Abstract
RATIONALE:
As a powerful ambient ion source, atmospheric pressure (AP)-MALDI enables direct analysis at atmospheric pressure/temperature and minimal sample preparation. With the increasing usage of AP-MALDI sources with Orbitrap instruments, systematic characterization of the extent of ion suppression effect (ISE) in AP-MALDI-Orbitrap mass spectrometry imaging (MSI) is desirable. Recently, a new low-pressure MALDI platform has been introduced that reportedly provided better sensitivity. While extensive research efforts have been devoted to improving spatial resolution, fewer studies focused on the characterization and sensitivity improvement of these MALDI platforms that, coupled with high-resolution Orbitraps, provide powerful strategy for MS imaging.
METHODS:
Here, we compared the analytical performance of AP and low-pressure (subatmospheric) MALDI sources to study the effect of pressure control in the ion source. Using a model peptide/protein mixture, we systematically evaluated the factors influencing ISE. Furthermore, the effect of laser spot size was evaluated through tissue imaging analysis of lipids and neuropeptides. The effects of ion suppression and laser spot size have also been examined by comparing the number of identified molecular species during MSI analysis.
RESULTS:
Several key operating parameters including source pressure, laser energy, laser repetition rate, and microscopic slide coating materials were optimized to minimize the ISE. Under the optimal conditions, the subatmospheric AP-MALDI-Orbitrap platform with high spatial and mass spectral resolution enabled significantly improved coverage of several lipid and neuropeptide families in the MS analysis of mouse brain tissue sections.
CONCLUSIONS:
The new SubAP-MALDI source coupled with an Orbitrap mass spectrometer was established as a viable platform for in situ endogenous biomolecular analysis with increased sensitivity compared to conventional AP-MALDI sources as evidenced by the confident identification of neuropeptides from mouse brain imaging analyses. The alleviated ISE was key to substantial performance improvement due to optimized intermediate pressure conditions and better ion collection by the ion funnel.
Keywords: Mass spectrometry imaging, ion suppression effect, ambient ionization, AP-MALDI, neuropeptide, lipid
INTRODUCTION
Electrospray ionization (ESI) and MALDI are two techniques extensively used to ionize a wide variety of biomolecules prior to mass spectrometry (MS) identification. While ion suppression effect (ISE) has been extensively studied in ESI,1–6 relatively little information has been provided to characterize and alleviate the ISE-induced sensitivity loss in MALDI.7 The information on the extent of ISE at different source parameters is especially critical for atmospheric pressure (AP)-MALDI because of the increasing usage of AP-MALDI MS methods for in situ molecular imaging of a variety of molecular species in biological tissue sections with high spatial resolution.8–9 Of note, AP-MALDI MSI enables sampling at semi-physiological conditions (i.e., at physiological temperature and pressure), which minimizes vacuum-induced sample alteration and the loss of volatile analytes.10 Compared to vacuum MALDI sources hyphenated with the Orbitrap XL platform equipped with a nitrogen laser (60 Hz, 337 nm),11–12 one of the distinct merits of the AP-MALDI-Orbitrap is the feasibility of achieving higher spatial resolution during tissue imaging analysis. Furthermore, the decreased laser spot size in AP MALDI source, together with collisional cooling under AP condition than that of under vacuum condition, helps to alleviate in-plume thermal and photochemical fragmentation, which might contribute to the gentle/soft nature of AP MALDI and better reproducibility compared to vacuum MALDI.13 Nevertheless, this softer ionization is not necessarily accompanied with higher sensitivity when compared with vacuum MALDI. Therefore, optimization of gas pressure in the MALDI source is a critical step to increase the ionization yield thus improving MALDI MS analysis.
To assess the ISE-induced sensitivity degradation, it is helpful to consider possible underlying ionization mechanisms in the MALDI source. There are two major models for the MALDI processes extensively discussed in the last 30 years, including the photochemical ionization model and the cluster ionization model.14–16 More recently, a thermal proton transfer model8, 17–18 has been proposed to explain the ionization processes in MALDI, while other experimental efforts have been devoted to the understanding of initial material ejection steps including the surface19–20 and bulk21–23 molecular evaporation assumptions. However, the common characteristics between the various models seem to be the secondary reactions, including ion-molecule reactions during in-plume charge transfer processes at a time scale (e.g., 10s-100s of ns) much longer than the laser pulse duration (e.g., 3–5 ns for N2 or Nd:YAG lasers).24 Numerous studies have been devoted to MALDI sensitivity improvement and ISE alleviation for a wide range of analytes. Previously, the development of a matrix with high salt-tolerance was reported to alleviate the ISE in protein and peptide analysis.25–26 Several studies compared the effects of matrix application techniques on several aspects of MALDI-MSI performance, including analyte diffusion, reproducibility, and sensitivity that are directly related to the ISE of a MALDI source.27–29 MALDI sensitivity was also reported to be improved through the rational design of matrices and the addition of glycerol which generates multiply-charged protein/peptide ions.30–32 Multiply-charged ions can also be produced with matrix and vacuum alone without the use of a laser,33 with a reflective geometry,34 or with a laser in a transmission geometry.34 A picosecond infrared laser system was recently used to extract and liberate highly-charged proteins in water.35–36 Ammonium formate37 and ammonium sulfate38 have been used to enhance the MALDI sensitivity for lipids and hydrophilic quaternary ammonium compounds, respectively. Addition of concentrated nitrilotriacetic acid to the matrix solution during sample preparation was also shown to effectively reduce the MALDI matrix-induced ISE and enhance the peptide and protein identification from peptide mass fingerprinting.39
Notably, differential and severe regional ISE during MALDI-MSI have recently been investigated.40 The sources for differential ISE during MSI include endogenous salt heterogeneity, cell density diversity, and inherent differences in ionization due to distinct chemical properties. It has thus further strengthened the necessity to systematically characterize and effectively alleviate the ISE of MALDI, especially for AP-MALDI. Recently, we have successfully applied an AP-MALDI-Orbitrap MSI platform for high spatial resolution MSI of various biomolecules.41 While previous studies were mainly focused on the improvement of spatial and mass resolution of AP-MALDI MSI, herein, we systematically evaluated the ISE-induced sensitivity degradation through optimization of several key operating parameters including source pressure, laser energy, laser firing repetition rate, and slide coating materials. In our experiments, a SubAP-MALDI source was coupled to a Q Exactive HF Orbitrap mass spectrometer to achieve both high spatial and mass spectral resolution. The new SubAP-MALDI-Orbitrap platform was found to offer increased sensitivity compared to that in the AP-MALDI due to the use of low-pressure conditions benefiting the ion yield during the desorption/ionization stage and better transmission of produced ions to the mass analyzer through the use of the ion funnel. By studying dried droplets containing mixtures of small and large peptides, we estimated the dependence of the ion yield on the laser spot size in the SubAP-MALDI source under conditions of optimal gas pressure and electric field. The effect of laser spot size on sensitivity was also studied in MSI experiments. In the latter, the extent of ISE has been evaluated for the AP and low-pressure MALDI sources by comparing both the number of identified molecular species and overall coverage. Collectively, we have demonstrated the utility of a new SubAP-MALDI platform for biomolecular analysis with reduced ISE. We established the limits of its detection and evaluated analytical performance for MSI with high spatial resolution.
EXPERIMENTAL SECTION
Chemical Reagents.
Methanol (MeOH), acetonitrile (ACN), acetic acid, and formic acid (FA) were purchased from Fisher Scientific (Pittsburgh, PA). α-cyano-4-hydroxycinnamic acid (CHCA) and 3-nitrobenzyl alcohol (3-NBA) were purchased from Sigma-Aldrich (St. Louis, MO). 2, 5-dihydroxybenzoic acid (DHB) was purchased from Acros Organics (Morris Plains, NJ). Microscope glass slides were purchased from VWR international, LLC (Radnor, PA). Conductive indium tin oxide (ITO)-coated glass slides were purchased from Bruker (Billerica, MA). Human insulin was purchased from Promega (Madison, WI, USA). Peptide standards (bradykinin, somatostatin II, angiotensin II, FMRF, FMRFamide-like peptide II (lobster)) were purchased from American Peptide Company (Sunnyvale, CA, USA). Amyloid beta peptides were purchased from Sangon Biotech Co., Ltd. (Shanghai, China). No further purifications were performed for any reagents. All solvents used in this study were of HPLC grade. Purified water (conductivity of 18.2 MΩ·cm) was obtained from Milli-Q Reference System (Millipore Corp., Bedford, MA, USA).
Matrix Preparation.
CHCA and DHB matrices were used in this study. Specifically, 10 mg/mL CHCA (ACN/H2O/FA = 49.95/49.95/0.1, v/v/v) and 150 mg/mL DHB (MeOH/H2O/FA = 49.95/49.95/0.1, v/v/v) were used for standard spot analysis. For MSI, the concentrations of CHCA and DHB were 5 mg/mL and 40 mg/mL, respectively.
Tissue Sample Preparation.
Animal experiments were conducted following institutional guidelines (University of Wisconsin-Madison, IACUC). Female C57BL/6J mice were anesthetized, perfused with chilled phosphate buffered saline, and decapitated, and the brains were removed. The brains were then cut along midline. Each hemisphere was embedded in gelatin (100 mg/mL in water), snap frozen in dry ice and stored at −80 °C. The gelatin-embedded tissue was cryo-sectioned into 12 μm slices on a cryostat (Thermo Scientific Microm HM 525, Thermo Scientific, Kalamazoo, MI) at −20 °C and thaw-mounted onto an ITO-coated slide or a microscope glass slide. The optical images of tissue sections were acquired by an Olympus SZX16 stereo microscope (Olympus, Center Valley, PA) with bright field illumination.
MSI Matrix Application.
The tissue slide was dried at room temperature for 30 minutes in a desiccator before matrix application. A robotic TM sprayer system (HTX Technologies, Carrobo, NC) was used for depositing DHB. The nozzle temperature was set to 80 °C with a moving velocity of 800 mm/min. Four or eight passes of matrix were deposited at a flow rate of 0.1 mL/min with 30 seconds drying time between each pass. The sprayed slide was dried at room temperature for 30 minutes and stored in a desiccator at −20 °C until analysis.
SubAP-MALDI.
A SubAP-MALDI (ng) UHR (MassTech, Columbia, MD) was used as a laser ion source. A similar laser module can be found in our previous study.41 Notably, the SubAP-MALDI source was equipped with a 355 nm Nd:YAG laser along with a two-stage ion funnel for improved ion collection and transmission (compared to the AP-MALDI source). The typical operating pressure range in a chamber containing the MALDI plate holder and the first funnel was 3–5 Torr, which makes it SubAP-MALDI, rather than AP-MALDI. The key laser parameters (unless otherwise specified) are: laser energy of 18.9% (of total 2 μJ) and repetition rate of 1 kHz for CHCA, and 18.9% and 200 Hz for DHB. The RF voltage used to confine the ions moving through the ion funnel was 250 V (peak-to-peak). Under optimal conditions, the voltage applied to the MALDI plate was approximately 250–300 V, and the voltage applied to the first electrode in the 15-mm long accelerating stack located 6 mm away from the MALDI plate was 180–200 V. Following that, a drop in the electric potential across the first funnel (60 mm long) was 80–100 V, with the voltage drop across the second funnel (110 mm long) being 10–20 V. The voltage drop between the exit from the first and the entrance to the second funnel was approximately 50–70 V.
Sample plate loading.
To load the sample plate into a SubAP-MALDI source, it requires a two-step process, vacuum breakage and vacuum build-up. Vacuum breakage: 1) With the ion funnel set to Standby mode, Eject the plate holder; 2) Turn the knob (on left side of source) clockwise until the vacuum gate valve is totally closed and the pressure gauge starts to show pressure increase. Then release the metal buckle (on right side of source) and wait ~2 min until the source automatically opens; 3) Carefully place the sample plate onto sample holder and ensure plate does not wiggle. Standard microscopic slides (typically 75mm × 25mm, 1 mm thick) need to be cut into a proper length (~50 mm) prior to loading so it stretches out the slide holder for less than 5 mm for each end or the glass might be crushed during the source initiation procedure. One is advised to use 25 mm ×50 mm slides to avoid the slide shortening. Vacuum built-up: 1) Close the source with buckle closed, 2) Turn on small roughing pump (first, make sure the handle on the pump hose is perpendicular to the hose) then flip handle to make it parallel to the hose to allow gas evacuation from the source. 3) When the vacuum gauge shows gas pressure drops below 40 Torr, start turning the knob counterclockwise and make 4 turns. 4) Turn handle 90 degrees on the hose to disconnect small vacuum pump from SubAP MALDI source then turn off the small pump. 5) When the pressure in the source drops below 10 Torr, fully open the gate valve by turning the knob counterclockwise until its full range is achieved. The pressure shown by gas gauge should drop again. The working pressure in the MALDI source should be between 3 and 5 Torr. Make sure that the vacuum pressure in the intermediate section shown in the Tune program is below 1.9 mbar.
Mass spectrometry.
The SubAP-MALDI source was coupled with a QE-HF mass spectrometer (Thermo Fisher Scientific, Bremen, Germany) for all data acquisition. A software tool (Target-ng, MassTech, Columbia, MD) was used to control the XY stage motion in the ion source, while iFunnel software (MassTech, Columbia, MD) was utilized to control the ion funnel operation. In the dried droplet sample preparation method, peptide standards were premixed with matrix solutions and then applied onto an ABI Opti-TOF 192 target plate (Applied Biosystems, Foster City, CA) and analyzed using a spiral motion mode for the laser spot travel across the sample surface. For MSI, constant speed raster motion was used. The mass spectrometer was operated in the MS mode with positive polarity. A mass range of m/z 400–4,000 was used, and the resolution was set at 120,000 unless otherwise specified. Automatic gain control (AGC) target was set to 3×106 with 300 ms maximum injection time. The AGC value was high enough to ensure that in every scan the ion accumulation took place exactly during 300 ms injection time which, with the constant scan speed of the MALDI target plate, ensured each pixel was the same size and prevented the Orbirap cell from being overloaded.
Data Analysis.
Xcalibur (Thermo Scientific, Bremen, Germany) was used to process spectra. ImageQuest (Thermo Scientific, Bremen, Germany) was used for image processing, including intensity normalization and ion image generation. LIPID Metabolites and Pathways Strategy (LIPID MAPS, University of California-San Diego, La Jolla, CA), Sweden peptide database (SwePep, Uppsala University, Uppsala, Sweden) and an in-house crustacean neuropeptide database (Li Lab, University of Wisconsin-Madison, Madison, WI) were used for accurate mass matching of lipids and neuropeptides with 5 ppm tolerance.
RESULTS AND DISCUSSION
Characterization of Ion Suppression Effects (ISE) in MALDI.
In order to characterize the ISE in MALDI analysis with the feasibility of source pressure control, we utilized a high spatial resolution SubAP-MALDI source with improved source pressure adjusting capability compared to the regular AP-MALDI source. Similar to the previous AP-MALDI setup developed by MassTech,41–42 SubAP-MALDI was interfaced with a QE-HF Orbitrap instrument (Scheme 1a). A representative mass spectrum of a peptide standard mixture recorded from the SubAP-MALDI-Orbitrap is shown in Figure 1. Large and small peptides, including FMRFamide (599 Da), Bradykinin (1060 Da), FMRF-like neuropeptide (1243 Da), somatostatin (1637 Da), Aβ (1–16) (1955 Da) and insulin (5807.6 Da) were tested. Insulin, for example, could be distinctly identified at the singly charge state with m/z 5808.7016, which is close to the mass limit of the QE-HF Orbitrap mass analyzer (6,000 Da). We then selected this peptide mixture whose molecular weights ranged from 600 Da to 5800 Da as a model system to characterize the ISE in SubAP-MALDI-Orbitrap. As Figure 1 shows that SubAP-MALDI spectrum is dominated by singly-charged analyte ions, we then assume the mechanism of the SubAP-MALDI ion production is similar to that of a conventional (vacuum) MALDI which also produces singly-charged analyte ions. The major difference from the vacuum source is that the SubAP-MALDI source produces ions under low-pressure (3–5 Torr) conditions in a separate housing (Scheme 1b) and contains the first (high pressure) section of the ion funnel. This housing is then interfaced with a second housing containing the low-pressure section of the ion funnel, and the latter is interfaced with the Orbitrap analyzer.
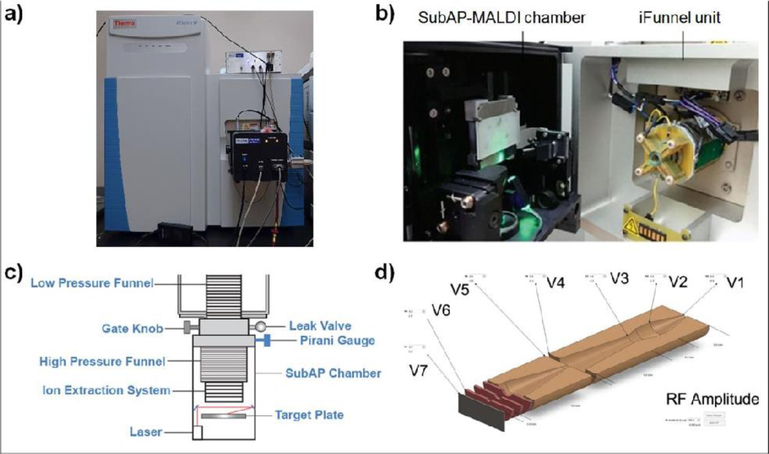
Scheme 1.
SubAP-MALDI-Orbitrap platform. a) A snapshot of the assembled SubAP-MALDI source interfaced with the QE-HF Orbitrap mass spectrometer. b) Sample loading platform and ion funnel. c) Schematic illustration for the SubAP-MALDI platform. d) RF and various voltages controlled by iFunnel software program.
Figure 1.
Representative SubAP-MALDI-Orbitrap mass spectrum of peptide standard mixture. The peptides were individually dissolved in water and then combined with matrix to the final amounts: FMRFamide, 71 nmol; FMRF-like neuropeptide, 0.019 nmol; somatostatin-14, 71 nmol; Bradykinin, 0.022 nmol; Aβ (1–16), 0.014 nmol and human insulin, 0.001 nmol. CHCA was taken as MALDI matrix in 10 mg/mL concentration. Laser fluence was 9.6% (100%, 2 μJ) and laser repetition rate was 1 kHz. Source pressure was kept at 3 Torr.
The unit containing the SubAP-MALDI plate holder and the laser was built as a monolithic housing. This unit was attached to the two-stage ion funnel which was installed into the mass spectrometer to replace the heated cone assembly comprising the capillary and S-Lens originally interfacing the Orbitrap and the ESI/AP-MALDI sources (Scheme 1c). All ions produced inside the unit were accelerated toward the head of the ion funnel and then to the mass spectrometer. The unit contained the following components: the UV laser, optics, XY-stages with the target plate holder, and control electronics. The MALDI samples were placed onto the surface of a stainless steel plate (target plate) and the latter was inserted into a target plate holder. The high voltage was applied across the target plate. Slightly lower voltage was applied across the first electrode in a short ion-accelerating stack placed in front of the ion funnel head to assist the transportation of the ions from the target plate toward the ion funnel and then to the mass spectrometer.
Ion suppression is often observed in MALDI analysis of mixtures containing multiple components, especially when analyzing mixtures of analytes with different ionization efficiencies/proton affinities/charge states. Herein, all species were singly-charged, therefore the charge effect could be ignored. However, each analyte exhibited distinct ionization efficiency as the molecular mass of these peptides varied from 600 Da to 5,800 Da. For example, peptides with higher mass, like insulin of 0.001 nmol, exhibited lower signal compared to those with lower mass such as FMRFamide of 71 nmol and bradykinin of 0.022 nmol. We then decided to use the ratio of higher mass peptide to lower mass peptide as an indicator for the ISE. The coexisting analyte-induced ion suppression effects have been evaluated using a similar concept in sonic spray ionization,6 nanoelectrospray ionization,43 and laser electrospray ionization44–45. To this end, a variety of source parameters have been tested to evaluate their effects on ISE, including laser energy, source pressure, laser firing frequency, and ion funnel voltages (Scheme 1d).
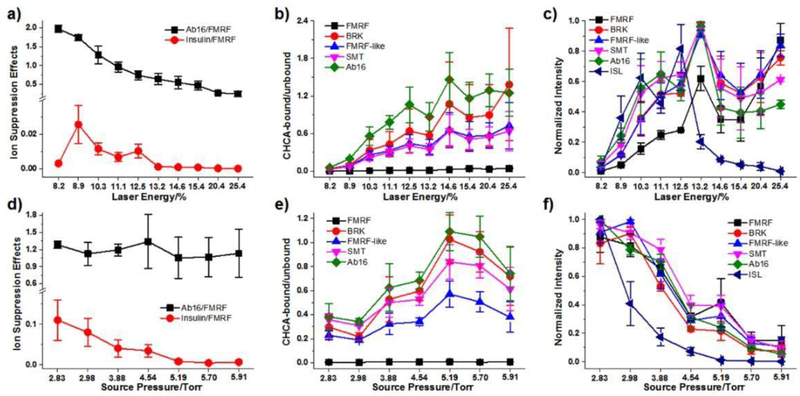
We first studied the effect of laser parameters, including laser energy and laser firing repetition rate. Three key indicators were employed to evaluate the performance of SubAP-MALDI, including intensity ratio-based ISE (through normalizing the intensity of Aβ (1–16) and insulin to FMRF), matrix bound degree (intensity ratio of CHCA-adducts to protonated peptide ions) and normalized analyte intensity (normalized to itself among parameter optimization). Figure 2a shows the dependency trend of intensity ratios of insulin to FMRF and Aβ (1–16) to FMRF as a function of laser energy. The lower intensity ratio of higher mass analytes to lower mass analytes indicates higher ISE during ionization. This intensity ratio drops when laser increases from 8.9%, as shown in Figure 2a which clearly indicated ISE gets more severe with the increase of laser energy. Meanwhile, Figure 2b shows increase of matrix adducts as the laser energy increases, which could be related to the thermo-regulated ionization and desorption processes as similar to normal MALDI regime. These observations indicate that a relatively low laser energy is desired to reduce ISE and matrix interferences in MALDI analysis. However, it should be noted that the intensity of analytes would decrease significantly when laser energy goes down from 13% to around 8.2%, as indicated in Figure 2c, suggesting the laser energy cannot be too low in order to obtain analyte signals of sufficient intensity. Using the same peptide mixture as well as matrix (CHCA), variation in laser repetition rate did not show significant influence on either ISE or formation of matrix adducts (Figure S1a-b) compared to laser energy, although the signal intensity varied when the laser repetition rate increased from 200 to 2,000 Hz (Figure S1c). The laser repetition rate was then optimized to be 1 kHz with laser energy 9–12% (100%, 2 μJ) when using CHCA as a matrix to analyze peptide standards. Of note, the laser frequency and energy for DHB matrix were optimized to be 200 Hz and 18–22% (Figure S2).
Figure 2.
Effects of laser energy and source pressure on ion suppression (a, d), matrix-binding (b, e) and intensity (c, f). All data were derived from SubAP-MALDI-Orbitrap spot analysis of peptide mixtures. Data in (c) and (f) were individually normalized to the highest peaks for each peptide throughout the tuning of laser energy and source pressure. Matrix, CHCA, 10 mg/mL. All error bars denote SD, n = 3.
In addition to laser energy, the gas pressure in the source is another important operating parameter that affects the performance of SubAP-MALDI. As shown in Figure 2d, the background gas pressure in the SubAP-MALDI source showed a significant effect on ISE. The gas pressure largely affects the process of co-ionization of analytes that possess larger size or higher mass, though such influence is not that severe for the analytes with smaller difference in mass. With the increase of source pressure, the suppression of insulin signal by those from smaller peptides was larger (Figure 2d). Meanwhile, the CHCA bound ratios of peptides showed high positive correlation with rising source pressure (Figure 2e). The signal intensity of all peptides tested herein drastically reduced with the increase in the source pressure (Figure 2f). The source pressure was thus optimized to be ~3.0 Torr. Furthermore, we also studied the effects of ion funnel voltages (Figure S1d-i), which indicated that relatively higher ion funnel voltages will produce higher intensities of individual peptides, but one should be also cautious about the resultant, more severe ISE induced by coexisting peptides. Taken together, the ion funnel RF voltage was optimized as 250 V and other voltages were adjusted as stated in the Experimental section.
In addition, coating material for the tissue loading slide was also evaluated as the effects on ISE during SubAP-MALDI-MSI. Figure S3a indicates that when using the ITO-coated glass slides, the number of neuropeptide identifications significantly surpassed that of using regular glass slides, although the overall lipid identifications remain almost constant. Further examinations of lateral distributions of neuropeptides and lipids in Figure S3c-f showed less diffusion of analytes when using ITO-coated slides. These observations suggest that ITO-coating might help alleviate the ISE in SubAP-MALDI-MSI, presumably benefiting from the enhanced electric field.
High-resolution SubAP-MALDI-Orbitrap MSI.
SubAP-MALDI is capable of high spatial resolution MSI with an adjustable laser spot size. After parameter optimization and systematic evaluation of the ISE, we investigated the utility of the SubAP-MALDI source for MS imaging with high spatial resolution while maintaining high sensitivity. Benefiting from the use of a low-pressure chamber and ion funnel setup, we found that the sensitivity was significantly improved compared to that of an AP-MALDI source interfaced with the same Orbitrap. This improvement could be attributed to higher ionization/desorption efficiency and increased efficiency of the ion transfer. In order to maximize the performance of the SubAP-MALDI source, the relationship between sensitivity and spatial resolution/raster step size has been investigated.
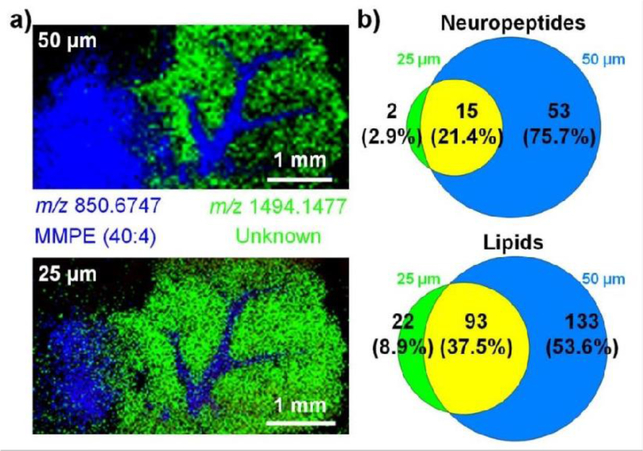
We first defined the actual spatial resolution of our SubAP-MALDI platform. A peptide mixture of FMRFamide-like neuropeptide, bradykinin, somatostatin and Aβ (1–16) was rastered under spiral motion mode with their respective images collected as shown in Figure S4 under laser energy 9.6% and repetition rate at 1 kHz with CHCA matrix. From the ion images of peptides, the trace could be distinguished within 20 μm without overlap, indicating that the actual spatial resolution at those parameters could be as high as 20 μm. The relationship between sensitivity and spatial resolution was then studied by varying step size of SubAP-MALDI source and comparing generated MSI ion images. The laser energy has been elevated from 9.6% to 18.9% to desorb peptides and lipids from tissue sections compared to the optimized value with pure analytes. Mouse brain tissues were tested with two step sizes (i.e., two laser spot diameters), 25 and 50 μm (Figure 3a). In order to make fair comparison, we plotted spatial distribution of two abundant ions (m/z 850.6747 and m/z 1494.1477) that were shared in both spectra. As illustrated in Figure 3a, the ion distribution images provided pixel-sharp separation between sub-regions within mouse cerebellum at step sizes of 25 and 50 μm without signal overlap. However, when spatial resolution was set at 25 μm, ion signals appreciably decreased and some signal blur or overlap appeared, which could be due to a larger variation of ion signals for two neighboring rows. This observation indicated that to obtain MS images of high quality (high signal-to-noise ratios), the laser spot size is critical and in this case the spatial resolution should be larger than 25 μm. In order to achieve higher spatial resolution, laser fluence and laser repetition rate optimization are recommended depending on the chosen matrix (CHCA requires lower laser fluence compared to DHB).
Figure 3.
Step size (laser spot size) affecting the performance of SubAP-MALDI-MSI on a mouse cerebellum tissue section. a) Ion images taken with two different step sizes. Two ions, m/z 850.6747 and m/z 1494.1477, are superimposed. b) The Venn diagram showing the comparisons of detected neuropeptides and lipid identification numbers with step size of 25 and 50 μm. Matrix, DHB, 40 mg/mL. Laser energy 18.9%, laser repetition rate 200 Hz.
After defining the actual resolution of SubAP-MALDI, the effect of step size on sensitivity and molecular species coverage were studied through comparisons of the numbers of identified neuropeptides and lipids. Figure 3b demonstrated the Venn diagrams for neuropeptide and lipid identifications at two different step sizes. At 25 μm step size, only 17 neuropeptides were identified from the mouse brain tissue section, while abundant lipids were detected. Notably, the neuropeptide identifications at step size of 50 μm increased dramatically to 68 (four-fold improvement compared to that obtained at step size of 25 μm). A similar trend was also observed in the lipid Venn diagram (Figure 3b). Importantly, our results present the effect of laser spot size on the overall quality of MS images which corroborates with previous findings.46
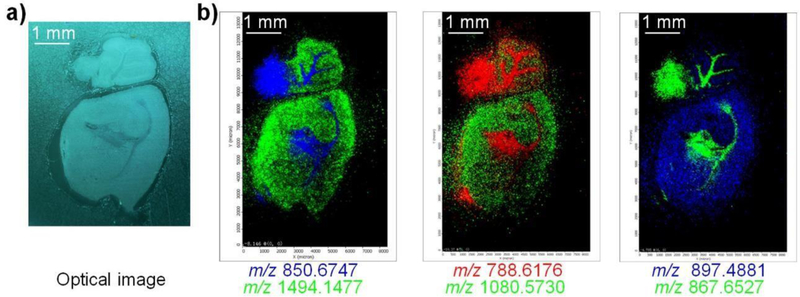
In addition to lateral resolution, image contrast is another important factor for discriminating specific compound distributions in biological tissues.10 Figure 4 showed the SubAP-MALDI-MSI results on a whole mouse brain tissue section. Both neuropeptides and lipids were simultaneously imaged with distinct image ion distributions in the cerebellum.
Figure 4.
Simultaneous imaging of neuropeptides and lipids on a whole mouse brain tissue section using SubAP-MALDI-MSI. a) Optical image of the mouse brain tissue section analyzed by MSI. b) SubAP-MALDI ion images for neuropeptides and lipids. For each image contrast, two ions were superimposed. Ion signals in b) were, m/z 850.6747 ([MMPE(40:4)+83+Li]+), m/z 1494.1477 (unknown), m/z 788.6176 ([PC(36:1)+H]+), m/z 1080.573 ([DGGRNFLRFamide+H]+), m/z 867.6527 ([TAG(52:9)+Na]+), and m/z 897.4881 ([IFVGGSRYamide+H]+). Step size, 50 μm. Matrix, DHB, 40 mg/mL. Laser energy 18.9%, firing frequency 200 Hz.
CONCLUSION
The SubAP-MALDI source interfaced with an Orbitrap mass spectrometer provides both high spatial and mass resolutions necessary for a high-throughput in situ biomolecular analysis. Based on the new SubAP-MALDI source hyphenated, by means of the ion funnel, to Orbitrap mass spectrometer, a model peptide/protein mixture was utilized to systematically characterize the factors influencing ISE such as source pressure, laser energy, laser repetition rate, ion funnel voltages, and glass slide coating materials. With the optimized parameters, the SubAP-MALDI-Orbitrap platform proves its feasibility of co-localizing numerous lipids and neuropeptides in a mouse brain section. The newly-assembled SubAP-MALDI-Orbitrap system features improved sensitivity compared to the AP-MALDI Orbitrap setup. It offers down to 25-micron spatial resolution while retaining relatively high ion signals for lipids/neuropeptides and faster imaging acquisition speed compared to conventional vacuum MALDI-LTQ-Orbitrap system. The enhanced spatial and mass spectral resolution for tissue imaging brought by this new SubAP-MALDI QE-HF platform offers an attractive alternative to the ever-expanding MSI toolbox.
Supplementary Material
ACKNOWLEDGEMENT
The authors would like to acknowledge Dr. Vladimir Doroshenko from MassTech Inc. for generous technical help and valuable discussions. The authors would also like to thank Dr. Chris Ikonomidou at the University of Wisconsin-Madison for providing mouse tissue samples. This work was supported in part by the National Institutes of Health (NIH) through grants R56MH110215 and R01 DK071801. LL acknowledges a Vilas Distinguished Achievement Professorship and Janis Apinis Professorship with funding provided by the Wisconsin Alumni Research Foundation and University of Wisconsin-Madison School of Pharmacy. KD acknowledges the National Institute of General Medical Sciences of the NIH under award number F31GM126870, as well as a predoctoral fellowship supported by the NIH, under Ruth L. Kirschstein National Research Service Award T32 HL 007936 from the National Heart Lung and Blood Institute to the University of Wisconsin-Madison Cardiovascular Research Center.
Footnotes
SUPPORTING INFORMATION
Additional supporting information as noted in the text can be found in the online version
REFERENCES
- 1.Liang HR; Foltz RL; Meng M; Bennett P, Ionization enhancement in atmospheric pressure chemical ionization and suppression in electrospray ionization between target drugs and stable-isotope-labeled internal standards in quantitative liquid chromatography/tandem mass spectrometry. Rapid Commun. Mass Spectrom 2003, 17 (24), 2815–2821. [DOI] [PubMed] [Google Scholar]
- 2.Wang S; Cyronak M; Yang E, Does a stable isotopically labeled internal standard always correct analyte response?: A matrix effect study on a LC/MS/MS method for the determination of carvedilol enantiomers in human plasma. J. Pharm. Biomed. Anal 2007, 43 (2), 701–707. [DOI] [PubMed] [Google Scholar]
- 3.Constantopoulos T; Jackson G; Enke C, Effects of salt concentration on analyte response using electrospray ionization mass spectrometry. J. Am. Soc. Mass Spectrom 1999, 10 (7), 625–634. [DOI] [PubMed] [Google Scholar]
- 4.King R; Bonfiglio R; Fernandez-Metzler C; Miller-Stein C; Olah T, Mechanistic investigation of ionization suppression in electrospray ionization. J. Am. Soc. Mass Spectrom 2000, 11 (11), 942–950. [DOI] [PubMed] [Google Scholar]
- 5.Hirabayashi A; Ishimaru M; Manri N; Yokosuka T; Hanzawa H, Detection of potential ion suppression for peptide analysis in nanoflow liquid chromatography/mass spectrometry. Rapid Commun. Mass Spectrom 2007, 21 (17), 2860–2866. [DOI] [PubMed] [Google Scholar]
- 6.Li G; Huang G, Alleviation of ion suppression effect in sonic spray ionization with induced alternating current voltage. J. Mass Spectrom 2014, 49 (7), 639–645. [DOI] [PubMed] [Google Scholar]
- 7.Qiao H; Spicer V; Ens W, The effect of laser profile, fluence, and spot size on sensitivity in orthogonal-injection matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom 2008, 22 (18), 2779–2790. [DOI] [PubMed] [Google Scholar]
- 8.Moskovets E; Misharin A; Laiko V; Doroshenko V, A comparative study on the analytical utility of atmospheric and low-pressure MALDI sources for the mass spectrometric characterization of peptides. Methods 2016, 104, 21–32. [DOI] [PubMed] [Google Scholar]
- 9.Laiko VV; Baldwin MA; Burlingame AL, Atmospheric Pressure Matrix-Assisted Laser Desorption/Ionization Mass Spectrometry. Anal. Chem 2000, 72 (4), 652–657. [DOI] [PubMed] [Google Scholar]
- 10.Kompauer M; Heiles S; Spengler B, Atmospheric pressure MALDI mass spectrometry imaging of tissues and cells at 1.4-mum lateral resolution. Nat. Methods 2017, 14 (1), 90–96. [DOI] [PubMed] [Google Scholar]
- 11.OuYang C; Chen B; Li L, High Throughput In Situ DDA Analysis of Neuropeptides by Coupling Novel Multiplex Mass Spectrometric Imaging (MSI) with Gas-Phase Fractionation. J Am. Soc. Mass Spectrom 2015, 26 (12), 1992–2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen B; Lietz CB; Li L, In Situ characterization of proteins using laserspray ionization on a high-performance MALDI-LTQ-Orbitrap mass spectrometer. J Am. Soc. Mass Spectrom 2014, 25 (12), 2177–2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gabelica V; Schulz E; Karas M, Internal energy build-up in matrix-assisted laser desorption/ionization. J. Mass Spectrom 2004, 39 (6), 579–593. [DOI] [PubMed] [Google Scholar]
- 14.Chang WC; Huang LC; Wang YS; Peng WP; Chang HC; Hsu NY; Yang WB; Chen CH, Matrix-assisted laser desorption/ionization (MALDI) mechanism revisited. Anal. Chim. Acta 2007, 582 (1), 1–9. [DOI] [PubMed] [Google Scholar]
- 15.Karas M; Kruger R, Ion formation in MALDI: the cluster ionization mechanism. Chem. Rev 2003, 103 (2), 427–440. [DOI] [PubMed] [Google Scholar]
- 16.Trimpin S; Wang B; Inutan ED; Li J; Lietz CB; Harron A; Pagnotti VS; Sardelis D; McEwen CN, A mechanism for ionization of nonvolatile compounds in mass spectrometry: considerations from MALDI and inlet ionization. J Am. Soc. Mass Spectrom 2012, 23 (10), 1644–1660. [DOI] [PubMed] [Google Scholar]
- 17.Bae YJ; Kim MS, A Thermal Mechanism of Ion Formation in MALDI. Annu. Rev. Anal. Chem 2015, 8, 41–60. [DOI] [PubMed] [Google Scholar]
- 18.Lu IC; Lee C; Lee YT; Ni CK, Ionization Mechanism of Matrix-Assisted Laser Desorption/Ionization. Annu. Rev. Anal. Chem 2015, 8, 21–39. [DOI] [PubMed] [Google Scholar]
- 19.Zhigilei LV; Garrison BJ, Microscopic mechanisms of laser ablation of organic solids in the thermal and stress confinement irradiation regimes. J. Appl. Phys 2000, 88 (3), 1281–1298. [Google Scholar]
- 20.Soltwisch J; Jaskolla TW; Dreisewerd K, Color matters--material ejection and ion yields in UV-MALDI mass spectrometry as a function of laser wavelength and laser fluence. J. Am. Soc. Mass. Spectrom 2013, 24 (10), 1477–88. [DOI] [PubMed] [Google Scholar]
- 21.Diologent L; Bolbach G; Focsa C; Ziskind M; Fournier I, Laser induced post-desolvation of MALDI clusters. Int. J. Mass spectrom 2017, 416, 29–36. [Google Scholar]
- 22.Rohlfing A; Leisner A; Hillenkamp F; Dreisewerd K, Investigation of the Desorption Process in UV Matrix-Assisted Laser Desorption/Ionization with a Liquid 3-Nitrobenzyl Alcohol Matrix by Photoacoustic Analysis, Fast-Flash Imaging, and UV-Laser Postionization. J. Phys. Chem. C 2009, 114 (12), 5367–5381. [Google Scholar]
- 23.Musapelo T; Murray KK, Particle production in reflection and transmission mode laser ablation: implications for laserspray ionization. J. Am. Soc. Mass. Spectrom 2013, 24 (7), 1108–15. [DOI] [PubMed] [Google Scholar]
- 24.Knochenmuss R; Zenobi R, MALDI ionization: the role of in-plume processes. Chem. Rev 2003, 103 (2), 441–452. [DOI] [PubMed] [Google Scholar]
- 25.Xu S; Ye M; Xu D; Li X; Pan C; Zou H, Matrix with high salt tolerance for the analysis of peptide and protein samples by desorption/ionization time-of-flight mass spectrometry. Anal. Chem 2006, 78 (8), 2593–2599. [DOI] [PubMed] [Google Scholar]
- 26.Wang S; Xiao Z; Xiao C; Wang H; Wang B; Li Y; Chen X; Guo X, (E)-Propyl alpha-Cyano-4-Hydroxyl Cinnamylate: A High Sensitive and Salt Tolerant Matrix for Intact Protein Profiling by MALDI Mass Spectrometry. J Am. Soc. Mass Spectrom 2016, 27 (4), 709–718. [DOI] [PubMed] [Google Scholar]
- 27.Gemperline E; Rawson S; Li L, Optimization and comparison of multiple MALDI matrix application methods for small molecule mass spectrometric imaging. Anal. Chem 2014, 86 (20), 10030–10035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Malys BJ; Owens KG, Improving the analyte ion signal in matrix-assisted laser desorption/ionization imaging mass spectrometry via electrospray deposition by enhancing incorporation of the analyte in the matrix. Rapid Commun. Mass Spectrom 2017, 31 (9), 804–812. [DOI] [PubMed] [Google Scholar]
- 29.Moskovets E; Chen HS; Pashkova A; Rejtar T; Andreev V; Karger BL, Closely spaced external standard: a universal method of achieving 5 ppm mass accuracy over the entire MALDI plate in axial matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom 2003, 17 (19), 2177–2187. [DOI] [PubMed] [Google Scholar]
- 30.Wiangnon K; Cramer R, Sample preparation: a crucial factor for the analytical performance of rationally designed MALDI matrices. Anal. Chem 2015, 87 (3), 1485–1488. [DOI] [PubMed] [Google Scholar]
- 31.Cramer R; Karas M; Jaskolla TW, Enhanced MALDI MS sensitivity by weak base additives and glycerol sample coating. Anal. Chem 2014, 86 (1), 744–751. [DOI] [PubMed] [Google Scholar]
- 32.Cramer R; Pirkl A; Hillenkamp F; Dreisewerd K, Liquid AP-UV-MALDI enables stable ion yields of multiply charged peptide and protein ions for sensitive analysis by mass spectrometry. Angew. Chem. Int. Ed 2013, 52 (8), 2364–2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Woodall DW; Wang B; Inutan ED; Narayan SB; Trimpin S, High-throughput characterization of small and large molecules using only a matrix and the vacuum of a mass spectrometer. Anal. Chem 2015, 87 (9), 4667–4674. [DOI] [PubMed] [Google Scholar]
- 34.Trimpin S; Inutan ED; Herath TN; McEwen CN, Matrix-assisted laser desorption/ionization mass spectrometry method for selectively producing either singly or multiply charged molecular ions. Anal. Chem 2010, 82 (1), 11–15. [DOI] [PubMed] [Google Scholar]
- 35.Lu Y; Pieterse CL; Robertson WD; Miller RJD, Soft Picosecond Infrared Laser Extraction of Highly Charged Proteins and Peptides from Bulk Liquid Water for Mass Spectrometry. Anal. Chem 2018, 90 (7), 4422–4428. [DOI] [PubMed] [Google Scholar]
- 36.Kwiatkowski M; Wurlitzer M; Omidi M; Ren L; Kruber S; Nimer R; Robertson WD; Horst A; Miller RJ; Schluter H, Ultrafast extraction of proteins from tissues using desorption by impulsive vibrational excitation. Angew. Chem. Int. Ed 2015, 54 (1), 285–288. [DOI] [PubMed] [Google Scholar]
- 37.Yang J; Caprioli RM, Matrix precoated targets for direct lipid analysis and imaging of tissue. Anal. Chem 2013, 85 (5), 2907–2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sugiyama E; Masaki N; Matsushita S; Setou M, Ammonium Sulfate Improves Detection of Hydrophilic Quaternary Ammonium Compounds through Decreased Ion Suppression in Matrix-Assisted Laser Desorption/Ionization Imaging Mass Spectrometry. Anal. Chem 2015, 87 (22), 11176–11181. [DOI] [PubMed] [Google Scholar]
- 39.Kim JS; Kim JY; Kim HJ, Suppression of matrix clusters and enhancement of peptide signals in MALDI-TOF mass spectrometry using nitrilotriacetic acid. Anal. Chem 2005, 77 (22), 7483–7488. [DOI] [PubMed] [Google Scholar]
- 40.Taylor A; Dexter A; Bunch J, Exploring ion suppression in mass spectrometry imaging of a heterogeneous tissue. Anal. Chem 2018, 90 (9), 5637–5645. [DOI] [PubMed] [Google Scholar]
- 41.Chen B; OuYang C; Tian Z; Xu M; Li L, A high resolution atmospheric pressure matrix-assisted laser desorption/ionization-quadrupole-orbitrap MS platform enables in situ analysis of biomolecules by multi-mode ionization and acquisition. Anal. Chim. Acta 2018, 1007, 16–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jackson SN; Muller L; Roux A; Oktem B; Moskovets E; Doroshenko VM; Woods AS, AP-MALDI Mass Spectrometry Imaging of Gangliosides Using 2,6-Dihydroxyacetophenone. J Am. Soc. Mass Spectrom 2018, 29 (7), 1463–1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li G; Yuan S; Pan Y; Liu Y; Huang G, Binding States of Protein-Metal Complexes in Cells. Anal. Chem 2016, 88 (22), 10860–10866. [DOI] [PubMed] [Google Scholar]
- 44.Flanigan PM; Perez JJ; Karki S; Levis RJ, Quantitative Measurements of Small Molecule Mixtures Using Laser Electrospray Mass Spectrometry. Anal. Chem 2013, 85 (7), 3629–3637. [DOI] [PubMed] [Google Scholar]
- 45.Perez JJ; Flanigan PM; Karki S; Levis RJ, Laser electrospray mass spectrometry minimizes ion suppression facilitating quantitative mass spectral response for multicomponent mixtures of proteins. Anal. Chem 2013, 85 (14), 6667–6673. [DOI] [PubMed] [Google Scholar]
- 46.Wiegelmann M; Dreisewerd K; Soltwisch J, Influence of the Laser Spot Size, Focal Beam Profile, and Tissue Type on the Lipid Signals Obtained by MALDI-MS Imaging in Oversampling Mode. J. Am. Soc. Mass. Spectrom 2016, 27 (12), 1952–1964. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.