Abstract
A central question in the circadian biology field concerns the mechanisms that translate ~24-hour oscillations of the molecular clock into overt rhythms. Drosophila melanogaster is a powerful system that provided the first understanding of how molecular clocks are generated and is now illuminating the neural basis of circadian behavior. The identity of ~150 clock neurons in the Drosophila brain and their roles in shaping circadian rhythms of locomotor activity have been described before. This review summarizes mechanisms that transmit time-of-day signals from the clock, within the clock network as well as downstream of it. We also discuss the identification of functional multisynaptic circuits between clock neurons and output neurons that regulate locomotor activity.
Keywords: Circadian rhythm, Rest:activity rhythms, Neuropeptide, Neuronal activity
Circadian pacemaking is a result of individually rhythmic clock neurons synchronized across a circuit. In the circadian system of Drosophila melanogaster, rhythmic neuronal activity also propagates downstream of the central clock neurons, s-LNvs, to output circuits that regulate behavioral rhythms.
Introduction
Circadian (~24 hour) rhythms allow animals to anticipate daily changes in their environment and coordinate their behavior and physiology with time of day. These rhythms are generated by an internal timing mechanism, which is synchronized to environmental cycles of light and temperature imposed by the rotation of earth. In its simplest form, a circadian system is modeled with three basic components: the clock, input pathways, and output pathways. The clock maintains ~24-hour rhythms even in constant darkness. Input pathways synchronize the clock to external signals such as light. Output pathways receive and translate circadian signals from the clock to produce biological rhythms.
Much of our molecular knowledge of circadian clocks came from genetic studies in the fruit fly, Drosophila melanogaster. In flies, circadian rhythms are typically studied using locomotor activity as the output. Under a 12-hour light:12-hour dark cycle, the fly exhibits a bimodal pattern in locomotor activity, with activity peaks anticipating the light-to-dark (evening) and dark-to-light (morning) transitions. Locomotor activity rhythms are dependent on endogenous clocks and persist in constant darkness (DD), albeit with a different pattern. In DD, the fly’s locomotor activity free-runs with the periodicity of the endogenous clock, which is about (but usually not exactly) 24-hours, such that activity each day generally occurs during the subjective day and rest occurs during the subjective night. Besides rest:activity rhythms, flies also exhibit rhythms in eclosion (emergence of adult flies from pupae), feeding, temperature preference, and sleep. Besides behavior, there are circadian rhythms at the cellular level, such as electrical activity of neurons, gene expression, and metabolic processes. A basic molecular clock mechanism regulates all output rhythms. Yet compared to our relatively advanced understanding of the molecular clock mechanism, the output mechanisms that turn molecular clock oscillations into diverse behavioral and physiological rhythms are not well understood.
Here, we review output mechanisms of the circadian clock in the Drosophila brain. We will start by describing the circadian clock network and the output mechanisms that occur within the network. Then, we will move beyond the circadian clock network and review recent work that identified output circuits regulating circadian rhythms of behavior and physiology.
The circadian clock network in Drosophila brain
The basic molecular oscillator in eukaryotes consists of transcriptional activators and repressors in a feedback loop. In Drosophila, the co-activator complex, CLOCK-CYCLE, drives transcription of the co-repressors, period (per) and timeless (tim). Accumulated PER and TIM proteins feed back to inhibit CLOCK-CYCLE activity. Delays are built into the basic molecular oscillator at multiple steps, and include post-transcriptional and post-translational mechanisms, which ensure 24-hr rhythms in PER and TIM expression [reviewed in (Zheng & Sehgal, 2012)].
Oscillations in the circadian clock are self-sustained. However, the clock is usually synchronized to external cues through a process called entrainment, which is crucial for adaption to the environment [reviewed in (Yoshii et al., 2016)]. Light is the primary entrainment cue, and in flies it involves a dedicated circadian photoreceptor, Cryptochrome (CRY). Upon light exposure, CRY binds TIM and targets TIM for ubiquitination by the E3 ligase, JETLAG, and then degradation. In addition to the CRY mechanism, light-input circuits from the visual system to the central clock neurons are also important for light entrainment.
In the Drosophila brain, there are ~150 clock neurons subdivided into six groups based on neuroanatomy. The six groups of PER-TIM-expressing neurons include the large and small ventral lateral neurons (l-LNvs and s-LNvs), the dorsal lateral neurons (LNds), the lateral posterior neurons (LPNs), and three groups of dorsal neurons (DN1s, DN2s and DN3s) (Figure 1) (Kaneko & Hall, 2000; Helfrich-Förster et al., 2007). Clock neurons between and within groups use a heterogenous set of neuropeptides and neurotransmitters for signaling [reviewed in (Beckwith & Ceriani, 2015)].
Figure 1: Circadian circuits in the fly brain.
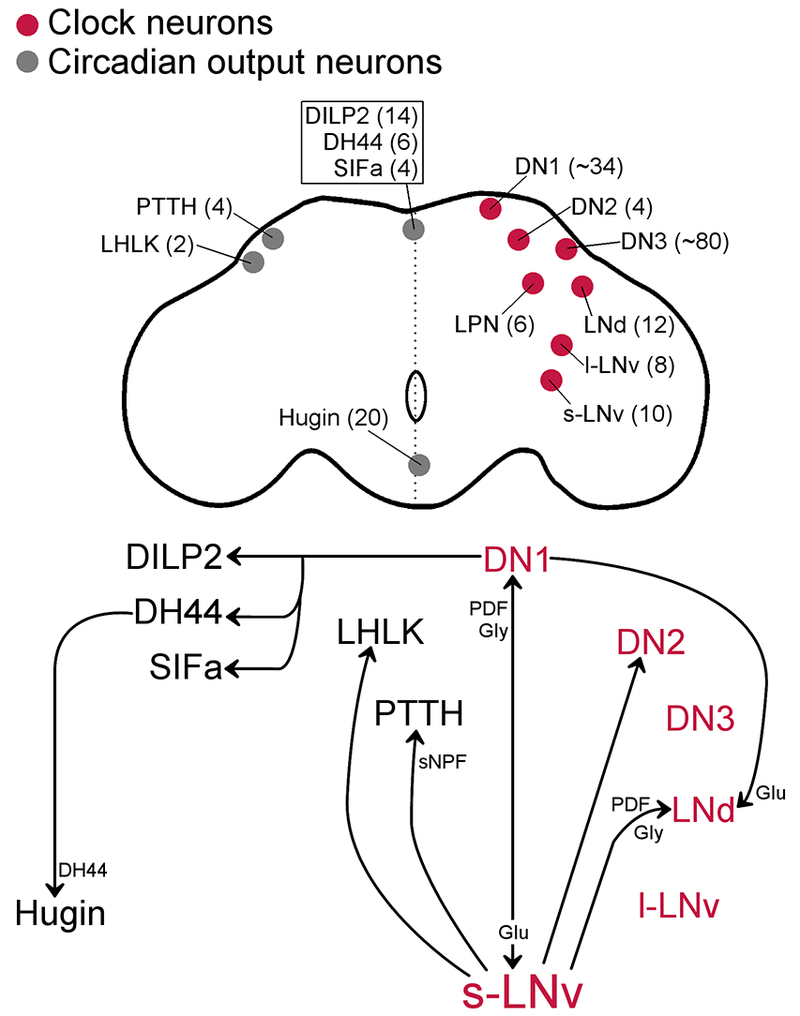
Top. Schematic representation of a fly brain with neuroanatomical locations of clock neurons (red, right hemisphere) and circadian output neurons (gray, left hemisphere or midline). Bilaterally represented neurons are labeled in only one of the hemispheres. Approximate total number of cells in the brain is indicated in parentheses. Bottom. Arrows represent the paths of communication between groups of circadian neurons. Circuits were mapped using neuronal activation and functional imaging and/or GRASP (GFP reconstitution across synaptic partners) methods. The neuropeptide/neurotransmitters that signal in the circuits were genetically identified by removing the peptide or neurotransmitter transporter in the presynaptic neuron and removing the receptor in the postsynaptic neuron. PDF mediates s-LNv communication to LNd, DN1, and LHLK (indirectly) (Leucokinin+ lateral horn). Glycine (Gly) also signals from s-LNv to DN1 and LNd. Short neuropeptide F (sNPF) signals in the s-LNv to PTTH circuit. Glutamate (Glu) signals from the DN1 to s-LNv and LNd. The molecules that signal between DN1 and PI neurons (DH44/SIFa/Dilp2) are unknown. DH44 neuropeptide signal from Dh44+ to hugin+ neurons.
The LNvs are comprised of two groups of neurons, s-LNvs and l-LNvs, and are genetically identified by expression of neuropeptide Pigment-Dispersing Factor (PDF) (Helfrich-Förster, 1995). There are four s-LNvs and four l-LNvs in each hemisphere of the fly brain. An additional pair of cells called the “5th s-LNvs” also has molecular clocks but is PDF-negative. The Pdf+ LNvs hold an important role in regulating rest:activity rhythms. Flies with ablated or electrically silenced LNvs have arrhythmic rest:activity behavior in DD (Renn et al., 1999; Nitabach et al., 2002; Depetris-Chauvin et al., 2011), and period null mutant flies with period restored in LNvs display normal rest:activity rhythms in DD (Grima et al., 2004).
Although the Pdf+ LNvs appear to have the primary role, robust rest:activity rhythms are a result of clock network coordination. When molecular clocks in the network are mismatched with one another, arrhythmicity, complex rhythms (comprised of multiple rhythmic components of different period lengths), or weak rest:activity rhythms emerge in the fly behavior (Yao and Shafer 2014). The network is often simply modeled as a system of dual oscillators, where oscillators in Pdf+ LNvs control the morning peak of locomotor activity, and oscillators in LNds and the 5th s-LNv control the evening peak (Grima et al., 2004; Stoleru et al., 2004; Rieger et al., 2006; Guo et al., 2014). The LNd group is comprised of six neurons per hemisphere. Blocking neurotransmission from a LNd subset results in a large proportion of arrhythmic flies in DD (Guo et al., 2014). In addition, molecular clocks in a subset of LNds drive transcriptional rhythms of a set of metabolic genes in the fat body, a peripheral tissue analogous to adipose/liver tissue, through Neuropeptide F signaling (Erion et al., 2016).
The DN1 group is comprised of 2 anterior (DN1a) and 15 posterior (DN1p) neurons. DN1ps serve diverse functions as integrators of light, temperature, and circadian cues as well as effectors of locomotor activity, sleep, and mating. DN1s have molecular clocks that can be entrained to temperature (Yoshii et al., 2010), and calcium (Ca2+) activity in DN1ps is regulated by temperature (Guo et al., 2016; Yadlapalli et al., 2018). DN1ps integrate temperature and light information to promote robust rest:activity rhythms (Zhang, Chung, et al., 2010; Zhang, Liu, et al., 2010) and also regulate sleep at specific times of day, through different circuits using either DH31 neuropeptide or glutamate signaling (Kunst et al., 2014; Guo et al., 2016). In addition, DN1ps mediate rhythms in male sex drive (Fujii et al., 2017).
The DN2s also have temperature-entrainable molecular clocks and regulate rhythms of temperature preference, namely the tendency of flies to seek different temperatures at different times of day (Yoshii et al., 2010; Kaneko et al., 2012). A circuit for temperature preference at dawn has been mapped from the thermosensory anterior cells to s-LNvs to DN2s (Tang et al., 2017). The molecular clocks in LPNs are also strongly synchronized to temperature cycles (Miyasako et al., 2007).
Finally, glial cells in the brain also express PER and TIM (Zerr et al., 1990). Astrocytes are important for rest:activity rhythms, although the molecular clock in these cells is dispensable (Ng et al., 2011). Glial cells are proposed to regulate outputs of clock neurons, but the signaling mechanisms remain to be uncovered (Ng & Jackson, 2015; Herrero et al., 2017). In the Drosophila blood-brain barrier (BBB), molecular clocks in glial cells drive circadian rhythms in BBB permeability (Zhang et al., 2018).
PDF is an important clock output factor in the clock network
In the clock network, pigment-dispensing factor (PDF) is an important clock output factor [reviewed in (Shafer & Yao, 2014)]. Loss or overexpression of Pdf causes arrhythmic rest:activity behavior (Renn et al., 1999; Helfrich-Förster et al., 2000), and mutations in the PDF receptor (PDFR) phenocopy Pdf mutants (Hyun et al., 2005; Lear, Merrill, et al., 2005; Mertens et al., 2005). PDFR is a G-protein coupled receptor that activates cAMP production upon binding of PDF peptide. An important function of PDF/PDFR signaling is to maintain coherent and synchronized molecular oscillations in the clock network (Lin et al., 2004; Yoshii et al., 2009). All the groups of clock neurons, except the l-LNvs, express PDFR and respond to PDF application (Shafer et al., 2008; Im & Taghert, 2010). Since PDFR in also expressed in Pdf+ s-LNv, PDF may feed back to cell-autonomously regulate the clock itself or output from the clock (Choi et al., 2012). Outside the clock network, PDFR expression is sparse in the brain (Im & Taghert, 2010), and while PDF does signal to non-clock neurons implicated in behavior (Pírez et al., 2013; Chen et al., 2016), it is unclear whether PDF signaling in these circuits confers circadian timing to behavior.
PDF levels cycle across the day at s-LNv terminals in the dorsal protocerebrum, indicating that PDF may be secreted in a circadian manner (Park et al., 2000). In addition, CLOCK regulates PDF expression at the post-transcriptional level through VRILLE, a repressor that functions in a second feedback loop interlocked with the core clock (Blau & Young, 1999; Gunawardhana & Hardin, 2017). CLOCK also regulates Pdf at the transcriptional level through a nuclear receptor called Hormone receptor-like 38 (Mezan et al., 2016). However, it is unclear whether rhythmic PDF levels or secretion are important for rest:activity rhythms (Kula et al., 2006). Instead, rhythmic PDF levels in the s-LNv terminals may be a secondary consequence of rhythmic neuronal firing or remodeling of the projections (discussed below). Furthermore, rhythmic PDF signaling may also occur through circadian-gated sensitivity to PDF in target neurons, mediated by PDFR and a small GTPase, Ral A (Klose et al., 2016). In summary, PDF is important for circadian rhythms, and its effect on circadian behavior is largely localized within the clock network.
Glycine and glutamate mediate reciprocal inhibition between the s-LNvs and DN1ps
Compared to neuropeptides, less is known about fast neurotransmitters in the clock network. However, within the s-LNv-DN1p circuit, the inhibitory neurotransmitter, glycine, is used in addition to PDF (Frenkel et al., 2017). Knockdown of the glycine transporter or disrupting glycine synthesis in the Pdf+ LNvs lengthens the period of rest:activity rhythms, suggesting LNvs are glycinergic. In addition, glycine application on DN1ps reduces their firing frequency, and knockdown of glycine receptors subunits in the DN1ps reduces the power of rest:activity rhythms in flies, confirming functional glycine signaling in the s-LNv to DN1 circuit.
In the reciprocal direction, DN1ps signal to the s-LNvs through glutamate, which appears to be an inhibitory neurotransmitter in this circuit (Hamasaka et al., 2007; Guo et al., 2016). A subset of the DN1ps expresses vesicular glutamate transporter (VGlut), and s-LNvs and LNds express the metabotropic glutamate receptor, mGluRA. Consistent with an inhibitory effect, glutamate application decreases Ca2+ levels in s-LNvs and LNds. Glutamate signaling is also relevant for behavioral rhythms—glutamate from non-LNv clock neurons is required for robust rest:activity rhythms and knockdown of mGluRA in Pdf+ LNvs lengthens the period of rest:activity rhythms (Hamasaka et al., 2007; Collins et al., 2012)
Neuropeptides sNPF and PDF set different phases of Ca2+ rhythms in clock network
Intercellular signaling is not only essential for synchronizing molecular clock rhythms but also coordinating neuronal activity rhythms in the clock network. It is thought that the molecular clock regulates the excitability of clock neurons, such that the neurons are more active at certain times of day than other times. Electrophysiological recordings from s-LNv, l-LNv, and DN1 have shown that the molecular clock drives these cells to be more active at dawn than at dusk (Table 1) (Cao & Nitabach, 2008; Sheeba, Gu, et al., 2008; Flourakis et al., 2015). Recent studies use genetically encoded Ca2+ sensors to perform longitudinal imaging of neuronal activity in the entire clock network over 24 hours, with the added advantages of obtaining more temporal information and precise determination of when clock neurons are most active (Liang et al., 2016, 2017). We review this work reported in a pair of papers by Liang, Holy, and Taghert.
Table 1:
Cycling in Circadian Circuits
| Neuronal group: | Cycling: | Highest at: (hours since lights-on) | Cycles in constant darkness? | Cycle lost in a clock mutant?: | Reference |
|---|---|---|---|---|---|
| Clock neurons | |||||
| s-LNv | Electrical activity | ~0 | N.d. | N.d. | (Cao & Nitabach, 2008) |
| Intracellular calcium (Ca2+) levels | 23-24 | Yes | Yes | (Liang et al., 2016) | |
| Complexity of projections | ~0 | Yes | Yes | (Fernández et al., 2008) | |
| Synapse contacts | ~2 | Yes | N.d. | (Gorostiza et al., 2014) | |
| Rho1 activity | ~12 | Yes | Yes | (Petsakou et al., 2015) | |
| PDF levels in projections | 0-6 | Yes | Yes | (Park et al., 2000) | |
| PDF and dopamine sensitivity | ~0 | Yes | N.d. | (Klose et al., 2016) | |
| l-LNv | Electrical activity | 1-6 | No (DD day 1); Yes (DD day 14) | Yes | (Cao & Nitabach, 2008; Sheeba, Gu, et al., 2008) |
| Ca2+ levels | 5-6 | Yes | Yes | (Liang et al., 2016) | |
| GABA sensitivity | Evening | N.d. | N.d. | (Li et al., 2017) | |
| LNd | Ca2+ levels | ~12 | Yes (highest at CT 8-9) | Yes | (Liang et al., 2016) |
| DN1 | Electrical activity | 0-4 or 20-24 | N.d. | Yes | (Flourakis et al., 2015) |
| Ca2+ levels | 18-20 | Yes | Yes | (Liang et al., 2016) | |
| DN2 | Synaptic contacts with s-LNvs | 22-24 | N.d. | N.d. | (Tang et al., 2017) |
| DN3 | Ca2+ levels | 17-18 | Yes | Yes | (Liang et al., 2016) |
| Circadian output neurons | |||||
| DILP2+ PI | Electrical activity | 0-4 | No | Yes | (Barber et al., 2016) |
| DH44+ PI | Ca2+ levels | 7-12 | Yes | Yes | (Cavey et al., 2016; Bai et al., 2018) |
| Hugin+ SEZ | Neuropeptide vesicle release | Night | N.d. | Yes | (King et al., 2017) |
| LK+ LH | Ca2+ levels | Night | Yes | Yes | (Cavey et al., 2016) |
| Carbachol sensitivity | Night | Yes | Yes | (Cavey et al., 2016) | |
| LK Receptor+ LH | Ca2+ levels | Day | Yes | Yes | (Cavey et al., 2016) |
| Carbachol sensitivity | Day | Yes | Yes | (Cavey et al., 2016) | |
N.d. = not determined
CT = circadian time
DD = constant darkness
LH = lateral horn
PI = pars intercerebralis
SEZ = subesophageal zone
Intracellular calcium (Ca2+) ions are important secondary messengers for many signaling pathways, and Ca2+ levels rise during electrical activity in neurons. In circadian regulation, Ca2+ signaling is both an input and output of the molecular clock (Ikeda, 2004; Harrisingh et al., 2007), with all groups of clock neurons displaying 24-hr Ca2+ rhythms. Despite synchrony of the molecular oscillator across the clock network, Ca2+ rhythms are asynchronous among the different groups of clock neurons (Table 1) (Liang et al., 2016). Ca2+ peaks in clock neurons occur at times that match with their roles in behavior. For example, s-LNvs control morning locomotor activity and have peak Ca2+ levels at dawn, and LNds control evening locomotor activity and have peak Ca2+ levels preceding the evening.
How does the clock network coordinate different phases of Ca2+ rhythms? To discover the mechanisms, Liang et al. (2017) focused on neuropeptides, such as PDF. In the absence of PDF, the Ca2+ peaks in LNds and DN3s are shifted from ~CT 8 and ~CT 16, respectively, to dawn (~CT 0). (CT is the circadian time is defined by an organism’s endogenous circadian clock in constant conditions; CT 0 corresponds to the start of subjective day and CT 12 to the start of subjective night). To determine if the shift in Ca2+ rhythms is a phase advance or delay, they applied synthetic PDF and found that Ca2+ levels decreased in LNds and DN3s, and importantly, the Ca2+ levels remained depressed for several hours. Therefore, PDF delays the Ca2+ peaks in LNds and DN3s and does so by staggering their Ca2+ peaks to two different times of the day. How one neuropeptide produces two different effects on phase is not known. The authors also determined that sNPF (short Neuropeptide F) inhibits Ca2+ and delays the Ca2+ peak in DN1s. sNPF in the clock network is required for rhythmic Ca2+ rhythms but not molecular clock oscillations in DN1s. Therefore, for certain clock neurons, circuit mechanisms may dominate over the cell-autonomous molecular clock in shaping Ca2+ rhythms. This study reported an inhibitory effect for PDF, while others have noted acute depolarization (Seluzicki et al., 2014; Vecsey et al., 2014) or increased Ca2+ levels in target cells (Seluzicki et al., 2014). In fact, PDF can apparently increase or decrease Ca2+ levels in different neurons within the DN1p cluster (Chatterjee et al., 2018). Opposing effects of PDF are also seen in the cockroach Rhyparobia maderae, such that responses to it vary across circadian clock neurons (Wei et al., 2014; Gestrich et al., 2018). Neuropeptides have complex roles in the clock network, as they synchronize the phases of molecular clocks and Ca2+ rhythms; in addition, acute and long-term effects of neuropeptides on target neurons may be different. How neuropeptides serve diverse functions is the clock network is still not well understood but likely involves divergent downstream signaling mechanisms (Duvall & Taghert, 2013; Seluzicki et al., 2014).
Circadian regulation of structural plasticity in s-LNvs
Circadian structural plasticity in the fly brain was first reported in the lamina, the first optic neuropil of the visual system [reviewed in (Górska-Andrzejak et al., 2015)]. In the lamina, many structures undergo circadian rhythms in morphological plasticity, including the retinal photoreceptor terminals, monopolar cells, and synapses (Pyza & Meinertzhagen, 1995; Weber et al., 2009; Górska-Andrzejak et al., 2013). Circadian plasticity of these structures is complex and involves multiple inputs from phototransduction pathways, clock neurons, and peripheral clocks in glia and photoreceptor cells.
Circadian structural plasticity has also been extensively studied in the terminal projections of s-LNvs in the dorsal protocerebrum. The s-LNv projections include both presynaptic and postsynaptic sites and are near most other clock neurons, implicating s-LNv projections as major sites for communication in the clock network (Helfrich-Förster et al., 2007; Yasuyama & Meinertzhagen, 2010). In the morning, the s-LNv terminals display greater complexity, with more arbors, branching, and volume, than at night (Fernández et al., 2008; Petsakou et al., 2015). Presumably, the increased terminal complexity indicates more synaptic connections. Indeed, using the GRASP (GFP reconstitution across synaptic partners) assay, which labels synaptic contacts between two populations of neurons, the number of contacts between s-LNv and their partners were found to be higher during the day than in the evening (Gorostiza et al., 2014; Tang et al., 2017). As such, the structural plasticity of s-LNv projections is a circadian output rhythm, regulated by the molecular clock and maintained in constant darkness (Fernández et al., 2008).
Circadian remodeling of s-LNv projections appears to be important for behavior, as disrupted remodeling of these projections is associated with compromised overt rest:activity rhythms. When the Pdf+ LNvs are acutely silenced, the s-LNv projections have reduced axonal complexity throughout the day (Depetris-Chauvin et al., 2011) and the flies display arrhythmic rest:activity behavior, although the s-LNv molecular clock still runs with a normal 24-hr period, suggesting that electrical activity is an output of the molecular clock and regulates remodeling of s-LNv projections. The circadian remodeling of s-LNv projections is also regulated by cell-autonomous expression of PDF and Mmp1, a matrix metalloproteinase that processes PDF (Depetris-Chauvin et al., 2014).
Other genes that affect rest:activity rhythms by dysregulating circadian remodeling of s-LNv projections, include Mef2 (Myocyte enhancer factor 2), a transcription factor expressed in all groups of clock neurons (Blanchard et al., 2010). Mef2 transcription is directly regulated by the CLOCK-CYCLE transcription factor complex, and Mef2, in turn, regulates transcription of many genes, including Fasciclin 2 (Fas2), the Drosophila ortholog of neural cell adhesion molecule, NCAM (Sivachenko et al., 2013). In s-LNvs, Mef2 promotes branching of the dorsal projections, while Fas2 represses branching through fasciculation, a phenomenon where axons stick to each other as they grow. From this study, a clock output mechanism emerges for circadian remodeling of s-LNv projections: CLOCK-CYCLE → Mef2 → Fas2 → s-LNv remodeling (Sivachenko et al., 2013). Dysregulation of Mef2 in Pdf+ LNvs leads to decreased power of rest:activity rhythms or complex rhythms (Blanchard et al., 2010; Sivachenko et al., 2013), but these behavioral changes are also correlated with altered molecular clocks in s-LNvs, suggesting Mef2 may also feedback onto the molecular clock (Blanchard et al., 2010).
Another pathway that regulates s-LNv projections involves Rho1, a member of the Rho family of GTPase signaling proteins. Rho1 activity cycles in the s-LNv projections and is highest in the evening (ZT12), when the projections are most condensed, which is consistent with the role of Rho1 in promoting retraction of projections (Petsakou et al., 2015). A Rho Guanine Nucleotide Exchange Factor (GEF), Puratrophin-1-like (Pura), activates Rho1 by promoting its association with GTP rather than GDP. Pura transcription cycles in s-LNvs and may be a direct target of CLOCK. Petsakou et al. (2015) proposed that clock-regulated Pura imposes rhythms in Rho1 activity, and the Rho-ROCK-myosin light chain pathway regulates actomyosin retraction of s-LNv projections in a circadian manner. When Rho1 is overexpressed in the Pdf+ LNv, the s-LNv projections do not branch in the morning, and flies display arrhythmic rest:activity behavior. At the molecular level, the s-LNv molecular clocks are normal, but in downstream DN1s, the molecular clocks are phase-shifted by up to 12 hours. Thus, remodeling of the s-LNvs has effects on other clock neurons.
The findings described above implicate two different pathways in circadian s-LNv remodeling, both of which are linked to the circadian clock. Both Mef2 and Pura are targets of CLOCK, but they are maximally expressed at different times of day, suggesting they regulate s-LNv projections in different ways (Blanchard et al., 2010; Sivachenko et al., 2013; Petsakou et al., 2015). Mechanisms of Mef2-Fas2 and Pura-Rho1 interaction are not known, and it is possible they act in parallel pathways, where Mef2-Fas2 regulate axon fasciculation cycles and Pura-Rho1 regulate axon growth- contraction cycles to together drive the circadian structural remodeling of s-LNv projections.
Clock output genes
Less is known about the circadian output pathways that transmit timekeeping signals from central clock cells to other parts of the brain to produce rest:activity rhythms. An output component is defined as a molecule or cell population that is regulated by the circadian clock but is not an intrinsic part of the clock mechanism. Several output genes have been implicated in behavioral rhythms, including na, slo, miR-279, Nf1, wake, and ebony. Dysregulation of these genes disrupts behavioral rhythms in animals but does so without affecting oscillations of the molecular clock. Many of these clock output genes exhibit clock-dependent diurnal variation in expression or function.
Na (narrow abdomen) encodes an ion channel with homology to the mammalian sodium leak channel, nonselective (NALCN), and is required broadly in the clock network for normal rest:activity rhythms (Lear, Lin, et al., 2005). In the posterior DN1 (DN1p) and l-LNv clock neurons, na is required for cycling of a sodium leak current, which contributes to oscillations in firing frequency and resting membrane potential (Flourakis et al., 2015). Nlf-1 (also known as Mid1) is a NA localization factor that is rhythmically expressed and clock-controlled. Nlf-1 is also required for robust rest:activity rhythms (Ghezzi et al., 2014; Flourakis et al., 2015). Together, NLF-1 and NA are part of a cell-autonomous clock output mechanism to ensure robust rhythms of neuronal activity.
The SLO (slowpoke) potassium channel was identified as an output factor, because its binding partner, SLOB (slowpoke binding protein) is coded by a clock-controlled gene with robust transcriptional rhythms (Claridge-Chang et al., 2001; McDonald & Rosbash, 2001; Ceriani et al., 2002). slo mutants are arrhythmic in constant darkness but have intact s-LNv molecular clocks. Instead, slo mutants have altered levels of PDF in s-LNv projections and desynchronized clocks in the DN1s (Fernández et al., 2007). slo may also have an important role outside the clock network, since clock neuron-specific rescue of slo only partially rescues rest:activity rhythms. Furthermore, dyschronic, a factor that regulates http://flybase.org/search/sloSLO expression, is required in non-clock neurons for rest:activity rhythms (Jepson et al., 2012).
miR-279 is a microRNA that regulates rest:activity rhythms by targeting and downregulating expression of Unpaired 1 (Upd1), a ligand of the JAK/STAT pathway (Luo & Sehgal, 2012). JAK/STAT signaling constitutes a critical pathway for development and immunity, but disrupting this pathway only in adulthood impairs rest:activity rhythms. miR-279 and Upd1 were found to be required in clock neurons for rest:activity rhythms, although their cellular requirements were not precisely mapped. Given findings that UPD1 is a fly analog of leptin and expressed in the Pdf+ LNvs, UPD1 could be an output of the s-LNvs (Beshel et al., 2017).
Wake (wide awake) is a clock output molecule that regulates the timing of sleep onset. wake mutants have a delayed sleep onset at night but normal rest:activity rhythms (Liu et al., 2014). WAKE levels cycle in the l-LNvs, peaking near dusk, when they are required to promote sleep. Previously, the l-LNvs were shown to promote arousal and respond to inhibition by GABA (Parisky et al., 2008; Shang et al., 2008; Sheeba, Fogle, et al., 2008). In l-LNvs, WAKE upregulates membrane localization of RDL, a GABA(A) receptor, which would inhibit the excitability of arousal-promoting l-LNvs. Indeed, in wake mutants, the l-LNvs show decreased GABA sensitivity and increased excitability (Liu et al., 2014). RDL also cycles in l-LNvs and is regulated by rhythmic degradation though the E3 ligase Fbxl4, whose transcription is clock-controlled. As expected, Fbxl4 mutants have the opposite phenotype of wake mutants, with a shorter latency to sleep onset at dusk (Li et al., 2017).
Nf1 (neurofibromatosis-1) encodes a Ras-specific GTPase activating protein required for rest:activity rhythms (Williams et al., 2001). Nf1 mutants have increased Ras/mitogen-activated protein kinase (MAPK) signaling, and loss-of-function mutations in the MAPK pathway can rescue rest:activity rhythms in Nf1 mutants. Restoring Nf1 in clock cells does not rescue the behavioral deficits. Instead, Nf1 is required broadly in the brain, presumably in multiple circadian neurons that regulate rest:activity rhythms (Bai et al., 2018). Not only does Nf1 regulate PDF levels in the s-LNv projections, it regulates Ca2+ and neuropeptide levels in circadian output neurons that are downstream of clock neurons (discussed below).
Ebony encodes a β-alanyl-biogenic amine synthase that controls the levels of free biogenic amines. EBONY is expressed exclusively in glial cells, where it functions to regulate rest:activity rhythms (Suh & Jackson, 2007). At least some of the glial expression of EBONY co-localizes with PER and TIM clock proteins, suggesting that ebony is an output molecule of glial clock cells.
All the clock output genes identified so far, except for WAKE, appear to broadly function in multiple groups of circadian clock neurons or glial cells. Expression of many of these genes, including nlf-1, slob, wake, and ebony, is controlled by the clock and so they directly link the circadian clock to neuronal activity or cell signaling. Despite arrhythmic behavior in the clock output mutants, molecular oscillations in the s-LNv central neurons remain unaffected. However, there may be examples where mechanisms that affect the electrical activity of clock neurons also feedback onto the molecular clock (Nitabach et al., 2002; Ruben et al., 2012), thus assigning output and input functions to some genes is not so clear-cut. Finally, since the output genes described above have only been studied in the context of the known circadian network, their additional action in as-yet unidentified circadian neurons cannot be excluded.
Circadian output circuits that regulate rhythms of behavior/physiology
In the last 5 years, with advances in circuit mapping tools, we have identified multisynaptic output circuits that regulate circadian rhythms (Figure 1). These circuits consist of non-clock neurons that convey circadian timing information from clock neurons to sites that control behavior or physiology. Output neurons receive inputs from clock neurons, either directly or indirectly through another group of output neurons. To date, assays of output neurons have revealed cycling of neural/cellular activity in a clock-dependent fashion (Table 1). Disruption of this neuronal activity disrupts the output rhythm without affecting the molecular clock. Therefore, most phenotypes from manipulating circadian output neurons are effects on rhythmicity of rest:activity rather than changes in circadian period, which is an intrinsic property of the clock. However, output neurons could feedback onto the clock to affect periodicity. As output circuits identified thus far are peptidergic and neuromodulatory in nature, and possibly also redundant, their disruption tends to weaken the amplitude of the rest:activity rhythm and not eliminate it altogether as would loss of molecular clock oscillations.
The pars intercerebralis (PI) has been proposed as a clock output region for many years. Ablation studies in cockroaches showed that the PI is required for locomotor activity rhythms (Nishiitsutsuji-Uwo et al., 1967; Matsui et al., 2009). Furthermore, in Drosophila, nearly all the circadian clock neurons, except for the l-LNvs, project to the PI (Helfrich-Förster, 1995; Kaneko & Hall, 2000; Helfrich-Förster et al., 2007). The PI is a major neurosecretory center with a high degree of neurochemical heterogeneity, as such functionally analogous to the mammalian hypothalamus (de Velasco et al., 2007). PI neurons regulate various behaviors in flies including sleep (Foltenyi et al., 2007; Crocker et al., 2010), feeding (Zhan et al., 2016), nutrient sensing (Dus et al., 2015), courtship (Terhzaz et al., 2007), and aggression (Davis et al., 2014). Thus, the PI may be a major output center for regulating circadian timing of behaviors.
Our group identified populations of PI neurons relevant for circadian rhythms. Three different PI groups, those that express DH44 (Diuretic hormone 44), SIFa (SIFamide), or DILP2 (Drosophila insulin-like peptide 2), synapse with DN1p clock neurons (Cavanaugh et al., 2014; Barber et al., 2016). The three PI groups are largely distinct from one another, with the exception that a pair of the Dh44+ neurons expresses low levels of DILP2 (Ohhara et al., 2018). Currently, it is not known whether s-LNvs or other clock neurons directly signal to the PI. Furthermore, we do not know the identity of the signaling molecules that mediate the DN1p to PI communication.
DH44→Hugin: A neuropeptidergic output circuit regulates rest:activity rhythms
The six Dh44+ neurons of the PI receive clock input through a multisynaptic circuit comprised of s-LNv → DN1 → Dh44+ PI (Cavanaugh et al., 2014). Activation or ablation of Dh44+ PI neurons reduces the power (or amplitude) of rest:activity rhythms without affecting the molecular oscillation of clock proteins in s-LNvs, demonstrating that Dh44+ PI neurons are output neurons downstream of the clock. In Dh44+ PI neurons, Ca2+ levels cycle across the 24-hr day, with peak activity occurring around evening and trough activity in the morning. Ca2+ cycling in Dh44+ neurons requires the Pdf+ LNvs, suggesting that rhythmic Ca2+ levels propagate from the s-LNvs to Dh44+ neurons (Cavey et al., 2016). In addition, the Nf1 circadian output gene cell-autonomously regulates Ca2+ cycling in Dh44+ neurons (Bai et al., 2018).
What about the role of the DH44 neuropeptide in rest:activity rhythms? DH44 and one of its receptors, DH44-R1, are required for strong rest:activity rhythms (Cavanaugh et al., 2014; King et al., 2017). Our group also mapped the circuit downstream of Dh44+ PI neurons to another set of neuropeptidergic neurons in the subesophageal zone. Knockdown of Dh44-R1 in hugin+ neurons reduces the power of rest:activity rhythms. In addition, hugin and its encoded neuropeptides, Hugin-γ and/or Prokynin-2, are required for robust rest:activity rhythms. hugin+ neurons themselves display clock-dependent cycling of neuropeptide vesicle release from their axon termini. A subset of hugin+ neurons projects back to the PI, potentially providing feedback regulation, while another subset of hugin+ neurons projects to the ventral nerve cord (VNC), where the circuit potentially modulates motor circuits driving locomotor activity (King et al., 2017). For the first time, we have a minimal, linear circuit between clock neurons and output neurons regulating locomotor activity.
SIFa+ PI neurons regulate rest:activity rhythms
In the same screen that identified Dh44+ PI neurons, the SIFa+ PI neurons were also found to regulate rest:activity rhythms (Cavanaugh et al., 2014). Ablation of all four SIFa+ neurons in the brain disrupts rest:activity rhythms but spares the s-LNv molecular clock. Loss of SIFa peptide itself produces a weaker effect on rest:activity rhythms than neuronal ablation, suggesting that other or co-neurotransmitters from SIFa+ neurons regulate rest:activity rhythms (Bai et al., 2018). Finally, circadian phenotypes in Nf1 mutants may be due to dysregulation of SIFa+ neurons. In Nf1 mutants with arrhythmic rest:activity behavior, both Ca2+ levels in SIFa+ neurons and mRNA levels of SIFa are elevated (Bai et al., 2018).
Dilp2+ PI neurons integrate circadian timing and metabolic signals
The fourteen Dilp2+ PI neurons and the insulin-like peptides have well-described roles in feeding and metabolism (Nässel et al., 2013). Similar to the Dh44+ and SIFa+ neurons, Dilp2+ neurons receive inputs from DN1p clock neurons (Barber et al., 2016). However, unlike their PI counterparts, Dilp2+ neurons do not appear to control rest:activity rhythms. Activation of Dilp2+ neurons in the adult fly is not sufficient to impair rest:activity rhythms (Cavanaugh et al., 2014). However, Dilp2+ neurons and insulin signaling may be important for development of circadian output circuits (Monyak et al., 2017). A set of Dilp2+ neurons project out of the brain and into the aorta, where circulating insulin-like peptides may be released to affect peripheral tissues, like the fat body. Dilp2+ neurons and insulin signaling regulate transcriptional rhythms of sxe2, a lipase in the fat body (Barber et al., 2016). As circadian output neurons, Dilp2+ neurons show cycling in electrical activity (Barber et al., 2016). Dilp2+ neurons exhibit higher electrical activity in the morning compared to the night, specifically increased firing frequency and burst firing events. These differences in electrical activity are lost in a period null mutant, demonstrating that cycling of Dilp2+ neuronal activity is clock-dependent. Cycling of electrical activity in Dilp2+ neurons is in phase with cycling in upstream clock neurons, DN1s and LNvs (Cao & Nitabach, 2008; Sheeba, Gu, et al., 2008; Flourakis et al., 2015). In addition to clock-regulation, firing in Dilp2+ neuron is regulated by feeding, since restricted feeding can shift the nighttime firing pattern of Dilp2+ neurons to the daytime firing pattern (Barber et al., 2016). Thus, Dilp2+ PI neurons integrate both circadian timing and metabolic signals.
Leucokinin regulates rest:activity rhythms
Leucokinin (Lk)-expressing neurons in the lateral horn are circadian output neurons that regulate sleep and rest:activity rhythms (Cavey et al., 2016). Both Lk and Lk receptor (Lk-R) mutants have reduced power of rest:activity rhythms. s-LNv clock neurons project to Lk+ lateral horn neurons, and firing of Pdf+ LNv neurons indirectly inhibits Ca2+ in Lk+ lateral horn neurons. LK-R is expressed broadly in the brain, including the lateral horn, ellipsoid body, and fan-shaped body, which are all areas implicated in locomotor control. The cellular requirement of LK-R for rest:activity rhythms has not been mapped. However, both Lk+ and Lk-R+ neurons in the lateral horn display cycling of Ca2+ levels that is dependent on the molecular clock and Pdf+ LNvs. Cycling of Ca2+ levels occurs with opposite phases in Lk+ and Lk-R+ neurons, since LK peptide decreases Ca2+ levels in Lk-R+ neurons. Lk+ and Lk-R+ lateral horn neurons also exhibit rhythms in excitability to carbachol, a cholinergic receptor agonist, that tracks with baseline Ca2+ rhythms. In summary, rhythmic neuronal activity can propagate to output neurons that are at least two synapses removed from clock neurons.
PTTH+ neurons regulate eclosion rhythms
Eclosion (adult emergence from pupae) occurs only once in the life of a fly, but rhythms of eclosion can be monitored in a population, with peaks of emerging flies typically observed around dawn. The prothoracic gland (PG) is an endocrine gland that produces ecdysone, the steroid hormone that controls molting. Eclosion rhythms are controlled by central brain clocks and peripheral clocks in the PG (Myers et al., 2003), but the brain clock has a dominant role over the PG clock (Selcho et al., 2017). The central clock transmits timing information to the PG clock through a s-LNv → PTTH+ neurons → PG circuit (Selcho et al., 2017). PTTH (prothoracicotropic hormone) is expressed in two pairs of brain neurons that receive input from s-LNvs via short Neuropeptide F. In turn, PTTH from the brain signals onto the PG through the PTTH receptor, torso. Knockdown of torso in the PG disrupts eclosion rhythms but has no effect on adult rest:activity rhythms (Selcho et al., 2017). These works highlight that s-LNvs control rest:activity rhythms and eclosion rhythms through different output circuits.
Conclusion
The molecular mechanism of the circadian oscillator has been worked out in detail. For many years, we knew much less about how oscillations of the molecular clock are translated into overt rhythms in behavior and physiology. Only recently has the field begun to identify functional connections within and downstream of the clock network, thus providing a neural basis for circadian rhythms. The primary focus of the field has been dissecting functional circuits that control rest:activity rhythms, and so the circuits that control other rhythmic behaviors in adult flies are underexplored. For all circadian circuits, clock-regulated cycling of neuronal activity appears to be the output mechanism for timekeeping and can propagate from clock neurons to output neurons along multisynaptic circuits. Longitudinal recording of neuronal activity over 24 hours remains a challenge in flies but will be informative for precisely studying how cycling of activity in circadian circuits is shaped by the molecular clock and neurotransmission.
The knowledge provided by dissection of circadian circuits will likely establish principles applicable to the function of circuits in general. The circadian model allows study of the neuromodulatory role of neuropeptides, the mechanisms coordinating release of a peptide and a fast neurotransmitter at the same synapse, and the use of different signaling mechanisms in response to the same neuromodulator, all in the context of robust behavioral outputs. Once the basic mechanisms governing these processes have been determined, their adaptation to more complex or nuanced behaviors can be ascertained. Moreover, while the emphasis in the circadian field is on 24-hour rhythms, the principles driving transmission of temporal signals may also be relevant for understanding those that occur on a shorter timescale.
Acknowledgements
The laboratory is supported by National Institutes of Health (NIH) grant R37 NS048471 (to A.S.) A.N.K. was supported in part by NIH T32 GM008216 and F31 NS100395.
Abbreviations
- BBB
blood-brain barrier
- Ca2+
intracellular calcium ion
- CRY
Cryptochrome
- CT
circadian time
- DD
constant darkness
- DH31
Diuretic Hormone 31
- DH44
Diuretic Hormone 44
- DH44-R1
diuretic hormone 44 receptor 1
- DILP2
Drosophila insulin-like peptide 2
- DN1
dorsal neuron (group 1)
- DN1p
posterior dorsal neuron (group 1)
- DN2
dorsal neuron (group 2)
- DN3
dorsal neuron (group 3)
- Fas2
Fasciclin 2
- GRASP
GFP reconstitution across synaptic partners
- LHLK
Leucokinin-expressing lateral horn neurons
- LK
Leucokinin
- LK-R
Leucokinin receptor
- l-LNv
large ventral lateral neuron
- LNd
dorsal lateral neuron
- LNv
ventral lateral neuron
- LPN
lateral posterior neuron
- Mef2
Myocyte enhancer factor 2
- mGluRA
metabotropic Glutamate Receptor
- na
narrow abdomen
- Nf1
Neurofibromatosis-1
Pigment-Dispersing Factor
- PDFR
Pigment-Dispersing Factor receptor
- PER
PERIOD
- PG
prothoracic gland
- PI
pars intercerebralis
- PTTH
prothoracicotropic hormone
- Pura
Puratrophin-1-like
- SEZ
subesophageal zone
- SIFa
SIFamide
- s-LNv
small ventral lateral neuron
- slo
slowpoke
- sNPF
short Neuropeptide F
- TIM
TIMELESS
- VGlut
Vesicular glutamate transporter
- VNC
ventral nerve cord
- wake
wide awake
- ZT
Zeitgeber Time
Footnotes
Conflict of Interest
The authors declare no conflict of interests.
References
- Bai L, Lee Y, Hsu CT, Williams JA, Cavanaugh D, Zheng X, Stein C, Haynes P, Wang H, Gutmann DH, & Sehgal A (2018) A Conserved Circadian Function for the Neurofibromatosis 1 Gene Cell Rep, 22, 3416–3426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber AF, Erion R, Holmes TC, & Sehgal A (2016) Circadian and feeding cues integrate to drive rhythms of physiology in Drosophila insulin-producing cells. Genes Dev, 30, 2596–2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckwith EJ & Ceriani MF (2015) Communication between circadian clusters: The key to a plastic network. FEBS Lett, 589, 3336–3342. [DOI] [PubMed] [Google Scholar]
- Beshel J, Dubnau J, & Zhong Y (2017) A Leptin Analog Locally Produced in the Brain Acts via a Conserved Neural Circuit to Modulate Obesity-Linked Behaviors in Drosophila. Cell Metab, 25, 208–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchard FJ, Collins B, Cyran SA, Hancock DH, Taylor MV, & Blau J (2010) The transcription factor Mef2 is required for normal circadian behavior in Drosophila. J. Neurosci, 30, 5855–5865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blau J & Young MW (1999) Cycling vrille Expression Is Required for a Functional Drosophila Clock. Cell, 99, 661–671. [DOI] [PubMed] [Google Scholar]
- Cao G & Nitabach MN (2008) Circadian control of membrane excitability in Drosophila melanogaster lateral ventral clock neurons. J. Neurosci, 28, 6493–6501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavanaugh DJ, Geratowski JD, Wooltorton JRA, Spaethling JM, Hector CE, Zheng X, Johnson EC, Eberwine JH, & Sehgal A (2014) Identification of a circadian output circuit for rest:activity rhythms in Drosophila. Cell, 157, 689–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavey M, Collins B, Bertet C, & Blau J (2016) Circadian rhythms in neuronal activity propagate through output circuits. Nat. Neurosci, 19, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceriani MF, Hogenesch JB, Yanovsky M, Panda S, Straume M, & Kay S. a (2002) Genome-wide expression analysis in Drosophila reveals genes controlling circadian behavior. J. Neurosci, 22, 9305–9319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee A, Lamaze A, De J, Mena W, Chélot E, Martin B, Hardin P, Kadener S, Emery P, & Rouyer F (2018) Reconfiguration of a Multi-oscillator Network by Light in the Drosophila Circadian Clock. Curr. Biol, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Reiher W, Hermann-Luibl C, Sellami A, Cognigni P, Kondo S, Helfrich-Förster C, Veenstra JA, & Wegener C (2016) Allatostatin A Signalling in Drosophila Regulates Feeding and Sleep and Is Modulated by PDF. PLOS Genet, 12, e1006346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi C, Cao G, Tanenhaus AK, McCarthy EV, Jung M, Schleyer W, Shang Y, Rosbash M, Yin JCP, & Nitabach MN (2012) Autoreceptor control of peptide/neurotransmitter corelease from PDF neurons determines allocation of circadian activity in Drosophila Cell Rep, 2, 332–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claridge-Chang A, Wijnen H, Naef F, Boothroyd C, Rajewsky N, & Young MW (2001) Circadian Regulation of Gene Expression Systems in the Drosophila Head. Neuron, 32, 657–671. [DOI] [PubMed] [Google Scholar]
- Collins B, Kane EA, Reeves DC, Akabas MH, & Blau J (2012) Balance of activity between LN(v)s and glutamatergic dorsal clock neurons promotes robust circadian rhythms in Drosophila. Neuron, 74, 706–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crocker A, Shahidullah M, Levitan IB, & Sehgal A (2010) Identification of a neural circuit that underlies the effects of octopamine on sleep:wake behavior. Neuron, 65, 670–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis SM, Thomas AL, Nomie KJ, Huang L, & Dierick HA (2014) Tailless and Atrophin control Drosophila aggression by regulating neuropeptide signalling in the pars intercerebralis. Nat. Commun, 5, 3177. [DOI] [PubMed] [Google Scholar]
- de Velasco B, Erclik T, Shy D, Sclafani J, Lipshitz H, McInnes R, & Hartenstein V (2007) Specification and development of the pars intercerebralis and pars lateralis, neuroendocrine command centers in the Drosophila brain. Dev. Biol, 302, 309–323. [DOI] [PubMed] [Google Scholar]
- Depetris-Chauvin A, Berni J, Aranovich EJ, Muraro NI, Beckwith EJ, & Ceriani MF (2011) Adult-specific electrical silencing of pacemaker neurons uncouples molecular clock from circadian outputs. Curr. Biol, 21, 1783–1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depetris-Chauvin A, Fernández-Gamba A, Gorostiza EA, Herrero A, Castaño EM, & Ceriani MF (2014) Mmp1 processing of the PDF neuropeptide regulates circadian structural plasticity of pacemaker neurons. PLoS Genet, 10, e1004700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dus M, Lai JS-Y, Gunapala KM, Min S, Tayler TD, Hergarden AC, Geraud E, Joseph CM, & Suh GSB (2015) Nutrient Sensor in the Brain Directs the Action of the Brain-Gut Axis in Drosophila. Neuron, 87, 139–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duvall LB & Taghert PH (2013) E and M Circadian Pacemaker Neurons Use Different PDF Receptor Signalosome Components in Drosophila. J. Biol. Rhythms, 28, 239–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erion R, King AN, Wu G, Hogenesch JB, & Sehgal A (2016) Neural clocks and Neuropeptide F/Y regulate circadian gene expression in a peripheral metabolic tissue. Elife, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández M. de la P., Chu J, Villella A, Atkinson N, Kay SA, & Ceriani MF (2007) Impaired clock output by altered connectivity in the circadian network. Proc. Natl. Acad. Sci. U. S. A, 104, 5650–5655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández MP, Berni J, & Ceriani MF (2008) Circadian remodeling of neuronal circuits involved in rhythmic behavior. PLoS Biol, 6, e69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flourakis M, Kula-Eversole E, Hutchison AL, Han TH, Aranda K, Moose DL, White KP, Dinner AR, Lear BC, Ren D, Diekman CO, Raman IM, & Allada R (2015) A Conserved Bicycle Model for Circadian Clock Control of Membrane Excitability. Cell, 162, 836–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foltenyi K, Greenspan RJ, & Newport JW (2007) Activation of EGFR and ERK by rhomboid signaling regulates the consolidation and maintenance of sleep in Drosophila. Nat. Neurosci, 10, 1160–1167. [DOI] [PubMed] [Google Scholar]
- Frenkel L, Muraro NI, Beltrán González AN, Marcora MS, Bernabó G, Hermann-Luibl C, Romero JI, Helfrich-Förster C, Castaño EM, Marino-Busjle C, Calvo DJ, & Ceriani MF (2017) Organization of Circadian Behavior Relies on Glycinergic Transmission. Cell Rep, 19, 72–85. [DOI] [PubMed] [Google Scholar]
- Fujii S, Emery P, & Amrein H (2017) SIK3-HDAC4 signaling regulates Drosophila circadian male sex drive rhythm via modulating the DN1 clock neurons. Proc. Natl. Acad. Sci. U. S. A, 114, E6669–E6677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gestrich J, Giese M, Shen W, Zhang Y, Voss A, Popov C, Stengl M, & Wei H (2018) Sensitivity to Pigment-Dispersing Factor (PDF) Is Cell-Type Specific among PDF-Expressing Circadian Clock Neurons in the Madeira Cockroach. J. Biol. Rhythms, 33, 35–51. [DOI] [PubMed] [Google Scholar]
- Ghezzi A, Liebeskind BJ, Thompson A, Atkinson NS, & Zakon HH (2014) Ancient association between cation leak channels and Mid1 proteins is conserved in fungi and animals. Front. Mol. Neurosci, 7, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorostiza EA, Depetris-Chauvin A, Frenkel L, Pírez N, & Ceriani MF (2014) Circadian pacemaker neurons change synaptic contacts across the day. Curr. Biol, 24, 2161–2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Górska-Andrzejak J, Damulewicz M, & Pyza E (2015) Circadian changes in neuronal networks. Curr. Opin. Insect Sci, 7, 76–81. [DOI] [PubMed] [Google Scholar]
- Górska-Andrzejak J, Makuch R, Stefan J, Görlich A, Semik D, & Pyza E (2013) Circadian expression of the presynaptic active zone protein Bruchpilot in the lamina of Drosophila melanogaster. Dev. Neurobiol, 73, 14–26. [DOI] [PubMed] [Google Scholar]
- Grima B, Chélot E, Xia R, & Rouyer F (2004) Morning and evening peaks of activity rely on different clock neurons of the Drosophila brain. Nature, 431, 869–873. [DOI] [PubMed] [Google Scholar]
- Gunawardhana KL & Hardin PE (2017) VRILLE Controls PDF Neuropeptide Accumulation and Arborization Rhythms in Small Ventrolateral Neurons to Drive Rhythmic Behavior in Drosophila. Curr. Biol, 27, 3442–3453.e4. [DOI] [PubMed] [Google Scholar]
- Guo F, Cerullo I, Chen X, & Rosbash M (2014) PDF neuron firing phase-shifts key circadian activity neurons in Drosophila. Elife, 2014, 1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo F, Yu J, Jung HJ, Abruzzi KC, Luo W, Griffith LC, & Rosbash M (2016) Circadian neuron feedback controls the Drosophila sleep--activity profile. Nature, 536, 292–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamasaka Y, Rieger D, Parmentier M-L, Grau Y, Helfrich-Förster C, & Nässel DR (2007) Glutamate and its metabotropic receptor in Drosophila clock neuron circuits. J. Comp. Neurol, 505, 32–45. [DOI] [PubMed] [Google Scholar]
- Harrisingh MC, Wu Y, Lnenicka GA, & Nitabach MN (2007) Intracellular Ca2+ regulates free-running circadian clock oscillation in vivo. J. Neurosci, 27, 12489–12499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfrich-Förster C (1995) The period clock gene is expressed in central nervous system neurons which also produce a neuropeptide that reveals the projections of circadian pacemaker cells within the brain of Drosophila melanogaster. Proc. Natl. Acad. Sci. U. S. A, 92, 612–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfrich-Förster C, Täuber M, Park JH, Mühlig-Versen M, Schneuwly S, & Hofbauer A (2000) Ectopic expression of the neuropeptide pigment-dispersing factor alters behavioral rhythms in Drosophila melanogaster. J. Neurosci, 20, 3339–3353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfrich-Förster C, Yoshii T, Wülbeck C, Grieshaber E, Rieger D, Bachleitner W, Cusamano P, & Rouyer F (2007) The lateral and dorsal neurons of Drosophila melanogaster: new insights about their morphology and function. Cold Spring Harb. Symp. Quant. Biol, 72, 517–525. [DOI] [PubMed] [Google Scholar]
- Herrero A, Duhart JM, & Ceriani MF (2017) Neuronal and Glial Clocks Underlying Structural Remodeling of Pacemaker Neurons in Drosophila. Front. Physiol, 8, 918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyun S, Lee Y, Hong S-T, Bang S, Paik D, Kang J, Shin J, Lee J, Jeon K, Hwang S, Bae E, & Kim J (2005) Drosophila GPCR Han is a receptor for the circadian clock neuropeptide PDF. Neuron, 48, 267–278. [DOI] [PubMed] [Google Scholar]
- Ikeda M (2004) Calcium Dynamics and Circadian Rhythms in Suprachiasmatic Nucleus Neurons. Neurosci, 10, 315–324. [DOI] [PubMed] [Google Scholar]
- Im SH & Taghert PH (2010) PDF receptor expression reveals direct interactions between circadian oscillators in Drosophila. J. Comp. Neurol, 518, 1925–1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jepson JEC, Shahidullah M, Lamaze A, Peterson D, Pan H, & Koh K (2012) dyschronic, a Drosophila homolog of a deaf-blindness gene, regulates circadian output and Slowpoke channels. PLoS Genet, 8, e1002671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko H, Head LM, Ling J, Tang X, Liu Y, Hardin PE, Emery P, & Hamada FN (2012) Circadian rhythm of temperature preference and its neural control in Drosophila. Curr. Biol, 22, 1851–1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko M & Hall JC (2000) Neuroanatomy of cells expressing clock genes in Drosophila: transgenic manipulation of the period and timeless genes to mark the perikarya of circadian pacemaker neurons and their projections. J. Comp. Neurol, 422, 66–94. [DOI] [PubMed] [Google Scholar]
- King AN, Barber AF, Smith AE, Dreyer AP, Sitaraman D, Nitabach MN, Cavanaugh DJ, & Sehgal A (2017) A Peptidergic Circuit Links the Circadian Clock to Locomotor Activity. Curr. Biol,. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose M, Duvall LB, Li W, Liang X, Ren C, Steinbach JHH, & Taghert PHH (2016) Functional PDF Signaling in the Drosophila Circadian Neural Circuit Is Gated by Ral A-Dependent Modulation. Neuron, 90, 781–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kula E, Levitan ES, Pyza E, & Rosbash M (2006) PDF cycling in the dorsal protocerebrum of the Drosophila brain is not necessary for circadian clock function. J. Biol. Rhythms, 21, 104–117. [DOI] [PubMed] [Google Scholar]
- Kunst M, Hughes ME, Raccuglia D, Felix M, Li M, Barnett G, Duah J, & Nitabach MN (2014) Calcitonin Gene-Related Peptide Neurons Mediate Sleep-Specific Circadian Output in Drosophila. Curr. Biol, 24, 2652–2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lear BC, Lin J-M, Keath JR, McGill JJ, Raman IM, & Allada R (2005) The ion channel narrow abdomen is critical for neural output of the Drosophila circadian pacemaker. Neuron, 48, 965–976. [DOI] [PubMed] [Google Scholar]
- Lear BC, Merrill CE, Lin J-M, Schroeder A, Zhang L, & Allada R (2005) A G protein-coupled receptor, groom-of-PDF, is required for PDF neuron action in circadian behavior. Neuron, 48, 221–227. [DOI] [PubMed] [Google Scholar]
- Li Q, Li Y, Wang X, Qi J, Jin X, Tong H, Zhou Z, Zhang ZC, & Han J (2017) Fbxl4 Serves as a Clock Output Molecule that Regulates Sleep through Promotion of Rhythmic Degradation of the GABAA Receptor. Curr. Biol, 27, 3616–3625.e5. [DOI] [PubMed] [Google Scholar]
- Liang X, Holy TE, & Taghert PH (2016) Synchronous Drosophila circadian pacemakers display nonsynchronous Ca2+ rhythms in vivo. Science, 351, 976–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X, Holy TE, & Taghert PH (2017) A Series of Suppressive Signals within the Drosophila Circadian Neural Circuit Generates Sequential Daily Outputs. Neuron, 94, 1173–1189.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Stormo GD, & Taghert PH (2004) The Neuropeptide Pigment-Dispersing Factor Coordinates Pacemaker Interactions in the Drosophila Circadian System. J. Neurosci, 24, 7951–7957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Lamaze A, Liu Q, Tabuchi M, Yang Y, Fowler M, Bharadwaj R, Zhang J, Bedont J, Blackshaw S, Lloyd TE, Montell C, Sehgal A, Koh K, & Wu MN (2014) WIDE AWAKE mediates the circadian timing of sleep onset. Neuron, 82, 151–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo W & Sehgal A (2012) Regulation of circadian behavioral output via a MicroRNA-JAK/STAT circuit. Cell, 148, 765–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui T, Matsumoto T, Ichihara N, Sakai T, Satake H, Watari Y, & Takeda M (2009) The pars intercerebralis as a modulator of locomotor rhythms and feeding in the American cockroach, Periplaneta americana. Physiol. Behav, 96, 548–556. [DOI] [PubMed] [Google Scholar]
- McDonald MJ & Rosbash M (2001) Microarray analysis and organization of circadian gene expression in Drosophila. Cell, 107, 567–578. [DOI] [PubMed] [Google Scholar]
- Mertens I, Vandingenen A, Johnson EC, Shafer OT, Li W, Trigg JS, De Loof A, Schoofs L, & Taghert PH (2005) PDF receptor signaling in Drosophila contributes to both circadian and geotactic behaviors. Neuron, 48, 213–219. [DOI] [PubMed] [Google Scholar]
- Mezan S, Feuz JD, Deplancke B, & Kadener S (2016) PDF Signaling Is an Integral Part of the Drosophila Circadian Molecular Oscillator. Cell Rep, 17, 708–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyasako Y, Umezaki Y, & Tomioka K (2007) Separate sets of cerebral clock neurons are responsible for light and temperature entrainment of Drosophila circadian locomotor rhythms. J. Biol. Rhythms, 22, 115–126. [DOI] [PubMed] [Google Scholar]
- Monyak RE, Emerson D, Schoenfeld BP, Zheng X, Chambers DB, Rosenfelt C, Langer S, Hinchey P, Choi CH, McDonald TV, Bolduc FV, Sehgal A, McBride SMJ, & Jongens TA (2017) Insulin signaling misregulation underlies circadian and cognitive deficits in a Drosophila fragile X model. Mol. Psychiatry, 22, 1140–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers EM, Yu J, & Sehgal A (2003) Circadian control of eclosion: interaction between a central and peripheral clock in Drosophila melanogaster. Curr. Biol, 13, 526–533. [DOI] [PubMed] [Google Scholar]
- Nässel DR, Kubrak OI, Liu Y, Luo J, & Lushchak OV (2013) Factors that regulate insulin producing cells and their output in Drosophila. Front. Physiol, 4, 252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng FS & Jackson FR (2015) The ROP vesicle release factor is required in adult Drosophila glia for normal circadian behavior. Front. Cell. Neurosci, 9, 256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng FS, Tangredi MM, & Jackson FR (2011) Glial cells physiologically modulate clock neurons and circadian behavior in a calcium-dependent manner. Curr. Biol, 21, 625–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiitsutsuji-Uwo J, Petropulos SF, & Pittendrigh CS (1967) Central nervous system control of circadian rhythmicity in the cockroach. I. Role of the pars intercerebralis. Biol. Bull, 133, 679–696. [Google Scholar]
- Nitabach MN, Blau J, & Holmes TC (2002) Electrical silencing of Drosophila pacemaker neurons stops the free-running circadian clock. Cell, 109, 485–495. [DOI] [PubMed] [Google Scholar]
- Ohhara Y, Kobayashi S, Yamakawa-Kobayashi K, & Yamanaka N (2018) Adult-specific insulin-producing neurons in Drosophila melanogaster. J. Comp. Neurol, 1–17. [DOI] [PubMed] [Google Scholar]
- Parisky KM, Agosto J, Pulver SR, Shang Y, Kuklin E, Hodge JJL, Kang K, Kang K, Liu X, Garrity PA, Rosbash M, & Griffith LC (2008) PDF cells are a GABA-responsive wake-promoting component of the Drosophila sleep circuit. Neuron, 60, 672–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JH, Helfrich-Förster C, Lee G, Liu L, Rosbash M, & Hall JC (2000) Differential regulation of circadian pacemaker output by separate clock genes in Drosophila. Proc. Natl. Acad. Sci, 97, 3608–3613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petsakou A, Sapsis TP, & Blau J (2015) Circadian Rhythms in Rho1 Activity Regulate Neuronal Plasticity and Network Hierarchy. Cell, 162, 823–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pírez N, Christmann BL, & Griffith LC (2013) Daily rhythms in locomotor circuits in Drosophila involve PDF. J. Neurophysiol, 110, 700–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyza E & Meinertzhagen IA (1995) Monopolar cell axons in the first optic neuropil of the housefly, Musca domestica L., undergo daily fluctuations in diameter that have a circadian basis. J. Neurosci, 15, 407–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renn SC, Park JH, Rosbash M, Hall JC, & Taghert PH (1999) A pdf neuropeptide gene mutation and ablation of PDF neurons each cause severe abnormalities of behavioral circadian rhythms in Drosophila. Cell, 99, 791–802. [DOI] [PubMed] [Google Scholar]
- Rieger D, Shafer OT, Tomioka K, & Helfrich-Förster C (2006) Functional analysis of circadian pacemaker neurons in Drosophila melanogaster. J. Neurosci, 26, 2531–2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruben M, Drapeau MD, Mizrak D, & Blau J (2012) A mechanism for circadian control of pacemaker neuron excitability. J. Biol. Rhythms, 27, 353–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selcho M, Millán C, Palacios-Muñoz A, Ruf F, Ubillo L, Chen J, Bergmann G, Ito C, Silva V, Wegener C, & Ewer J (2017) Central and peripheral clocks are coupled by a neuropeptide pathway in Drosophila. Nat. Commun, 8, 15563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seluzicki A, Flourakis M, Kula-Eversole E, Zhang L, Kilman V, & Allada R (2014) Dual PDF signaling pathways reset clocks via TIMELESS and acutely excite target neurons to control circadian behavior. PLoS Biol, 12, e1001810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafer OT, Kim DJ, Dunbar-Yaffe R, Nikolaev VO, Lohse MJ, & Taghert PH (2008) Widespread receptivity to neuropeptide PDF throughout the neuronal circadian clock network of Drosophila revealed by real-time cyclic AMP imaging. Neuron, 58, 223–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafer OT & Yao Z (2014) Pigment-Dispersing Factor Signaling and Circadian Rhythms in Insect Locomotor Activity. Curr. Opin. insect Sci, 1, 73–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang Y, Griffith LC, & Rosbash M (2008) Light-arousal and circadian photoreception circuits intersect at the large PDF cells of the Drosophila brain. Proc. Natl. Acad. Sci. U. S. A, 105, 19587–19594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheeba V, Fogle KJ, Kaneko M, Rashid S, Chou Y-T, Sharma VK, & Holmes TC (2008) Large ventral lateral neurons modulate arousal and sleep in Drosophila. Curr. Biol, 18, 1537–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheeba V, Gu H, Sharma VK, O’Dowd DK, & Holmes TC (2008) Circadian- and light-dependent regulation of resting membrane potential and spontaneous action potential firing of Drosophila circadian pacemaker neurons. J. Neurophysiol, 99, 976–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivachenko A, Li Y, Abruzzi KC, & Rosbash M (2013) The transcription factor Mef2 links the Drosophila core clock to Fas2, neuronal morphology, and circadian behavior. Neuron, 79, 281–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoleru D, Peng Y, Agosto J, & Rosbash M (2004) Coupled oscillators control morning and evening locomotor behaviour of Drosophila. Nature, 431, 862–868. [DOI] [PubMed] [Google Scholar]
- Suh J & Jackson FR (2007) Drosophila ebony activity is required in glia for the circadian regulation of locomotor activity. Neuron, 55, 435–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X, Roessingh S, Hayley SE, Chu ML, Tanaka NK, Wolfgang W, Song S, Stanewsky R, & Hamada FN (2017) The role of PDF neurons in setting the preferred temperature before dawn in Drosophila. Elife, 6, e23206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terhzaz S, Rosay P, Goodwin SF, & Veenstra JA (2007) The neuropeptide SIFamide modulates sexual behavior in Drosophila. Biochem. Biophys. Res. Commun, 352, 305–310. [DOI] [PubMed] [Google Scholar]
- Vecsey CG, Pírez N, & Griffith LC (2014) The Drosophila neuropeptides PDF and sNPF have opposing electrophysiological and molecular effects on central neurons. J. Neurophysiol, 111, 1033–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber P, Kula-Eversole E, & Pyza E (2009) Circadian control of dendrite morphology in the visual system of Drosophila melanogaster. PLoS One, 4, e4290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei H, Yasar H, Funk NW, Giese M, Baz E-S, & Stengl M (2014) Signaling of Pigment-Dispersing Factor (PDF) in the Madeira Cockroach Rhyparobia maderae. PLoS One, 9, e108757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams JA, Su HS, Bernards A, Field J, & Sehgal A (2001) A circadian output in Drosophila mediated by neurofibromatosis-1 and Ras/MAPK. Science, 293, 2251–2256. [DOI] [PubMed] [Google Scholar]
- Yadlapalli S, Jiang C, Bahle A, Reddy P, Meyhofer E, & Shafer OT (2018) Circadian clock neurons constantly monitor environmental temperature to set sleep timing. Nature, 555, 98–102. [DOI] [PubMed] [Google Scholar]
- Yasuyama K & Meinertzhagen IA (2010) Synaptic connections of PDF-immunoreactive lateral neurons projecting to the dorsal protocerebrum of Drosophila melanogaster. J. Comp. Neurol, 518, 292–304. [DOI] [PubMed] [Google Scholar]
- Yoshii T, Hermann-Luibl C, & Helfrich-Förster C (2016) Circadian light-input pathways in Drosophila. Commun. Integr. Biol, 9, e1102805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshii T, Hermann C, & Helfrich-Förster C (2010) Cryptochrome-positive and -negative clock neurons in Drosophila entrain differentially to light and temperature. J. Biol. Rhythms, 25, 387–398. [DOI] [PubMed] [Google Scholar]
- Yoshii T, Wülbeck C, Sehadova H, Veleri S, Bichler D, Stanewsky R, & Helfrich-Förster C (2009) The neuropeptide pigment-dispersing factor adjusts period and phase of Drosophila’s clock. J. Neurosci, 29, 2597–2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerr DM, Hall JC, Rosbash M, & Siwicki KK (1990) Circadian fluctuations of period protein immunoreactivity in the CNS and the visual system of Drosophila. J. Neurosci, 10, 2749–2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan YP, Liu L, & Zhu Y (2016) Taotie neurons regulate appetite in Drosophila. Nat. Commun, 7, 13633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Chung BY, Lear BC, Kilman VL, Liu Y, Mahesh G, Meissner R-AA, Hardin PE, & Allada R (2010) DN1p Circadian Neurons Coordinate Acute Light and PDF Inputs to Produce Robust Daily Behavior in Drosophila. Curr. Biol, 20, 591–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang SL, Yue Z, Arnold DM, Artiushin G, & Sehgal A (2018) A Circadian Clock in the Blood-Brain Barrier Regulates Xenobiotic Efflux. Cell, 173, 130–139.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu Y, Bilodeau-Wentworth D, Hardin PE, & Emery P (2010) Light and temperature control the contribution of specific DN1 neurons to Drosophila circadian behavior. Curr. Biol, 20, 600–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X & Sehgal A (2012) Speed control: cogs and gears that drive the circadian clock. Trends Neurosci, 35, 574–585. [DOI] [PMC free article] [PubMed] [Google Scholar]