Abstract
Skeletal muscle macrophages participate in repair and regeneration following injury. However, their role in physiological adaptations to exercise is unexplored. We determined whether endurance exercise training (EET) alters macrophage content and characteristics in response to resistance exercise (RE), and whether macrophages are associated with other exercise adaptations. Subjects provided vastus lateralis biopsies before and after one bout of RE, after 12 weeks of EET (cycling), and after a final bout of RE. M2 macrophages (CD11b+/CD206+) did not increase with RE, but increased in response to EET (P < 0.01). Increases in M2 macrophages were positively correlated with fiber hypertrophy (r = 0.49) and satellite cells (r = 0.47). M2c macrophages (CD206+/CD163+) also increased following EET (P < 0.001), and were associated with fiber hypertrophy (r = 0.64). Gene expression was quantified using NanoString. Following EET, the change in M2 macrophages was positively associated with changes in HGF, IGF1, and extracellular matrix genes. EET decreased expression of IL6 (P < 0.05), C/EBPβ (P < 0.01), and MuRF (P < 0.05), and increased expression of IL-4 (P < 0.01), TNFα (P < 0.01) and the TWEAK receptor FN14 (P < 0.05). The change in FN14 gene expression was inversely associated with changes in C/EBPβ (r = −0.58) and MuRF (r = −0.46) following EET. In cultured human myotubes, siRNA inhibition of FN14 increased expression of C/EBPβ (P < 0.05) and MuRF (P < 0.05). Our data suggest that macrophages contribute to the muscle response to EET, potentially including modulation of TWEAK-FN14 signaling.
Introduction
Both endurance and resistance exercise promote maintenance of muscle mass and function1,2. Understanding how exercise exerts beneficial effects could provide strategies for mimicking or improving exercise responses. Macrophages participate in muscle repair and regeneration by modulating inflammation, stem cells, cytokines, growth factors, and extracellular matrix. However, their role in the physiological adaptation to exercise is relatively unexplored. Macrophages exhibit phenotypic variability and plasticity, occupying a spectrum from “M1” (inflammatory) to “M2” (anti-inflammatory)3.
Macrophages exert effects on myogenic stem cells, satellite cells (SCs), which give rise to myogenic progenitor cells (MPCs). SCs and MPCs express monocyte chemoattractants4, and macrophages promote in vitro MPC proliferation and differentiation5,6. In damaged muscle, M1 macrophages produce inflammatory cytokines (TNFα, IL1β) that signal through canonical NFκB and other pathways to promote SC proliferation7,8. In later stages of repair, macrophages shift toward M2 activation, and produce anti-inflammatory cytokines (TGFβ, IL10)9,10, driving non-canonical NFκB signaling11 and promoting MPC differentiation. Muscle macrophages also produce growth factors, including HGF12, which promote SC activation and proliferation13. Macrophage depletion impairs recovery from muscle damage caused by contusion14, unloading15 or neurotoxin delivery9.
Macrophages may participate in the regulation of muscle mass by helping to balance catabolic and anabolic signaling. Macrophage-derived inflammatory cytokines, including IL1β, TNFα, and TWEAK, are observed in various disease states, and drive muscle atrophy via canonical NFκB signaling. Canonical NFκB then drives transcription of MuRF, a muscle-specific E3 ubiquitin ligase, leading to protein degradation7,8,16. On the other hand, non-canonical NFκB signaling promotes PGC1α production and mitochondrial biogenesis11,17. Additionally, M2 macrophages produce IGF1, which supports damage repair, protein synthesis, and maintenance of muscle mass15,18–20.
While the macrophage response to muscle injury is well described, their role in adaptation to exercise is largely unstudied. It is clear that the macrophage response to damaging exercise mimics the response to injury, as evidenced by macrophage infiltration following forced lengthening contractions21, electrical stimulation22, downhill running23, and synergist ablation surgery24. Macrophage regulation of ECM remodeling is documented in lung25, liver26, and kidney27, where mechanistic studies have shown macrophage regulation of fibroblasts and pericytes. Similar pathways are likely operative in muscle since repair and hypertrophy require ECM remodeling. Accordingly, ECM and M2 macrophage genes are concurrently up-regulated following resistance exercise (RE) and/or endurance exercise training (EET) in humans28,29. However, EET caused decreased muscle macrophage content in mice30, and did not affect muscle macrophage content in rats31.
Muscle macrophage function may be impaired with obesity, aging, and sedentary lifestyle. Our group previously reported higher CD68+ muscle macrophages in obese versus lean humans32. We also reported decreased macrophage content, and blunted macrophage response to acute RE, in old versus young men33. We and others have shown increased SCs following EET in humans34–36. In middle aged women (a subset of this cohort), we have further shown that EET modulates the transcriptional and SC response to RE37. We therefore sought to determine whether increased M2 macrophages would be correlated to increased SC content and transcriptional alterations following EET. Since EET alleviates some of the muscle deficits associated with a sedentary lifestyle, we further hypothesized that increasing physical activity via EET would alter the macrophage response to acute RE in humans. Toward this end, we collected muscle from human vastus lateralis at baseline, following a single bout of RE, following 12 weeks of EET, and again after a final bout of RE. Immunohistochemistry was used to assess muscle macrophage and SCs and muscle fiber size, and the NanoString nCounter system was used to measure transcription of related genes at each time point.
Methods
Human subjects
All procedures were performed according to the principles set forth in the Declaration of Helsinki, and approved by the Institutional Review Board of the University of Kentucky. All research participants were informed of the purpose and design of the study, and they provided written consent prior to participation. Exclusion criteria included: smoking, coronary disease, congestive heart failure, chronic inflammatory diseases, or orthopedic problems that could limit the ability to perform the exercise protocols. Participants were sedentary and were asked to maintain consistent dietary and lifestyle habits throughout the study. The cohort includes subjects with a wide range of insulin sensitivity (determined by frequently sampled intravenous glucose tolerance testing, previously described38), age, BMI, and aerobic fitness (N = 23, Table 1). Using a small subset (N = 7) of this study cohort, we have previously reported that satellite cell and transcriptional responses to RE were modulated by EET37. In this cohort, we have also reported on satellite cell proliferation, muscle fiber hypertrophy (N = 23)39 and angiogenesis (N = 26)38 in response to EET. In this report, we investigate changes in M2 macrophage response to RE and the effects of EET on that response. We further assess relationships among fiber hypertrophy, muscle macrophages, and gene expression. Figure 1 provides an overview of the study design.
Table 1.
Characteristics of study participants.
| Variable | Mean ± SEM (range) |
|---|---|
| Immunohistochemstry using all four biopsy time points | |
| N = 14, 78% female | |
| Age | 49.5 ± 3.2 (29–68) |
| BMI | 30.8 ± 1.3 (24.1–39.25) |
| VO2 max | 27.5 ± 2.7 (13.8–53.2) |
| SI | 3.26 ± 0.53 (0.92–6.81) |
| Immunohistochemstry using baseline and post-EET biopsies | |
| N = 23, 78% female | |
| Age | 47.5 ± 2.7 (26–68) |
| BMI | 30.8 ± 1.2 (23.3–41.8) |
| VO2 max | 27.8 ± 2.0 (13.8–53.2) |
| SI | 3.21 ± 0.42 (0.92–7.12) |
| NanoString mRNA | |
| N = 20, 75% female | |
| Age | 49.8 ± 2.3 (29–64) |
| BMI | 31.5 ± 1.2 (24.3–41.8) |
| VO2 max | 27.1 ± 1.8 (13.8–53.2) |
| SI | 3.04 ± 0.48 (0.65–7.12) |
Figure 1.
Overview of the study design. At baseline, subjects underwent a vastus lateralis biopsy, as well as a fitness assessment by graded exercise test for VO2max. Subjects next performed one bout of damaging RE, with a second muscle biopsy taken 3 days later. Subjects then underwent 12 weeks of EET on a cycle ergometer. Exercise intensity was gradually increased and rest periods were gradually decreased so that subjects exercised non-stop for 45 minutes at a heart rate corresponding to approximately 65% of VO2max throughout the training period. At the end of the EET protocol, VO2max was measured, and a third muscle biopsy was obtained 3 days after the final endurance bout. Participants performed one more bout of RE, with a fourth muscle biopsy obtained 3 days later.
Biopsies
At baseline, we obtained the first of four vastus lateralis biopsies using a 5 mm Bergstrom needle with suction. For immunohistochemistry, approximately 100 mg of each biopsy was mounted with fibers perpendicular to a cork using tragacanth gum and snap frozen in liquid nitrogen-cooled isopentane. For isolation of primary myogenic precursor cells (MPCs), approximately 75 mg of tissue was placed in phosphate buffered saline with 1 g/L glucose and penicillin/streptomycin overnight prior to tissue dissociation. For gene expression, the remaining portion of each biopsy was snap-frozen in liquid nitrogen.
Graded exercise testing
Subjects next performed maximal graded exercise testing (GXT), with assessment of VO2max and integrated electrocardiogram on a calibrated exercise bicycle ergometer. Subjects maintained a pedaling rate of 60–70 rpm, with a workload intensity beginning at 20 watts and increasing by 20 watts every 2 minutes until VO2max was achieved. Initial resistance was based on participant fitness. Continuous measures of oxygen consumption and CO2 production were obtained using the Vmax 229 system (Viasys Healthcare, Yorba Linda, CA). Respiratory exchange ratio, heart rate, blood pressure, and rate of perceived exertion were recorded in the final 30 seconds of each work watts stage.
Resistance exercise
Subjects performed an acute bout of RE using the leg contralateral to the baseline biopsy leg. RE consisted of 5 sets of 8 repetitions of leg extensions at 80% of 1 repetition max, with a sixth set performed to exhaustion. Seventy-two hours after RE, a second muscle biopsy was obtained from the exercised leg.
Endurance exercise training (EET)
Participants underwent 12 weeks of EET using a stationary cycle ergometer (Monark 828E Ergometer Upright Bike), and a target intensity corresponding to 65% of VO2max and approximately 75–80% of maximum heart rate, as determined by baseline maximal GXT measures. Training intensity was monitored using Polar A3 heart rate monitors (Polar Electro Inc., Woodbury, NY). Subjects were required to exercise three days/week for 45 minutes and were allowed to take intermittent breaks if they were unable to maintain constant exercise for the entire session. Throughout the training period, exercise intensity was gradually increased and rest periods were gradually decreased so that during the eighth through twelfth weeks of training, subjects were able to exercise for 45 minutes consecutively without rest at a heart rate corresponding to 65% of VO2max. Average power output per workout was calculated in excel [Power output = (work/time)/6.12].
Following 12 weeks of EET, subjects underwent a third muscle biopsy. In order to detect chronic adaptations to EET, biopsy 3 was performed 72 hours after the last bout of exercise. Next, subjects repeated the RE protocol that was performed at the beginning of the study, and a fourth muscle biopsy was obtained 72 hours later.
Immunohistochemistry (IHC)
We performed IHC on vastus lateralis from 23 subjects, chosen based on the availability and quality of muscle mounts. All staining was performed on 7 µm sections of frozen muscle. Human muscle macrophage IHC has been described in detail and validated in our previous publication40. Primary macrophage antibodies include: mouse anti-human CD11b/CR3, clone Bear-1 (Cell Sciences, MON1019-1), mouse anti-human CD163 (Hycult Biotech, HM2157), and goat anti-Human MMR/CD206 (R&D, AF2534). Secondary antibodies include: biotinylated goat anti-mouse IgG1 (Jackson ImmunoResearch, 115-065-205) and biotinylated rabbit anti-goat IgG (Vector Laboratories, BA-5000). Thermo Scientific AlexaFlour systems were used to add fluorescent tags to secondary antibodies. IHC images were obtained using 20× magnification with a Zeiss AxioImager M1 upright microscope and analyzed using Zen Lite software (Carl Zeiss AG, Oberkochen, Germany). Macrophages, fibers, and fiber size were analyzed manually. Macrophages were counted as M2 if they co-stained for CD11b and CD206. For M2 macrophage counting, the smallest section contained 123 cross-sectional fibers and the mean fiber number was 275 ± 15.9 SEM. M2c macrophages were identified by co-staining for CD206 and CD163. For M2c macrophage counting, the smallest section contained 91 cross-sectional fibers and the mean fiber number was 256 ± 22.3 SEM. Macrophage counts were normalized to fiber number. Muscle fiber size was assessed by quantifying cross sectional area (CSA) using IHC against laminin (Abcam, AB14055) to distinguish fiber borders. Satellite cell content was determined using IHC against Pax7 (DSHB), as described in our previous publication39.
Human primary myotube culture, FN14 knock-down, and FN14 overexpression
Human vastus lateralis tissue was finely minced in PBS with 1 g/L glucose, dispase II (2.4 U/ml; Roche Applied Science), and collagenase D (1 mg/ml; Sigma-Aldrich). Tissue was then incubated at 37 °C for 60 minutes with gentle rotation, then passed through a 40 μm cell strainer. Cells were washed and pre-plated in in growth medium (HAM’s F-10, 20% fetal bovine serum and 5 ng/ml basic fibroblast growth factor) to allow adherent cells (fibroblasts) to attach. Media containing non-adherent cells (MPCs) was gently moved to a fresh culture dish. MPCs were passaged in growth medium approximately 4 times, allowed to become approximately 90% confluent, then differentiated into myotubes in DMEM with 1 g/L glucose and 2.5% fetal bovine serum. On the fifth day of differentiation, cells were treated with 90 pM scramble RNA or small interfering RNA targeted against FN14 (siFN14) (Invitrogen Stealth RNAi siRNA, ThermoFisher), delivered using Lipofectamine RNAiMAX Reagent (ThermoFisher) according to the manufacturer’s protocol. Forty eight hours after siFN14 treatment, cells were harvested for RNA extraction. For FN14 overexpression, human myotubes were differentiated for 5 days, and then transfected with 500 ng/mL of pCMVScript control (vector) or pCMVScript encoding the full length human FN14 (FN14-FL)41. Myotubes were transfected for 24 hours using Lipofectamine 3000 reagent (ThermoFisher), and then harvested for RNA.
Gene expression
RNA extraction
Approximately 30–40 mg of frozen vastus lateralis was homogenized in QIAzol Lysis Reagent (QIAGEN, Hilden, Germany, 79306). To obtain RNA from cultured myotubes, cells were washed with PBS and QIAzol was added. Cells were then scraped off of culture dishes and submitted to brief bead homogenization. For both biopsies and cells, RNA was precipitated and washed using the RNeasy kit (QIAGEN, 74104). RNA quantity and quality was assessed using the Agilent 2100 Bioanalyser (Agilent Technologies, Santa Clara, CA).
NanoString nCounter
In human muscle biopsies, all genes except TWEAK were measured using the nCounter analysis system (NanoString Technologies, Seattle, WA)42–44. The nCounter system allowed us to design a hypothesis-driven custom probe set containing 6 housekeeping genes and 102 genes of interest related to muscle hypertrophy, extracellular matrix remodeling, macrophages, and inflammation37. The custom code set was then hybridized with 100 ng of RNA from each biopsy. Gene expression was normalized to the geometric mean of six housekeeping genes (β-actin, Cyclophilin A, Cyclophilin B, TATA binding protein, Tubulin-β, and Ubiquitin C), and the mean of eight negative controls was subtracted. The data are presented as normalized counts. Eight genes in the code set were not detectable, leaving us with 94 genes available for analysis. Subjects used for NanoString analysis were the first 20 subjects who completed the study and had sufficient RNA quality (RIN ≥ 7.0) at each time point.
Real-time PCR
Real-time PCR was used to measure changes in gene expression in cultured human myotubes following FN14 knock-down, as well as TWEAK expression in muscle biopsies (TWEAK was not included in the NanoString code set). RNA was reverse transcribed with SuperScript™ IV VILO™ (ThermoFisher) and quantitative real-time rtPCR was performed using PowerUp™ SYBR™ Green Real-Time PCR Master Mix (ThermoFisher). Gene expression was calculated using the 2^ΔΔCt method, with genes of interest normalized against the geometric mean of three housekeeping genes (18S RNA, β2 microglobulin (B2M), and phosphoglycerate kinase (PGK)). Primer sequences are given in Supplementary Table 1.
Statistics
All data are expressed as mean ± SEM. Significance was predetermined to be P ≤ 0.05. Exact P values for biologically relevant trends are reported. 2-way ANOVA was used to determine whether RE or EET affected skeletal muscle macrophage content, with RE, EET, and RE x EET included in the model as independent variables. Paired t-tests were used to determine the effects of endurance training (biopsy 1 versus biopsy 3) on gene expression and macrophages/fiber. Paired t-tests were also used to determine the transcriptional effects of FN14 knock-down and overexpression in cultured myotubes. Linear regressions employed the Pearson product-moment correlation coefficient when the two continuous variables were normally distributed (including all figures depicting regressions). Because we designed a hypothesis-driven custom code set, NanoString gene expression analyses were not corrected for multiple comparisons. Statistical analyses were performed with JMP v. 12 (SAS Institute, Cary, NC).
Results
Baseline subject characteristics are provided in Table 1. The study consisted of mostly women with a broad range in age (29–68 years), BMI (23.3–41.8) and fitness (VO2max 13.8–53.2). Menopause status and medications are provided in Supplementary Table 2. All four biopsies were available for macrophage immunohistochemistry (IHC) from 14 subjects, with baseline and post-EET biopsies available for 23 of the 26 subjects who completed the trial. RNA was isolated for NanoString gene expression analyses on biopsies from baseline and post EET from 20 subjects. Both IHC and NanoString results were available at baseline and post EET on biopsies from 17 subjects. There were no statistically significant differences between the subgroups of subjects used for differing analyses (Table 1).
M2 macrophages increase following EET and are associated with fiber hypertrophy and satellite cells
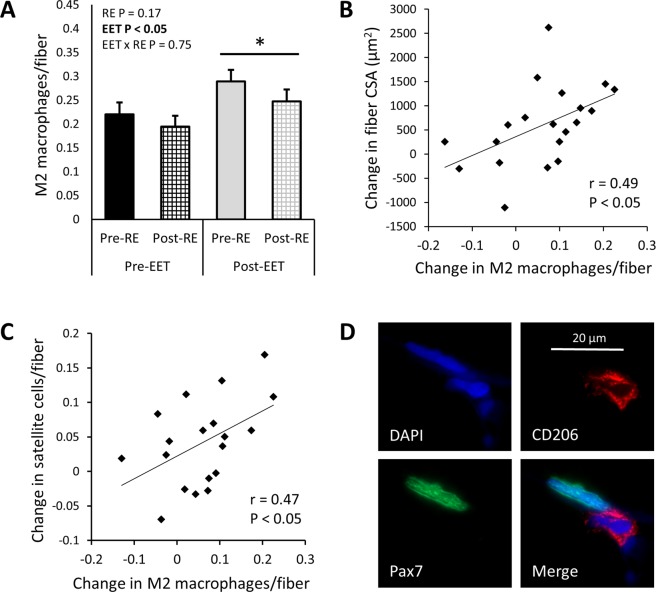
Since macrophages are known to participate in the response to muscle injury45,46, we hypothesized that a single bout of high intensity RE in sedentary humans would induce muscle macrophage infiltration within 3 days. We further hypothesized that EET in the sedentary participants would modify the subsequent response to RE. We therefore performed IHC staining for CD11b (a pan macrophage marker) and CD206 (an M2 macrophage marker) on muscle biopsies taken at baseline, 3 days after RE, 3 days after the last EET session, and 3 days after a second bout of RE (Fig. 1 shows an overview of the study design). Representative macrophage staining is shown in Fig. 2. The M2 (CD11b+/CD206+) macrophage response to the exercise protocols is shown in Fig. 3A. Contrary to our original hypothesis and differing from our findings in a previous cohort33, macrophages/fiber were not increased 3 days after performing RE (leg extensions) to exhaustion (RE P = 0.17; Fig. 3A). However, 12 weeks cycle ergometer EET elicited a significant increase in M2 macrophages/fiber (EET P < 0.05; Fig. 3A), and EET did not alter macrophage numbers in response to a final bout of RE (RE x EET P = 0.75; Fig. 3A). We were able to perform IHC for M2 macrophages in 9 additional subjects with available biopsies from baseline and following EET, which confirmed the significant increase in M2 macrophages following EET (P < 0.01, N = 23; Supplementary Fig. 1). Finally, we counted cells that were CD11b+/CD206−. In human skeletal muscle, these cells are rare and may constitute an M1-like population. However we did not observe sufficient numbers of these cells to perform statistical analyses.
Figure 2.
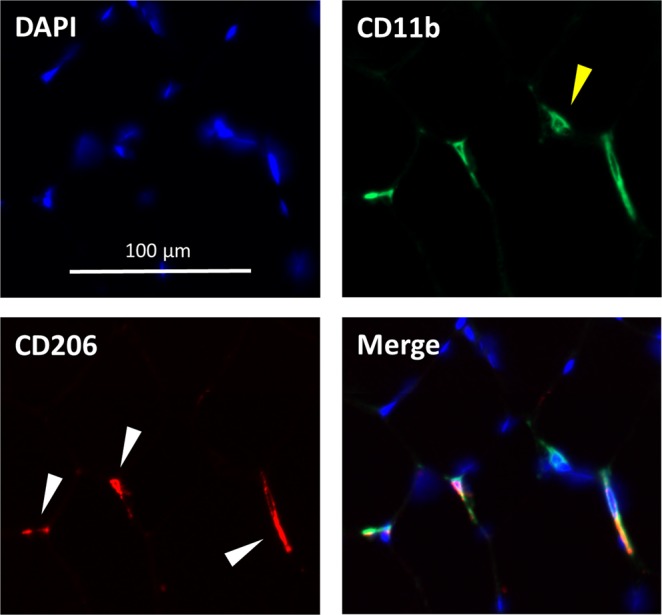
Representative skeletal muscle macrophage immunohistochemistry. M2 macrophages were identified via co-stain for CD11b and CD206. Blue DAPI stain identifies nuclei. Green indicates CD11b, used as a pan-macrophage marker. Yellow carrot indicates a CD11b+/CD206- macrophage. Red indicates CD206, used to identify M2 macrophages. White carrots indicate cells that express both markers (CD11b+/CD206+). A merged image shows CD11b, CD206, and DAPI.
Figure 3.
In human vastus lateralis, macrophages increase following 12 weeks EET and are associated with muscle fiber hypertrophy. (A) M2 (CD11b+/CD206+) macrophages do not increase 3 days after a single bout of RE. However, M2 macrophages are increased following EET (P < 0.05) and EET does not modulate the macrophage response 3 days after a final bout of RE (RE x EET NS, N = 14, 2-way ANOVA). (B) Following EET, the increase in macrophages per fiber is associated with muscle fiber hypertrophy, assessed by change in fiber cross-sectional area (CSA) (r = 0.49, P < 0.05, N = 20). (C) The change in M2 macrophages per fiber is also positively associated with increased satellite cells per fiber (assessed by Pax7 IHC) following EET (r = 0.47, P < 0.05, N = 18). (D) M2 macrophages are sometimes observed in close proximity to Pax7+ satellite cells, as in this 100x z-stack image.
In this cohort, we previously reported muscle fiber hypertrophy (assessed by fiber cross -sectional area (CSA) using IHC for laminin) and increased satellite cell content (assessed by IHC for Pax7) following EET39. Because macrophages are known to participate in tissue remodeling47, we investigated whether the EET-induced increase in macrophages was associated with increased fiber size and satellite cells. Indeed, the change in M2 macrophages/fiber was positively associated with the change in fiber CSA following EET (r = 0.49, P < 0.05; Fig. 3B). Additionally, the change in M2 macrophages/fiber was also positively associated with the change in satellite cells/fiber resulting from EET (r = 0.47, P < 0.05; Fig. 3C). On six tissue sections, we performed IHC for both Pax7 and CD206, and we observed several instances of M2 macrophages in close proximity to satellite cells (Fig. 3D). Although most participants showed gains in average power output per 45 minute workout (watts) following EET (P < 0.01; Supplementary Fig. 2), there was no relationship between gains in average power and increased muscle macrophage content. Similarly, most participants showed increased VO2max following EET (P < 0.01; Supplementary Fig. 3), but changes in VO2max were not correlated with changes in muscle macrophage content. Moreover, there were no relationships among age, BMI, or change in BMI and baseline M2 macrophages or change in M2 macrophages with EET.
Changes in M2 macrophages are associated with changes in gene expression in response to EET
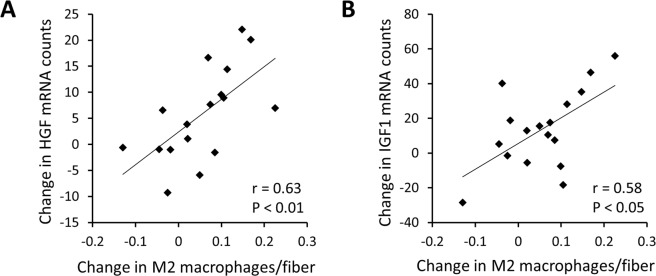
Using a hypothesis-driven, custom-designed NanoString code set, we measured expression of genes related to macrophages/inflammation, growth and ECM expansion, at baseline and following EET. Changes in M2 macrophage abundance, assessed by IHC, were significantly correlated to changes in CD11b (ITGAM gene) and CD206 (MRC1 gene) mRNA counts, as well as other macrophage markers (Table 2). Consistent with the observation that macrophages are associated with fiber hypertrophy and satellite cell number, the training-induced increase in macrophages was also positively associated with increased transcription of two growth factor genes: HGF, which is up-regulated during surgical overload-induced hypertrophy48,49 (r = 0.63, P < 0.01; Fig. 4A), and IGF1 (isoform 4; class IA), whose splice variants are known to drive various stages of the hypertrophic response to exercise50,51 (r = 0.58, P < 0.05; Fig. 4B). Furthermore, the change in M2 macrophages following EET was positively associated with changes in numerous extracellular matrix related genes (Table 2). Other growth, ECM and inflammation genes that were not correlated to changes in macrophages are shown in Supplementary Table 3. Several genes whose changes were significantly correlated to the change in M2 macrophage number were also significantly altered by EET, including: HGF, IGF1, CD11b (ITGAM), SPARC (osteonectin), LOX (lysyl oxidase), COL6A1, COL5A1, and SERPINE1 (PAI-1) (See Table 2 and Supplementary Table 4).
Table 2.
Correlation between changes in M2 macrophages/fiber and changes in ECM and inflammation gene expression following EET.
| Gene name | Alternate name | r | P |
|---|---|---|---|
| Inflammation | |||
| CCL18 | C-C motif chemokine ligand 18 | 0.73 | 0.001** |
| ITGAX | Integrin subunit alpha X, CD11c | 0.58 | 0.014* |
| ITGAM | Integrin subunit alpha M, CD11b | 0.58 | 0.014* |
| CCL5 | C-C motif chemokine ligand 5, RANTES | 0.56 | 0.019* |
| CD68 | CD68 molecule | 0.56 | 0.019* |
| HMOX1 | Heme oxygenase 1 | 0.51 | 0.035* |
| MRC1 | Mannose receptor C-type 1, CD206 | 0.49 | 0.045* |
| Extracellular matrix | |||
| MMP14 | Matrix metallopeptidase 14 | 0.69 | 0.002* |
| SERPINE1 | Plasminogen activator inhibitor-1, PAI-1 | 0.64 | 0.006* |
| SPARC | Osteonectin | 0.63 | 0.007* |
| ELN | Elastin | 0.56 | 0.018* |
| COL5A1 | Collagen type V α-1 chain | 0.56 | 0.021* |
| COL6A1 | Collagen type VI α-1 chain | 0.54 | 0.026* |
| TGFB1 | Transforming growth factor β-1 | 0.50 | 0.040* |
| LOX | Lysyl oxidase | 0.49 | 0.047* |
M2 (CD11b+/CD206+) macrophages were assessed by IHC. Gene expression was quantified using a custom-designed NanoString nCounter system code set. r = Pearson correlation coefficient. **P < 0.01, *P < 0.05. P values are not adjusted for multiple comparisons. N = 20.
Figure 4.
Following EET, the change in M2 macrophages per fiber is positively associated with gene expression changes in the hypertrophic growth factors (A). HGF (r = 0.63, P < 0.01), and (B). IGF1 (r = 0.58, P < 0.05) in human vastus lateralis. Gene expression changes were measured using the NanoString nCounter system. P values are not adjusted for multiple comparisons. N = 17.
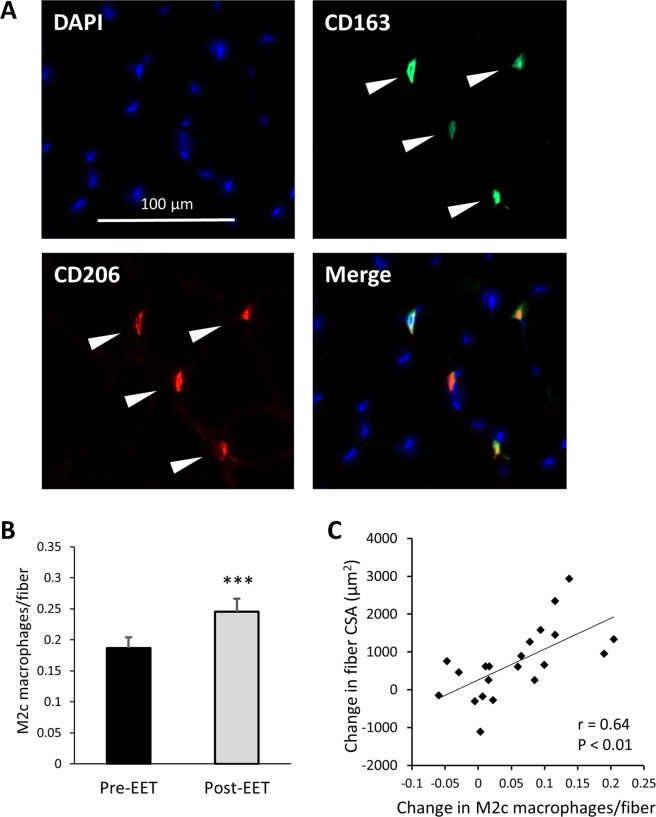
CD163+ M2c macrophages increase following EET and are associated with fiber hypertrophy
M2 macrophages are increasingly understood to include a functionally diverse set of cells, many of which express CD163, and are known as M2c macrophages. M2c macrophages are believed to be involved in extracellular matrix remodeling, since they are stimulated by TGFβ and they produce TGFβ52. We therefore sought to determine whether EET increases CD163+ M2c macrophages. Representative CD163 staining is shown in Fig. 5A. CD163 was expressed on 67.1% (±4.9% SEM) of CD206+ macrophages prior to training, and on 70.0% (±4.7% SEM) after training (not significant). Although EET did not influence the ratio of CD163+ to total M2 macrophages, it resulted in a highly significant increase in M2c macrophages per fiber (P < 0.001; Fig. 5B). As with total CD206+ M2 macrophages, the increase in M2c macrophages was tightly correlated to muscle fiber hypertrophy following EET (r = 0.64, P < 0.01; Fig. 5C). However, change in M2c macrophages was not associated with change in satellite cells, HGF mRNA, or IGF1 mRNA. Whereas we observed numerous associations between M2 macrophages and gene expression, changes in M2c macrophages were only associated with changes in 6 genes: TIMP2 (r = 0.63, P < 0.01), MMP2 (r = 0.54, P < 0.05), TNFα (r = 0.55, P < 0.05), CD68 (r = 0.54, P < 0.05), and tissue factor (F3 gene) (r = 0.53, P < 0.05). Lastly, neither baseline M2c macrophages nor change in M2c macrophages were associated with age, BMI, VO2max, or change in BMI or VO2max.
Figure 5.
In human vastus lateralis, M2c (CD206+/CD163+) macrophages increase following 12 weeks EET and are associated with muscle fiber hypertrophy. (A) Representative skeletal muscle M2c macrophage immunohistochemistry: Nuclei were stained with DAPI (blue). CD163 stain (green) was used as a specific M2c marker, while CD206 (red) was used as a pan M2 marker. A merged image shows M2c macrophages that co-stain for CD206 and CD163. White carrots indicate CD206+/CD163+ M2c macrophages. (B) EET results in increased M2c macrophages per fiber (P < 0.001, paired t-test), and (C) the increase in M2c macrophages per fiber is associated with muscle fiber hypertrophy, as assessed by change in fiber cross-sectional area (CSA, µm2) (r = 0.64, P < 0.01). N = 20.
EET alters inflammation-related gene expression
Expression of several inflammatory and anti-inflammatory genes were altered with EET, but were not directly correlated to changes in M2 or M2c macrophage number. The anti-inflammatory cytokine IL4 exhibited increased gene expression following EET (P < 0.01; Fig. 6A), whereas the inflammatory cytokine IL6 exhibited decreased gene expression after training (P < 0.05; Fig. 6A). Moreover, EET led to decreased expression of other known NFκB interacting genes, including the transcription factor C/EBPβ (P < 0.01; Fig. 6B) and the muscle specific E3 ubiquitin ligase MuRF (P < 0.05; Fig. 6B). C/EBPβ can heterodimerize with NFκB subunits to either antagonize or synergistically promote gene transactivation53–55, while MuRF is a known NFκB p50/65 target gene in skeletal muscle56–58. In spite of reduced expression of IL6, C/EBPβ, and MuRF, other known NFκB interacting genes were induced by EET, including TNFα (P < 0.01; Fig. 6C), which participates in MPC activation59,60, and FN14 (P < 0.05; Fig. 6C), a member of the TNF receptor superfamily. Supplementary Table 4 shows the effect of EET on expression of other inflammation and macrophage related genes. We were unable to determine whether IL10 mRNA was affected by EET because it was below the detectable limit of our NanoString code set.
Figure 6.
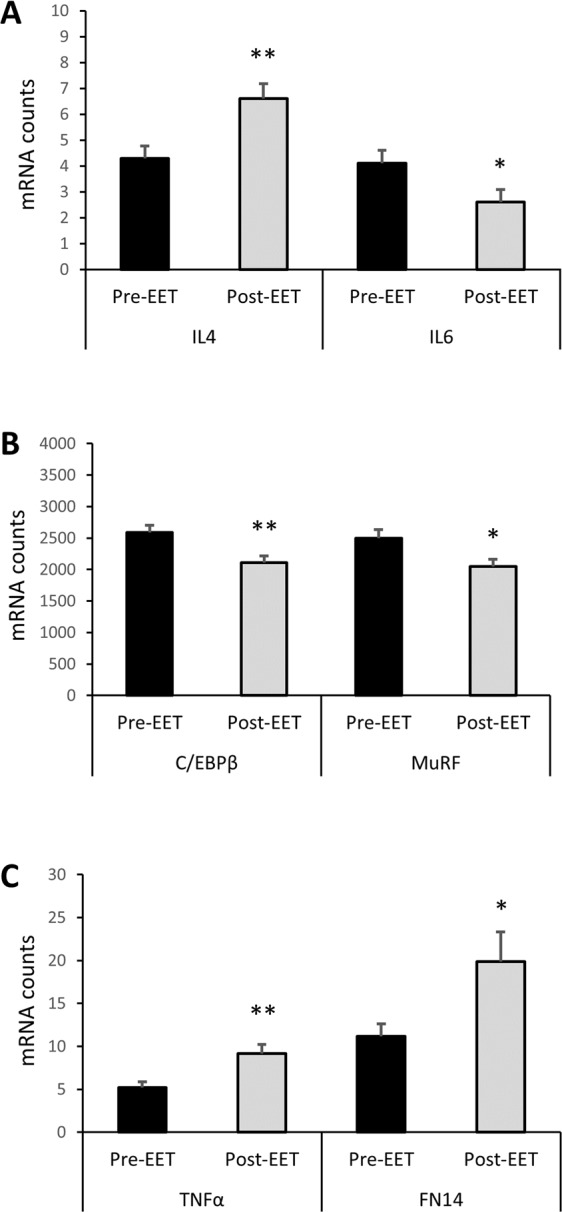
Effect of 12 weeks of EET on inflammation-related gene expression in human vastus lateralis. Gene expression was measured using the NanoString nCounter system. (A) The anti-inflammatory cytokine IL4 is increased (P < 0.01), while the pleiotropic cytokine IL6 is decreased (P < 0.05) following EET. (B) EET results in decreased expression of the NFκB target genes C/EBPβ (P < 0.01) and MuRF (P < 0.05). (C) EET causes increased expression of TNFα (P < 0.01) and the TNF receptor super family member FN14 (P < 0.05). Paired t-tests. P values are not adjusted for multiple comparisons. N = 20.
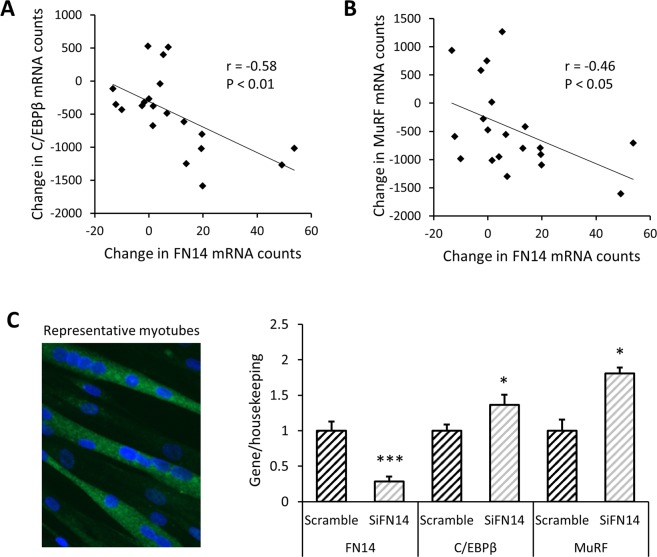
FN14 expression is inversely related to C/EBPβ and MuRF gene expression
Fn14 and its endogenous ligand TWEAK are known to exert paradoxical effects in skeletal muscle61. Furthermore, FN14 transcription has been shown to increase acutely following exercise62,63 and to be associated with fiber hypertrophy64. In our cohort, EET increased FN14 gene expression, but did not affect TWEAK gene expression (Supplementary Fig. 4). Since FN14 signaling can activate both canonical and non-canonical NFκB pathways65, we hypothesized that the change in FN14 expression would relate to changes in other inflammation related genes that were modulated by EET. After training, the change in FN14 expression was not correlated to changes in IL4, IL6, or TNFα. However, the training-induced change in FN14 was positively associated with the change in myogenin (r = 0.58, P < 0.05; Supplementary Fig. 5), a transcription factor that is required for muscle differentiation and has been reported to be inhibited by the NFκB p65 subunit11. Moreover, the EET-induced increase in FN14 mRNA was inversely correlated to changes in C/EBPβ (r = −0.58, P < 0.01; Fig. 7A) and MuRF (r = −0.46, P < 0.05; Fig. 7B). Given these observations, we hypothesized that when TWEAK remains constant, FN14 may inhibit transcription of C/EBPβ and MuRF. To test this hypothesis, we delivered small interfering RNA against FN14 (siFN14) to cultured primary human myotubes. Forty eight hours after siFN14 delivery, expression of FN14 was reduced by 71% versus scramble control (P < 0.001; Fig. 7C). TWEAK gene expression is readily detectible in human myotubes, with an average Ct of 26.1 ± 0.16 SEM, or approximately 15% of the level of FN14 in scramble cells. Neither TWEAK nor myogenin gene expression were affected by FN14 knock-down. However, siFN14 caused increased gene expression of both C/EBPβ (P < 0.05; Fig. 7C) and MuRF (P < 0.05; Fig. 7C). However, overexpression of FN14 in human myotubes did not have the opposite effect. Transfection with FN14-FL induced a 35.5-fold (±3.45 SEM) increase in FN14 gene expression (P < 0.001), without affecting expression of C/EBPβ or MuRF (Supplementary Fig. 6).
Figure 7.
FN14 gene expression is inversely correlated to expression of C/EBPβ and MuRF. In human vastus lateralis, change in FN14 expression is inversely correlated to change in (A). C/EBPβ expression (r = −0.58, P < 0.01), and (B) MuRF expression (r = −0.46, P < 0.05) resulting from 12 weeks endurance exercise training. P values are not adjusted for multiple comparisons. N = 20. (C) In cultured primary human myotubes, siRNA was used to knock down FN14 (siFN14), leading to a 71% decrease in FN14 expression (P < 0.001), together with increased expression of C/EBPβ (P < 0.05) and MuRF (P < 0.05) compared to scramble control cells. Paired t-tests were used to analyze data using 2 wells of each treatment in 2 individual cell lines and 1 well of each treatment in a third cell line (N = 5 data points per condition).
Discussion
We tested the hypothesis that EET would enhance the macrophage response to an acute bout of RE. We were surprised to observe that muscle macrophage content was increased following EET, but not RE. To our knowledge, we are the first to show that human muscle M2 and M2c macrophages increase following 12 weeks EET. In rodent studies, EET appears to reduce or have no effect on muscle macrophage content30,31. Our results in humans may differ from those observed in rodents because each of our endurance workouts contained some resistance stimulus and led to increased fiber CSA. Furthermore, we incrementally increased the cycle ergometer resistance throughout the study, contributing to increased average power output/workout as the study progressed.
The EET-elicited increase in M2 macrophage content was associated with increased satellite cell content. HGF and IGF1 may mediate the satellite cell response, as mRNAs encoding both growth factors are increased in proportion to M2 macrophages following EET, consistent with the idea that macrophages are an important source of these growth factors and play an active role in modulating the satellite cell niche12,18. Accordingly, we observed instances of macrophages in close proximity to satellite cells. This is consistent with reports indicating that macrophage-mediated SC activation requires proximal positioning66, and that the two cell types may share areas where intercellular space is less than 20 nm wide, termed “tight surface appositions”67.
Changes in genes associated with ECM remodeling were also positively correlated to changes in M2 macrophages with training, which is consistent with previous gene expression reports28,29 and is also consistent with the supposition that macrophages are involved in the hypertrophic response to EET and contribute to ECM allostasis. We did not perform statistical corrections for multiple tests because our NanoString code set was specifically designed to augment our overarching hypothesis that changes in macrophages would be associated with changes in growth-related, ECM, and inflammatory genes. Therefore, all correlational data should be interpreted as hypothesis generating rather than causal or mechanistic in the context of whole animal exercise responses.
EET caused changes in inflammation-related gene expression, and some of these changes were not directly correlated to changes in macrophage abundance. Nonetheless, increased expression of IL4, along with decreased expression of IL6, C/EBPβ, and MuRF, is consistent with a shift toward anti-inflammatory, pro-hypertrophic signaling following EET, and may result from changes in macrophage function. However, we were intrigued by the observation that EET resulted in up-regulation of TNFα and FN14, the receptor for TWEAK. Both TNFα and TWEAK exert pleiotropic effects on skeletal muscle. While TNFα is thought to promote satellite cell/MPC proliferation59,60, TWEAK has been shown to either promote68,69 or inhibit70 satellite cell/MPC proliferation. When chronically elevated, both TNFα and TWEAK drive canonical NFκB p50/p65-mediated protein catabolism and muscle loss16,59,65,71–73. MuRF is a known transcriptional target of p50/p6558,74, and C/EBPβ and p50/p65 can synergistically transactivate numerous gene promoters55. We therefore hypothesize that C/EBPβ may act synergistically with p50/p65 to drive MuRF transcription. Indeed, the MuRF promoter contains 8 predicted p65 response elements and 4 predicted CCAAT (C/EBPβ interacting) motifs within 2000 bp of its transcription start site75,76.
FN14-TWEAK signaling is primarily regulated through FN14 abundance, as TWEAK did not change with EET. Although FN14 has primarily been studied within the context of muscle damage and regeneration61, FN14 transcription has been associated with hypertrophic stimuli. FN14 is acutely elevated following a single bout of either endurance or resistance exercise62,63. It is associated with type II fiber hypertrophy following taper from training64. Furthermore, FN14 was induced approximately 500-fold in an individual champion sprint runner following a single bout of resistance exercise77. These associations between FN14 and hypertrophic stimuli are consistent with our observation that transcription of C/EBPβ and MuRF genes are inversely associated with FN14 in our training cohort, and that C/EBPβ and MuRF mRNAs are increased following FN14 knock-down in primary human myotubes. The apparent disconnect between ligand (TWEAK) and receptor (FN14) effects may be explained by TWEAK-independent effects of FN14. For example, FN14 has been shown to promote myogenin expression regardless of TWEAK levels in C2C12 cells78. Additionally, TWEAK expression is higher in macrophages compared to other cell types79 and increased levels of membrane bound TWEAK relative to soluble TWEAK promotes canonical NFκB signaling80. Thus, relatively low levels of TWEAK in our human myotube cultures may favor non-canonical NFκB signaling via FN14 and the subsequent suppression of C/EBPβ and MuRF transcription. Furthermore, overexpression of FN14 in cultured myotubes did not affect C/EBPβ or MuRF gene expression, indicating that basal levels of TWEAK-independent FN14 signaling may be sufficient to suppress C/EBPβ and MuRF transcription.
Here we report a highly significant increase in CD163+ M2c macrophages following EET. CD163 is highly expressed on tissue resident macrophages81 and is associated with the M2c macrophage transcriptional program in vitro82. Membrane bound CD163 decreases TWEAK bioactivity by binding and internalizing it83, while soluble CD163 functions as a decoy receptor for TWEAK84,85. We hypothesize that when CD163 is low and TWEAK-FN14 interactions high, canonical NFκB signaling may dominate, favoring C/EBPβ and MuRF transcription and atrophy. Conversely, when CD163 is abundant and TWEAK-FN14 interactions are decreased, C/EBPβ and MuRF transcription may be suppressed, and hypertrophic pathways predominate. We did not expect, nor did we observe, a correlation between increased M2c macrophage content and increased FN14 expression since FN14 is expressed by myofibers and participates in both hypertrophic and atrophic signaling. Thus, optimal hypertrophic responses may include alterations in cytokine signaling, FN14 inhibition of C/EBPβ and MuRF, which may be augmented by increased CD163 abundance, and increased M2 macrophage signaling via HGF and IGF1.
In the current cohort, we were surprised to observe no macrophage response to RE. This may be due to the heterogeneity of the cohort, which included individuals with a broad range of age and metabolic health. We previously reported a macrophage response 3 days after RE in young healthy men, which was reduced in older men33. Nonetheless, our current results might also be explained by the timing of our biopsy collection. While we performed vastus lateralis biopsies 72 hours after RE, an elegant study by Mackey and Kjaer performed biopsies 7 days after a highly damaging electrical stimulation protocol and reported dramatic macrophage infiltration in humans21. We also suspect that changes in macrophage morphology could account for inconsistent detection before and after RE; “amoeboid” (contracted, spherical) macrophages have been associated with inflammatory signaling while “ramified” (flattened, with multiple processes) macrophages have been associated with quiescence, surveillance, and/or anti-inflammatory signaling86,87. If muscle macrophages are more amoeboid following RE, they may be less likely to appear in a 7 µm tissue slice and therefore more difficult to quantify.
Taken together, these results indicate that in the absence of severe tissue damage, muscle resident macrophages are more polarized toward M2 than previously appreciated. Following endurance exercise training, M2 macrophages are associated with fiber hypertrophy and satellite cell accumulation, and may reduce inflammation, potentially via suppression of TWEAK signaling. Further elucidation of the beneficial functions of skeletal muscle macrophages following exercise could lead to novel approaches to overcome muscular deficits observed in obesity, aging, and other pathological conditions.
Supplementary information
Acknowledgements
The authors would like offer our sincere gratitude to Ms. Samantha Michaelis and Mr. Kyle Polley for counting macrophages. We would also like to thank Ms. Donna Wall and Dr. Kuey Chen of the University of Kentucky Genomics Core Laboratory for assistance with NanoString nCounter processing. We are grateful to Dr. Jeffrey A. Winkles of the University of Maryland for kindly providing FN14 and vector control plasmids. The authors also offer our sincere thanks to the many participants who were a part of this study. This work was supported by the following NIH grants: DK071349 and AG046920 (C.A.P. and P.A.K.); UL1TR001998; P20 GM103527.
Author Contributions
Study conception and design: R.G.W., K.K., B.S.F., P.AK., C.A.P. Data collection: R.G.W., K.K., J.M., C.S.F., B.D.P., J.S.G., B.Z. Data analysis and interpretation: R.G.W., K.K., C.S.F., B.S.F., P.A.K., C.A.P. Figure preparation: R.G.W., K.K., Drafting the manuscript: R.G.W. Critical revision of the article: R.G.W., K.K., B.D.P., B.S.F., P.A.K., C.A.P. All authors reviewed and approved the manuscript prior to submission.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-37187-1.
References
- 1.Way KL, Hackett DA, Baker MK, Johnson NA. The Effect of Regular Exercise on Insulin Sensitivity in Type 2 Diabetes Mellitus: A Systematic Review and Meta-Analysis. Diabetes Metab J. 2016;40:253–271. doi: 10.4093/dmj.2016.40.4.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grace A, Chan E, Giallauria F, Graham PL, Smart NA. Clinical outcomes and glycaemic responses to different aerobic exercise training intensities in type II diabetes: a systematic review and meta-analysis. Cardiovasc Diabetol. 2017;16:37. doi: 10.1186/s12933-017-0518-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Martinez FO, Gordon S. The M1 and M2 paradigm of macrophage activation: time for reassessment. F1000Prime Rep. 2014;6:13. doi: 10.12703/P6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chazaud B, et al. Satellite cells attract monocytes and use macrophages as a support to escape apoptosis and enhance muscle growth. J Cell Biol. 2003;163:1133–1143. doi: 10.1083/jcb.200212046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cantini M, et al. Macrophage-secreted myogenic factors: a promising tool for greatly enhancing the proliferative capacity of myoblasts in vitro and in vivo. Neurol Sci. 2002;23:189–194. doi: 10.1007/s100720200060. [DOI] [PubMed] [Google Scholar]
- 6.Massimino ML, et al. ED2+ macrophages increase selectively myoblast proliferation in muscle cultures. Biochem Biophys Res Commun. 1997;235:754–759. doi: 10.1006/bbrc.1997.6823. [DOI] [PubMed] [Google Scholar]
- 7.Sakuma K, Aoi W, Yamaguchi A. Molecular mechanism of sarcopenia and cachexia: recent research advances. Pflugers Arch. 2017;469:573–591. doi: 10.1007/s00424-016-1933-3. [DOI] [PubMed] [Google Scholar]
- 8.Peterson JM, Bakkar N, Guttridge DC. NF-kappaB signaling in skeletal muscle health and disease. Curr Top Dev Biol. 2011;96:85–119. doi: 10.1016/B978-0-12-385940-2.00004-8. [DOI] [PubMed] [Google Scholar]
- 9.Arnold L, et al. Inflammatory monocytes recruited after skeletal muscle injury switch into antiinflammatory macrophages to support myogenesis. J Exp Med. 2007;204:1057–1069. doi: 10.1084/jem.20070075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saclier M, et al. Differentially activated macrophages orchestrate myogenic precursor cell fate during human skeletal muscle regeneration. Stem cells. 2013;31:384–396. doi: 10.1002/stem.1288. [DOI] [PubMed] [Google Scholar]
- 11.Bakkar N, et al. IKK/NF-kappaB regulates skeletal myogenesis via a signaling switch to inhibit differentiation and promote mitochondrial biogenesis. J Cell Biol. 2008;180:787–802. doi: 10.1083/jcb.200707179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sawano S, et al. Supplementary immunocytochemistry of hepatocyte growth factor production in activated macrophages early in muscle regeneration. Anim Sci J. 2014;85:994–1000. doi: 10.1111/asj.12264. [DOI] [PubMed] [Google Scholar]
- 13.Villena J, Brandan E. Dermatan sulfate exerts an enhanced growth factor response on skeletal muscle satellite cell proliferation and migration. J Cell Physiol. 2004;198:169–178. doi: 10.1002/jcp.10422. [DOI] [PubMed] [Google Scholar]
- 14.Liu X, et al. Macrophage depletion impairs skeletal muscle regeneration: The roles of regulatory factors for muscle regeneration. Cell Biol Int. 2017;41:228–238. doi: 10.1002/cbin.10705. [DOI] [PubMed] [Google Scholar]
- 15.Dumont N, Frenette J. Macrophages protect against muscle atrophy and promote muscle recovery in vivo and in vitro: a mechanism partly dependent on the insulin-like growth factor-1 signaling molecule. Am J Pathol. 2010;176:2228–2235. doi: 10.2353/ajpath.2010.090884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sato S, Ogura Y, Kumar A. TWEAK/Fn14 Signaling Axis Mediates Skeletal Muscle Atrophy and Metabolic Dysfunction. Front Immunol. 2014;5:18. doi: 10.3389/fimmu.2014.00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bakkar N, et al. IKKalpha and alternative NF-kappaB regulate PGC-1beta to promote oxidative muscle metabolism. J Cell Biol. 2012;196:497–511. doi: 10.1083/jcb.201108118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tonkin J, et al. Monocyte/Macrophage-derived IGF-1 Orchestrates Murine Skeletal Muscle Regeneration and Modulates Autocrine Polarization. Mol Ther. 2015;23:1189–1200. doi: 10.1038/mt.2015.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pelosi L, et al. Local expression of IGF-1 accelerates muscle regeneration by rapidly modulating inflammatory cytokines and chemokines. FASEB J. 2007;21:1393–1402. doi: 10.1096/fj.06-7690com. [DOI] [PubMed] [Google Scholar]
- 20.Lu H, Huang D, Ransohoff RM, Zhou L. Acute skeletal muscle injury: CCL2 expression by both monocytes and injured muscle is required for repair. FASEB J. 2011;25:3344–3355. doi: 10.1096/fj.10-178939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mackey AL, Kjaer M. The breaking and making of healthy adult human skeletal muscle in vivo. Skelet Muscle. 2017;7:24. doi: 10.1186/s13395-017-0142-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hu L, et al. Low-frequency electrical stimulation attenuates muscle atrophy in CKD–a potential treatment strategy. J Am Soc Nephrol. 2015;26:626–635. doi: 10.1681/ASN.2014020144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zuo, Q., Wang, S. C., Yu, X. K. & Chao, W. W. Response of macrophages in rat skeletal muscle after eccentric exercise. Chin J Traumatol, 10.1016/j.cjtee.2017.12.001 (2018). [DOI] [PMC free article] [PubMed]
- 24.DiPasquale DM, et al. Urokinase-type plasminogen activator and macrophages are required for skeletal muscle hypertrophy in mice. Am J Physiol Cell Physiol. 2007;293:C1278–1285. doi: 10.1152/ajpcell.00201.2007. [DOI] [PubMed] [Google Scholar]
- 25.Duffield JS, Lupher M, Thannickal VJ, Wynn TA. Host responses in tissue repair and fibrosis. Annu Rev Pathol. 2013;8:241–276. doi: 10.1146/annurev-pathol-020712-163930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krenkel O, Tacke F. Liver macrophages in tissue homeostasis and disease. Nat Rev Immunol. 2017;17:306–321. doi: 10.1038/nri.2017.11. [DOI] [PubMed] [Google Scholar]
- 27.Huen SC, Cantley LG. Macrophages in Renal Injury and Repair. Annu Rev Physiol. 2017;79:449–469. doi: 10.1146/annurev-physiol-022516-034219. [DOI] [PubMed] [Google Scholar]
- 28.Gordon PM, et al. Resistance exercise training influences skeletal muscle immune activation: a microarray analysis. J Appl Physiol (1985) 2012;112:443–453. doi: 10.1152/japplphysiol.00860.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pourteymour S, et al. Global mRNA sequencing of human skeletal muscle: Search for novel exercise-regulated myokines. Mol Metab. 2017;6:352–365. doi: 10.1016/j.molmet.2017.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Samaan, M. C. et al. Endurance interval training in obese mice reduces muscle inflammation and macrophage content independently of weight loss. Physiol Rep2, 10.14814/phy2.12012 (2014). [DOI] [PMC free article] [PubMed]
- 31.Kurosaka M, Naito H, Ogura Y, Machida S, Katamoto S. Satellite cell pool enhancement in rat plantaris muscle by endurance training depends on intensity rather than duration. Acta Physiol (Oxf) 2012;205:159–166. doi: 10.1111/j.1748-1716.2011.02381.x. [DOI] [PubMed] [Google Scholar]
- 32.Varma V, et al. Muscle inflammatory response and insulin resistance: synergistic interaction between macrophages and fatty acids leads to impaired insulin action. Am J Physiol Endocrinol Metab. 2009;296:E1300–1310. doi: 10.1152/ajpendo.90885.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Przybyla B, et al. Aging alters macrophage properties in human skeletal muscle both at rest and in response to acute resistance exercise. Exp Gerontol. 2006;41:320–327. doi: 10.1016/j.exger.2005.12.007. [DOI] [PubMed] [Google Scholar]
- 34.Joanisse S, et al. Evidence for the contribution of muscle stem cells to nonhypertrophic skeletal muscle remodeling in humans. FASEB J. 2013;27:4596–4605. doi: 10.1096/fj.13-229799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Charifi N, Kadi F, Feasson L, Denis C. Effects of endurance training on satellite cell frequency in skeletal muscle of old men. Muscle Nerve. 2003;28:87–92. doi: 10.1002/mus.10394. [DOI] [PubMed] [Google Scholar]
- 36.Hoedt A, et al. Satellite cell response to erythropoietin treatment and endurance training in healthy young men. J Physiol. 2016;594:727–743. doi: 10.1113/JP271333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Murach, K. A. et al. Cycle training modulates satellite cell and transcriptional responses to a bout of resistance exercise. Physiol Rep4, 10.14814/phy2.12973 (2016). [DOI] [PMC free article] [PubMed]
- 38.Walton, R. G. et al. Insulin-resistant subjects have normal angiogenic response to aerobic exercise training in skeletal muscle, but not in adipose tissue. Physiol Rep3, 10.14814/phy2.12415 (2015). [DOI] [PMC free article] [PubMed]
- 39.Fry CS, et al. Fibre type-specific satellite cell response to aerobic training in sedentary adults. J Physiol. 2014;592:2625–2635. doi: 10.1113/jphysiol.2014.271288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kosmac, K. et al. Immunohistochemical Identification of Human Skeletal Muscle Macrophages. Bio Protoc8, 10.21769/BioProtoc.2883 (2018). [DOI] [PMC free article] [PubMed]
- 41.Brown SA, Cheng E, Williams MS, Winkles JA. TWEAK-independent Fn14 self-association and NF-kappaB activation is mediated by the C-terminal region of the Fn14 cytoplasmic domain. PLoS One. 2013;8:e65248. doi: 10.1371/journal.pone.0065248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Geiss GK, et al. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat Biotechnol. 2008;26:317–325. doi: 10.1038/nbt1385. [DOI] [PubMed] [Google Scholar]
- 43.Northcott PA, et al. Rapid, reliable, and reproducible molecular sub-grouping of clinical medulloblastoma samples. Acta Neuropathol. 2012;123:615–626. doi: 10.1007/s00401-011-0899-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Veldman-Jones MH, et al. Reproducible, Quantitative, and Flexible Molecular Subtyping of Clinical DLBCL Samples Using the NanoString nCounter System. Clin Cancer Res. 2015;21:2367–2378. doi: 10.1158/1078-0432.CCR-14-0357. [DOI] [PubMed] [Google Scholar]
- 45.Tidball JG, Villalta SA. Regulatory interactions between muscle and the immune system during muscle regeneration. Am J Physiol Regul Integr Comp Physiol. 2010;298:R1173–1187. doi: 10.1152/ajpregu.00735.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chazaud B, et al. Dual and beneficial roles of macrophages during skeletal muscle regeneration. Exerc Sport Sci Rev. 2009;37:18–22. doi: 10.1097/JES.0b013e318190ebdb. [DOI] [PubMed] [Google Scholar]
- 47.Wynn TA, Vannella KM. Macrophages in Tissue Repair, Regeneration, and Fibrosis. Immunity. 2016;44:450–462. doi: 10.1016/j.immuni.2016.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tanaka Y, et al. Expression of mRNA for specific fibroblast growth factors associates with that of the myogenic markers MyoD and proliferating cell nuclear antigen in regenerating and overloaded rat plantaris muscle. Acta Physiol (Oxf) 2008;194:149–159. doi: 10.1111/j.1748-1716.2008.01866.x. [DOI] [PubMed] [Google Scholar]
- 49.Yamaguchi A, Ishii H, Morita I, Oota I, Takeda H. mRNA expression of fibroblast growth factors and hepatocyte growth factor in rat plantaris muscle following denervation and compensatory overload. Pflugers Arch. 2004;448:539–546. doi: 10.1007/s00424-004-1282-5. [DOI] [PubMed] [Google Scholar]
- 50.Philippou A, Barton ER. Optimizing IGF-I for skeletal muscle therapeutics. Growth Horm IGF Res. 2014;24:157–163. doi: 10.1016/j.ghir.2014.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schiaffino S, Dyar KA, Ciciliot S, Blaauw B, Sandri M. Mechanisms regulating skeletal muscle growth and atrophy. FEBS J. 2013;280:4294–4314. doi: 10.1111/febs.12253. [DOI] [PubMed] [Google Scholar]
- 52.Roszer T. Understanding the Mysterious M2 Macrophage through Activation Markers and Effector Mechanisms. Mediators Inflamm. 2015;2015:816460. doi: 10.1155/2015/816460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang K. Molecular mechanisms of hepatic apoptosis regulated by nuclear factors. Cell Signal. 2015;27:729–738. doi: 10.1016/j.cellsig.2014.11.038. [DOI] [PubMed] [Google Scholar]
- 54.Newton, K. & Dixit, V. M. Signaling in innate immunity and inflammation. Cold Spring Harb Perspect Biol4, 10.1101/cshperspect.a006049 (2012). [DOI] [PMC free article] [PubMed]
- 55.Stein B, Cogswell PC, Baldwin AS., Jr. Functional and physical associations between NF-kappa B and C/EBP family members: a Rel domain-bZIP interaction. Mol Cell Biol. 1993;13:3964–3974. doi: 10.1128/MCB.13.7.3964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li H, Malhotra S, Kumar A. Nuclear factor-kappa B signaling in skeletal muscle atrophy. J Mol Med (Berl) 2008;86:1113–1126. doi: 10.1007/s00109-008-0373-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fry, C. S. et al. Glucocorticoids increase skeletal muscle NF-kappaB inducing kinase (NIK): links to muscle atrophy. Physiol Rep4, 10.14814/phy2.13014 (2016). [DOI] [PMC free article] [PubMed]
- 58.Wu CL, Kandarian SC, Jackman RW. Identification of genes that elicit disuse muscle atrophy via the transcription factors p50 and Bcl-3. PLoS One. 2011;6:e16171. doi: 10.1371/journal.pone.0016171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li YP. TNF-alpha is a mitogen in skeletal muscle. Am J Physiol Cell Physiol. 2003;285:C370–376. doi: 10.1152/ajpcell.00453.2002. [DOI] [PubMed] [Google Scholar]
- 60.Alvarez B, et al. Tumor necrosis factor-alpha exerts interleukin-6-dependent and -independent effects on cultured skeletal muscle cells. Biochim Biophys Acta. 2002;1542:66–72. doi: 10.1016/S0167-4889(01)00167-7. [DOI] [PubMed] [Google Scholar]
- 61.Tajrishi MM, Zheng TS, Burkly LC, Kumar A. The TWEAK-Fn14 pathway: a potent regulator of skeletal muscle biology in health and disease. Cytokine Growth Factor Rev. 2014;25:215–225. doi: 10.1016/j.cytogfr.2013.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Raue U, Jemiolo B, Yang Y, Trappe S. TWEAK-Fn14 pathway activation after exercise in human skeletal muscle: insights from two exercise modes and a time course investigation. J Appl Physiol (1985) 2015;118:569–578. doi: 10.1152/japplphysiol.00759.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dickinson, J. M. et al. Transcriptome response of human skeletal muscle to divergent exercise stimuli. J Appl Physiol (1985), 10.1152/japplphysiol.00014.2018 (2018). [DOI] [PubMed]
- 64.Murach K, et al. Single muscle fiber gene expression with run taper. PLoS One. 2014;9:e108547. doi: 10.1371/journal.pone.0108547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Enwere EK, Lacasse EC, Adam NJ, Korneluk RG. Role of the TWEAK-Fn14-cIAP1-NF-kappaB Signaling Axis in the Regulation of Myogenesis and Muscle Homeostasis. Front Immunol. 2014;5:34. doi: 10.3389/fimmu.2014.00034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Du H, et al. Macrophage-released ADAMTS1 promotes muscle stem cell activation. Nature communications. 2017;8:669. doi: 10.1038/s41467-017-00522-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ceafalan, L. C. et al. Skeletal muscle regeneration involves macrophage-myoblast bonding. Cell Adh Migr, 1–8, 10.1080/19336918.2017.1346774 (2017). [DOI] [PMC free article] [PubMed]
- 68.Dogra C, Changotra H, Mohan S, Kumar A. Tumor necrosis factor-like weak inducer of apoptosis inhibits skeletal myogenesis through sustained activation of nuclear factor-kappaB and degradation of MyoD protein. J Biol Chem. 2006;281:10327–10336. doi: 10.1074/jbc.M511131200. [DOI] [PubMed] [Google Scholar]
- 69.Girgenrath M, et al. TWEAK, via its receptor Fn14, is a novel regulator of mesenchymal progenitor cells and skeletal muscle regeneration. EMBO J. 2006;25:5826–5839. doi: 10.1038/sj.emboj.7601441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ogura Y, Mishra V, Hindi SM, Kuang S, Kumar A. Proinflammatory cytokine tumor necrosis factor (TNF)-like weak inducer of apoptosis (TWEAK) suppresses satellite cell self-renewal through inversely modulating Notch and NF-kappaB signaling pathways. J Biol Chem. 2013;288:35159–35169. doi: 10.1074/jbc.M113.517300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sakuma K, Aoi W, Yamaguchi A. Current understanding of sarcopenia: possible candidates modulating muscle mass. Pflugers Arch. 2015;467:213–229. doi: 10.1007/s00424-014-1527-x. [DOI] [PubMed] [Google Scholar]
- 72.Sakuma, K. & Yamaguchi, A. In Skeletal Muscle-From Myogenesis to Clinical Relations (InTech, 2012).
- 73.Dogra C, et al. TNF-related weak inducer of apoptosis (TWEAK) is a potent skeletal muscle-wasting cytokine. FASEB J. 2007;21:1857–1869. doi: 10.1096/fj.06-7537com. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cai D, et al. IKKbeta/NF-kappaB activation causes severe muscle wasting in mice. Cell. 2004;119:285–298. doi: 10.1016/j.cell.2004.09.027. [DOI] [PubMed] [Google Scholar]
- 75.Dreos R, Ambrosini G, Groux R, Cavin Perier R, Bucher P. The eukaryotic promoter database in its 30th year: focus on non-vertebrate organisms. Nucleic Acids Res. 2017;45:D51–D55. doi: 10.1093/nar/gkw1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dreos R, Ambrosini G, Perier RC, Bucher P. The Eukaryotic Promoter Database: expansion of EPDnew and new promoter analysis tools. Nucleic Acids Res. 2015;43:D92–96. doi: 10.1093/nar/gku1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Trappe S, et al. Skeletal muscle signature of a champion sprint runner. J Appl Physiol (1985) 2015;118:1460–1466. doi: 10.1152/japplphysiol.00037.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Dogra C, Hall SL, Wedhas N, Linkhart TA, Kumar A. Fibroblast growth factor inducible 14 (Fn14) is required for the expression of myogenic regulatory factors and differentiation of myoblasts into myotubes. Evidence for TWEAK-independent functions of Fn14 during myogenesis. J Biol Chem. 2007;282:15000–15010. doi: 10.1074/jbc.M608668200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Maecker H, et al. TWEAK attenuates the transition from innate to adaptive immunity. Cell. 2005;123:931–944. doi: 10.1016/j.cell.2005.09.022. [DOI] [PubMed] [Google Scholar]
- 80.Roos C, et al. Soluble and transmembrane TNF-like weak inducer of apoptosis differentially activate the classical and noncanonical NF-kappa B pathway. J Immunol. 2010;185:1593–1605. doi: 10.4049/jimmunol.0903555. [DOI] [PubMed] [Google Scholar]
- 81.Lau SK, Chu PG, Weiss LM. CD163: a specific marker of macrophages in paraffin-embedded tissue samples. Am J Clin Pathol. 2004;122:794–801. doi: 10.1309/QHD6-YFN8-1KQX-UUH6. [DOI] [PubMed] [Google Scholar]
- 82.Lurier EB, et al. Transcriptome analysis of IL-10-stimulated (M2c) macrophages by next-generation sequencing. Immunobiology. 2017;222:847–856. doi: 10.1016/j.imbio.2017.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Moreno JA, et al. The CD163-expressing macrophages recognize and internalize TWEAK: potential consequences in atherosclerosis. Atherosclerosis. 2009;207:103–110. doi: 10.1016/j.atherosclerosis.2009.04.033. [DOI] [PubMed] [Google Scholar]
- 84.Akahori H, et al. CD163 interacts with TWEAK to regulate tissue regeneration after ischaemic injury. Nat Commun. 2015;6:7792. doi: 10.1038/ncomms8792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bover LC, et al. A previously unrecognized protein-protein interaction between TWEAK and CD163: potential biological implications. J Immunol. 2007;178:8183–8194. doi: 10.4049/jimmunol.178.12.8183. [DOI] [PubMed] [Google Scholar]
- 86.McWhorter FY, Wang T, Nguyen P, Chung T, Liu WF. Modulation of macrophage phenotype by cell shape. Proc Natl Acad Sci USA. 2013;110:17253–17258. doi: 10.1073/pnas.1308887110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Durafourt BA, et al. Comparison of polarization properties of human adult microglia and blood-derived macrophages. Glia. 2012;60:717–727. doi: 10.1002/glia.22298. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.