Abstract
Mouse embryonic stem cells (mESCs) are pluripotent and can differentiate into cells belonging to the three germ layers of the embryo. However, mESC pluripotency and genome stability can be compromised in prolonged in vitro culture conditions. Several factors control mESC pluripotency, including Wnt/β-catenin signaling pathway, which is essential for mESC differentiation and proliferation. Here we show that the activity of the Wnt/β-catenin signaling pathway safeguards normal DNA methylation of mESCs. The activity of the pathway is progressively silenced during passages in culture and this results into a loss of the DNA methylation at many imprinting control regions (ICRs), loss of recruitment of chromatin repressors, and activation of retrotransposons, resulting into impaired mESC differentiation. Accordingly, sustained Wnt/β-catenin signaling maintains normal ICR methylation and mESC homeostasis and is a key regulator of genome stability.
Introduction
The evolutionarily conserved Wnt/β-catenin signaling pathway controls many cellular and developmental processes, including cell proliferation, cell fate determination and tissue homeostasis1. Mutations affecting the Wnt/β-catenin pathway often lead to disease, cancer progression and developmental defects.
The canonical Wnt/β-catenin-dependent pathway integrates membrane, cytoplasmic and nuclear components, such as Wnt ligands, Frizzled (FZD) receptors and co-receptors, AXIN/glycogen synthase kinase 3 (GKS3)/Adenomatosis polyposis coli (APC)/Casein Kinase I (CKI) destruction complex, β-catenin protein and several transcription factors1,2. In the absence of Wnt ligands, cytoplasmic β-catenin is constantly degraded by the action of the AXIN/GSK3/APC/CKI destruction complex. On the contrary, the destruction complex is disassembled when Wnt ligands bind to the FZD receptors. As a consequence, β-catenin translocates to the nucleus where it associates with TCF/LEF (T-cell factor/lymphoid enhancing factor) nuclear complex and activates Wnt targeted gene expression3.
During embryogenesis Wnt/β-catenin signaling plays a fundamental role in the establishment of both dorso-ventral and anterior-posterior axis and its role is essential for normal gastrulation. Indeed, β-catenin knockout embryos are lethal since they fail to develop the mesodermal and endodermal germ layers4,5. Accordingly, Wnt/β-catenin represents a key pathway for mouse embryonic stem cell (mESC) identity and homeostasis.
Mouse ESCs, derived from the inner cell mass (ICM) of the blastocyst6,7 are pluripotent stem cells, which are able to generate the three germ layers and can be expanded in vitro indefinitely. Their long-term self-renewal ability has been attributed to the protein regulatory network that includes several pluripotency factors, such as Nanog, Oct4 and Rex1, among others8–11. In this context, the role of β-catenin during mESC differentiation has been shown to be indispensable, since β-catenin null mESCs undergo apoptosis at the onset of the differentiation process12–14. However β-catenin function in ESC self-renewal has been largely debated14–20. The dual role of β-catenin can be attributed to its capacity to form complexes with many downstream factors, including key pluripotency genes, such as Oct421,22.
In parallel to the core pluripotency factors, several epigenetic mechanisms control mESC identity through chromatin remodeling processes23. In particular, reversible changes on DNA methylation, followed by histone modifications, control both pluripotency and differentiation genes in mESCs, recapitulating the epigenetic profile of the pre-implantation embryo stage24,25. While developmental genes are characterized by flexible and reversible epigenetic regulation mechanisms to allow their dynamic expression, stable DNA methylation ensures silencing and protection of retrotransposons from moving around in the genome and causing potential mutations26. The same applies to imprinted genes, which reside in clusters27 and are regulated from one major cis-acting element called the imprinting control region, ICR. In mammals, DNA methylation is maintained stable and it can be propagated through cell division28–30 by a mechanism of DNA methylation maintenance coupled to DNA replication, which involves the action of different enzymes including DNA methyltransferase I (DNMT1). Along with DNA methylation, other epigenetic factors, such as ZFP57, KAP1, DNMT1, H3K9me3 and others, are involved in marking the ICRs and in protecting the methylated DNA. Indeed, loss of ZFP57, KAP1, DNMT1 or other repressors, leads to loss of imprinting and genomic instability in mESCs, and, thereby, to embryonic lethality27,31–34.
Epigenetic instability in imprinted genes and transposons has been observed in several mESC lines and can be attributed to culture conditions, sex of the cells, isolation procedures35 or prolonged in vitro culture of mESCs36–39. In particular, mESCs with global loss of methylation at the ICRs have been shown to contribute to chimeras, but mice developed several types of tumors by one year of age40. The mechanisms causing genomic aberrations and destabilization are still debated. However, downregulation of several epigenetic factors, such as DNMT1, KAP1, G9a, has been correlated with the epigenetic instability of the cells34,41–46.
Mouse embryonic stem cells represent an essential model to study in vitro the mechanisms that regulate embryo development. Therefore, it is important to fully understand the mechanisms that control cell identity, genomic stability and cell homeostasis. Wnt/β-catenin signaling has been investigated to be crucial for gene transcriptional regulation of mESCs, including pluripotency genes. Though, the connection between Wnt signaling and the epigenetic regulatory mechanisms has not been elucidated up to now. Here we investigated a novel role of Wnt/β-catenin signaling as a key player involved in epigenetic changes that preserve mESC identity and genome stability. We found that mESCs cultured in vitro for prolonged time showed loss of Wnt activity and downregulation of β-catenin protein, which correlated with a general loss of DNA methylation, affecting the ICRs, and leading to impaired mESC differentiation. On the contrary, sustained levels of Wnt/β-catenin ensure ICR methylation maintenance over time, suggesting a possible role for this signaling pathway in the protection of silent genomic regions and, therefore, in the maintenance of the genomic stability.
Results
Wnt/β-catenin activity is downregulated in mESCs after prolonged in vitro culture
The functional role of the Wnt/β-catenin pathway has been widely investigated in pluripotent stem cells. While the activation of Wnt pathway is indispensable for mouse embryonic stem cell (mESC) differentiation, its role in self-renewal and cell identity maintenance has been largely debated. Thus, we decided to analyze the activity of the Wnt/β-catenin pathway in mESCs cultured for a prolonged time, in particular its influence on pluripotency and homeostasis, including cell proliferation, differentiation potential and epigenetic stability.
To this aim we cultured E14 mESCs for several passages in the Serum + LIF medium. We observed that E14 mESCs cultured for many passages, around seventy, (old passage mESCs, henceforth called OP-mESCs), showed homogeneous morphology, characterized prevalently by flat clones, when compared to the same mESCs that were kept in culture for only fourteen passages (young passage mESCs, henceforth called YP-mESCs) (Fig. 1a,b). Similar results were obtained with GS1 mESCs, derived from a different strain, that were grown in prolonged culture conditions (around fifty passages, OP-mESCs) (Fig. S1a,b). In contrast, both E14 and GS1 YP-mESC cultures displayed heterogeneous morphology, including both round shaped and flat morphology clones (Figs 1b and S1b, upper panels). Pluripotent cell heterogeneity is due to fluctuation of pluripotency marker expression within the cell population. Even though cell heterogeneity can be found in almost all pluripotent stem cells, including induced pluripotent stem cells, the mechanisms causing gene expression variability and changes in morphology are still under investigation47–53.
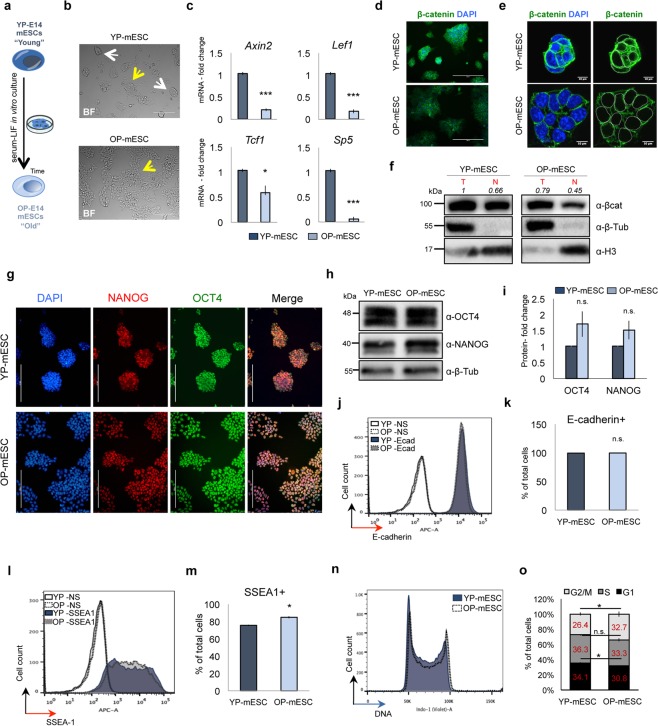
Figure 1.
Prolonged in vitro cell culture of E14 mouse embryonic stem cells (mESCs) correlates with low Wnt/β-catenin activity. (a) Schematic representation of Young (YP) and Old passage (OP) E14 mESCs. (b) Representative bright field images of YP- and OP-mESCs. Round-shaped and flat colonies are indicated by white and yellow arrow, respectively. (c) Quantitative real-time PCR showing the expression profiles of Axin2, Lef1, Tcf1, Sp5 in YP- and OP- mESCs. The transcriptional levels are normalized to Gapdh as reference gene. Data are represented as fold change (2−ΔΔCt) relative to the YP-E14 mESCs and means of n = 3 independent experiments ± SE. (d,e) Representative immunofluorescence (d) and confocal microphotographs (e) of β-catenin. Nuclear demarcation is indicated by white circles (right panel). (f) Western blot analysis showing total and nuclear β-catenin protein in YP- and OP-mESCs and its quantification (n = 1) relative to total β-catenin in YP-mESCs. For quantification, densitometric analysis was performed with ImageJ software. The quantification reflects the relative amounts as a ratio of each protein band relative to their loading control. (g) Representative immunofluorescence images showing OCT4 (green), NANOG (red) and their merge in YP- and OP-E14 mESCs. (h,i) Representative western blot analysis of OCT4 and NANOG in YP- and OP-mESCs (h) and its quantification represented as fold change over the protein amount in YP-mESCs and means of n = 3 independent experiments ± SE (i). (f,h,i) Full scan blots are available in Supplementary Fig. 6. (j–m) FACS-plot showing the percentage of E-cadherin +(j) and SSEA1 +cells (l) in YP- and OP- mESCs and its quantification (k,m) as means of 3 technical replicates ± SE (NS: non stained). (n,o) Representative cell cycle FACS profile analyzed with Flowjo software (n) and its quantification (o) represented as percentage of total cells and means of n = 3 independent experiments ± SE. Scale bar is 200 (b,d,g) and 10 μm (e). (d,e left panel, and g) Nuclei were stained with DAPI. (f,h) β-tubulin and H3 were used as loading controls. (c,i,k,m,o) Asterisks indicate statistical significance calculated by unpaired two-tailed t test analysis (n.s. not significant; *p-value < 0.05; ***p-value < 0.001).
Interestingly, both E14 and GS1 OP-mESCs showed low Wnt activity, since the Wnt targets Axin2, Lef1, Tcf1 and Sp5 were significantly downregulated, when compared to YP-mESCs (Figs 1c and S1c). In addition, total β-catenin protein was downregulated in E14 and GS1 OP-mESCs, as indicated by the microscope fluorescence intensity (Figs 1d–e and S1d,e). Additionally, by western blot analysis we observed lower amount of both total and nuclear β-catenin protein in OP-mESCs when compared to YP-mESCs (Figs 1f and S1f), suggesting again a reduction in the canonical Wnt/β-catenin signaling activity in OP-mESCs.
The difference in morphology of OP-mESCs did not correspond to significantly altered pluripotency gene expression. In particular, we compared the expression of NANOG and OCT4 protein levels among OP-mESCs and YP-mESCs for both E14 and GS1 strains (Figs 1g–i and S1g,i). We did not find any relevant difference in the expression pattern (Figs 1g and S1g) or significant changes in the protein level of NANOG and OCT4 (Figs 1h,i and S1h,i) among YP- and OP-mESCs. Thus, YP- and OP-mESCs expressed comparable levels of NANOG and OCT4.
Moreover, we performed FACS analysis to detect protein expression of the pluripotency cell membrane markers E-cadherin and SSEA154–59. The expression of E-cadherin was similar between YP- and OP-mESCs in both cell lines (Figs 1j,k and S1j,k). The percentage of cells expressing SSEA1 was higher in OP-mESC E14 (Fig. 1l,m), but did not change in GS1 mESCs (Fig. S1l,m). These data further confirmed that the pluripotency genes were not downregulated after prolonged culturing or even they were slightly upregulated in OP cells, as in the case of E14 mESCs.
Since prolonged in vitro culturing39,60 and Wnt/β-catenin activity61 can affect cell proliferation, we compared cell cycle progression in both YP- and OP-mESCs. Both E14 and GS1 OP-mESCs displayed a significantly lower percentage of cells in G1 phase in comparison with the YP-mESCs (Figs 1n,o and S1n,o). Moreover, E14 OP-mESCs also showed a significant increase in the number of cells in G2/M phase, with respect to the YP-mESCs that on the contrary, displayed a higher number of cells in G1 and S phases (Fig. 1n,o). Overall, these data suggest that OP-mESCs are characterized by higher proliferation rate along with Wnt signaling down-regulation.
Wnt signaling and β-catenin protein are essential for differentiation and cell fate determination12–14,20,62. Thus, we induced embryoid body (EB) formation from both E14 and GS1 YP- and OP-mESCs (Figs 2a and S2a) to evaluate their differentiation capacity. EBs can recapitulate in vitro many aspects of cell differentiation that occur during early embryogenesis63. Interestingly, even though all the mESCs that we used could form normal aggregates in suspension (Figs 2b and S2b, left panels) only YP-mESCs were able to generate beating EBs (Movies S1, S2) and to form large three-dimensional multicellular structures (Figs 2b and S2b, right and upper panel). In contrast, OP-mESCs gave rise to small structures, and we did not observe any beating EBs (Movies S3, S4) up to day 9 (Figs 2b and S2b, right and lower panels). We further characterized the EBs by analyzing transcriptional levels of genes corresponding to the three germ layers at day 6 (D6) and 12 (D12) of differentiation. For this we used undifferentiated YP- and OP-mESCs as controls (ESC). In particular, EBs derived from E14 or GS1 OP-mESCs showed lower level of the mesodermal marker Nkx2.5 already at day 6 with respect to the YP-mESCs, while the endoderm marker Gata6 was significantly downregulated only at day 12 of EB differentiation (Figs 2c and S2c). These results were consistent with already published studies, which reported that Wnt/β-catenin pathway activity is essential for mesoendoderm specification12,62. On the other hand, the ectodermal marker Otx2 was significantly upregulated in OP cells already at the mESC stage and it further increased with differentiation, being expressed at much higher level in OP-EBs with the respect to YP-EBs. This result suggests that the differentiation could be biased toward ectoderm (Figs 2c and S2c), which was confirmed when we induced neural differentiation64,65 of both YP- and OP-mESCs (Figs 2d and S2d). Indeed, OP-mESCs expressed higher levels of Sox1, when compared to YP-mESCs and this was also the case at day 3 of neural differentiation (Figs 2e and S2e). We confirmed these results by analyzing the expression of other neural markers at day 8 (D8) of differentiation, such as Nestin and III β –tubulin (TUJ1) and Pax6 (Figs 2f–g and S2f,g). OP-mESCs expressed higher level of Nestin and III β –tubulin (TUJ1) protein at day 8 (D8) of neural differentiation, with respect to YP-mESCs, in both cell lines (Figs 2f and S2f). Moreover, both Nestin and III β –tubulin (TUJ1) positive cells obtained from OP-mESCs were characterized by a more branched and elongated morphology, suggesting a faster neural differentiation (Figs 2f and S2f). Pax6 was upregulated in OP-mESC, in both E14 and GS1 (Figs 2g and S2g, left plot) and although Fgf5 was upregulated in E14 OP- neural precursors, it showed lower levels in differentiated GS1 OP-mESCs compared to their YP counterpart (Figs 2g and S2g, right plot). Surprisingly, although similar levels of pluripotency genes were expressed in both YP- and OP-mESCs, during differentiation the levels of Rex1, Oct4 and Nanog, remained much higher in OP-EBs with respect to the YP-EBs (Figs 2c and S2c, lower plots). These data suggest that the differentiation potential of mESCs, in particular toward the meso-endodermal germ layers, is strongly impaired in prolonged culture condition, and correlates with high level of pluripotency genes in both OP-mESCs and OP-EBs.
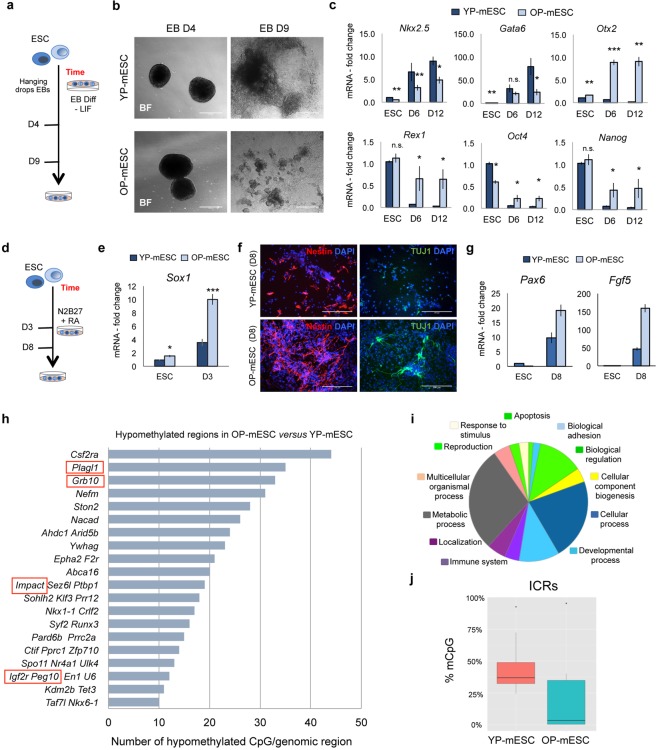
Figure 2.
Old passage E14 mESCs show differentiation defects and loss of methylation at ICRs. (a) Schematic representation of embryoid body (EB) differentiation protocol of YP- and OP- mESCs. (b) Representative bright field images showing EBs at day 4 (D4) and 9 (D9) obtained from both YP- and OP- E14 mESCs. Scale bar is 400 μm. (c) Quantitative real-time PCR showing the expression profiles of differentiation genes (Nkx2.5, Gata6, Otx2) and pluripotency genes (Rex1, Oct4, Nanog) in YP- and OP- E14 mESCs (ESC) and during EB differentiation at day 6 (D6) and day 12 (D12). (d) Schematic representation of neural differentiation protocol of YP- and OP- mESCs. (e) Quantitative real-time PCR showing the expression profiles of Sox1 at day 3 (D3) of N2B27 + retinoic acid (RA) treatment in YP- and OP-E14 mESCs (ESC). (f) Representative immunofluorescence images showing Nestin (left panels) and III β-tubulin (TUJ1, right panels) protein expression in YP- and OP- mESCs at day 8 (D8) of neural differentiation. (g) Quantitative real-time PCR experiment showing the expression profiles of Pax6 and Fgf5 at day 8 (D8) of N2B27 + retinoic acid (RA) treatment in YP- and OP- E14 mESCs (ESC). (c,e,g) The transcriptional levels are normalized to Gapdh as a reference gene. Data are represented as fold change (2−ΔΔCt) relative to the YP-E14 mESCs and the results are means of n = 3 independent experiments ± SE (c,e) and means of n = 3 technical replicated for SD (g). (c,e) Asterisks indicate statistical significance calculated by unpaired two-tailed t test analysis (n.s. not significant; *p < 0.05; **p < 0.01; ***p-value < 0.001). (h) Number of hypomethylated common CpGs in OP- versus YP- E14 mESCs analyzed by RRBS covered by at least 10 reads and showed at least 25% of methylation reduction. Red rectangle indicates imprinted regions. (i) Gene ontology of hypomethylated regions in OP-E14 mESCs analyzed by PANTHER (www.pantherdb.org). (j) Box-plot, from min–max values, showing the distribution of mCpG levels at ICRs in YP- and OP-E14 mESCs determined by RRBS analysis. The plots indicate the first quartile, median (black line) and third quartile. Data are obtained from the average of n = 2 biological replicates.
Reduced activity of Wnt/β-catenin signaling in OP-mESCs correlates with loss of DNA methylation, affecting also ICRs
Several epigenetic alterations causing DNA methylation changes have been associated with prolonged in vitro cell culture36. In pluripotent stem cells, DNA methylation regulates many cellular processes including cell differentiation. Altered DNA methylation has often been associated with impaired differentiation capacity of mESCs44. In this context, Wnt/β-catenin pathway regulates pluripotent stem cell differentiation, however its involvement in DNA methylation has not been explored. We, therefore, wondered whether OP-mESCs, which express low level of β-catenin protein and Wnt target transcripts, could show any change in DNA methylation levels. Thus, we performed Reduced Representation Bisulfite Sequencing (RRBS), a technique that combines restriction enzymes and bisulfite sequencing to enrich for the genomic areas with high CpG content66, using both YP- and OP-mESCs. Only 3 genomic regions, localized close to Repin1, Nkx2-1 and Gm11846 genes, were hypermethylated in E14 OP-mESCs after prolonged culture, when compared to the YP-mESCs (Table S1). In contrast, we found a general hypomethylation in E14 OP-mESCs. Indeed, about 100 genomic CpG-rich regions displayed reduced DNA methylation in OP-mESCs with respect to YP-mESCs (Fig. 2h and Table S1). The coding genes nearby these hypomethylated genomic regions belong to different gene families and they control several biological processes, including metabolic and developmental processes, as analyzed by PANTHER functional classification (Fig. 2i). Among the list of hypomethylated regions we found several imprinted genes. Imprinted genes have been previously described to be crucial for metabolic and developmental process regulation67,68. In particular, the ICRs corresponding to Plagl1, Grb10, Impact, Igf2r and Peg10 loci showed reduced DNA methylation in many CpGs in OP-mESCs but not in YP-mESCs (Fig. 2h). ICRs control many elements within the imprinted clusters and they have been shown to be generally stable in the pre-implantation embryo, and to be methylated in only one allele27,32. By analyzing only ICRs, we observed that YP-mESCs showed a normal profile of DNA methylation (around 50%, corresponding to one specific allele), while methylation was lost in several ICRs in OP-mESCs (Fig. 2j). Our results indicate that impairment of the ICR status, such as loss of DNA methylation, can affect pluripotency and differentiation potential of mESCs40.
To validate the RRBS data and to examine the methylation profile of additional ICRs, we performed Combined Bisulfite Restriction Analysis (COBRA) coupled to pyrosequencing analysis. Methylation analysis by COBRA includes the use of methylation sensitive restriction enzymes that can digest DNA only when methylated. As expected, some of the ICRs were hypomethylated in OP-mESCs and not in YP-mESCs, such as Airn, Rasgrf1, Peg10 and Grb10, as shown by the enzymatic digestion pattern (Fig. S2h). On the contrary Ig-DMR and Gnas XL did not show differences in DNA methylation (Fig. S2h). In parallel, we also included genomic DNA extracted from a wild type mouse (Ctrl gDNA) as a positive control, carrying a normal methylation profile, and from Zfp57 knockout mESCs (Zfp57 KO) that have been shown to lose methylation at several ICRs34.
Finally, to quantify the DNA methylation changes observed by COBRA, we performed pyrosequencing analysis. Even in this case, E14 YP-mESCs showed normal levels of DNA methylation (around 50%) at the analyzed ICRs (Airn, Rasgrf1, Peg10, Grb10, Ig-DMR and Gnas XL) that was comparable to the mouse genomic DNA (Ctrl gDNA) (Fig. S2i). On the contrary, OP-mESC methylation profile was similar with that of Zfp57 KO mESCs, as Airn, Rasgrf1, Peg10 and Grb10 had less than 20–30% of methylation. However, no changes were detected with passages in Ig-DMR and Gnas XL ICRs (Fig. S2i). In addition, by performing bisulfite-PCR sequencing we also observed loss of methylation at several CG dinucleotides in both KvDMR (also called Kcnq1) and Inpp5fV2 ICRs in E14 OP- but not in YP-mESCs (Fig. S2j). The KvDMR ICR was hypomethylated also in the GS1 OP-mESCs when compared to YP-mESCs (Fig. S2j,k left). However, the Inpp5fV2 ICR was already hypomethylated in the GS1 YP-mESCs and did not show any further loss of methylation with passages (Fig. S2k, right).
Overall these data indicate that downregulation of endogenous Wnt signaling and of β-catenin protein level occurs in prolonged mESC cultures and this correlates with loss of DNA methylation. In particular, we observed loss of methylation at several ICRs, including both maternally and paternally methylated loci that, in turn, control the expression of a variety of coding and non-coding genes within the imprinted clusters.
Wnt/β-catenin activity “protects” the ICRs against de-methylation
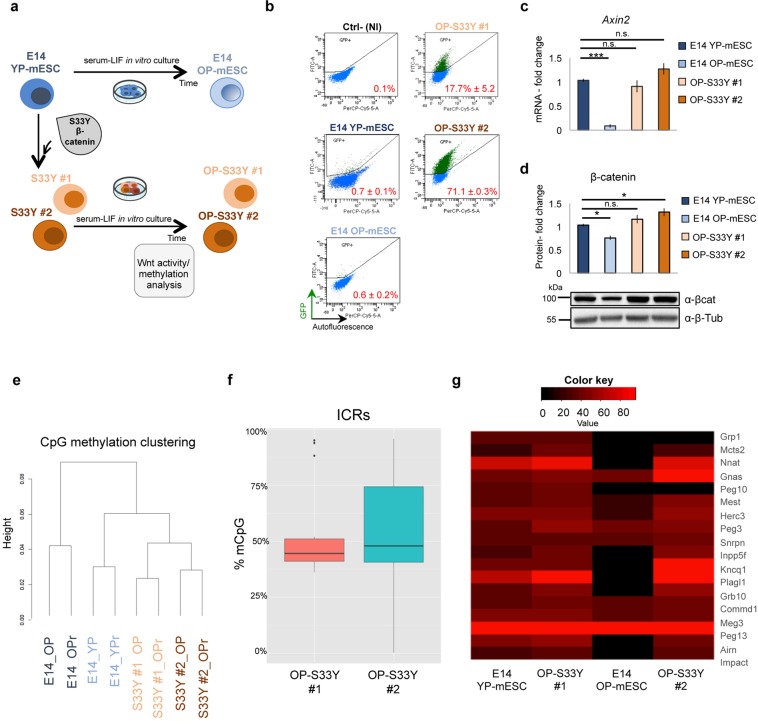
To further investigate the implication of Wnt/β-catenin signaling in the control and protection of the ICRs against de-methylation, we generated two Wnt signaling mutant mESC clones overexpressing the S33Y-mutated β-catenin protein in E14 YP-mESCs. This β-catenin protein mutant is stable since it is not recognized and degraded by the AXIN/GSK3β/APC/CKI destruction complex and it is retained in the nucleus69,70. We named the two E14-derived mESC clones, carrying the S33Y-mutant β-catenin protein, S33Y-β-cat #1 and S33Y-β-cat #2. To test if sustained Wnt/β-catenin signaling activity could preserve methylation at ICRs, we cultured the clones in the conditions that were used to derive E14 OP-mESCs (Fig. 3a). We next tested the level of Wnt signaling by analyzing the activity of the topflash reporter (7TGP)71 in the old passage ESC clones (OP-S33Y-β-cat #1 and OP-S33Y-β-cat #2) and we compared them with both E14 YP- and OP-mESCs. OP-S33Y-β-cat #1 and OP-S33Y-β-cat #2 retained high level of Wnt activity as indicated by the top-flash reporter (Fig. 3b) and by high expression of Axin2 (Fig. 3c). Since the 7TGP lentiviral vector carries a puromycin resistance cassette, prior to the Wnt activity analysis we selected the infected cells to ensure homogeneous expression of the reporter within the cell population in each condition. In parallel, to test the topflash reporter reliability we treated E14 mESCs with either DMSO or CHIR (3 μM) for 24 hours as previously reported15,72. As expected, the 7TGP reporter was activated upon CHIR but not DMSO treatment (Fig. S3a). Moreover, total β-catenin protein was maintained at similar level as in E14 YP-mESCs, rather than being downregulated as in E14 OP-mESCs (Fig. 3d).
Figure 3.
β-catenin overexpressing mESC clones maintain high Wnt/β-catenin activity and normal ICR methylation level after several in vitro passages. (a) Scheme showing how β-catenin overexpressing mESC clones (S33Y #1, #2) were obtained and grown. (b) Representative FACS-plot showing the percentage of positive mESCs for 7TGP topflash reporter activity in YP-mESCs, OP-mESCs, OP-S33Y #1 and OP-S33Y #2 mESC clones. The non infected (NI) cells were used as negative control (Ctrl-). The FITC and the Per-CP-Cy5.5-A detectors were used to identify GFP +(y axis) and autofluorescence (false positive) cells (x axis). The number of recorded events spans from 12000 (OP-mESC) to 18000 (YP-mESCs). Data are represented as means of n = 3 independent experiments ± SD. (c) Quantitative real-time PCR experiments showing the expression profiles of Axin2 in YP-mESCs, OP-mESCs, OP-S33Y #1 and OP-S33Y #2 mESC clones. The transcriptional levels are normalized to Gapdh as a reference gene. Data are represented as fold change (2−ΔΔCt) relative to the YP-E14 mESCs and the results are means of n = 3 independent experiments ± SE. Asterisks indicate statistical significance calculated by unpaired two-tailed t test analysis (n.s. not significant; ***p < 0.001). (d) Western blot analysis showing total β-catenin protein levels in E14 YP-mESCs, OP-mESCs, OP-S33Y #1 and OP-S33Y #2 mESC clones and its quantification. Data are represented as fold change over the protein amount in YP-mESCs and means of n = 3 independent experiments ± SE. β-tubulin was used as loading control. For western-blot quantification densitometric analysis was carried out by using ImageJ software. The quantification reflects the relative amounts as a ratio of each protein band relative to their loading control. (e) Cluster analysis of the four different mESCs. For each line 2 different biological replicates were represented. (f) Box-plot, from min–max values, showing the distribution of mCpG levels at ICRs in OP-S33Y #1 and OP-S33Y #2 mESC clones determined by RRBS analysis. The plots indicate the first quartile, median (black line) and third quartile. Data are obtained from the average of n = 2 biological replicates. (g) Heat-map representation of ICR methylation levels in YP-mESCs, OP-mESCs, OP-S33Y #1 and OP-S33Y #2 mESC clones.
Next, we tested the methylation status of OP-S33Y-β-cat #1 and OP-S33Y-β-cat #2 mutant clones by performing RRBS analysis and compared the mutant clones with both E14 YP- and OP-mESCs. The clusters and PCA analysis grouped together E14 YP-mESCs, OP-S33Y-β-cat #1 and OP-S33Y-β-cat #2 (Figs 3e and S3b), but not E14 OP-mESCs, which clusterized apart. In addition, the two replicates belonging to each sample perfectly correlated between them (Fig. S3c). As expected, OP-S33Y-β-cat #1 and OP-S33Y-β-cat #2 clones maintained the methylation at the ICRs at around 50%, indicating allele-specific methylation (Fig. 3f). Moreover, by plotting together all the conditions it was clear that E14 OP-mESCs showed loss of methylation at many ICRs if compared to YP-mESCs and to the mutant clones (Fig. 3g).
To further validate the RRBS data and to analyze additional ICRs, we performed COBRA analysis and pyrosequencing quantification (Fig. S3d,e). Methylation at Airn, Rasgrf1, Grb10 and Ig-DMR ICRs was maintained normal, at around 50%. However, in the OP-S33Y-β-cat #2 clone Gnas XL and Peg10 were hypermethylated and hypomethylated respectively, suggesting other possible and unpredictable cellular mechanisms, or perturbations occurring in specific clones that could be induced by selection effects. Finally, OP-S33Y-β-cat #1 and OP-S33Y-β-cat #2 mESC clones maintained a round shaped morphology also after several passages in vitro (Fig. S3f).
Overall these data indicate that sustained Wnt/β-catenin activity in mESCs cultured for several passages in Serum + LIF medium prevents the loss of methylation at ICRs and at other genomic regions. Accordingly, loss of Wnt activity can affect DNA methylation and, as a consequence, mESC differentiation potential.
DNA hypomethylation results in loss of chromatin repressor recruitment at the ICRs
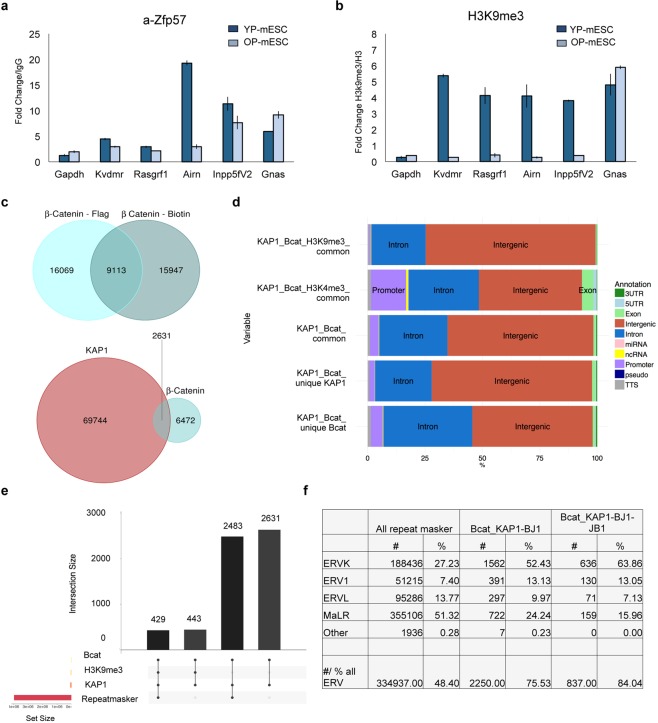
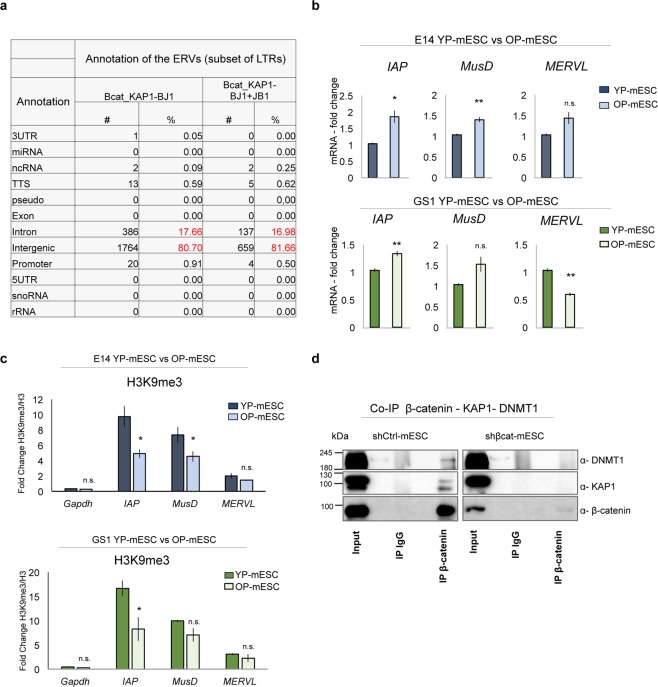
To protect the methylated allele from de-methylation, several chromatin repressors are recruited at the ICRs. In particular, ZFP57 is highly expressed in mESCs and can directly bind to a methylated hexanucleotide DNA motif within imprinted control regions34. After binding, ZFP57 recruits KAP1, which in turn interacts with other heterochromatin-associated histone marks, including H3K9me333,34. Thus, we decided to investigate whether ZFP57 and H3K9me3 levels changed during mESC passages along with the loss of DNA methylation at the ICRs. To this aim, we performed chromatin immunoprecipitation (ChIP)-qPCR experiments for both ZFP57 and H3K9me3 on some known ICR target regions34. Both ZFP57 and H3K9me3 binding decreased in KvDMR, Rasgrf1, Airn and Inpp5fV2 ICRs, reflecting their methylation status. On the contrary, no reduction of ZFP57 and H3K9me3 binding occurs at the Gnas ICR, which did not show loss of DNA methylation in OP-mESCs (Figs 4a,b and S2h,i). Gapdh was used as negative control region and showed low levels of enrichment across all conditions as expected (Fig. 4a,b). Similar results were obtained in GS1 mESCs. GS1 OP-mESCs were characterized by loss of ZFP57 and H3K9me3 recruitment at KvDMR, Rasgrf1, Airn, but not at Inpp5fV2, which was already de-methylated in GS1 YP-mESCs (Figs S4a,b and S2k). These data show that ZFP57 and H3K9me3 ChIP levels are decreased at hypomethylated ICRs, consistently with the methylation profiles shown previously (Figs 2h–j, 3g and S2h,k).
Figure 4.
β-catenin and KAP1 share intergenic common DNA binding sites localized mainly on LTRs and ERVs. (a,b) Representative ChIP-qPCR experiment (out of n = 2 independent experiments) of ZFP57 (a) and H3K9me3 (b) recruitment at several ICRs. The data are represented as fold change (2−ΔΔCt) over IgG (a) or H3 (b) and means ± SD. (c) Venn diagram showing overlapping regions between ChIP-sequencing profiles of β-catenin and KAP1. The peaks between Flag- (β-catenin-Flag) and Biotin- (β-catenin-Biotin) tagged endogenous β-catenin published by Zhang and colleagues73 were intersected among them. The common bound regions were then overlapped with KAP1 ChIP-sequencing peaks performed in BJ1 mESCs by Anvar and colleagues31. (d) Bar chart showing genomic distribution of unique and common peaks among β-catenin, KAP1, H3K9me3, H3K4me3. (e) Bar chart showing ChIP-sequencing peaks intersection among KAP1, β-catenin, KAP1, H3K9me3 and Repeat masker. The number of common overlapping peaks is indicated on the top of the bars. (f) Table showing the different LTR and ERV families represented as number (#) and percentage (%) over Repeat maskers (column 2, 3), the total number of common overlapping peaks between β-catenin and KAP1 in BJ1 (column 4, 5), the total number of common overlapping peaks among β-catenin, KAP1 in BJ1 and KAP1 in JB1 mESCs (columns 6, 7).
β-catenin and KAP1 proteins share common genomic bound regions
Having observed the effect of the Wnt/β-catenin signaling downregulation, we wondered whether β-catenin could play a direct or indirect role in the control of the ICR methylation and epigenetic changes. By analyzing published ChIP-sequencing datasets we observed that several β-catenin binding sites localized close to KAP1, which is responsible for the recruitment of repressors in silent genomic regions. In particular, we compared published β-catenin ChIP-sequencing profiles73 with those of KAP131. We first overlapped ChIP-sequencing profiles between Biotin-tagged and Flag-tagged β-catenin, and we found 9113 common peaks (almost 69% of all peaks) (Fig. 4c, upper diagram), confirming the previously published results73. Next, we intersected these 9113 peaks with KAP1 ChIP-sequencing profiles31 carried out using two different mESC strains (BJ1 and JB1 mESCs) and we found 2631 common overlapping regions between β-catenin and KAP1 in BJ1 mESCs (Fig. 4c, lower diagram). Since the number of total bound regions was much lower in KAP1 ChIP-sequencing dataset in JB1 mESCs (38713 versus 72535 for KAP1 in BJ1 mESCs), we found less common regions bound by both β-catenin and KAP1 (Table S2). As expected, most of the common regions bound from both β-catenin and KAP1, were intergenic or within introns, around 64% and 30% respectively considering both KAP1 ChIP-sequencing data sets (KAP1 in BJ1 and JB1 mESCs) (Fig. 4d and Table S2).
In parallel, we observed that, among the overlapping peaks, around 400–500 regions were also enriched for either H3K9me3 or H3K4me3, which are associated to silent or active chromatin, respectively (Fig. S4c,d and Table S2). For these analysis, we used the already published ENCODE ChIP-sequencing data for H3K9me3 (GSM1000147) and H3K4me3 (GSM769008)74. As expected, the H3K9me3 peaks were located mostly within intergenic genomic regions, while the H3K4me3 bound sites were distributed among intergenic, introns and promoters, including CpG islands (Figs 4d, S4e and Table S2). Nevertheless, in the overlapping regions among β-catenin, KAP1 and H3K9me3 we could find only two ICRs (Grb10 and Meg3), as potential targets of β-catenin. Notably, the same two ICRs were also bound by ZPF5731, which specifically binds to methylated ICRs, as we previously observed in the overlapping regions between β-catenin and ZFP57. However, in this case, the total number of common binding sites was very low (Table S2).
Interestingly, almost all common genomic regions, bound by β-catenin and KAP1 were enriched in repeats (94% and 97% considering KAP1 datasets carried out in BJ1 and JB1 mESCs, respectively). This was also true when we overlapped the data with H3K9me3 peaks (Fig. 4e and Table S2). Among the different repeats the most enriched were the LTRs, which constitute 40% of the common regions bound by β-catenin and KAP1 (Fig. S4f and Table S3). Accordingly, 78–79% of the LTRs bound by both β-catenin and KAP1 were located within intergenic regions (Fig. S4g), confirming the previous data, showed in Fig. 4d and Supplementary Table S2.
Long-terminal repeat (LTR) elements belong to the third repeat class and represent 10% of all mammalian transposable elements75,76. All mammalian LTRs derive from the vertebrate-specific endogenous retroviral elements (ERVs), which can be grouped into sub-classes (I–III) and subfamilies, such as murine retroviral-related sequences (MURRSs, class I), ERVK, class II, ERVL and mammalian apparent LTR retrotransposons (MaLR, (class III), and others75–77. The expression of retroelements has been previously detected in different developmental stages. However, in most tissues and during embryo development the transcription of ERVs is counteracted by several epigenetic mechanisms, including DNA methylation, chromatin repressors (i.e. KAP1) and heterochromatin-associated histone marks, such as H3K9me346,78–80.
As expected most of the LTRs were found in the overlapping common regions between β-catenin and KAP1 corresponded to endogenous retroviral elements, with the ERVK family (including Intracisternal A-particle (IAP) and early transposons (ETn//MusD)) being the most represented one, followed by MaLR and ERV1 families (Fig. 4f). Accordingly, almost 80% and 17% of ERVs were located within intergenic genomic regions and introns, respectively (Fig. 5a).
Figure 5.
β-catenin interacts with the chromatin repressive complex. (a) Table showing genomic annotation of common ERVs bound by β-catenin and KAP1 in BJ1 (columns 2, 3), and among β-catenin, KAP1 in BJ1 and KAP1 in JB1 mESCs (columns 4, 5), represented as number (#) and percentage (%) over the total common peaks. (b) Quantitative real-time PCR experiments showing the expression profiles of ERVs (IAP, MusD, MERVL) in E14 (upper charts) and GS1 (lower charts) YP- and OP- mESCs. The transcriptional levels are normalized to Gapdh as a reference gene. Data are represented as fold change (2−ΔΔCt) relative to the YP-mESCs and the results are means of n = 3 independent experiments ± SE. (c) ChIP-qPCR analysis of H3K9me3 recruitment at IAP, MusD and MERVL ERVs in E14 (upper charts) and GS1 (lower charts) YP- and OP- mESCs. The data are represented as fold change (2−ΔΔCt) over H3 and means of n = 3 independent experiments ± SE. (b,c) Asterisks indicate statistical significance calculated by unpaired two-tailed t test analysis (n.s. not significant; *p < 0.05; **p < 0.01). (d) Co-immunoprecipitation of β-catenin with either KAP1 or DNMT1 followed by western-blot analysis, in shCtrl -or shβcat- transduced E14 mESCs. 10% of input was used for DNMT1, KAP1 and β-catenin IP. IgG were used as negative control. An empty well was included between each experimental condition (Input, IgG and IP) to avoid cross-contamination.
β-catenin can directly interact with KAP1 protein and regulate the expression of retrotransposons
To further investigate the epigenetic changes in YP- and OP-mESCs, we also analyzed the expression of some endogenous retroviruses, along with ICR DNA methylation. IAP and MusD subsets of ERVs were significantly upregulated in the E14 OP-mESCs (Fig. 5b, upper panel) but not MERVL, when compared to YP-mESCs. Interestingly, IAP expression increases also in GS1 OP-mESCs (Fig. 5b, lower panel), suggesting that these endogenous retroviral elements could be more active in OP- than in YP-mESCs.
Retrotransposons (such as IAP and MusD) are located within repressed genomic regions and they have been associated with induction and spreading of heterochromatin marks, such as H3K9me381. Furthermore, they are controlled by KAP1-mediated repressive complexes, which prevent their activation and genomic spreading46,79,80. Therefore, to analyze whether the transcriptional up-regulation of the ERVs in OP-mESCs correlates with loss of heterochromatin marks, we performed ChIP- qPCR analysis for H3K9me3 in E14 and GS1 YP- and OP-mESCs. The recruitment of H3K9me3 decreased at IAP LTR in both E14 and GS1 OP-mESCs, and at MusD in E14 OP-mESCs (Fig. 5c upper and lower panels), which was consistent with their expression profile. However, the ChIP-qPCR profile did not follow MERVL expression changes, suggesting that other repressive mechanisms control specifically these elements in the OP-mESCs (Fig. 5b,c). In mESCs the expression of ERVs is controlled by a number of chromatin repressive factors, including KAP1, DNMT1, among others82. In particular, KAP1 acts as a co-repressor by facilitating the recruitment of repressive complexes at ERVs, ICRs or other silenced genomic regions46. Since, in our analysis almost all of the common overlapping regions corresponded to intergenic regions enriched in LTRs, we reasoned that β-catenin could interact with the chromatin repressive complex, in particular with KAP1. To confirm this hypothesis, we performed co-immunoprecipitation (CoIP) experiments in E14 YP-mESCs carrying either a short hairpin against a control sequence with no predicted genomic target (shCtrl) or against β-catenin (shβcat). In shCtrl-transduced mESCs, β-catenin was co-immunoprecipitated with KAP1 and DNMT1, suggesting that it can interact with each one of the components of the epigenetic repressive complex (Fig. 5d, left panel). As control, we performed CoIP experiments in the shβcat-infected mESCs. In absence of β-catenin neither KAP1 nor DNMT1 could be immunoprecipitated as expected (Fig. 5d, right panel). Importantly, the amount of total (as observed by the input band) and the immunoprecipitated β-catenin was much lower in the shβcat-infected mESCs, showing the high silencing efficiency (Fig. 5d, right panel).
Inhibition of β-catenin causes impaired mESC differentiation and changes in retrotransposon expression, but it does not affect ICR methylation
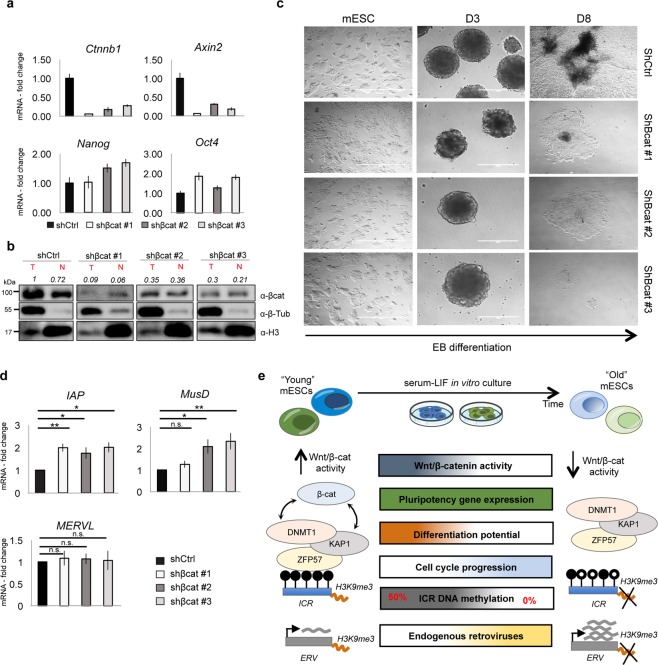
To further investigate whether β-catenin plays a direct role in protecting ICRs and retrotransposons, thereby safegarding genomic stability, we knocked down β-catenin in E14 YP-mESCs by using three different pLKO-based silencing constructs83,84. We tested β-catenin silencing efficiency by performing qPCR and by western blot analysis. β-catenin transcript and protein (both total and nuclear) were efficiently downregulated (Fig. 6a, upper panel, and 6b), as well as the downstream target Axin2 (Fig. 6a, upper panel). Neither the expression of pluripotency markers, Nanog and Oct4 (Fig. 6a, lower panel), nor the self-renewal capacity (Fig. 6c, left micrographs) were impaired after β-catenin silencing in mESCs as expected14,85. Nevertheless, mESCs carrying β-catenin silencing shRNAs could not properly differentiate. The embryoid bodies started to disaggregate already at day 3 (Fig. 6c), displaying a phenotype similar with the one reported in β-catenin knockout mESCs thus confirming the already published data14,85.
Figure 6.
β-catenin silencing impairs mESC differentiation. (a) Quantitative real-time PCR analysis showing β-catenin silencing efficiency, Axin2 and pluripotency marker (Nanog, Oct4) levels. The transcriptional levels are normalized to Gapdh as a reference gene. Data are represented as fold change (2−ΔΔCt) relative to the shCtrl-infected mESCs and the results are means of n = 3 technical replicated ± SD. (b) Western blot analysis showing protein levels of total and nuclear β-catenin in shCtrl-, shβcat#1-, shβcat#2- and shβcat#3- transduced E14 mESCs (n = 1). Quantification of total and nuclear β-catenin is represented as relative to total β-catenin in shCtrl-transduced mESCs. β-tubulin and H3 were used as loading controls. For western-blot quantification densitometric analysis was carried out by using ImageJ software. The quantification reflects the relative amounts as a ratio of each protein band relative to their loading control. (c) Representative bright field images of mESCs and embryoid bodies (EBs) at day 3 (D3), 8 (D8) after β-catenin silencing (shβcat #1, #2, #3) versus the control condition (shCtrl). Scale bar is 400 μm. (d) Quantitative real-time PCR experiments showing the expression profiles of ERVs (IAP, MusD, MERVL) in shCtrl-, shβcat#1-, shβcat#2- and shβcat#3- transduced E14 mESCs. The transcriptional levels are normalized to Gapdh as a reference gene. Data are represented as fold change (2−ΔΔCt) relative to shCtrl-infected mESCs and are means of n = 3 independent experiments ± SE. Asterisks indicate statistical significance calculated by unpaired two-tailed t test analysis (n.s. not significant; *p < 0.05; **p < 0.01). (e) Schematic representation showing cellular and epigenetic changes occurring in prolonged in vitro mESC cultures along with Wnt/β-catenin pathway downregulation.
When we analyzed DNA methylation profile by RRBS, we did not observe a drastic loss of genome-wide DNA methylation at CpG islands and ICRs, in contrast to what we observed for OP-mESCs (Fig. S5a,b, and Table S4). We performed RRBS analysis on both shCtrl- and shβcat- transduced mESCs and EBs at day 8 of differentiation. We focused on the mESCs carrying shβcat #1 construct, which showed the highest silencing efficiency. Shβcat-infected mESCs displayed lower methylation at Peg10, Peg3, Inpp5fV2, Airn (Fig. S5b, black arrows), whereas Snrpn, Commd1 and Impact had less methylation in the shβcat-transduced EBs, when compared to control (shCtrl) (Fig. S5b, blue arrows). Moreover, we validated the RRBS data by performing COBRA analysis on several ICRs, such as Airn, Grb10, Rasgrf1, Ig-DMR, Peg10 and Gnas XL. The methylation profile (as showed by the enzymatic digestion pattern) did not almost change between shCtrl- and shβcat-transduced mESCs (Fig. S5c), suggesting that the methylation differences observed in the RRBS analysis were small and not comparable to the ones observed in OP-mESCs. Nevertheless, we observed an upregulation of IAP, MusD, but not of MERVL expression after β-catenin silencing (Fig. 6d) suggesting that β-catenin silencing might affect the epigenetic regulation of silent genomic regions, likely acting together with repressive complexes. In conclusion, the observation that β-catenin removal did not cause relevant changes in DNA methylation might indicate that the endogenous β-catenin amount is already limiting and further reduction does not affect the methylation level.
Discussion
Since mouse embryonic stem cells (mESCs) were isolated, in the early 80’s6,7, many groups started to investigate the mechanisms that define their pluripotency. Mouse ESCs present many comparable properties with the inner cell mass (ICM) of the blastocyst, such as the expression of pluripotency genes and the capacity to contribute to the formation of the three germ layers. Up to now, many studies have investigated embryonic developmental mechanisms by exploiting the capacity of mESCs to self-renew in vitro indefinitely. Mouse ESCs are usually cultured in vitro in the presence of LIF86,87, which directly controls the expression of the pluripotency gene-network that includes Oct4, Nanog and other factors88.
Nevertheless, whether the in vitro cultured mESCs are identical to the ICM has been a topic of debate of the last years. With the aim to obtain mESCs with features as much as possible similar to the ICM, the 2i-LIF culturing medium was designed to drive mESCs toward a naïve pluripotent state, characterized by high expression of several pluripotency genes, including Rex189,90. This has been attributed to the activity of two small drug inhibitors (2i) of MEK1/2 and GSK3β, the latter leading to Wnt/β-catenin signaling activation. However, the identity of mESCs is not only dependent on the pluripotency gene network but also on the maintenance of a correct epigenetic state. Indeed, many groups previously observed genomic instability of mouse and human ESCs when they were cultured in vitro for a long time36. Recently, it has been shown that the constant inhibition of MEK1/2 is detrimental for mESC homeostasis after prolonged in vitro culture, leading to chromosomal aberrations, severe global hypomethylation and impaired differentiation capacity39. However, this study did not addressed whether Wnt/β-catenin activity also had an epigenetic effect in prolonged mESC cultures.
The essential role of Wnt/β-catenin signaling in mESC differentiation is widely accepted. Loss of β-catenin12–14 or Wnt3a17,91 causes embryo cell death when gastrulation starts. Likewise, we observed that OP-mESCs, which showed decreased endogenous Wnt/β-catenin activity, could not generate beating embryoid bodies with the same efficiency as YP-mESCs. On the contrary, they expressed high levels of ectodermal markers, such as Otx2, Sox1, Pax6, Nestin and III β-Tubulin, during both EB and neural differentiation. E14 OP-mESCs also expressed high levels of Fgf5 during differentiation, but GS1 OP-mESCs did not, suggesting a possible epigenetic dysregulation of this gene in this strain. Since the differentiation potential of OP-mESCs was biased toward the ectodermal fate, these data might suggest that OP-mESCs could be more primed with respect to the YP-mESCs, and they could share some common features with epiblast stem cells (EpiSCs)92,93. It is important to note however that EpiSCs were shown as able to generate the three germ layers in vitro and maintain their genomic integrity93. In our study, the expression level of pluripotency genes was maintained high in OP-EBs, indicating that OP-mESCs could not properly exit from pluripotency state, in agreement with what was previously observed in β-catenin knockout mESCs85. Indeed, β-catenin knockout–derived teratomas displayed high level of pluripotency markers and features comparable to germ cell tumors.
In addition to molecular pathways alterations, several epigenetic changes have been investigated in both mouse and human ESCs. These epigenetic changes can occur both at the level of DNA methylation and histone modifications and they affect the chromatin structure and ESC identity. In particular, DNA methylation is sensitive to external stimuli and cellular stress. DNA methylation pattern is maintained stable during cell replication through the action of DNA methyltransferases (DNMTs) and other repressors, since tuned methylation levels are indispensable for mESC stability and differentiation. Loss of DNMTs leads to severe global epigenetic deregulation, which causes developmental defects and embryonic lethality, as previously reported39,44,94–96. Accordingly, in our study, we observed changes in DNA methylation at several CpG enriched genomic regions, which were prevalently hypomethylated upon prolonged culturing, and consequently, impaired mESC differentiation potential. Moreover, the hypomethylated CpG regions appeared to control several biological processes as indicated by the gene ontology analysis, with the metabolic processes as the most represented. Additionally, some of the analyzed hypomethylated regions were found nearby genes that have been previously reported to interact with Wnt pathway and its downstream components in different cellular contexts97–101. Some of these genes have been described to inhibit Wnt/β-catenin signaling pathway, such as Grb10, Kdm2b, Tet3, Pcdhgc5, Mir148a97–101. On the contrary, other hypomethylated regions corresponded to genes that act as activators or inhibitors of Wnt signaling, depending on the molecular and cellular context, such as Runx3, En1, Nkx6-1, Epha2, Nr4a102–107.
In particular, the OP-mESCs showed high hypomethylation at the level of imprinted genes, which commonly show stable allele-specific methylation pattern in the pre-implantation embryo. It is important to take into consideration that the imprinted genes are organized in clusters with a common ICR. Methylation changes of one ICR can affect the expression of many imprinted genes within the cluster (around 4–5). In this study, we observed that the YP-mESCs show 50% of methylation at many ICRs, indicating that allele specific methylation was lost in prolonged cultures, extended to both maternally and paternally imprinted loci. Additionally, the OP-mESCs were characterized by higher retrotransposon expression, such as IAP, when compared to YP-mESCs. Both imprinted genes and retrotransposons are tightly epigenetically controlled27,31–34,45,46,79–81 since they play essential roles during embryonic and extra-embryonic tissue formation. Loss of imprinting or transcriptional changes in retrotransposons, in particular in IAP, could indeed be detrimental to genomic stability and embryo development26,36,39,40,94,96,108,109. Accordingly, both E14 and GS1 OP-mESCs could not properly differentiate when compared to YP cells.
OP-mESCs were characterized by significantly faster cell cycle progression with respect to YP-mESCs, suggesting that the epigenetic changes could also affect cell cycle check-points. Interestingly, loss of methylation at the KvDMR ICR has been associated with lower expression of the cell cycle inhibitor cyclin-dependent kinase inhibitor 1 C, (Cdkn1c)110. In addition, Wnt/β-catenin pathway has been reported to have an anti-proliferative effect, by directly regulating the expression of cell cycle repressor genes61, thus safeguarding mESC identity. In line with previous published studies, we observed that both E14 and GS1 mESCs downregulated the expression of Wnt downstream targets (Axin2, Lef1, Tcf1, Sp5) and β-catenin protein at late passages. Wnt activity reduction was not associated with altered pluripotency gene expression, but it was translated into impaired differentiation potential, faster cell cycle progression and genomic instability, thus loss of mESC homeostasis. The factors causing Wnt/β-catenin pathway downregulation in prolonged cell cultures still remain unknown. Among possible triggering events, oxidative stress could be a putative inducing factor. Indeed, increased oxidative stress has been reported to antagonize Wnt signaling and has been previously described to induce genomic aberrations and loss of cell homeostasis in long-term in vitro cell culture111–114. In particular, increased oxidative stress can antagonize Wnt signaling by inducing expression of Forkhead box-O (FOXO) transcription factor115. FOXO competes with TCFs for its interaction with β-catenin, thus inhibiting TCF transcriptional activity and the canonical Wnt/β-catenin signaling pathway116,117. Additionally, among the hypomethylated regions analyzed by RRBS we found some inhibitors of Wnt/β-catenin pathway, including Kdm2b, which inhibits the stability of β-catenin protein98, thereby creating a feedback loop that could accelerate β-catenin degradation.
It is important to note that different mESC lines can display disparate endogenous levels of Wnt/β-catenin activity, due to either the diverse in vitro culturing conditions or different timing of mESC isolation from the embryo20,62. YP-GS1 mESCs showed low levels of nuclear β-catenin protein, if compared to the YP-E14 mESCs. Interestingly, ICRs, such as Inpp5fV2 region were hypomethylated in GS1 mESCs already at early passages, suggesting that this cell line was less stable with respect to E14. The role of Wnt activity in homeostasis maintenance has been largely described in the adult stem cells. For instance, long-term hematopoietic stem cells show a decrease in the canonical Wnt/β-catenin signaling activity during ageing118. In parallel, other studies has reported that many epigenetic changes, including loss of imprinting, occur in hematopoietic and other adult stem cell compartments119. The observation that mESCs carrying gain of function Wnt mutants (S33Y-β-cat #1 and S33Y-β-cat #2) maintained normal or even higher level of methylation at the ICRs also after prolonged in vitro culturing, further strengthen the hypothesis of a possible “protective” role due to Wnt/β-catenin pathway, which can control epigenetic stability. Even after prolonged culturing, these clones retained Wnt/β-catenin activity and methylation at similar levels of YP-mESCs. However, it is important to take into account that additional epigenetic changes might occur in some of the clones, due to stochastic perturbations. Indeed, some of the ICRs, such as Peg10, Gpr1 were hypomethylated in the OP-S33Y-β-cat #2 mESC clone, whereas others showed an increase in DNA methylation level in both β-catenin mutant clones. However, in both β-catenin overexpressing clones the overall DNA methylation profile was similar to the YP-mESCs.
Finally, we observed that β-catenin interacts with KAP1 and DNMT1 repressors, therefore it can be considered as a regulator of epigenetic stability maintenance. By analyzing published ChIP-sequencing datasets we observed that β-catenin and KAP1 share common target regions, located within intergenic regions and overlapping with endogenous retroviral elements. These data further strengthen the conclusion that β-catenin could mediate the action of repressors recruited by KAP1 on silent genomic regions, even though the potential mechanism still remains unknown. Interestingly, β-catenin protein has been previously described to interact with DNMT1 in colorectal cancer cells120 or with other chromatin factors in mESCs121. However, it is important to note that, when we silenced β-catenin in mESCs we did not detect drastic methylation changes at the ICRs, though shβcat- transduced cells upregulated the transcriptional levels of IAP and MusD endogenous retroviruses. These data suggest that either the endogenous levels of β-catenin are already limiting and its further decrease does not impair methylation maintenance, or, alternatively, that β-catenin does not directly act on the ICRs but it acts as a mediator for the repressive complexes. Up to date, the factors causing genomic instability in mESCs are not clear, even though many chromatin modifiers have been described to act on repressed genomic regions.
All in all, our data suggest that Wnt/β-catenin activity need to be maintained constantly active in mESCs during passages to ensure correct cell identity and epigenetic stability. Loss of Wnt activity results in global hypomethylated DNA, loss of chromatin repressor recruitment and activation of silent genomic regions, resulting in impaired mESC differentiation and altered cell cycle progression (Fig. 6e). Sustained activation of the Wnt signaling pathway results in maintenance of methylation at most of the ICRs after prolonged in vitro culture. In conclusion, Wnt/β-catenin pathway, mediates a large number of molecular and biological processes including DNA methylation at ICRs to ensure correct cell and tissue homeostasis.
Materials and Methods
Cell lines and differentiation protocols
GS-1 (129 Sv) and E14 (129/Ola) mouse embryonic stem cells (mESCs) were obtained from Merrill’s laboratory122 and purchased from ATCC, respectively. Both mESC cell lines were maintained in 0.1% gelatin (Millipore ES-006-B)-coated plates mESC medium, which consisted of DMEM supplemented with 15% fetal bovine serum (FBS), L– glutamine (2 mM), penicillin (100 U/ml), streptomycin (100 µg/ ml), sodium pyruvate (1 mM), non-essential amino acid (NEAA) (0,1 mM), 2-mercaptoethanol (0,5 mM) and ESGRO mLif (1000 U/ml). E14 mESCs were thawed at passage 8 and expanded up to passage 14 for the analysis of young passage (henceforth called YP-E14 mESCs) E14 mESCs. GS1 were thawed from passage 18 and expanded up to passage 22 for the analysis of young passage (henceforth called YP-GS1 mESCs) GS1 mESCs. To obtain the old passage (henceforth called OP-mESCs) E14 and GS1 mESCs were kept in culture for ~70 and ~50 passages, respectively, in mESC medium. At each passage cells were detached by using trypsin (0.025% trypsin and 0.04% EDTA, SIGMA 25300-054) at 37 °C, centrifuged for 5 minutes and 300 rcf and plated with a dilution ratio of 1:15–1:20 at each passage.
The differentiation medium for the production of embryoid bodies (EBs) consisted of mESC culture medium without LIF. The cells were harvested by trypsinisation, counted, and propagated in hanging drops (400 single mESCs/30 µl initial drop) for 2 days, before being transferred to 10 cm2 bacterial dishes, where the cells grow in suspension. On day 5, the embryoid bodies were transferred onto gelatinized p100 dishes always in differentiation medium, which consisted of mESC culture medium without the LIF. The medium was changed every 2 days and the beating embryoid bodies were observed starting from day 8 of the differentiation process. For expression profile analysis the cells were harvested and pelleted at day 0 (ESC), 6 and 12 of the differentiation process.
For neural differentiation in monolayer, undifferentiated mESCs were gently dissociated using trypsin (0.025% trypsin and 0.04% EDTA, SIGMA 25300-054) at 37 °C and plated onto 0.1% gelatin-coated tissue culture plastic at a density of 0.5–1.5 × 104/cm2 in N2B27 medium [1:1mix of DMEM/F12 (GIBCO) supplemented with N2 (GIBCO) and Neurobasal medium (GIBCO) supplemented with B27 (GIBCO)], L–glutamine (0,5 mM), 2-mercaptoethanol (0,1 mM) and retinoic acid (1 μM). The medium was refreshed every other day64,65. For expression profile analysis cells were detached by using Accutase (A1110501, GIBCO) and pelleted at 300 rcf for 5 minutes.
Total protein extraction
Cells were trypsinized (0.025% trypsin and 0.04% EDTA, SIGMA 25300-054) at 37 °C, pelleted at 300 rcf and washed twice with PBS. During each wash cells were pelleted at 300 rcf for 5 min 4 °C. Cell lysis was performed on ice for 25 min, in RIPA buffer (150 mM NaCl, 1% Nonidet P40, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulphate, 50 mM Tris-HCl, pH 8.0) containing protease (SIGMA P8340) and phosphatase inhibitors (SIGMA P2850). Insoluble material was pelleted by centrifugation at 16,000 rcf for 30 min at 4 °C. Protein concentrations were determined using the Bradford assay (Bio-Rad 500-0006). Western blot was performed as specified in the apposite section with the antibodies indicated in Table S6.
Nuclear protein extraction
For nuclear protein extraction cells were trypsinized (0.025% trypsin and 0.04% EDTA, SIGMA 25300-054) at 37 °C, pelleted at 300 rcf and washed twice with cold PBS. During each wash cells were pelleted at 300rcf for 5 min at 4 °C. Cells were incubated in hypotonic buffer (10 mM Tris-HCl pH 7.8, 5 mM KCl, 2 mM MgCl2 DTT 1 mM) containing protease inhibitors (SIGMA P8340) for 10 min at 4 °C. Cells were pelleted at 300 rcf for 5 min at 4 °C and plasma membrane lysis was performed in 0,25% NP-40 hypotonic buffer on ice for 15 min. Nuclei were pelleted at 300 rcf for 15 min at 4 °C and washed twice in hypotonic buffer. Isolated nuclei were incubated in RIPA buffer (150 mM NaCl, 1% Nonidet P40, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulphate, 50 mM Tris-HCl, pH 8.0) containing protease (SIGMA P8340) and phosphatase inhibitors (SIGMA P2850). Insoluble material was pelleted by centrifugation at 16,000 rcf for 30 min at 4 °C. Protein concentrations were determined using the Bradford assay (Bio-Rad 500-0006). Western blot was performed as specified in the apposite section with the antibodies indicated in Table S6.
Protein immunoprecipitation
For each immunoprecipitation condition 50 µl of Dynabeads (Thermo scientific 10004D) were washed 3 times in 500 µl of cold CHAPS buffer (50 mM TrisHCl pH 7.5, 150 mM NaCl, 0.15% CHAPS) containing protease inhibitors (SIGMA P8340). To separate the beads from the wash solution the tubes were placed on the magnet. The isolated beads were re-suspended in 500 µl of antibody solution containing 8 µg of antibody (DNMT1 (Abcam, ab87656) β-catenin (Millipore, 06-734) KAP1 (Abcam, ab10483),) or IgG (Abcam, ab46540) in cold CHAPS buffer containing protease inhibitors) and incubated O/N at 4 °C on a rotating wheel.
Cell fractionation and nuclei isolation was performed as described in the previous paragraph. For co-immunoprecipitation experiments, nuclei were lysed in CHAPS buffer (50 mM TrisHCl pH 7.5, 150 mM NaCl, 0.15% CHAPS containing protease inhibitors) for 15 min at 4 °C and were immerged in liquid nitrogen for 2 min and successively thawed on ice to perform a freeze-thaw lysis. Insoluble material was pelleted by centrifugation at 16,000 rcf for 30 min at 4 °C and the supernatants (100 µg of the nuclear protein extract) were incubated with antibody-coupled dynabeads overnight O/N at 4 °C. Beads were washed three times with CHAPS buffer containing protease inhibitors and elution was performed by boiling beads in Laemmli buffer (1x) at 95 °C for 10 min. Western blot was performed as specified in the apposite section with the antibodies indicated in Table S6.
Western blot
Either total protein extract or nuclear protein extract was mixed with 4 × Laemmli buffer (40% glycerol, 240 mM Tris/HCl, pH 6.8, 8% SDS, 0.04% bromophenol blue, 5% β-mercaptoethanol) and denatured at 99 °C for 10 minutes. Either total protein extract or nuclear protein extract, or co-immunoprecipitation eluate was separated by SDS-PAGE, and transferred to poly vinylidene difluoride membrane (BIO-RAD 162-0177). The membranes were blocked with 5% non-fat dry milk (SIGMA 70166) in TBS-Tween 20 (0,1%) (SIGMA P1379) for 60 min, incubated with primary antibodies (β-catenin (BD, 610153), NANOG (Calbiochem, #SC1000), OCT-4 (Santa Cruz, sc-5279), β-tubulin (SIGMA, T0198), DNMT1 (Abcam, ab87656), KAP1 (Abcam, ab10483)) overnight at 4 °C. The working dilution of each antibody is listed in Table S6. The poly vinylidene difluoride membrane was then washed three times with TBS-T for 15 min, incubated with the peroxidase-conjugated secondary antibody (1:2000, Amersham Biosciences NA931 (Mouse IgG) and NA934 (Rabbit IgG)) in TBS-T with 5% non-fat dry milk for 60 min, and washed three times with TBS-T for 10 min. Immunoreactive proteins were detected using Pierce ECL Western Blotting Substrate (Thermo Scientific 32106). Densitometric analysis was carried-out by using ImageJ software. The quantification reflects the relative amounts as a ratio of each protein band relative to their loading control.
Reduced representation bisulfite sequencing (RRBS) data analysis
Reads were processed by adaptor trimming (Illumina Pipeline Casava v1.8.2), filtered for low quality reads (Trim Galore v0.2.8) and subjected to quality control (FastQC). Reads were aligned using Bismark v0.7.9124 to the Mus musculus genome (assembly NCBI37/mm9). CpG methylation calls were extracted using the Bismark methylation extractor v0.7.9. The methylation level of a DNA region was defined using SeqMonk v0.32.1 pipeline (Simon Andrews, Babraham Institute, UK) considering at least 2 CpGs covered by at least 10 reads. Hypomethylated DNA regions were identified by searching for sequences with common symmetric CpGs (at least 10 CpGs covered by at least 10 reads that were less than 2 kb apart) with a decrease in methylation of >25%. Clustering and correlation analysis were performed using R package methylKit125. The RRBS data are available under the GSE109417 accession number.
Analysis of published ChIP-seq data sets
The CpG islands data was downloaded from the UCSC genome annotation data-base for the July 2007 assembly of the mouse genome (mm9, NCBI build 37). The repeat types and coordinates were extracted from the RepeatMasker file (UCSC table browser). We used bedtools (v2.25.0)126 to overlap ChIP-seq peak data coordinates. The annotatePeaks tools from HOMER suite of programs127 was used to annotate the resulting peak overlaps (using mm9 version of Mus musculus genome assembly).
Relevant figures were produced in the R environment using mainly ggplot2128, reshape2129 VennDiagram130 and UpSetR131 packages.
Supplementary information
Acknowledgements
We are grateful to Bernhard Payer, Frederic Lluis, Ruben Sebastian Perez and other members of the Cosma laboratory for helpful suggestions and discussions. We thank Hapreet Kukreja and Vincenzo Riso for technical suggestions. We thank the ENCODE Consortium for generating the ChiP sequencing datasets. We acknowledge the support from Spanish Ministry of Economy and Competitiveness and FEDER funds (SAF2011-28580, BFU2014-54717-P, BFU2017-86760-P and BFU2015-71984-ERC to M.P.C.), as well as ‘Centro de Excelencia Severo Ochoa 2013–2017’, Secretaria d’Universitats i Recerca del Departament d’Economia i Coneixement de la Generalitat de Catalunya grant (2014 SGR1137 to M.P.C.), AGAUR (SGR 2017-2019 to M.P.C) and the CERCA Programme/Generalitat de Catalunya (to M.P.C.), the European Union’s Horizon 2020 research and innovation programme under grant agreement CellViewer No 686637 (to M.P.C.), People Programme Marie Curie Actions of the European Union’s Seventh Framework Programme (FP7/2007–2013/, n° 290123 to I.T.), La Caixa international PhD fellowship (to F.S.) and Ministerio de Ciencia e Innovació FPI (to F.A.) and People Program (Marie Curie Actions) FP7/2007–2013 under REA grant (608959 to M.V.N.).
Author Contributions
M.P.C. and I.T. designed the experiments and performed the data analysis. I.T. performed the majority of the experiments and data analysis. F.S. performed western-blot, co-immunoprecipitation analysis and flow cytometry analysis. M.C. performed RRBS data analysis. S.B. performed ChIP-sequencing data analysis. M.S.D. performed pyrosequencing analysis. U.D.V. and M.V.N. contributed to cell culture and helped with in vitro experiments. K.A. generated the S33Y-β-cat #1 and S33Y-β-cat #2 mutant mESC clones. F.A. generated the silencing constructs. D.M. supervised pyrosequencing and COBRA analysis. A.R. supervised RRBS data analysis. M.P.C. and I.T. wrote the manuscript. M.P.C. supervised the project.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Marco Cammisa and Sarah Bonnin contributed equally.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-37442-5.
References
- 1.Rao TP, Kuhl M. An updated overview on Wnt signaling pathways: a prelude for more. Circulation research. 2010;106:1798–1806. doi: 10.1161/CIRCRESAHA.110.219840. [DOI] [PubMed] [Google Scholar]
- 2.Katoh M, Katoh M. WNT signaling pathway and stem cell signaling network. Clin Cancer Res. 2007;13:4042–4045. doi: 10.1158/1078-0432.CCR-06-2316. [DOI] [PubMed] [Google Scholar]
- 3.Staal FJ, Clevers H. Tcf/Lef transcription factors during T-cell development: unique and overlapping functions. The hematology journal: the official journal of the European Haematology Association / EHA. 2000;1:3–6. doi: 10.1038/sj/thj/6200001. [DOI] [PubMed] [Google Scholar]
- 4.Haegel H, et al. Lack of beta-catenin affects mouse development at gastrulation. Development. 1995;121:3529–3537. doi: 10.1242/dev.121.11.3529. [DOI] [PubMed] [Google Scholar]
- 5.Huelsken J, et al. Requirement for beta-catenin in anterior-posterior axis formation in mice. The Journal of cell biology. 2000;148:567–578. doi: 10.1083/jcb.148.3.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Evans MJ, Kaufman MH. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981;292:154–156. doi: 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- 7.Martin GR. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proceedings of the National Academy of Sciences of the United States of America. 1981;78:7634–7638. doi: 10.1073/pnas.78.12.7634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boyer LA, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chambers I, et al. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell. 2003;113:643–655. doi: 10.1016/S0092-8674(03)00392-1. [DOI] [PubMed] [Google Scholar]
- 10.Jaenisch R, Young R. Stem cells, the molecular circuitry of pluripotency and nuclear reprogramming. Cell. 2008;132:567–582. doi: 10.1016/j.cell.2008.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Niwa H, Miyazaki J, Smith AG. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nature genetics. 2000;24:372–376. doi: 10.1038/74199. [DOI] [PubMed] [Google Scholar]
- 12.Blauwkamp TA, Nigam S, Ardehali R, Weissman IL, Nusse R. Endogenous Wnt signalling in human embryonic stem cells generates an equilibrium of distinct lineage-specified progenitors. Nature communications. 2012;3:1070. doi: 10.1038/ncomms2064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bone HK, Nelson AS, Goldring CE, Tosh D, Welham MJ. A novel chemically directed route for the generation of definitive endoderm from human embryonic stem cells based on inhibition of GSK-3. Journal of cell science. 2011;124:1992–2000. doi: 10.1242/jcs.081679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lyashenko N, et al. Differential requirement for the dual functions of beta-catenin in embryonic stem cell self-renewal and germ layer formation. Nature cell biology. 2011;13:753–761. doi: 10.1038/ncb2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Anton R, Kestler HA, Kuhl M. Beta-catenin signaling contributes to stemness and regulates early differentiation in murine embryonic stem cells. FEBS letters. 2007;581:5247–5254. doi: 10.1016/j.febslet.2007.10.012. [DOI] [PubMed] [Google Scholar]
- 16.Kielman MF, et al. Apc modulates embryonic stem-cell differentiation by controlling the dosage of beta-catenin signaling. Nature genetics. 2002;32:594–605. doi: 10.1038/ng1045. [DOI] [PubMed] [Google Scholar]
- 17.Liu P, et al. Requirement for Wnt3 in vertebrate axis formation. Nature genetics. 1999;22:361–365. doi: 10.1038/11932. [DOI] [PubMed] [Google Scholar]
- 18.Ogawa K, Nishinakamura R, Iwamatsu Y, Shimosato D, Niwa H. Synergistic action of Wnt and LIF in maintaining pluripotency of mouse ES cells. Biochemical and biophysical research communications. 2006;343:159–166. doi: 10.1016/j.bbrc.2006.02.127. [DOI] [PubMed] [Google Scholar]
- 19.Singla DK, Schneider DJ, LeWinter MM, Sobel B. E. wnt3a but not wnt11 supports self-renewal of embryonic stem cells. Biochemical and biophysical research communications. 2006;345:789–795. doi: 10.1016/j.bbrc.2006.04.125. [DOI] [PubMed] [Google Scholar]
- 20.ten Berge D, et al. Embryonic stem cells require Wnt proteins to prevent differentiation to epiblast stem cells. Nature cell biology. 2011;13:1070–1075. doi: 10.1038/ncb2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kelly KF, et al. beta-catenin enhances Oct-4 activity and reinforces pluripotency through a TCF-independent mechanism. Cell stem cell. 2011;8:214–227. doi: 10.1016/j.stem.2010.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thomson M, et al. Pluripotency factors in embryonic stem cells regulate differentiation into germ layers. Cell. 2011;145:875–889. doi: 10.1016/j.cell.2011.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell research. 2011;21:381–395. doi: 10.1038/cr.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bernstein BE, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 25.Azuara V, et al. Chromatin signatures of pluripotent cell lines. Nature cell biology. 2006;8:532–538. doi: 10.1038/ncb1403. [DOI] [PubMed] [Google Scholar]
- 26.Bourc’his D, Bestor TH. Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking Dnmt3L. Nature. 2004;431:96–99. doi: 10.1038/nature02886. [DOI] [PubMed] [Google Scholar]
- 27.Edwards CA, Ferguson-Smith AC. Mechanisms regulating imprinted genes in clusters. Current opinion in cell biology. 2007;19:281–289. doi: 10.1016/j.ceb.2007.04.013. [DOI] [PubMed] [Google Scholar]
- 28.Bostick M, et al. UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science. 2007;317:1760–1764. doi: 10.1126/science.1147939. [DOI] [PubMed] [Google Scholar]
- 29.Song J, Teplova M, Ishibe-Murakami S, Patel DJ. Structure-based mechanistic insights into DNMT1-mediated maintenance DNA methylation. Science. 2012;335:709–712. doi: 10.1126/science.1214453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Arita K, Ariyoshi M, Tochio H, Nakamura Y, Shirakawa M. Recognition of hemi-methylated DNA by the SRA protein UHRF1 by a base-flipping mechanism. Nature. 2008;455:818–821. doi: 10.1038/nature07249. [DOI] [PubMed] [Google Scholar]
- 31.Anvar Z, et al. ZFP57 recognizes multiple and closely spaced sequence motif variants to maintain repressive epigenetic marks in mouse embryonic stem cells. Nucleic acids research. 2016;44:1118–1132. doi: 10.1093/nar/gkv1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barlow, D. P. & Bartolomei, M. S. Genomic imprinting in mammals. Cold Spring Harbor perspectives in biology6, 10.1101/cshperspect.a018382 (2014). [DOI] [PMC free article] [PubMed]
- 33.Ideraabdullah FY, Bartolomei MS. ZFP57: KAPturing DNA methylation at imprinted loci. Mol Cell. 2011;44:341–342. doi: 10.1016/j.molcel.2011.10.008. [DOI] [PubMed] [Google Scholar]
- 34.Quenneville S, et al. In embryonic stem cells, ZFP57/KAP1 recognize a methylated hexanucleotide to affect chromatin and DNA methylation of imprinting control regions. Mol Cell. 2011;44:361–372. doi: 10.1016/j.molcel.2011.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zvetkova I, et al. Global hypomethylation of the genome in XX embryonic stem cells. Nature genetics. 2005;37:1274–1279. doi: 10.1038/ng1663. [DOI] [PubMed] [Google Scholar]
- 36.Garitaonandia I, et al. Increased risk of genetic and epigenetic instability in human embryonic stem cells associated with specific culture conditions. PloS one. 2015;10:e0118307. doi: 10.1371/journal.pone.0118307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bai Q, et al. Temporal analysis of genome alterations induced by single-cell passaging in human embryonic stem cells. Stem cells and development. 2015;24:653–662. doi: 10.1089/scd.2014.0292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maitra A, et al. Genomic alterations in cultured human embryonic stem cells. Nature genetics. 2005;37:1099–1103. doi: 10.1038/ng1631. [DOI] [PubMed] [Google Scholar]
- 39.Choi J, et al. Prolonged Mek1/2 suppression impairs the developmental potential of embryonic stem cells. Nature. 2017;548:219–223. doi: 10.1038/nature23274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Holm TM, et al. Global loss of imprinting leads to widespread tumorigenesis in adult mice. Cancer cell. 2005;8:275–285. doi: 10.1016/j.ccr.2005.09.007. [DOI] [PubMed] [Google Scholar]
- 41.Zhang T, et al. G9a/GLP Complex Maintains Imprinted DNA Methylation in Embryonic Stem Cells. Cell Rep. 2016;15:77–85. doi: 10.1016/j.celrep.2016.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen T, Ueda Y, Dodge JE, Wang Z, Li E. Establishment and maintenance of genomic methylation patterns in mouse embryonic stem cells by Dnmt3a and Dnmt3b. Molecular and cellular biology. 2003;23:5594–5605. doi: 10.1128/MCB.23.16.5594-5605.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tsumura A, et al. Maintenance of self-renewal ability of mouse embryonic stem cells in the absence of DNA methyltransferases Dnmt1, Dnmt3a and Dnmt3b. Genes Cells. 2006;11:805–814. doi: 10.1111/j.1365-2443.2006.00984.x. [DOI] [PubMed] [Google Scholar]
- 44.Jackson M, et al. Severe global DNA hypomethylation blocks differentiation and induces histone hyperacetylation in embryonic stem cells. Molecular and cellular biology. 2004;24:8862–8871. doi: 10.1128/MCB.24.20.8862-8871.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Karimi MM, et al. DNA methylation and SETDB1/H3K9me3 regulate predominantly distinct sets of genes, retroelements, and chimeric transcripts in mESCs. Cell stem cell. 2011;8:676–687. doi: 10.1016/j.stem.2011.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rowe HM, et al. KAP1 controls endogenous retroviruses in embryonic stem cells. Nature. 2010;463:237–240. doi: 10.1038/nature08674. [DOI] [PubMed] [Google Scholar]
- 47.Cahan P, Daley GQ. Origins and implications of pluripotent stem cell variability and heterogeneity. Nat Rev Mol Cell Biol. 2013;14:357–368. doi: 10.1038/nrm3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chambers I, et al. Nanog safeguards pluripotency and mediates germline development. Nature. 2007;450:1230–1234. doi: 10.1038/nature06403. [DOI] [PubMed] [Google Scholar]
- 49.Godwin S, et al. An extended model for culture-dependent heterogenous gene expression and proliferation dynamics in mouse embryonic stem cells. NPJ Syst Biol Appl. 2017;3:19. doi: 10.1038/s41540-017-0020-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Guo G, et al. Serum-Based Culture Conditions Provoke Gene Expression Variability in Mouse Embryonic Stem Cells as Revealed by Single-Cell Analysis. Cell Rep. 2016;14:956–965. doi: 10.1016/j.celrep.2015.12.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hayashi K, de Sousa Lopes SMC, Tang F, Lao K, Surani MA. Dynamic equilibrium and heterogeneity of mouse pluripotent stem cells with distinct functional and epigenetic states. Cell stem cell. 2008;3:391–401. doi: 10.1016/j.stem.2008.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kumar RM, et al. Deconstructing transcriptional heterogeneity in pluripotent stem cells. Nature. 2014;516:56–61. doi: 10.1038/nature13920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Toyooka Y, Shimosato D, Murakami K, Takahashi K, Niwa H. Identification and characterization of subpopulations in undifferentiated ES cell culture. Development. 2008;135:909–918. doi: 10.1242/dev.017400. [DOI] [PubMed] [Google Scholar]
- 54.Knowles BB, Aden DP, Solter D. Monoclonal antibody detecting a stage-specific embryonic antigen (SSEA-1) on preimplantation mouse embryos and teratocarcinoma cells. Curr Top Microbiol Immunol. 1978;81:51–53. doi: 10.1007/978-3-642-67448-8_8. [DOI] [PubMed] [Google Scholar]
- 55.Redmer T, et al. E-cadherin is crucial for embryonic stem cell pluripotency and can replace OCT4 during somatic cell reprogramming. EMBO Rep. 2011;12:720–726. doi: 10.1038/embor.2011.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Solter D, Knowles BB. Monoclonal antibody defining a stage-specific mouse embryonic antigen (SSEA-1) Proceedings of the National Academy of Sciences of the United States of America. 1978;75:5565–5569. doi: 10.1073/pnas.75.11.5565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Soncin F, Ward CM. The function of e-cadherin in stem cell pluripotency and self-renewal. Genes (Basel) 2011;2:229–259. doi: 10.3390/genes2010229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sottile F, Aulicino F, Theka I, Cosma MP. Mesenchymal stem cells generate distinct functional hybrids in vitro via cell fusion or entosis. Sci Rep. 2016;6:36863. doi: 10.1038/srep36863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhao W, Ji X, Zhang F, Li L, Ma L. Embryonic stem cell markers. Molecules. 2012;17:6196–6236. doi: 10.3390/molecules17066196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Izadpanah R, et al. Long-term in vitro expansion alters the biology of adult mesenchymal stem cells. Cancer Res. 2008;68:4229–4238. doi: 10.1158/0008-5472.CAN-07-5272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.De Jaime-Soguero A, et al. Wnt/Tcf1 pathway restricts embryonic stem cell cycle through activation of the Ink4/Arf locus. PLoS Genet. 2017;13:e1006682. doi: 10.1371/journal.pgen.1006682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.ten Berge D, et al. Wnt signaling mediates self-organization and axis formation in embryoid bodies. Cell stem cell. 2008;3:508–518. doi: 10.1016/j.stem.2008.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kurosawa H. Methods for inducing embryoid body formation: in vitro differentiation system of embryonic stem cells. Journal of bioscience and bioengineering. 2007;103:389–398. doi: 10.1263/jbb.103.389. [DOI] [PubMed] [Google Scholar]
- 64.Ying QL, Stavridis M, Griffiths D, Li M, Smith A. Conversion of embryonic stem cells into neuroectodermal precursors in adherent monoculture. Nat Biotechnol. 2003;21:183–186. doi: 10.1038/nbt780. [DOI] [PubMed] [Google Scholar]
- 65.Xu J, et al. Retinoic acid promotes neural conversion of mouse embryonic stem cells in adherent monoculture. Mol Biol Rep. 2012;39:789–795. doi: 10.1007/s11033-011-0800-8. [DOI] [PubMed] [Google Scholar]
- 66.Meissner A, et al. Reduced representation bisulfite sequencing for comparative high-resolution DNA methylation analysis. Nucleic acids research. 2005;33:5868–5877. doi: 10.1093/nar/gki901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Smith FM, Garfield AS, Ward A. Regulation of growth and metabolism by imprinted genes. Cytogenet Genome Res. 2006;113:279–291. doi: 10.1159/000090843. [DOI] [PubMed] [Google Scholar]
- 68.Tycko B, Morison IM. Physiological functions of imprinted genes. J Cell Physiol. 2002;192:245–258. doi: 10.1002/jcp.10129. [DOI] [PubMed] [Google Scholar]
- 69.Mao J, et al. Low-density lipoprotein receptor-related protein-5 binds to Axin and regulates the canonical Wnt signaling pathway. Mol Cell. 2001;7:801–809. doi: 10.1016/S1097-2765(01)00224-6. [DOI] [PubMed] [Google Scholar]
- 70.Zeng X, et al. Initiation of Wnt signaling: control of Wnt coreceptor Lrp6 phosphorylation/activation via frizzled, dishevelled and axin functions. Development. 2008;135:367–375. doi: 10.1242/dev.013540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fuerer C, Nusse R. Lentiviral vectors to probe and manipulate the Wnt signaling pathway. PloS one. 2010;5:e9370. doi: 10.1371/journal.pone.0009370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Faunes F, et al. A membrane-associated beta-catenin/Oct4 complex correlates with ground-state pluripotency in mouse embryonic stem cells. Development. 2013;140:1171–1183. doi: 10.1242/dev.085654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang X, Peterson KA, Liu XS, McMahon AP, Ohba S. Gene regulatory networks mediating canonical Wnt signal-directed control of pluripotency and differentiation in embryo stem cells. Stem Cells. 2013;31:2667–2679. doi: 10.1002/stem.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Consortium EP. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mouse Genome Sequencing C, et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 76.Stocking C, Kozak CA. Murine endogenous retroviruses. Cell Mol Life Sci. 2008;65:3383–3398. doi: 10.1007/s00018-008-8497-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Guallar D, et al. Expression of endogenous retroviruses is negatively regulated by the pluripotency marker Rex1/Zfp42. Nucleic acids research. 2012;40:8993–9007. doi: 10.1093/nar/gks686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Maksakova IA, Mager DL, Reiss D. Keeping active endogenous retroviral-like elements in check: the epigenetic perspective. Cell Mol Life Sci. 2008;65:3329–3347. doi: 10.1007/s00018-008-8494-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rowe HM, et al. TRIM28 repression of retrotransposon-based enhancers is necessary to preserve transcriptional dynamics in embryonic stem cells. Genome Res. 2013;23:452–461. doi: 10.1101/gr.147678.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rowe HM, Trono D. Dynamic control of endogenous retroviruses during development. Virology. 2011;411:273–287. doi: 10.1016/j.virol.2010.12.007. [DOI] [PubMed] [Google Scholar]
- 81.Rebollo R, et al. Retrotransposon-induced heterochromatin spreading in the mouse revealed by insertional polymorphisms. PLoS Genet. 2011;7:e1002301. doi: 10.1371/journal.pgen.1002301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Groh S, Schotta G. Silencing of endogenous retroviruses by heterochromatin. Cell Mol Life Sci. 2017;74:2055–2065. doi: 10.1007/s00018-017-2454-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Aulicino F, Theka I, Ombrato L, Lluis F, Cosma MP. Temporal perturbation of the Wnt signaling pathway in the control of cell reprogramming is modulated by TCF1. Stem Cell Reports. 2014;2:707–720. doi: 10.1016/j.stemcr.2014.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Theka I, Sottile F, Aulicino F, Garcia AC, Cosma MP. Reduced expression of Paternally Expressed Gene-3 enhances somatic cell reprogramming through mitochondrial activity perturbation. Sci Rep. 2017;7:9705. doi: 10.1038/s41598-017-10016-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Okumura N, et al. beta-catenin functions pleiotropically in differentiation and tumorigenesis in mouse embryo-derived stem cells. PloS one. 2013;8:e63265. doi: 10.1371/journal.pone.0063265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Smith AG, et al. Inhibition of pluripotential embryonic stem cell differentiation by purified polypeptides. Nature. 1988;336:688–690. doi: 10.1038/336688a0. [DOI] [PubMed] [Google Scholar]
- 87.Williams RL, et al. Myeloid leukaemia inhibitory factor maintains the developmental potential of embryonic stem cells. Nature. 1988;336:684–687. doi: 10.1038/336684a0. [DOI] [PubMed] [Google Scholar]
- 88.Niwa H. How is pluripotency determined and maintained? Development. 2007;134:635–646. doi: 10.1242/dev.02787. [DOI] [PubMed] [Google Scholar]
- 89.Nichols J, Smith A. Naive and primed pluripotent states. Cell stem cell. 2009;4:487–492. doi: 10.1016/j.stem.2009.05.015. [DOI] [PubMed] [Google Scholar]
- 90.Ying QL, et al. The ground state of embryonic stem cell self-renewal. Nature. 2008;453:519–523. doi: 10.1038/nature06968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Takada S, et al. Wnt-3a regulates somite and tailbud formation in the mouse embryo. Genes & development. 1994;8:174–189. doi: 10.1101/gad.8.2.174. [DOI] [PubMed] [Google Scholar]
- 92.Tosolini M, Jouneau A. From Naive to Primed Pluripotency: In Vitro Conversion of Mouse Embryonic Stem Cells in Epiblast Stem Cells. Methods Mol Biol. 2016;1341:209–216. doi: 10.1007/7651_2015_208. [DOI] [PubMed] [Google Scholar]
- 93.Tesar PJ, et al. New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature. 2007;448:196–199. doi: 10.1038/nature05972. [DOI] [PubMed] [Google Scholar]
- 94.Jackson-Grusby L, et al. Loss of genomic methylation causes p53-dependent apoptosis and epigenetic deregulation. Nature genetics. 2001;27:31–39. doi: 10.1038/83730. [DOI] [PubMed] [Google Scholar]
- 95.Lei H, et al. De novo DNA cytosine methyltransferase activities in mouse embryonic stem cells. Development. 1996;122:3195–3205. doi: 10.1242/dev.122.10.3195. [DOI] [PubMed] [Google Scholar]
- 96.Takebayashi S, Tamura T, Matsuoka C, Okano M. Major and essential role for the DNA methylation mark in mouse embryogenesis and stable association of DNMT1 with newly replicated regions. Molecular and cellular biology. 2007;27:8243–8258. doi: 10.1128/MCB.00899-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Tezuka N, Brown AM, Yanagawa S. GRB10 binds to LRP6, the Wnt co-receptor and inhibits canonical Wnt signaling pathway. Biochemical and biophysical research communications. 2007;356:648–654. doi: 10.1016/j.bbrc.2007.03.019. [DOI] [PubMed] [Google Scholar]
- 98.Lu L, et al. Kdm2a/b Lysine Demethylases Regulate Canonical Wnt Signaling by Modulating the Stability of Nuclear beta-Catenin. Dev Cell. 2015;33:660–674. doi: 10.1016/j.devcel.2015.04.006. [DOI] [PubMed] [Google Scholar]
- 99.Li X, et al. Tet proteins influence the balance between neuroectodermal and mesodermal fate choice by inhibiting Wnt signaling. Proceedings of the National Academy of Sciences of the United States of America. 2016;113:E8267–E8276. doi: 10.1073/pnas.1617802113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Mah KM, Houston DW, Weiner JA. The gamma-Protocadherin-C3 isoform inhibits canonical Wnt signalling by binding to and stabilizing Axin1 at the membrane. Sci Rep. 2016;6:31665. doi: 10.1038/srep31665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Jiang Q, et al. MicroRNA-148a inhibits breast cancer migration and invasion by directly targeting WNT-1. Oncol Rep. 2016;35:1425–1432. doi: 10.3892/or.2015.4502. [DOI] [PubMed] [Google Scholar]
- 102.Ju X, et al. Context-dependent activation of Wnt signaling by tumor suppressor RUNX3 in gastric cancer cells. Cancer Sci. 2014;105:418–424. doi: 10.1111/cas.12356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Alves dos Santos MT, Smidt MP. En1 and Wnt signaling in midbrain dopaminergic neuronal development. Neural Dev. 2011;6:23. doi: 10.1186/1749-8104-6-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Nouri N, et al. Excessive Wnt/beta-catenin signaling promotes midbrain floor plate neurogenesis, but results in vacillating dopamine progenitors. Mol Cell Neurosci. 2015;68:131–142. doi: 10.1016/j.mcn.2015.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Huang J, et al. EphA2 promotes epithelial-mesenchymal transition through the Wnt/beta-catenin pathway in gastric cancer cells. Oncogene. 2014;33:2737–2747. doi: 10.1038/onc.2013.238. [DOI] [PubMed] [Google Scholar]
- 106.Wu L, Amarachintha S, Xu J, Oley F, Jr., Du W. Mesenchymal COX2-PG secretome engages NR4A-WNT signalling axis in haematopoietic progenitors to suppress anti-leukaemia immunity. Br J Haematol. 2018 doi: 10.1111/bjh.15548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zhou F, et al. Nuclear receptor NR4A1 promotes breast cancer invasion and metastasis by activating TGF-beta signalling. Nature communications. 2014;5:3388. doi: 10.1038/ncomms4388. [DOI] [PubMed] [Google Scholar]
- 108.Hutnick LK, Huang X, Loo TC, Ma Z, Fan G. Repression of retrotransposal elements in mouse embryonic stem cells is primarily mediated by a DNA methylation-independent mechanism. J Biol Chem. 2010;285:21082–21091. doi: 10.1074/jbc.M110.125674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Yoder JA, Walsh CP, Bestor TH. Cytosine methylation and the ecology of intragenomic parasites. Trends Genet. 1997;13:335–340. doi: 10.1016/S0168-9525(97)01181-5. [DOI] [PubMed] [Google Scholar]
- 110.Diaz-Meyer N, et al. Silencing of CDKN1C (p57KIP2) is associated with hypomethylation at KvDMR1 in Beckwith-Wiedemann syndrome. J Med Genet. 2003;40:797–801. doi: 10.1136/jmg.40.11.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Allen RG, Tresini M. Oxidative stress and gene regulation. Free Radic Biol Med. 2000;28:463–499. doi: 10.1016/S0891-5849(99)00242-7. [DOI] [PubMed] [Google Scholar]
- 112.Tower J. Stress and stem cells. Wiley Interdiscip Rev Dev Biol. 2012;1:789–802. doi: 10.1002/wdev.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Gu Y, et al. Changes in mesenchymal stem cells following long-term culture in vitro. Mol Med Rep. 2016;13:5207–5215. doi: 10.3892/mmr.2016.5169. [DOI] [PubMed] [Google Scholar]
- 114.Halliwell B. Oxidative stress in cell culture: an under-appreciated problem? FEBS letters. 2003;540:3–6. doi: 10.1016/S0014-5793(03)00235-7. [DOI] [PubMed] [Google Scholar]
- 115.Almeida M, Han L, Martin-Millan M, O’Brien CA, Manolagas SC. Oxidative stress antagonizes Wnt signaling in osteoblast precursors by diverting beta-catenin from T cell factor- to forkhead box O-mediated transcription. J Biol Chem. 2007;282:27298–27305. doi: 10.1074/jbc.M702811200. [DOI] [PubMed] [Google Scholar]
- 116.Hoogeboom D, et al. Interaction of FOXO with beta-catenin inhibits beta-catenin/T cell factor activity. J Biol Chem. 2008;283:9224–9230. doi: 10.1074/jbc.M706638200. [DOI] [PubMed] [Google Scholar]
- 117.Iyer S, et al. FOXOs attenuate bone formation by suppressing Wnt signaling. J Clin Invest. 2013;123:3409–3419. doi: 10.1172/JCI68049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Florian MC, et al. A canonical to non-canonical Wnt signalling switch in haematopoietic stem-cell ageing. Nature. 2013;503:392–396. doi: 10.1038/nature12631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Oittinen, M. et al. PRC2 Enacts Wnt Signaling in Intestinal Homeostasis and Contributes to the Instigation of Stemness in Diseases Entailing Epithelial Hyperplasia or Neoplasia. Stem Cells, 10.1002/stem.2479 (2016). [DOI] [PubMed]
- 120.Song J, et al. A Protein Interaction between beta-Catenin and Dnmt1 Regulates Wnt Signaling and DNA Methylation in Colorectal Cancer Cells. Mol Cancer Res. 2015;13:969–981. doi: 10.1158/1541-7786.MCR-13-0644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Yakulov T, Raggioli A, Franz H, Kemler R. Wnt3a-dependent and -independent protein interaction networks of chromatin-bound beta-catenin in mouse embryonic stem cells. Mol Cell Proteomics. 2013;12:1980–1994. doi: 10.1074/mcp.M112.026914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Pereira L, Yi F, Merrill BJ. Repression of Nanog gene transcription by Tcf3 limits embryonic stem cell self-renewal. Molecular and cellular biology. 2006;26:7479–7491. doi: 10.1128/MCB.00368-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Xiong Z, Laird PW. COBRA: a sensitive and quantitative DNA methylation assay. Nucleic acids research. 1997;25:2532–2534. doi: 10.1093/nar/25.12.2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Krueger F, Andrews SR. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics. 2011;27:1571–1572. doi: 10.1093/bioinformatics/btr167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Akalin A, et al. methylKit: a comprehensive R package for the analysis of genome-wide DNA methylation profiles. Genome biology. 2012;13:R87. doi: 10.1186/gb-2012-13-10-r87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Heinz S, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Wickham, H. ggplot2: Elegant Graphics for Data Analysis. Springer-Verlag New YorkVIII, 213, 10.1007/978-0-387-98141-3 (2009).
- 129.Zhang Z. Reshaping and aggregating data: an introduction to reshape package. Ann Transl Med. 2016;4:78. doi: 10.3978/j.issn.2305-5839.2016.01.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Chen H, Boutros PC. VennDiagram: a package for the generation of highly-customizable Venn and Euler diagrams in R. BMC Bioinformatics. 2011;12:35. doi: 10.1186/1471-2105-12-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Conway JR, Lex A, Gehlenborg N. UpSetR: an R package for the visualization of intersecting sets and their properties. Bioinformatics. 2017;33:2938–2940. doi: 10.1093/bioinformatics/btx364. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.