The relationship between the microbiome and the immune system is significant in human health and disease. Aberrations in both the gut microbiome and the immune system are associated with diseases such as inflammatory bowel disease [1], rheumatoid arthritis [2], asthma [3], HIV [4] and anti-PD-1 immunotherapy response failures in cancer patients [5–7]. There is an increasing understanding that microbiome-immune system interplay is a major factor driving immune phenotypes in the context of health and disease [8]. Microbiome-immune system interplay is of interest to the immunotherapy community and others in two ways. First, as suggested by the cancer immunotherapy research, we might be better able to stage or treat patients with a more refined understanding of which microbes interact with which immune cells to drive an immune response to cancer. In this scenario, a microbe–immune cell relationship could serve as a biomarker for response to treatment. Second, through improved understanding of particular mechanisms of interaction that impact disease and its progression, we may be able to identify particular microbes or sets of microbes that can be used as therapies [6]. In this commentary, we build the case for assessing both the microbiome and the immune system with high-dimensional technologies, performing integrative analysis of these datasets, and supporting visual analysis of the results with a goal of prioritizing relationships for follow-up experiments.
High-dimensional ‘omic’ techniques measure a large number of analytes that might differ with disease or response to treatment. These analytes can serve as biomarkers that predict response to therapy [7], or allow for the generation of testable hypotheses that lead to mechanistic insights [9]. Several mechanistic relationships between immune cells, gut microbes, and disease are already known. For example, Bacteroides fragilis, a common gut commensal, produces zwitterionic polysaccharides (ZPS), including polysaccharide A, which can induce IL-10 production by regulatory-T cells and has conferred protection from pathologic inflammation in mouse models [10]. Faecalibacterium prausnitzii, which comprises about 5% of the fecal microbiome in healthy adults, interacts with dendritic cells causing them to produce IL-10 [11]. Segmented filamentous bacteria induce the accumulation of CD4+ T cells that produce IL-17 and IL-22 [12]. We might reasonably speculate that there are a number of other bacteria:immune cell:cytokine mechanisms that we have not yet discovered, and that aberrations of these mechanisms might be associated with disease.
The case for integrative analysis
Techniques like 16S ribosomal RNA (rRNA)-targeted, shotgun metagenomic and transcriptomic sequencing, and metabolomics allow researchers to deeply interrogate microbiome composition and function. Similarly, omic technologies such as time of flight mass cytometry (CyTOF) (immunophenotyping), single cell RNASeq, and multiplex cytokine arrays (e.g., Luminex or MSD) support in depth characterization of immune cell repertoire and milieu. However, analyzing the resulting data as an integrated whole to identify microbiome-immune system interplay remains a challenge. Yet this integrative analysis, coupled with experimental follow-up, can facilitate the development of a mechanistic understanding of the contribution of the microbiome to disease [13]. Some authors report that integrative analysis can provide greater predictive accuracy for biomarkers of disease [14,15] than analyzing the data from a single assay in isolation. For example, combing methylation and gene expression offers better predictive power in The Cancer Genome Atlas glioblastoma multiforme dataset than does gene expression alone. The authors treat gene expression as a downstream event regulated by methylation, which is aligned with conventional mechanistic understanding [14]. Zhang et al. use this same data to show that combining gene expression, methylation, copy number and microRNA yields a better predictive model than does any one of these assays alone [16]. Importantly, interpreting the results of integrative analysis may require collaboration among experts in multiple domains such as microbiology and immunology. These thoughts are aligned with those of Manzoni and colleagues, who argue that fragmented niche groups, each with their own analytical programs, limit the flow of communication and data interpretation [17].
Commonly, 16S microbiome data may be presented as principal coordinate plots with one point per sample [6]; normalized bar charts of taxa per sample [6]; or relative abundance heat maps, with one square per sample per taxa [6,7]. Immunological readouts are often presented as scatterplots by group, with one point per sample [5,7]. A common approach for combining microbiome and immunological data is a heat map with one square per microbe-immune readout pair, with colors encoding the magnitude and direction of correlations [5,6]. We suggest that additional approaches for analyzing and visualizing this microbiome–immune system interplay are likely to provide additional insight.
At a high level, each omic assay yields data for a large number of name-value pairs. For example, we might collect data that the bacterial genus Bacteroides makes up 0.4 of the sequence reads from a microbial sample and that CD4+ T cells constitute 30% of the total T cells from a blood or tissue sample. Tersely, we can note that Bacteroides = 0.4 and CD4+ T cells = 30, with Bacteroides and CD4+ T cells being analyte names. We can combine analytes from multiple assays into a single dataset, and apply a variety of existing machine learning algorithms to these integrated data. These algorithms can support classification (e.g., decision trees, random forests and artificial neural networks), pattern finding (e.g., principal component analysis, association rule mining, hierarchical clustering and self-organizing maps) and numeric prediction (e.g., high-dimensional penalized regression such as LASSO, ridge and elastic net). We discuss many of these approaches in a review [18]. However, such machine learning approaches do not necessarily identify relationships that span ‘omes’, which is important for developing a better understanding of microbiome-immune system interplay.
Pairwise cross-omic regressions
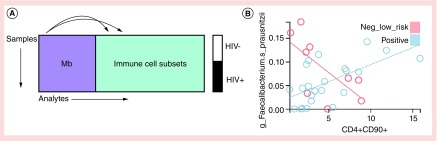
One approach for integrative analysis that spans omic data are pairwise regressions across the datasets, as illustrated in Figure 1A, with every microbe compared with every immune cell subset. In a dataset with 50 microbes and 100 immune cell subsets, this would generate 5000 regression results. On a contemporary personal computer, this processing takes less than 1 min. A representative regression result is shown in Figure 1B. In this example, the specified regression model allows the relationship between an immune cell population (as assessed with CyTOF technology) and a microbial taxon (as assessed with 16S rRNA sequencing) to differ based on disease state. The detailed plot illustrates the fitted relationship for each group with a colored line, and includes one point for the specific pair of microbe:immune cell measurements from samples drawn from one person, allowing us to visually assess the fit of the model.
Figure 1. . Pairwise cross-omic regressions.
(A) Pairwise linear regressions across two omic assays. Samples from HIV- or HIV+ individuals are assayed for microbiome relative abundance and for immune cell subsets. Values for each microbe are compared with values for each immune cell subset using a regression model of the form Mb ∝ HIV + IC + IC × HIV, where Mb is the microbe and IC is the immune cell and HIV is HIV status. This model allows the fitted relationships to differ by group. (B) A detailed plot shows the relationship between Faecalibacterium prausnitzii and immune cell subset CD4+CD90+. The relationship is positive for the HIV positive cohort, but negative for the HIV negative cohort.
The emerging technologies mentioned above give us greater ability than ever to begin to interrogate the complexity of microbe–immune interactions, at a level of detail that supports follow-up laboratory experiments with specific microbe strains and specific immune cell subsets. Although we hope to recover known interactions, such as associations between B. fragilis, IL-10 expression and regulatory-T cells in the gut mucosa, other findings may be highly exploratory. For example, we may identify novel immune cell phenotypes based on well-known populations (e.g., CD4 T cells) coupled with a variety of functional markers (e.g., CTLA4, CXCR5 or CD90), associated with microbes for which we have limited functional understanding. Given the likelihood that we may identify relationships that have not been previously reported in the literature, other information, such as correlation with disease phenotypes, may aid in prioritizing the most promising relationships for mechanistic follow-up.
The flexibility of the regression framework, coupled with the fine-grained resolution of the analytes, provides an approach that is fundamentally different from an approach that computes correlations for a few well-known immune cell populations to a few bacterial orders and families. For example, in Figure 1B, we show a positive relationship between F. prausnitzii and CD4+CD90+ T cells in HIV+ participants, but a negative relationship in HIV-participants. Conceptually, this observed relationship between a microbial species and an immune cell population that differs by disease state is sufficiently detailed as to suggest possibilities for follow-up experiments. For example, assuming the species classification is sufficiently specific, and that the microbe is culturable, we could combine the microbe with immune cells in an ex vivo experiment to see if the microbe could induce immune cell differentiation.
Furthermore, the regression framework allows us to explore microbe-immune system interplay in a wide variety of experimental designs. For example, we can inspect the relationships between microbes and immune cells, testing for a differential relationship between responders and nonresponders. This type of analysis is also well suited to our goal of understanding immune interactions that occur in the context of HIV infection (Figure 1). Since HIV infects and kills CD4+ T cells, a central regulator of the immune system, it is important to understand how microbe:immune relationships may change in the context of this disease. We might see a positive association in one group and a negative association in another. We can also account for multiple treatment arms, and for responders and nonresponders in each arm. We can account for longitudinal samples from the same person. We can include covariates like treatment center, sex and age.
Visual analysis of the top table
Of course, not all of the thousands of regression results are interesting. Therefore, we must first filter them by statistical significance to yield a ‘top table’ of candidate relationships similar to those derived from gene expression analysis. This top table gives us our next challenge: prioritizing the results for follow-up. To address this challenge, our team is developing a visual analysis environment for exploring regression results that characterize microbiome–immune system interplay [19]. Our approach involves close collaboration among experts in bioinformatics, microbiology and immunology. Hereafter, we refer to the microbiologists and immunologists as domain experts.
Given a statistically significant relationship, domain experts would like to inspect the underlying detail, vetting the result for scientific relevance. The domain experts may be interested in goodness of fit of the model, the magnitude of the readouts (are they large enough to be credible and measured with precision), the dynamic range of the readouts (are the changes from low to high large enough to correspond to changes in biological processes), the presence of outliers, and evidence of bimodal distributions. Importantly, both microbiome data and immunophenotyping data include analytes that are not well studied. Thus, another vetting criterion is that at least one of the analytes is of interest to the research community. If one of the analytes is well-known, a strong association with a lesser-studied analyte from another assay may generate interest in the novel analyte. Although the above discussion focuses on data generated by 16S sequencing and CyTOF immunophenotyping, these concepts apply to data generated by other techniques, such as transcriptomics, metabolomics, single cell RNA-Seq and multiplex cytokine arrays.
Visual analysis supports the refinement of a shared mental model among representatives of diverse scientific disciplines. The ability to interact with the data creates both interest and understanding. Interactive visual analysis also allows the domain experts to better communicate their data analysis needs and data nuances to the bioinformaticists. In our work-to-date, we have observed that interactive visual analysis allows domain experts to screen hundreds of candidate relationships quickly. It allows these experts to discuss their respective knowledge about particular microbes and immune readouts in the context of disease. It further allows them to group regression models based on metapatterns, such as ‘positive association in healthy patients but no association in disease’ or ‘negative association in disease but not in health’. Promising results can be further studied in the lab with ex vivo stimulation experiments and in mouse studies. We also anticipate that analyzing the top table of regression results with network-based approaches will provide additional understanding [20,21].
In conclusion, better understanding of microbiome–immune system interplay may provide insight into microbe-based immunotherapies, and into patient responses to therapies. Technologies such as 16S sequencing, shotgun metagenomics, transcriptomics, CyTOF, single cell RNASeq and multiplex cytokine arrays allow us to simultaneously characterize large numbers of microbes, immune cell subsets and cytokines. Pairwise regression models across these data collections generate fine-grained results for visual analysis and vetting. These regression models also support a wide variety of experimental designs. Interactive visual analysis allows bioinformaticists, immunologists and microbiologists to refine a shared mental model, thereby strengthening the collaboration. The ability to interactiveIy explore results allow researchers to quickly prioritize results for follow-up, increasing opportunities for discovery.
Footnotes
Financial & competing interests disclosure
The authors have funding from NIH Grant 5 R01 LM009254 11. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
- 1.Sartor RB. Microbial influences in inflammatory bowel diseases. Gastroenterology. 2008;134(2):577–594. doi: 10.1053/j.gastro.2007.11.059. [DOI] [PubMed] [Google Scholar]
- 2.Chen J, Wright K, Davis JM, et al. An expansion of rare lineage intestinal microbes characterizes rheumatoid arthritis. Genome Med. 2016;8:43. doi: 10.1186/s13073-016-0299-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huang YJ, Boushey HA. The microbiome in asthma. J. Allergy Clin. Immunol. 2015;135(1):25–30. doi: 10.1016/j.jaci.2014.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lozupone CA, Li M, Campbell TB, et al. Alterations in the gut microbiota associated with HIV-1 infection. Cell Host Microbe. 2013;14(3):329–339. doi: 10.1016/j.chom.2013.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Routy B, Chatelier EL, Derosa L, et al. Gut microbiome influences efficacy of PD-1–based immunotherapy against epithelial tumors. Science. 2018;359(6371):91–97. doi: 10.1126/science.aan3706. [DOI] [PubMed] [Google Scholar]
- 6.Gopalakrishnan V, Spencer CN, Nezi L, et al. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science. 2018;359(6371):97–103. doi: 10.1126/science.aan4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Matson V, Fessler J, Bao R, et al. The commensal microbiome is associated with anti-PD-1 efficacy in metastatic melanoma patients. Science. 2018;359(6371):104–108. doi: 10.1126/science.aao3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pabst O. Correlation, consequence, and functionality in microbiome-immune interplay. Immunol. Rev. 2017;279(1):4–7. doi: 10.1111/imr.12584. [DOI] [PubMed] [Google Scholar]
- 9.Arneson D, Shu L, Tsai B, Barrere-Cain R, Sun C, Yang X. Multidimensional integrative genomics approaches to dissecting cardiovascular disease. Front. Cardiovasc. Med. 2017;4 doi: 10.3389/fcvm.2017.00008. www.frontiersin.org/articles/10.3389/fcvm.2017.00008/full [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Neff CP, Rhodes ME, Arnolds KL, et al. Diverse intestinal bacteria contain putative zwitterionic capsular polysaccharides with anti-inflammatory properties. Cell Host Microbe. 2016;20(4):535–547. doi: 10.1016/j.chom.2016.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martín R, Bermúdez-Humarán LG, Langella P. Searching for the bacterial effector: the example of the multi-skilled commensal bacterium faecalibacterium prausnitzii. Front. Microbiol. 2018;9 doi: 10.3389/fmicb.2018.00346. www.frontiersin.org/articles/10.3389/fmicb.2018.00346/full [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ivanov II, Atarashi K, Manel N, et al. Induction of intestinal Th17 Cells by segmented filamentous bacteria. Cell. 2009;139(3):485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lozupone CA. Unraveling interactions between the microbiome and the host immune system to decipher mechanisms of disease. mSystems. 2018;3(2):e00183–17. doi: 10.1128/mSystems.00183-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang W, Baladandayuthapani V, Morris JS, Broom BM, Manyam G, Do K-A. iBAG: integrative Bayesian analysis of high-dimensional multiplatform genomics data. Bioinformatics. 2013;29(2):149–159. doi: 10.1093/bioinformatics/bts655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li Y, Wu F-X, Ngom A. A review on machine learning principles for multi-view biological data integration. Brief Bioinform. 2018;19(2):325–340. doi: 10.1093/bib/bbw113. [DOI] [PubMed] [Google Scholar]
- 16.Zhang Y, Li A, Peng C, Wang M. Improve glioblastoma multiforme prognosis prediction by using feature selection and multiple kernel learning. IEEE/ACM Trans. Comput. Biol. Bioinform. 2016;13(5):825–835. doi: 10.1109/TCBB.2016.2551745. [DOI] [PubMed] [Google Scholar]
- 17.Manzoni C, Kia DA, Vandrovcova J, et al. Genome, transcriptome and proteome: the rise of omics data and their integration in biomedical sciences. Brief Bioinform. 2018;19(2):286–302. doi: 10.1093/bib/bbw114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Siebert JC, Wagner BD, Juarez-Colunga E. Integrating and mining diverse data in human immunological studies. Bioanalysis. 2014;6(2):209–223. doi: 10.4155/bio.13.309. [DOI] [PubMed] [Google Scholar]
- 19.Siebert J, Neff CP, Schneider JM, Palmer B, Lozupone C, Gorg C. VOLARE: visual analysis of disease-associated microbiome-immune system interplay. bioRxiv. 2018:431379. doi: 10.1186/s12859-019-3021-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yabu JM, Siebert JC, Maecker HT. Immune profiles to predict response to desensitization therapy in highly HLA-sensitized kidney transplant candidates. PLoS ONE. 2016;11(4):e0153355. doi: 10.1371/journal.pone.0153355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Whiting CC, Siebert J, Newman AM, et al. Large-scale and comprehensive immune profiling and functional analysis of normal human aging. PLoS ONE. 2015;10(7):e0133627. doi: 10.1371/journal.pone.0133627. [DOI] [PMC free article] [PubMed] [Google Scholar]