on the Markov random field model, in which not only the unigrams but also the ordered and unordered bi-grams in a posed query are considered in the retrieval score computation. The term frequency score is
where qi is a query term, D is the document, θD is a language model built using D, tfqi,D is the term frequency of qi in D, cfqi is the collection frequency of qi, |C| is the total number of terms across all the documents, |D| is the document length, and μ is the Dirichlet prior for the smoothing effect. Likewise, the functions for the ordered and unordered bi-grams are defined in a similar way:
where and indicate the frequencies of the terms qi and qi+1 within an ordered window of N word positions and within a unordered window of M word positions respectively. The final scoring function is the weighted sum of the the three constituent functions
where Q = q1, … , q|Q| is the query and λT, λO, and λU are weights for the unigram, ordered bigram, and unordered bigram components respectively. This SDM scoring function is the baseline throughout all our experiments where we measure the effectiveness of our matching score feature and other auxiliary features.
B. Question-Answer Matching (QAMat) Model
Our QA matching (QAMat) model is an attention-based neural network based on prior efforts on siamese networks in NLP [5]. However, the main difference is that we use separate parameters for encoding the question and candidate sentences while the original siamese network uses the same parameters until the final distance layer. Given the linguistic (lexical and syntactic) layout of a question and the importances of various words in it are different in nature from the relevance of different tokens observed in a candidate answer sentence, different parameter sets for encoding them separately are necessary. Due to this, we see our network as “matching” sentences instead of computing similarity between them.
As outlined earlier, the BioASQ training datasets provide a list of human adjudicated text snippets that are relevant to each question. As such, we train the QAMat model with the pairs of questions and relevant sentences in the ground truth training text snippets. Thus we expect the model to score sentences in a document with regards to their potential for containing an answer to a specific question. We first outline the architecture and subsequently elaborate on training dataset generation.
1). Architecture:
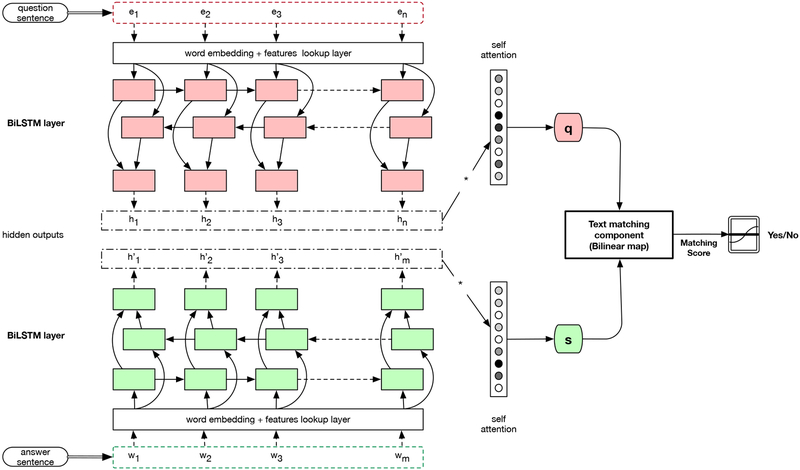
Beyond simple averaging of word embeddings in a sentence, researchers have attempted to build neural models that encode a phrase [6], a sentence, or a document [7] into a discriminative low dimensional vector representation. For QA in particular, a paragraph can be matched to a question sentence to find an answer phrase span in that paragraph [8]. We follow a similar approach where given a question sentence and a candidate answer sentence, the neural net estimates the probability that the answer sentence contains information pertinent to answer the question. We train two bidirectional long short-term memory networks (BiLSTMs [9]), one for encoding a question sentence and the other for encoding a candidate answer sentence as shown in Figure 1.
Fig. 1:
Question-Answering Text Matching (QAMat) Model Architecture
Question Sentence Encoding–
All the tokens in a question sentence Q are mapped to corresponding word embeddings. The word embeddings are then fed into the question BiLSTM to produce hidden node outputs
where ei are embeddings of words in the question and hj are concatenations of the forward and backward LSTM hidden outputs for the j-th position. All hi are subsequently combined into a single fixed-size vector specifically in the form a weighted sum with weights
determined via self-attention and where αj quantifies the attention that needs to be put on the corresponding question word and w is the attention parameter vector learned as part of training. To this weighted sum representation of the question, we concatenate a one-hot 4-bit vector indicating the type of a question to encode the set {yes/no, factoid, list, summary} given the question type may affect the matching process.
Answer Sentence Encoding–
Similarly, we encode a candidate answer sentence representation using a second BiLSTM using word embeddings for the answer sentence tokens. The hidden outputs of the candidate answer sentence are combined using another attention layer just like for the question sentence. Then, the resulting two sentence representations are compared to each other in the next text matching component.
Semantic Matching Model–
Our matching component is based on well known metric learning constructs to measure relatedness or similarity between two vectors [10]. We tested approaches ranging from simple dot product to bilinear maps and recent neural tensor networks [11]. Based on experiments, we finalized the bilinear map metric g(s, q) = sT Wq where s and q are candidate answer sentence and question embeddings respectively as defined in the previous two paragraphs and W is the parameter matrix for the bilinear transformation. In the end, the output scores g(s, q) are passed to the logistic function. The network in Figure 1 is trained with the binary cross-entropy loss function to evaluate the prediction quality.
2). Building Datasets for the QAMat model:
Each instance to train the QAMat model takes the form of a pair of sentences, one representing the question and the other representing the candidate answer sentence. An instance is positive if the second sentence in the pair is relevant to answering the question represented by the first sentence. We use the BioASQ data from previous years for training this. Specifically, all sentences of human-curated text snippets in BioASQ data are labeled as the relevant group. To populate the irrelevant group, we randomly select from the relevant documents those sentences that do not appear in the relevant text snippets. Since the examples are from the relevant documents, we expect the context to be related to the topic of the document but not directly containing content to glean the answer. We also sample irrelevant examples from the entire document collection given the chance of the random samples from over 27 million documents being relevant to the question is extremely low. The proportions for training are as follows:
50% of the sentences are relevant examples, and the other half are irrelevant examples.
Among irrelevant examples, half are sampled from the relevant documents (but outside snippets that contain answers) and half are from the rest of the corpus (irrelevant documents).
C. MeSH Distribution across Questions and Journals
QAMat component from Section II–B is our main explicit feature directly comparing question and document contents. Here we discuss an auxiliary feature involving thematic overlap between question contents and the journal in which a candidate document is published. The medical subject headings (MeSH) is a well-known standardized hierarchical vocabulary used to tag biomedical articles (just like keywords) to facilitate future thematic search by researchers who use NLM’s PubMed search engine. Besides individual articles, a journal name is also assigned a set of MeSH terms. The MeSH terms for an article or journal can be treated as a thematic abstraction of the content in them. MeSH terms can also be extracted using NLM’s medical text indexer (MTI) tool that outputs MeSH terms for any piece of text. Our intuition is that if we can build a distribution of MeSH terms occurring across questions and journals, we can use it to design a feature that takes as input the question and candidate document (thus its journal) and output a score for it based on thematic overlap.
We build a distribution matrix M where the rows are MeSH terms from questions in the training data and the columns are MeSH terms of the journals of the corresponding relevant training documents. Here M[mi][m′j] contains the number of times in the training data we encountered a question with MeSH term mi with a corresponding answer document whose journal has the associated MeSH term m′j. More specifically, let is the set of questions in the training data. Let R(Q) be the set of relevant documents for . Let t(Q) be the MeSH terms mentioned in Q and let t(D) be the set of MeSH terms for the journal of document D. We fill the table M via
where ‘+= k’ refers to increment-by-k operation. We subsequently normalize each row by dividing each cell value with the sum of all elements in that row. With this, M[mi][m′j} now represents P(m′j|mi) – the probability estimate of encountering an answer document whose journal has MeSH term m′j given the question contains term mi. With this setup, given a new question Q, for a candidate document D, the score is
It is straightforward to note μ ∈ [0,1] given the normalization step in building M and the 1/t(Q) factor in computing μ.
D. Semantic Predications in SemMedDB
SemMedDB [2] is a repository of semantic predications (subject-predicate-object triples) that are extracted from the biomedical scientific literature indexed by PubMed using rule-based NLP techniques. The NLM provides an updated SemMedDB every year to include predications from newer articles. In each predication, the subject and object are biomedical entities (e.g., diseases, drugs, and procedures) represented by concepts from the unified medical language system (UMLS). The predicates (e.g., treats and causes) that connect the subject and object come from an extended semantic network [12]. For example, for a PubMed document sentence “We conclude that tamoxifen therapy is more effective for early stage breast cancer patients”, SemMedDB would contain the predication (Tamoxifen Citrate [C0079589], treats, Breast Carcinoma [C0678222]) where the C codes in square braces represent UMLS unique concept identifiers for the entities. We note that relations in SemMedDB have corresponding provenance information of particular sentences (in PubMed citations) they came from. Given the BioASQ search corpus is also PubMed citations, we design features that capture semantic links between concept mentions in the question. Specifically, from a question sentence, we use NLM’s MetaMap software to extract UMLS concepts C(Q) for question Q. For a candidate document D, let C(D) be all UMLS concepts that participated (either as subject or object) in at least one predication in D and let R(D) be set of all predications in D. Our first binary feature π1(Q,D) is set to 1 if and only if |{(i, j) : i,j ∈ C(Q) and (i,p,j) ∈ R(D)}| > 0 for some predicate p. That is, π1 fires only if there exists at least one SemMedDB triple in D whose subject and object are both present in Q. The second feature π2(Q,D) = (|C(Q) ∩ C(D)|)/|C(Q)| is a numerical feature (∈ [0,1]) that measures the proportion of number of concepts present in both Q and semantic predication based concept set C (D) to the total number of concepts in Q.
E. Feature Weighting Methods
Finally, to rerank the top few documents returned by the SDM model, we need a way to combine all the five scores derived from the (1). preliminary SDM retrieval (Section II–A), (2). QAMat (Section II–B), (3). MeSH distribution (Section II–C), (4). SemMedDB relation match, and (5). SemMedDB concept proportion (Section II–D). We note that we scale features to [0,1] range before combining them for final document ranking. Except for the QAMat score, all other features score the entire document. For QAMat, we produce a score for each sentence in the candidate document. To arrive at the final document-level score, we can consider the average of all QAMat scores for all sentences in it, just the maximum value among sentences, or both the average and max scores. Based on our experiments, we chose the simpler maximum score option as involving the average score did not improve the validation set performances.
1). Adaptive Random Search:
The adaptive random search (ARS) method is a particular instance of a class of stochastic optimization methods where a weighted sum of feature scores is used as the final score for ranking documents. In this case, we have five weights α1, … , α5 such that ∑i αi = 1, so the final score is also in [0,1] since all constituent scores are in that range too. ARS starts with a random configuration of αis and incrementally updates them as it proceeds to explore the search space. It does not require derivatives when performing updates. Instead of using a fixed step size, ARS dynamically increases or decreases the step size based on the observed difference between the performances on a validation dataset. Karnopp [13] discusses the details of the ARS algorithm, which we incorporated in our system to optimize the weights for the ranking features.
2). Learning-to-Rank Algorithms:
Learning-to-rank [14] (L2R) has emerged from the machine learning community as an automated way of learning functions that can rank a list of documents in response to an input query based on different query-specific features extracted from the documents. We also compare ARS against a variety of L2R algorithms as implemented in the RankLib library2. For the training data, we use all five feature scores and a binary judgment (‘relevant’ or ‘irrelevant’) for each item. Whether we use ARS or an L2R algorithm, the feature weighting model is built solely from the training dataset.
III. EXPERIMENTS AND RESULTS
We perform experiments on the BioASQ QA datasets (years 2014 through 2017) focusing on the past two years for testing scenarios to examine the efficacy of the proposed approaches. Before we get into our results, we outline some system configuration details for experiments.
SDM component (Section II–A): For this initial document retrieval component, we used its implementation by the Galago search engine [3]. Indexing of the documents was done by the Krovetz stemmer (https://sourceforge.net/p/lemur/wiki/KrovetzStemmer/), included in the Galago system. The window width for the ordered query tokens (N in Section II–A) is increased from the default setting of 1 to 3. The unordered width is increased from the default setting of 4 to 8 (M in Section II–A). Empirically, this setting improved the recall scores. We choose the default settings in the Galago implementation of SDM and set unigram score weight λT = 0.8, ordered distance score weight λO = 0.15, and unordered window weight λU = 0.05. Finally, the maximum number of documents to be retrieved using SDM is set to 30.
QAMat component (Section II–B): For the neural matching component, we use pre-trained word embeddings with 300 dimensions trained on Wikipedia using fastText [15]. The dimensionality of the BiLSTM hidden layers is set to 256 (determined via experiments). For regularization, we apply a dropout to the inputs of the LSTM layers with the dropout rate of 0.3. The attention layer output is 512 dimensional given the hidden layer output is 256 dimensions in each direction in the BiLSTM. In order to indicate the type of the given question, four additional bits are appended to the question representation; hence the parameter matrix W of the following bilinear matching function is set to (512 × 516). The maximum number of epochs is set to 30 with early stopping enabled, and batch size is fixed at 128. We train the model using Adamax optimizer with an initial learning rate of 0.005 and a weight decay of 0.0005. Gradient clipping is set to 10 to avoid the exploding gradient problem. All other network weights are based on default initializations in PyTorch [16].
A. Experiments for the QAMat Feature
In Table II, we show the counts of datasets created for training the QAMat model as discussed in Section II-B2. We chose the datasets to be balanced given we do not want to compromise too much on recall and because we have other evidences (SDM, MeSH distribution, SemMedDB match scores) to alleviate precision trade-off concerns. For each question, the positive examples in the datasets were based on those found in the BioASQ datasets and negative examples were generated randomly from the rest of the corpus. We achieved test set accuracies of ≈ 87% for the QAMat component. Next, we look at a sample question and QAMat scores (before they are passed to the sigmoid function) for answer sentences.
TABLE II:
Training dataset counts for QAMat training
| dataset | relevant | irrelevant |
| train (2014–2015) | 23,466 | 23,466 |
| test (2016) | 16,706 | 16,706 |
| (a) datasets for testing on year 2016 | ||
| dataset | relevant | irrelevant |
| train (2014–2016) | 33,075 | 33,075 |
| test (2017) | 9,582 | 9,582 |
| (b) datasets for testing on year 2017 | ||
Table III shows how the QAMat model scores the sentences of an example relevant document and also the ones of another random irrelevant document for the question “Orteronel was developed for treatment of which cancer?”. As we can see, the relevant document sentences that succinctly discuss treatment of cancer with orteronel have scored high. Other sentences in the document that contain a lot more information do not have as high a score as smaller sentences that pointedly talk about orteronel drug therapy for cancer. All the sentences in the irrelevant document attain negative scores, all of which are worse than the lowest score achieved by the relevant sentences.
TABLE III:
QAMat scores for sentences of a relevant and an irrelevant document for an example question
| Question: Orteronel was developed for treatment of which cancer? | |
|---|---|
| Score | Sentence of a relevant document |
| 3.3877 | Orteronel also known as TAK-700 is a novel hormonal therapy that is currently in testing for the treatment of prostate cancer. |
| −0.2706 | Orteronel inhibits the 17,20 lyase activity of the enzyme CYP17A1, which is important for androgen synthesis in the testes, adrenal glands and prostate cancer cells. |
| −2.2917 | Preclinical studies demonstrate that orteronel treatment suppresses androgen levels and causes shrinkage of androgen-dependent organs, such as the prostate gland. |
| 0.2731 | Early reports of clinical studies demonstrate that orteronel treatment leads to reduced prostate-specific antigen levels, a marker of prostate cancer tumor burden, and more complete suppression of androgen synthesis than conventional androgen deprivation therapies that act in the testes alone. |
| −2.2761 | Treatment with single-agent orteronel has been well tolerated with fatigue as the most common adverse event, while febrile neutropenia was the dose-limiting toxicity in a combination study of orteronel with docetaxel. |
| −0.3830 | Recently, the ELM-PC5 Phase III clinical trial in patients with advanced-stage prostate cancer who had received prior docetaxel was unblinded as the overall survival primary end point was not achieved. |
| 2.2541 | However, additional Phase III orteronel trials are ongoing in men with earlier stages of prostate cancer. |
| Score | Sentence of a random (irrelevant) document |
| −7.0492 | The dynamics of antibody response in guinea pigs infected with Coxiella burnetii was investigated by microagglutination MA and complement-fixation CF tests with different preparations of C. burnetii antigens. |
| −4.5142 | At the onset of antibody response the highest antibody titres were detected by the MA test with natural antigen 2, later on by the MA test with artificial antigen 2. |
| −7.1499 | Throughtout the 1-year period of observation, the CF antibody levels were usually lower and, with the exception of the highest infectious doses, the CF antibodies appeared later than agglutinating antibodies. |
| −7.1499 | There was no difference in the appearance of agglutinating and CF antibodies directed to antigen 1. |
| −4.2314 | Inactivation of the sera caused a marked decrease in antibody titres when tested with artificial antigen 2, whereas the antibody levels remained unchanged when tested with natural antigen 2. |
B. L2R Vs ARS for Feature Weighting
Table IV shows the mean average precision (MAP) results when using different feature weighting methods. Surprisingly, ARS outperforms all other methods except for one out of ten batches considered. MART, Coordinate Ascent, and Random Forests more or less perform at the same level but trail behind ARS. We believe L2R algorithms may perform better in situations where features used have non-trivial correlations. In this case, it appears the features considered may be contributing complementary evidence.
TABLE IV:
Feature weighting method comparison based on MAP
| BioASQ test datasets | MAP for learning-to-rank algorithms | ||||||
|---|---|---|---|---|---|---|---|
| ARS | MART | RankBoost | AdaRank | CoordAscent | LambdaMART | RandForests | |
| year 2016, batch 1 | 0.4438 | 0.4181 | 0.3731 | 0.3792 | 0.4296 | 0.4025 | 0.4175 |
| year 2016, batch 2 | 0.4780 | 0.4625 | 0.3698 | 0.4396 | 0.4493 | 0.4497 | 0.4736 |
| year 2016, batch 3 | 0.4534 | 0.4198 | 0.3366 | 0.4009 | 0.4274 | 0.4026 | 0.4417 |
| year 2016, batch 4 | 0.4388 | 0.4036 | 0.3490 | 0.3813 | 0.4127 | 0.4022 | 0.4296 |
| year 2016, batch 5 | 0.3722 | 0.3563 | 0.2869 | 0.3263 | 0.3551 | 0.3314 | 0.3729 |
| year 2017, batch 1 | 0.4075 | 0.3843 | 0.2616 | 0.1233 | 0.3786 | 0.3517 | 0.3975 |
| year 2017, batch 2 | 0.4363 | 0.4334 | 0.3300 | 0.1457 | 0.4299 | 0.4227 | 0.4263 |
| year 2017, batch 3 | 0.4534 | 0.4377 | 0.3223 | 0.1536 | 0.4456 | 0.4105 | 0.4434 |
| year 2017, batch 4 | 0.3891 | 0.3693 | 0.2598 | 0.1193 | 0.3763 | 0.3362 | 0.3791 |
| year 2017, batch 5 | 0.2316 | 0.2068 | 0.1226 | 0.0793 | 0.2170 | 0.1887 | 0.2216 |
C. Ablation Study
We perform a feature ablation study to measure the contributions of different features discussed in Section II. We first build a full model consisting of all features and subsequently drop each component, one at a time, to note the dip in performance (here MAP). Table V shows the results of these experiments for test sets from 2016 and 2017. The first rows in Table V(a) and Table V(b) have results from our full model and the last rows are based on the baseline SDM model (Section II–A). Rows 2–4 indicate dropped components from Sections II–B–II–D respectively. The bold scores indicate the values that had the biggest drop from the corresponding full featured model score in the first row. We also note that the blue colored scores (1st rows) indicate the best performance achieved in each test batch. That is, in all batches, our fully featured model obtained the best scores.
TABLE V:
Ablation study - Bold entries indicate biggest drop in MAP and blue entries correspond to best MAP values
| Models | Optimized ARS weights | MAP | ||||||||
| SDM score | QAMat | MeSH Dist. | SemMedDB1 | SemMedDB2 | batch1 | batch2 | batch3 | batch4 | batch5 | |
| All | 0.3043 | 0.4747 | 0.0846 | 0.0102 | 0.1264 | 0.4438 | 0.4780 | 0.4534 | 0.4388 | 0.3722 |
| – QAMat | 0.4515 | - | 0.2848 | 0.0718 | 0.1919 | 0.4202 | 0.4721 | 0.4227 | 0.4145 | 0.3604 |
| – MeSH Dist. | 0.3279 | 0.5771 | - | 0.0309 | 0.0642 | 0.4410 | 0.4659 | 0.4476 | 0.4307 | 0.3614 |
| – SemMedDB | 0.2896 | 0.5365 | 0.1739 | - | - | 0.4352 | 0.4680 | 0.4329 | 0.4161 | 0.3521 |
| Baseline | 1.0000 | - | - | - | - | 0.4279 | 0.4709 | 0.4306 | 0.4219 | 0.3505 |
| (a) Results on year 2016 datasets | ||||||||||
| Models | Optimized ARS weights | MAP | ||||||||
| SDM score | QAMat | MeSH Dist. | SemMedDB1 | SemMedDB2 | batch1 | batch2 | batch3 | batch4 | batch5 | |
| All | 0.1665 | 0.7298 | 0.0411 | 0.0062 | 0.0564 | 0.4075 | 0.4363 | 0.4534 | 0.3891 | 0.2316 |
| – QAMat | 0.5243 | - | 0.1832 | 0.2406 | 0.0520 | 0.3782 | 0.4190 | 0.4372 | 0.3690 | 0.2181 |
| – MeSH Dist. | 0.3121 | 0.5494 | - | 0.0638 | 0.0747 | 0.3956 | 0.4221 | 0.4471 | 0.3772 | 0.2144 |
| – SemMedDB | 0.2970 | 0.5220 | 0.1811 | - | - | 0.3808 | 0.4316 | 0.4459 | 0.3758 | 0.2154 |
| Baseline | 1.0000 | - | - | - | - | 0.3959 | 0.4176 | 0.4378 | 0.3746 | 0.2133 |
| (b) Results on year 2017 datasets | ||||||||||
We display the optimized [0, 1] ARS weights in Table V in columns 2–6. We observe that QAMat score takes the highest weight by a large margin compared to other feature weights. Furthermore, QAMat’s weight increases in 2017 compared with its weight in 2016 potentially due to the availability of more training data for 2017. However, the baseline SDM model (last rows) by itself does reasonably well but scores around 2% below our full model’s MAP. Moreover, our model can highlight sentences based on high QAMat scores that are expected to contain crucial information pertinent for answering the question. Coming to ablation results, from rows 2–4, we notice that dropping the QAMat component causes the biggest drop in MAP in most of the cases. Although the MeSH distribution and SemMedDB features were useful, the ablation results show that their contribution is much less than that of the baseline SDM scores and QAMat scores.
IV. RELATED WORK
Our main contribution here is the retrieval of relevant documents with an end goal of finding answers to specific questions in biomedicine. Unlike other ad hoc IR tasks, the BioASQ IR task is unique in the sense that it is part of a more complex set of tasks including snippet retrieval and QA. In this section, we briefly discuss other efforts related to this paper.
Biomedical information retrieval has benefited from multiple shared tasks including TREC genomics [17], clinical decision support [18], and precision medicine [19] tracks, the CLEF user-centered health information retrieval task [20], and the BioASQ retrieval and QA task [1]. The use of neural approaches for IR is on the rise in general [21], also for question-answer matching [22] and biomedical QA [23], [24]. However, classical non-neural IR approaches especially those that employ pseudo-relevance feedback and extensions of SDM model are topping the BioASQ IR task during recent years [25]. Our immediate goal is to combine the best of both worlds to build a superior IR system as elaborated in future research directions in Section V.
Our sentence matching component is mainly derived from recent research in machine reading comprehension (MRC). Over the past few years, researchers made significant progress with end-to-end MRC models by utilizing various input embeddings and attention mechanisms. Seo et al. [26] combined character embeddings along with pre-trained word embeddings with an attention flow mechanism to model the context for a query. In the transformer architecture proposed by Vaswani et al. [27], the multi-head attention mechanism allows the model to jointly attend to information from different representation subspaces at different positions. These relatively more complex architectures may be useful in our matching task too.
V. CONCLUSION
In this paper, we demonstrated the effectiveness of the three different relevance measures for a biomedical document retrieval task where the query takes the form of a question. The first involves computing matching scores via dense neural representations of both the question sentence and candidate answer sentences. The second one utilizes thematic overlap between a document and the question based on distributional information of MeSH terms in questions and journals of corresponding answer documents. The third prioritizes documents that contain relations between concepts found in the question. We showed that our proposed features help improve the retrieval quality consistently, and the official results in the 2018 BioASQ task (Table I) confirm this finding. Next we discuss some future research directions.
Based on the SDM model in Section II–A, we limit the number of documents to retrieve for reranking to 30. Although it is important to limit the size of the candidate document set to be reranked, additional experiments where pseudo-relevance feedback is employed on top of SDM might be beneficial. That is, based on the top scoring (using the QAMat model) sentences in the top 30 documents, we may be able to expand the query to obtain more highly relevant documents with a second SDM fetch operation. The expansion can be in the form of new query terms or entities that ought to be included in the query.
We used the type of question (yes/no, factoid, list, or summary) as part of the question representation matching process in Section II–B. However, the 4-bit vector that represents the question type is added after the attention mechanism is applied to form a weighted vector for the question. It would be interesting to see how the scoring would change if the question type information is used as part of the attention mechanism. This can be accomplished by choosing a different attention parameter vector for each question type. Although this would be more time consuming, it might help the attention mechanism to focus more on words that might matter based on the question type.
Also, for factoid and list question types, we may be able to ascertain the semantic type of the entities that constitute the answer. For the example for the question in Table III, through NLP methods involving dependency parsing, we might be able to determine that the answer entity is a disease (cancer, specifically). We can then parametrize the attention mechanism for the answer sentence and also the matching process based on this additional piece of information about the answer type. For instance, a candidate sentence that has more entities of the answer type detected in the question ought to be scored higher than other sentences that do not contain answer type entities.
ACKNOWLEDGEMENT
This research is supported by the U.S. National Library of Medicine through grant R21LM012274. We also gratefully acknowledge the support of the NVIDIA Corporation for its donation of the Titan X Pascal GPU used for this research. We thank anonymous reviewers for their helpful comments.
Footnotes
2018 17th IEEE International Conference on Machine Learning and Applications
The MAP values in the table are much smaller than what they ought to be due to the special way BioASQ organizers compute AP, for which they always divide the p@k sum by 10 instead of the actual number of relevant documents (given the maximum number of relevant items they allow for a system is ten). This makes the MAP value much smaller given many questions have < 10 relevant documents. In our experiments in the rest of the paper, we use the standard MAP formula to give realistic scores.
Open source collection of learning-to-rank implementations part of the Lemur project: http://sourceforge.net/p/lemur/wiki/RankLib/
Contributor Information
Jiho Noh, Department of Computer Science, University of Kentucky, Lexington KY..
Ramakanth Kavuluru, Div. of Biomedical Informatics (Internal Medicine), University of Kentucky, Lexington KY..
REFERENCES
- [1].Tsatsaronis G, Balikas G, Malakasiotis P, Partalas I, Zschunke M, Alvers MR, Weissenborn D, Krithara A, Petridis S, Polychronopoulos D et al. , “An overview of the BIOASQ large-scale biomedical semantic indexing and question answering competition,” BMCbioinformatics, vol. 16, no. 1, p. 138, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].National Library of Medicine. (2016) Semantic MEDLINE Database. http://skr3.nlm.nih.gov/SemMedDB/.
- [3].Croft WB, Metzler D, and Strohman T, Search engines: Information retrieval in practice. Addison-Wesley Reading, 2010. [Google Scholar]
- [4].Metzler D and Croft WB, “A Markov random field model for term dependencies,” in Proceedings of the 28th annual international ACM SIGIR conference. ACM, 2005, pp. 472–479. [Google Scholar]
- [5].Mueller J and Thyagarajan A, “Siamese recurrent architectures for learning sentence similarity” in AAAI, vol. 16, 2016, pp. 2786–2792. [Google Scholar]
- [6].Yin W and Schutze H, “Discriminative phrase embedding for paraphrase identification,” in Proceedings of the 2015 Conference of the North American Chapter of the ACL, 2015, pp. 1368–1373. [Google Scholar]
- [7].Le Q and Mikolov T, “Distributed representations of sentences and documents,” in International Conference on Machine Learning, 2014, pp. 1188–1196. [Google Scholar]
- [8].Chen D, Fisch A, Weston J, and Bordes A, “Reading wikipedia to answer open-domain questions,” in Proceedings of the 55th Annual Meeting of the ACL ), vol. 1, 2017, pp. 1870–1879. [Google Scholar]
- [9].Hochreiter S and Schmidhuber J, “Long short-term memory,” Neural computation, vol. 9, no. 8, pp. 1735–1780, 1997. [DOI] [PubMed] [Google Scholar]
- [10].Kulis B et al. , “Metric learning: A survey,” Foundations and Trends® in Machine Learning, vol. 5, no. 4, pp. 287–364, 2013. [Google Scholar]
- [11].Socher R, Chen D, Manning CD, and Ng A, “Reasoning with neural tensor networks for knowledge base completion,” in Advances in neural information processing systems, 2013, pp. 926–934. [Google Scholar]
- [12].National Library of Medicine. (2016) Current Relations in the Semantic Network. https://www.nlm.nih.gov/research/umls/META3_current_\relations.html.
- [13].Karnopp DC, “Random search techniques for optimization problems,” Automatica, vol. 1, no. 2–3, pp. 111–121, 1963. [Google Scholar]
- [14].Liu T-Y, “Learning to rank for information retrieval,” Foundations and Trends in Information Retrieval, vol. 3, no. 3, pp. 225–331, 2009. [Google Scholar]
- [15].Bojanowski P, Grave E, Joulin A, and Mikolov T, “Enriching word vectors with subword information,” Transactions of the Association of Computational Linguistics, vol. 5, no. 1, pp. 135–146, 2017. [Google Scholar]
- [16].Paszke A, Gross S, Chintala S, Chanan G, Yang E, DeVito Z, Lin Z, Desmaison A, Antiga L, and Lerer A, “Automatic differentiation in PyTorch,” NIPS 2017 Workshop on Autodiff, 2017. [Google Scholar]
- [17].Roberts PM, Cohen AM, and Hersh WR, “Tasks, topics and relevance judging for the TREC genomics track: five years of experience evaluating biomedical text information retrieval systems,” Information Retrieval, vol. 12, no. 1, pp. 81–97, 2009. [Google Scholar]
- [18].Roberts K, Simpson M, Demner-Fushman D, Voorhees E, and Hersh W, “State-of-the-art in biomedical literature retrieval for clinical cases: a survey of the TREC 2014 CDS track,” Information Retrieval Journal, vol. 19, no. 1–2, pp. 113–148, 2016. [Google Scholar]
- [19].Roberts K, Demner-Fushman D, Voorhees EM, Hersh WR, Bedrick S, Lazar AJ, and Pant S, “Overview of the TREC 2017 precision medicine track,” in Proceedings of TREC Conference, 2017, pp. 1–13. [PMC free article] [PubMed] [Google Scholar]
- [20].Zuccon G, Palotti J, Goeuriot L, Kelly L, Lupu M, Pecina P,Mueller H, Budaher J, and Deacon A, “The IR task at the CLEF eHealth evaluation lab 2016: user-centered health information retrieval,” in CLEF 2016-Conference and Labs of the Evaluation Forum, vol. 1609, 2016, pp. 15–27. [Google Scholar]
- [21].Onal KD, Zhang Y, Altingovde IS, Rahman MM, Karagoz P,Braylan A, Dang B, Chang H-L, Kim H, McNamara Q et al. , “Neural information retrieval: At the end of the early years,” Information Retrieval Journal, vol. 21, no. 2–3, pp. 111–182, 2018. [Google Scholar]
- [22].Tran NK and Niedereée C, “Multihop attention networks for question answer matching,” in The 41st International ACM SIGIR Conference. ACM, 2018, pp. 325–334. [Google Scholar]
- [23].Mollá D, “Macquarie university at BioASQ 5B-query-based summarisation techniques for selecting the ideal answers,” BioNLP 2017, pp. 67–75, 2017. [Google Scholar]
- [24].Wiese G, Weissenborn D, and Neves M, “Neural question answering at BioASQ 5B,” BioNLP 2017, pp. 76–79, 2017. [Google Scholar]
- [25].Jin Z-X, Zhang B-W, Fang F, Zhang L-L, and Yin X-C, “A multistrategy query processing approach for biomedical question answering: USTB_PRIR at BioASQ 2017 Task 5B,” BioNLP, pp. 373–380, 2017. [Google Scholar]
- [26].Seo M, Kembhavi A, Farhadi A, and Hajishirzi H, “Bidirectional attention flow for machine comprehension,” in Proceedings of the International Conference on Learning Representations, 2017. [Google Scholar]
- [27].Vaswani A, Shazeer N, Parmar N, Uszkoreit J, Jones L, Gomez AN, Kaiser L, and Polosukhin I, “Attention is all you need,” in Advances in Neural Information Processing Systems, 2017, pp. 5998–6008. [Google Scholar]