Abstract
Immunotherapy represents one of the most promising therapeutic approaches in lung cancer, however 50% of lung cancer patients will not respond to this treatment, while others will have transitory or durable responses. Because side effects may be life threatening and treatment costs remain very high, the identification of predictive markers is mandatory and actually extensively studied. Factors that determine response to immune checkpoint inhibitors (ICI) are numerous including tumor microenvironment, immune tumor infiltrates, expression of immune checkpoint proteins (PD-1/PD-L1), gene expression signatures and molecular tumor profiles. Based on high impact factor publications and recent literature this review focuses on the potential predictive value of tumor molecular alterations and tumor mutation burden as predictive markers of response or resistance to ICI. We also discuss the role of circulating tumor DNA (ctDNA) to monitor ICI responses and propose an algorithm that integrates molecular markers upcoming recommendations for first line treatment.
Keywords: Lung cancer, immunotherapy, next generation sequencing (NGS), oncogene drivers, tumor mutation load (TML)
Introduction
Lung cancer remains the most common cause of cancer death worldwide with more than a million deaths per year. In non-smokers, carcinogenesis is often linked to the presence of somatic molecular alterations in specific oncogenic drivers. The use of selective inhibitors such as anti-EGFR, anti-ALK/ROS1 therapies can lead to tumor shrinkage and prolonged survival. In smokers and in patients without druggable target, immunotherapy is a very promising approach. It was first shown that survival was increased in patients receiving an immune checkpoint inhibitor (ICI) in monotherapy as compared to standard of care in second line treatment (1), in first line for subgroups of patients (2) and more recently clinical benefit was observed in patients receiving combination therapies in first or second lines (3). Indeed, combining ICIs with other anticancer strategies such as targeted therapy, chemotherapy, radiation therapy as well as combinations of different ICIs increases effectiveness. Regardless of treatment regimen, nearly half of lung cancer patients will not respond to ICIs and determinants are still being investigated to better select responders. Sensitivity to ICIs is multifactorial involving tumor genetics background, immune cell infiltrates and the level of immune-modulators such as PD-L1 or PD1 expression. The expression of ICI targets PD1-PD-L1 was investigated and related to increased response to ICIs although it was shown that patients with PD-L1 positive tumors could be non-responders whereas patients with PD-L1 negative tumors could have clinical benefits. Arguments linked to differences between antibodies used to test PD-L1 expression and to tumor heterogeneity were the most convincing. PD-L1 immunohistochemistry (IHC) despite its drawbacks remains the only validated marker use in clinical practice to select responders for first line pembrolizumab monotherapy (2).
For patients with lung cancer, tumor molecular testing is important, to allow individual treatment decisions and is part of patients’ care. Enlarged molecular testing is now feasible in routine thanks to the implementation, in clinical practice, of targeted next generation sequencing (NGS) cancer gene panels to investigate genetic tumor profiles. This easy access to somatic mutations in tumors raised the question of the predictive value of mutation types and mutation load on response to ICIs. The predictive value of oncogene drivers, as EGFR or ALK fusion and mutations such as TP53, STK11/LKB1, KRAS, was analyzed in ICI series of patients. It was demonstrated that immune cell infiltrates depend upon mutation types driving various response levels to ICI. Recently, tumor mutation load (TML), that is the number of non-synonymous somatic point mutations defined by whole-exome sequencing (WES) was linked to ICIs responses in all tumor types, any treatment lines and any type of treatment (combo or monotherapy) (4). However, exome sequencing is time consuming and not yet cost effective for TML evaluation, which hampers its use in clinical settings. For that reason, large targeted NGS panels have been evaluated as surrogate methods to identify potential drivers and to evaluate TML. The accuracy of targeted panels to evaluate TML is a current subject of research. Performance evaluation of different panels showed correct concordance with WES however the definition of high and low TML groups and the choice of a clinically relevant cut-off remain to be validated. We will discuss these different points in line with data from ICIs clinical trials.
As observed for targeted therapy, patients receiving ICIs will ultimately relapse. Studies are on going to identify molecular markers associated to secondary resistance and circulating tumor DNA (ctDNA) is also being evaluated as part of treatment monitoring.
This review focuses on the predictive value and the implementation of molecular markers in clinical practice to help select and monitor patients receiving immunotherapy.
Genetic determinants of response to ICI
One hypothesis is that tumor immune response is activated by antigenic peptides arising from tumor mutations that act as neoantigens, however not all mutations are efficiently turned into neoantigens. As an example, driver oncogenes often lack immunogenicity (5,6). Interaction between molecular profiles and response to ICIs can be analyzed at gene level. Are specific gene alterations related to response to ICIs? Or at a global level, how is TML linked to response to ICIs?
First trials identified smoking as a predictive marker of response to ICIs (1) and rapidly it was showed that non-smokers with EGFR mutated or ALK rearranged tumors do not do well with ICIs and should not receive first line ICIs even though tumor cells may express high PD-L1 levels [see Miura et al. (7) for review]. Indeed up-regulation of PD-L1 is not rare in EGFR mutated or ALK rearranged lung tumors and linked to oncogene induced up-regulation, activation of ERK or mTOR signaling (8,9). Most trials have excluded EGFR, ALK and ROS1 positive tumors and literature is scarce on the subject and relies on small subgroups of patients. In second line, the use of ICIs remains controversial. The CheckMate 057 and KEYNOTE-010 trials, which tested the benefit of nivolumab and pembrolizumab over docetaxel chemotherapy in second line did not show any differences between study arms among EGFR-mutant or ALK-fusion patients (1,10).
Patients with EGFR mutated tumors and no identified secondary resistance mechanism should be offered chemotherapy rather than an ICIs (11). It seems clear that for patients with oncogene drivers or druggable secondary resistance mechanisms, targeted therapy is the first choice. This lack of efficacy could be due to low levels of CD8+ tumor-infiltrating lymphocytes, and to non-inflamed microenvironment that limits the efficacy of ICIs (12). However recent results from the IMpower150 trial may change treatment option for patients with tyrosine kinase inhibitor (TKI) relapse. Indeed, this trial included EGFR and ALK positive patients that had progressed after TKI and showed that progression-free survival (PFS) benefit was observed with atezolizumab + chemotherapy + bevacizumab vs. chemotherapy + bevacizumab in this subgroup of patients, with a mean PFS of 9.7 versus 6.1 months, HR (95% CI) 0.59 (0.37–0.94) (13). Combo using TKIs and ICIs are on-going to evaluated clinical benefits of the association. Tables 1-3 summarized the main phase 2/3 clinical trials which evaluated pembrolizumab (Table 1) (2,3,14-16), atezolizumab (Table 2) (13,17-20) and nivolumab (Table 3) (1,10,21-23) in lung cancer.
Table 1. Summary of the main phase 2/3 clinical trials evaluating pembrolizumab in lung cancer.
| Study | Reference | Phase | N | Purposes | Results |
|---|---|---|---|---|---|
| KEYNOTE-021: NCT02039674 | Langer et al. 2016 | 2 | 123 | Carboplatin and pemetrexed with or without pembrolizumab | An objective response was achieved in 55% (33/60) of patients in the pembrolizumab plus chemotherapy group compared with 29% (18/63) in the chemotherapy alone group (P=0.0016) |
| NCT02879994 | Lisberg et al. 2018 | 2 | 11 | Pembrolizumab in EGFR-mutant, PD-L1 positive (>1%), TKI naïve patients | Lack of efficacy of pembrolizumab in TKI naïve, EGFR-mutant, PD-L1 patients even in case of PD-L1 expression >50% |
| KEYNOTE-010: NCT01905657 | Herbst et al. 2016 | 2/3 | 1,034 | Pembrolizumab vs. docetaxel for previously treated PD-L1 positive (>1%) NSCLC | OS was significantly longer for pembrolizumab vs. docetaxel (HR P=0.0008) |
| No significant difference was achieved in PFS between pembrolizumab and docetaxel groups | |||||
| In the subgroup of patients with PD-L1 positive tumours (expression >50%), OS and PFS were significantly longer with pembrolizumab than with docetaxel (P<0.001) | |||||
| NCT02142738 | Reck et al. 2016 | 3 | 305 | Pembrolizumab vs. chemotherapy for PD-L1 positive (>50%) previously untreated advanced NSCLC | Median PFS was 10.3 months with pembrolizumab vs. 6 months with chemotherapy (P<0.001) |
| Estimated OS at 6 months was 80.2% in the pembrolizumab group vs. 72.4% in the chemotherapy group (P=0.005) | |||||
| The RR was 44.8% with pembrolizumab vs. 27.8% with chemotherapy | |||||
| NCT02578680 | Gandhiet al. 2018 | 3 | 616 | Pembrolizumab plus chemotherapy in previously untreated advanced non-squamous NSCLC | Estimated OS rate at 12 months was 69.2% in the pembrolizumab plus chemotherapy group vs. 49.4% in the chemotherapy group (P<0.001) |
| An increased expression of PD-L1 significantly improved OS | |||||
| Median PFS was significantly longer in pembrolizumab plus chemotherapy group compared with chemotherapy group (8.8 vs. 4.9 months respectively, P<0.001) |
OS, overall survival; TKI, tyrosine kinase inhibitor; PFS, progression-free survival; NSCLC, non-small cell lung cancer; RR, response rate.
Table 2. Summary of the main phase 2/3 clinical trials evaluating atezolizumab in lung cancer.
| Study | Reference | Phase | N | Purposes | Results |
|---|---|---|---|---|---|
| BIRCH: NCT02031458 | Peters et al. 2017 | 2 | 659 | Atezolizumab as first line or subsequent therapy for PD-L1 positive (>5%) advanced NSCLC | ORR was 22%, 19% and 18% for first-, second- and third-line therapy respectively. ORR was increased in PD-L1 >50% subgroup |
| Median OS was 23.5, 15.5 and 13.2 months for the three groups respectively | |||||
| POPLAR: NCT01903993 | Fehrenbacher et al. 2016 | 2 | 144 | Atezolizumab vs. docetaxel for patients who progressed on post-platinum chemotherapy | OS was 12.6 months for atezolizumab vs. 9.7 months for docetaxel (P=0.04) |
| Increasing improvement in OS was associated with increasing PD-L1 expression | |||||
| OAK: NCT02008227 | Rittmeyer et al. 2017 | 3 | 1,225 | Atezolizumab vs. docetaxel in patients with previously treated NSCLC | OS was 13.8 months with atezolizumab vs. 9.6 months with docetaxel (P=0.0003) |
| OS in the PD-L1 positive tumors (>1%) was 15.7 months with atezolizumab vs. 10.3 months with docetaxel (P=0.0102) | |||||
| OS in the PD-L1 low (<1%) or negative tumors was 12.6 months with atezolizumab vs. 8.9 months with docetaxel | |||||
| NCT02366143 | Socinski et al. 2018 | 3 | 1,202 | Atezolizumab for first line treatment of metastatic non-squamous NSCLC | The addition of atezolizumab to BCP (bevacizumab + carboplatin + paclitaxel) for first line treatment improved PFS (8.3 vs. 6.8 months respectively, P<0.001) and OS (19.2 vs. 14.7 months respectively, P=0.02), regardless of PD-L1 expression and EGFR or ALK genetic alteration status |
| Pooled POPLAR, OAK: NCT01903993, NCT02008227 | Gandara et al. 2018 | 3 | 273, 797 | Atezolizumab vs. chemotherapy | In patients recruited in OAK with tumors showing a TML >16 mut/Mb (TML plasma test): PFS HR 0.65, 95% CI (0.47–0.92) favors atezolizumab |
| In patients recruited in OAK with tumors showing a TML<16 mut/Mb (TML plasma test): PFS HR 0.98, 95% CI (0.80–1.20) |
NSCLC, non-small cell lung cancer; OS, overall survival; PFS, progression-free survival; TML, tumor mutation load; HR, hazard ratio; ORR, objective response rate; mut, mutations.
Table 3. Phase 2/3 clinical trials evaluating nivolumab in lung cancer.
| Study | Reference | Phase | N | Purposes | Results |
|---|---|---|---|---|---|
| CheckMate 032: NCT01928394 | Antonia et al. 2016 | 1/2 | 216 | Nivolumab alone vs. nivolumab plus ipilimumab in small-cell lung carcinoma which had progressed after platinum-based chemotherapy | OR was achieved in 10% (10/98) of patients receiving nivolumab alone (3 mg/kg), 23% (14/61) of patients receiving nivolumab (1 mg/kg) and ipilimumab (3 mg/kg) and 19% (10/54) of patients receiving nivolumab (3 mg/kg) and ipilimumab (1 mg/kg) |
| CheckMate 057: NCT01673867 | Borghaei et al. 2015 | 3 | 582 | Nivolumab vs. docetaxel in patients with non-squamous NSCLC that had progressed after platinum-based doublet chemotherapy | OS was 12.2 months with nivolumab vs. 9.4 months with docetaxel (P=0.002) |
| OS rate at 12 months was 51% in the nivolumab group vs. 39% in the docetaxel group. OS at 18 months was 39% vs. 23% respectively | |||||
| RR was 19% with nivolumab vs. 12% with docetaxel (P=0.02) | |||||
| Median PFS was 2.3 months with nivolumab vs. 4.2 months with docetaxel | |||||
| CheckMate 017: NCT01642004 | Brahmer et al. 2015 | 3 | 272 | Nivolumab vs. docetaxel in patients with squamous NSCLC that had progressed after first-line chemotherapy | Median OS was 9.2 months with nivolumab vs. 6 months with docetaxel |
| OS rate at 12 months was 42% with nivolumab vs. 24% with docetaxel | |||||
| RR was 20% with nivolumab vs. 9% with docetaxel (P=0.008) | |||||
| Median PFS was 3.5 vs. 2.8 months respectively (P<0.001) | |||||
| All of these results were independent of PD-L1 expression level | |||||
| CheckMate 026: NCT02041553 | Carbone et al. 2017 | 3 | 541 | First-line nivolumab vs. chemotherapy in PD-L1 positive (>1%) NSCLC | Median PFS was 4.2 months with nivolumab vs. 5.9 months with chemotherapy (P=0.25) among patients with a PD-L1 tumor-expression >5% |
| Median OS was 14.4 vs. 13.2 months | |||||
| In patients with high TML, PFS was 9.7 vs. 5.3 months | |||||
| In patients with low-medium TML, PFS was 4.1 vs. 6.9 months | |||||
| CheckMate 227: NCT02477826 | Hellmann et al. 2018 | 3 | 75 | Multipart phase 3; different nivolumab based regimen vs. chemotherapy; nivolumab-ipilimumab vs. chemotherapy in patients with TML high | In all patients PFS was 4.9 months (95% CI, 4.1 to 5.6) with nivolumab plus ipilimumab and 5.5 months (95% CI, 4.6 to 5.6) with chemotherapy |
| In patients with tumors showing a TML>10 mut/Mb (FoundationOne): PFS was 7.2 months (95% CI, 5.5 to 13.2) vs. 5.5 months (95% CI, 4.4 to 5.8) irrespective of tumor PD-L1 expression level | |||||
| In patients with tumors showing TML >13 mut/Mb: no difference in PFS was shown between nivolumab monotherapy vs. chemotherapy |
NSCLC, non-small cell lung cancer; TML, tumor mutation load; OS, overall survival; PFS, progression-free survival; RR, response rate. OR, odds ratio.
In smokers, KRAS and TP53 are frequently mutated and their predictive value on response to ICIs was studied in different publications but results are inconsistent. It was suggested that KRAS-TP53 co-mutation could predict response to immunotherapy. Indeed KRAS/TP53 mutated samples demonstrated a favorable immune infiltrate and a higher mutation burden (24). However, in another publication TP53 mutations were modestly associated with increased response and were related to high TML (24) whereas KRAS had no predictive value. Finally, TP53 mutated, STK11 and EGFR wild type tumors were associated with a higher density of CD8 T-cell and PD-L1 expression. This subgroup was associated with a high TML and a longer PFS was reported in immunotherapy treated patients (25). At this point, the predictive value of KRAS and TP53 is not robust enough to be used in clinical practice to select patients for ICIs.
Due to the low frequency of BRAF-mutant non-small cell lung cancer (NSCLC), the immune response and immunological characteristics of tumors has not been extensively studied. A recent paper based on a retrospective analysis of 39 patients with BRAF mutated cancer that received ICIs showed that BRAF mutant NSCLC is associated with high level of PD-L1 expression and that ICIs have favorable activity with an objective response rate of 25% and 33% in BRAF V600E and BRAF non-V600E mutant respectively (26).
At the opposite, LKB1/STK11 mutations in association or not with KRAS were related to a lack of response to immunotherapy (27). This could be related to specific immune environment linked to LKB1/STK11 mutated tumors (28,29). Hellmann et al. showed that none of the tumors with STK11/LKB1 alterations in their series responded to treatment (23). They made similar observation with PTEN mutations although it did not reach significance. Consistent with these findings another group has compared patients with durable clinical benefit (DCB) versus no durable benefit (NDB) based on targeted NGS data (MSK-IMPACT). Authors showed that KRAS had no predictive value but that STK11 mutations were related to NDB. No other association was mentioned (30).
Altogether tumor cells molecular profiles can significantly impact the tumor immune microenvironment (i.e., T-cells, macrophages and neutrophils density) however large series are needed to validate the predictive value of individual gene mutation as markers of response to ICIs. Among frequently mutated genes, STK11/LKB1 has the strongest predictive value with consistent observations of a lack of response to ICIs.
Tumor mutational load (TML) as a predictive marker
Determination of TML
Tumor mutation load or tumor mutation burden (TML, TMB), is the number of non-synonymous somatic point mutations based on exome sequencing. It is expressed as a number of mutations per mega base (Mb). When TML is estimated by smaller sequencing panels using targeted NGS, the total number of alterations is reported to panel coverage. Therefore, results can be compared to WES.
WES sequencing is actually the goal standard; however, it remains expensive and time-consuming, which hampers its clinical practice applications. Different strategies have been tested and compared to WES to validate TML determination by targeted NGS panels. Results showed that methods were concordant when large comprehensive panel of over 1 to 1.5 Mb corresponding to more than 350 genes were used. However smaller panels were reported as surrogate methods, they are used in association with specific bioinformatics pipelines, algorithms to adjust mutation value per gene (31-33). If all studies agree on the positive predictive value of high TML, the validation of a clinically meaningful cut-off and the definition of “high” remains debated and differs between series or clinical trials (30,34). One difficulty is that TML is a continuous variable with an intermediate TML group. Cutoffs were first chosen to optimize specificity and sensitivity retrospectively in clinical trials. However, those used in prospective trials could be different from those used in retrospective trials. As examples, in Rizvi et al. (30) the cut-off was set at 178 mutations per exome (approximately 6/Mb), in the CheckMate 026 ancillary study to 243 mutations (7–8/Mb), in Chalmers et al. a study demonstrating that TML measurements from comprehensive NGS using the FoundationOne assay (Cambridge, MA, USA) was correlated to WES, TML high was defined as 20 mutations per/Mb (35).
Any TML quantification methods, WES or NGS panels can accurately identify low, intermediate and high TML samples however the definition of a precise threshold may not fit clinical practice. Indeed, mutation count could slightly differ due to the use of different methods, threshold may differ between situations first or second line, combination or monotherapy, differ between tumor types or subtypes and could be heterogeneous between primary and metastasis. The identification of a precise cut-off will be very difficult to validate or will be linked to a companion test which could ultimately limit the access to immunotherapy for patients with lung cancer. At the opposite any laboratory can validate a global TML classification as high/intermediate/low according to internal controls and to its own series of patients allowing large use of this maker to predict response to ICIs. Moreover, TML classification should be integrated to therapeutic options: monotherapy, combination therapy, first or second line and associated with other markers as PD-L1 expression, immune infiltrates to refine treatment selection. Indeed combination of markers may be more accurate as compared to TML only (24).
A few publications evaluated TML on cell free DNA. It was done mainly using comprehensive targeted panels. However due to the frequent low mutation allele frequencies (MAF) in ctDNA, it requires sequencing at high depth, the development of specific analysis pipelines and increases costs. However, for samples known to have high MAF in plasma, TML measurements should be possible using standard pipelines and may be an option. CtDNA is often described as a surrogate marker of tumor heterogeneity and could measure the sum of alterations in different tumor or metastatic sites. The predictive impact of TML measured in circulating DNA remains to be validated however it was recently evaluated as an ancillary study of the OAK trial using targeted NGS and 1.125 Mb of coding region corresponding to 394 genes. The authors showed that 59% of variants were shared between tumor and plasma DNA, that plasma and tissue TML were correlated and that the best cutoff to selected responders to atezolizumab was 16 mutations/Mb (35).
TML and response
In lung cancer, somatic mutation load was related to tobacco exposure and to a specific molecular smoking signature (36). Despite a great variability, lung cancers frequently exhibit high TML related to tobacco exposure. Rizvi et al. demonstrated on a small series of patients using WES that TML was predictive for treatment response. TML high tumors are predicted to have more neoantigens expressed. It was shown that neoantigen load correlated with TML, high PD-L1, CTLA4 and CD8+ infiltrates (33).
TML was explored in clinical trials and in different treatment combinations. In first line monotherapy, the ancillary study of CheckMate 026 explored its predictive value in a population of lung cancer patients with a PD-L1 expression of 5% or more. Main result from this phase 3 trial was that nivolumab was not associated with significantly longer PFS as compared to chemotherapy. However, TML was assessed in a sub group of patients using WES. PFS was longer in the subgroup of patients with high TML defined as >243 mutations per exome corresponding to about 8 mutations/Mb (median, 9.7 vs. 5.8 months; hazard ratio for disease progression or death, 0.62; 95% CI, 0.38 to 1.00). There was no difference of PFS between the low and medium TML groups. The absence of difference observed for overall survival (OS) was attributed to treatment crossover. No overlap was found between PD-L1 expression and TML however patients with both PD-L1 >50% and TML high experienced longer PFS (22). A retrospective study evaluating patients recruited in the MKS-IMPACT trial that received ICIs, tested the value of TML to predict clinical benefit defined as complete, partial or stable disease that lasted more than 6 months. In patients with clinical benefit TML was significantly higher, 8.5 vs. 6.6/Mb and odd ratios for clinical benefit increased with TML. The rate of clinical benefit and PFS were improved for patients with TML above the 50th percentile and even more for those in the top decile of TML (30). These results are consistent with the hypothesis that a high TML enhance tumor immunogenicity and increase clinical benefit of immunotherapy (36).
In patients receiving combination therapy (nivolumab and ipilimumab) in the CheckMate 012 trial, TML determined by WES was the strongest marker of response using the median (158 mutations/exome) as cut-off. However a continuous effect of TML on PFS was demonstrated (24). The impact of TML was also described with atezolizumab, a cut-off of 16/Mb discriminates patients with favorable outcome with atezolizumab as compared to chemotherapy (37). Similar observations were made in studies combining ICIs to chemotherapy. Subgroup analyses from CheckMate 227 reported that patients with high TML (>10 mutations per Mb) demonstrated clinical benefits with the nivolumab + ipilimumab or the nivolumab + chemotherapy associations versus chemotherapy alone. No difference was observed in the group of patients with low TML (38). Tables 1-3 reports main trials in NSCLC and potential associated biomarkers.
Based on different studies, high TML predicts response to ICIs used in monotherapy or in combinations, however some patients with low TML respond to treatment and some with high TML have short PFS.
TML is the surrogate marker of tumor neoantigen load (TNagL). Different algorithms taking into account various parameters including peptide binding to patients’ specific HLA isoforms have been developed to estimate neoantigen load. It was shown that TNagL is much lower than TML with only a few neoantigens present even when TML is high (39). High TML increases the chance that, at random, neoantigens are synthetized by tumor cells. Due to the importance of neoantigens in activating immune responses, TNagL is an attractive biomarker to better identify responders to ICIs.
Repair pathway defects and TML
Repair deficiencies are often associated to cancer and linked to high TML. It is especially true for mismatch repair (MMR) and replicative polymerases (POLE, POLD1) deficiencies. MMR deficiency is associated to lynch syndrome and occurs in sporadic cancers mainly colorectal, endometrial, ovarian and gastric tumors. MMR deficiency causes replication errors to accumulate, is linked to a microsatellite instable (MSI) phenotype and to high TML. The MSI phenotype is a predictive marker of response to ICIs in various cancer types and can easily be assessed by standard routine methods (40). Data from the AACR GENIE database shows that 2.1%, 1.09%, 1.65% and 1.87% of lung tumors have mutations in the MMR genes MSH2, MLH1, PMS2 and MSH6 respectively (41). However, the identification of an MSI phenotype is rare in lung cancer (<0.5%) and is often associated to other predictors (TML and PD-L1) so the identification of the MMR status in lung is not justified (42,43). Mutations in replicative polymerases have been described in patients treated by ICIs. In the Rizvi series, patients with POLE and POLD1 mutations had long term responses. The identification of POLE and POLD1 exonuclease domain mutation is accessible using targeted NGS in routine diagnostics. To conclude, the identification of repair pathway defects such as MMR deficiency which is rare considering lung cancer and mutation in DNA polymerases POLE and POLD1 are surrogate markers of TML (30).
Resistance to immunotherapy
Three different resistance mechanisms to immunotherapy were described: (I) the primary resistance is defined as an absence of response to immunotherapy; (II) the adaptive immune resistance mechanism, in which cancer cells are recognized by the immune system but escape to immune attack; and (III) the acquired resistance, in which a tumor initially responds to immunotherapy but relapses after a period of time. These mechanisms may be related to tumor-cell intrinsic or extrinsic factors such as the absence of antigenic proteins, the absence of antigen presentation, a genetic T-cell exclusion or an insensibility to T-cells… (44). Some of these factors could be sustained by different molecular alterations.
Molecular features of resistance to immunotherapy
Activation of Wnt/β-catenin pathway
Several oncogenic signaling pathways have been associated with resistance mechanisms to immunotherapy. For instance, an activation of the Wnt/β-catenin pathway was described as a mechanism of primary or acquired resistance to immunotherapy. Activation of this pathway through stabilization of β-catenin modulates formation of transcriptional complexes, leading to suppression of CCL4 transcription and secretion. CCL4 is a chemokine implicated in the recruitment of immune cells like NK cells and monocytes. CCL4 expression is associated with improved response to immunotherapy. The lack of chemokine secretion decreases immune cells recruitment, activation and may be responsible for immunotherapy resistance. In murine melanoma models, the lack of Wnt/β-catenin signaling is associated with a good response to immunotherapy whereas an activation results in resistance (45). Molecular alterations such as CTNNB1 mutations especially in exon 3 or copy number variation (i.e., deletions) are known to promote β-catenin stabilization and are found in lung cancer. In the same way, adenomatous polyposis coli (APC) mutations also decrease ubiquitin-proteasome degradation of β-catenin (46).
Constitutive expression of PD-L1 through Akt/mTOR pathway activation
The constitutive expression of PD-L1 by cancer cells leads to an active inhibition of immunotherapy response. In this context, different molecular alterations have been pointed out to trigger PD-L1 constitutive expression. For example, activation of the Akt-mTOR signaling pathway regulates PD-L1 expression in vitro and in vivo (47) and promotes immunotherapy resistance. Loss of PTEN through deletion or inactivating mutations is commonly found in different cancers including lung and is associated with an increased PD-L1 expression (48). In the same pathway, activating mutations of PIK3CA or AKT could also enhance Akt/mTOR signaling and tightly regulate PD-L1 expression in lung cancer (47). Moreover, PI3K-AKT pathway inhibitors were shown to improve the efficacy of immunotherapy in murine models (49).
Impairment of the interferon-gamma (IFN-γ) signaling pathway
The IFN-γ signaling pathway plays a key role of the immune response. IFN-γ is produced and released by T-cells and induces anti-tumor immune response. The binding of IFN-γ on cell surface IFN-γ receptors (IFNGR1 and 2) induces receptor dimerization allowing activation of the interferon receptor-associated JAK (Janus kinase 1 or 2). JAKs then activate STATs (signal transducer and activator of transcription proteins) which translocate to the nucleus to induce transcription of IFN-γ targeted genes (50). Both primary and acquired resistances to immunotherapy may be mediated through JAK/STAT pathway impairment (51). For instance, inactivating mutations of JAK1 p.Gln503* or JAK2 p.Phe547_splice c.1641+2T>G result in a lack of response to IFN-γ stimulation in melanoma (52). Inactivating mutations in genes of the IFN-γ signaling pathway protect tumor cells from IFN-γ mediated anti-tumor immunity (53).
Loss of MHC class I expression in tumor cells
Beta-2-microglobulin (β2m) is a component of MHC class I complex widely implicated in processing and antigen presentation to the cell surface. Downregulation of β2m through acquired mutation of the B2M gene leads to a lack of MHC class I expression in tumor cells and also provide resistance to immunotherapy in lung cancer mouse model (54).
CtDNA concentration as a predictive marker of immunotherapy response
CtDNA is now commonly used to manage TKI treatment in EGFR mutated NSCLC. In a cohort of nivolumab treated NSCLC patients, low ctDNA concentration at first evaluation (<2 ng/mL) is associated with clinical benefit and increased overall response rate. Moreover, a decrease of ctDNA concentration between diagnosis and first evaluation is also correlated with PFS, clinical benefit and tumor response (55). CtDNA concentration may also be used in association with medical imaging for the assessment of lung cancer immunotherapy response. A >50% decrease in mutant allele fraction from baseline seems to be correlated with radiological response to immunotherapy (56). Quantification of ctDNA in patients receiving ICIs is feasible in clinics thanks to the development of targeted NGS panel in routine diagnostics.
Discussion/conclusions
ICIs are widely used and evaluated in lung cancer. Immunotherapy is a therapeutic option for patients with metastatic lung cancer, in monotherapy or in combinations and is also extensively evaluated in patients with localized disease. However not all patients will benefit from these treatments, toxicities may be life threatening and treatment costs are important. It is therefore mandatory to refine patients’ selection and to personalize the use of ICIs.
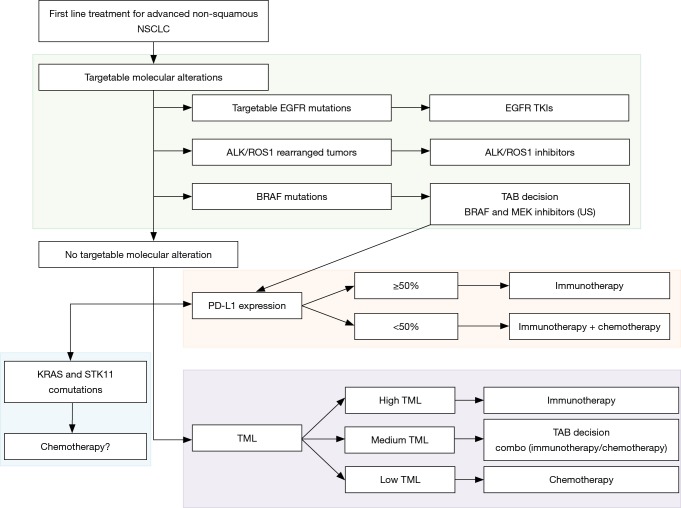
PD-L1 expression level is currently the only validated marker in first line for NSCLC. Immunotherapy is recommended in first line for patients with an EGFR or ALK wild type, PD-L1 >50% tumor. There is no validated recommendation for combination therapies or second line treatments although trials try to point out groups of patients with a higher clinical benefit. In this context, will molecular testing, mutation profiling and TML evaluation help refine patient selection? Based on the recent consensus statement on immunotherapy for the treatment of NSCLC published by the society for immunotherapy of cancer (57), Figure 1 summarized first-line treatment options and associated potential markers. Regardless of PD-L1 expression patients with targetable alterations are to receive targeted therapy as first line treatment. Last year, the FDA approved the association of dabrafenib and trametinib in first line treatment of BRAF mutated advanced NSCLC. However, for these patients, immunotherapy could be also considered as an option in tumors that expressed high levels of PD-L1. To our knowledge, no study compared immunotherapy vs. BRAF and MEK targeted therapy in patients with BRAF mutated tumors. A case-by-case TAB decision could be suitable in these cases. Activating KRAS mutations are the most commonly found oncogenic driver in tobacco induced NSCLC. Although studies are inconsistent concerning the predictive value of KRAS mutations, different studies underlined the value of STK11 mutations with or without KRAS alterations. Indeed, patients showed decreased ORR, shorter PFS, shorter OS and poor clinical outcomes regardless of PD-L1 or KRAS status or compared to KRAS-only (27). Therefore, chemotherapy may be considered as first line treatment for patients with STK11 mutated tumors.
Figure 1.
First-line treatment algorithm of advanced non-squamous NSCLC. NSCLC, non-small cell lung cancer; TML, tumor mutation load; TKI, tyrosine kinase inhibitor.
TML and PD-L1 expression are two independent predictors of response to immunotherapy. Their combined use will rely on the development of TML testing. Both may be important to support the decision of immunotherapy or combine therapies in patients without druggable drivers. One challenge will be to select the best treatment for patients with TML low/PD-L1 high or TML high/PD-L1 low cancers for which combination strategies might be more efficient than monotherapies.
The recent development of immunotherapy has greatly improved the management of patients diagnosed with lung cancer. Optimizing treatment and combination therapies remains a real challenge to detect primary or acquired resistance and to monitor treatment during follow-up. There are data supporting the role of molecular testing and mainly TML as a meaningful biomarker of response to immunotherapy. However, response to ICIs depends upon numerous parameters including tumor cells and immune infiltrates and it is likely that patient selection will not rely on a unique biomarker. It is time to develop standardized testing algorithms allowing comparison between drugs and efforts should be done to identify new biomarkers especially in long responder patients. Molecular screening and TML evaluation will be useful to detect molecular alterations involved in ICIs responses. However, to reach clinical value it may first be important to define the population of patients to be selected. Are we expecting tumor stabilization, transitory responses or long-time responses? Biomarkers will probably be different. To optimize immunotherapy and truly show impact on survival we have to learn how to combine biomarkers and identify the best combination. In daily practice, EGFR, ALK, ROS1 and PD-L1 can help, ICIs should not be proposed to patients with STK11 mutated tumors at least in first line as there are consistent results showing no response in this situation and TML needs to be implemented in clinics but interpretation guidelines are still to be validated. Real progress has been made in the management of lung cancer patients thanks to the development of ICIs, we have to progress in monitoring their use to maximize responses, to minimize side effects and rationalize costs.
Acknowledgements
None.
Footnotes
Conflicts of Interest: H Blons declares occasional lectures and boards for Roche, AstraZeneca, Boehringer Ingelheim, Pfizer, BMS, and MSD. Pierre Laurent-Puig declares participation lectures and boards for Roche, AstraZeneca, Boehringer Ingelheim, Pfizer, BMS, MSD, MDS Serrono, Amgen, and Biocartis. The other authors have no conflicts of interest to declare.
References
- 1.Borghaei H, Paz-Ares L, Horn L, et al. Nivolumab versus Docetaxel in Advanced Nonsquamous Non-Small-Cell Lung Cancer. N Engl J Med 2015;373:1627-39. 10.1056/NEJMoa1507643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reck M, Rodríguez-Abreu D, Robinson AG, et al. Pembrolizumab versus Chemotherapy for PD-L1–Positive Non-Small-Cell Lung Cancer. N Engl J Med 2016;375:1823-33. 10.1056/NEJMoa1606774 [DOI] [PubMed] [Google Scholar]
- 3.Gandhi L, Rodríguez-Abreu D, Gadgeel S, et al. Pembrolizumab plus Chemotherapy in Metastatic Non–Small-Cell Lung Cancer. N Engl J Med 2018;378:2078-92. 10.1056/NEJMoa1801005 [DOI] [PubMed] [Google Scholar]
- 4.Goodman AM, Kato S, Bazhenova L, et al. Tumor Mutational Burden as an Independent Predictor of Response to Immunotherapy in Diverse Cancers. Mol Cancer Ther 2017;16:2598-608. 10.1158/1535-7163.MCT-17-0386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dong ZY, Zhang JT, Liu SY, et al. EGFR mutation correlates with uninflamed phenotype and weak immunogenicity, causing impaired response to PD-1 blockade in non-small cell lung cancer. OncoImmunology 2017;6:e1356145. 10.1080/2162402X.2017.1356145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schumacher TN, Schreiber RD. Neoantigens in cancer immunotherapy. Science 2015;348:69-74. 10.1126/science.aaa4971 [DOI] [PubMed] [Google Scholar]
- 7.Miura Y, Sunaga N. Role of Immunotherapy for Oncogene-Driven Non-Small Cell Lung Cancer. Cancers 2018;10:245. 10.3390/cancers10080245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yoneshima Y, Ijichi K, Anai S, et al. PD-L1 expression in lung adenocarcinoma harboring EGFR mutations or ALK rearrangements. Lung Cancer 2018;118:36-40. 10.1016/j.lungcan.2018.01.024 [DOI] [PubMed] [Google Scholar]
- 9.Ota K, Azuma K, Kawahara A, et al. Induction of PD-L1 Expression by the EML4-ALK Oncoprotein and Downstream Signaling Pathways in Non-Small Cell Lung Cancer. Clin Cancer Res 2015;21:4014-21. 10.1158/1078-0432.CCR-15-0016 [DOI] [PubMed] [Google Scholar]
- 10.Brahmer J, Reckamp KL, Baas P, et al. Nivolumab versus Docetaxel in Advanced Squamous-Cell Non–Small-Cell Lung Cancer. N Engl J Med 2015;373:123-35. 10.1056/NEJMoa1504627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hanna N, Johnson D, Temin S, et al. Systemic Therapy for Stage IV Non–Small-Cell Lung Cancer: American Society of Clinical Oncology Clinical Practice Guideline Update. J Clin Oncol 2017;35:3484-515. 10.1200/JCO.2017.74.6065 [DOI] [PubMed] [Google Scholar]
- 12.Gainor JF, Shaw AT, Sequist LV, et al. EGFR Mutations and ALK Rearrangements Are Associated with Low Response Rates to PD-1 Pathway Blockade in Non-Small Cell Lung Cancer: A Retrospective Analysis. Clin Cancer Res 2016;22:4585-93. 10.1158/1078-0432.CCR-15-3101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Socinski MA, Jotte RM, Cappuzzo F, et al. Atezolizumab for First-Line Treatment of Metastatic Nonsquamous NSCLC. N Engl J Med 2018;378:2288-301. 10.1056/NEJMoa1716948 [DOI] [PubMed] [Google Scholar]
- 14.Langer CJ, Gadgeel SM, Borghaei H, et al. Carboplatin and pemetrexed with or without pembrolizumab for advanced, non-squamous non-small-cell lung cancer: a randomised, phase 2 cohort of the open-label KEYNOTE-021 study. Lancet Oncol 2016;17:1497-508. 10.1016/S1470-2045(16)30498-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lisberg A, Cummings A, Goldman JW, et al. A Phase II Study of Pembrolizumab in EGFR-Mutant, PD-L1+, Tyrosine Kinase Inhibitor Naïve Patients With Advanced NSCLC. J Thorac Oncol 2018;13:1138-45. 10.1016/j.jtho.2018.03.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Herbst RS, Baas P, Kim DW, et al. Pembrolizumab versus docetaxel for previously treated, PD-L1-positive, advanced non-small-cell lung cancer (KEYNOTE-010): a randomised controlled trial. Lancet 2016;387:1540-50. 10.1016/S0140-6736(15)01281-7 [DOI] [PubMed] [Google Scholar]
- 17.Peters S, Gettinger S, Johnson ML, et al. Phase II Trial of Atezolizumab As First-Line or Subsequent Therapy for Patients With Programmed Death-Ligand 1–Selected Advanced Non–Small-Cell Lung Cancer (BIRCH). J Clin Oncol 2017;35:2781-9. 10.1200/JCO.2016.71.9476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fehrenbacher L, Spira A, Ballinger M, et al. Atezolizumab versus docetaxel for patients with previously treated non-small-cell lung cancer (POPLAR): a multicentre, open-label, phase 2 randomised controlled trial. Lancet 2016;387:1837-46. 10.1016/S0140-6736(16)00587-0 [DOI] [PubMed] [Google Scholar]
- 19.Rittmeyer A, Barlesi F, Waterkamp D, et al. Atezolizumab versus docetaxel in patients with previously treated non-small-cell lung cancer (OAK): a phase 3, open-label, multicentre randomised controlled trial. Lancet 2017;389:255-65. 10.1016/S0140-6736(16)32517-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gandara DR, Paul SM, Kowanetz M, et al. Blood-based tumor mutational burden as a predictor of clinical benefit in non-small-cell lung cancer patients treated with atezolizumab. Nat Med 2018;24:1441-8. 10.1038/s41591-018-0134-3 [DOI] [PubMed] [Google Scholar]
- 21.Antonia SJ, López-Martin JA, Bendell J, et al. Nivolumab alone and nivolumab plus ipilimumab in recurrent small-cell lung cancer (CheckMate 032): a multicentre, open-label, phase 1/2 trial. Lancet Oncol 2016;17:883-95. 10.1016/S1470-2045(16)30098-5 [DOI] [PubMed] [Google Scholar]
- 22.Carbone DP, Reck M, Paz-Ares L, et al. First-Line Nivolumab in Stage IV or Recurrent Non–Small-Cell Lung Cancer. N Engl J Med 2017;376:2415-26. 10.1056/NEJMoa1613493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hellmann MD, Nathanson T, Rizvi H, et al. Genomic Features of Response to Combination Immunotherapy in Patients with Advanced Non-Small-Cell Lung Cancer. Cancer Cell 2018;33:843-52.e4. 10.1016/j.ccell.2018.03.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dong ZY, Zhong WZ, Zhang XC, et al. Potential Predictive Value of TP53 and KRAS Mutation Status for Response to PD-1 Blockade Immunotherapy in Lung Adenocarcinoma. Clin Cancer Res 2017;23:3012-24. 10.1158/1078-0432.CCR-16-2554 [DOI] [PubMed] [Google Scholar]
- 25.Biton J, Mansuet-Lupo A, Pécuchet N, et al. TP53, STK11 and EGFR Mutations Predict Tumor Immune Profile and the Response to anti-PD-1 in Lung Adenocarcinoma. Clin Cancer Res 2018;24:5710-23. 10.1158/1078-0432.CCR-18-0163 [DOI] [PubMed] [Google Scholar]
- 26.Dudnik E, Peled N, Nechushtan H, et al. BRAF Mutant Lung Cancer: Programmed Death Ligand 1 Expression, Tumor Mutational Burden, Microsatellite Instability Status, and Response to Immune Check-Point Inhibitors. J Thorac Oncol 2018;13:1128-37. 10.1016/j.jtho.2018.04.024 [DOI] [PubMed] [Google Scholar]
- 27.Skoulidis F, Goldberg ME, Greenawalt DM, et al. STK11/LKB1 Mutations and PD-1 Inhibitor Resistance in KRAS -Mutant Lung Adenocarcinoma. Cancer Discov 2018;8:822-35. 10.1158/2159-8290.CD-18-0099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koyama S, Akbay EA, Li YY, et al. STK11/LKB1 Deficiency Promotes Neutrophil Recruitment and Proinflammatory Cytokine Production to Suppress T-cell Activity in the Lung Tumor Microenvironment. Cancer Res 2016;76:999-1008. 10.1158/0008-5472.CAN-15-1439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mansuet-Lupo A, Alifano M, Pécuchet N, et al. Intratumoral Immune Cell Densities Are Associated with Lung Adenocarcinoma Gene Alterations. Am J Respir Crit Care Med 2016;194:1403-12. 10.1164/rccm.201510-2031OC [DOI] [PubMed] [Google Scholar]
- 30.Rizvi H, Sanchez-Vega F, La K, et al. Molecular Determinants of Response to Anti–Programmed Cell Death (PD)-1 and Anti–Programmed Death-Ligand 1 (PD-L1) Blockade in Patients With Non–Small-Cell Lung Cancer Profiled With Targeted Next-Generation Sequencing. J Clin Oncol 2018;36:633-41. 10.1200/JCO.2017.75.3384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roszik J, Haydu LE, Hess KR, et al. Novel algorithmic approach predicts tumor mutation load and correlates with immunotherapy clinical outcomes using a defined gene mutation set. BMC Med 2016;14:168. 10.1186/s12916-016-0705-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lyu GY, Yeh YH, Yeh YC, et al. Mutation load estimation model as a predictor of the response to cancer immunotherapy. NPJ Genom Med 2018;3:12. 10.1038/s41525-018-0051-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Campesato LF, Barroso-Sousa R, Jimenez L, et al. Comprehensive cancer-gene panels can be used to estimate mutational load and predict clinical benefit to PD-1 blockade in clinical practice. Oncotarget 2015;6:34221-7. 10.18632/oncotarget.5950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Steuer CE, Ramalingam SS. Tumor Mutation Burden: Leading Immunotherapy to the Era of Precision Medicine? J Clin Oncol 2018;36:631-2. 10.1200/JCO.2017.76.8770 [DOI] [PubMed] [Google Scholar]
- 35.Chalmers ZR, Connelly CF, Fabrizio D, et al. Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden. Genome Med 2017;9:34. 10.1186/s13073-017-0424-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rizvi NA, Hellmann MD, Snyder A, et al. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 2015;348:124-8. 10.1126/science.aaa1348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Legrand F, Gandara DR, Mariathasan S, et al. Association of high tissue TMB and atezolizumab efficacy across multiple tumor types. J Clin Oncol 2018;36:abstr 12000.
- 38.Borghaei H, Hellmann MD, Paz-Ares L, et al. Nivolumab (Nivo) + platinum-doublet chemotherapy (Chemo) vs chemo as first-line (1L) treatment (Tx) for advanced non-small cell lung cancer (NSCLC) with <1% tumor PD-L1 expression: Results from CheckMate 227. J Clin Oncol 2018;36:abstr 9001.
- 39.Brown SD, Warren RL, Gibb EA, et al. Neo-antigens predicted by tumor genome meta-analysis correlate with increased patient survival. Genome Res 2014;24:743-50. 10.1101/gr.165985.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Le DT, Durham JN, Smith KN, et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science 2017;357:409-13. 10.1126/science.aan6733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.AACR Project GENIE Consortium AACR Project GENIE: Powering Precision Medicine through an International Consortium. Cancer Discov 2017;7:818-31. 10.1158/2159-8290.CD-17-0151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vanderwalde A, Spetzler D, Xiao N, et al. Microsatellite instability status determined by next-generation sequencing and compared with PD-L1 and tumor mutational burden in 11,348 patients. Cancer Med 2018;7:746-56. 10.1002/cam4.1372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Warth A, Körner S, Penzel R, et al. Microsatellite instability in pulmonary adenocarcinomas: a comprehensive study of 480 cases. Virchows Arch 2016;468:313-9. 10.1007/s00428-015-1892-7 [DOI] [PubMed] [Google Scholar]
- 44.Sharma P, Hu-Lieskovan S, Wargo JA, et al. Primary, Adaptive, and Acquired Resistance to Cancer Immunotherapy. Cell 2017;168:707-23. 10.1016/j.cell.2017.01.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Spranger S, Bao R, Gajewski TF. Melanoma-intrinsic β-catenin signalling prevents anti-tumour immunity. Nature 2015;523:231-5. 10.1038/nature14404 [DOI] [PubMed] [Google Scholar]
- 46.Wang B, Tian T, Kalland KH, et al. Targeting Wnt/β-Catenin Signaling for Cancer Immunotherapy. Trends Pharmacol Sci 2018;39:648-58. 10.1016/j.tips.2018.03.008 [DOI] [PubMed] [Google Scholar]
- 47.Lastwika KJ, Wilson W, Li QK, et al. Control of PD-L1 Expression by Oncogenic Activation of the AKT–mTOR Pathway in Non–Small Cell Lung Cancer. Cancer Res 2016;76:227-38. 10.1158/0008-5472.CAN-14-3362 [DOI] [PubMed] [Google Scholar]
- 48.Song M, Chen D, Lu B, et al. PTEN Loss Increases PD-L1 Protein Expression and Affects the Correlation between PD-L1 Expression and Clinical Parameters in Colorectal Cancer. PLoS One 2013;8:e65821. 10.1371/journal.pone.0065821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Peng W, Chen JQ, Liu C, et al. Loss of PTEN Promotes Resistance to T Cell-Mediated Immunotherapy. Cancer Discov 2016;6:202-16. 10.1158/2159-8290.CD-15-0283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Marabelle A, Aspeslagh S, Postel-Vinay S, et al. JAK Mutations as Escape Mechanisms to Anti–PD-1 Therapy. Cancer Discov 2017;7:128-30. 10.1158/2159-8290.CD-16-1439 [DOI] [PubMed] [Google Scholar]
- 51.Shin DS, Zaretsky JM, Escuin-Ordinas H, et al. Primary Resistance to PD-1 Blockade Mediated by JAK1/2 Mutations. Cancer Discov 2017;7:188-201. 10.1158/2159-8290.CD-16-1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zaretsky JM, Garcia-Diaz A, Shin DS, et al. Mutations Associated with Acquired Resistance to PD-1 Blockade in Melanoma. N Engl J Med 2016;375:819-29. 10.1056/NEJMoa1604958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sucker A, Zhao F, Pieper N, et al. Acquired IFNγ resistance impairs anti-tumor immunity and gives rise to T-cell-resistant melanoma lesions. Nat Commun 2017;8:15440. 10.1038/ncomms15440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gettinger S, Choi J, Hastings K, et al. Impaired HLA Class I Antigen Processing and Presentation as a Mechanism of Acquired Resistance to Immune Checkpoint Inhibitors in Lung Cancer. Cancer Discov 2017;7:1420-35. 10.1158/2159-8290.CD-17-0593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Giroux Leprieur E, Herbretau G, Dumenil C, et al. Circulating tumor DNA evaluated by Next-Generation Sequencing is predictive of tumor response and prolonged clinical benefit with nivolumab in advanced non-small cell lung cancer. OncoImmunology 2018;7:e1424675. 10.1080/2162402X.2018.1424675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Goldberg SB, Narayan A, Kole AJ, et al. Early Assessment of Lung Cancer Immunotherapy Response via Circulating Tumor DNA. Clin Cancer Res 2018;24:1872-80. 10.1158/1078-0432.CCR-17-1341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Brahmer JR, Govindan R, Anders RA, et al. The Society for Immunotherapy of Cancer consensus statement on immunotherapy for the treatment of non-small cell lung cancer (NSCLC). J Immunother Cancer 2018;6:75. 10.1186/s40425-018-0382-2 [DOI] [PMC free article] [PubMed] [Google Scholar]