Abstract
Objective:
The functional polarization of CD4 T cells determines their antimicrobial effector profile, but may also impact the susceptibility to infection with HIV-1. Here, we analyzed the susceptibility of CD4 T cells with different functional polarization to infection with X4- and R5-tropic HIV-1.
Methods:
CD4 T cells with a Th1, Th2, Th17 and Th9 polarization were subjected to in vitro infection assays with X4, R5 or VSV-G-pseudotyped HIV-1. In addition, we sorted differentially-polarized CD4 T cell subsets from individuals treated with antiretroviral therapy and analyzed the tropism of viral env sequences.
Results:
Th9-polarized CD4 T cells and, to a lesser extent, Th2-polarized CD4 T cells expressed higher surface levels of CXCR4, and are more permissive to X4-tropic infection in vitro. In contrast, Th1 and Th17 CD4 T cells exhibited stronger surface expression of CCR5, and were more susceptible to infection with R5-tropic viruses. Correspondingly, the distribution of X4-tropic viral sequences in ART-treated HIV-1-infected patients was biased towards Th9/Th2 cells, while R5-tropic sequences were more frequently observed in Th17 cells.
Conclusion:
CD4 T cell polarization is associated with a distinct susceptibility to X4- and R5-tropic HIV-1 infection.
Keywords: HIV-1, viral tropism, CD4 T cell polarization
Introduction
CD4 T cells represent the predominant target cell population for HIV-1, but the susceptibility to viral infection varies considerably among individual cells. These differences are likely related to the profound diversity among different CD4 T cell subpopulations, each of which exhibits a discrete phenotypic, functional and developmental profile [1]. Originally, CD4 T cells were dichotomized into Th1-polarized cells, responsible for antiviral immune defense and characterized by expression of IFN-γ and the master transcription factor t-bet, and Th2-polarized cells, which are involved in anti-helminthic and anti-parasitic immune protection and are typically identified by secretion of IL-4 and the transcription factor Gata-3 [2]. More recently, significant progress has been made in defining a range of alternative T helper cell subpopulations that were not previously recognized as distinct entities. These cells include Th9 cells, which contribute to anti-tumor immune defense through secretion of interleukin-9 [3], Th17 cells with important roles for anti-bacterial and anti-fungal immune protection [4], Th22 cells, T regulatory cells and T follicular helper cells [5].
An important aspect of CD4 T cell polarization is the expression of distinct phenotypic surface markers that determine the cellular trafficking and tissue homing behavior. Indeed, prior studies have defined specific clusters of chemokine receptors in functionally-polarized CD4 T cells that impact the cellular responsiveness to microenvironmental chemokine milieus and organize the distribution of cells in individual tissue compartments [6]. Since HIV-1 uses the chemokine receptors CXCR4 and CCR5 as co-receptors for viral entry, differences in chemokine receptor expression among cells with discrete functional polarization may profoundly affect the tropism of viral species a given cell is susceptible to [7–9]. In this study, we show opposing patterns in CXCR4/CCR5 surface expression between Th1 and Th17 cells on the one side, and Th2 or Th9 cells on the opposite side, and identify characteristic differences in the distribution of R5-tropic and X4-tropic viral species in these CD4 T cell subsets in vivo.
Methods
Patients
HIV-1 positive and negative study participants were recruited from the Massachusetts General Hospital and the Brigham and Women’s Hospital (both in Boston, MA, USA). PBMC samples were used according to protocols approved by the respective Institutional Review Boards. Study subjects gave written informed consent to participate in accordance with the Declaration of Helsinki.
Cell sorting and flow cytometry
Cryopreserved whole PBMC were thawed and cultured in RPMI medium supplemented with 10% FCS. Cells were stimulated for 12 hours with CD3/CD28 beads (Miltenyi Biotec) and for additional 3 hour with PMA/ionomycin (100ng/ml and 1ug/ml respectively), in the presence of Golgistop and Brefeldin A (both at 1 μg/ml) to inhibit cytokine release. Cells were surface stained with monoclonal antibodies to CXCR4, CCR5 (during initial stimulation), CD4, CD3, CD45RO (during the last 3 hours of stimulation), and with LIVE/DEAD Fixable Blue (Thermofisher) as a viability dye. Cells were then fixed and permeabilized using commercially available buffers (BioLegend), followed by intracellular cytokine staining with antibodies directed against IFN-γ, IL-4, IL-9, and IL-17. Afterwards, cells were washed and indicated cell populations were analyzed by flow cytometry on BD LSR Fortessa or sorted in a specifically designated biosafety cabinet (Baker Hood), using a FACS Aria cell sorter (BD Biosciences) at 70 pounds per square inch. Cell sorting was performed by the Ragon Institute Imaging Core Facility at Massachusetts General Hospital, and resulted in isolation of lymphocytes with the defined phenotypic characteristics of >95% purity.
In vitro HIV-1 infection assays
Freshly isolated whole PBMC from HIV-1 negative donors were cultured in RPMI medium (supplemented with 10% FCS and 50 U/ml of rhIL-2) and infected (MOI=1, unless otherwise indicated) with a GFP-encoding VSV-G-pseudotyped virus or GFP-encoding R5-tropic (Ba-L) or X4-tropic (NL4–3) viral strains. After two hours, cells were washed with PBS and cultured at 200,000 cells/well in 96-well round-bottom plates for 2 days (VSV-G pseudotyped HIV-1) or 4 days (R5/X4-tropic HIV-1). Afterwards, cells were stimulated and subjected to surface and intracellular antibody staining as described above, and analyzed on a LSRII flow cytometer.
Viral sequencing and tropism analysis
Cell lysates from 5 sorted CD4 T cell subpopulations from 7 HIV-1 positive individuals were used for single-genome HIV-1 envelope sequence analysis encompassing the V3 region. HIV-1 DNA concentrations in each CD4 T cell population were measured by ddPCR. In order to obtain single genome HIV-1 sequences, PCR plates were set up with cellular lysate dilutions yielding <30% positive PCR reactions for each cell population, according to a protocol described before [10]. Two sequential PCR reactions were performed using outer primers envA/LA11 and inner primers LA12 and LA13 [11]. Amplification products were then purified and sequenced using standard Sanger sequencing procedures. Sequences were aligned with an HXB2 reference sequence using MEGA6.06 [12]. A neighbor-joining method was used to construct phylogenetic trees with phylogenetically informative HIV-1 nucleotide sequences. Viral tropism was determined using the geno2pheno program (http://coreceptor.bioinf.mpi-inf.mpg.de/), using the original g2p coreceptor prediction method and 10% false positive rate (FPR).
Statistics
Data are summarized as individual data plots with horizontal lines reflecting the median. Differences were tested for statistical significance using Fisher’s exact test, or ANOVA followed by Dunn’s test for multiple comparisons, as appropriate.
Results
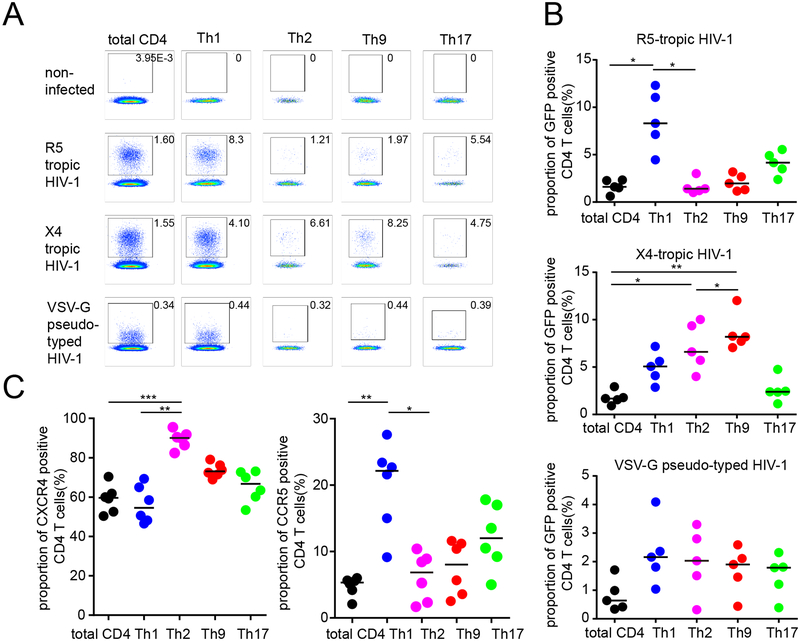
To investigate the influence of viral tropism on the susceptibility of differentially-polarized CD4 T cell populations to HIV-1, we initially conducted ex-vivo infection experiments. For this purpose, CD4 T cells from freshly isolated whole PBMC samples were in vitro infected with GFP-encoding R5-tropic, X4-tropic or VSV-G pseudotyped HIV-1 strains. At day 4, the cells were subjected to sequential stimulations with CD3/CD28 beads and PMA/Ionomycin, and processed to intracellular cytokine staining for IFN-γ, IL-4, IL-9 and IL-17, the signature cytokines for Th1, Th2, Th9 and Th17 cells, respectively; cells simultaneously secreting more than one cytokine, indicative of mixed functional polarizations, were not considered for this analysis. Subsequently, the proportion of GFP+ events in CD4 T cells secreting any of the selected cytokines, was determined by flow cytometry (Figure 1A); proportions of GFP+ events in total CD4 T cells were assessed for comparative purposes. CD4 T cells secreting multiple combinations of these cytokines, indicative of mixed functional polarizations, were not considered for this study. Overall, these investigations showed that among all CD4 T helper cell populations tested, Th1 cells had highest levels of susceptibility to R5-tropic viral infection, followed by Th17 cells; Th9 and Th2 cells were poorly susceptible to R5-tropic HIV-1 (Figure 1B). In contrast, the cellular susceptibility to X4-tropic infection was most pronounced in Th9 cells and, to a lesser extent, in Th2 cells; Th1 and Th17 cells exhibited very limited permissiveness to X4-tropic infection (Figure 1B). Consistent with the differential susceptibility to infection with R5-tropic and X4-tropic viruses, we noted that the cell surface expression of CCR5 was 4–5-fold higher in Th1 cells compared to Th9 and Th2 cells; CCR5 expression in Th17 cells was also elevated although not to the same extent as in Th1 cells (Figure 1C). Differences in CXCR4 expression among cell subsets were more limited, although a trend for increased expression of this marker was visible for Th2 and Th9 cells. Interestingly, we observed roughly equal levels of cellular susceptibility to infection with VSV-G-pseudotyped viruses among all analyzed subsets (Figure 1B), strongly suggesting that the observed differences between viral replicative activity in individual cell subsets was unrelated to cell-intrinsic restriction of viral replication steps during the post-entry phase of the viral life-cycle.
Figure 1: Increased susceptibility of Th9 and Th2 cells to X4-tropic HIV-1 in vitro.
(A-B): Proportions of GFP-positive CD4 T cells within indicated T helper cell subpopulations after in vitro infection with GFP-encoding X4-tropic, R5-tropic or VSV-G pseudotyped HIV-1. (A) shows representative flow cytometry dot plots from one study person, (B) demonstrates cumulative data from viral infections in PBMC from five HIV-1 negative study subjects. (C): Proportions of cells with CCR5 and CXCR4 surface expression in indicated CD4 T cell subpopulations. Data from six HIV-1 negative study subjects are shown. Data were tested for statistical significance using one-way ANOVA, followed by Dunn’s test for multiple comparison.
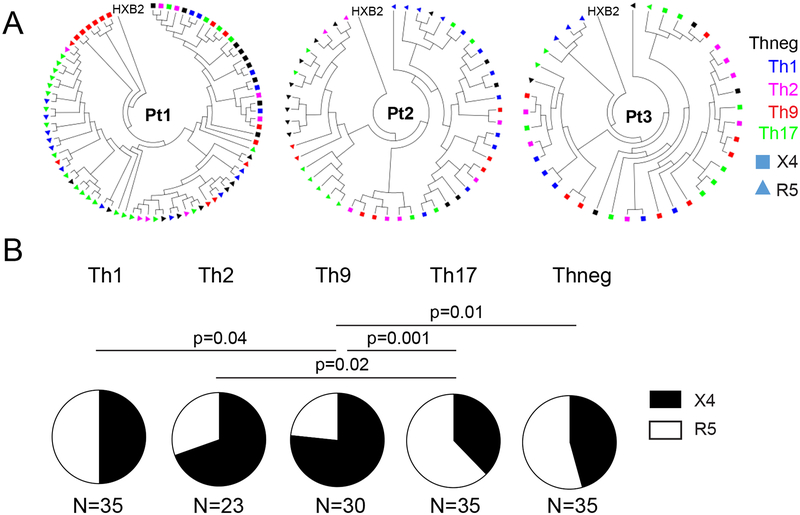
To determine whether viral tropism influences the susceptibility of CD4 T cells infected in vivo, we sorted the described polarized CD4 T cell populations, as well as a control CD4 T cell population secreting none of these cytokines (termed Thneg in this manuscript), from 7 individuals (all male, median age of 56 (range:33–60), median CD4 T cell count/ul of 1033 (range: 516–1554)), who received suppressive antiretroviral therapy, as described previously [13]. Proviral DNA was subjected to single-genome sequencing of HIV-1 env. In four of these study subjects, all sequences retrieved from the different cell populations were R5-tropic, consistent with homogenous infection with an R5-tropic virus. However, in three study persons, we identified a mix of X4- and R5-tropic sequences. Notably, in Th1 cells, as well as in Thneg, the frequencies of X4- and R5-tropic viral sequences were well balanced, and accounted for roughly equal proportions of viral sequences (Figure 2). In contrast, viral sequences in Th9, and to a lesser degree in Th2 cells, were heavily biased towards X4-tropic sequences. Th17 cells exhibited a disproportionate enrichment with R5-tropic viral sequences. Together, these data suggest distinct differences between the susceptibility of differentially-polarized CD4 T cells to R5- and X4-tropic HIV-1 in vivo.
Figure 2: Increased frequencies of X4-tropic HIV-1 in Th9 and Th2 cells in vivo.
(A): Circular phylogenetic trees indicating X4-tropic (squares) and R5-tropic (triangles) HIV-1 env sequences in indicated CD4 T cell subsets sorted from three different patients. (B): Relative proportions of X4- and R5-tropic sequences in indicated CD4 T cell populations in vivo. Pooled data from three study subjects are shown. Differences were tested for statistical significance using Fisher’s exact test.
Discussion
Few examples highlight recent advances in immunology as profoundly as the discovery of the complexity and diversity of CD4 T cell subpopulations with distinct functional polarizations. While the specific roles and functions of differentially-polarized CD4 T cells for antimicrobial immune defense is continuously increasing, relatively little is known about how this functional lineage commitment affects the role of CD4 T cells as target cells for HIV-1. This is particularly true for recently discovered CD4 T cell populations such as Th9 cells, a population of long-lived CD4 T helper cells that plays a distinct role for anti-tumor immune defense [14], but may also promote allergic inflammation and be involved in the pathogenesis of several inflammatory or autoimmune diseases [15]. Here, we show that Th9 cells and, to a lesser extent Th2 cells, have a remarkably elevated susceptibility to X4-tropic viruses in vitro, and more frequently harbor X4-tropic HIV-1 env DNA in vivo. Notably, these studies were limited to the analysis of HIV-1 env DNA segments, which in many cases are parts of defective provirus that do not encode for replication-competent virus [16]. Therefore, it is currently unclear whether Th9 cells also represent a specific viral reservoir niche for X4-tropic viruses that supports long-term persistence of X4-tropic, replication-competent viruses; however, massive single-genome sequencing of large numbers of viral sequences retrieved from Th9-/Th2-positive cells may in the future allow to address that question. The identification of specific CD4 T cell subpopulations that serve as preferred sites for viral persistence of X4-tropic virus may be relevant in the context of current strategies aiming at inducing a functional cure of HIV-1 through therapeutic manipulation of CCR5 surface expression [17], and in the setting of treatment with pharmacological CCR5 inhibitors as part of antiretroviral combination regimens.
Interestingly, we observed that HIV-1 env sequences in Th1 cells were relatively evenly balanced between X4- and R5-tropic sequences in the three patients we analyzed, despite a considerably higher surface expression of CCR5 and a higher susceptibility to R5-tropic virus in Th1 cells in vitro. Since we observed a similar susceptibility of all analyzed cell populations to VSV-G pseudotyped virus, it is unlikely that viral restriction mechanisms inside CD4 T cells are responsible for this observation. Rather, these results may reflect a more dynamic expression of CCR5 in vivo, which is typically low in resting naïve and central-memory CD4 T cells but increases upon cellular activation, particularly in effector-memory CD4 T cells [18]. In contrast, surface expression of CXCR4 appears to be more stable and less dependent on immune activation and cellular differentiation.
The present studies were restricted to an analysis of Th1, Th2, Th9 and Th17 cells, but these cell populations are far from representing the entirety of the functionally-committed CD4 T cell pool. As such, future studies will be necessary to determine if functional lineage commitment towards a profile of e. g. regulatory T cells, of T follicular helper cells or of Th22 cells is associated with altered permissiveness to X4- or R5-tropic HIV-1. Continuous progress in understanding the developmental and functional characteristics of individual CD4 T cell subsets to HIV-1 may help to delineate a more comprehensive understanding of the CD4 T cell populations susceptible to HIV-1.
Acknowledgments
ML is supported by NIH grants AI098487, AI106468, AI114235, AI117841, AI120008, AI124776. XGY is supported by NIH grants AI116228, AI078799, HL134539.
Footnotes
The authors declare that conflicts of interest do not exist.
References
- 1.Zhu J, Yamane H, Paul WE. Differentiation of effector CD4 T cell populations (*). Annu Rev Immunol 2010,28:445–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Glimcher LH, Murphy KM. Lineage commitment in the immune system: the T helper lymphocyte grows up. Genes Dev 2000,14:1693–1711. [PubMed] [Google Scholar]
- 3.Kaplan MH. Th9 cells: differentiation and disease. Immunol Rev 2013,252:104–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Muranski P, Restifo NP. Essentials of Th17 cell commitment and plasticity. Blood 2013,121:2402–2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schmitt N, Ueno H. Regulation of human helper T cell subset differentiation by cytokines. Curr Opin Immunol 2015,34:130–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morita R, Schmitt N, Bentebibel SE, Ranganathan R, Bourdery L, Zurawski G, et al. Human blood CXCR5(+)CD4(+) T cells are counterparts of T follicular cells and contain specific subsets that differentially support antibody secretion. Immunity 2011,34:108–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gosselin A, Monteiro P, Chomont N, Diaz-Griffero F, Said EA, Fonseca S, et al. Peripheral blood CCR4+CCR6+ and CXCR3+CCR6+CD4+ T cells are highly permissive to HIV-1 infection. J Immunol 2010,184:1604–1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vicenzi E, Panina-Bodignon P, Vallanti G, Di Lucia P, Poli G. Restricted replication of primary HIV-1 isolates using both CCR5 and CXCR4 in Th2 but not in Th1 CD4(+) T cells. J Leukoc Biol 2002,72:913–920. [PubMed] [Google Scholar]
- 9.Moonis M, Lee B, Bailer RT, Luo Q, Montaner LJ. CCR5 and CXCR4 expression correlated with X4 and R5 HIV-1 infection yet not sustained replication in Th1 and Th2 cells. AIDS 2001,15:1941–1949. [DOI] [PubMed] [Google Scholar]
- 10.Josefsson L, Eriksson S, Sinclair E, Ho T, Killian M, Epling L, et al. Hematopoietic precursor cells isolated from patients on long-term suppressive HIV therapy did not contain HIV-1 DNA. J Infect Dis 2012,206:28–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sharkey M, Babic DZ, Greenough T, Gulick R, Kuritzkes DR, Stevenson M. Episomal viral cDNAs identify a reservoir that fuels viral rebound after treatment interruption and that contributes to treatment failure. PLoS Pathog 2011,7:e1001303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol 2007,24:1596–1599. [DOI] [PubMed] [Google Scholar]
- 13.Lee GQ, Orlova-Fink N, Einkauf K, Chowdhury FZ, Sun X, Harrington S, et al. Clonal expansion of genome-intact HIV-1 in functionally polarized Th1 CD4+ T cells. J Clin Invest 2017,127:2689–2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vegran F, Apetoh L, Ghiringhelli F. Th9 cells: a novel CD4 T-cell subset in the immune war against cancer. Cancer Res 2015,75:475–479. [DOI] [PubMed] [Google Scholar]
- 15.Schmitt E, Klein M, Bopp T. Th9 cells, new players in adaptive immunity. Trends Immunol 2014,35:61–68. [DOI] [PubMed] [Google Scholar]
- 16.Bruner KM, Murray AJ, Pollack RA, Soliman MG, Laskey SB, Capoferri AA, et al. Defective proviruses rapidly accumulate during acute HIV-1 infection. Nat Med 2016,22:1043–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stone D, Kiem HP, Jerome KR. Targeted gene disruption to cure HIV. Curr Opin HIV AIDS 2013,8:217–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Buzon MJ, Sun H, Li C, Shaw A, Seiss K, Ouyang Z, et al. HIV-1 persistence in CD4+ T cells with stem cell-like properties. Nat Med 2014,20:139–142. [DOI] [PMC free article] [PubMed] [Google Scholar]