Abstract
Mutations in the β-catenin gene (CTNNB1) have been implicated in the pathogenesis of some cancers. The recent development of cancer genome databases has facilitated comprehensive and focused analyses on the mutation status of cancer-related genes. We have used these databases to analyze the CTNNB1 mutations assembled from different tumor types. High incidences of CTNNB1 mutations were detected in endometrial, liver, and colorectal cancers. This finding agrees with the oncogenic role of aberrantly activated β-catenin in epithelial cells. Elevated frequencies of missense mutations were found in the exon 3 of CTNNB1, which is responsible for encoding the regulatory amino acids at the N-terminal region of the protein. In the case of metastatic colorectal cancers, inframe deletions were revealed in the region spanning exon 3. Thus, exon 3 of CTNNB1 can be considered to be a mutation hotspot in these cancers. Since the N-terminal region of the β-catenin protein forms a flexible structure, many questions arise regarding the structural and functional impacts of hotspot mutations. Clinical identification of hotspot mutations could provide the mechanistic basis for an oncogenic role of mutant β-catenin proteins in cancer cells. Furthermore, a systematic understanding of tumor-driving hotspot mutations could open new avenues for precision oncology.
Keywords: β-catenin, cancer genome database, hotspot mutations
INTRODUCTION
β-Catenin is an important co-activator downstream of the oncogenic Wnt signaling pathway, so mutations in the β-catenin gene (CTNNB1) have been implicated in oncogenesis (Korinek et al., 1997; Morin et al., 1997; Polakis, 2012b). Recently, large-scale cancer databases, such as The Cancer Genome Atlas (TCGA) pan-cancer analysis project, have leveraged systemic analyses on genome, exome, and transcriptome data from all types of cancers (Blum et al., 2018; Hutter and Zenklusen, 2018; Tomczak et al., 2015). Multidimensional cancer genome data are available on cBioPortal, an open platform for cancer genome analysis and visualization (Cerami et al., 2012; Gao et al., 2013). In this review, we have employed pan-cancer genome databases to analyze the current status of β-catenin gene (CTNNB1) mutations to identify mutation hotspots and to re-evaluate the oncogenic roles of specific β-catenin mutant proteins. An extensive review on the clinical aspects of the β-catenin protein is beyond the scope of this mini review, so we have provided a brief introduction regarding the basic biology of the β-catenin protein.
A BRIEF INTRODUCTION TO THE β-CATENIN PROTEIN
β-Catenin is a multitasking protein involved in transcription and cell adhesion (Hur and Jeong, 2013; Kumar and Bashyam, 2017; Valenta et al., 2012). In particular, β-catenin is an important co-activator of Wnt target genes, such as cyclin D1 and c-myc (Korinek et al., 1997; Morin et al., 1997). However, in differentiated cells, where Wnt signaling is off, the central regulatory mechanism for β-catenin is sequential phosphorylation in the N-terminal region followed by ubiquitin-mediated proteolysis (Fig. 1A). Casein Kinase-1α phosphorylates the S45 residue and primes subsequent phosphorylation on T41/S37/S33 by GSK-3β, leading to the binding of ubiquitin E3 ligase β-transducin repeats-containing proteins (β-TrCP) at the N-terminal region (D32 to S37) in a phosphorylation-dependent manner (Hart et al., 1998; Liu et al., 2002). Specific phosphorylation and ubiquitination occur in the APC/Axin complex, termed as the β-catenin destruction complex (Stamos and Weis, 2013). In contrast, the destruction complex functions no more, so the level of the β-catenin protein in the cytoplasm increases following Wnt activation (Fig. 1B). The mechanism by which Wnt signaling stabilizes β-catenin needs to be better understood in the aspect of the β-catenin destruction complex (Kim et al., 2013; 2015; Li et al., 2012; Taelman et al., 2010). Finally, Wnt-stimulated β-catenin is translocated into the nucleus, where it acts as transcriptional co-activator with DNA binding TCF/LEF proteins and activates many developmentally important, cancer-related and pathogenic genes (Nusse and Clevers, 2017).
Fig. 1. A schematic diagram of the Wnt signaling pathway.
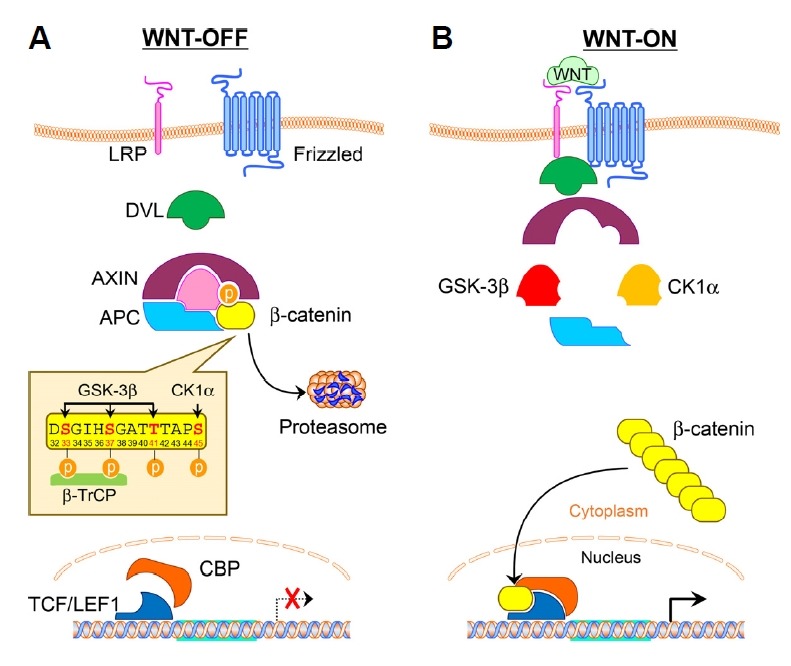
(A) Wnt-off. In the absence of Wnt stimulation, β-catenin is phosphorylated by CK1α and GSK3β followed by ubiquitin-proteasome mediated proteolysis. (B) Wnt-on. Upon Wnt stimulation, the destruction complex is not functional, so the β-catenin protein is translocated into the nucleus and acts as a transcriptional co-activator to regulate oncogenic target genes. APC, Adenomatous polyposis; DVL, Disheveled.
FREQUENCY OF GENOMIC ALTERATIONS IN THE CTNNB1 GENE IN CANCERS
Small-scale targeted gene analysis demonstrates mutations in the β-catenin gene (CTNNB1 ) in some cancers (Polakis, 2007; 2012b). Large-scale β-catenin mutational landscape was revealed from clinical sequencing of 10,000 prospective cancer patients by the Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT) (Zehir et al., 2017). Figure 2 shows the frequency of CTNNB1 alterations across tumor types. A high frequency of CTNNB1 mutations are found in endometrial (16%), hepatobiliary (12%), melanoma (7%), and colorectal (6%) cancers. It is noteworthy that the frequency of APC mutations is much higher (approximately 70%) than that of CTNNB1 in colorectal cancer, but CTNNB1 and APC mutations exist in the exclusive manner. The alteration frequency of CTNNB1 in endometrial, liver, and colorectal cancer from other genomic analysis networks is compiled in Table 1. In addition, cancer cell lines with mutations in CTNNB1 are summarized in Table 2.
Fig. 2. The alteration frequency of CTNNB1 and APC across cancer types.
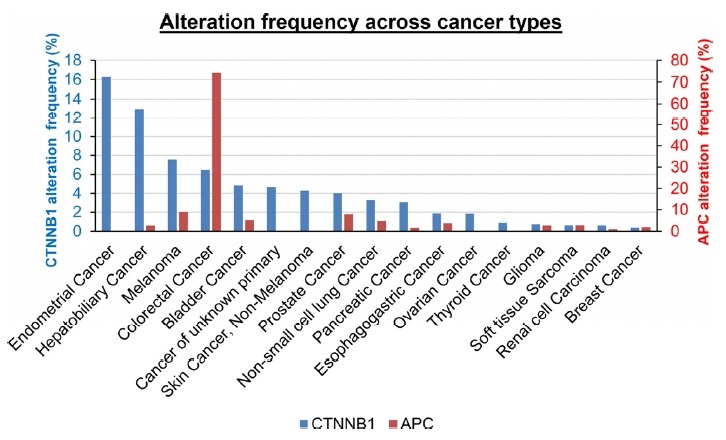
Data obtained from the MSK-IMPACT pan-cancer study on cBioportal (www.cbioportal.org).
Table 1.
The alteration frequency of CTNNB1 in endometrial, liver, and colorectal cancer
| Cancer type | Sequencing data source | No. Sequenced | No. Alteration (%) | No. Exon3-mut (%) | Reference |
|---|---|---|---|---|---|
| Endometrial cancer | Endometrial Cancer (MSK, 2018) | 187 | 27 (14.4) | 25 (13.4) | Soumerai et al., 2018 |
| Uterine Corpus Endometrial Carcinoma (TCGA, Nature 2013) | 240 | 71 (29.6) | 63 (26.3) | Cancer Genome Atlas Research et al., 2013a | |
| Uterine Carcinosarcoma (TCGA, PanCancer Atlas) | 56 | 1 (1.8) | 0 (0.0) | Cancer Genome Atlas Research et al., 2013b | |
| Uterine Clear Cell Carcinoma (NIH, Cancer 2017) | 16 | 0 (0.0) | 0 (0.0) | Le Gallo et al., 2017 | |
| Liver cancer | Liver Hepatocellular Carcinoma (TCGA, PanCancer Atlas) | 353 | 95 (26.9) | 78 (22.1) | Cancer Genome Atlas Research et al., 2013b |
| Liver Hepatocellular Carcinoma (AMC, Hepatology 2014) | 231 | 53 (22.9) | 46 (19.9) | Ahn et al., 2014 | |
| Liver Hepatocellular Carcinoma (RIKEN, Nat Genet 2012) | 25 | 3 (12.0) | 3 (12.0) | Fujimoto et al., 2012 | |
| Hepatocellular Carcinomas (Inserm, Nat Genet 2015) | 243 | 87 (35.8) | 76 (31.3) | Schulze et al., 2015 | |
| Hepatocellular Adenoma (Inserm, Cancer Cell 2014) | 30 | 13 (43.3) | 11 (36.7) | Pilati et al., 2014 | |
| Colorectal cancer | Colorectal Adenocarcinoma (TCGA, Nature 2012) | 212 | 11 (5.2) | 1 (0.5) | Cancer Genome Atlas, N. et al., 2012 |
| Colorectal Adenocarcinoma (Genentech, Nature 2012) | 72 | 5 (6.9) | 2 (2.8) | Seshagiri et al., 2012 | |
| Colorectal Adenocarcinoma (DFCI, Cell Reports 2016) | 619 | 31 (5.0) | 8 (1.3) | Giannakis et al., 2016 | |
| Metastatic colorectal cancer (MSK, Cancer Cell 2018) | 1099 | 84 (7.6) | 19 (1.7) | Yaeger et al., 2018 | |
| Colon Adenocarcinoma (TCGA, PanCancer Atlas) | 389 | 27 (6.9) | 15 (3.9) | Cancer Genome Atlas Research et al., 2013b | |
| Rectum Adenocarcinoma (TCGA, PanCancer Atlas) | 137 | 8 (5.8) | 0 (0.0) | Cancer Genome Atlas Research et al., 2013b |
Data obtained from the listed cancer studies on cBioportal (www.cbioportal.org)
Table 2.
Status of mutations in cancer cell lines harboring activating mutations of CTNNB1
| Cancer type | Cell Line | Mutations | ||||
|---|---|---|---|---|---|---|
|
| ||||||
| CTNNB1 | APC | TP53 | BRAF | KRAS | ||
| Colorectal cancer | SW48 | S33Y | R2714C | |||
| CCK81 | T41A | Y159C | P278H | S273N | ||
| SNU407 | T41A | R726C | G12D | |||
| HCT116 | S45del | G13D | ||||
| LS180 | S45F | R1788C | D211G | G12D | ||
| Gastric cancer | KE39 | D32N | V272L | |||
| AGS | G34E | G12D | ||||
| SNU719 | G34V | |||||
| OCUM1 | S45C | |||||
| Endometrial cancer | HEC265 | D32V, X561_splice | P1233L | |||
| HEC6 | D32V | V160A | ||||
| HEC108 | S37P, D207G | S678G, A2388V, T2514I | P151H | |||
| JHUEM2 | S37C | |||||
| SNGM | S37P | A2V | G12V | |||
| Lung cancer | MORCPR | S33L | P865L, A2122dup | P152Rfs*18 | G13C | |
| SW1573 | S33F | G12C | ||||
| LXF289 | T41A | R248W | ||||
| HCC15 | S45F, Y670* | D2796G | D259V | |||
| Liver cancer | HUH6 | G34V | N239D, A159D | |||
| SNU398 | S37C | |||||
| Melanoma | SKMEL1 | S33C | V600E | |||
| COLO783 | S45del | P27L | V600E | |||
Mutation data obtained from Cancer Cell Line Encyclopedia (Novartis/Broad, Nature, 2012) on cBioportal (www.cbioportal.org).
Abbreviation: del, deletion; dup, duplication; fs, frame shift; splice, splice site mutation;
stop codon
Endometrial cancer
Wnt/β-catenin pathway has been linked to endometrial cancer. Loss of APC function in the mouse endometrium induces nuclear β-catenin accumulation in uterine hyperplasia and squamous cell metaplasia. Although APC loss alone does not lead to malignant transformation, APC loss enhances endometrial tumorigenesis driven by PTEN loss (van der Zee et al., 2013). The majority of CTNNB1 alterations occur in endometrial carcinoma, but not in carcinosarcoma or clear cell carcinoma as indicated in Table 1 (Cancer Genome Atlas Research et al., 2013a; Cancer Genome Atlas Research et al., 2013b; Le Gallo et al., 2017; Soumerai et al., 2018). In endometrial cancer cases from TCGA, the alterations of CTNNB1 or APC genes are 30% or 12% of 240 patients, respectively (Cancer Genome Atlas Research et al., 2013a). Loss of APC function arises from truncation of the gene, but the frequency is only 6% in endometrial cancer. Thus, in endometrial cancer, CTNNB1 mutations, rather than APC mutations, might be direct driver mutations. Recently, 245 endometrial cancer patient samples were sequenced using 46–200 gene panels. CTNNB1 mutations appear more frequently in low-grade (grades 1–2) and early-stage (stages I–II) patients. More significantly, the patients harboring CTNNB1 mutations are associated with worse recurrence-free survival (Kurnit et al., 2017).
Hepatocellular carcinoma (HCC)
Liver cancer is the seventh most common cancer and the fourth leading cause of cancer mortality worldwide (Bray et al., 2018). However, treatment options are still limited for patients with advanced HCC due to the heterogeneity of genome alterations. Genome-wide studies have been carried out to identify driver genes responsible for tumorigenesis. SNP array analysis of 125 HCC cases have identified that four genes (CTNNB1 (32.8%), TP53 (20.8%), ARID1A (16.8%), and AXIN1 (15.2%)) are altered in more than 10% of the samples (Guichard et al., 2012). Whole exome sequencing analysis with 231 early-stage HCC Korean patient samples identified recurrent somatic mutations in CTNNB1 (23%) and TP53 (32%) (Ahn et al., 2014). In addition, CTNNB1 and TP53 were found to be frequently altered in a large cohort of HCC patient samples (Cancer Genome Atlas Research et al., 2013b; Fujimoto et al., 2012; Schulze et al., 2015). The CTNNB1 gene is also frequently altered in hepatocellular adenoma (HCA) (Pilati et al., 2014). β-catenin transgenic mouse models have been used to define a function of β-catenin in HCC tumorigenesis (Nejak-Bowen and Monga, 2011). Ectopic expression of either wild-type or mutant β-catenin is not sufficient to induce tumorigenesis (Harada et al., 2002; Nejak-Bowen et al., 2010). In some cases, β-catenin may accelerate tumorigenesis in cooperating with activated Ha-Ras (Harada et al., 2004) or heterozygote deleted Lkb1 (Miyoshi et al., 2009).
Colorectal cancer (CRC)
Wnt/β-cat signaling plays an important role in the tumorigenesis of CRC (Polakis, 2012b). In particular, alteration of APC, a negative regulator in Wnt signaling, is found in approximately 70% of CRC patients. Most APC alterations are truncation mutations, which cannot facilitate the proteolysis of β-catenin. In addition, loss of heterozygosity is frequently found in colorectal cancers. As shown in Table 1, genetic alterations also occurred in the CTNNB1 gene (5% of TCGA, 5% of DFCI, 6.9% of Genentech) (Giannakis et al., 2016; Seshagiri et al., 2012). Several studies reported that β-catenin has oncogenic activity in CRC cells, so the inhibition of β-catenin by gene targeting or knockdown resulted in growth inhibition of colorectal cancer cells (Cancer Genome Atlas, 2012; Green et al., 2001; Kim et al., 2002; Roh et al., 2001).
MUTATION HOTSPOTS IN EXON 3 (ENCODING THE N-TERMINAL REGION) OF THE β-CATENIN GENE
The β-catenin protein is composed of three domains: an N-terminal domain (~130 aa), a central domain (residue 141-664) made of 12 Armadillo (Arm) repeats and a C-terminal domain (~100 aa) (Fig. 3A). The central domain of the protein, the Arm repeats domain, forms a rigid rod-like structure and interacts with many binding proteins (Xu and Kimelman, 2007). However, it has been difficult to determine the structure of the terminal regions (N- and C-terminals) of β-catenin, so they are likely to be flexible and could be intrinsically disordered (Xing et al., 2008). Interestingly, the N-terminal region of the β-catenin protein is encoded by exon 3 (amino acid residues 5–80) of CTNNB1, so the N-terminal mutations can also be referred to as exon 3 mutations. CTNNB1 mutation hotspots were statistically analyzed by the Sorting Intolerant From Tolerant (SIFT) and the Polymorphism Phenotyping (PolyPhen) (Adzhubei et al., 2010; Naus, 1982; Sim et al., 2012).
Fig. 3. Diagram of β-catenin protein domains and hotspot mutations.
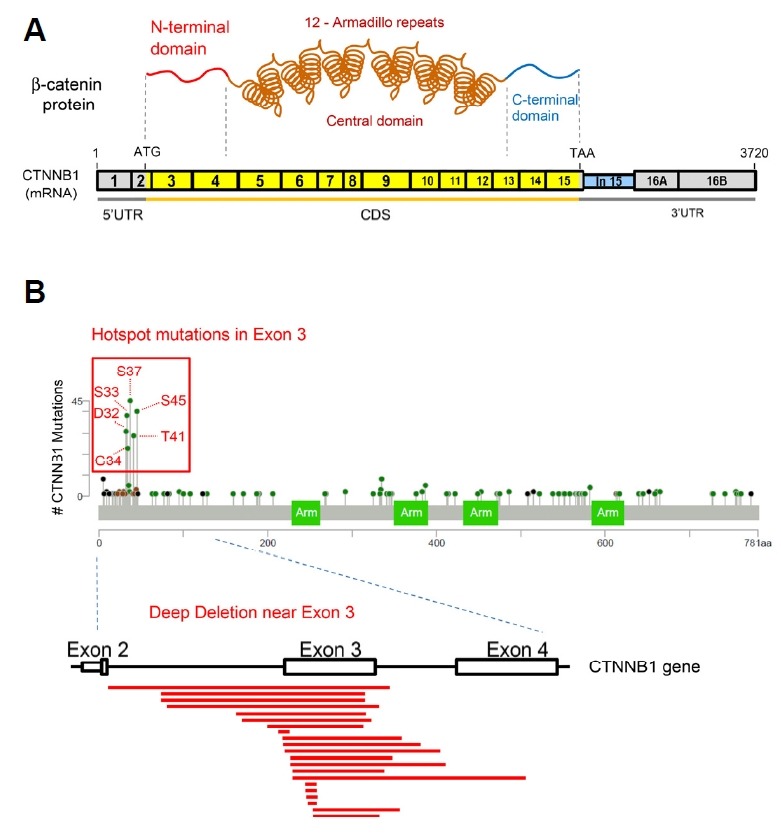
(A) A schematic diagram of the β-catenin protein and mRNA. UTR, untranslated region; CDS, coding sequence; ATG, translation start codon; TAA, translation stop codon. (B) Exon 3 hotspot mutations of CTNNB1 are marked on the lollipop plot downloaded from the MSK-IMPACT pan-cancer study on cBioportal. Deep deletions near Exon 3 of CTNNB1 pre-mRNA are indicated as red lines. Deletion data were obtained from metastatic colorectal cancer study (MSK) on cBioportal.
Missense mutations affecting the N-terminal region of the β-catenin protein
In most cancers, mutations are found in the N-terminal region of β-catenin, especially in exon 3 of β-catenin mRNA (Fig. 3B). In endometrial cancers, integrated analysis showed that exon 3 mutations in β-catenin mRNA are associated with an aggressive phenotype of low-grade and low-stage in younger women (Liu et al., 2014). These studies suggest that β-catenin mutations can be a prognostic marker for aggressive endometrial cancer. Additionally, in liver cancer, hotspot mutations in CTNNB1 were deeply analyzed in a large cohort of patients from HCA to carcinoma (HCC). S45, K335, and N387 mutations result in weak activation of β-catenin and are frequently found in HCA. T41 mutations show relatively moderate activation. Exon 3 deletion and β-TrCP binding site (D32-S37) mutations show strong activation and are enriched in HCA/HCC borderline and HCC, respectively. Highly activated β-catenin is associated with malignant tumors, as evaluated by glutamine synthase staining. Although S45 mutations show weak activation, most S45 mutant alleles in HCC are duplicated, resulting in strong activation of β-catenin. This study suggests that HCA harboring high/moderate mutants or S45 mutants may be associated with malignant transformation (Rebouissou et al., 2016). Accelerated liver regeneration and hepatocarcinogenesis was also observed in mouse overexpressing S45 mutant β-catenin (Nejak-Bowen et al., 2010). In colorectal cancers, most somatic mutations are observed at D32, S33, G34, S37, T41, and S45 in exon 3 of β-catenin mRNA. These hotspot mutations have been shown to stabilize β-catenin by disturbing the phosphorylation-dependent ubiquitination, leading to tumorigenesis. S45 is a priming-phosphorylation site for Casein Kinase I alpha (CK1α) (Liu et al., 2002). S33, S37, and T41 are further phosphorylated by GSK3β. D32 and G34 is required to bind with β-TrCP, a component of ubiquitin E3 ligase for phosphorylated β-catenin (Aberle et al., 1997; Hart et al., 1998).
Exon 3-spanning in-frame deletion in metastatic colorectal cancers
Recently, prospective targeted sequencing was reported with metastatic and early-stage colorectal cancer patients of a large cohort study (Yaeger et al., 2018). In this MSK study, the frequency of CTNNB1 alterations (8%) is slightly higher than that in TCGA cohort (5% of TCGA pan-cancer atlas), but in-frame deletion is highly enriched in the MSK cohort. This difference may be due to the distinct features between MSK and TCGA cohorts. The MSK cohort includes 47% of metastases that were not included in TCGA cohort, representing more aggressive and advanced disease. Activating hotspot mutations of β-catenin are more frequently occurred in microsatellite instability-high (MSI-H) tumors than in microsatellite stable (MSS) tumors (25% of MSI-H, 6% of MSS). Interestingly, however, exon 3-spanning in-frame deletions were identified only in MSS tumors and the nuclear staining of β-catenin was observed in tumors harboring inframe deletions in CTNNB1 (Yaeger et al., 2018).
CONCLUSION
Large-scale analysis of pan-cancer genomic database revealed a high frequency of CTNNB1 mutations in endometrial, liver, and colorectal cancers. In addition, mutations are frequently located near exon 3 of CTNNB1, which encode for the regulatory amino acids (D32, S33, G34, S37, T41, and S45) at the N-terminal region of the protein. Since the N-terminal region is highly unstructured and flexible, the contributions of N-terminal hotspot mutations from a structural perspective are not easy to comprehend (Dar et al., 2017; Gottardi and Peifer, 2008; Xing et al., 2008). Rather, their contribution to cancer development should be understood in terms of their roles in normal and pathogenic epithelial cell states.
FUTURE PERSPECTIVES
Re-evaluating hotspot mutations
The high frequency of mutations affecting the GSK3β and β-TrCP-binding sites (D32, S33, G34, S37) can be explained by their roles in the β-catenin destruction complex (Megy et al., 2005; Stamos and Weis, 2013). However, higher frequencies of S45 and T41 mutations cannot be easily explained in terms of the residues for priming and relay kinases, respectively. In fact, recent study suggested the uncoupling of CK1α phosphorylation on S45 residue to GSK3β phosphorylation on S37/S33 residues. The phosphorylations on the T41/S45 residues of β-catenin were spatially uncoupled from the phosphorylated S33/S37/T41 (Maher et al., 2010). In addition, a previous study reported that the phosphorylations on S33/S37/T41 can occur in the absence of the phospho-S45 in colon cancer cells (Wang et al., 2003). In desmoid-type fibromatosis, protein stability and target genes for the S45F mutant are different from those of the wild-type β-catenin (Colombo et al., 2017). Moreover, the S45F mutation is associated with low efficacy of a cyclooxygenase-2 inhibitor in desmoid tumors (Hamada et al., 2014). It will be important to determine the oncogenic role of the S45 mutant β-catenin protein, as a type of mutation distinct from other mutant β-catenin proteins.
β-catenin in multiple protein complexes
β-Catenin protein was first discovered as a component of the adherens junction (Ozawa et al., 1989). Later, it is considered as a multitasking protein involved in transcription as well as in cell adhesion (Hur and Jeong, 2013; Kumar and Bashyam, 2017; Valenta et al., 2012). However, it should be noted that most β-catenin proteins reside in the adhesion complex near the plasma membrane in which it interacts with E-cadherin and α-catenin with high affinities (Huber and Weis, 2001). Multiple roles of β-catenin protein may come from multiprotein assembly forming distinct complexes in different intracellular locations (Xu and Kimelman, 2007). In the nucleus, β-catenin associates with DNA binding proteins, such as TCF/LEF and BCL9 (Graham et al., 2001; Sampietro et al., 2006). Collectively, the N-terminal region of β-catenin is critical for regulating the adhesion and transcription functions of the protein. Thus, the regulatory mechanism of phosphorylation may differ in distinct β-catenin complexes (Dar et al., 2016). Therefore, many questions arise as to whether the specific mutant β-catenin proteins can form a previously unknown complex, in addition to the adhesion, destruction, and transcription complexes (Fig. 4). We hope that the clinical information gained from the large cancer genome databases could facilitate the study of novel functions of β-catenin in RNA metabolism as an RNA-binding protein (Hur and Jeong, 2013; Kim et al., 2009; Kim et al., 2012; Lee and Jeong, 2006). To enhance our understanding of such novel functions, a systematic mutant β-catenin library could be developed to link the differential functional impacts to specific mutations in cancer. More functional studies on specific mutant β-catenin proteins will open up new avenues for elucidating the mechanisms underlying mutant β-catenin-mediated oncogenesis.
Fig. 4. Proposed model for the role of mutant β-catenin proteins in distinct complexes.
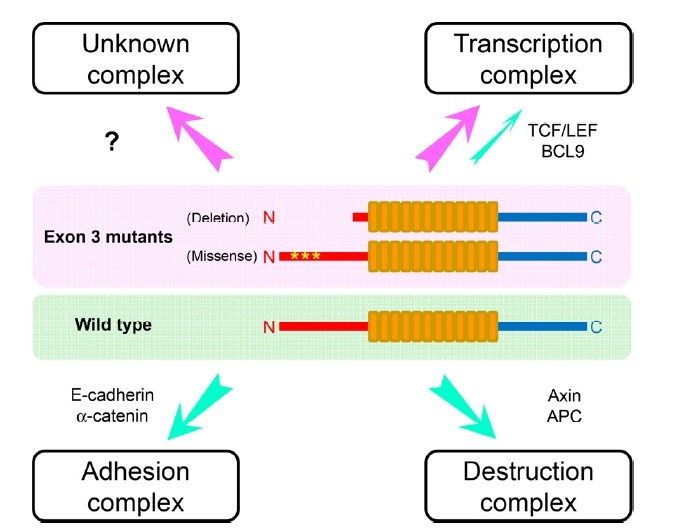
The green indicates wild-type β-catenin protein and the pink indicates exon 3-mutated β-catenin proteins. In addition to the adhesion, destruction and transcription complexes with the indicated proteins, additional unknown protein complexes are likely to be formed by the mutant β-catenin proteins.
Novel therapeutic approach for mutant β-catenin proteins
β-Catenin protein has been a prime target for anti-cancer drug development, but some limitations may suspend successful drug development. In most cases, wild-type β-catenin protein have been utilized as a target protein and Wnt signaling activated transcription is used as a screening read-out (Cui et al., 2018; Krishnamurthy and Kurzrock, 2018; Polakis, 2012a). As a novel strategy, the information obtained for mutant β-catenin can be implemented for mutant-specific anti-cancer therapeutics, as utilized for mutant p53 proteins (Bykov et al., 2018; Kotler et al., 2018). Large-scale clinical analysis could provide important information on the functions of cancer-related proteins and cancer signaling, as shown here (Hyman et al., 2017). Therefore, future research should be directed toward a precision oncology strategy by identifying the molecular signature of cancer-related genes and exploiting cancer genome databases (Zehir et al., 2017).
ACKNOWLEDGMENTS
We apologize to the researchers whose works are not cited because of the space limitations. This study was supported by a grant from Dankook University (2016), Korea Research Foundation (NRF-2013R1A1A2057796 and NRF-2016R1A2B2007879) and the National R&D Program for Cancer Control, Ministry for Health and Welfare, Republic of Korea (1320170).
REFERENCES
- Aberle H., Bauer A., Stappert J., Kispert A., Kemler R. β-catenin is a target for the ubiquitin-proteosome pathway. EMBO J. 1997;16:3797–3804. doi: 10.1093/emboj/16.13.3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adzhubei I.A., Schmidt S., Peshkin L., Ramensky V.E., Gerasimova A., Bork P., Kondrashov A.S., Sunyaev S.R. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn S.M., Jang S.J., Shim J.H., Kim D., Hong S.M., Sung C.O., Baek D., Haq F., Ansari A.A., Lee S.Y., et al. Genomic portrait of resectable hepatocellular carcinomas: implications of RB1 and FGF19 aberrations for patient stratification. Hepatology. 2014;60:1972–1982. doi: 10.1002/hep.27198. [DOI] [PubMed] [Google Scholar]
- Blum A., Wang P., Zenklusen J.C. SnapShot: TCGA-analyzed tumors. Cell. 2018;173:530. doi: 10.1016/j.cell.2018.03.059. [DOI] [PubMed] [Google Scholar]
- Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- Bykov V.J.N., Eriksson S.E., Bianchi J., Wiman K.G. Targeting mutant p53 for efficient cancer therapy. Nat Rev Cancer. 2018;18:89–102. doi: 10.1038/nrc.2017.109. [DOI] [PubMed] [Google Scholar]
- Cancer Genome Atlas N. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research N. Kandoth C., Schultz N., Cherniack A.D., Akbani R., Liu Y., Shen H., Robertson A.G., Pashtan I., Shen R., et al. Integrated genomic characterization of endometrial carcinoma. Nature. 2013a;497:67–73. doi: 10.1038/nature12113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research N. Weinstein J.N., Collisson E.A., Mills G.B., Shaw K.R., Ozenberger B.A., Ellrott K., Shmulevich I., Sander C., Stuart J.M. The cancer genome atlas pan-cancer analysis project. Nat Genet. 2013b;45:1113–1120. doi: 10.1038/ng.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerami E., Gao J., Dogrusoz U., Gross B.E., Sumer S.O., Aksoy B.A., Jacobsen A., Byrne C.J., Heuer M.L., Larsson E., et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo C., Belfiore A., Paielli N., De Cecco L., Canevari S., Laurini E., Fermeglia M., Pricl S., Verderio P., Bottelli S., et al. β-catenin in desmoid-type fibromatosis: deep insights into the role of T41A and S45F mutations on protein structure and gene expression. Mol Oncol. 2017;11:1495–1507. doi: 10.1002/1878-0261.12101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui C., Zhou X., Zhang W., Qu Y., Ke X. Is β-catenin a druggable target for cancer therapy? Trends Biochem Sci. 2018;43:623–634. doi: 10.1016/j.tibs.2018.06.003. [DOI] [PubMed] [Google Scholar]
- Dar M.S., Singh P., Mir R.A., Dar M.J. β-catenin N-terminal domain: An enigmatic region prone to cancer causing mutations. Mutat Res. 2017;773:122–133. doi: 10.1016/j.mrrev.2017.06.001. [DOI] [PubMed] [Google Scholar]
- Dar M.S., Singh P., Singh G., Jamwal G., Hussain S.S., Rana A., Akhter Y., Monga S.P., Dar M.J. Terminal regions of β-catenin are critical for regulating its adhesion and transcription functions. Biochim Biophys Acta. 2016;1863:2345–2357. doi: 10.1016/j.bbamcr.2016.06.010. [DOI] [PubMed] [Google Scholar]
- Fujimoto A., Totoki Y., Abe T., Boroevich K.A., Hosoda F., Nguyen H.H., Aoki M., Hosono N., Kubo M., Miya F., et al. Whole-genome sequencing of liver cancers identifies etiological influences on mutation patterns and recurrent mutations in chromatin regulators. Nat Genet. 2012;44:760–764. doi: 10.1038/ng.2291. [DOI] [PubMed] [Google Scholar]
- Gao J., Aksoy B.A., Dogrusoz U., Dresdner G., Gross B., Sumer S.O., Sun Y., Jacobsen A., Sinha R., Larsson E., et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannakis M., Mu X.J., Shukla S.A., Qian Z.R., Cohen O., Nishihara R., Bahl S., Cao Y., Amin-Mansour A., Yamauchi M., et al. Genomic correlates of immune-cell infiltrates in colorectal carcinoma. Cell Rep. 2016;15:857–865. doi: 10.1016/j.celrep.2016.03.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottardi C.J., Peifer M. Terminal regions of β-catenin come into view. Structure. 2008;16:336–338. doi: 10.1016/j.str.2008.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham T.A., Ferkey D.M., Mao F., Kimelman D., Xu W. Tcf4 can specifically recognize β-catenin using alternative conformations. Nat Struct Biol. 2001;8:1048–1052. doi: 10.1038/nsb718. [DOI] [PubMed] [Google Scholar]
- Green D.W., Roh H., Pippin J.A., Drebin J.A. β-catenin antisense treatment decreases β-catenin expression and tumor growth rate in colon carcinoma xenografts. J Surg Res. 2001;101:16–20. doi: 10.1006/jsre.2001.6241. [DOI] [PubMed] [Google Scholar]
- Guichard C., Amaddeo G., Imbeaud S., Ladeiro Y., Pelletier L., Maad I.B., Calderaro J., Bioulac-Sage P., Letexier M., Degos F., et al. Integrated analysis of somatic mutations and focal copy-number changes identifies key genes and pathways in hepatocellular carcinoma. Nat Genet. 2012;44:694–698. doi: 10.1038/ng.2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada S., Futamura N., Ikuta K., Urakawa H., Kozawa E., Ishiguro N., Nishida Y. CTNNB1 S45F mutation predicts poor efficacy of meloxicam treatment for desmoid tumors: a pilot study. PloS one. 2014;9:e96391. doi: 10.1371/journal.pone.0096391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada N., Miyoshi H., Murai N., Oshima H., Tamai Y., Oshima M., Taketo M.M. Lack of tumorigenesis in the mouse liver after adenovirus-mediated expression of a dominant stable mutant of β-catenin. Cancer Res. 2002;62:1971–1977. [PubMed] [Google Scholar]
- Harada N., Oshima H., Katoh M., Tamai Y., Oshima M., Taketo M.M. Hepatocarcinogenesis in mice with β-catenin and Ha-ras gene mutations. Cancer Res. 2004;64:48–54. doi: 10.1158/0008-5472.can-03-2123. [DOI] [PubMed] [Google Scholar]
- Hart M., Concordet J-P, Lassot I., A;bert O, Santos E., Durand H., Perret C., Rubinfeld B., Margottin F., Benarous R., et al. The F-box protein β-TrCP associates with phosphorylated β-catenin and regulates its activity in the cell. Curr Biol. 1998;9:207–210. doi: 10.1016/s0960-9822(99)80091-8. [DOI] [PubMed] [Google Scholar]
- Huber A.H., Weis W.I. The structure of the β-catenin/E-cadherin complex and the molecular basis of diverse ligand recognition by β-catenin. Cell. 2001;105:391–402. doi: 10.1016/s0092-8674(01)00330-0. [DOI] [PubMed] [Google Scholar]
- Hur J., Jeong S. Multitasking β-catenin: from adhesion and transcription to RNA regulation. Anim Cells Syst. 2013;17:299–305. [Google Scholar]
- Hutter C., Zenklusen J.C. The Cancer Genome Atlas: Creating Lasting Value beyond Its Data. Cell. 2018;173:283–285. doi: 10.1016/j.cell.2018.03.042. [DOI] [PubMed] [Google Scholar]
- Hyman D.M., Taylor B.S., Baselga J. Implementing Genome-Driven Oncology. Cell. 2017;168:584–599. doi: 10.1016/j.cell.2016.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H., Vick P., Hedtke J., Ploper D., De Robertis E.M. Wnt signaling translocates Lys48-linked polyubiquitinated proteins to the lysosomal pathway. Cell Rep. 2015;11:1151–1159. doi: 10.1016/j.celrep.2015.04.048. [DOI] [PubMed] [Google Scholar]
- Kim I., Kwak H., Lee H.K., Hyun S., Jeong S. β-Catenin recognizes a specific RNA motif in the cyclooxygenase-2 mRNA 3′-UTR and interacts with HuR in colon cancer cells. Nucleic Acids Res. 2012;40:6863–6872. doi: 10.1093/nar/gks331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.S., Crooks H., Foxworth A., Waldman T. Proof-of-principle: oncogenic β-catenin is a valid molecular target for the development of pharmacological inhibitors. Mol Cancer Ther. 2002;1:1355–1359. [PubMed] [Google Scholar]
- Kim M.Y., Hur J., Jeong S. Emerging roles of RNA and RNA-binding protein network in cancer cells. BMB Rep. 2009;42:125–130. doi: 10.5483/bmbrep.2009.42.3.125. [DOI] [PubMed] [Google Scholar]
- Kim S.E., Huang H., Zhao M., Zhang X., Zhang A., Semonov M.V., MacDonald B.T., Zhang X., Garcia Abreu J., Peng L., et al. Wnt stabilization of β-catenin reveals principles for morphogen receptor-scaffold assemblies. Science. 2013;340:867–870. doi: 10.1126/science.1232389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korinek V., Barker N., Morin P.J., van Wichen D., de Weger R., Kinzler K.W., Vogelstein B., Clevers H. Constitutive transcriptional activation by a β-catenin-Tcf complex in APC−/− colon carcinoma. Science. 1997;275:1784–1787. doi: 10.1126/science.275.5307.1784. [DOI] [PubMed] [Google Scholar]
- Kotler E., Shani O., Goldfeld G., Lotan-Pompan M., Tarcic O., Gershoni A., Hopf T.A., Marks D.S., Oren M., Segal E. A systematic p53 mutation library links differential functional impact to cancer mutation pattern and evolutionary conservation. Mol Cell. 2018;71:178–190 e178. doi: 10.1016/j.molcel.2018.06.012. [DOI] [PubMed] [Google Scholar]
- Krishnamurthy N., Kurzrock R. Targeting the Wnt/β-catenin pathway in cancer: Update on effectors and inhibitors. Cancer Treat Rev. 2018;62:50–60. doi: 10.1016/j.ctrv.2017.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar R., Bashyam M.D. Multiple oncogenic roles of nuclear β-catenin. J Biosci. 2017;42:695–707. doi: 10.1007/s12038-017-9710-9. [DOI] [PubMed] [Google Scholar]
- Kurnit K.C., Kim G.N., Fellman B.M., Urbauer D.L., Mills G.B., Zhang W., Broaddus R.R. CTNNB1 (β-catenin) mutation identifies low grade, early stage endometrial cancer patients at increased risk of recurrence. Mod Pathol. 2017;30:1032–1041. doi: 10.1038/modpathol.2017.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Gallo M., Rudd M.L., Urick M.E., Hansen N.F., Zhang S., Program N.C.S., Lozy F., Sgroi D.C., Vidal Bel A., Matias-Guiu X., et al. Somatic mutation profiles of clear cell endometrial tumors revealed by whole exome and targeted gene sequencing. Cancer. 2017;123:3261–3268. doi: 10.1002/cncr.30745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H.K., Jeong S. β-Catenin stabilizes cyclooxygenase-2 mRNA by interacting with AU-rich elements of 3′-UTR. Nucleic acids Res. 2006;34:5705–5714. doi: 10.1093/nar/gkl698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li V.S., Ng S.S., Boersema P.J., Low T.Y., Karthaus W.R., Gerlach J.P., Mohammed S., Heck A.J., Maurice M.M., Mahmoudi T., et al. Wnt signaling through inhibition of β-catenin degradation in an intact Axin1 complex. Cell. 2012;149:1245–1256. doi: 10.1016/j.cell.2012.05.002. [DOI] [PubMed] [Google Scholar]
- Liu C., Li Y., Semenov M., Han C., Baeg G-H, Tan Y., Zhang Z., Lin X., He X. Control of β-cateninphosphorylation/degradation by a dual-kinase mechanism. Cell. 2002;108:837–847. doi: 10.1016/s0092-8674(02)00685-2. [DOI] [PubMed] [Google Scholar]
- Liu Y., Patel L., Mills G.B., Lu K.H., Sood A.K., Ding L., Kucherlapati R., Mardis E.R., Levine D.A., Shmulevich I., et al. Clinical significance of CTNNB1 mutation and Wnt pathway activation in endometrioid endometrial carcinoma. J Natl Cancer Inst. 2014;106 doi: 10.1093/jnci/dju245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maher M.T., Mo R., Flozak A.S., Peled O.N., Gottardi C.J. β-catenin phosphorylated at serine 45 is spatially uncoupled from β-catenin phosphorylated in the GSK3 domain: implications for signaling. PloS one. 2010;5:e10184. doi: 10.1371/journal.pone.0010184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megy S., Bertho G., Gharbi-Benarous J., Baleux F., Benarous R., Girault J.P. Solution structure of a peptide derived from the oncogenic protein β-Catenin in its phosphorylated and nonphosphorylated states. Peptides. 2005;26:227–241. doi: 10.1016/j.peptides.2004.09.021. [DOI] [PubMed] [Google Scholar]
- Miyoshi H., Deguchi A., Nakau M., Kojima Y., Mori A., Oshima M., Aoki M., Taketo M.M. Hepatocellular carcinoma development induced by conditional β-catenin activation in Lkb1+/− mice. Cancer Sci. 2009;100:2046–2053. doi: 10.1111/j.1349-7006.2009.01284.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin P.J., Sparks A.B., Korinek V., Barker N., Clevers H., Vogelstein B., Kinzler K.W. Activation of β-catenin-Tcf signaling in colon cancer by mutations in β-catenin or APC. Science. 1997;275:1787–1790. doi: 10.1126/science.275.5307.1787. [DOI] [PubMed] [Google Scholar]
- Naus J.I. Approximations for distributions of scan statistics. J Am Stat Assoc. 1982;77:177–183. [Google Scholar]
- Nejak-Bowen K.N., Monga S.P. β-catenin signaling, liver regeneration and hepatocellular cancer: sorting the good from the bad. Semin Cancer Biol. 2011;21:44–58. doi: 10.1016/j.semcancer.2010.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nejak-Bowen K.N., Thompson M.D., Singh S., Bowen W.C., Dar M.J., Khillan J., Dai C., Monga S.P.S. Accelerated liver regeneration and hepatocarcinogenesis in mice overexpressing serine-45 mutant β-catenin. Hepatology. 2010;51:1603–1613. doi: 10.1002/hep.23538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nusse R., Clevers H. Wnt/β-Catenin Signaling, Disease, and Emerging Therapeutic Modalities. Cell. 2017;169:985–999. doi: 10.1016/j.cell.2017.05.016. [DOI] [PubMed] [Google Scholar]
- Ozawa M., Baribault H., Kemler R. The cytoplasmic domain of the cell adhesion molecule uvomorulin associates with three independent proteins structurally related in different species. EMBO J. 1989;8:1711–1717. doi: 10.1002/j.1460-2075.1989.tb03563.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilati C., Letouze E., Nault J.C., Imbeaud S., Boulai A., Calderaro J., Poussin K., Franconi A., Couchy G., Morcrette G., et al. Genomic profiling of hepatocellular adenomas reveals recurrent FRK-activating mutations and the mechanisms of malignant transformation. Cancer cell. 2014;25:428–441. doi: 10.1016/j.ccr.2014.03.005. [DOI] [PubMed] [Google Scholar]
- Polakis P. The many ways of Wnt in cancer. Curr Opin Genet Dev. 2007;17:45–51. doi: 10.1016/j.gde.2006.12.007. [DOI] [PubMed] [Google Scholar]
- Polakis P. Drugging Wnt signalling in cancer. EMBO J. 2012a;31:2737–2746. doi: 10.1038/emboj.2012.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polakis P. Wnt signaling in cancer. Cold Spring Harbor perspectives in biology. 2012b;4 doi: 10.1101/cshperspect.a008052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebouissou S., Franconi A., Calderaro J., Letouze E., Imbeaud S., Pilati C., Nault J.C., Couchy G., Laurent A., Balabaud C., et al. Genotype-phenotype correlation of CTNNB1 mutations reveals different ss-catenin activity associated with liver tumor progression. Hepatology. 2016;64:2047–2061. doi: 10.1002/hep.28638. [DOI] [PubMed] [Google Scholar]
- Roh H., Green D.W., Boswell C.B., Pippin J.A., Drebin J.A. Suppression of β-catenin inhibits the neoplastic growth of APC-mutant colon cancer cells. Cancer Res. 2001;61:6563–6568. [PubMed] [Google Scholar]
- Sampietro J., Dahlberg C.L., Cho U.S., Hinds T.R., Kimelman D., Xu W. Crystal structure of a β-catenin/BCL9/Tcf4 complex. Mol Cell. 2006;24:293–300. doi: 10.1016/j.molcel.2006.09.001. [DOI] [PubMed] [Google Scholar]
- Schulze K., Imbeaud S., Letouze E., Alexandrov L.B., Calderaro J., Rebouissou S., Couchy G., Meiller C., Shinde J., Soysouvanh F., et al. Exome sequencing of hepatocellular carcinomas identifies new mutational signatures and potential therapeutic targets. Nat Genet. 2015;47:505–511. doi: 10.1038/ng.3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seshagiri S., Stawiski E.W., Durinck S., Modrusan Z., Storm E.E., Conboy C.B., Chaudhuri S., Guan Y., Janakiraman V., Jaiswal B.S., et al. Recurrent R-spondin fusions in colon cancer. Nature. 2012;488:660–664. doi: 10.1038/nature11282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sim N.L., Kumar P., Hu J., Henikoff S., Schneider G., Ng P.C. SIFT web server: predicting effects of amino acid substitutions on proteins. Nucleic Acids Res. 2012;40:W452–457. doi: 10.1093/nar/gks539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soumerai T.E., Donoghue M.T.A., Bandlamudi C., Srinivasan P., Chang M.T., Zamarin D., Cadoo K.A., Grisham R.N., O’Cearbhaill R.E., Tew W.P., et al. Clinical utility of prospective molecular characterization in advanced endometrial cancer. Clin Cancer Res. 2018;24:5939–5947. doi: 10.1158/1078-0432.CCR-18-0412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamos J.L., Weis W.I. The β-catenin destruction complex. Cold Spring Harb Perspect Biol. 2013;5:a007898. doi: 10.1101/cshperspect.a007898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taelman V.F., Dobrowolski R., Plouhinec J.L., Fuentealba L.C., Vorwald P.P., Gumper I., Sabatini D.D., De Robertis E.M. Wnt signaling requires sequestration of glycogen synthase kinase 3 inside multivesicular endosomes. Cell. 2010;143:1136–1148. doi: 10.1016/j.cell.2010.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomczak K., Czerwinska P., Wiznerowicz M. The cancer genome atlas (TCGA): an immeasurable source of knowledge. Contemp Oncol (Pozn) 2015;19:A68–77. doi: 10.5114/wo.2014.47136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenta T., Hausmann G., Basler K. The many faces and functions of β-catenin. EMBO J. 2012;31:2714–2736. doi: 10.1038/emboj.2012.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Zee M., Jia Y., Wang Y., Heijmans-Antonissen C., Ewing P.C., Franken P., DeMayo F.J., Lydon J.P., Burger C.W., Fodde R., et al. Alterations in Wnt-β-catenin and Pten signalling play distinct roles in endometrial cancer initiation and progression. J Pathol. 2013;230:48–58. doi: 10.1002/path.4160. [DOI] [PubMed] [Google Scholar]
- Wang Z.H., Vogelstein B., Kinzler K.W. Phosphorylation of β-catenin at S33, S37, or T41 can occur in the absence of phosphorylation at T45 in colon cancer cells. Cancer Res. 2003;63:5234–5235. [PubMed] [Google Scholar]
- Xing Y., Takemaru K., Liu J., Berndt J.D., Zheng J.J., Moon R.T., Xu W. Crystal structure of a full-length β-catenin. Structure. 2008;16:478–487. doi: 10.1016/j.str.2007.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W., Kimelman D. Mechanistic insights from structural studies of β-catenin and its binding partners. J Cell Sci. 2007;120:3337–3344. doi: 10.1242/jcs.013771. [DOI] [PubMed] [Google Scholar]
- Yaeger R., Chatila W.K., Lipsyc M.D., Hechtman J.F., Cercek A., Sanchez-Vega F., Jayakumaran G., Middha S., Zehir A., Donoghue M.T.A., et al. Clinical sequencing defines the genomic landscape of metastatic colorectal cancer. Cancer cell. 2018;33:125–136 e123. doi: 10.1016/j.ccell.2017.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zehir A., Benayed R., Shah R.H., Syed A., Middha S., Kim H.R., Srinivasan P., Gao J., Chakravarty D., Devlin S.M., et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med. 2017;23:703–713. doi: 10.1038/nm.4333. [DOI] [PMC free article] [PubMed] [Google Scholar]


