Abstract
In many insects, X-linked inversions fix at a higher rate and are much less polymorphic than autosomal inversions. Here we report that in Drosophila, X-linked inversions also capture 67% more genes. We estimated the number of genes captured through an Approximate Bayesian Computational (ABC) analysis of gene orders in 9 species of Drosophila. X-linked inversions fixed with a significantly larger gene content. Further, X-linked inversions of intermediate size enjoy highest fixation rate, while the fixation rate of autosomal inversions decreases with size. A less detailed analysis in Anopheles suggests a similar pattern holds in mosquitoes. We develop a population genetic model that assumes the fitness effects of inversions scale with the number of genes captured. We show that the same conditions that lead to a higher fixation rate also produce a larger size for inversions on the X.
Keywords: Sex chromosomes, evolution, population genetics
1. INTRODUCTION
The evolution of inversions on sex chromosomes is of interest for two reasons. First, chromosomal inversions play a key role in the evolution of sex chromosomes of many groups of animals and plants (Charlesworth & Charlesworth 2005; Bachtrog et al. 2011; Bachtrog 2013; Charlesworth 2013). The non-recombining sex determination region can expand by fixation of inversions that suppress recombination between the X and Y (or Z and W) chromosomes. In mammals and other taxa, the fixation of multiple inversions has generated “evolutionary strata” that show different levels of divergence between the sex chromosomes (Lahn & Page 1999; Liu et al. 2004; Handley et al. 2004). Second, sex chromosomes have unique genetic properties (Bachtrog et al. 2011). X-linked genes are hemizygous in males, they have fewer copies in the population than do autosomal genes, and they spend more of their evolutionary lives in females. These characteristics are expected to impact the properties of chromosome rearrangements that fix on sex chromosomes (Charlesworth et al. 1987; Pennell et al. 2015). Understanding differences between inversions on sex chromosomes and autosomes may give general insights into how inversions evolve throughout the genome.
Previous studies of inversions in Orthoptera and Diptera have found a “faster X” effect -- higher fixation rates on the X chromosome than on autosomes (White 1973; Charlesworth et al. 1987; Bhutkar et al. 2008; von Grotthuss et al. 2010). The X chromosome in marsupials also shows more rearrangements by inversions than do autosomes (Charlesworth et al. 1987; Deakin et al. 2012). Likewise in birds, the Z chromosome is subject to much more extensive intra-chromosomal rearrangements than are autosomes (Griffin et al. 2007; Nanda et al. 2008). A second intriguing observation from Orthoptera and Diptera is that there is a striking deficiency of inversion polymorphism on the X compared to autosomes (Kitzmiller 1977; Charlesworth et al. 1987; Pombi et al. 2008; Neafsey et al. 2015).
Charlesworth et al. (1987) developed a series of models to explain the faster X effect. In their analysis of inversions, they assumed that heterozygotes suffer a disadvantage (e.g., because of meiotic problems). They found that the fixation rates can be higher on the X than the autosomes if inversions that are homozygous or hemizygous have a fitness advantage that is sufficiently large. They also showed that inversions (or any other kind of mutation) have higher fixation rates on the X when they are beneficial and partly or completely recessive. The conditions that maintain polymorphic inversions have also been studied theoretically (Pamilo 1979; Avery 1984; Curtsinger 1980). Consistent with the deficit of polymorphisms seen on the X, those models show that there is a reduced parameter space for polymorphism on the X relative to the autosomes. To date, other distinctive properties of inversions on the X have not been studied.
In this paper, we address the sizes of inversions. This focus is motivated by studies in Drosophila and Anopheles that show several patterns. Polymorphic inversions that are common and geographically widespread tend to be larger than rare inversions with localized distributions (Wallace 1954; Olvera et al. 1979; Brehm & Krimbas 1991; Cáceres et al. 1997; Pombi et al. 2008). A comparison between D. melanogaster and D. yakuba suggested that the fixation rates of inversions vary with their size (York et al. 2007). Inversions can also impact patterns of gene expression in the genome (Cassone et al. 2011; Fuller et al. 2016), and the number of differentially expressed genes might scale with inversion size. There are also theoretical reasons to suspect that the sizes of inversions affect how likely they are to become established. If inversions fix because they link together locally adapted alleles, the probability that a new inversion spreads increases with the number of locally-adapted loci that it captures (Kirkpatrick & Barton 2006). In their models for the fixation of inversions, Nei et al. (1967) and Kimura and Ohta (1969) hypothesized that the size of an inversion affects its fitness through the number of deleterious mutations that it is likely to capture.
We use gene order across the genomes of nine Drosophila species (von Grotthuss et al. 2010) to study the evolution of inversions. Our approach uses a novel scheme based on Approximate Bayesian computation (ABC) to estimate their sizes. Our analysis focuses on fixed inversions that differ between species, and we do not attempt to explain patterns of polymorphism. We measure size in terms of the number of genes that an inversion captures. We find that inversions fixed on X are larger on average. The distribution of sizes on the X is also distinctive. On autosomes, the smallest inversions fix most frequently, while on the X it is inversions of intermediate size that are most frequent. Less detailed analyses of two species of Anopheles mosquitoes suggest that inversions are also larger on the X in those taxa.
We develop a population genetic model to explain this “bigger-on-the-X” pattern. The key assumption is that the fitness effects of inversions are proportional to the number of genes they carry. We find the conditions regarding fitness effects and dominance that result in larger inversions becoming fixed on the X. We show that our model also explains other salient features of inversions observed in natural populations.
2. MATERIALS AND METHODS
Our analyses are based on the gene order for nine species of flies (Drosophila ananassae, D. erecta, D. grimshawi, D. mojavensis, D. pseudoobscura, D. virilis, D. willistoni, D. yakuba and D. melanogaster) as determined by von Grotthuss et al. (2010). The gene orders, in turn, are based on the reference genomes for those species (Drosophila 12 Genomes Consortium 2007). We used the phylogeny of these species estimated by Powell and DeSalle (1995) and shown in Figure 1. This phylogeny is consistent with that estimated from whole genomes (Drosophila 12 Genomes Consortium 2007).
Figure 1.
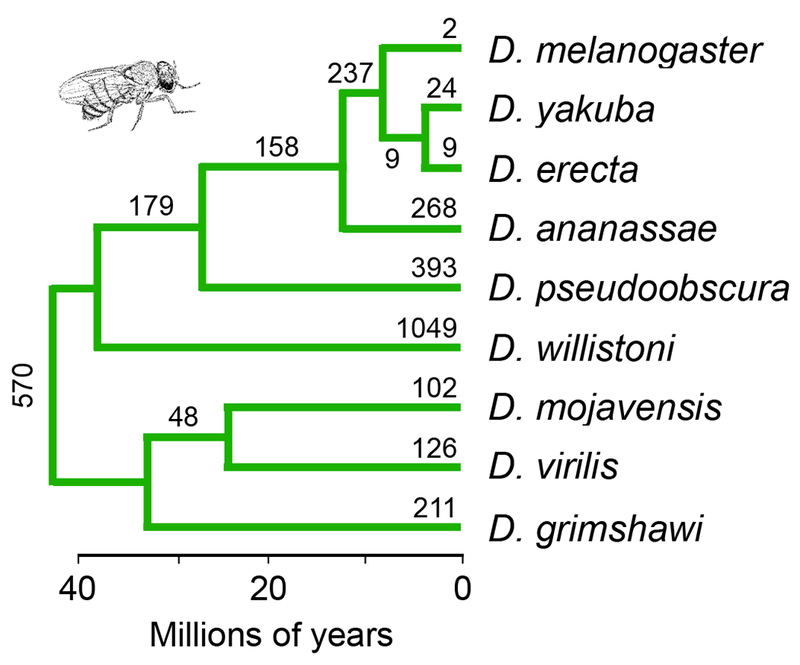
The phylogeny of the Drosophila species analyzed in this study (Powell and DeSalle 1995; Drosophila 12 Genomes Consortium 2007; Crosby et al. 2007). The estimated numbers of inversions fixed along each branch are from von Grotthuss et al. (2010).
We do not account for polymorphism caused by inversions that are currently segregating. We do not, however, expect that to affect the results noticeably. Inversions in Drosophila melanogaster, whose ages are the best characterized of any species in the genus, are typically only about 105 years old (Corbett-Detig & Hartl 2012). The branches on the phylogeny are 10 to 100 times longer, and so (assuming that inversions in melanogaster are representative) polymorphisms will little impact on estimates of differences between species. For semantic simplicity, we refer to the inversions found in the reference genomes as “fixed”, but in reality, some of them are certain to be polymorphic.
One approach to estimate the sizes of inversions fixed in different species would be to reconstruct their breakpoints using parsimony, then count the number of genes between the breakpoints. However, we found using simulations that this strategy greatly underestimates the sizes of inversions. This bias results because the breakpoints of older inversions are covered by younger ones (Bourque & Pevzner 2002), making the older inversions seem smaller.
We therefore devised the following strategy based on Approximate Bayesian Computation, or ABC (Beaumont et al. 2002). In Step 1, we used parsimony to estimate the distribution of inversion sizes with the software package GRIMM (Tesler 2002) under default parameter settings, considering the sign of the genes. In Step 2, we simulate the fixation of inversions on the phylogeny of the nine species. The number of inversions fixed along each branch is drawn from a Poisson distribution with a mean given by the product of the branch length and the fixation rate. This rate is sampled from a uniform prior distribution, with limits of 0 and 1.2 times the number of inversions estimated for that branch in Step 1. (These limits were chosen for optimal convergence by preliminary analyses.) For each inversion, a breakpoint is randomly chosen between two adjacent genes. The second breakpoint is randomly chosen such that the number of genes between the two breakpoints follows a gamma distribution with given size (µ) and shape (γ) parameters. Note that a new inversion can overlap with or be nested within an inversion that fixed previously. In Step 3, we again use the parsimony method of Step 1 to estimate the distribution of inversion sizes in the simulated data set. In Step 4, we compare the distribution estimated from the simulated data (Step 3) to the distribution estimated from the real data (Step 1). We measure the fit of the simulated data to the real data using the difference in the numbers of inversions that fix and the difference in their mean sizes. We repeat Steps 2 to 4, adjusting the parameters of the gamma distribution (µ and γ) until the simulated data converge on the real data in Step 4.
We repeated this entire procedure 108 times. In each run, the parameters for the gamma distribution were sampled from log-uniform distributions, with µ sampled in the range [1,1000] and γ sampled in the range [0.1, 10]. The posterior distribution was obtained by a rejection algorithm in which the 104 simulations with the smallest Euclidean distances to the real data were retained. The posterior distributions of µ and γ were estimated from those simulations.
This procedure was carried out separately for each chromosome arm (Muller element). This allows us to compare the X chromosome with the autosomes, and to compare the different autosomal arms. We excluded the small dot chromosome (Muller element F) for two reasons: it has only 5% of the genes carried by the other chromosomes, which strongly skews the sizes of inversions downwards, and the quality of this chromosome’s assemblies are lower than those for the other chromosomes (Leung et al. 2015).
To test the taxonomic generality of the results, we also studied two species of Anopheles mosquitoes. The quality of the genome assemblies for the mosquitoes are inferior to those in the flies, so these results should be treated with caution. We compared 3,958 orthologous genes in An. gambiae and An. stephensi, the two mosquitoes with the best reference genomes (Neafsey et al. 2015).
3. RESULTS
3.1. The sizes of inversions in Drosophila
We find that inversions in Drosophila that have fixed on the X are on average 67% larger than those on autosomes (Figure 2). The maximum a posteriori (MAP) estimate for the average size of inversions that fix on X is 496 genes (95% credible interval = [382, 575]), while on autosomes it is 297 genes (c.i. = [198, 378]). This difference is significant at the 0.5% level. The mean size of inversions that fix on the X is also significantly larger than the means of the four autosomal arms when each of the latter are treated separately (Table 1). These trends are also seen when inversion size is measured as a fraction of the genes on its chromosome arm that were captured. On the X, on average inversions capture 30% of the genes, while on autosomes they capture only 12% of the genes (p < 10−15, Wilcoxon test).
Figure 2.
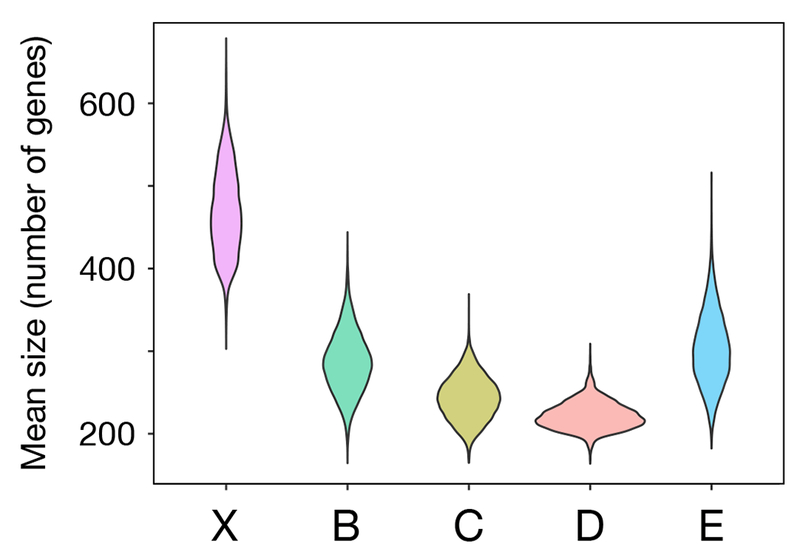
Posterior distributions of the mean sizes of inversion on the X chromosome and the arms of the autosomes (Muller elements B, C, D, and E).
Table 1.
The maximum a posteriori (MAP) estimates and 95% credible intervals for the mean sizes of inversions on the five Muller elements (major chromosomal arms) in Drosophila. Muller element A is the X chromosome, while the others comprise the autosomes.
| Muller element | MAP estimate | 95% c.i. |
|---|---|---|
| X (A) | 496 | [382,575] |
| B | 283 | [226,371] |
| C | 258 | [199,299] |
| D | 240 | [195,293] |
| E | 336 | [224,412] |
The shapes of the distributions of inversion sizes also differ between the X and autosomes (Figure 3). The mode of the distribution on the X is 422 genes, which is significantly greater than 0. In contrast, the mode for autosomes is 0. (In reality, inversions of size 0 do not exist. This result is a minor artifact of the gamma distribution that we fit to the data. We interpret this result to mean that inversions with very few genes are most likely to fix.) In sum, the most frequent inversions to fix on the X are intermediate in size, while on autosomes it is the smallest inversions that have the highest fixation rate.
Figure 3.
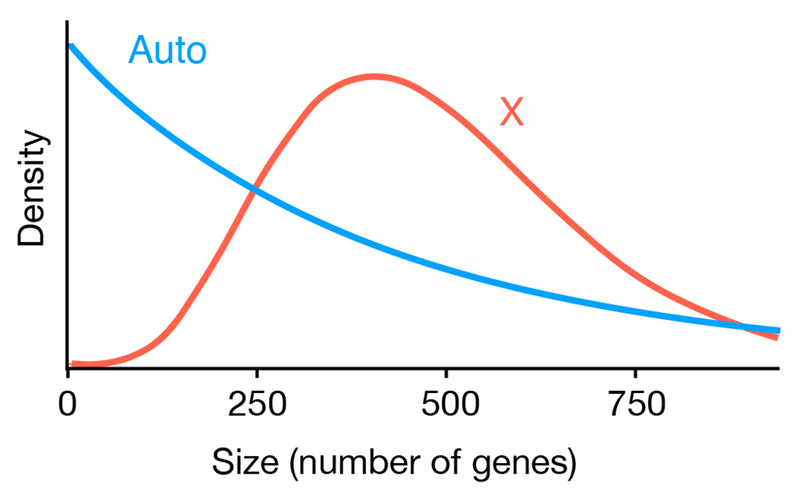
The distributions of inversion sizes on the X and the autosomes estimated by the ABC analysis. The mode of inversion size on the X is 422, significantly larger than 0. The mode on autosomes is not significantly larger than 0, however, and so the larger the size of inversion, the less likely it will be fixed.
One of the autosomal arms provides an interesting natural experiment to test the effect of sex linkage on inversion size. Muller Element D is fused to the X chromosome in D. willistoni and D. pseudoobscura. We found that the mean size of inversions on Element D when fused is 263 genes (c.i. = [182, 379]), while when it is not fused the mean is 233 genes (c.i. = [170, 284]). Although the trend is consistent with what we found in the comparison of the X and autosomes, the difference is not statistically significant.
3.2. Inversions in mosquitoes
The results for inversions in mosquitoes are consistent with those from the flies. In Anopheles, the average size of inversions fixed on the X is much larger than those on the autosomes: 26 vs. 1 marker gene (p < 0.01, one way Wilcoxon test). The result remains significant when the inversion sizes are scaled relative to chromosome size. The pattern is all the more striking when one considers that the autosomes in An. gambiae have 3 to 4 times more genes than the X (Neafsey et al. 2015), and so inversions on autosomes have the potential to span many more genes.
We emphasize that the result is much less robust than those from Drosophila because of the quality of the genome assemblies. Nevertheless, they suggest that the bigger-on-the-X pattern may be general.
3.3. A population genetic model
Next, we used a population genetic model to develop a hypothesis to explain why inversions fixed on the X might be larger. The key assumptions are that an inversion’s fitness effects are proportional to its size. Our analysis is a minor extension of models developed by Charlesworth et al. (1987).
On autosomes, let the relative fitnesses of standard (that is, uninverted) homozygotes, heterozygotes, and inverted homozygotes be
| WSS | WSI | WII |
|---|---|---|
| 1 | 1+s1y | 1+s2y |
where y is the size of the inversion. On X chromosomes, we assume full dosage compensation and no sex differences in fitness. Consequently, the relative fitness of males that carry the inverted X chromosome is (1 + s2y) relative to those with the standard chromosome. The selection parameters s1 and s2 can be positive or negative, allowing for cases in which inversions are either deleterious or beneficial, and for arbitrary patterns of dominance.
Following Charlesworth et al. (1987), we calculated the fixation rates of inversions on autosomes and the X using Kimura’s (1962) diffusion approximation. Assuming weak selection, the fixation rate for inversions of size y on autosomes is
| (1) |
where µ(y) is the rate that inversions of that size originate by mutation and N is the population size. The fixation rate on the X chromosome is
| (2) |
We assume that the mutation rates for inversions on the X and autosomes are the same.
We denote the relative fixation rate for inversions on the X compared to those on autosomes as R = KX/KA. Inspection of Equations (1) and (2) shows that R is greater than 1, meaning that inversions have a higher fixation rate on the X, whenever
| (3) |
An analogous result was derived previously by Charlesworth et al. (1987).
This condition can be satisfied when inversion heterozygotes are deleterious and when they are advantageous. When heterozygotes are beneficial, the condition requires that the fitness effects are partly recessive, such that homozygotes are more than twice as fit as heterozygotes. When heterozygotes are deleterious, the condition is met when the inversion is partly dominant (2s1 < s2 < 0), and when it is underdominant (s1 < 0, s2 > 0).
All else equal, if R increases with the size of inversions, then the mean size of inversions that fix will be larger on the X than on the autosomes. To show that this condition is met, we linearize R in terms of s1N y and s2N y, which gives
| (4) |
Thus R increases with y, and inversions that fix on the X will be larger on average than those on autosomes, whenever condition (3) is met.
An example of the distributions of inversion sizes predicted by this model is shown in Figure 4. Here we assumed that new inversions generated by mutation have an exponential distribution with a mean of 200 genes. Inversions are beneficial, with Nes2 = 0.1, and slightly underdominant, with Nes1 = −0.04. Under those assumptions, inversions fix more frequently on the X, and their mean size is larger (60 genes on autosomes vs. 309 genes on the X).
Figure 4.
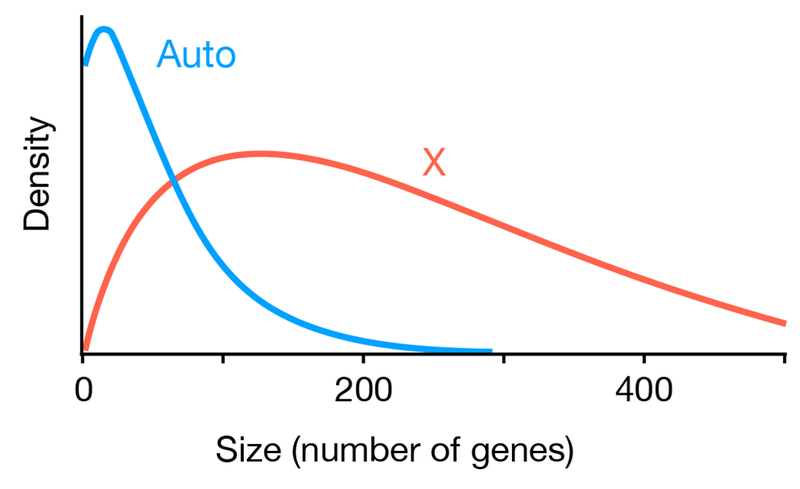
Distributions of sizes for inversions that fix on autosomes and the X chromosome predicted by the model. In this example, inversions are assumed to be beneficial (Nes2 = 0.1) and slightly under dominant (Nes1 = −0.04), and the sizes of new mutant inversions is exponential with a mean of 200 genes. With those parameters, the model predicts the mean size of inversions that fix will be 60 genes on autosomes and 309 genes on the X.
4. DISCUSSION
The evolutionary genetics of inversions has a rich history dating back to the laboratory studies of Sturtevant and the work on natural populations by Dobzhansky (Hoffmann & Rieseberg 2008; Kirkpatrick 2010). Much of the research has focused on the inversion polymorphisms that are abundant in some species, such as Drosophila pseudoobscura (Dobzhansky 1981) and Anopheles mosquitoes (Coluzzi et al. 2002). Another important research theme has been the important role that inversions play in blocking recombination between the X and Y (or Z and W) sex chromosomes (Bachtrog 2013; van Doorn & Kirkpatrick 2007).
This paper focuses on somewhat less studied aspects: the differences between inversions that have fixed on the X chromosome and the autosomes, and how those differences can inform us about the fitness effects of inversions. Our key finding is that inversions in Drosophila that have fixed on the X are larger than those on autosomes. Males are achiasmatic in these flies, and so this contrast cannot involve blocking recombination between the X and the Y. Instead, it must trace back to differences in how selection or mutation acts on those chromosomes. The same pattern is seen in comparisons between two species of Anopheles mosquitoes, which have comparable recombination rates in females and males (Zheng et al. 1996).
These results are consistent with a population genetic model that assumes the fitness effects of an inversion are proportional to its size. When that is true, the bigger-on-the-X pattern is expected under the same fairly general conditions that cause inversions to fix more frequently on the X, an empirical pattern that has been documented previously (Charlesworth et al. 1987; Bhutkar et al. 2008; Neafsey et al. 2015). These conditions are satisfied in several situations: when inversions are beneficial as heterozygotes and more than twice as beneficial as homozygotes, when inversions are underdominant, and when inversions are deleterious (both heterozygotes and homozygotes) and partly or wholly dominant. Thus the bigger-on-the-X pattern does not require that the inversion be deleterious when heterozygous, as assumed in some other models (Lande 1979; Charlesworth et al. 1987).
The model suggests what fitness effects can lead to the pattern, but provides no biological insight about what might produce those effects. Several mechanisms can be hypothesized. In one scenario that our model predicts will lead to the bigger-on-the-X effect, inversions are underdominant, and the deleterious fitness effects in heterozygotes increase with the size of the inversion. In many organisms, inversions are underdominant because single crossovers within the inverted region lead to aneuploid gametes (White 1973). Because the probability of a crossover increases with the size of an inversion, this would cause fitness loss in heterozygotes to increase with the size of the inversion, as required by the model. This scenario may not apply to Drosophila, however, which have mechanisms that largely suppress the deleterious effects of inversion heterozygotes (White 1973). Alternatively, inversions could be underdominant simply because of their genetic content, rather than their effects on recombination.
In a second scenario compatible with the predictions of our model, inversions increase fitness and are partly recessive. One situation in which this can occur is when inversions spread because of their effects in suppressing recombination between loci carrying locally adapted alleles (Kirkpatrick & Barton 2006; Charlesworth et al. 2017). The selective benefit of suppressing recombination scales with the initial recombination rate and with the number of loci involved. All else equal, larger inversions will span more of the linkage map, so they will have larger fitness advantage from suppressing recombination. Further, inversions on X chromosomes will have greater effects on decreasing recombination than those on autosomes: the X spends two-thirds of its evolutionary life in females, where it can recombine, while autosomes spend only half of their lives in females. Thus if locally-adapted loci are partly recessive, we might expect inversions on the X to fix more frequently.
In sum, several biological mechanisms could create the conditions causing inversions that fix on the X to be larger and more frequent than those on autosomes, as predicted by our model. The shapes of the size distributions estimated for the X and autosomes by the ABC analysis also differ: the mode is at the smallest size for inversions on autosomes, but at an intermediate size for those on the X (Figure 3). These shapes are determined by the distribution of sizes of new inversions generated by mutation as well as the fixation probabilities for mutations of different sizes. Figure 4 shows an example of the distributions predicted by the model assuming that the distribution of sizes of inversions arising by mutation is exponential; that is, the smallest inversions are most frequent. Further, in this example inversions are slightly underdominant, and so there is stronger selection against them as heterozygotes when they first appear. A result of these two factors is that the frequency of inversions that fix on autosomes declines with inversion size. In contrast, inversions that fix most frequently on the X are intermediate in size. That is because they have a selective advantage in males (as hemizygotes) even when rare, and that advantage grows with the size of the inversion.
Our model is highly simplified in several regards. Perhaps the most extreme is that we assume all inversions of the same size have the same fitness effects. This means that there is no allowance for the possibility that some inversions are overdominant, for example, or that their fitnesses can vary in space and time. At best, our model hopes to capture some average features of inversions. Inversions polymorphisms maintained (for example) by overdominance or local adaptation are a fascinating but likely very small subset of all inversions generated by mutation and fixed by selection and drift. Our model does not seek to understand how that set of inversions evolves.
The bigger-on-the-X effect may contribute to patterns involving sterility and other reproductive incompatibilities between populations and species. Inversions can contribute to incompatibilities. When they do, it is plausible that larger inversions will be more likely to carry alleles responsible for incompatibilities. The bigger-on-the-X effect will then cause the X chromosome to contribute to incompatibilities more often than autosomes. This pattern, called the “large X effect”, is seen in some taxa (reviewed in Charlesworth et al. (1987), Coyne and Orr (1989), and Presgraves (2008)). Consistent with that trend, segments of X chromosomes introgress between species less often than do segments of autosomes in Drosophila flies (Kulathinal et al. 2009) and Anopheles mosquitoes (Fontaine et al. 2015). Likewise, the X chromosome in mice (Mus) (Macholan et al. 2007) and the Z chromosome in flycatchers (Ficedula) (Saetre et al. 2003) show less introgress than do autosomes.
Two other hypotheses might also explain the bigger-on-the-X pattern seen in flies. First, the genetic content is often quite different on the X chromosome. The large X effect mentioned earlier might result from these differences. Genes with male-biased expression are significantly underrepresented on the X chromosome in flies (Parisi et al. 2003), and sexually antagonistic loci may be enriched on the X in D. melanogaster (Innocenti & Morrow 2010). The expression levels of genes on the X diverge faster than those on the autosome in flies (Meisel et al. 2012). It is plausible that one or more of these genetic differences between the X and the autosomes drives the pattern.
Second, new inversions generated by mutation might tend to be larger on the X than the autosomes. Transposons and repetitive sequences have been implicated in the mutational origin of inversions in several organisms (Cáceres et al. 1999; Goidts et al. 2004; Coulibaly et al. 2007). Perhaps differences between the X and autosomes in the distributions of those (and possibly other) genomic elements biases the mutational spectrum towards larger inversions on the X.
Regardless of why bigger inversions establish on the X, our results suggest inversions been fixed may affect more genes on the X chromosome and may have larger evolutionary impacts than those on autosomes.
ACKNOWLEDGEMENTS
We are grateful to four reviewers for constructive comments. This work was supported by the National Institutes of Health grant R01-GM116853 and Swiss National Science Foundation grant CRS113-147625.
Footnotes
DATA ACCESSIBILITY STATEMENT
We used publicly available data for our analyses. Orthologous marker genes in Drosophila were taken from von Grotthuss et al. (2010) (DOI: 10.1101/gr.103713.109) and FlyBase (flybase.org). Orthologous marker genes in Anopheles were taken from Neafsey et al. (2015) (DOI: 10.1126/science.1258522) and VectorBase (https://www.vectorbase.org/).
References
- Avery PJ (1984) The population genetics of haplo-diploids and X-linked genes. Genetical Research 44, 321–341. [Google Scholar]
- Bachtrog D (2013) Y-chromosome evolution: emerging insights into processes of Y-chromosome degeneration. Nature Reviews Genetics 14, 113–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachtrog D, Kirkpatrick M, Mank JE, et al. (2011) Are all sex chromosomes created equal? Trends in Genetics 27, 350–357. [DOI] [PubMed] [Google Scholar]
- Beaumont MA, Zhang WY, Balding DJ (2002) Approximate Bayesian computation in population genetics. Genetics 162, 2025–2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhutkar A, Schaeffer SW, Russo SM, et al. (2008) Chromosomal rearrangement inferred from comparisons of 12 Drosophila genomes. Genetics 179, 1657–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourque G, Pevzner PA (2002) Genome-scale evolution: Reconstructing gene orders in the ancestral species. Genome Research 12, 26–36. [PMC free article] [PubMed] [Google Scholar]
- Brehm A, Krimbas CB (1991) Inversion polymorphism in Drosophila obscura. Journal of Heredity 82, 110–117. [DOI] [PubMed] [Google Scholar]
- Cáceres M, Barbadilla A, Ruiz A (1997) Inversion length and breakpoint distribution in the Drosophila buzzatii species complex: Is inversion length a selected trait? Evolution 51, 1149–1155. [DOI] [PubMed] [Google Scholar]
- Cáceres M, Ranz JM, Barbadilla A, Long M, Ruiz A (1999) Generation of a widespread Drosophila inversion by a transposable element. Science 285, 415–418. [DOI] [PubMed] [Google Scholar]
- Charlesworth B, Coyne JA, Barton NH (1987) The relative rates of evolution of sex-chromosomes and autosomes. American Naturalist 130, 113–146. [Google Scholar]
- Charlesworth D (2013) Plant sex chromosome evolution. Journal of Experimental Botany 64, 405–420. [DOI] [PubMed] [Google Scholar]
- Charlesworth D, Barton NH, Charlesworth B (2017) The sources of adaptive variation. Proceedings of the Royal Society B 284, 20162864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth D, Charlesworth B (2005) Sex chromosomes: Evolution of the weird and wonderful. Current Biology 15, R129–R131. [DOI] [PubMed] [Google Scholar]
- Drosophila 12 Genomes Consortium (2007) Evolution of genes and genomes on the Drosophila phylogeny. Nature 450, 203–218. [DOI] [PubMed] [Google Scholar]
- Coulibaly MB, Lobo NF, Fitzpatrick MC, et al. (2007) Segmental duplication implicated in the genesis of inversion 2Rj of Anopheles gambiae. Plos One 2, e849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne JA, Orr HA (1989) Patterns of speciation in Drosophila. Evolution 43, 362–381. [DOI] [PubMed] [Google Scholar]
- Curtsinger JW (1980) On the opportunity for polymorphism with sex-linkage or haplodiploidy. Genetics 96, 995–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deakin JE, Bender HS, Pearse A- M, et al. (2012) Genomic restructuring in the Tasmanian Devil facial tumour: Chromosome painting and gene mapping provide clues to evolution of a transmissible tumour. PLoS Genetics 8, e1002483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontaine MC, Pease JB, Steele A, et al. (2015) Extensive introgression in a malaria vector species complex revealed by phylogenomics. Science 347, 1258524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goidts V, Szamalek JM, Hameister H, Kehrer-Sawatzki H (2004) Segmental duplication associated with the human-specific inversion of chromosome 18: a further example of the impact of segmental duplications on karyotype and genome evolution in primates. Human Genetics 115, 116–122. [DOI] [PubMed] [Google Scholar]
- Griffin DK, Robertson LBW, Tempest HG, Skinner BM (2007) The evolution of the avian genome as revealed by comparative molecular cytogenetics. Cytogenetic and Genome Research 117, 64–77. [DOI] [PubMed] [Google Scholar]
- Handley LL, Ceplitis H, Ellegren H (2004) Evolutionary strata on the chicken Z chromosome: Implications for sex chromosome evolution. Genetics 167, 367–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann AA, Rieseberg LH (2008) Revisiting the impact of inversions in evolution: From population genetic markers to drivers of adaptive shifts and speciation? Annual Review of Ecology, Evolution and Systematics 39, 21–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innocenti P, Morrow EH (2010) The sexually antagonistic genes of Drosophila melanogaster. PLoS Biology 8, e1000335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M (1962) On probability of fixation of mutant genes in a population. Genetics 47, 713–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick M (2010) How and why chromosome inversions evolve. PLoS Biology 8, e1000501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick M, Barton N (2006) Chromosome inversions, local adaptation, and speciation. Genetics 173, 419–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitzmiller JB (1977) Chromosomal differences among species of Anopheles mosquitoes. Mosquito Systematics 9, 113. [Google Scholar]
- Kulathinal RJ, Stevison LS, Noor MAF (2009) The genomics of speciation in Drosophila: Diversity, divergence, and introgression estimated using low-coverage genome sequencing. PLoS Genetics 5, e1000550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahn BT, Page DC (1999) Four evolutionary strata on the human X chromosome. Science 286, 964–967. [DOI] [PubMed] [Google Scholar]
- Lande R (1979) Effective deme sizes during long-term evolution estimated from rates of chromosomal rearrangement. Evolution 33, 234–251. [DOI] [PubMed] [Google Scholar]
- Liu ZY, Moore PH, Ma H, et al. (2004) A primitive Y chromosome in papaya marks incipient sex chromosome evolution. Nature 427, 348–352. [DOI] [PubMed] [Google Scholar]
- Macholan M, Munclinger P, Sugerkova M, et al. (2007) Genetic analysis of autosomal and X-linked markers across a mouse hybrid zone. Evolution 61, 746–771. [DOI] [PubMed] [Google Scholar]
- Nanda I, Schlegelmilch K, Haaf T, Schartl M, Schmid M (2008) Synteny conservation of the Z chromosome in 14 avian species (11 families) supports a role for Z dosage in avian sex determination. Cytogenetic and Genome Research 122, 150–156. [DOI] [PubMed] [Google Scholar]
- Neafsey DE, Waterhouse RM, Abai MR, et al. (2015) Highly evolvable malaria vectors: The genomes of 16 Anopheles mosquitoes. Science 347, 1258522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olvera O, Powell JR, Rosa M, et al. (1979) Population genetics of Mexican Drosophila. 6. Cytogenetic aspects of the inversion polymorphism in Drosophila pseudoobscura. Evolution 33, 381–395. [DOI] [PubMed] [Google Scholar]
- Pamilo P (1979) Genic variation at sex-linked loci: Quantification of regular selection models. Hereditas 91, 129–133. [DOI] [PubMed] [Google Scholar]
- Parisi M, Nuttall R, Naiman D, et al. (2003) Paucity of genes on the Drosophila X chromosome showing male-biased expression. Science 299, 697–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennell MW, Kirkpatrick M, Otto SP, et al. (2015) Y Fuse? Sex chromosome fusions in fishes and reptiles. PLoS Genetics 11, e1005237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pombi M, Caputo B, Simard F, et al. (2008) Chromosomal plasticity and evolutionary potential in the malaria vector Anopheles gambiae sensu stricto: insights from three decades of rare paracentric inversions. BMC Evolutionary Biology 8, 309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell JR, DeSalle R (1995) Drosophila molecular phylogenies and their uses. Evolutionary Biology 38, 87–138. [Google Scholar]
- Tesler G (2002) GRIMM: genome rearrangements web server. Bioinformatics 18, 492–493. [DOI] [PubMed] [Google Scholar]
- van Doorn GS, Kirkpatrick M (2007) Turnover of sexual chromosomes induced by sexual conflict. Nature 449, 909–912. [DOI] [PubMed] [Google Scholar]
- von Grotthuss M, Ashburner M, Ranz JM (2010) Fragile regions and not functional constraints predominate in shaping gene organization in the genus Drosophila. Genome Research 20, 1084–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace B (1954) Coadaptation and the gene arrangements of Drosophila pseudoobscura. International Union of Biological Sciences Symposium 15, 67–94. [Google Scholar]
- White MJD (1973) Animal Cytology and Evolution, 3rd edn Cambridge University Press, Cambridge. [Google Scholar]
- York TL, Durrett R, Nielsen R (2007) Dependence of paracentric inversion rate on tract length. BMC Bioinformatics 8, 115. [DOI] [PMC free article] [PubMed] [Google Scholar]


