Abstract
Chronic lymphocytic leukaemia is the most prevalent leukaemia in Western countries. It is an incurable disease characterized by a highly variable clinical course. Chronic lymphocytic leukaemia is an ideal model for studying clonal heterogeneity and dynamics during cancer progression, response to therapy and/or relapse because the disease usually develops over several years. Here we report an analysis by deep sequencing of sequential samples taken at different times from the affected organs of two patients with 12- and 7-year disease courses, respectively. One of the patients followed a linear pattern of clonal evolution, acquiring and selecting new mutations in response to salvage therapy and/or allogeneic transplantation, while the other suffered loss of cellular tumoral clones during progression and histological transformation.
Introduction
Chronic Lymphocytic Leukaemia (CLL) is the most prevalent leukaemia in Western countries. It is an incurable disease characterized by a highly variable clinical course that results from expansion of clonal CD5 + B-lymphocytes growing in bone marrow (BM), blood, lymph nodes (LN) or other lymphoid compartments. In the last years, multiple studies have reported that CLL disease is not just heterogeneous between patients but also individual CLL samples are genetically heterogeneous and contain subclonal cellular populations1–3. It is broadly accepted that this diversity evolves under selective treatment pressures, such as chemotherapy, that favor selection of resistant clones that emerge after treatment1,4. Two major patterns for clonal evolution have been described: linear and branched evolution. In the linear evolution model, a linear sequence of mutational events takes place within a single clone while branched evolution presents spatial and temporal heterogeneity and two or more clone could coexist and evolve in parallel5.
CLL is an ideal model for studying clonal heterogeneity and dynamics during cancer progression, response to therapy and/or relapse because the disease usually develops over several years, it could affect different haematological compartments and its presentation is fairly heterogeneous among patients3,6,7. Here we report an analysis by deep sequencing of sequential samples taken at different times from the affected organs of two patients with 12- and 7-year disease courses, respectively.
Results
Whole exome sequencing in index samples
Patient 1 (P1) was a 50-year-old male diagnosed with stage A/II CLL in 2000 (see Patient 1 description in “Methods” section). He was CD38-negative, ZAP70-positive, had unmutated IgHV, and a normal karyotype with an 11q22-23 deletion (ATM) in 67% of interphase nuclei, as revealed by fluorescence in situ hybridization (FISH) analysis. In March 2004 the patient was treated with fludarabine achieving partial remission (PR); two years later, in March 2006, he came to our institution with symptomatic disease, generalized lymphadenopathy and splenomegaly. Before he started treatment with FCR (fludarabine, cyclophosphamide, rituximab) plus rituximab maintenance for 6 months, we collected sample P1.1 of peripheral blood mononuclear cells (PBMCs). In September 2012 (aged 61 years, 12 years after diagnosis) the patient died of disease progression and samples were obtained from different compartments: P1.13 (PBMCs), P1.14 (lymph node) and P1.15 (spleen). Whole exome sequencing (WES) was performed in these index samples as well as in DNA from oral mucosa (P1.N). Our analysis identified 46 non-synonymous variants (non-sense, missense, frameshift and splicing) as being present in at least one sample.
Patient 2 (P2) was a 62-year-old male diagnosed with stage B/II CLL in September 2006 (see Patient 2 description) the patient presented symptomatic disease. The first sample (P2.1, PBMCs) was taken in January 2007, and it was CD38-negative, ZAP-70-positive, IgHV unmutated, and had a normal karyotype with deletions of 11q22-23 and 13q14 detected by FISH in 50% and 25% of interphase nuclei, respectively. In March 2008 was treated with 6 cycles of FCR, and rituximab maintenance for 3 years (REM [Rituximab in maintenance] clinical trial) and in October 2012 (P2.4, PBMCs), P2 rapidly developed an increase in lymphocytosis with generalized lymphadenopathy and was treated with anti-CD37 (TRU-016) plus bendamustine and attained PR. In July 2013, a Richter transformation was diagnosed in a lymph node (LN) biopsy (P2.5), the patient was treated with salvage chemotherapy plus bortezomib, but had no clinical response and died in January 2014, aged 68, 7 years after diagnosis. WES was performed in these index samples and oral mucosa (P2.N) and we identified 32 non-synonymous variants present in at least one of them.
Clonal heterogeneity and evolution
With the whole set of non-synonymous variants identified by WES, we designed a custom panel to analyse every available sample by deep sequencing (median coverage 4125×), including the index samples (Table S1). This, together with the karyotype and FISH information, allowed us to follow the genetic changes on 13 points over 6 years in P1 and in 5 samples over 7 years in P2.
Patient 1
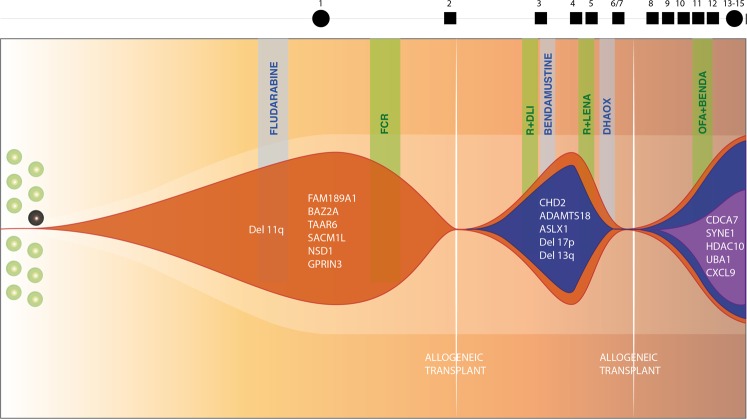
In P1, FISH revealed an 11q22-23 (ATM) deletion in 90% of tumoral cells when diagnosed in September 2000 (see Patient Description). In March 2004, P1 was treated with fludarabine achieving PR; two years later, in January 2006, P1 showed symptomatic disease with lymphadenopathy and splenomegaly and was treated with FCR and rituximab maintenance. In sample P1.1 (March 2006, before treatment with FCR)), ATM and 17p13.1 (TP53) deletions were detected in 85% and 3% of cells, respectively, as well as somatic mutations in seven genes (TP53, FAM189A1, SACM1L, TAAR6, GPRIN3, NSD1 and BAZ2A) with variant allelic frequencies (VAFs) ranging from 17% to 50% (Table S2 and Fig. S1A–C in SI). In June 2007 the patient received a non-myeloablative allogenic stem cell transplant (SCT). Later on, in February 2009, the disease progressed and the patient was treated with rituximab and donor lymphocyte infusion, attaining PR; in the sample analyzed in September 2009 (P1.3) we were able to detect new sub-clonal mutations in CHD2, ASXL1 and ADAMTS18, among other genes, with VAFs ranging from 3% to 6% (Table S2 and Fig. S1A,B and C in SI). At this point, 11q22-23, 17p13.1 and 13q14 deletions (66%, 23% and 30% respectively) and MYC amplification (15%) were detected by FISH. P1 was treated with R-bendamustine (September 2009) without response, and subsequently was given R-lenalidomide (March 2010, P1.4), which yielded a short-lived response, with the patient progressing in October 2010. The patient rapidly developed a generalised growth of lymph nodes and lymphocytosis that were refractory to two subsequent lines of treatment (R-bendamustine and R-lenalidomide). Mutational data validated that tumoral cells did not respond to treatment with R-bendamustine (median VAF increased from P1.3 to P1.4) and only partially to R-lenalidomide (slight decrease in VAFs in sample P1.5, September 2010). In February 2011 (P1.6), cytogenetic and mutational data indicated an increase in the tumoral cell population. The patient was subjected to salvage chemotherapy with four cycles of dexamethasone, high-dose cytarabine and oxaliplatin, which produced a PR (P1.7), and consolidated in August 2011 with a second non-myeloablative allogeneic SCT (sample P1.8 taken before transplant). The same mutations, although with lower VAF than in P1.6, were detected in P1.7, reflecting the PR to salvage chemotherapy. Cytogenetic data were consistent with mutation changing patterns provided by the sequencing analysis data. In February 2012, after treatment with salvage chemotherapy and allogeneic SCT the patient progressed and sample P1.9 was found to have new mutated genes (CDCA7, SYNE1, HDAC10, UBA1, CXCL9, among others), with VAFs from 7% to 15%. FISH revealed deletions of 11q22-23 (in 95% of cells), 13q14 (57%) and 17p13 (76%), in addition to a complex karyotype (see Patients’ description in the Methods section). During the period from February to September 2012 (exitus), P1 was treated with ofatumumab-bendamustine without clinical response; samples P1.10 to P1.13 were also sequenced, and showed increase in the VAFs of the mutations and in the percentage of cells with cytogenetic alterations (Fig. 1, Table S2A,B and Fig. S1A–C in SI).
Figure 1.
Schema of the inferred tumour clonal evolution path of the CLL patient 1. Sequential acquisition of somatic alterations in patient 1 leads to its genomic diversification. Exposure to successive chemotherapy, immunochemotherapy and allogeneic transplants would be associated with acquisition of new alterations and may profile the CLL landscape. FCR: fludarabine, cyclophosphamide, rituximab; R + LENA: rituximab plus lenalidomide; OFA-Benda: ofatumumab-bendamustine; DLI: donor lymphocyte infusion; DHA0X: high-dose cytarabine and oxaliplatin.
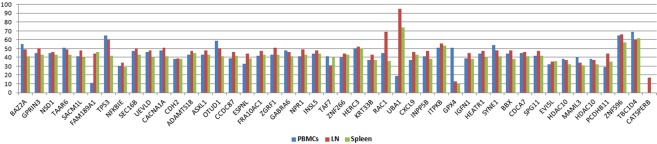
We also analysed the differences in genetic alterations in samples from different anatomical locations, LN, spleen and PBMCs at the end-stage of the disease (P1.13-15), and found that the three compartments shared almost the same variants, although some had different VAFs (Figs 2, S1D and Table S2A,B in SI), such as CASTPERB, which was exclusively mutated in LN or GPX4 with higher VAF in PBMCs.
Figure 2.
Variant Allelic Frequency of the non-synonymous somatic mutations in peripheral blood lymphocytes (PBMCs), lymph node (LN) and spleen at the end of the disease from Patient 1.
Patient 2
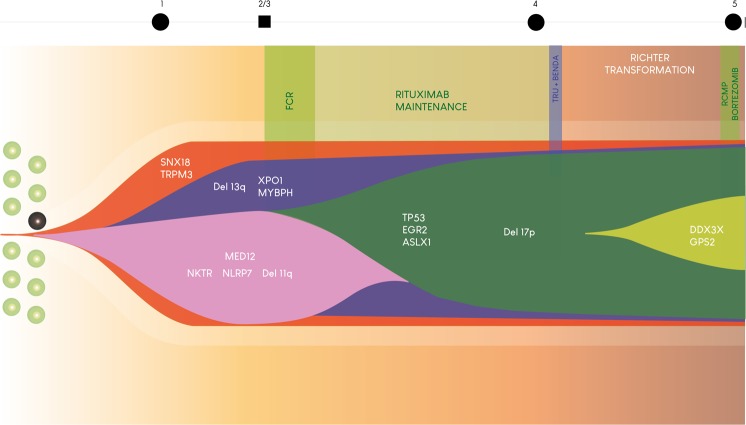
P2 showed at three months after diagnosis (sample P2.1, January 2007) concomitant 11q22-23 and 13q14 deletions in 50% and 25% of tumoral cells, respectively. The landscape of somatic mutations in this patient was consistent with the existence of two clones derived from an ancestral one with mutations in SNX18 and TRPM3 (VAF of 50%, meaning around 100% of cells) (Table S3A,B and Fig. S2 in SI). At diagnosis, one clone encompassed the 11q22-23 deletion and mutations in MED12, NLRP7 and NKTR (50% of cells), whose VAF decreased over the course of the disease; the second clone featured the 13q14 deletion (in 25% of the cells) and mutations in XPO1, among others, whose median VAF increased over time, as may be seen by comparing the patterns of mutations in samples P2.1 and P2.2. No treatment was given to P2 until March 2008 when he was treated with FCR and rituximab maintenance for 3 years, in the context of a clinical trial, and achieved complete response (samples P2.2 [PBMCs] and P2.3 [bone marrow] were taken in February 2008, before treatment). In September 2012 (sample P2.4) the patient developed general lymphadenopathy and exhibited a complex karyotype along with 11q22-23 (9%), 17p13 (55%) and 13q14 deletions (63%), and MYC amplification (19%) detected by FISH; a new tumoral clone with mutations in TP53, EGR2 and ASXL1 emerged while the clone with 11q22-23 deletion was almost completely lost, in parallel with an increase of the subclone with an 13q14 deletion and XPO1 mutation. The patient was treated with TRU-016 (anti-CD37) and bendamustine (October 2012) achieving PR. In July 2013, P2 developed a generalized lymphadenopathy along with malaise, weight loss, daily fever and night sweats. A Richter transformation was diagnosed after a LN biopsy (sample P2.5; see Patient Description in SI). In this sample, neither the 11q22-23 deletion, nor the variants in MED12, NLRP7, NKTR, PTPRG or PRDM6 could be detected, but new mutated genes emerged, DDX3 X and GPS2 being the most relevant. In the case of the mutational landscape of PBMCs (P2.2) and bone marrow (P2.3), the same pattern and VAFs in mutated genes were found. Although samples P2.4 and P2.5 came from different organs (PBMCs and LN, respectively), a Richter transformation occurred between the two time-points. It was not possible to determine whether these differences were due to the transformation (although this was the most likely cause) or to the different organs involved, due to the absence of a PBMC sample obtained simultaneously with the LN sample, because the patient did not have lymphocytosis (Fig. 3, Table S3A,B and Fig. S2 in SI). Patient was treated with salvage chemotherapy and bortezomib with two cycles without clinical response and died in January 2014 of disease progression.
Figure 3.
Schema of the inferred tumour clonal evolution path of the CLL patient 2. CLL therapy could induce novel mutagenesis and/or accelerate clonal evolution by “killing” a clone and selecting other containing mutations in genes that might confer resistance. Sustained treatment with targeted antigen-specific immunotherapy with anti-CD20 and subsequent anti-CD37 monoclonal antibodies might interfere with stem cell plasticity and provoke tumor escape through clonal evolution with acquisition of additional genetic alterations responsible for histological transformation. FCR: fludarabine, cyclophosphamide, rituximab; TRU-Benda: TRU-016-bendamustine; RCMP: rituximab, liposomal doxorubicin and prednisone.
Discusion
In this study, we described results from sequencing sequential samples taken at different time-points of disease evolution from two CLL patients with 12- and 7-year disease courses.
Patient P1 followed a linear pattern of clonal evolution, acquiring and selecting new mutations in response to salvage therapy and/or allogeneic SCT (Fig. 1). The founder alteration was the 11q22-23 deletion and the acquisition of the TP53 biallelic alteration, one of the clearest predictive factors of treatment resistance, appeared to be the inflection point. The successive treatments received by P1 not only failed to eliminate the initial clones but also contributed linearly to, or at least did not prevent, the acquisition of new alterations, which did not respond to any subsequent treatment. This fact was more evident after the two non-myeloablative allogenic SCT, when a higher number of new mutations appeared (Table S2A). After the first SCT, mutations in CDH2, ADAMTS18 and ASXL1 were detected with low VAFs (P1.3) that increased during disease evolution (P1.4 and successive samples). After the second one, genes such as ITPKB, IGFN1, SYNE1, CDCA7 and HDAC10, among others, were also found mutated, probably giving a survival advantage to the tumoral clone.
When we compared the mutational landscape in PBMCs, LN and spleen, we did not detect any significant differences between the three compartments; previous studies analysing different parts of the same tumour or single cells, have described differences in their mutational landscapes8,9. In our case, we were not able to perform this type of single cell or spatial analysis, and, therefore, putative minor clones, with low VAFs, may be underrepresented. The clones we have detected were similar in the three compartments and may be attributed to predominant clones with a broader somatic mutational landscape.
We draw attention to two points regarding patient P2: the displacement of the clone with 11q22-23 deletion and mutations in driver genes such as MED12 by that with the 13q14 deletion and mutation in XPO1, and the presence of a new tumoral clone that emerged after FCR-Rm treatment, with a TP53 biallelic alteration, MYC amplification and mutations in EGR2, AXL1 and RPS15 (Fig. 3). XPO1 mutation has been described as a recurrent and initiating event in CLL and other B cell-derived neoplasias10,11. EGR2 mutations have been associated with clinically aggressive CLL12,13. Eventually, after anti-CD37 plus bendamustine therapy, the Richter transformation was diagnosed. The TP53 biallelic alteration is associated with a higher risk of transformation as well as NOTCH1 activation and MYC abnormalities14,15. A previous study identified mutations in DDX3X, RPS15 and GPS2 (which were all mutated in P2) as part of a driver gene profile associated with 17p deletion in CLL16. RNA processing and splicing alterations play central roles in this patient: four genes (MED12, XPO1, DDX3X , RPS15) involved in these biological processes were found to be mutated, as well as chromatin remodelling genes (PRDM6, ASXL1, MSX2) (for a review see17).
ASXL1 was found mutated in the two patients after FCR treatment. ASXL1 mutations have been associated with shorter overall survival and worse response to immunosuppression18. A case report of a CLL patient19 also identified a mutation in ASXL1 gene after FCR treatment, and other studies focused on acute myeloid leukaemia and myelodysplastic syndrome have identified mutations in ASXL1 after treatment with chemotherapy20–22. We hypothesize that the appearance of ASXL1 mutations in the two patients could be a consequence of the CIT received by both patients.
In summary, this study provides complementary information about clonal evolution and heterogeneity in CLL. We show here two different patterns of clonal dynamics, P1 acquiring new alterations at relapse, after allogeneic SCT and salvage chemotherapy, and P2 loss cellular tumoral clones after treatment and during progression. We also hypothesize that there might be two mechanisms involved in the pathogenesis of non-germinal centre DLBCL transformation in CLL. First, cell-cycle dysregulation induced by acquired MYC rearrangements, along with the impairment of the DNA damage response through acquired TP53 loss, may play a role in the transformation event. Second, sustained treatment with targeted antigen-specific immunotherapy with anti-CD20 and subsequent anti-CD37 monoclonal antibodies might interfere with stem cell plasticity and provoke tumour escape through acquisition of additional genetic alterations, responsible for histological transformation, some of them associated to 17p deletion in CLL.
Methods
Patients Description
This study was approved by the Hospital Universitario Puerta de Hierro-Majadahonda institutional ethical committee. Patients provided informed consent in accordance with local institutional review board requirements and the Declaration of Helsinki before enrolment. All research methods were performed in accordance with Institutional Guidelines and regulations. Treatment protocols were according to the Haematology Department guidelines and patient 2 also participated in two research phase II clinical trials (REM: EudraCT No.: 2007-002733-36 and Otlertuzumab plus Bendamustine: NCT01188681) approved by the Institutional ethics committee.
Patient 1, a physically fit 50-year-old male was diagnosed in September 2000 with stage A/II CLL, with splenomegaly of 4 cm below the left costal margin (b.l.c.m.), a lymphocyte count of 112,000/µL, a haemoglobin count of 14.5 g/dl, and a platelet count of 150,000/µL. He was CD38-negative, ZAP70-positive, had unmutated IgHV, and a normal karyotype with an 11q22-23 del (ATM del in 67% of interphase nuclei), as revealed by fluorescence in situ hybridization (FISH).
In May 2004, 45 months after diagnosis, the patient presented with increasing fatigue, a lymphocyte doubling time of 6 months and splenomegaly of 6 cm b.l.c.m. He was treated with fludarabine (25 mg/m2 × 5 days × 6 cycles) and achieved partial remission.
He presented at 21 months since first treatment (64 months from diagnosis) with symptomatic disease, generalized lymphadenopathy and splenomegaly of 4 cm b.l.c.m. (Sample P1.1, February 2006) and in March 2006 (67 months from diagnosis) was treated with FCR (fludarabine, cyclophosphamide, rituximab) plus rituximab maintenance for 6 months, achieving complete clinical remission, and was consolidated with a non-myeloablative allogeneic stem cell transplant (SCT) of HLA-identical unrelated adult donor in June 2007 (82 months after diagnosis) (Sample P1.2, before transplant).
The patient relapsed 20 months after transplantation (102 months, February 2009) with rapidly progressive lymphocytosis and generalized lymphadenopathy of 2–4 cm and retroperitoneal adenopathy of 6 cm, along with acquisition of a 17p1.3 deletion, as revealed by FISH, and a TP53 mutation, as identified by cDNA Sanger sequencing (inframe deletion: c.376_396del21; p.Y126_K132delYSPALNK). He was treated with rituximab and increasing monthly doses of donor lymphocyte infusion (4 cycles), and achieved partial remission (PR). The patient progressed in October 2010, developing rapidly growing generalised lymph nodes and lymphocytosis that were refractory to two subsequent lines of treatment (R-bendamustine and R-lenalidomide).
In February 2011 (126 months after diagnosis), the patient received four cycles of salvage chemotherapy with dexamethasone, high-dose cytarabine and oxaliplatin, achieving PR, and was consolidated in August 2011 with a second non-myeloablative allogeneic SCT of HLA-identical unrelated cord-blood. Patient remained in partial remission until February 2012 (138 months after diagnosis), when progressed with a 7-cm bulky retroperitoneal lymphadenopathy and was treated with ofatumumab-bendamustine for 3 cycles, but without clinical response. In September 2012, the patient died of disease progression without histopathological evidence of Richter transformation.
Patient 2, a physically fit 62-year-old male was diagnosed in September 2006 with stage B/II CLL with generalized lymphadenopathy of 1.5–2.0 cm without hepatomegaly or splenomegaly, a lymphocyte count of 16.750/µL, a haemoglobin count of 14.8 g/dl and a platelet count of 159,000/µL. His sample was CD38-negative, ZAP-70-positive, IgHV unmutated, and had a normal karyotype with deletions of 11q22-23 (ATM) and 13q14 (D13S319) detected by FISH in 50% and 25% of interphase nuclei, respectively.
The patient was treated in March 2008 (15 months after diagnosis) with 6 cycles of FCR, and rituximab maintenance for 3 years (REM clinical trial), and he achieved a complete response.
In October 2012 (70 months after diagnosis) the patient developed rapidly increasing lymphocytosis with generalized lymphadenopathy of 3-4 cm and molecular cytogenetic studies showed a complex acquired karyotype [45–46, XY, −10, del(11)(q14;q21), del(11)(q11), del(13)(q14;q21), i(17)(q10)] with deletions of 17p13 (p53) (55%), 13q14 (63%), C-MYC amplification (19%) and a notably low percentage of ATM deletion (9%) in the 11q22-23 region. The patient was treated with anti-CD37 plus bendamustine for 6 cycles (clinical trial) and achieved partial remission. In July 2013 (79 months) presented with rapidly growing cervical and axillary lymphadenopathy of 3–5 cm along with progressive malaise, weight loss, night sweats and fever with no documented active infection. A lymph node biopsy showed Richter transformation into a diffuse large B cell lymphoma (DLBCL) (Ki67: 70%, CD20+ high, CD5−, CD10−, BCL-6−, MUM-1+, FOXp1+, CD38+ and EBERs−). The bone marrow had no infiltration by DLBCL, but showed diffuse infiltration of small and medium size lymphocytes. CD20+, CD5+, CD23+, BCL2+ and TP53 overexpression and Ki67+ (40%). FISH data showed 17p13 deletion (75%), homozygous 13q14 deletion (83%), and CMYC amplification/translocation (19%). P2 was treated with salvage immunochemotherapy (rituximab, liposomal doxorubicin and prednisone) and bortezomib for 2 cycles without clinical response and died in January 2014 (86 months after diagnosis), 7 months after diagnosis of transformation to non-germinal centre DLBCL with the same IgHV mutational pattern as the original CLL clone.
Fluorescence In Situ Hybridization (FISH)
Cytogenetic aberrations were analysed by FISH with the VysisCLL FISH Probe Kit, following the manufacturer’s recommendations for detecting deletions of TP53 (17p13.1), ATM (11q22.3), D13S319 (13q14.3), MYC rearrangements/amplification (8q24.12-q24.13) and gain of the D12Z3 sequence (trisomy 12) in peripheral blood specimens from CLL patients.
Cut-off values for a positive FISH result were 3% and 10% for gains and deletions, respectively. Routine FISH testing is generally performed for patients with CLL prior to starting treatment. Follow-up FISH testing is not routinely performed at defined intervals but may be requested by the treating physician when there is disease progression and treatment is considered.
Samples
Fifteen samples were collected from peripheral blood of Patient 1 between diagnosis and exitus (P1.1 to P1.13) as well as biopsies from lymph node (P1.14) and spleen (P1.15) from necropsy and buccal mucosa (P1.N). We analysed three samples from peripheral blood mononuclear cells of Patient 2 (P2.1, P2.2 and P2.3) one from bone marrow (P2.4), a lymph node (P2.5) and buccal mucosa for germinal DNA (P2.N).
DNA was obtained from peripheral blood mononuclear cells separated by Ficoll (Rafer, Zaragoza, Spain). Tumour cell purity was calculated based on the CD19/CD5 ratio, measured by FACS.
DNA was extracted with DNAzol Genomic DNA Isolation Reagent (Molecular Research Center, OH, USA) according to the manufacturer’s instructions. The quality and quantity of purified DNA was assessed by both fluorimeter (Qubit, Invitrogen, MA, USA) and gel electrophoresis.
Whole-Exome Sequencing
Whole-exome sequencing (WES) was performed in DNA from mononuclear cells of index samples (P1.1, P1.13, P1.14, P1.15 and P1.N from Patient 1, and P2.1, P2.4, P2.5 and P2.N from Patient 2).
Whole-exome capture and sequencing were performed at the Spanish National Centre for Genomic Analysis (CNAG, Barcelona, Spain). Tumour and germline DNA were enriched for coding regions in the genome using the SureSelect-XT Human exon capture 50 Mb kits V4 and V5 (Agilent Technologies, Santa Clara, CA, USA). According to the manufacturer’s protocol, these include the coding exons of c.20,000 genes, covering > 50 Mb of the genome. Paired-end sequencing, resulting in 76 bases from each end of the fragments, was performed using a HiSeq. 2000 Genome Analyzer (Illumina Inc., San Diego, CA, USA). Targeted NGS data was demultiplexed with HiSeq Software (Illumina). Base calling and quality control were carried out with the Illumina RTA sequence analysis pipeline (Illumina) (Supplementary Table S4).
We performed two parallel pipelines for alignment and variant calling. In the first, the sequence reads were trimmed until the first base was read with a quality of more than 10. Mapping to human genome build hg19 (GRCh37) was done with the GEM23, allowing up to four mismatches. Reads not mapped by the GEM (approximately 4%) were subjected to a final round of mapping with BFAST24. Results were merged and only uniquely mapping non-duplicate read pairs were used for further analyses. The SAMtools suite25, with default settings, was used to call single nucleotide variants and short insertions and deletions. Identified variants in regions with low mappability, a read depth of <10 or a strand bias with a value of P < 0.001 were filtered out. Variant annotation and effect prediction were performed with Annovar26 and snpEff27, including information from the Single Nucleotide Polymorphism Database (dbSNP, build 135)28, the 1000 Genomes Project, the Exome Variant Server (NHLBI GO Exome Sequencing Project, Seattle, WA, USA; http://evs.gs.washington.edu/EVS/) and an internal database of sequence variants identified in a set of more than 100 control samples. To compare tumour and normal cells, the significance of Fisher’s exact test was calculated for positions with different genotypes in the two samples based on alternative allele frequencies.
For the second pipeline, quality was assessed using the FASTQC tool (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/), and alignment performed using Burrows-Wheeler Aligner25 using GRCh37/hg19 assembly. The samples were realigned with the GATK tool29 to improve variant calling, which was done with Mutect 1.1.730. Annotation was carried out with Variant Effect Predictor (VEP)31.
The sequence data have been deposited in the Sequence Read Archive at http://www.ncbi.nlm.nih.gov/sra/, with accession number (SUB2978993).
Target Sequencing
Based on mutations identified by WES, a TruSeq Custom Amplicon v1.5 panel was designed. The target regions are provided in Table S1 in SI. The probes for this targeted custom panel were designed with DesignStudio (Illumina) and consisted of 266 amplicons with an average size of 180 bp and a cumulative targeted region of 20 kb. Polymorphisms were avoided in the design of primers.
DNA target enrichment was performed using the protocol described in the TruSeq Amplicon—Cancer Panel Library Preparation Guide (March 2016; Illumina). The total amount of input DNA for capture ranged from 30 to 100 ng. After preparation, indexing and bead purification, the libraries were quantified by Qubit analysis to assess successful enrichment and amplification. Libraries were then normalized on beads and pooled for sequencing in accordance with the TruSeq Amplicon protocol. The pooled libraries were paired-end (2 × 151), and sequenced with V2 chemistry on a MiSeq instrument (Illumina), as described in the manufacturer’s protocol. Alignment and variant calling were performed using MiSeq reporter and variants identified in previous analyses were verified by visualization in IGV tool32 (Integrative Genomics Viewer).
Somatic single nucleotide variants (SNVs) identified by deep sequencing were used as the input into PyClone (v0.13.0) to identify those with similar VAFs and to infer the different clones and their patterns of evolution33–35.
Supplementary information
Acknowledgements
We are indebted to the patients who contributed to this study. We also thank E. Ramil (Genomics Unit) from IDIPHISA, and C. González-Rincón for her help drawing the figures. This work was supported by the Spanish Ministry of Economy and Competence (MINECO): SAF2013-47416-R; ISCIII-MINECO AES-FEDER (Plan Estatal de I + D + I 2008–2011 and 2013–2016) (PI14/00221 (MSB), PIE14/0064 (MSB), PIE15/0081 (MAP), PI16/01294 (MAP), CIBERONC CB16/12/00291 (MAP)), Madrid Autonomous Community, B2017/BMD3778 (MAP, MSB, FA-S), F Hoffmann-La Roche (JAGM); CNIO Bioinformatics Unit work has been supported by Marie-Curie Career Integration Grant CIG334361. J.G.-R. is a recipient of a i-PFIS predoctoral fellowship (IFI14/00003); NM was supported by AECC Scientific Foundation; S.D. was supported by the Torres Quevedo subprogramme (MICINN) under grant agreement PTQ-12-05391. K.T. and J.P.-P. are supported by Severo Ochoa FPI grant doctoral fellowship by the Spanish MINECO. MSB currently holds a Miguel Servet II contract (CPII16/00024) supported by ISCIII-MINECO AES-FEDER (Plan Estatal I + D + I 2013–2016) and the Fundación de Investigación Biomédica Puerta de Hierro.
Author Contributions
J.G.-R. performed experiments, analysed the data and wrote the manuscript; S.G., B.F.-C., N.P.-S., S.N.-G. processed samples and performed experiments; N.M., processed samples, analysed data; K.V., J.P.-P., S.D., S.B., I.G., F.A.-S. analysed the data, M.A.P. analysed data and supervised the study; J.A.G.M. performed the diagnosis and/or collected the samples and clinical and pathological data, supervised the project and wrote the manuscript; M.S.-B. designed and supervised the project, analysed the data and wrote the manuscript. All the authors revised and approved the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
José A. García-Marco and Margarita Sánchez-Beato jointly supervised this work.
Contributor Information
José A. García-Marco, Email: jagarciam@aehh.org
Margarita Sánchez-Beato, Email: msbeato@idiphim.org.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-37389-7.
References
- 1.Ding L, et al. Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing. Nature. 2012;481:506–510. doi: 10.1038/nature10738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Burgess DJ. Leukaemia: targeted therapy re-enABLed? Nat. Rev. Cancer. 2011;11:460. doi: 10.1038/nrc3099. [DOI] [PubMed] [Google Scholar]
- 3.Schuh A, et al. Monitoring chronic lymphocytic leukemia progression by whole genome sequencing reveals heterogeneous clonal evolution patterns. Blood. 2012;120:4191–4196. doi: 10.1182/blood-2012-05-433540. [DOI] [PubMed] [Google Scholar]
- 4.Guièze R, Wu CJ. Genomic and epigenomic heterogeneity in chronic lymphocytic leukemia. Blood. 2015;126:445–453. doi: 10.1182/blood-2015-02-585042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Greaves M, Maley CC. Clonal evolution in cancer. Nature. 2012;481:306–313. doi: 10.1038/nature10762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Del Giudice I, et al. Inter- and intra-patient clonal and subclonal heterogeneity of chronic lymphocytic leukaemia: evidences from circulating and lymph nodal compartments. Br. J. Haematol. 2016;172:371–383. doi: 10.1111/bjh.13859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sutton L-A, Rosenquist R. Clonal evolution in chronic lymphocytic leukemia: impact of subclonality on disease progression. Expert Rev. Hematol. 2015;8:71–78. doi: 10.1586/17474086.2015.972930. [DOI] [PubMed] [Google Scholar]
- 8.Martelotto LG, et al. Whole-genome single-cell copy number profiling from formalin-fixed paraffin-embedded samples. Nat. Med. 2017;23:376–385. doi: 10.1038/nm.4279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhao, Z. et al. Evolution of multiple cell clones over a 29-year period of a CLL patient. Nat. Commun. 7 (2016). [DOI] [PMC free article] [PubMed]
- 10.Puente XS, et al. Whole-genome sequencing identifies recurrent mutations in chronic lymphocytic leukaemia. Nature. 2011;475:101–105. doi: 10.1038/nature10113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Takahashi, K. et al. Clinical implications of cancer gene mutations in patients with chronic lymphocytic leukemia treated with lenalidomide. Blood, 10.1182/blood-2017-11-817296 (2018). [DOI] [PMC free article] [PubMed]
- 12.Damm F, et al. Acquired Initiating Mutations in Early Hematopoietic Cells of CLL Patients. Cancer Discov. 2014;4:1088–1101. doi: 10.1158/2159-8290.CD-14-0104. [DOI] [PubMed] [Google Scholar]
- 13.Young E, et al. EGR2 mutations define a new clinically aggressive subgroup of chronic lymphocytic leukemia. Leukemia. 2017;31:1547–1554. doi: 10.1038/leu.2016.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rossi D, et al. The genetics of Richter syndrome reveals disease heterogeneity and predicts survival after transformation. Blood. 2011;117:3391–3401. doi: 10.1182/blood-2010-09-302174. [DOI] [PubMed] [Google Scholar]
- 15.Fabbri G, et al. Analysis of the chronic lymphocytic leukemia coding genome: role of NOTCH1 mutational activation. J. Exp. Med. 2011;208:1389–1401. doi: 10.1084/jem.20110921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu L, et al. Survival of Del17p CLL Depends on Genomic Complexity and Somatic Mutation. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2017;23:735–745. doi: 10.1158/1078-0432.CCR-16-0594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lazarian G, Guièze R, Wu CJ. Clinical Implications of Novel Genomic Discoveries in Chronic Lymphocytic Leukemia. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2017;35:984–993. doi: 10.1200/JCO.2016.71.0822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ferrando AA, López-Otín C. Clonal evolution in leukemia. Nat. Med. 2017;23:1135–1145. doi: 10.1038/nm.4410. [DOI] [PubMed] [Google Scholar]
- 19.Ge, H. et al. A case report of extranodal NK/T-cell lymphoma in patient with chronic lymphocytic leukemia. Medicine (Baltimore)97 (2018). [DOI] [PMC free article] [PubMed]
- 20.Larsson CA, Cote G, Quintás-Cardama A. The Changing Mutational Landscape of Acute Myeloid Leukemia and Myelodysplastic Syndrome. Mol. Cancer Res. MCR. 2013;11:815–827. doi: 10.1158/1541-7786.MCR-12-0695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Abdel-Wahab O, et al. ASXL1 Mutations Promote Myeloid Transformation Through Loss of PRC2-Mediated Gene Repression. Cancer Cell. 2012;22:180–193. doi: 10.1016/j.ccr.2012.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gelsi-Boyer V, et al. Mutations of polycomb-associated gene ASXL1 in myelodysplastic syndromes and chronic myelomonocytic leukaemia. Br. J. Haematol. 2009;145:788–800. doi: 10.1111/j.1365-2141.2009.07697.x. [DOI] [PubMed] [Google Scholar]
- 23.Marco-Sola S, Ribeca P. Efficient Alignment of Illumina-Like High-Throughput Sequencing Reads with the GEnomic Multi-tool (GEM) Mapper. Curr. Protoc. Bioinforma. 2015;50(11):13.1–20. doi: 10.1002/0471250953.bi1113s50. [DOI] [PubMed] [Google Scholar]
- 24.Homer N, Merriman B, Nelson SF. BFAST: an alignment tool for large scale genome resequencing. PloS One. 2009;4:e7767. doi: 10.1371/journal.pone.0007767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinforma. Oxf. Engl. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cingolani P, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strainw1118; iso-2; iso-3. Fly (Austin) 2012;6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smigielski EM, Sirotkin K, Ward M, Sherry ST. DbSNP: a database of single nucleotide polymorphisms. Nucleic Acids Res. 2000;28:352–355. doi: 10.1093/nar/28.1.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McKenna A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cibulskis K, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat. Biotechnol. 2013;31:213–219. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McLaren W, et al. The Ensembl Variant Effect Predictor. Genome Biol. 2016;17:122. doi: 10.1186/s13059-016-0974-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thorvaldsdóttir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief. Bioinform. 2013;14:178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roth A, et al. PyClone: statistical inference of clonal population structure in cancer. Nat. Methods. 2014;11:396–398. doi: 10.1038/nmeth.2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ulintz PJ, Greenson JK, Wu R, Fearon ER, Hardiman KM. Lymph Node Metastases in Colon Cancer Are Polyclonal. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2018;24:2214–2224. doi: 10.1158/1078-0432.CCR-17-1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kridel R, et al. Histological Transformation and Progression in Follicular Lymphoma: A Clonal Evolution Study. PLoS Med. 2016;13:e1002197. doi: 10.1371/journal.pmed.1002197. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.