Abstract
The polymerase complex proteins (PB2, PB1, and PA) are responsible primarily for the replication of avian influenza virus and play an important role in virus virulence, mammalian adaptation, and interspecies transmission. In this study; eight Egyptian LPAI-H9N2 viruses isolated from apparent healthy chickens and quails from 2014 to 2016. Characterization of complete nucleotide sequences, phylogenetic and mutation analysis were carried out. The measurement of thermodynamic stability of the H9N2 polymerase protein in comparison to human H3N2 and H1N1 proteins was carried out using in silico method. Phylogenetic analysis of these viruses revealed a close relationship to viruses isolated from neighboring Middle Eastern countries with an average of 96–99% homology. They are sharing the common ancestor A/quail/Hong Kong/G1/1997 (G1-Like) without any evidence for genetic reassortment. In addition, eight markers related to virulence were identified, including the combination of 627V and 391E in the PB2 gene with full-length PB1-F2 and PA-X proteins were observed in all viruses and the substitution N66S in PB1-F2 which suggest increasing virus virulence. Moreover, six markers that may affect the virus replication and transmission in mammalian hosts were identified. Five mutations related to mammalian adaptation show a structural stabilizing effect on LPAI-H9N2 polymerase complex protein according to the free-energy change (ΔΔG). Three out of those six adaptive mutations shown to increase polymerase complex protein stability were found in Egyptian LPAI-H9N2 viruses similar to Human H3N2 and H1N1 (661 in PB2, 225 and 409 in PA genes). Our results suggested that the stabilizing mutations in the polymerase complex protein have likely affected the protein structure and induced favorable conditions for avian virus replication and transmission in mammalian hosts. Indeed, the study reports the mutational analysis of the circulating LPAI-H9N2 strains in Egypt.
Keywords: Polymerasebasic1, 2 (PB1PB2), Polymerase acid (PA), H9N2, G1-LIKE, Free-energy change, H3N2 and H1N1
1. Introduction
The low pathogenic avian influenza LPAI-H9N2 subtype is classified into two major genetic lineages in both poultry and wild birds into North American and the Eurasian LPAI-LPAI-H9N2 lineages [26]. The Eurasian lineages consist of at least three sublineages represented by their prototype strains: A/Quail/Hong Kong/G1/1997(H9N2) (G1-like), A/Duck/Hong Kong/Y280/7 (Y280-like), A/Chicken/Beijing/1/94 (BJ94-like), and A/Chicken/Korea/38349P96323/96 (Korean-like). The LPAI-LPAI-H9N2 viruses have four distinct groups (A, B, C and D) co-circulating in the Middle East and Central-Asian countries [68]. Groups A and B are circulating extensively in the Middle East countries [44]. According to polymerases genes, all Egyptian isolates clustered in group A with isolates from Israel, Saudi Arabia, and Jordan [33].
LPAI-H9N2/G1 belong to Orthomyxoviridae family, influenza A virus, which possesses eight negative-sense single-stranded RNA segments [1]. 11–14 major proteins are encoded. three polymerase proteins polymerase basic protein 2 (PB2), polymerase basic protein 1 (PB1), polymerase acidic protein (PA), nucleoprotein (NP), NA and HA, matrix proteins (M1 and M2) and non-structural proteins (NS1 and NS2) [65]. The LPAI-H9N2 viruses share (PB2, PB1, PA, NP, M, and NS) genes with the lethal H5N1 viruses causing human disease in 1997. This stain crossed the species barrier to transmitted to humans in Hong Kong and Guangzhou, China [25].
The RNA-dependent RNA polymerase (RdRP) of influenza virus is composed of three viral proteins (PB2, PB1, and PA) and involved in both transcription and replication of the RNA genome [28], with about ∼250 KD molecular weight [16], [29]. Associated with the nucleoprotein (NP) encapsidated viral gene segments forming what is called polymerase complex [35].
The PB1 subunit plays a key role in both the assembly of three Polymerase subunits and the catalytic function of RNA polymerization [28]. while PA and PB2 would be involved in RNA replication and transcription [5]. The position 627 in PB2 gene is critical for pathogenicity and LPAI-LPAI-H9N2 viral replication in mice [37]. It was reported that PA mutations in the Amino acids T157A and T162A weaken the proteolysis Function of PA [48] but some PA mutation enhances the polymerase replication activity at higher temperatures to that influence the virus adaptations [7].
The alternative open reading frame (ORF) of segment 2 resulting from the encoding of the auxiliary proteins: PB1-F2 and PB1-N40 [14]. Which associating increase virus pathogenicity by causing cells apoptosis [23]. The N66S and L82S mutations in the PB1-F2 protein are important for increasing viral pathogenicity and mammalian adaptation respectively [15]. Furthermore, the segment 3 encodes several additional isoforms of PA proteins such as PA-X, PA-N55, and PA-N182 [54]. 252 amino acids are the full-length of a PA-X protein [4]. Viruses encoding full-length PA-X were more virulent and cause more severe inflammatory responses in mice. [22]. PA-N55 and PA-N182 proteins are highly conserved among IAV strains [2]. It is suggesting that these minor viral proteins are involved in host restriction as well as pathogenicity [42].
This study was carried out to determine the in-depth genetic analysis of polymerase genes and study the molecular markers related to virus replication and pathogenicity of recent viruses isolated from Egypt. We investigate the extent to which the effects of individual mutations on protein stability change as other residues diverge.
2. Materials and methods
2.1. Virus isolation and propagation
As a part of routine avian influenza surveillance program conducted in Egypt; Cloacal and oropharyngeal swab samples were collected from chickens and quails and were characterized at the National Laboratory for Veterinary Quality Control on Poultry Production (NLQP, Egypt). Extraction of Viral RNA using a QIAamp viral RNA Mini Kit (QIAGEN, Hilden, Germany) according to the manufacturer’s protocol. RNA extracts were tested for influenza type A by real-time RT-PCR to amplify 244 bp of the M segment [60], then Sub-typing of the (H5, H9, H7) was done using specific sets of primers and probes to determine the HA and NA subtypes (Supplementary Table 2) [56], [58] with One-Step RRT-PCR Kit (QIAGEN, Hilden, Germany). The real-time PCR reactions have run on Stratagene MX3005P real-time PCR machine (Stratagene, Amsterdam, and the Netherlands).
The extracted RNAs were tested further for IBV and NDV by real-time RT-PCR to ensure freedom of isolates from other pathogens affecting respiratory system [39], [66]. The positive influenza A samples with RT-PCR were initially isolated in 10-day-old specific pathogen free embryonated chicken eggs according to standard protocols [55]. The allantoic fluid was harvested, tested for Hemagglutination, and then stored at −80C until use.
According to HA phylogenetic grouping, eight viruses were selected representing LPAI-H9N2 virus groups circulating in Egypt. Isolates were collected from January 2014 to July 2016 from backyard poultry holdings and commercial farms representing Upper and Lower Egypt and including chicken and quails. The epidemiological data of the selected samples is shown in (Supplementary Table 1).
2.2. Sequencing and Phylogenetic analysis of the Pb1, PB2 and PA genes
The first-strand cDNA was synthesized using Superscript III Reverse Transcriptase (Invitrogen, Carlsbad, CA) and Uni-12 primer as per the manufacturer’s protocol. The full length of each gene was amplified using gene-specific forward and reverse primer (Table S2). Using a platinum®Taq DNA polymerase high fidelity (Invitrogen, Carlsbad, CA). The electrophoresis of PCR products was done on ethidium bromide stained Agarose gel and the amplified products of expected correct size visualized by gel documentation system BDA digital – Image capture (Biometra, Germany). Amplicons of the appropriate sizes were subsequently gel purified using a QIAquick Gel Extraction Kit (QIAGEN, Hilden, Germany). The purified PCR products were used directly for sequencing reactions using a Big Dye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA) according to manufacturer’s, the reaction product was purified by exclusion chromatography in CentriSep columns (Princeton Separations, Adelphia, NJ). The recovered materials sequenced using a 3500xl DNA Analyzer (Applied Biosystems). Sequences were assembled using seqscape and BioEdit 7.0 were used for multiple sequence alignment [6], [27]. Phylogenetic analysis was conducted by using the neighbor-joining algorithm with the Kimura 2-parameter model MEGA 6 [63]. Strain A/Turkey/Wisconsin/1/1966 was used as the root of the tree and the reliability of Phylogenetic inference at each branch node was estimated by the bootstrap method with 1000 replications. The trees included the most Egyptian LPAI-LPAI-H9N2 virus sequences available in the GenBank database, closely related LPAI-H9N2 viruses from other Middle Eastern countries, representative viruses from the groups A-D [41]. Using the SeqMan DNA Lasergene 6 software (DNASTAR, Madison, WI, USA) [17] to determine the identity Percent matrices comparing the genes under study to each other.
2.3. Measurement of the effect of mutation on protein stability
We selected eighteen substitutions which affect the ability of virus replication and/or the pathogenicity. In order to determine the effect of these selected mutations on the protein stability, we compared results with the human influenza A strains from 2015 and included in the 2017/2018 flu vaccine of the WHO [45]. These mutations are (PB1-13, 317), (PB1-F2-66, 82), (PB2-318, 391, 504, 647, 661), and (PA-57, 127, 225, 328, 400, 404, 409, 550, 672). Using the iStable® web site: (http://predictor.nchu.edu.tw/iStable/) [11], integrates the results from several stability prediction programs, and with the support vector machine (SVM)- like Mutant2.0 [9]. The Predictions are performed starting from the protein sequence, stability calculations were done with the polymerase subunits structure at 37 °C and pH = 7.0. I-Mutant 2.0 can be used both as a classifier for predicting the sign of the protein stability change upon mutation and as a regression estimator for predicting the related free energy change value (DDG). When predicting DDG values associated with mutations, the correlation of predicted with expected/experimental values is 0.71 (with a standard error of 1.30 kcal/mol) and 0.62 (with a standard error of 1.45 kcal/mol). [11].
2.4. Modeling of the protein structure
For understanding the significance of a single amino acid substitution on protein function based on 3D structure, we simulate the protein from the SWISS-PDB viewer, (https://swissmodel.expasy.org). The mutation analysis was performed using the PyMOL version 1.5.0.4 (http://www.pymol.org) [53] to identify the protein using the PDB file format obtained by Swiss model. Selected mutations were mapped in the structure of the polymerase subunits from one selected isolate representing the isolates under study (A/chicken/Egypt/1455V/2014 LPAI-H9N2 virus). The figures were prepared with the “Mutagenesis” wizard and the ray trace renderer of PyMol.
3. Results
3.1. Virus isolation and confirmation
Egyptian LPAI-LPAI-H9N2 viruses were isolated from sick and healthy broiler chickens and quail in Egypt. The samples were collected from January 2014 to July 2016; the samples were representing five Governorates of Upper and Lower Egypt. According to the type of breeding the selected viruses represent backyard poultry holdings, live bird markets and commercial farms, including chicken and quails. The details of epidemiological information, GenBank accession numbers, collection area, date of collection and type of breeding of these viruses are provided in (Supplementary Table 1). The Real-time RT-PCR results for eight isolates (CT values) for H9 subtype ranged from 12 to 18. The appropriate expected size of each gene was confirmed by gel electrophoresis. The electrophoresis of PCR products was done on ethidium bromide stained Agarose gel and the amplified products of expected correct size.
3.2. Phylogenetic analysis
3.2.1. Polymerase basic two
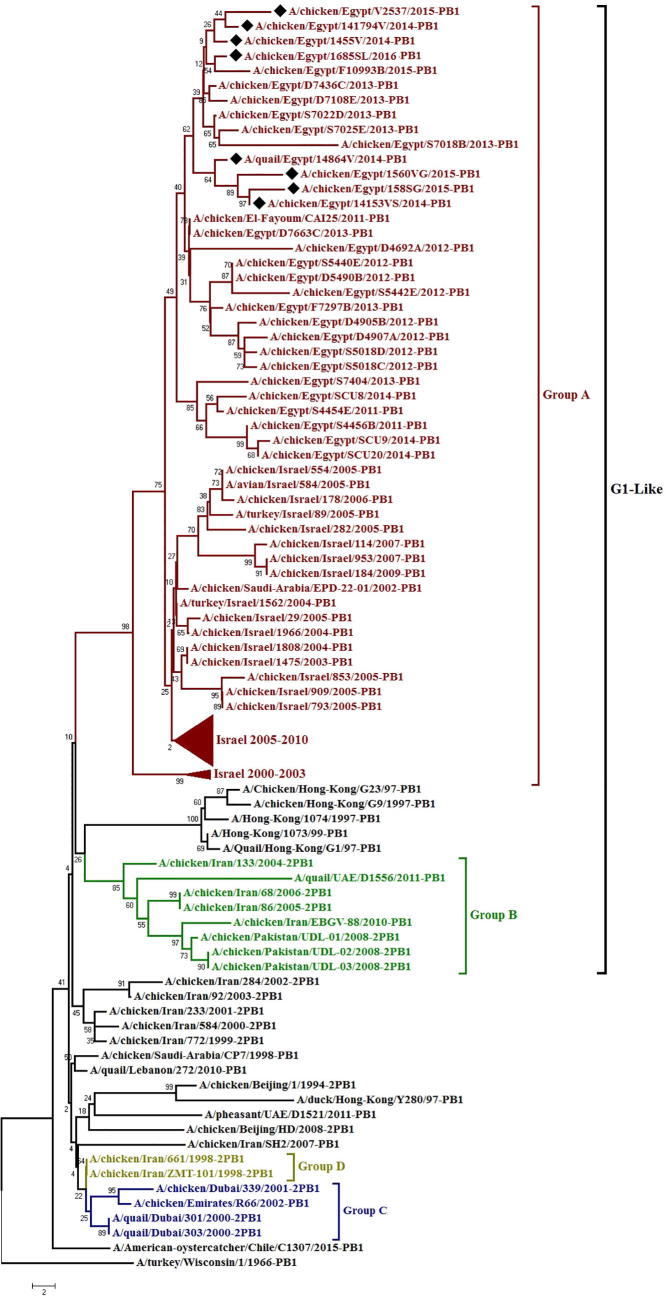
Phylogenetic analyses showed that all the PB2 genes of LPAI-H9N2 viruses isolated from Egypt are part of the lineage represented by A/QUAIL/Hong Kong/G1/1997(G1), which is the most dominant lineage worldwide, clustered with isolates from Israel, Iran in group A (Fig. 1). Egyptian viruses clustered in several minor sub-groups all viruses under study cluster together except for Q/14864V which different and take a different branch. The nucleotide and deduced amino acid sequence similarities among Egyptian strains ranged from 95 to 99% and 96 to 100%; respectively, and showing higher similarity (97–99%) to A/turkey/Israel/311/2009 rather than to LPAI-LPAI-H9N2 progenitor viruses such as G1 (93–94%) (Table 1), K627 and N701 substitutions in PB2 protein were not found in any of the LPAI-H9N2 viruses sequences, Instead all isolates possessed Egyptian viruses that displayed aspartate at position 701D as shown in valine at position 627V (Fig. 4), 391E, K318R, A661T, I504V and K526R were recognized in all viruses under study (Table 3). The mutations 355I and 453T became dominant in Egyptian LPAI-LPAI-H9N2 viruses. In addition, 41 distinct mutations were identified in comparison to ancestor strain Q/HK/G1/97 that characteristic to the Egyptian viruses and some of the recent Israeli strains from other LPAI-H9N2 strains (Supplementary Table 3).
Fig. 1.
Phylogenetic tree of the PB2gene of avian influenza subtype LPAI-H9N2 viruses isolated in Egypt during 2014–2016 and reference isolates from Gen Bank. A black rhomboid indicates isolates sequenced specifically for this study Boldface indicates avian viruses. Group A is shown in red, group B is shown in green, group C is shown in blue and group D are shown in olive.
Table 1.
Nucleotide and amino acid identity% of polymerase genes of Egyptian H9N2 viruses, Middle Eastern viruses, and reference strains.
| Average | Egypt 2011–2013 | Egypt 2006 | G1-Like | Human H9N2 | Y280-Like | Korea-Like | American-like | Israel 2009 | Saudi Arabia 2006 | |
|---|---|---|---|---|---|---|---|---|---|---|
| Identity% | ||||||||||
| Pb2 gene | NT | 95–100 | 87–88 | 86–87 | 86–87 | 83–84 | 86–87 | 82 | 96–97 | 85–86 |
| AA | 96–100 | 94–95 | 93–94 | 93–94 | 93–94 | 93–94 | 94–95 | 97–99 | 94–95 | |
| Pb1 gene | NT | 97–100 | 89–90 | 88–89 | 88–89 | 87–88 | 61–63 | 86–87 | 97–98 | 88–89 |
| AA | 98–100 | 97–98 | 96–97 | 96–97 | 95–96 | 57 | 95–96 | 98–99 | 96–97 | |
| PA gene | NT | 97–99 | 90 | 86–87 | 86–87 | 86 | 89–90 | 84–85 | 97–98 | 89–90 |
| AA | 97–100 | 95–96 | 93–94 | 93–94 | 93–95 | 94–95 | 93–95 | 97–98 | 94–96 | |
FN: The comparison was done between of the eight Egyptian H9N2 viruses in this study, H9N2 viruses from The Middle East and other reference strains.
NT = nucleotide identity%,AA = amino acid identity%, Egyptian H9N2 viruses: from 2011till 2016, Egypt2006: A/avian/Egypt/920431/2006/H9N2, G1-Like: A/Q/HK/G1/97, Human H9N2: A/HK/1073/99, Y280-Like: A/duck/HK/Y280/97, Korea-Like: A/Ck/Korea/38349-p96323/96, American-like: A/turkey/Wisconsin/1/66, Israel2009:A/turkey/Israel/311/09, SA/2006:A/avian/SA/910136/2006.
Fig. 4.
the three-dimensional structure showing Crystal structure of the polymerase subunits of the LPAI-H9N2 influenza virus. Shows the distribution of different substitutions on each gene, selected mutations were mapped in the structure of the polymerase subunits from the influenza A/chicken/Egypt/1455V/2014 LPAI-H9N2 virus using PyMol viewer (PyMOL 1.1 program (DeLano Scientific LLC)).
Table 3.
Mutations determinants in polymerase genes that correlated to the replication, spread or pathogenicity of H9N2 viruses.
| Protein | Amino acid position | Egyptian H9N2 under study | Avian preference | Mammalian preference | Reference |
|---|---|---|---|---|---|
| PB2 | 44 | A | A | S | [44] |
| 64 | M | M | T | [44] | |
| 81 | T | T | M | [47] | |
| 199 | A | A | S | [44] | |
| 271 | T | T | A | [44] | |
| 256 | D | D | G | [48] | |
| 318 | R(4), K(4) | K | R | [48] | |
| 333 | T | T | I | [40] | |
| 355 | M(6), I(2) | K | Q | [44] | |
| 475 | L | L | M | [40] | |
| 482 | K | K | R | [40] | |
| 504 | V | I | V | [45] | |
| 588 | A | A | I | [44] | |
| 613 | V | V | T | [40] | |
| 627 | V | E | K | [40] | |
| 647 | I | I | I | [40] | |
| 661 | A(7), T(1) | A | T | [40] | |
| 674 | A | A/S | T | [44] | |
| 701 | D | D | N | [40] | |
| 702 | K | K | R | [65] | |
| 714 | S(2),G(6) | S | R | [44] | |
| PB1 | 13 | P | L | P | [44] |
| 327 | R | R | K | [48] | |
| 336 | V | V | I | [48] | |
| 375 | N | N | S | [44] | |
| 538 | D | D | G | [44] | |
| 578 | K | K | Q | [44] | |
| 678 | S | S | N | [44] | |
| PB1-F2 | 68 | T | T | I | [48] |
| 73 | K | K | R | [48] | |
| 76 | V | V | A | [48] | |
| 79 | R | R | Q | [48] | |
| 82 | S | L | S | [48] | |
| 87 | E | E | G | [48] | |
| PA | 28 | P | P | L | [48] |
| 55 | D | D | N | [48] | |
| 57 | R(6),K(2) | R | Q | [48] | |
| 65 | S | S | L/Y | [44] | |
| 100 | V | V | A | [44] | |
| 133 | E | E | G | [44] | |
| 225 | S(7),G(1) | S | C | [48] | |
| 241 | C | C | Y | [48] | |
| 268 | L | L | I | [48] | |
| 312 | K | K | R | [48] | |
| 356 | K | K | R | [48] | |
| 382 | E | E | D | [48] | |
| 400 | S | Q/T/S | L | [48] | |
| 404 | A | A | S | [48] | |
| 409 | S(7),N(1) | S | N | [48] | |
| 552 | T | T | S | [44] | |
| 556 | Q | Q | R | [44] | |
| 615 | K | K | N | [44] | |
3.2.2. Polymerase basic one
Phylogenetic analysis showed that The PB1 genes of Egyptian isolates clustered with isolates from Israel and Saudi Arabia in group A sharing the same progenitor virus G1 closely related to Israel viruses (2005–07) (Fig. 2). Like PB2; the nucleotide and amino acid similarity among Egyptian viruses to each other, Israel viruses, Middle East viruses and G1virus shown (Table 1), All Egyptian viruses under study had Isoleucine at position 317, L13P, V14A, and A473V (Fig. 4). The PB1-F2 encoded a full-length protein of 90 AA and showing the substitutions N66S and L82S in all Egyptian LPAI-LPAI-H9N2 viruses, while the substitution T68I was not present, further mutations were described to foster replication in mammals and influence virulence was mentioned in (Table 2). Egyptian viruses differed from the G1 strain in the PB1protein by twenty-one mutations (Supplementary Table 3). Six substitutions recorded in some viruses under study (K176R, I181V, R350K, R386K, E398D and S515P) which first recorded in Egyptian LPAI-H9N2 viruses.
Fig. 2.
Phylogenetic tree of the PB1gene of avian influenza subtype LPAI-H9N2 viruses isolated in Egypt during 2014–2016 and reference isolates from Gen Bank. A black rhomboid indicates isolates sequenced specifically for this study Boldface indicates avian viruses. Group A is shown in red, group B is shown in green, group C is shown in blue and group D is shown in olive.
Table 2.
Virulence determinants in polymerase subunits of the Egyptian H9N2 viruses.
3.2.3. Polymeric acid
Phylogenetic analysis showed that the PA gene of Egyptian isolates belonged to the G1-lineage and clustered with isolates from Israel in group A (Fig. 3) the nucleotide and amino acid similarity of PA gene among Egyptian viruses to each other, Israel viruses, Middle East viruses and G1virus shown (Table 1). Analysis of the PA gene showed that all Egyptian isolates possessed previously recognized ribosomal frameshifting responsible for viral protein PA-X 252 amino acid. Amino acid substitutions V127, L672, L550 and S409N residues were observed in all Egyptian LPAI-LPAI-H9N2 viruses (Fig. 4). Additional substitutions that have been described to affect virulence in avian and/or mammalian hosts were found in PA encoding sequences are shown in (Table 2, Table 3); all Egyptian viruses under study show G186S. Egyptian LPAI-H9N2 viruses PA protein differed from the G1-like strain by 37 mutations (Supplementary Table 3).
Fig. 3.
Phylogenetic tree of the PA gene of avian influenza subtype LPAI-H9N2 viruses isolated in Egypt during 2014–2016 and reference isolates from Gen Bank. A black rhomboid indicates isolates sequenced specifically for this study Boldface indicates avian viruses. Group A is shown in red, group B is shown in green, group C is shown in blue and group D are shown in olive.
3.3. Effect of mutation on protein stability
The Free-energy analysis revealed that out of the eighteen selected substitutions which known to affect the ability of virus replication and/or the pathogenicity, six mutations (PB1-F2 L82, PB2-K318R, and A661 and PA-S225, I400S, S409N) have a structural stabilizing effect on polymerase complex protein. The free-energy change upon mutation (ΔΔG) showed Positive values (0.11, 0.58, 0.23, 0.14, 0.08, 0.39) while the other selected substitutions showed negative values (Table 4), Three out of those adaptive mutations shown to increase polymerase complex protein stability similar to Human H3N2 and H1N1 (661 in PB2, 225 and 409 in PA genes). Despite that, the free-energy analysis of the substitutions (PB2-318, T661, and PA-S409N) suggested a destabilizing effect; the Predicting stability with i-Mutant2.0 SEQ® suggesting stabilizing effect. The 3D structure of a protein is simulating the protein obtained from the SWISS-PDB viewer (Fig. 4).
Table 4.
Predicting stability changes of polymerase protein complex upon selected mutations in polymerase genes.
| Gene | Amino Acid Position | Vaccine strain for (H3N2-H1N1) according to WHO | Free-energy change (ΔΔG, in kcal/mol)a with I Stable | Predicting stability b with i-Mutant2.0 SEQ | Egyptian H9N2 Under Study | Free-energy change (ΔΔG, in kcal/mol)a with I Stable |
Predicting stabilityb with i-Mutant2.0 SEQ |
|---|---|---|---|---|---|---|---|
| PB1 | *317 | M | −1.04 | Decrease | I | −0.75 | Decrease |
| 13 | I/V | 0.59 | Decrease | P | −0.78 | Decrease | |
| PB1-F2 | *66 | N | −2.11 | Decrease | S | 0.52 | Increase |
| 82 | S | −1.34 | Decrease | (8)L | 0.11 | Increase | |
| PB2 | 318 | R | −0.07 | Decrease | (4)R | 0.58 | Increase |
| (4)K | −0.03 | Increase | |||||
| *391 | D | −0.09 | Decrease | E | 0.23 | Increase | |
| *504 | V | −1.80 | Decrease | V | −1.53 | Decrease | |
| 647 | I | −0.85 | Decrease | I | −0.78 | Decrease | |
| 661 | T | 0.05 | Increase | (7)A | −0.03 | Increase | |
| (1)T | −0.63 | Decrease | |||||
| PA | 57 | R | −0.53 | Decrease | R(6),K(2) | −1.08 | Decrease |
| *127 | V | 0.6 | Decrease | V | −0.6 | Decrease | |
| 225 | S | 0.17 | Increase | (6)S | 0.14 | Increase | |
| (1)G | −1.33 | Decrease | |||||
| *328 | K | 0.27 | Increase | R | −0.47 | Decrease | |
| 400 | S | −1.20 | Decrease | S | 0.08 | Increase | |
| 404 | A | −0.26 | Decrease | S | −0.33 | Decrease | |
| 409 | N | 0.64 | Increase | (1)N | −0.01 | Increase | |
| (7)S | 0.39 | Increase | |||||
| *550 | L | −1.52 | Decrease | L | −1.55 | Decrease | |
| *672 | L | −0.96 | Decrease | L | −1.52 | Decrease | |
Vaccine strain for (H3N2-H1N1) according to WHO; WHO recommends that trivalent vaccines for use in the 2017/18 northern hemisphere influenza season contain the following: an A/Michigan/45/2015 (H1N1)pdm09-like virus; an A/Hong Kong/4801/2014 (H3N2)-like virus; and a B/Brisbane/60/2008-like virus [38].
Abbreviations: ΔΔG, free-energy change upon mutation, polymerase subunits subunit.
Positive values indicate a stabilizing effect, while negative values indicate a destabilizing effect; polymerase subunits.
Calculated Free-energy change with I Stable server [39].
Predicting stability changes upon mutation from the protein sequence or structure with i-Mutant2.0 SEQ [40].
Refer to the residue related to virus virulence.
4. Discussion
Since 2011 The H9N2 viruses have a widespread in Egypt [18]. in recent times there were many human cases with H9N2 viruses were reported in Egypt [19]. In this study we attempted to correlate the mutations affect the avian H9N2 virus virulence and mammalian adaptation with their effect on protein stability, we isolated eight LPAI-LPAI-H9N2 viruses from apparent healthy chicken and quail flocks from five Egyptian governorates. Blast analysis of the nucleotide sequences from the polymerases subunits showed that the isolated LPAI-H9N2 viruses were closely related G1/ LPAI-LPAI-H9N2 strains. The virus shared the common ancestor A/QUAIL/HONG KONG/G1/1997isolate which has contributed the internal genes of the H5N1 virus circulating in Asia [25]. The phylogenetic trees of the polymerases subunits (PB2, PB1 and PA) revealed that the corresponding alleles of each viral segments of LPAI-H9N2 viruses were of Eurasian in origin (G1-like), suggesting that all the three polymerases subunits were closely related to Israeli isolates (2005–2010) especially A/turkey/Israel/311/2009 virus, with an average percent pairwise nucleotide and amino acid distances less than (<4%). Accordingly, they possess the same grouping of Israeli isolates based on the genome constellations of the LPAI-LPAI-H9N2 viruses described by [20]. They were related to group A the same like other LPAI-H9N2 strains circulated in the Middle East countries. That confirms the previous study on Egyptian LPAI-LPAI-H9N2 viruses that there is no obvious change or any evidence of reassortment recorded in chicken or quail viruses [32].
Understanding the genetic characteristics and mutation trend of the Egyptian LPAI-H9N2 virus polymerases subunits is important to recognize the behavior of H9N2. Identification of virulence markers of (PB2, PB1, and PA) genes of Egyptian LPAI-LPAI-H9N2 viruses under study revealed Seven Virulence markers, The substitutions (PB2-D391E and I504V), (PB1-M/V317I and N66S of PB1-F2), and (PA -127V, 550 L, and 672 L) which tend to increase virus virulence [13], [15], [30], [34], [43], [52]. As previously reported by in Egyptian LPAI-H9N2 Virus [33]. Substitution M/V317I, similar to the precursor strain Q/Hk/G1/97 and to the human virus A/Hong Kong/1073/99, furthermore it showed to be characteristic for LPAI-H9N2 and H5N1 of human origin [8]. The Egyptian LPAI-H9N2 viruses of this study .were encoded PB1-F2 and PA-X full-length proteins (90aa and 252aa respectively), which have the potential importance in virulence determinant of influenza virus [14]. It is well known that the full-length PA-X in human cells conferred 10- to 100-fold increase in viral replication and 5–8% increase in apoptosis [22].
Transmission of influenza viruses from aquatic birds to mammals is promoted by the viral proteins adaptation to the new host which includes the PB2 subunit of the viral polymerase complex. This protein has been described as an important host range factor [51]. In the present study, the majority of adaptive mutations found in the polymerases subunits identified within PB2. The naturally occurring substitutions (L89V, G309A, R477G and A676T) [36] were detected in Egyptian LPAI-H9N2 viruses. The substitutions (K318R, I504V, 647I, E627V, and A661 T) Also, were detected suggesting increased polymerase activity and virus replication concluding that the PB2 subunit of the LPAI-H9N2 viruses[71]. Two avian-like amino acids detected in few LPAI-H9N2 viruses under study at positions 318 and 661 lysine (K) and alanine (A) respectively located at the functional domain that is responsible for the polymerase subunits interaction [47], the substitution 647I detected in most of The Egyptian LPAI-H9N2 and other human and avian influenza virus strains. on the other hand, a single amino acid substitution is insufficient to retrieve the permissive tropism [70]. The PB2 protein displayed valine at position 627 (E627V) is typical of G1 viruses lineage, group A [20]. This mutation (E627V) suggested increasing H5N1 virus replication in mammalian cells and virulence in mice compared with PB2-627E [62].the three mutations (L13P, V14A, 317I, and 473V) in PB1 protein affect the virus replication in mammalian hosts [21], [38]. The substitutions L13P, 317I, and 473V suggested to enhancing polymerase activity of LPAI-H9N2 in mammalian hosts and in mice [43], [67]. it is recorded that the PB1-V14A substitution reduced polymerase activity, viral shedding, and transmissibility of H5N1 but does not always correlate with pathogenicity in chickens [61]. The mammalian-host-associated substitution PB1-F2 L82S was also identified in all of the viruses under study; whereas T68I was not detected in any of Egyptian viruses under study. The deduced PA amino acid sequence shows mammalian-host associated substitution at residues K328R it was recorded that K328R PA enhances the ability of a human H7N9 virus to replicate and cause severe disease in mice [72]. The substitution S409N determined in our study is known to be important for changing host range from avian to human [12].
The mutation G186S was found recently in PA of Egyptian viruses for the first time by [33], and also found in all viruses under this study. It is located in the N-terminal region of PA and it is found to be necessary to enhance avian virus polymerase activity in mammalian host cells and produced higher levels of viral protein which involved in mammalian adaptation and pathogenicity [7]. The substitution N383D in PA found in all viruses under study and may play an important role in the activity of the polymerase and in the accumulation of the PA and PB1 subunits in the nucleus of infected cells [59]. The substitutions K355M and P453T in PB2 became dominant in Egyptian viruses; also the substitutions 355I-PB2 and S516P-PB1 which was not previously described. The substitution S516P cited in The catalytic domain for endonuclease (508 to 522) [46]. Additional substitutions that have been described to affect virulence in avian and/or mammalian hosts were found in polymers genes encoding sequences [12], [52].
Stable folding is essential for a protein to perform its function, the stability results from cooperative interactions of the protein amino acids constituents [31], Not every substitution at a given site is predicted to be stabilizing [57]. the balance between stabilizing and destabilizing mutations will affect the protein function [24], [64]. There was study suggested that the pandemic influenza A virus of 1918 is closely related to the avian influenza virus than are other human viruses [50]. But, it is difficult to predict the number of the mutations which would make an avian influenza virus able to infect the humans efficiently.
The Free-energy analysis showed that (PB1-F2-L82, PB2-D391E, and A661 and PA-S225, I400S, S409N) have a structural stabilizing effect on polymerase complex protein, the free-energy change upon mutation (ΔΔG) showing Positive values indicate a stabilizing effect, while the others selected substitutions showing negative values to indicate a destabilizing effect on polymerase subunits [40], according to the result the residue A is consistently more stabilizing than T at site PB2-661, and S is consistently more stabilizing than G at site 225-PA; This “stability-function” predicts that it usually possible to replace residues known to be important for function, reducing protein activity but concomitantly increasing its stability [57]. The positions (PB1-F2-82, PB2-318 and PA 409) have stabilizing effect on protein whatever the residue found in those locations.
The S225, PB1-F2-82, PB2-318, and A661 those are affecting the virus replication in mammalian hosts besides the stabilizing effect on protein [34]. Pollock et al. Have used computer simulations to suggest that there are widespread evolutionary shifts in mutational effects, where for instance a mutation is destabilizing in one homolog but stabilizing in another homolog due to interactions with other covarying sites [49]. We agree with [3] whose find that mutational effects on stability are highly conserved, with relatively little interdependence. The above-mentioned mutations are found to be conserved among Egyptian LPAI-H9N2 viruses, so they can increase proteins stability through a generation of favorable intra- and inter monomer interactions, and it may reflect the continuous adaptation of the influenza virus. While other selected substitutions show destability of polymerase complex protein however that needs further studies to investigate that. We suggest that interactions that are lost with the mutation are compensated by interactions with other amino acids, leading to a more stable protein.
Evidently, the presence of mutations in several sites can lead to a rapid decrease in efficacy of protein stability if changes which affect the protein function [10]. Our results suggest that many mutations arising since 2011 have likely stabilized the structure of polymerase complex by introducing favorable interactions within and between monomers. Single point mutations seem to have little impact on protein stability [69], but recruitment of several mutations could have a great impact on the overall stability of the protein.
5. Conclusion
Genetic characterization of LPAI-H9N2/G1 viruses circulating in Egypt has remarkably important to grasp the key features for the emergence of LPAI-H9N2 among poultry and as a potential pandemic virus. Phylogenetically no significant changes versus the previous 2014–16 were observed. LPAI-H9N2 viruses are shown to be under the acquisition of mutations accumulation numbers of new substituting mutations which detected in polymerase subunits that correlated to virus virulence and mammalian adaptation. Some of these mutations described here can be considered to improve the protein stability.
Acknowledgements
Thanks to the National Laboratory for Quality Control on Poultry Production (NLQP), Animal Health Research Institute. Special thanks for entire gene analysis unit staff especially Drs. Amany Adel and Naglaa Hagag for their great help during the study.
Compliance with Ethics Guidelines
The authors declare that they have no conflict of interest. This article does not contain any studies with human or animal subjects performed by any of the authors.
Authors’ contributions
The study was designed by Abdelsatar Arafa, Hussein A. Hussein, Mohamed A. Shalaby; data collecting, analyzed and manuscript preparation by Zeinab Mosaad; data interpretation and manuscript reviewed by Abdelsatar Arafa, Hussein A. Hussein, and Mohamed A. Shalaby.
Footnotes
Peer review under responsibility of National Research Center, Egypt.
Supplementary data associated with this article can be found, in the online version, at https://doi.org/10.1016/j.jgeb.2018.02.008.
Appendix A. Supplementary material
Reference
- 1.Alexander D.J. Vaccine. 2007;25:5637–5644. doi: 10.1016/j.vaccine.2006.10.051. [DOI] [PubMed] [Google Scholar]
- 2.Alexander D.J. Sixth Edition. W.B. Saunders; Edinburgh: 2008. Chapter 26 - Orthomyxoviridae – avian influenza, Poultry Diseases; pp. 317–332. [Google Scholar]
- 3.Ashenberg O., Gong L.I., Bloom J.D. Proc Natl Acad Sci. 2013;110:21071–21072. doi: 10.1073/pnas.1314781111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bavagnoli L., Cucuzza S., Campanini G., Rovida F., Paolucci S., Baldanti F., Maga G. Nucleic Acids Res. 2015;43:9405–9417. doi: 10.1093/nar/gkv926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Biswas S.K., Nayak D.P. J Virol. 1994;68:1819–1826. doi: 10.1128/jvi.68.3.1819-1826.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burland T.G. Bioinformatics methods and protocols. 1999:71–91. [Google Scholar]
- 7.Bussey K.A., Desmet E.A., Mattiacio J.L., Hamilton A., Bradel-Tretheway B., Bussey H.E., Kim B., Dewhurst S., Takimoto T. J Virol. 2011;85:7020–7028. doi: 10.1128/JVI.00522-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cameron K., Gregory V., Banks J., Brown I., Alexander D., Hay A., Lin Y. Virology. 2000;278:36–41. doi: 10.1006/viro.2000.0585. [DOI] [PubMed] [Google Scholar]
- 9.Capriotti E., Fariselli P., Casadio R. Nucleic Acids Res. 2005;33:W306–W310. doi: 10.1093/nar/gki375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Castelán-Vega J.A., Magaña-Hernández A., Jiménez-Alberto A., Ribas-Aparicio R.M. Advances and applications in bioinformatics and chemistry: AABC. 2013;7:37–44. doi: 10.2147/AABC.S68934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen C.-W., Lin J., Chu Y.-W. BMC Bioinf. 2013;14:S5. doi: 10.1186/1471-2105-14-S2-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen G.-W., Chang S.-C., Mok C.-K., Lo Y.-L., Kung Y.-N., Huang J.-H., Shih Y.-H., Wang J.-Y., Chiang C., Chen C.-J. Emerg Infect Dis. 2006;12:1353–1360. doi: 10.3201/eid1209.060276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen H., Bright R.A., Subbarao K., Smith C., Cox N.J., Katz J.M., Matsuoka Y. Virus Res. 2007;128:159–163. doi: 10.1016/j.virusres.2007.04.017. [DOI] [PubMed] [Google Scholar]
- 14.Chen W., Calvo P.A., Malide D., Gibbs J., Schubert U., Bacik I., Basta S., O'Neill R., Schickli J., Palese P. Nat Med. 2001;7:1306–1312. doi: 10.1038/nm1201-1306. [DOI] [PubMed] [Google Scholar]
- 15.Conenello G.M., Tisoncik J.R., Rosenzweig E., Varga Z.T., Palese P., Katze M.G. J Virol. 2011;85:652–662. doi: 10.1128/JVI.01987-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Detjen B.M., St Angelo C., Katze M.G., Krug R.M. J Virol. 1987;61:16–22. doi: 10.1128/jvi.61.1.16-22.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dnastar I. Wisconsin; USA: 1999. Madison. [Google Scholar]
- 18.El-Zoghby E.F., Arafa A.-S., Hassan M.K., Aly M.M., Selim A., Kilany W.H., Selim U., Nasef S., Aggor M.G., Abdelwhab E. Arch Virol. 2012;157:1167–1172. doi: 10.1007/s00705-012-1269-z. [DOI] [PubMed] [Google Scholar]
- 19.F. EMPRES, 2015.
- 20.Fusaro A., Monne I., Salviato A., Valastro V., Schivo A., Amarin N.M., Gonzalez C., Ismail M.M., Al-Ankari A.-R., Al-Blowi M.H. J Virol. 2011;JVI 00219-00211. [Google Scholar]
- 21.Gabriel G., Dauber B., Wolff T., Planz O., Klenk H.-D., Stech J. Proc Natl Acad Sci USA. 2005;102:18590–18595. doi: 10.1073/pnas.0507415102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gao H., Sun H., Hu J., Qi L., Wang J., Xiong X., Wang Y., He Q., Lin Y., Kong W. J Gen Virol. 2015;96:2036–2049. doi: 10.1099/vir.0.000143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.García-Sastre A., Biron C.A. Science. 2006;312:879–882. doi: 10.1126/science.1125676. [DOI] [PubMed] [Google Scholar]
- 24.Gong L.I., Suchard M.A., Bloom J.D. Elife. 2013;2:e00631. doi: 10.7554/eLife.00631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guan Y, Shortridge KF, Krauss S, Webster RG. In: Proceedings of the National Academy of Sciences, 96 (1999) 9363–9367%@ 0027–8424. [DOI] [PMC free article] [PubMed]
- 26.Guo Y.J., Krauss S., Senne D.A., Mo I.P., Lo K.S., Xiong X.P., Norwood M., Shortridge K.F., Webster R.G., Guan Y. Virology. 2000;267:279–288. doi: 10.1006/viro.1999.0115. [DOI] [PubMed] [Google Scholar]
- 27.Hall T. North Carolina State University, Raleigh; 2004.
- 28.Honda A., Mizumoto K., Ishihama A. Proc Natl Acad Sci. 2002;99:13166–13171. doi: 10.1073/pnas.152456799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Honda A., Mukaigawa J., Yokoiyama A., Kato A., Ueda S., Nagata K., Krystal M., Nayak D.P., Ishihama A. J Biochem. 1990;107:624–628. doi: 10.1093/oxfordjournals.jbchem.a123097. [DOI] [PubMed] [Google Scholar]
- 30.Iqbal M., Yaqub T., Reddy K., McCauley J.W. PLoS One. 2009;4:e5788. doi: 10.1371/journal.pone.0005788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jaenicke R. Biochemistry. 1991;30:3147–3161. doi: 10.1021/bi00227a001. [DOI] [PubMed] [Google Scholar]
- 32.Kandeil A., El-Shesheny R., Maatouq A., Moatasim Y., Cai Z., McKenzie P., Webby R., Kayali G., Ali M.A. J Gen Virol. 2017;98:548–562. doi: 10.1099/jgv.0.000657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kandeil A., El-Shesheny R., Maatouq A.M., Moatasim Y., Shehata M.M., Bagato O., Rubrum A., Shanmuganatham K., Webby R.J., Ali M.A. Arch Virol. 2014;159:2861–2876. doi: 10.1007/s00705-014-2118-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Katz J.M., Lu X., Tumpey T.M., Smith C.B., Shaw M.W., Subbarao K. J Virol. 2000;74:10807–10810. doi: 10.1128/jvi.74.22.10807-10810.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lamb R.A., Krug R., Knipe D. Fields Virol. 2001;1 [Google Scholar]
- 36.Li J., Ishaq M., Prudence M., Xi X., Hu T., Liu Q., Guo D. Virus Res. 2009;144:123–129. doi: 10.1016/j.virusres.2009.04.008. [DOI] [PubMed] [Google Scholar]
- 37.Li Q.W., X, He J., Ning Z., Hu Y. PLoS Biol, One. 2012;6 [Google Scholar]
- 38.Lycett S., Ward M., Lewis F., Poon A., Pond S.K., Brown A.L. J Virol. 2009;83:9901–9910. doi: 10.1128/JVI.00608-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Meir R., Maharat O., Farnushi Y., Simanov L. J Virol Methods. 2010;163:190–194. doi: 10.1016/j.jviromet.2009.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miyazawa S., Jernigan R.L. Macromolecules. 1985;18:534–552. [Google Scholar]
- 41.Monne I., Hussein H.A., Fusaro A., Valastro V., Hamoud M.M., Khalefa R.A., Dardir S.N., Radwan M.I., Capua I., Cattoli G. Influenza Other Respir Viruses. 2013;7:240–243. doi: 10.1111/j.1750-2659.2012.00399.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Muramoto Y., Noda T., Kawakami E., Akkina R., Kawaoka Y. J Virol. 2013;87:2455–2462. doi: 10.1128/JVI.02656-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Naffakh N., Tomoiu A., Rameix-Welti M.-A., van der Werf S. Annu Rev Microbiol. 2008;62:403–424. doi: 10.1146/annurev.micro.62.081307.162746. [DOI] [PubMed] [Google Scholar]
- 44.Naguib MM, Arafa AS, El-Kady MF, Selim AA, Gunalan V, Maurer-Stroh S, et al. Infection, Genetics Evolut: J Mol Epidemiol. Evolut Genetics Infect Dis 2015;34:278–91. [DOI] [PubMed]
- 45.W.H. Organization, March 2017.
- 46.Palese P., Shaw M.L. Fields Virol. 2007;2:1647–1689. [Google Scholar]
- 47.Perales B., de la Luna S., Palacios I., Ortín J. J Virol. 1996;70:1678–1686. doi: 10.1128/jvi.70.3.1678-1686.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Perales B., Sanz-Ezquerro J.J., Gastaminza P., Ortega J., Santarén J.F., Ortín J., Nieto A. J Virol. 2000;74:1307–1312. doi: 10.1128/jvi.74.3.1307-1312.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pollock D.D., Thiltgen G., Goldstein R.A. Proc Natl Acad Sci. 2012;109:E1352–E1359. doi: 10.1073/pnas.1120084109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Reid A.H., Taubenberger J.K., Fanning T.G. Nat Rev Microbiol. 2004;2:909. doi: 10.1038/nrmicro1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rodriguez-Frandsen A., Alfonso R., Nieto A. Virus Res. 2015;209:23–38. doi: 10.1016/j.virusres.2015.03.017. [DOI] [PubMed] [Google Scholar]
- 52.Rolling T., Koerner I., Zimmermann P., Holz K., Haller O., Staeheli P., Kochs G. J Virol. 2009;83:6673–6680. doi: 10.1128/JVI.00212-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schrödinger L. There is no corresponding record for this reference; 2010.
- 54.Sediri H. 2015.
- 55.Setiawaty V., Dharmayanti N.L., Misriyah H.A., Pawestri M., Azhar G., Tallis L., Schoonman G. Samaan. Zoonoses and public health. 2015;62:381–387. doi: 10.1111/zph.12158. [DOI] [PubMed] [Google Scholar]
- 56.Shabat M.B., Meir R., Haddas R., Lapin E., Shkoda I., Raibstein I., Perk S., Davidson I. J Virol Methods. 2010;168:72–77. doi: 10.1016/j.jviromet.2010.04.019. [DOI] [PubMed] [Google Scholar]
- 57.Shoichet B.K., Baase W.A., Kuroki R., Matthews B.W. Proc Natl Acad Sci. 1995;92:452–456. doi: 10.1073/pnas.92.2.452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Slomka M., Pavlidis T., Banks J., Shell W., McNally A., Essen S., Brown I. Avian Dis. 2007;51:373–377. doi: 10.1637/7664-060906R1.1. [DOI] [PubMed] [Google Scholar]
- 59.Song J., Feng H., Xu J., Zhao D., Shi J., Li Y., Deng G., Jiang Y., Li X., Zhu P. J Virol. 2011;85:2180–2188. doi: 10.1128/JVI.01975-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Spackman E., Senne D.A., Myers T.J., Bulaga L.L., Garber L.P., Perdue M.L., Lohman K., Daum L.T., Suarez D.L. J Clin Microbiol. 2002;40:3256–3260. doi: 10.1128/JCM.40.9.3256-3260.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Suzuki Y., Uchida Y., Tanikawa T., Maeda N., Takemae N., Saito T. J Virol. 2014;JVI doi: 10.1128/JVI.01564-14. 01564-01514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Taft A.S., Ozawa M., Fitch A., Depasse J.V., Halfmann P.J., Hill-Batorski L., Hatta M., Friedrich T.C., Lopes T.J., Maher E.A., Ghedin E., Macken C.A., Neumann G., Kawaoka Y. Nat Commun. 2015;6:7491. doi: 10.1038/ncomms8491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Taverna D.M., Goldstein R.A. Proteins: Struct, Function, Bioinform. 2002;46:105–109. doi: 10.1002/prot.10016. [DOI] [PubMed] [Google Scholar]
- 65.von Itzstein M., Wu W.-Y., Kok G.B., Pegg M.S., Dyason J.C., Jin B., Van Phan T., Smythe M.L., White H.F., Oliver S.W. Nature. 1993;363:418–423. doi: 10.1038/363418a0. [DOI] [PubMed] [Google Scholar]
- 66.Wise M.G., Suarez D.L., Seal B.S., Pedersen J.C., Senne D.A., King D.J., Kapczynski D.R., Spackman E. J Clin Microbiol. 2004;42:329–338. doi: 10.1128/JCM.42.1.329-338.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Xu C., Hu W.-B., Xu K., He Y.-X., Wang T.-Y., Chen Z., Li T.-X., Liu J.-H., Buchy P., Sun B. J Gen Virol. 2012;93:531–540. doi: 10.1099/vir.0.036434-0. [DOI] [PubMed] [Google Scholar]
- 68.Xu K.M., Li K.S., Smith G.J., Li J.W., Tai H., Zhang J.X., Webster R.G., Peiris J.S., Chen H., Guan Y. J Virol. 2007;81:2635–2645. doi: 10.1128/JVI.02316-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yang H., Chang J.C., Guo Z., Carney P.J., Shore D.A., Donis R.O., Cox N.J., Villanueva J.M., Klimov A.I., Stevens J. J Virol. 2014;88:4828–4838. doi: 10.1128/JVI.02278-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yao Y., Mingay L.J., McCauley J.W., Barclay W.S. J Virol. 2001;75:5410–5415. doi: 10.1128/JVI.75.11.5410-5415.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhao F., Tian J., Lin T., Shao J., Zhang Y., Chen Y., Chang H. J Infect. 2016;73:95–97. doi: 10.1016/j.jinf.2016.02.004. [DOI] [PubMed] [Google Scholar]
- 72.Zhao Y., Yu Z., Liu L., Wang T., Sun W., Wang C., Xia Z., Gao Y., Zhou B., Qian J., Xia X. Vet Microbiol. 2016;187:8–14. doi: 10.1016/j.vetmic.2016.02.027. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.