SUMMARY
In colorectal cancer patients, a high density of cytotoxic CD8+ T cells in tumors is associated with better prognosis. Using a Stat3 loss-of-function approach in two wnt/β-catenin-dependent autochthonous models of sporadic intestinal tumorigenesis, we unravel a complex intracellular process in intestinal epithelial cells (IECs) that controls the induction of a CD8+ T cell based adaptive immune response. Elevated mitophagy in IECs causes iron(II)-accumulation in epithelial lysosomes, in turn, triggering lysosomal membrane permeabilization. Subsequent release of proteases into the cytoplasm augments MHC class I presentation and activation of CD8+ T cells via cross-dressing of dendritic cells. Thus, our findings highlight a so-far-unrecognized link between mitochondrial function, lysosomal integrity, and MHC class I presentation in IECs and suggest that therapies triggering mitophagy or inducing LMP in IECs may prove successful in shifting the balance toward anti-tumor immunity in colorectal cancer.
In Brief
Enhanced mitophagy in intestinal epithelial cells promotes anti-tumor immunity through increasing lysosomal membrane permeabilization that augments MHC I presentation and CD8+ T cell activation.
Graphical Abstract

INTRODUCTION
Colorectal cancer (CRC) belongs to a group of the most commonly diagnosed cancers in both men and women in the Western world (Siegel et al., 2016). Activating mutations in the Wnt pathway that mostly affect APC or CTNNB1 are found in the majority of cases; however, apart from these, only a small number of additional genes are significantly mutated (Cancer Genome Atlas, 2012). Instead, colorectal carcinogenesis depends on the close interaction of mutated tumor cells with their microenvironment (Grivennikov et al., 2010). The presence of tumor-infiltrating T cells and, in particular, CD8+ T cells and increased interferon-gamma (IFNγ) expression has prognostic relevance and is associated with prolonged survival, whereas a T helper 17 (Th17) T-cell-dominated immune response is associated with a worse outcome (Fridman et al., 2012). While the clinical correlation between T cell infiltration and prognosis is undisputed and has led to the development of an immune score that can be employed to predict survival of CRC patients (Fridman et al., 2012), less is known about the underlying cellular and molecular mechanisms that drive T cell polarization during intestinal carcinogenesis.
Activation of the transcription factor STAT3 has been documented in a wide range of tumors, including CRC (Bollrath and Greten, 2009). Multiple pathways, including receptor engagement by interleukin-6 (IL-6) family members, G-protein-coupled receptors, Toll-like receptors, and microRNAs, have been identified to activate JAK-STAT3 signaling in tumor cells and infiltrating immune cells (Yu et al., 2014). This results in Y705 phosphorylation of STAT3 and nuclear translocation of STAT3 to drive transcription of genes involved in cell-cycle regulation, cell survival, cell migration, and, importantly, immunosuppression. In addition to phosphorylation of the Y705 residue of STAT3, phosphorylation can occur at serine-727 (S727) as a result of receptor engagement by various growth factors, such as epidermal growth factor (EGF), and signals that are transduced via the ERK-, p38-, JNK-, or PKCδ-pathways (Decker and Kovarik, 2000). Apart from STAT3’s classical role as a transcription factor, non-canonical STAT3-dependent functions also have been proposed (Demaria et al., 2014; Gough et al., 2009; Wegrzyn et al., 2009).
Autophagy represents an intracellular degradation process that encloses ubiquitinated proteins in vesicles termed “autophagosomes,” which subsequently fuse with lysosomes (Levine and Kroemer, 2008). Besides its role as a response mechanism to cellular stress (e.g., during nutrient deprivation), autophagy also serves as the specific degradation mechanism of aged and/or damaged mitochondria, a process known as “mitophagy.” The kinase PINK1 binds to mitochondria with decreased membrane potential, the driving force of mitochondrial respiration, and thereby marks these to degradation via the autophagosomal-lysosomal pathway (Youle and Narendra, 2011). Mitochondria are rich in iron-containing macromolecules; therefore, mitophagy, which leads to transfer of mitochondrial proteins into lysosomes, can contribute to lysosomal iron content, which is critical for their resistance to reactive oxygen species (Terman et al., 2010).
The adaptive immune system relies on the presentation of cellular antigens by other cells on their surface via major histo-compatibility complex (MHC) molecules. Whereas MHC class II molecules are present only on specialized antigen-presenting cells (mainly dendritic cells, DCs), MHC class I molecules are present on virtually all cells of mammalian organisms and can be recognized by CD8+ T cells. To bind to nascent MHC molecules, antigens have to be transported into the endoplasmic reticulum (ER) by the transmembrane transport system transporter associated with antigen presentation (TAP).
In addition to a large body of evidence indicating that the proteasome is the origin of most antigens presented via MHC class I molecules, some evidence indicates that other proteases can contribute to antigen processing (Cruz et al., 2017; Münz, 2016; Rock et al., 2010). Most notably, the lysosomal protease cathepsin S has been found to facilitate antigen generation in DCs in the endosomal-lysosomal compartment before being presented by MHC class I molecules (Hari et al., 2015; Shen et al., 2004). In addition, in intestinal epithelial cells (IECs) as well as bone-marrow-derived antigen-presenting cells, cathepsin S is involved in antigen-presentation by MHC class II molecules, whereas cathepsin L is required for this in thymic cortical epithelial cells (Beers et al., 2005; Nakagawa et al., 1998).
Apart from the ability of DCs to present peptide antigens from their cytoplasm, these cells are also involved in the uptake and processing of extracellular antigens with MHC class I molecules to CD8+ T cells by cross presentation (Cruz et al., 2017). Recently, it was noted that, in addition to foreign protein, DCs can also take up complete antigen-MHC class I (and II) complexes and present them to T cells, a process termed “cross-dressing” (Nakayama, 2015). This process has been implicated in anti-tumor immunity as well as anti-viral host defense (Nakayama, 2015).
RESULTS
Loss of Stat3 in IECs Prevents Sporadic Tumorigenesis and Causes Infiltration of CD8+ T Cells during Tumor Initiation
Stat3flox and Stat3ΔIEC mice were subjected to 6 weekly injections of the somatic mutagen azoxymethane (AOM) and tumor development was examined 18 weeks later. While AOM induces oncogenic point mutations in β-catenin and led to development of tubular adenomas in the colons of 100% of Stat3-proficient (Stat3flox) animals, tumor formation was completely abolished in mice where Stat3 was deleted in IECs (Figures 1A–1C). Strikingly, the absence of Stat3 also prevented formation of pre-neoplastic foci which still occurred in these mice when challenged with AOM in the context of chronic inflammation during CAC (Bollrath et al., 2009), suggesting the complete loss of mutagenized Stat3-deficient IECs during initiation of sporadic tumorigenesis. Histological examination of AOM challenged Stat3ΔIEC mice revealed a pronounced accumulation of lamina propria CD3+ T cells when compared to Stat3flox controls (Figures 1D–1F). T cell accumulation in Stat3ΔIEC mice was preceded by an elevated number of CD11c+, but not F4/80+ or Gr1+, cells (Figure 1H) and increased Il12 mRNA expression (Figure 1I) in the colonic lamina propria at an early time point in the model when first pre-neoplastic aberrant crypt foci (ACF) were detectable in both genotypes (Figure 1G; data not shown). This raised the possibility that AOM exposure triggered an adaptive immune response against mutagenized cell in Stat3ΔIEC mice, thereby preventing their subsequent outgrowth into macroscopic colonic tumors. This finding is in line with a recent report demonstrating that CD8+ T cells can be stimulated by retinoic acid in vivo and, in doing so, can contribute to controlling AOM/DSS-induced tumor growth (Bhattacharya et al., 2016).
Figure 1. Loss of Stat3 in IEC Blocks Initiation of Sporadic Intestinal Tumorigenesis.
(A) Tumor incidence in Stat3F/F (n = 10) and Stat3ΔIEC (n = 9) mice 18 weeks after initial AOM challenge. Data are mean ± SEM, **p < 0.01 by one-sample t test.
(B–E) Representative H&E-stained sections (B and C) and CD3 immunohistochemistry (IHC) (D and E) of AOM-challenged colons. Scale bars, 1 mm (B and C) and 50 μm (D and E).
(F) Quantification of infiltrating CD3+ T cells in colonic non-tumor areas. Data are mean ± SEM, ***p < 0.001 by Student’s t test.
(G) Representative H&E-stained colonic aberrant crypt focus from a Stat3ΔIEC mouse 8 weeks after the first AOM injection. Scale bar, 100 μm.
(H) Flow cytometric analysis of different myeloid cell subpopulations in colonic lamina propria of Stat3F/F and Stat3ΔIEC mice 1 week after the last AOM injection Data are mean ± SEM; n = 5 per genotype; **p < 0.01 by Student’s t test.
(I) Relative mRNA expression 1 week after the last AOM injection. Data are mean ± SEM; n = 5 per genotype.
(J–M) IHC of p-Stat3S727 (J and K) and p-Stat3Y705 (L and M) in β-catc.a. mice. Scale bars, 100 μm (J and L) and 50 μm (K and M).
(N) Survival of β-catc.a. (n = 15), β-catc.a./Stat3ΔIEC (n = 10) and β-catc.a./Stat3+/Δ mice (n = 11). ***p < 0.001 by log-rank test.
See also Figure S1.
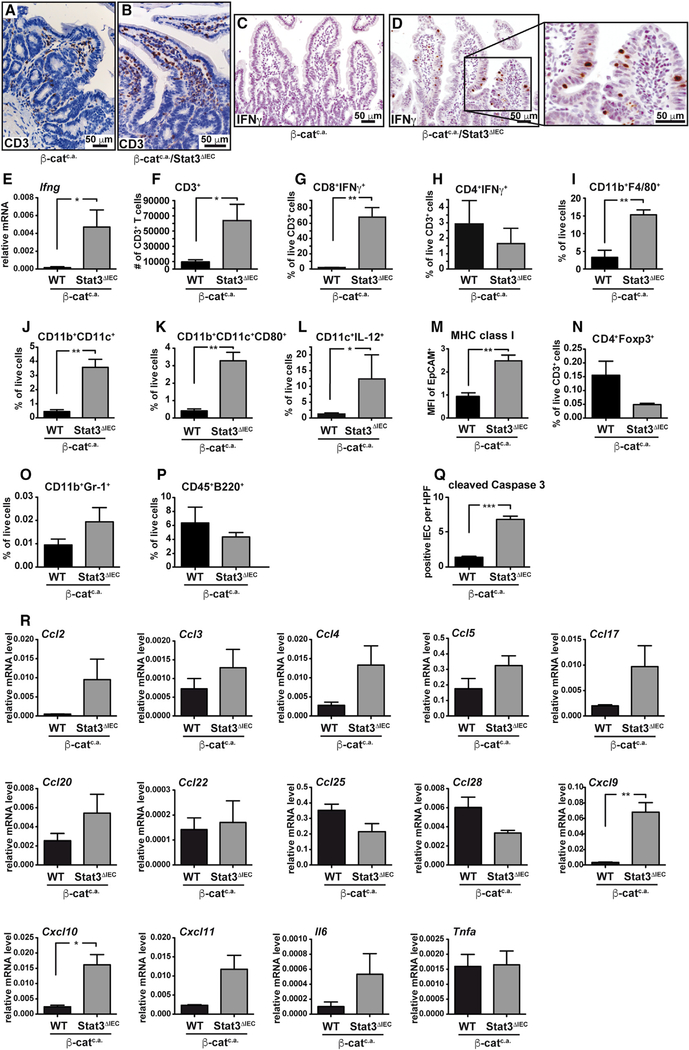
To distinguish whether an underlying mechanism is dependent on the mutagenic effect of AOM in IECs or the influence of mutated IECs on their cellular environment, we utilized villin-creERT2/CtnnbloxEx3/WT mice, termed “β-catc.a.,” as a defined genetic model for oncogenic activation of the wnt pathway. This model confers IEC-specific and tamoxifen-induced expression of an exon-3-deleted—and thereby stabilized form of— β-catenin. Accordingly, β-catc.a. mice mimic the AOM-induced missense mutation in exon 3 of Ctnnb that results in stabilization of the corresponding protein and aberrant activation of the wnt/β-catenin pathway (Greten et al., 2004) observed in >80% of human colorectal cancer (CRC) (Cancer Genome Atlas, 2012). Mice expressing stabilized β-catenin are characterized by rapid expansion of proliferative intestinal crypt stem cells and loss of absorptive enterocytes causing death of β-catc.a. mice within ~4 weeks after oral tamoxifen application (Schwitalla et al., 2013). IEC-specific expression of the serine 727- and tyrosine 705-phosphorylated form of Stat3 indicated marked Stat3 activation in transformed crypts of β-catc.a. mice (Figures 1J–1M). By contrast, IEC-restricted homozygous ablation of Stat3 significantly prolonged survival of the corresponding β-catc.a./Stat3ΔIEC mice (median survival of β-catc.a. mice 23 days versus 31 days in β-catc.a./Stat3ΔIEC mice; median survival of heterozygous β-catc.a./Stat3+/ΔIEC mice: 22 days; n ≥ 10; p < 0.001) (Figure 1N). We did not detect any evidence for improved differentiation or changes in proliferation of Stat3-deficient β-catc.a. IECs (Figures S1A–S1H), yet a higher villus/crypt ratio indicated a block in crypt expansion in β-catc.a./Stat3ΔIEC mice compared to Stat3-proficient controls (Figure S1G). Moreover, β-catc.a./Stat3ΔIEC mice were characterized by a marked accumulation of CD3+ cells, similar to our observations in AOM-challenged Stat3ΔIEC mice, as well as a high number of IFNγ-expressing infiltrating intraepithelial lymphocytes (Figures 2A–2D). Real-Time PCR confirmed elevated Ifng expression in the mucosa of β-catc.a./Stat3ΔIEC mice when compared to their Stat3 proficient counterparts (Figure 2E). These findings were further corroborated by fluorescence-activated cell sorting (FACS) analysis, which revealed a marked increase of CD3+ T cells in the lamina propria of β-catc.a./Stat3ΔIEC mice and CD8+ cells of the αβ lineage, rather than CD4+ T cells as the main source of IFNγ (Figures 2F–2H; data not shown). This was accompanied by a significantly increased the number of cleaved caspase3-positive apoptotic IECs in β-catc.a./Stat3ΔIEC mice (Figure 2Q), supporting cytotoxic activity of CD8+ T cells. Furthermore, β-catc.a./Stat3ΔIEC mice showed an elevated frequency of CD11b+F4/80+ macrophages (Figure 2I), activated CD11b+/CD11c+ dendritic cells (DC) that expressed CD80 and IL-12 (Figures 2J–2L), and elevated expression of MHC class I in EpCAM+ IECs (Figure 2M). In contrast, other notable immune cell populations were unchanged (Figures 2N–2P). In addition, we could detect a number of T cell related chemokines, most notably the IFNγ-inducible chemokines CXCL-9, CXCL-10, and CXCL-11, to be upregulated in Stat3-deficient IECs during tumorigenesis (Figure 2R). Collectively, these data suggest that loss of Stat3 in IECs triggered an adaptive immune response leading to IEC death, which delayed expansion of β-catenin mutant crypt cells associated with prolonged survival of β-catc.a./Stat3ΔIEC mice.
Figure 2. Stat3ΔIEC Mice Are Characterized by Increased Accumulation of CD8+/IFNγ+ T Cells in the Intestine upon Activation of wnt Signaling.
(A–D) IHC of CD3 (A and B) and IFNγ (C and D) 15 days after starting tamoxifen.
(E) Relative Ifng mRNA expression in small intestinal mucosa of β-catc.a. and β-catc.a./Stat3ΔIEC mice on day 15; n = 6/genotype.
(F) Quantification of infiltrating CD3+ T cells.
(G–L) Flow cytometric analysis of IFNγ in CD8+ (G) and CD4+ (H) T cells, CD11b+F4/80+ (I) and CD11b+CD11c+ (J) cells as well as CD80 (K) and IL-12 (L) in CD11c+ cells on day 15; n = 3/genotype.
(M) Surface expression (mean fluorescence intensity, MFI) of MHC class I 3 days after first tamoxifen; n = 4/genotype.
(N–P) Flow cytometric analysis of CD4+Foxp3+ (N), CD11b+Gr1+ (O), and CD45+B220+ (P) cells on day 15; n = 3/genotype.
(Q) Number of cleaved caspase 3-pos. IEC; n = 4/genotype; R 20 high-power fields (HPF)/animal.
(R)Relative gene expression of the indicated cytokines or chemokines in IECs on day 15; n = 3/genotype.
In (E)–(R), data are mean ± SEM. Scale bars, 50 μm. *p < 0.05, **p < 0.01 by Student’s t test.
Prolonged Survival of β-Catc.a. /Stat3ΔIEC Mice Depends on Presence of CD8+ T Cells and IFNγ
Genetically modified mouse models are usually not very immunogenic and do not express many neo-antigens because of their low mutational burden. Therefore, we aimed to confirm a T cell contribution to this model and sequenced TCRα- and β-chain CDR3 regions in sorted CD8+ T cells from intestinal mucosa of β-catc.a. and β-catc.a./Stat3ΔIEC mice. Indeed, loss of Stat3 in β-catenin mutant IECs led to clonal expansion of CD8+ T cells (Figure 3A) and less diverse T cell receptors (TCRs) (Figures 3B, 3C, and S2A–S2H). These findings are in line with an enhanced antigen-dependent immune response in these mice. This was further corroborated when we introduced a model antigen by employing ROSAOVA mice that contain an immunogenic peptide from ovalbumin (ova, amino acids [aa] 246–353) flanked by inversely oriented loxP sites and targeted into the ubiquitously active ROSA26 locus (Sandhu et al., 2011). ROSAOVA mice were crossed with both β-catc.a. mice (β-catc.a./OVAIEC) as well as β-catc.a./Stat3ΔIEC mice (β-catc.a./Stat3ΔIEC/OVAIEC) and resultant offspring were treated with tamoxifen. Expectedly, flow cytometry on day 15 using a Kb/OVA (SIINFEKL) pentamer confirmed a significantly increased number of SIINFEKL-specific CD8+ T cells in the intestinal mucosa in β-catc.a./Stat3ΔIEC/OVAIEC mice (Figure 3D). Moreover, staining of IECs using the 25-D1.16 monoclonal antibody that specifically reacts with ovalbumin-derived peptide SIINFEKL bound to H-2Kb of MHC class I revealed increased binding on Stat3-deficient IECs, confirming enhanced antigen processing in IECs from β-catc.a./Stat3ΔIEC/OVAIEC mice (Figure 3E). Similarly, the number of SIINFEKL-specific CD8+ T cells was elevated in AOM-challenged Stat3ΔIEC/OVAIEC mice when compared to OVAIEC mice (Figure 3F).
Figure 3. Suppression of Intestinal Tumor Initiation in Stat3ΔIEC Mice Depends on CD8+/IFNγ+ T Cells.
(A–C) Statistical analysis for clonality (A) and diversity of T cell receptor (TCRα and TCRβ) (B and C) in sorted mucosal CD8+ T cells on day 15; n = 3 and 4/genotype, respectively; *p < 0.05 by Mann-Whitney U test. (D and E) Flow cytometric quantification of SIINFEKLMHC pentamer binding CD3+CD8+ T cells (D) in mucosa and surface expression of SIINFEKL-binding MHC class I on IEC (E) on day 15. n = 5 mice/genotype.
(F) Flow cytometric quantification of SIINFEKL-MHC pentamer binding CD3+CD8+ T cells in AOM-challenged mice (week 18); n ≥ 4/genotype.
(G) Survival of β-catc.a. and β-catc.a./Stat3ΔIEC mice injected with CD8+ T-cell-depleting antibody; n ≥ 5/genotype.
(H) Survival of β-catc.a./Stat3ΔIEC mice that had been transplanted with bone marrow (BM) from CD11c-DTR (n = 6) or wild-type (WT) mice (n = 6) 8 weeks before tamoxifen administration and diphteria toxin injection to deplete CD11c+ cells.
(I) Survival of β-catc.a./Ifng (n = 9) and β-catc.a./Stat3ΔIEC/Ifng mice (n = 11).
*p < 0.05 by Student’s t test (D–F). The dashed gray lines in (A)–(C) represent survival curves of untreated β-catc.a. and β-catc.a./Stat3ΔIEC mice as reproduced from Figure 1N for better comparison.
In (A)–(F), data are mean ± SEM. See also Figure S2.
To functionally confirm that the survival advantage of β-catc.a./Stat3ΔIEC mice was indeed dependent on the observed adaptive immune response rather than on IEC-specific cell autonomous effects such as proliferation, we either depleted CD8+ T cells using a neutralizing antibody or CD11c+ DCs using CD11c-DTR mice (Jung et al., 2002). Expectedly, antibody-mediated loss of CD8+ T cells or diphtheria toxin-induced ablation of CD11c+ DCs reduced survival of β-catc.a./Stat3ΔIEC mice (median survival 24 days in CD8+ T cell or CD11c+ DCs depleted β-catc.a./Stat3ΔIEC animals, Figure 3G and 3H). A comparable reduction of survival was also observed in β-catc.a./Stat3ΔIEC/Ifng−/− compound mutants, with a median survival of 23 days (Figure 3I). Collectively, these data strongly suggest that IEC-specific Stat3 activation precludes the generation of an effective CD8+ T cell-dependent immune response during the tumor initiation phase.
CD8+ T Cell Activation Is a Consequence of Lysosomal Membrane Permeabilization
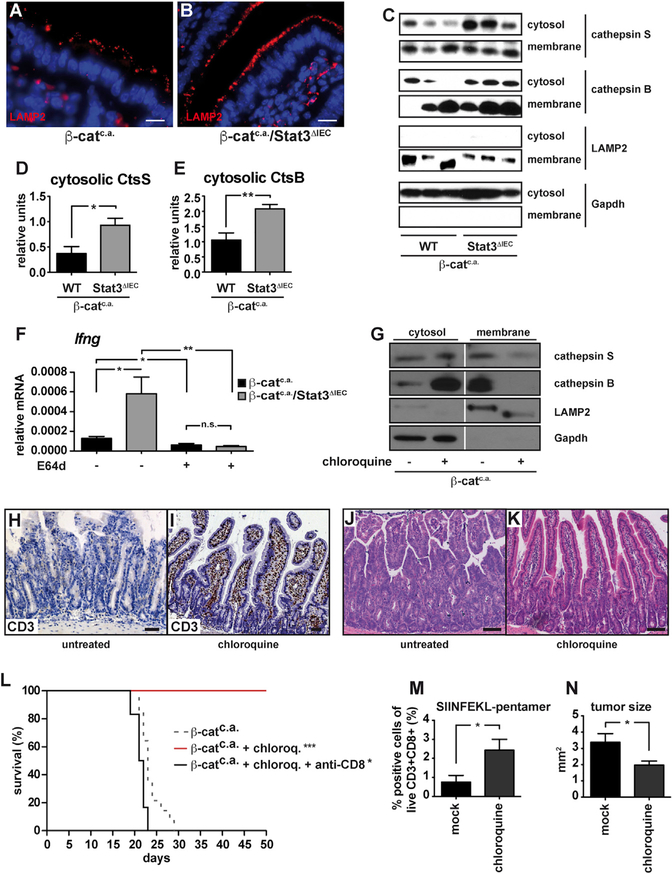
Having established the consequence of Stat3 deficiency in IECs during intestinal tumor initiation, we aimed to identify the responsible mechanism that prevented CD11c+ DC and T cell activation. In fully malignant murine colon cancer cells or B16 melanoma cells expression of dominant negative Stat3 increases expression of pro-inflammatory cytokines, including tumor necrosis factor alpha (TNF-α), IL-6, CCL5, CXCL10, and IFNβ, which triggers CD8+ T cell activation (Wang et al., 2004). Surprisingly, we did not observe marked expression differences for these cytokines and chemokines between primary IECs of Stat3 wild-type (WT) β-catc.a. and Stat3-deficient β-catc.a./Stat3ΔIEC mice 3 days after the first tamoxifen application (Figure S3A). Instead, careful histomorphological examination revealed enlarged lysosomal structures in IECs of β-catc.a./Stat3ΔIEC mice when stained for LAMP2 (Figures 4A and 4B), which prompted us to examine lysosomal function. Lysosomal membrane permeabilization (LMP) in response to the acute cellular stress commonly elicited in mutagenized cells, leads to the release of cathepsins from lysosomes into the cytosol (Boya and Kroemer, 2008). While total expression of cathepsins S, L, and B and of the lysosomal protease inhibitor cystatin C remained unaffected in IECs of β-catc.a./Stat3ΔIEC mice (Figures S3B and S3C), proteases accumulated in cytoplasmic fractions of these cells presumably in response to increased release from lysosomes (Figures 4C–4E). Importantly, administration of the membrane permeable pan-cysteine protease inhibitor E64d prevented upregulation of Ifng gene expression in both β-catc.a. and β-catc.a./Stat3ΔIEC mice (Figure 4F), as well as IEC apoptosis (Figure S3D), supporting the notion that lysosomal proteases play an important role in CD8+ T cell activation. To examine whether LMP was essential in this context and whether such immune cell activation could also be elicited in Stat3-proficient IECs, we treated β-catc.a. mice with chloroquine, a well-known inducer of LMP (Boya et al., 2003). Indeed, chloroquine triggered release of cathepsins from lysosomes in β-catenin mutant IECs (Figure 4G) and caused accumulation of CD3+ T cells in the lamina propria of β-catc.a. mice (Figures 4H and4I) as well as upregulation of Ifng mRNA (Figure S3E). Furthermore, cleaved caspase-3+ apoptotic IECs were markedly increased in chloroquine-treated β-catc.a. mice (Figure S3F) in line with a cytotoxic T cell activation. Consequently, chloroquine prevented expansion of β-catenin mutant crypts and death of β-catc.a. mice in a CD8+ T-cell-dependent manner (Figures 4J–4L and S3G). In agreement with this notion, we could observe an increased number of SIINFEKL-specific CD8+ T cells in the intestinal mucosa AOM-challenged OVAIEC mice (Figure 4M). Consequently, chloroquine treatment over a period of 4 weeks reduced tumor load of established AOM-induced adenomas in WT mice (Figure 4N), strongly supporting the stimulation of an anti-tumor immune response upon LMP induction.
Figure 4. Loss of Stat3 in IECs Induces Lysosomal Membrane Permeabilizaton.
(A and B) Immunofluorescent LAMP2 staining visualizing lysosomes in the mucosa of β-catc.a. (A) and β-catc.a./Stat3ΔIEC (B) mice on day 3 after first tamoxifen. Scale bar, 10 μm.
(C–E) Immunoblot analysis of cathepsin S, cathepsin B, LAMP2, and Gapdh in cytosolic and membrane fractions (C), and densitometric quantification of cytosolic cathepsin S (D) and cytosolic cathepsin B (E) normalized to Gapdh on day 3; Data are mean ± SEM; n ≥ 6/genotype.
(F) Ifng-mRNA in β-catc.a. and β-catc.a./Stat3ΔIEC mice treated with E64d on day 15; Data are mean ± SEM; n ≥ 5/genotype.
(G) Immunoblot analysis of cathepsin S, cathepsin B, Lamp2, and Gapdh in IECs isolated from untreated or chloroquine-treated (60 mg/kg) β-catc.a. mice on day 3.
(H and I) Immunohistochemical analysis of CD3+ T cells in unchallenged (H) and chloroquine-treated (I) β-catc.a. mice on day 15.
(J and K) H&E staining of untreated (J) or chloroquine-treated (K) β-catc.a. mice on day 15.
(L) Survival of chloroquine-treated (n = 8), chloroquine- and anti-CD8-antibody-treated (n = 6), or untreated β-catc.a. mice (n = 15); *p < 0.05, ***p < 0.001 by log-rank test. The dashed line (for β-catc.a. mice) has been reproduced from Figure 1N for better comparison.
(M) SIINFEKL-MHC pentamer binding CD3+CD8+ T cells in OVAIEC mice injected with chloroquine or saline 13 weeks after first AOM injection; Data are mean ± SEM; n = 6/genotype.
(N) Mean tumor size in AOM-treated mice that received daily chloroquine (n = 5) or saline (n = 8) intraperitoneally (i.p.) for 4 weeks starting 10 weeks after the last AOM injection.
*p < 0.05, **p < 0.01 by Student’s t test. Data are mean ± SEM.
See also Figure S3.
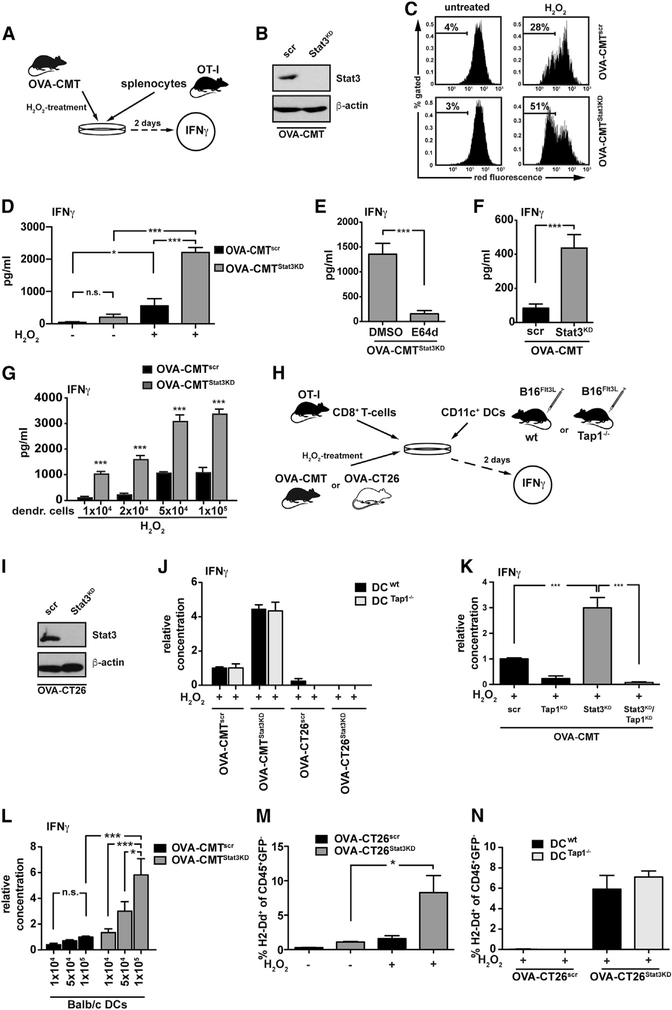
To further examine the link between LMP in IECs and CD8+ T cell recruitment, we established an ex vivo co-culture system. The system comprises an ovalbumin-expressing murine (C57BL/6) colon carcinoma cell line (OVA-CMT93) stably expressing a Stat3 miRNA or a scrambled control (OVA-CMTStat3KD or OVA-CMTscr, respectively) that were co-cultured together with splenocytes from OT-I transgenic mice harboring a T cell receptor (TCR) that recognizes the ovalbumin-derived peptide (SIINFEKL) on cytotoxic T cells. The level of T cell activation was measured by the release of IFNγ by OT-I cells (Figure 5A). After confirmation of successful Stat3 knockdown (KD) (Figure 5B), we verified increased LMP upon stimulation with H2O2 in OVA-CMTStat3KD cells by flow cytometric monitoring of the reduction in acidophilic acridine orange staining (Figure 5C). While unchallenged OVA-CMTStat3KD or OVA-CMTscr cells resulted in similar OT-I T cell activation, induction of LMP in tumor cells by H2O2-treatment prior to their co-culture with OT-I splenocytes significantly elevated IFNγ production (Figure 5D). Similar to the in vivo situation and consistent with a higher degree of LMP, IFNγ release by OT-I cells was greatly enhanced when Stat3 was absent from ovalbumin expressing tumor cells and markedly reduced upon treatment of OVA-CMTStat3KD with the pan-cysteine protease inhibitor E64d (Figure 5E), further supporting the notion that LMP-dependent protease release may be involved in antigen processing in epithelial cells. Indeed, when H2O2 challenged OVA-CMTStat3KD or OVA-CMTscr cells were co-cultured with MACS-purified OT-I CD8+ T cells in the absence of CD11c+ DCs, this was sufficient to elicit IFNγ secretion (Figure 5F), yet IFNγ production was greatly enhanced when increasing numbers of CD11c+ DCs were added (Figure 5G) suggesting the transfer and presentation of MHC class I molecules via cross-dressing (Nakayama, 2015) rather than cross-presentation. To examine this possibility, we co-cultured ovalbumin-expressing CT26 colon tumor cells of BALB/c origin (OVA-CT26Stat3KD and OVA-CT26scr) or haplo-identical OVA-CMTStat3KD and OVA-CMTscr together with C57BL/6-derived DCs from WT or Tap1-deficient mice along with OT-I CD8 T cells (Figure 5H). In the presence of BALB/c CT26 cells no OT-I T cell activation could be observed regardless of whether or not CT26 cells had been pre-treated with H2O2. Instead, T cell activation was only triggered by OVA-CMT cells, yet absence of Tap1 in DCs did not affect IFNγ release (Figure 5J), supporting the notion that T cell activation occurred independently of cross presentation. In contrast, knockdown of Tap1 in tumor cells prevented IFNγ production by OT-I cells (Figures 5K and S4A), providing further evidence that antigen-processing can occur directly in tumor cells. This was further supported when addition of DCs derived from BALB/c mice enhanced the stimulation of OT-I T cells in a dose-dependent manner when cultured together with OVA-CMTStat3KD cells but not with OVA-CMTscr cells (Figure 5L). Moreover, when we co-cultured BALB/c CT26 cells with C57BL/6 splenocytes, we could detect the presence of the BALB/c MHC class I allele Dd on C57BL/6 DCs. The MHC transfer was enhanced by H2O2 pretreatment of tumor cells, which, again, occurred independent of Tap1 expression in DCs (Figures 5M and 5N). Collectively, these results further strongly supported our hypothesis that Stat3-deficient tumor cells can directly process antigens in a LMP-dependent manner and cross-dress DCs to induce an effective CD8+ T cell response.
Figure 5. T Cell Activation by Antigen-Expressing Tumor Cells Is Enhanced by LMP and DC Cross-Dressing In Vitro.
For a Figure360 author presentation of Figure 5, see the figure legend at https://10.1016/j.cell.2018.05.028.
(A) Experimental setup of co-culture experiment.
(B) Immunoblot analysis of STAT3.
(C) Representative flow cytometric analysis of acridine-orange-stained OVA-CMTscr and OVA-CMTStat3KD cells.
(D) IFNγ released by OT-I splenocytes after co-culture with OVA-CMTscr or OVA-CMTStat3KD cells for 2 days; n = 6 from 2 independent experiments.
(E) IFNγ released by OT-I splenocytes that had been co-cultured for 2 days with E64d-pre-treated OVA-CMTStat3KD cells; n = 6 from 2 independent experiments. Note that protease inhibitor was present during H2O2 stimulation but absent during co-culture with OT-I cells.
(F) IFNγ released by CD8+ OT-I T cells co-cultured for 2 days with H2O2-pre-treated OVA-CMTscr or OVA-CMTStat3KD cells; n = 6 from 2 independent experiments.
(G)IFNγ released by CD8+ OT-I T cells with DCs co-cultured with H2O2-pre-treated OVA-CMTscr or OVA-CMTStat3KD cells; n ≥ 3 from 2 independent experiments.
(H) Experimental setup of co-culture experiments.
(I) Immunoblot analysis of STAT3.
(J) IFNγ released by OT-I T cells co-cultured with WT or Tap1−/− CD11c+ DCs and with H2O2-stimulated OVA-CMT or OVA-CT26 cells; n = 6 from 2 independent experiments.
(K) IFNγ released by OT-I splenocytes after co-culture with H2O2-stimulated OVA-CMT cells with knockdown of Tap1 and/or Stat3; n = 6 from 2 independent experiments. ***p < 0.001 by Student’s t test.
(L) IFNγ released by CD8+ OT-I T cells co-cultured with OVA-CMTscr or OVA-CMTStat3KD cells and DCs from BALB/c donors as indicated; n = 12 from 4 independent experiments. *p < 0.05, ***p < 0.001 by one-way ANOVA.
(M)Quantification of H2-Dd on splenocytes from WT C57BL/6 donor mice co-cultured with OVA-CT26scr or OVA-CT26Stat3KD cells pre-treated as indicated; n = 6 from 2 independent experiments.
*p < 0.05 by Student’s t test.
(N) Quantification of H2-Dd on splenocytes from WT or Tap1−/− C57BL/6 donor mice co-cultured with H2O2 pretreated OVA-CT26scr or OVA-CT26Stat3KD cells; n = 6 from two independent experiments.
*p < 0.05, ***p < 0.001 by Student’s t test (D–G). In (D)–(G) and (J)–(N), data are mean ± SEM. See also Figure S4.
Previously, it had been suggested that cathepsin S is engaged in MHC class II-mediated antigen presentation in IECs in vivo, whereas cathepsin L enables this in thymic cortical epithelial cells (Beers et al., 2005; Nakagawa et al., 1998; Shen et al., 2004). To functionally examine whether cathepsin S, cathepsin L, or alternative cathepsins might be responsible for MHC-class-I-mediated antigen presentation in Stat3-deficient tumor cells, we individually knocked down expression of single cathepsins B, D, F, H, L or S in OVA-CMTStat3KD cells and examined IFNγ release by OT-I T cells upon H2O2 treatment prior to the co-culture. Indeed, reduced expression of either cathepsin S or cathepsin L OVA-CMTStat3KD cells markedly reduced IFNγ release, while knockdown of the other cathepsins did not (Figure S4B). To confirm the contribution of cathepsin S and cathepsin L to the observed phenotype in Stat3-deficient/β-catenin mutant IECs in vivo, we crossed Ctss−/− whole-body mutants to β-catc.a./Stat3ΔIEC animals. Surprisingly, the loss of cathepsin S in the corresponding Ctss−/−/β-catc.a./Stat3ΔIEC mice did not affect Ifng expression on day 15. Similarly, IECs restricted deletion of cathepsin L (CtslΔIEC/β-catc.a./Stat3ΔIEC) and even simultaneous loss of cathepsin S and L neither prevented IFNγ upregulation (Figure S4C) nor affected LMP in intestinal epithelia of Ctss−/−/CtslΔIEC/β-catc.a./Stat3ΔIEC mice (Figure S4D). Importantly, we could not detect reduced protease activity in IECs of these mice when using the cathepsin substrate z-Leu-Arg-AMC (Figure S4E), indicating that in vivo the absence of cathepsin S and L could be compensated by other cysteine proteases as suggested in a murine pancreatic neuroendocrine tumor model (Akkari et al., 2016).
Enhanced Mitophagy Leads to Iron(II) Accumulation in Lysosomes, thereby Triggering the Fenton Reaction
Next, we aimed to address the mechanism causing LMP. LMP can be triggered by various stimuli, the most common being ROS-mediated lysosomal destabilization in the event of intralysosomal iron(II) accumulation (Boya and Kroemer, 2008). During the Fenton reaction iron(II) catalyzes the conversion of H2O2 into highly reactive hydroxyl and hydroperoxyl radicals (Winterbourn, 1995). Oncogenic wnt activation triggers ROS production in IECs (Myant et al., 2013), and, using the fluorescent Fe2+-specific probe IP-1 (Au-Yeung et al., 2013), we confirmed enhanced iron(II) accumulation in Stat3-deficient CMT93 cells (Figures 6A–6D). The signal of the iron probe IP-1 co-localized with LysoTracker fluorescence, confirming lysosomes as the predominant intracellular site of iron(II) accumulation in OVA-CMT cells (Figures S5A–S5F). However, we were not able to detect significant alterations in expression of genes coding for proteins involved in iron uptake (Figure S5G). Instead, we observed an increased presence of components of the mitochondrial oxidative phosphorylation system (COXII) in lysosomes of OVA-CMTStat3KD cells (Figures 6E–6H, S5H, and S5I). This was suggestive of enhanced mitophagy, a process selectively eliminating damaged or excessive mitochondria by the autophagic pathway as the transfer of iron-containing electron transporting complexes to lysosomes results in intralysosomal iron(II) accumulation (Terman et al., 2006; Youle and Narendra, 2011). Interestingly, serine-phosphorylated Stat3 had been suggested to be involved in mitochondrial function to preserve oxidative phosphorylation in cardiac and nerve cells as well as RAS-transformed tumor cells (Gough et al., 2009; Wegrzyn et al., 2009). However, a transcriptionally constitutive active Stat3 (Stat3C) induces a metabolic switch toward aerobic glycolysis (Demaria et al., 2010) in a Hif1α-dependent manner, thereby eliciting the Warburg effect commonly described in cancers (Warburg et al., 1924). In Stat3-deficient CMT93 cells we observed an increased basal and maximal oxygen consumption rate, decreased glucose-uptake and increased ATP levels (Figures 6I–6L and S5J), suggesting enhanced oxidative phosphorylation. In line with this notion, OVA-CMTStat3KD cells proliferated significantly more in carbohydrate-deficient medium, or in the presence of the glycolysis inhibitors bromopyruvate or deoxyglucose, or when cultured in galactose, which all require mitochondrial activity to yield a positive energy balance but proliferated indifferently in the presence of glucose (Figure S5K). Staining with MitoTracker indicated an elevated number of mitochondria in OVA-CMTStat3KD cells while at the same time the mitochondrial membrane potential was decreased in a larger proportion of these cells (Figures 6M and 6N). Collectively, these data suggested a higher turnover of mitochondria in Stat3-deficient cells, which was supported by flow cytometry after labeling OVA-CMTscr and OVA-CMTStat3KD cells with Mito-Timer (Hernandez et al., 2013) (Figure 6O). To confirm that LMP induction was dependent on mitochondrial Stat3, we triggered LMP in Stat3-deficient fibroblasts by H2O2 treatment. Retroviral reconstitution of Stat3−/− cells with either WT Stat3, a mitochondrial-targeted version of Stat3 (MLS-Stat3), a dominant negative tyrosine-to-phenylalanine mutant (MLS-Stat3Y705F), or a DNA binding mutant (MLS-Stat3E434A/435A) all prevented increased LMP. Because the additional inactivation of the S727 phosphorylation site (MLS-Stat3Y705F/S727A) blocked induction of LMP to a lesser extent than the other mutants, we speculate LMP was prevented by S727 phosphorylated Stat3 in mitochondria rather than Stat3-dependent transcription (Figure 6P). Accordingly, expression levels of Stat3-dependent genes involved in glycolysis or mitochondrial respiration (Demaria et al., 2010) were not markedly changed in OVA-CMTStat3KD cells (Figure S5L).
Figure 6. Stat3 Deficiency Induces Mitophagy and Increased Lysosomal Iron Load.
(A–D) Confocal live cell microscopy of the fluorescent Fe2+-specific probe IP-1 in OVA-CMTscr (A and B) or OVA-CMTStat3KD (C and D) cells. LUT as depicted on the right-hand side.
(E–H) Mitophagy in OVA-CMTscr (E and F) and OVA-CMTStat3KD (G and H) was visualized by co-staining for the mitochondrial protein COXII (green) and the lysosomal membrane protein LAMP2 (red).
(I and J) Respirometry of intact cells. Basal rate was measured after an initial stabilization time (position a in Figure S5J) (I) and the maximal rate after addition of a saturating amount of FCCP (positions b and c for OVA-CMTscr and OVA-CMTStat3KD, respectively, in Figure S5J) (J); n = 10 from 10 independent experiments.
(K) Relative glucose uptake over 12 hr; n = 7 from 2 independent experiments.
(L) ATP content as measured by luciferase activity; n = 12 from 4 independent experiments.
(M) MFI of cells incubated with MitoTracker; n = 11 from 3 independent experiments.
(N) Relative fraction of cells showing low TMRM staining; n = 9 from 3 independent experiments.
(O) Ratio of cells showing green or red fluorescence of MitoTimer, indicating new or old mitochondria, respectively; n = 18 from 3 independent experiments.
(P) Relative amount of pale bodies in MEFs reconstituted with the indicated variant of Stat3a after treatment with H2O2; n = 7 of 3 independent experiments.
*p < 0.05, **p < 0.001 by one-way ANOVA against not-reconstituted Stat3−/− MEFs. *p < 0.05,**p < 0.01, ***p < 0.001 by Student’s t test (J–O).
In (I)–(P), data are mean ± SEM. See also Figure S5.
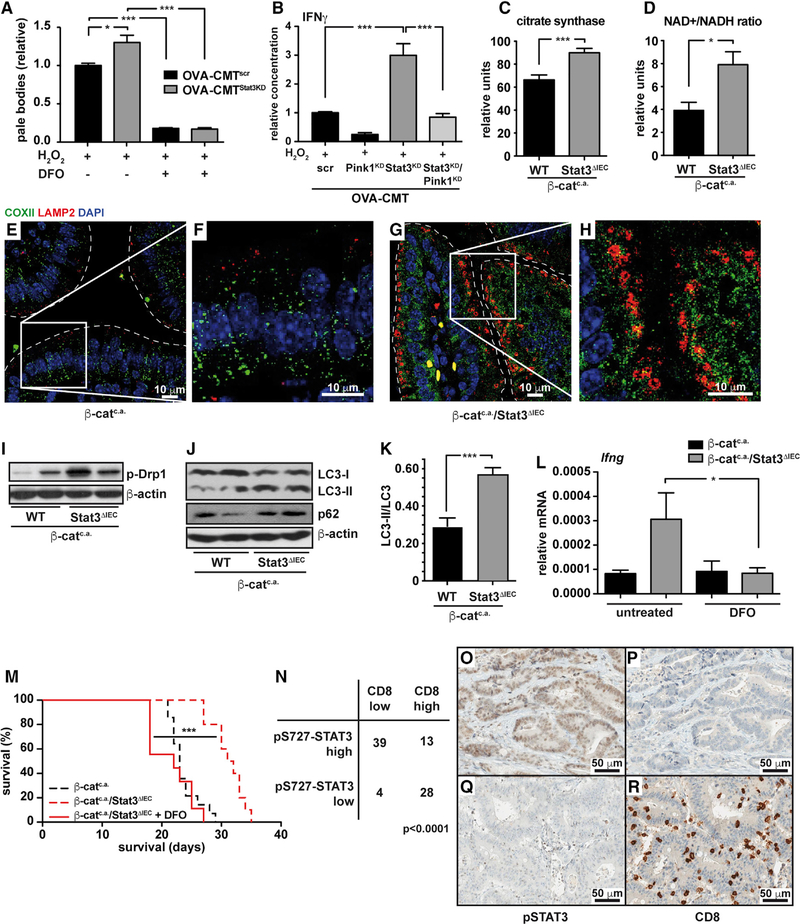
To confirm the functional relevance of intralysosomal iron(II) accumulation for enhanced LMP in OVA-CMTStat3KD cells, we treated CMT cells with the iron chelator deferoxamine (DFO) prior to the H2O2 challenge, and this was indeed sufficient to prevent LMP (Figures 7A and S6A–S6H). The direct involvement of mitophagy in lysosomal iron accumulation, as well as T cell activation, was demonstrated by the finding, that RNAi-mediated gene knockdown of Pink1, a kinase responsible for the degradation of damaged mitochondria in the Pink1-Parkin pathway, in OVA-CMTStat3KD cells markedly reduced lysosomal iron levels (Figures S6I–S6M), and prevented IFNγ production in the OT-I co-culture system (Figure 7B). Moreover, pre-treatment of CMT93-cells with antimycin A or rotenone, which inhibit complex III and I of the respiratory chain, respectively, induce mitophagy (Fang et al., 2014) and increase the lysosomal iron(II) load (Figures S6N–S6P), thus sensitizing CMT93 cells to H2O2-induced LMP, which could be prevented by DFO (Figure S6Q).
Figure 7. Lysosomal Iron Enhances Anti-tumor Immune Reaction In Vivo.
(A) Relative fraction of OVA-CMTscr and OVA-CMTStat3KD cells with low red acridine orange fluorescence (pale bodies); n = 4 of two independent experiments.
(B) IFNγ released by OT-I splenocytes after co-culture with H2O2 stimulated OVA-CMT cells with knockdown of Pink1 and/or Stat3; n = 6 from 2 independent experiments.
(C) Citrate synthase activity in mitochondrial isolates of IECs of β-catc.a. and β-catc.a./Stat3ΔIEC 3 days after the start of tamoxifen. Samples were normalized to total protein content and measured in triplicates of 6 mice each.
(D) NAD+/NADH-ratio in β-catc.a. and β-catc.a./Stat3ΔIEC mice on day 3; n ≥ 4/genotype.
(E–H) Co-staining for the mitochondrial protein COXII (green) and the lysosomal membrane protein LAMP2 (red) in β-catc.a. (E and F) and β-catc.a./Stat3ΔIEC (G and H) mice on day 3.
(I) Immunoblot analysis of phospho-Drp1 protein in lysates of IECs of β-catc.a. and β-catc.a./Stat3ΔIEC mice on day 3.
(J and K) Immunoblot analysis of LC3 and p62 (J) and quantification of LC3-II/LC3 conversion ratio (K) in β-catc.a. and β-catc.a./Stat3ΔIEC mice on day 15.
(L) Relative Ifng-mRNA in β-catc.a. and β-catc.a./Stat3ΔIEC mice treated with the iron-chelator DFO (400 mg/kg bodyweight) or control; n ≥ 5/genotype.
(M) Survival of β-catc.a./Stat3ΔIEC mice (n = 9) treated with DFO. Dashed lines have been reproduced from Figure 1N for better comparison. ***p < 0.001 by log-rank test.
(N) Correlation of pS727-STAT3 expression and presence of CD8+ T cells in human colorectal cancer (CRC, N = 84; χ2 test, p < 0.0001).
(O–R) Representative immunohistochemical analysis of pS727-STAT3 (O and Q) and CD8 (P and R) in human CRC. Scale bars, 50 μm.
See also Figure S6.
*p < 0.05, ***p < 0.001 by Student’s t test (A–D). In (A)–(D) and (J)–(L), data are mean ± SEM.
In line with improved oxidative phosphorylation in Stat3-deficient CMT cells in culture, in IECs of β-catc.a./Stat3ΔIEC we also detected elevated citrate synthase activity and an increased NAD+/NADH ratio (Figures 7C and 7D). Expression of genes controlling glycolysis was unchanged (Figure S6R) and immunofluorescent staining indicated the presence of COXII in lysosomes of β-catc.a./Stat3ΔIEC IECs (Figures 7E–7H). Electron microscopy confirmed morphological changes in mitochondrial ultrastructure, such as cristaelysis (Figures S6S and S6T) and immunoblot analysis revealed elevated activation of Drp1 in β-catc.a./Stat3ΔIEC IECs (Figure 7I). Furthermore, conversion of LC3-I to LC3-II in β-catc.a./Stat3ΔIEC IECs was elevated, suggesting enhanced autophagosome formation, yet simultaneous accumulation of p62 suggested decreased autophagic flux (Figures 7J and 7K) in agreement with induction of mitophagy but impaired lysosomal function. Importantly, DFO administration to β-catc.a. and β-catc.a./Stat3ΔIEC mice prevented Ifng upregulation in β-catc.a./Stat3ΔIEC mice (Figure 7L) and prevented prolonged survival of β-catc.a./Stat3ΔIEC mice (Figure 7M), thus further supporting the notion that accumulation of intralysosomal iron(II) was essential for the observed anti-tumor cell immune phenotype in these animals. In contrast, pharmacological suppression of autophagy using either 3-methyladenine or genetic ablation of Atg7 did not prevent IFNγ upregulation in lamina propria of β-catc.a./Stat3ΔIEC mice but, instead, increased it even further (Figures S6U and S6V).
A newly established immune score based on the presence of different T cell subsets can efficiently predict tumor recurrence in CRC patients (Fridman et al., 2012). In particular the presence of CD8+ T cells and high IFNγ expression are associated with a better prognosis. Therefore, we examined whether STAT3 activation was involved in this phenomenon. Indeed, when we analyzed CRC biopsies, we found a significant inverse correlation between the accumulation of S727-phosphorylated STAT3 in tumor epithelia and the frequency of infiltrating CD8+ T cells (Figures 7N–7R) suggesting that STAT3 may also be functionally involved in the suppression of CD8+ T cell recruitment in human CRC.
DISCUSSION
Here, we have identified a so far unrecognized complex multistep process linking altered mitochondrial function and antigen presentation in IECs. By triggering mitophagy and increasing the amount of intralysosomal ferrous iron lysosomes are sensitized for the induction of LMP via the Fenton reaction. As a consequence cathepsins are released into the cytoplasm, enabling antigen processing directly in IECs. While we have discovered this connection in the context of Stat3 deletion, this mechanism is not strictly Stat3-dependent considering that LMP can be pharmacologically induced by chloroquine in a Stat3 WT background, thus suggesting a previously unrecognized role for lysosomes in regulating the immunogenicity of malignant transformed epithelial cells. Knockdown of both cathepsin S and cathepsin L could prevent OT-I T cell activation ex vivo, yet the absence of cathepsin S and cathepsin L could be compensated by other proteases in vivo. This suggests that not only one single cathepsin is involved in antigen presentation in IECs but also several proteases have the capacity to process the relevant antigen in vivo. Nevertheless, our data underscore the importance of LMP in IECs for CD8+ T cell activation since prevention of LMP by iron chelation was equally effective in suppressing IFNγ upregulation as blockade of protease activity using E64d. Whether only the release of cathepsins to the cytoplasm or also cell death of IECs, which is usually associated with LMP induction (Boya and Kroemer, 2008), contributes to enhanced T cell activation still has to be determined.
Our data support the concept of cross-dressing (Nakayama, 2015) and suggest that the capacity of DCs to cross present is not essential for T cell activation in this scenario. Most likely, however, DCs are still required to provide essential co-stimulatory signals and indeed in vivo proper T cell activation by cross-dressing depends on the presence of DCs (Cerovic et al., 2015). Interestingly, chloroquine-induced LMP in β-catc.a. mice IECs extended survival more efficiently than loss of Stat3 in IECs. Chloroquine can enhance human CD8+ T cell responses against soluble antigens by inhibiting endosomal acidification in DCs (Accapezzato et al., 2005). Therefore, we cannot rule out that apart from affecting LMP in IECs some of the substantial tumor suppression is mediated by chloroquine’s ability to influence endosomes in DCs as well.
In addition to controlling the transcription of a wide range of genes involved in cell proliferation and survival (Bollrath and Greten, 2009), Stat3 has been suggested to confer transcription-independent functions including the regulation of mitochondrial activity (Meier and Larner, 2014). Indeed, mitochondrial Stat3 has been associated with decreased ETC activity, mainly complexes I, II, and V (Gough et al., 2009; Wegrzyn et al., 2009). Further studies showed that Stat3 deficiency decreased activity in complexes III and IV (Meier and Larner, 2014). In contrast, a constitutively active form of Stat3 (Stat3-C) supports a shift from oxidative phosphorylation toward glycolysis (Warburg effect) in mouse embryonic fibroblasts (MEFs) (Demaria et al., 2010). That the latter phenotype is at least in part dependent on Y705 phosphorylation and nuclear functions of Stat3 highlights the pleiotropic role of Stat3 in mitochondrial activity and illustrates that actions of Stat3 in the mitochondria greatly vary between types of cells and experimental systems. The relative contribution of mitochondrial Stat3 to a given response probably most likely depends on the relative contribution of nuclear Stat3 in the particular context. Cancer-cell-generated lactate is now considered a critical immune suppressive metabolite and reducing lactate production by blocking glycolysis or direct inhibition of LDH-A has been suggested to restore physiological T cell function (Choi et al., 2013). Our data suggest that in addition to inhibiting lactate production enhanced oxidative phosphorylation may trigger mitophagy, which would stimulate antigen processing thus further contributing to improved intestinal tumor suppressive immune function. Interestingly, the mitochondrial phenotype of Stat3-deficient cells comprising more mitochondria and enhanced oxidative phosphorylation does not match those typically of cells undergoing mitophagy. However, the fact that knockdown of Pink1 prevents LMP and CD8+ T cell activation strongly supports a contribution of mitophagy even though also mitophagy-independent functions of PINK1 have been described (Matheoud et al., 2016). Thus, it will be of great interest to define the exact molecular alterations in Stat3-deficient mitochondria causing this particular phenotype.
Our findings may provide the basis for novel therapeutic strategies that exploit anti-tumor immunity in CRC patients. These could encompass therapies interfering with mitochondrial function, thereby triggering mitophagy and the release of LMP-inducing compounds. Compounds directed against Stat3 or presumably its upstream Jak tyrosine kinases or chloroquine might be good candidates to examine in this context.
STAR★METHODS
KEY RESOURCES TABLE
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Florian R. Greten (greten@gsh.uni-frankfurt.de).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice
Stat3ΔIEC mice have been described previously (Bollrath et al., 2009; Takeda et al., 1998). CtnnbloxEx3/wt mice (Harada et al., 1999) were crossed to villin-creERT2 (el Marjou et al., 2004) mice to obtain β-catc.a. mice. These were intercrossed with Stat3F/F, Ctss−/− (Shi et al., 1999), CtslF/F (Tholen et al., 2014), Atg7F/F (Komatsu et al., 2005) or ROSAOVA (Sandhu et al., 2011) animals to obtain the respective compound mutants. All mice were kept on a mixed genetic background. OT-I, Tap1−/−, Ifng−/− mice and CD11c-DTR mice were obtained from Jackson Laboratories (Bar Harbor, USA). Appropriate littermate controls were used in all experiments.
Age and sex-matched cohorts comprised of male and female mice at 6 to 12 weeks of age were used for experiments. All mice were kept on a 12-hour light/dark cycle in individually ventilated cages in specific pathogen-free facilities at Klinikum rechts der Isar, Munich and the Georg-Speyer-Haus, Frankfurt/Main, with chow (standard formulation) and water supply ad libitum. All procedures were reviewed and approved by the Regierung of Oberbayern or the Regierungspräsidium Darmstadt, respectively. In general, mice were housed in groups of 3–5 mice per cage, unless experimental procedures (e.g., supplemented chow) or individual behavior (e.g., fighting) required single housing.
Human Samples
Human tumor samples were obtained from the tumor bank at the Institute of Pathology at Klinikum rechts der Isar, Munich or the Victoria Cancer Biobank, Melbourne. Samples have been analyzed per case and no assignment to groups (e.g., sex, age) have been performed. All procedures were performed with the approval of the respective ethics committees of the Technical University of Munich and the Victoria Cancer Biobank on samples from subjects that had given their written informed consent.
METHOD DETAILS
In vivo analyses
To induce colon tumors mice were injected i.p. with azoxymethane (Sigma-Aldrich, St. Louis, USA) once a week for 6 weeks at 10 mg/kg. In villin-creERT2 mice deletion was induced by five daily oral administrations of 1 mg tamoxifen (Sigma-Aldrich) dissolved in an ethanol/oil mixture, or, in the case of AOM-treated OVAVil or OVAVil/Stat3ΔIEC mice, administered by tamoxifen-containing chow (Genobios, Laval, France, dosage 400 mg/kg). Depletion of CD8+ T cells in β-catc.a. and β-catc.a./Stat3ΔIEC mice was achieved by i.p. injection of 150 μg anti-CD8-antibody (clone 2.43, BE0061, BioXCell, Lebanon, USA) in 100 μl PBS for 3 consecutive days prior to oral administration of tamoxifen for 5 days. After completion of tamoxifen-administration mice were again injected for 3 consecutive days with 150 μg of antibody. Then mice were injected every 7 days for 3 consecutive days. CD8-cell depletion was confirmed by flow cytometric analysis of splenocytes. For depletion of dendritic cells, bone marrow of CD11c-DTR-mice was isolated, cleared of CD3+ T cells and tail-vein injected into 9 Gy irradiated recipients. Mice were allowed to recover for 8 weeks and depletion of DC was carried out by i.p. injection 4 μg/kg bodyweight diphtheria toxin (Merck, Darmstadt, Germany) every second day. Chloroquine (Sigma-Aldrich) was i.p. injected at 60 mg/kg bodyweight in 0.9% NaCl-solution, 3-MA (Sigma-Aldrich) at 30 mg/kg bodyweight in PBS, daily starting upon induction of β-cat-activation. Deferoxamine (DFO) (Sigma-Aldrich) was dissolved in 0.9% NaCl-solution and injected i.p. at 400 mg/kg every other day starting upon induction of β-cat-activation, E64d (Santa Cruz Biotechnology, Dallas, USA) was dissolved in an ethanol/oil-mixture and applied as oral gavage at 5 mg/kg bodyweight daily starting upon induction of β-cat-activation. ACC (Sigma-Aldrich) was dissolved in the drinking water at 0.5% (w/v) and applied continously starting upon induction of β-cat-activation.
Protein Analysis and Immunohistochemistry
Paraffin sections (3 μm) were stained using standard immunohistochemical procedures. In order to isolate intestinal epithelial cells intestines have been opened longitudinally, cleared of mucus, cut into small pieces (approx. 5 mm) and incubated in PBS containing 5 mM EDTA and 2 mM DTT with gentle agitation at 37°C for 10 min. Epithelial cells were then separated from underlying tissue by heavy vortexing and subsequent centrifugation.
For extraction of cytosolic proteins pelleted cells were dissolved in extraction buffer (25 μg/ml digitonin, 250 mM sucrose, 20 mM HEPES, 10 mM KCl, 1.5 mM MgCl2, 1 mM EDTA, 1 mM EGTA, pH 7.5 (all purchased from Carl Roth, Karlsruhe, Germany or Sigma-Aldrich) and incubated for 15 min on ice. Heavy membrane particles were pelleted at 10.000 g for 10 min. and dissolved in modified RIPA buffer (50 mM Tris-HCl, 250 mM NaCl, 25 mM sodium pyrophosphate, 3 mM EDTA, 3 mM EGTA, 1% Triton-X, 0.5%, Igepal CA-630, 10% Glycerol, pH 7.5, all from Carl Roth or Sigma-Aldrich) and cleared of lipids by additional centrifugation at 17.000 g for 12 min. Protease inhibitor cocktail (Roche Diagnostics, Mannheim, Germany) was added according to the manufacturer’s instructions. For whole cell extracts cells pelleted, dissolved in modified RIPA buffer as above and cleared of lipids by additional centrifugation at 17.000 g for 12 min in presence of protease inhibitors.
Antibodies used were anti-CD3 (IS503, Dako, Hamburg, Germany), IFNγ (BAF485, R&D Systems, Minneapolis, USA), Y705-phosphorylated Stat3 (9145), S727-phosphorylated Stat3 (9134S), LC3 (2775) p-Drp1 (4494S) and cleaved Caspase-3 (9579, all Cell Signaling Technologies, Danvers, USA), p62 (GP62-C, Progen, Heidelberg, Germany), Stat3 (610190, BD, Franklin Lakes, USA), MHC class I (sc-59199), Cathepsin S (sc-6505), Gapdh (sc-32233, Santa Cruz), β-Actin (A4700, Sigma-Aldrich), Cathepsin B (ab58802), LAMP2 (ab13524, Abcam, Cambridge, UK) and COXII (55070–1-AP, Proteintech, Rosemont, USA). Biotinylated secondary antibodies were purchased from Vector Labs (Burlingame, USA). For human tissue, anti-CD8 (108M-96, Cell Marque, Rocklin, USA) and S727-phosphorylated-Stat3 (ab30647, Abcam) were used.
Immunofluorescent staining was performed on paraffin sections or Lab-Tek II Chamber slides (Thermo Fisher Scientific, Waltham, USA) using standard procedures and indicated primary antibodies. Secondary antibodies were fluorochrome-conjugated anti-rabbit IgG-Alexa Fluor 488 (A-21206, Thermo Fisher Scientific) and anti-rat IgG-Cy3 (Jackson Immunoresearch, West Grove, USA). For detection of mouse primary antibodies, the M.O.M. Fluorescein Kit (Vector Labs) was used according to the manufacturer’s instructions. Where indicated slides were analyzed using a SP5 confocal laser scanning microscope (Leica, Wetzlar, Germany). Size of tumors was quantified by measuring the largest area of each tumor in serial sections (approx. 250 μm distance between single sections) using a AxioImager microscope and AxioVision software (Zeiss, Jena, Germany).
For electron microscopy tissue samples from small intestines were excised, fixed with 2.5% glutaraldehyde solution and processed according to standard procedures.
Isolation and analysis of lamina propria cells
For the isolation of lamina propria cells intestines of mice were opened longitudinally, cleared of mucus, chopped into small pieces and digested in supplemented RPMI (Thermo Fisher Scientific) containing 1 mg/ml Collagenase I (C0130, Sigma-Aldrich), 1 mg/ml Dispase II (Roche), 20 μg/ml DNase I, 10 mM HEPES, non-essential amino acids, Glutamine (all Sigma-Aldrich or Thermo Fisher Scientific) and 2% fetal calf serum (Thermo Fisher Scientific) for 15 – 20 min. at 37°C while gently shaking. Liberated cells were collected and placed on ice. The digestion has been repeated for 2 additional times using fresh digestion medium. Following the third digestion step all fractions were combined and passed through a 70 μm strainer (BD). Flow cytometric analysis was performed on a Gallios flow cytometer (Beckman-Coulter, Brea, USA), FACSCalibur, FACSCantoII or LSRFortessa (all BD). The following fluorochrome-conjugated antibodies were used: CD3-Alexa Fluor 700 (56–0032), CD4-eFluor 450 (48–0042), CD8α-APC (17–0081), IFNγ-PerCP-Cy5.5 (45–7311), CD11c-FITC (11–0114), CD11b-APC-eFluor 780 (47–0112), CD80-PE-Cy7 (25–0801-82), Gr-1-Alexa Fluor 700 (56–5931-82), F4/80-APC (17–4801-82), EpCAM-eFluor 450 (48–5791-82), Foxp3-PE (12–5773), B220-efluor 450 (48–0452), SIINFEKL-H2Kb (12–5743-82, all Thermo Fisher Scientific, Santa Cruz, USA), CD45-BV786 (564225) and IL-12-PE (554479, all BD). Appropriate isotype control antibodies were purchased from Thermo Fisher Scientific. For intracellular staining cells were ex vivo restimulated in supplemented RPMI containing either 1 μg/ml ionomycin and 20 ng/ml phorbol-myristate-acetate (for T cells) or 0.1 μg/ml LPS (for DC, all Sigma-Aldrich) in presence of Golgi-Plug (BD) for 5 hr at 37°C. Cells were prestained with 0.5 μg/ml ethidium bromide monoazide (Sigma-Aldrich) for 15 min in bright light or Live/Dead Fixable Blue Dead Cell Stain Kit, fixed in IC-fixation buffer for 30 min on ice and stained in permeabilization buffer (all Thermo Fisher Scientific). SIINFEKL-MHCI-pentamer was purchased from ProImmune (Oxford, UK) and used according to the manufacturer’s instructions.
FACS-sort has been performed on a FACSAria or FACSAria Fusion (all BD).
Cell culture and T cell activation assay
Stat3−/− mouse embryonic fibroblasts were provided by Thomas Decker and reconstituted with the indicated plasmids by retroviral transduction as described previously (Wegrzyn et al., 2009). The murine colon carcinoma cell lines CMT93 was obtained from ATCC. Cells were cultured under standard conditions using Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% FCS and 1% Penicillin-Streptomycin. CMT93 cells were transfected with an expression vector encoding chicken ovalbumin cDNA (pCI-OVA). The vector pCI-OVA has been generated by inserting the OVA-cassette amplified from DEC205-OVA (Loschko et al., 2011) into the pCI-neo vector (Promega #TB215) at the NheI-NotI-site. CT26 cells were transfected with an expression vector encoding ovalbumin along with EGFP (pCI-OVA-IRES-GFP) and described previously (Gamrekelashvili et al., 2007). Stable clones were selected with medium containing 1.5 mg/ml geneticin (Thermo Fisher Scientific) and termed OVA-CMT or OVA-CT26. Stable knockdown of Stat3 was achieved by transfection of OVA-CMT or OVA-CT26 cells with pcDNA6.2-Stat3-miRNA (containing a synthetic Stat3-binding pre-miRNA: TCGTGAAACGTGAGCGACTCAAACTGGTTTTGGCCACTGACTGACCAGTTTGACGCTCACGTTT; target sequence: CAGTTTGAGTCGCTCACGTTT) or pcDNA6.2-scrambled from BLOCK-it Pol II miR RNAi Expression Vector Kit (Thermo Fisher Scientific #K493600), after removal of the EmGFP-expression cassette according to the manufacturer’s instructions. Clones were selected using supplemented DMEM containing 10 μg/ml blasticidin S (Thermo Fisher Scientific) and successful knockdown of Stat3 protein was confirmed by immunoblot analysis. For siRNA-mediated knockdown-experiments a pool of 4 pre-validated siRNAs (“ON-TARGETplus-smart pool”) per gene was purchased from Dharmacon/GE Healthcare (Buckinghamshire, UK) and transfected using Viromer green (Lipocalyx, Halle, Germany) or DharmaFECT 4 (Dharmacon) according to manufacturer’s instructions.
In T cell activation assays OVA-CMT cells were pretreated with H2O2 at 1 mM, E64d (10 μg/ml) or DMSO (all Sigma-Aldrich) for 2 hr at 37°C and washed in cell culture medium. Then 5×104 OVA-CMT cells were added to 5×105 OT-I splenocytes into a 96-well plate and incubated at 37°C for 2 days. IFNγ levels in supernatants were determined by ELISA (DY485, R&D Systems) according to the manufacturer’s instructions.
In co-culture experiments using purified DC and CD8+ T cells, DC were prepared from the spleen of C57B/6 wild-type or Tap1−/− mice 14 days after s.c. injection of 5×106 Flt3L-expressing B16 melanoma cells (Mach et al., 2000) or from BALB/c wild-type mice. DC were labeled with a biotinylated CD11c-antibody (553800, BD) and sorted using MACS-separation columns (Miltenyi, Bergisch-Gladbach, Germany) according to the manufacturer’s instructions. Splenic OT-I CD8+ T cells were magnetically purified by negative selection using CD8a+ T cell isolation kit II (130–095-236, Miltenyi). Purity was confirmed by flow cytometry and found to be greater than 95%. 5×104 CMT-OVA or OVA-CT26 cells were pretreated for 2 hr with 1 mM H2O2, washed and incubated with 2×105 OT-I CD8+-T cells with indicated amount of DC. Experiments were performed at least two times in triplicates.
For MHC class I transfer studies purified DC from C57B/6J or Tap1−/− (on C57B/6J background) mice were incubated with OVA-CT26 cells for 12–16 hr at 37°C. The cells then were stained with anti-CD45-APC-efluor 780 (47–0451-82), anti-CD11c-efluor 450 (48–0114-82, all Thermo Fisher Scientific), anti-H-2Kb–Percp-Cy5.5 (562831) and anti-H-2Dd-PE antibodies (553580, all BD) and analyzed by flow cytometry. The cells CD45+CD11c+GFP- were identified as DC and analyzed for (surface) expression of H-2Dd.
For quantification of LMP, cells were incubated in DMEM (Thermo Fisher Scientific) supplemented with 10% FCS and 1% Penicillin-Streptomycin in presence or absence of H2O2 (Sigma-Aldrich) at 1 mM for 2 hr. Then the cells were washed and cultured in supplemented DMEM for another 6 hr. Then cells were detached using Trypsin-EDTA (Thermo Fisher Scientific), and incubated with 50 ng/ml acridine orange (Calbiochem/Merck, Darmstadt, Germany) in supplemented DMEM for 15 min. at 37°C. Then cells were washed in PBS and analyzed by flow cytometry at 488 nm excitation and 620 ± 30 nm emission to quantify the red fluorescence of the lysosomal compartment. Where indicated cells were pre-incubated with rotenone (25 μM), antimycin A (100 μM) or DMSO overnight and with DFO (1 mM, all Sigma-Aldrich) for 1 hr prior to H2O2-stimulation.
All cell lines have been maintained at 37°C in 5% CO2.
Metabolic analyses
Cathepsin activity was measured using cytosolic fractions generated as stated above in presence of pefabloc (1 μM). Assay was performed in triplicates in a reaction buffer (50 mM sodium acetate, 8 mM EDTA, 1 mM DTT, 1 μM pefabloc, all Sigma-Aldrich) using 2 μg of protein lysate and 10 μM protease substrate z-LR-AMC (ES008, R&D Systems). Increase of fluorescence was measured at 390/485 nm on a spectrophotometer (Fluostar Omega, BMG Labtech, Ortenberg, Germany) and slope (ΔE/t) was calculated at linear phase.
Iron content was assessed by confocal live cell microscopy of cells grown on chambered optical coverslips (μ-slides, Ibidi, Martinsried, Germany) incubated in phenol-red free medium in presence of the Fe2+-sensitive probe IP-1 at 20 μM and Hoechst 33342 (Sigma-Aldrich) at 1 μg/ml and, where indicated, with LysoTracker red DND-99 (Thermo Fisher Scientific) at 25 nM for 30 min. Where indicated cells were pre-treated overnight with rotenone (25 μM), antimycin A (100 μM) or DMSO (all Sigma-Aldrich).
The oxygen consumption of trypsinized OVA-CMT93scr and OVA-CMT93STAT3KD cells was determined at 37°C using an Oxygraph-2k (Oroboros, Innsbruck, Austria). In each case, 3×106 cells were suspended in 2 mL pre-warmed FCS-supplemented DMEM medium and transferred into one of the two Oxygraph-chambers. 10 synchronous measurements with both cell lines were performed. Oxygen consumption rates (OCR) were determined after stabilization of the flux signal (1st deviation of the change in oxygen concentration) by using the ‘mark tool’ of the DatLab5 control software (Oroboros). After recording the basal respiration (basal OCR), oligomycin (Sigma-Aldrich) (2 μg/ml) was added to inhibit mitochondrial adenosine-triphosphate (ATP) synthesis. Subsequently carbonyl cyanide-p-trifluoromethoxyphenylhydrazone (FCCP, Sigma-Aldrich) was added stepwise to assess maximal capacity of the mitochondrial electron-transfer chain (maximal OCR) that occurred usually at 4.5 – 5.5 μM of the uncoupler. Finally, 2 mM KCN (Sigma-Aldrich) was added to determine the mitochondria-independent oxygen consumption of the cells.
In order to quantify the glucose uptake 5×105 cells were incubated in supplemented DMEM in presence of 5 mg/ml glucose for 24 hr. Then glucose concentration in the supernatant was assayed using the Glucose Assay Kit (Sigma-Aldrich) according to the manfacturer’s instructions and substracted from initial concentration. Total cellular adenosine-triphosphate (ATP) content was measured using the kit CellTiter-Glo (Promega, Madison, USA) according to the manufacturer’s instructions using 5×104 cells. Cell proliferation was assayed by seeding 104 OVA-CMT93 cells and counting the cell number using an automated cell counter (CASY, Roche Innovatis, Reutlingen, Germany) after 84h culture in glucose and pyruvate-free DMEM (Thermo Fisher Scientific) supplemented with 10% FCS and glucose (5 mg/ml), galactose (5 mg/ml), 2-deoxyglucose (1,5 mg/ml) or 3-bromopyruvate (100 μM, all Sigma-Aldrich) as indicated.
Mitochondrial content of cells was measured by flow cytometry following incubation of cells with MitoTracker deep red (250 nM, Thermo Fisher Scientific) in conventional cell culture medium for 30 min at 37°C. To estimate the mitochondrial membrane potential cells were incubated with tetramethylrhodamin-methylester (TMRM, 50 nM) in HBSS supplemented with 10 mM HEPES (all Thermo Fisher Scientific) for 30 min at 37°C and subjected to flow cytometry.
MitoTimer was expressed in OVA-CMTscr or OVA-CMTStat3KD cells by co-transfecting cells with pTRE-Tight-MitoTimer (Hernandez et al., 2013) and pES.1–2(M2N)p (containing reverse tetracycline-responsive transactivator M2) (Urlinger et al., 2000) using Lipofect-amine 2000 (Thermo Fisher Scientific) according to the manufacturer’s instructions. 2 days after transfection cells were seeded in triplicates, treated with doxycycline (Sigma-Aldrich) at 2 μg/ml for 1 hr, then incubated for additional 48 hr in conventional cell culture medium and treated with doxycycline for 2 hr. Cells were then analyzed by flow cytometry for fluorescence at 530/30 nm and 585/ 42 nm at excitation of 488 nm for new and old mitochondria, respectively.
NAD+/NADH ratio was measured using fresh tissue of small intestine and the NAD/NADH Assay Kit (ab65348, Abcam) according to the manufacturer instructions.
Citrate synthase activity was assayed using mitochondria isolated from small intestine epithelial cells resuspended in 0.25 M sucrose, 10 mM Tris, 1 mM EDTA, ph 7.2 passed 15 times through a 21G-needle. Unbroken cells were pelleted at 750 g for 5’ and discarded. The supernatant was subjected to centrifugation at 9,000 g for 15’ and the pellet resuspended in 0.25 M sucrose, 2 mM HEPES, 0.1 mM EGTA, pH 7.4 (with KOH) and used as crude mitochondrial fraction. To release matrix enzymes mitochondria were pelleted at 10,000 g for 15’ and resuspended in 25 mM potassium phosphate buffer, pH 7.2, 5 mM MgCl2 and subjected to 3 freeze-thaw cycles. The assay was performed adding acetyl CoA (0.1 mM) and oxaloacetic acid (0.1 mM) in buffer containing 50 mM KPi, pH 7.4, 0.1 mM 5,5’-dithio-bis-(2-nitrobenzoic acid) (DTNB, all Sigma-Aldrich or Carl Roth) and slope (ΔE/t) was measured at 412 nm using a spectrophotometer (SpectraMax 340, Molecular Devices, Sunnyvale, USA).
RNA Analysis
Total RNA was extracted from snap-frozen tissue, isolated enterocytes or cultured cells using a RNeasy Mini Kit (QIAGEN, Hilden, Germany) according to the manufacturer’s instructions. SuperScript II Reverse Transcriptase (Thermo Fisher Scientific) was used for synthesis of cDNA. Real-time PCR analysis using Fast-Start Universal SYBR Green PCR Master Mix (Roche Diagnostics) was performed on a StepOne Plus Real-Time PCR System (Thermo Fisher Scientific). Data has been normalized to the mRNA level of a housekeeping gene (cclophilinA) using the delta CT method (2CTcyclo-CTgene). Primer sequences are listed in Table S1.
NGS-based TCR repertoire analysis
NGS-based TCR repertoire analysis was performed by Repertoire Genesis (Osaka, Japan). Total RNA was converted to complementary DNA (cDNA) with Superscript III reverse transcriptase (Thermo Fisher Scientific). BSL-18E primer containing polyT18 (see below) and a SphI site was used for cDNA synthesis. After cDNA synthesis, double strand (ds)-cDNA was synthesized with E. coli DNA polymerase I, E. coli DNA Ligase, and RNase H. ds-cDNAs were blunted with T4 DNA polymerase (all Thermo Fisher Scientific). P10EA/ P20EA adaptor was ligated to the 5’ end of the ds-cDNA and then cut with SphI restriction enzyme. After removal of adaptor and primer with MinElute Reaction Cleanup kit (QIAGEN), PCR was performed with KAPA HiFi DNA Polymerase (Kapa Biosystems, Woburn, USA) using either TCR α chain constant region-specific (mCA1) or TCR β-chain constant region-specific primers (mCB1) and P20EA (Table S1). PCR conditions were as follows: 98°C (20 s), 65°C (30 s), and 72°C (1 min) for 20 cycles. The second PCR was performed with either mCA2 or mCB2 and P20EA primers using the same PCR conditions. Amplicons were prepared by amplification of the second PCR products using P22EA-ST1-R and either mCA-ST1-R or mCB-ST1-R. After PCR amplification, index (barcode) sequences were added by amplification with Nextera XT index kit v2 setA (illumina, San Diego, USA). The indexed products were mixed in an equal molar concentration and quantified by a Qubit 2.0 Fluorometer (Thermo Fisher Scientific). Sequencing was done with the Illumina Miseq paired-end platform (2 × 300bp). Assignment of sequences was performed by determining sequences with the highest identity in a dataset of reference sequences from the international ImMunoGeneTics information system® (IMGT) database (http://www.imgt.org). Data processing, assignment, and data aggregation were automatically performed using repertoire analysis software originally developed by our group (Repertoire Genesis, RG). RG implemented a program for sequence homology searches using BLATN, an automatic aggregation program, a graphics program for TRV and TRJ usage, and CDR3 length distribution. Sequence identities at the nucleotide level between query and entry sequences were automatically calculated. Nucleotide sequences of CDR3 regions ranged from conserved Cysteine at position 104 (Cys104) of IMGT nomenclature to conserved Phenylalanine at position 118 (Phe118) and the following Glycine (Gly119) was translated to deduced amino acid sequences. Sequence read having identical TRV, TRJ and deduced amino acid sequence of CDR3 was defined as unique read. The copy number of unique reads were automatically counted by RG software in each sample and then ranked in order of the copy number. Percentage occurrence frequencies of sequence reads with TRAV, TRAJ, TRBV and TRBJ genes in total sequence reads were calculated.
QUANTIFICATION AND STATISTICAL ANALYSIS
Please refer to the Figure Legends or the Experimental Details for description of sample size and statistical details. Cell culture assays have been performed in triplicates and in two independent experiments, unless stated otherwise. Data are expressed as mean ± SEM. Differences were analyzed by log-rank, two-tailed Student’s t test, Mann-Whitney U test, one-way ANOVA (using Bonferroni post test), one-sample t test or χ2-test using Prism 5 (GraphPad Software, La Jolla, USA) or Excel (Microsoft, Redmond, USA), p values ≤ 0.05 were considered significant.
Supplementary Material
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| CD8 (mouse, for in vivo depletion) | BioXCell | Cat# BE0061; RRID:AB_1125541 |
| CD3 (IHC) | Dako | Cat# IS503; RRID:AB_2732001 |
| IFNγ (IHC) | R&D Systems | Cat# BAF485; RRID:AB_356516 |
| pY705-STAT3 | Cell Signaling | Cat# 9145; RRID:AB_2491009 |
| pS727-STAT3 (IHC, mouse) | Cell Signaling | Cat# 9134S; RRID:AB_331589 |
| LC3 | Cell Signaling | Cat# 2775; RRID: AB_915950 |
| phospho-DRP1 | Cell Signaling | Cat# 4494S; RRID:AB_11178659 |
| cleaved Caspase-3 | Cell Signaling | Cat# 9579; RRID:AB_10897512 |
| STAT3 | BD | Cat# 610190; RRID:AB_397589 |
| p62 | Progen | Cat# GP62-C; RRID:AB_2687531 |
| MHC class I | Santa Cruz | Cat# sc-59199; RRID:AB_1126186 |
| Cathepsin S | Santa Cruz | Cat# sc-6505; RRID:AB_2245620 |
| Gapdh | Santa Cruz | Cat# sc-32233; RRID:AB_627679 |
| β-actin | Sigma | Cat# A4700; RRID:AB_476730 |
| LAMP2 | Abcam | Cat# ab13524; RRID:AB_2134736 |
| Cathepsin B | Abcam | Cat# ab58802; RRID:AB_940824 |
| COXII | Proteintech | Cat# 55070–1-AP; RRID: AB_10859832 |
| CD3-Alexa Fluor 700 (flow cytometry) | Thermo Fisher Scientific | Cat# 56–0032; RRID:AB_529508 |
| CD4-eFluor 450 | Thermo Fisher Scientific | Cat# 48–0042; RRID:AB_1272194 |
| CD8α-APC | Thermo Fisher Scientific | Cat# 17–0081; RRID:AB_469334 |
| IFNγ-PerCP-Cy5.5 (flow cytometry) | Thermo Fisher Scientific | Cat# 45–7311; RRID:AB_1107020 |
| CD11c-FITC (flow cytometry) | Thermo Fisher Scientific | Cat# 11–0114; RRID:AB_464941 |
| CD11b-APC-eFluor 780 | Thermo Fisher Scientific | Cat# 47–0112; RRID:AB_1603193 |
| CD80-PE-Cy7 | Thermo Fisher Scientific | Cat# 25–0801; RRID:AB_2573369 |
| Gr-1-Alexa Fluor 700 | Thermo Fisher Scientific | Cat# 56–5931; RRID:AB_494007 |
| F4/80-APC | Thermo Fisher Scientific | Cat# 17–4801; RRID:AB_469452 |
| EpCAM-eFluor 450 | Thermo Fisher Scientific | Cat# 48–5791; RRID:AB_10717090 |
| Foxp3-PE | Thermo Fisher Scientific | Cat# 12–5773; RRID:AB_465936 |
| B220-efluor 450 | Thermo Fisher Scientific | Cat# 48–0452; RRID:AB_1548761 |
| SIINFEKL-H2Kb | Thermo Fisher Scientific | Cat# 12–5743; RRID:AB_925774 |
| CD45-BV786 | BD | Cat# 564225; RRID:AB_2716861 |
| IL-12-PE | BD | Cat# 554479; RRID:AB_395420 |
| biotinylated CD11c (MACS) | BD | Cat# 553800; RRID:AB_395059 |
| CD45-APC-efluor 780 | Thermo Fisher Scientific | Cat# 47–0451; RRID:AB_1548790 |
| CD11c-efluor 450 (flow cytometry) | Thermo Fisher Scientific | Cat# 48–0114; RRID:AB_1548665 |
| H-2Kb-Percp-Cy5.5 | BD | Cat# 562831; RRID:AB_2732002 |
| H-2Dd-PE | BD | Cat# 553580; RRID:AB_394938 |
| pS727-STAT3 (IHC, human) | Abcam | Cat# ab30647; RRID:AB_779085 |
| CD8 (IHC, human) | Cell Marque | 108M-96; RRID:AB_1158208 |
| Biological Samples | ||
| Human CRC specimen | TUM-MRI-Tissue Biobank at the Department of Pathology of the Technical University Munich and the Victorian Cancer Biobank, Melbourne. | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Tamoxifen | Sigma | Cat# T5648 |
| Tamoxifen-containing chow | Genobios | Cat# 2847 |
| Azoxymethane | Sigma | Cat# A5486 |
| Diphtheria toxin | Merck | Cat# 322326 |
| Chloroquine | Sigma | Cat# C6628 |
| 3-methyladenine | Sigma | Cat# M9281 |
| Deferoxamine | Sigma | Cat# D9533 |
| E64d | Santa Cruz | Cat# sc-201280 |
| N-Acetyl-L-cysteine | Sigma | Cat# A7250 |
| Collagenase I | Sigma | Cat# C0130 |
| Dispase II | Roche | Cat# 04942078001 |
| Fetal bovine serum | Thermo Fisher Scientific | Cat# 10500056 |
| Phorbol 12-myristate 13-acetate | Sigma | Cat# P8139 |
| Ionomycin | Sigma | Cat# I3909 |
| Lipopolysaccharide | Sigma | Cat# L2630 |
| Golgi Plug | BD | Cat# 555029 |
| Ethidium bromide monoazide | Sigma | Cat# E2028 |
| Live/Dead Fixable Blue Dead Cell Stain Kit | Thermo Fisher Scientific | Cat# L23105 |
| IC-fixation buffer | Thermo Fisher Scientific | Cat# 00–8222 |
| Geneticin | Thermo Fisher Scientific | Cat# 10131027 |
| Blasticidin S | Thermo Fisher Scientific | Cat# R21001 |
| SIINFEKL-pentamer (PE) | ProImmune | Cat# F093–2A-E |
| Hydrogen peroxide | Sigma | Cat# H1009 |
| Anti-biotin microbeads | Miltenyi Biotech | Cat# 130–090-485 |
| CD8a+ T cell isolation kit II | Miltenyi Biotech | Cat# 130–095-236 |
| MACS magnetic separation columns | Miltenyi Biotech | Cat# 130–042-401 |
| Acridine orange | Merck | Cat# 113000 |
| Rotenone | Sigma | Cat# R8875 |
| Antimycin A | Sigma | Cat# A8674 |
| z-LR-AMC | R&D | Cat# ES008 |
| IP-1 | Au-Yeung et al., 2013 | N/A |
| LysoTracker red DND-99 | Thermo Fisher Scientific | Cat# L7528 |
| Oligomycin | Sigma | Cat# O4876 |
| carbonyl cyanide-p- trifluoromethoxyphenylhydrazone (FCCP) | Sigma | Cat# C2920 |
| Potassium cyanide | Sigma | Cat# 60178 |
| 2-deoxyglucose | Sigma | Cat# D8375 |
| 3-bromopyruvate | Sigma | Cat# 16490 |
| MitoTracker deep red | Thermo Fisher Scientific | Cat# M22426 |
| tetramethylrhodamin-methylester (TMRM) | Thermo Fisher Scientific | Cat# I34361 |
| Lipofectamine 2000 | Thermo Fisher Scientific | Cat# 11668019 |
| Viromer green | Lipocalyx | Cat# VG-01LB |
| Dharmafect 4 | Dharmacon/GE Healthcare | Cat# T-2004 |
| Critical Commercial Assays | ||
| M.O.M. detection kit, fluorescein | Vector Labs. | Cat# FMK-2201 |
| IFNg ELISA kit | R&D Systems | Cat# DY485 |
| NAD+/NADH assay kit | Abcam | Cat# ab65348 |
| Glucose assay kit | Sigma | Cat# GAGO20 |
| Cell titer glow | Promega | Cat# G7570 |
| RNeasy Mini Kit | QIAGEN | Cat# 74106 |
| SuperScript II Reverse Transcriptase | Thermo Fisher Scientific | Cat# 18064–071 |
| Fast-Start Universal SYBR Green PCR Master Mix | Roche Diagnostics | Cat# 4913914001 |
| Experimental Models: Cell Lines | ||
| OVA-CMT | This paper | N/A |
| OVA-CT26 | Gamrekelashvili et al., 2007 | N/A |
| wt MEFs | provided by Thomas Decker | N/A |
| Stat3−/− MEFs | provided by Thomas Decker | N/A |
| B16-FLT3L | Mach et al., 2000 | N/A |
| Experimental Models: Organisms/Strains | ||
| mouse: Stat3F/F: B6;129P2-Stat3tm1Aki | Takeda et al., 1998 | RRID:IMSR_APB:2010 |
| mouse: CtnnbloxEx3/wt: Ctnnb1tm1Mmt | Harada et al., 1999 | MGI:1858008 |
| mouse: villin-creERT2: Tg(Vil-cre/ERT2)23Syr | el Marjou et al., 2004 | MGI:3053826 |
| mouse: Ctss−/−: Ctsstm1Hap | Shi et al., 1999 | RRID:MGI:3573791 |
| mouse: CtslF/F: Ctsltm1.1Thre | Tholen et al., 2014 | RRID:MGI:5810299 |
| mouse: Atg7F/F: Atg7tm1.1Tchi | Komatsu et al., 2005 | RRID:MGI:3590136 |
| mouse: ROSAOVA: Gt(ROSA)26Sortm2(OVA/EGFP)Dwir | Sandhu et al., 2011 | MGI:5056498 |
| mouse: OT-I: Tg(TcraTcrb)1100Mjb | The Jackson Laboratories | strain#003831; RRID:MGI:5292730 |
| mouse: Tap1: Tap1tm1Arp | The Jackson Laboratories | strain# 002944; RRID:MGI:3621930 |
| mouse: CD11c-DTR: Tg(Itgax-DTR/GFP)57Lan | The Jackson Laboratories | strain# 002944; MGI:3057163 |
| mouse: Ifng−/−: Ifngtm1Ts | The Jackson Laboratories | strain# 002287; RRID:MGI:4838444 |
| Oligonucleotides | ||
| ON-TARGETplus-smart pool non-target siRNA | Dharmacon/GE Healthcare | Cat# D-001810–10 |
| ON-TARGETplus-smart pool mouse Stat3 siRNA | Dharmacon/GE Healthcare | Cat# L-040794–01 |
| ON-TARGETplus-smart pool mouse Tap1 siRNA | Dharmacon/GE Healthcare | Cat# L-057436–01 |
| ON-TARGETplus-smart pool mouse Pink1 siRNA | Dharmacon/GE Healthcare | Cat# L-044666–00 |
| ON-TARGETplus-smart pool mouse Ctsb siRNA | Dharmacon/GE Healthcare | Cat# L-044712–00 |
| ON-TARGETplus-smart pool mouse Ctsd siRNA | Dharmacon/GE Healthcare | Cat# L-051673–01 |
| ON-TARGETplus-smart pool mouse Ctsf siRNA | Dharmacon/GE Healthcare | Cat# L-040442–01 |
| ON-TARGETplus-smart pool mouse Ctsh siRNA | Dharmacon/GE Healthcare | Cat# L-040410–01 |
| ON-TARGETplus-smart pool mouse Ctsl siRNA | Dharmacon/GE Healthcare | Cat# L-060456–01 |
| ON-TARGETplus-smart pool mouse Ctss siRNA | Dharmacon/GE Healthcare | Cat# L-043220–01 |
| For primer sequences used in PCR experiments, please refer to Table S1. | ||
| Recombinant DNA | ||
| pCI-OVA | this paper | N/A |
| pCI-OVA-IRES-GFP | Gamrekelashvili et al., 2007 | N/A |
| DEC205-OVA | Loschko et al., 2011 | N/A |
| pcDNA6.2-Stat3-miRNA | this paper | N/A |
| pcDNA6.2-non target control miRNA | Thermo Fisher Scientific | Cat# K493600 |
| pTRE-Tight-MitoTimer | Hernandez et al., 2013 | Addgene plasmid # 50547 |
| pES.1–2(M2N)p | Urlinger et al., 2000 | N/A |
| MSCV-Stat3a | Wegrzyn et al., 2009 | N/A |
| MLS-Stat3 | Wegrzyn et al., 2009 | N/A |
| MLS-Stat3-Y705F | Wegrzyn et al., 2009 | N/A |
| MLS-Stat3-Y705F/S727A | Wegrzyn et al., 2009 | N/A |
| MLS-Stat3-E434A/E435A | Wegrzyn et al., 2009 | N/A |
| Software and Algorithms | ||
| Excel for Mac 2016 | Microsoft | https://www.microsoft.com/en-us/; RRID:SCR_016137 |
| Prism 5 | GraphPad | https://www.graphpad.com/; RRID:SCR_002798 |
| AxioVision 4.9.1 | Zeiss | https://www.zeiss.com |
Highlights.
Increased mitophagy in IECs triggers lysosomal membrane permeabilization
Lysosomal membrane permeabilization enhances antigen presentation in IECs
Enhanced antigen presentation in IECs augments MHC class I presentation
Mitophagy and lysosomal integrity in IECs regulate anti-tumor immunity
ACKNOWLEDGMENTS
We thank Kerstin Burmeister, Saskia Ettl, Kathleen Mohs, Hana Kunkel, Christin Danneil, Natalia Delis, and Eva Rudolf for technical assistance, as well as the Histology and Flow Cytometry Core Facilities at the Georg-Speyer-Haus. We are grateful to Johanna Joyce and Masaaki Komatsu for generously providing Ctss−/− and floxed Atg7 mice, respectively, and we thank Chris Chang for generously providing IP-1. The plasmid pTRE-Tight-MitoTimer was a kind gift from Roberta Gottlieb (Addgene plasmid # 50547), DEC205-OVA was a kind gift from Anne Krug, and pES.1–2(M2N)p was kindly provided by Rainer Loew. We gratefully acknowledge the TUM-MRI-Tissue Biobank at the Department of Pathology, Klinikum rechts der Isar, and the Victorian Cancer Biobank for providing colon cancer specimens. We would like to thank Lisa Sevenich, Firouzeh Korangy, Ari Waisman, and Thomas Blankenstein for helpful discussions. This work was supported in part by the LOEWE Center for Cell and Gene Therapy Frankfurt (CGT, III L 4–518/17.004) and institutional funds from the Georg-Speyer-Haus, as well as grants from the Deutsche Forschungsgemeinschaft (DFG) (SFB 850 Project B7 to T.R.; Gr1916/5–1, FOR2438 Gr1916/11–1, SFB 815, 1177, and 1292 to F.R.G.).
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
SUPPLEMENTAL INFORMATION
Supplemental Information includes six figures and one table and can be found with this article online at https://doi.org/10.1016/j.cell.2018.05.028.
REFERENCES
- Accapezzato D, Visco V, Francavilla V, Molette C, Donato T, Paroli M, Mondelli MU, Doria M, Torrisi MR, and Barnaba V (2005). Chloroquine enhances human CD8+ T cell responses against soluble antigens in vivo. J. Exp. Med 202, 817–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akkari L, Gocheva V, Quick ML, Kester JC, Spencer AK, Garfall AL, Bowman RL, and Joyce JA (2016). Combined deletion of cathepsin protease family members reveals compensatory mechanisms in cancer. Genes Dev. 30, 220–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Au-Yeung HY, Chan J, Chantarojsiri T, and Chang CJ (2013). Molecular imaging of labile iron(II) pools in living cells with a turn-on fluorescent probe. J. Am. Chem. Soc 135, 15165–15173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beers C, Burich A, Kleijmeer MJ, Griffith JM, Wong P, and Rudensky AY (2005). Cathepsin S controls MHC class II-mediated antigen presentation by epithelial cells in vivo. J. Immunol 174, 1205–1212. [DOI] [PubMed] [Google Scholar]
- Bhattacharya N, Yuan R, Prestwood TR, Penny HL, DiMaio MA, Reticker-Flynn NE, Krois CR, Kenkel JA, Pham TD, Carmi Y, et al. (2016). Normalizing microbiota-induced retinoic acid deficiency stimulates protective CD8(+) T cell-mediated immunity in colorectal cancer. Immunity 45, 641–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollrath J, and Greten FR (2009). IKK/NF-kappaB and STAT3 pathways: central signalling hubs in inflammation-mediated tumour promotion and metastasis. EMBO Rep. 10, 1314–1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollrath J, Phesse TJ, von Burstin VA,Putoczki T, Bennecke M, Bateman T, Nebelsiek T, Lundgren-May T, Canli O, Schwitalla S, et al. (2009). gp130-mediated Stat3 activation in enterocytes regulates cell survival and cell-cycle progression during colitis-associated tumorigenesis. Cancer Cell 15, 91–102. [DOI] [PubMed] [Google Scholar]
- Boya P, and Kroemer G (2008). Lysosomal membrane permeabilization in cell death. Oncogene 27, 6434–6451. [DOI] [PubMed] [Google Scholar]
- Boya P, Gonzalez-Polo RA, Poncet D, Andreau K, Vieira HL, Roumier T, Perfettini JL, and Kroemer G (2003). Mitochondrial membrane permeabilization is a critical step of lysosome-initiated apoptosis induced by hydroxychloroquine. Oncogene 22, 3927–3936. [DOI] [PubMed] [Google Scholar]
- Cancer Genome Atlas, N.; Cancer Genome Atlas Network (2012). Comprehensive molecular characterization of human colon and rectal cancer. Nature 487, 330–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerovic V, Houston SA, Westlund J, Utriainen L, Davison ES, Scott CL, Bain CC, Joeris T, Agace WW, Kroczek RA, et al. (2015). Lymph-borne CD8α+ dendritic cells are uniquely able to cross-prime CD8+ T cells with antigen acquired from intestinal epithelial cells. Mucosal Immunol. 8, 38–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi SY, Collins CC, Gout PW, and Wang Y (2013). Cancer-generated lactic acid: a regulatory, immunosuppressive metabolite? J. Pathol 230, 350–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz FM, Colbert JD, Merino E, Kriegsman BA, and Rock KL (2017). The biology and underlying mechanisms of cross-presentation of exogenous antigens on MHC-I molecules. Annu. Rev. Immunol 35, 149–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker T, and Kovarik P (2000). Serine phosphorylation of STATs. Oncogene 19, 2628–2637. [DOI] [PubMed] [Google Scholar]
- Demaria M, Giorgi C, Lebiedzinska M, Esposito G, D’Angeli L, Bartoli A, Gough DJ, Turkson J, Levy DE, Watson CJ, et al. (2010). A STAT3-mediated metabolic switch is involved in tumour transformation and STAT3 addiction. Aging (Albany N.Y.) 2, 823–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demaria M, Camporeale A, and Poli V (2014). STAT3 and metabolism: how many ways to use a single molecule? Int. J. Cancer 135, 1997–2003. [DOI] [PubMed] [Google Scholar]
- elMarjou F,Janssen KP,Chang BH,Li M,Hindie V,Chan L,Louvard D, Chambon P, Metzger D, and Robine S (2004). Tissue-specific and inducible Cre-mediated recombination in the gut epithelium. Genesis 39, 186–193. [DOI] [PubMed] [Google Scholar]
- Fang EF, Scheibye-Knudsen M, Brace LE, Kassahun H, SenGupta T, Nilsen H, Mitchell JR, Croteau DL, and Bohr VA (2014). Defective mitophagy in XPA via PARP-1 hyperactivation and NAD(+)/SIRT1 reduction. Cell 157, 882–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridman WH, Pages F, Sautes-Fridman C, and Galon J (2012). The immune contexture in human tumours: impact on clinical outcome. Nat. Rev. Cancer 12, 298–306. [DOI] [PubMed] [Google Scholar]
- Gamrekelashvili J, Krüger C, von Wasielewski R, Hoffmann M, Huster KM, Busch DH, Manns MP, Korangy F, and Greten TF (2007). Necrotic tumor cell death in vivo impairs tumor-specific immune responses. J. Immunol 178, 1573–1580. [DOI] [PubMed] [Google Scholar]
- Gough DJ, Corlett A, Schlessinger K, Wegrzyn J, Larner AC, and Levy DE (2009). Mitochondrial STAT3 supports Ras-dependent oncogenic transformation. Science 324, 1713–1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greten FR, Eckmann L, Greten TF, Park JM, Li ZW, Egan LJ, Kagnoff MF, and Karin M (2004). IKKbeta links inflammation and tumorigenesis in a mouse model of colitis-associated cancer. Cell 118, 285–296. [DOI] [PubMed] [Google Scholar]
- Grivennikov SI, Greten FR, and Karin M (2010). Immunity, inflammation, and cancer. Cell 140, 883–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada N, Tamai Y, Ishikawa T, Sauer B, Takaku K, Oshima M, and Taketo MM (1999). Intestinal polyposis in mice with a dominant stable mutation of the beta-catenin gene. EMBO J. 18, 5931–5942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hari A, Ganguly A, Mu L, Davis SP, Stenner MD, Lam R, Munro F, Namet I, Alghamdi E, Fürstenhaupt T, et al. (2015). Redirecting soluble antigen for MHC class I cross-presentation during phagocytosis. Eur. J. Immunol 45, 383–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez G, Thornton C, Stotland A, Lui D, Sin J, Ramil J, Magee N, Andres A, Quarato G, Carreira RS, et al. (2013). MitoTimer: a novel tool for monitoring mitochondrial turnover. Autophagy 9, 1852–1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung S, Unutmaz D, Wong P, Sano G, De los Santos K, Sparwasser T, Wu S, Vuthoori S, Ko K, Zavala F, et al. (2002). In vivo depletion of CD11c+ dendritic cells abrogates priming of CD8+ T cells by exogenous cell-associated antigens. Immunity 17, 211–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu M, Waguri S, Ueno T, Iwata J, Murata S, Tanida I, Ezaki J, Mizushima N, Ohsumi Y, Uchiyama Y, et al. (2005). Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J. Cell Biol 169, 425–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B, and Kroemer G (2008). Autophagy in the pathogenesis of disease. Cell 132, 27–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loschko J, Heink S, Hackl D, Dudziak D, Reindl W, Korn T, and Krug AB (2011). Antigen targeting to plasmacytoid dendritic cells via Siglec-H inhibits Th cell-dependent autoimmunity. J. Immunol 187, 6346–6356. [DOI] [PubMed] [Google Scholar]
- Mach N, Gillessen S, Wilson SB, Sheehan C, Mihm M, and Dranoff G (2000). Differences in dendritic cells stimulated in vivo by tumors engineered to secrete granulocyte-macrophage colony-stimulating factor or Flt3-ligand. Cancer Res. 60, 3239–3246. [PubMed] [Google Scholar]
- Matheoud D, Sugiura A, Bellemare-Pelletier A, Laplante A, Rondeau C, Chemali M, Fazel A, Bergeron JJ, Trudeau LE, Burelle Y, et al. (2016). Parkinson’s disease-related proteins PINK1 and Parkin repress mitochondrial antigen presentation. Cell 166, 314–327. [DOI] [PubMed] [Google Scholar]
- Meier JA, and Larner AC (2014). Toward a new STATe: the role of STATs in mitochondrial function. Semin. Immunol 26, 20–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Münz C (2016). Autophagy beyond intracellular MHC class II antigen presentation. Trends Immunol. 37, 755–763. [DOI] [PubMed] [Google Scholar]
- Myant KB, Cammareri P, McGhee EJ, Ridgway RA, Huels DJ, Cordero JB, Schwitalla S, Kalna G, Ogg EL, Athineos D, et al. (2013). ROS production and NF-κB activation triggered by RAC1 facilitate WNT-driven intestinal stem cell proliferation and colorectal cancer initiation. Cell Stem Cell 12, 761–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T, Roth W, Wong P, Nelson A, Farr A, Deussing J, Villadangos JA, Ploegh H, Peters C, and Rudensky AY (1998). Cathepsin L: critical role in Ii degradation and CD4 T cell selection in the thymus. Science 280, 450–453. [DOI] [PubMed] [Google Scholar]
- Nakayama M (2015). Antigen presentation by MHC-dressed cells. Front. Immunol 5, 672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rock KL, Farfán-Arribas DJ, and Shen L (2010). Proteases in MHC class I presentation and cross-presentation. J. Immunol 184, 9–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandhu U, Cebula M, Behme S, Riemer P, Wodarczyk C, Metzger D, Reimann J, Schirmbeck R, Hauser H, and Wirth D (2011). Strict control of transgene expression in a mouse model for sensitive biological applications based on RMCE compatible ES cells. Nucleic Acids Res. 39, e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwitalla S, Fingerle AA, Cammareri P, Nebelsiek T, Göktuna SI, Ziegler PK, Canli O, Heijmans J, Huels DJ, Moreaux G, et al. (2013). Intestinal tumorigenesis initiated by dedifferentiation and acquisition of stem-cell-like properties. Cell 152, 25–38. [DOI] [PubMed] [Google Scholar]
- Shen L, Sigal LJ, Boes M, and Rock KL (2004). Important role of cathepsin S in generating peptides for TAP-independent MHC class I cross-presentation in vivo. Immunity 21, 155–165. [DOI] [PubMed] [Google Scholar]
- Shi GP, Villadangos JA, Dranoff G, Small C, Gu L, Haley KJ, Riese R, Ploegh HL, and Chapman HA (1999). Cathepsin S required for normal MHC class II peptide loading and germinal center development. Immunity 10, 197–206. [DOI] [PubMed] [Google Scholar]
- Siegel RL, Miller KD, and Jemal A (2016). Cancer statistics, 2016. CA Cancer J. Clin 66, 7–30. [DOI] [PubMed] [Google Scholar]
- Takeda K, Kaisho T, Yoshida N, Takeda J, Kishimoto T, and Akira S (1998). Stat3 activation is responsible for IL-6-dependent T cell proliferation through preventing apoptosis: generation and characterization of T cell-specific Stat3-deficient mice. J. Immunol 161, 4652–4660. [PubMed] [Google Scholar]
- Terman A, Kurz T, Gustafsson B, and Brunk UT (2006). Lysosomal labilization. IUBMB Life 58, 531–539. [DOI] [PubMed] [Google Scholar]
- Terman A, Kurz T, Navratil M, Arriaga EA, and Brunk UT (2010). Mitochondrial turnover and aging of long-lived postmitotic cells: the mitochondrial-lysosomal axis theory of aging. Antioxid. Redox Signal 12, 503–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tholen M, Hillebrand LE, Tholen S, Sedelmeier O, Arnold SJ, and Reinheckel T (2014). Out-of-frame start codons prevent translation of truncated nucleo-cytosolic cathepsin L in vivo. Nat. Commun 5, 4931. [DOI] [PubMed] [Google Scholar]
- Urlinger S, Baron U, Thellmann M, Hasan MT, Bujard H, and Hillen W (2000). Exploring the sequence space for tetracycline-dependent transcriptional activators: novel mutations yield expanded range and sensitivity. Proc. Natl. Acad. Sci. USA 97, 7963–7968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Niu G, Kortylewski M, Burdelya L, Shain K, Zhang S, Bhattacharya R, Gabrilovich D, Heller R, Coppola D, et al. (2004). Regulation of the innate and adaptive immune responses by Stat-3 signaling in tumor cells. Nat. Med 10, 48–54. [DOI] [PubMed] [Google Scholar]
- Warburg O, Poesner K, and Negelein E (1924). On the metabolism of carcinoma cells. Biochem. Z 9, 309–344. [Google Scholar]
- Wegrzyn J, Potla R, Chwae YJ, Sepuri NB, Zhang Q, Koeck T, Derecka M, Szczepanek K, Szelag M, Gornicka A, et al. (2009). Function of mitochondrial Stat3 in cellular respiration. Science 323, 793–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winterbourn CC (1995). Toxicity of iron and hydrogen peroxide: the Fenton reaction. Toxicol. Lett. 82-83, 969–974. [DOI] [PubMed] [Google Scholar]
- Youle RJ, and Narendra DP (2011). Mechanisms of mitophagy. Nat. Rev. Mol. Cell Biol 12, 9–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Lee H, Herrmann A, Buettner R, and Jove R (2014). Revisiting STAT3 signalling in cancer: new and unexpected biological functions. Nat. Rev. Cancer 14, 736–746. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.