The transcription factor SlMYC1 regulates mono- and sesquiterpene biosynthesis differentially in leaf and stem trichomes and is involved in the formation of type VI glandular trichomes in tomato.
Abstract
Tomato (Solanum lycopersicum) glandular trichomes function as biochemical factories that synthesize a diverse array of specialized metabolites. Terpenoids are the most diverse class of plant specialized metabolites, with volatile mono- and sesquiterpenes playing important roles in plant defense. Although the biosynthetic pathways of volatile terpenes in tomato glandular trichomes have been well described, little is known about their regulation. Here, we demonstrate that SlMYC1, a basic helix-loop-helix transcription factor, differentially regulates mono- and sesquiterpene biosynthesis in the type VI glandular trichomes of tomato leaves and stems. SlMYC1 functions as a positive regulator of monoterpene biosynthesis in both leaf and stem trichomes but as a negative regulator of sesquiterpene biosynthesis in stem trichomes. SlMYC1 is also essential for type VI glandular trichome development, as knocking down SlMYC1 led to the production of smaller type VI glandular trichomes at lower densities, and knocking out this gene led to their absence. Our findings reveal a role for SlMYC1 not only in type VI glandular trichome development but also in the regulation of terpene biosynthesis in tomato.
INTRODUCTION
Trichomes are epidermal outgrowths on the stem and leaf surfaces of plants that serve as physical barriers to herbivorous arthropods. These structures also function as biochemical factories, as they synthesize, store, and secrete a wide range of specialized metabolites, such as terpenoids, phenylpropanoids, flavonoids, alkaloids, and acyl sugars (Schilmiller et al., 2008). Cultivated tomato (Solanum lycopersicum) and its wild relatives have eight types of trichomes, including both glandular (types I, IV, VI, and VII) and non-glandular (types II, III, V, and VIII) trichomes (Glas et al., 2012). However, our knowledge of the development of glandular trichomes in tomato is very limited. The Woolly (Wo) gene, encoding a homeodomain-Leu zipper (HD-ZIP) protein and the Hair gene, encoding a Cys2-His2 zinc finger protein that interacts with the Wo gene product, are essential for type I trichome development together with the CyclinB2 gene (Yang et al., 2011; Gao et al., 2017; Chang et al., 2018). The hairless gene from tomato, encoding a subunit of a regulatory complex that controls the nucleation of actin filaments, was recently mapped and cloned, indicating that actin plays an important role in the proper development of several types of trichomes (Kang et al., 2010a, 2016). The dialytic and odorless-2 loci, which affect trichome development, await further characterization (Kang et al., 2010b; Chang et al., 2016). Ectopic expression of SlMIXTA1, encoding a MYB transcription factor in tomato, led to higher numbers of glandular and non-glandular trichomes (Ewas et al., 2016, 2017). It has become clear that jasmonic acid (JA) also plays an important role in regulating the development of type VI glandular trichomes, as JA-deficient plants have reduced numbers of these trichomes (Yan et al., 2013; Bosch et al., 2014), and there is evidence that the JA-Ile receptor plays a role in this process as well (Li et al., 2004).
Type I and IV glandular trichomes are sources of acyl sugars (McDowell et al., 2011). Type IV trichomes are ubiquitous in some wild tomato relatives (Simmons and Gurr, 2005), but in S. lycopersicum, they are present only on embryonic and juvenile leaves (Vendemiatti et al., 2017). Type VI trichomes can produce terpenoids, flavonoids, and methyl ketones (Williams et al., 1980; Fridman et al., 2005; Besser et al., 2009; Schilmiller et al., 2010b; Schmidt et al., 2011; Kang et al., 2014). In the wild tomato Solanum habrochaites, type VI glands can produce large amounts of volatile mono- and sesquiterpenes, whereas type VI trichomes in cultivated tomato mainly accumulate monoterpenes (Besser et al., 2009; Sallaud et al., 2009; Schilmiller et al., 2009; Bleeker et al., 2011; Falara et al., 2011). The glandular heads of type VI trichome in these two species have distinct features (Bergau et al., 2015).
Volatile terpenes, representing the largest and most diverse class of plant volatile metabolites, contribute to plant defense responses, as they are repellent or toxic to herbivores or attract predators or parasitoids of the attacking insects (Gershenzon and Dudareva, 2007; Tholl, 2015). In S. lycopersicum, 45 terpene synthases (TPSs) have been identified, 30 of which appear to be functional (Falara et al., 2011, 2014). Transcripts for 14 of these genes have been detected in stem trichomes (Spyropoulou et al., 2014a), and some are induced by JA (van Schie et al., 2007; Bleeker et al., 2011; Falara et al., 2011). SlTPS5 and SlTPS9 are specifically expressed in the glandular heads of type VI trichomes (Spyropoulou et al., 2014a; Kortbeek et al., 2016).
In S. lycopersicum, only three transcription factors that are potentially involved in regulating volatile terpene biosynthesis in trichomes, SlEOT1 (Expression of Terpenoids 1), SlWRKY73, and SlMYC1, have thus far been identified (Spyropoulou et al., 2014a, 2014b). SlEOT1 and SlWRKY73 transactivate the SlTPS5 promoter in Nicotiana benthamiana leaves, and SlMYC1 transactivates the promoters of several TPS genes (Spyropoulou et al., 2014a, 2014b). SlMYC1 is a basic helix-loop-helix (bHLH) transcription factor. A number of bHLHs from several species have been shown to act as positive regulators of terpenoid biosynthesis: in Catharanthus roseus (Madagascar periwinkle) for monoterpenoid indole alkaloid biosynthesis (Zhang et al., 2011; Van Moerkercke et al., 2015, 2016); in Medicago truncatula (barrelclover) for triterpene saponin biosynthesis (Mertens et al., 2016); in Arabidopsis (Arabidopsis thaliana) for sesquiterpene synthase gene expression (Hong et al., 2012); in Artemisia annua (sweet wormwood) for artemisinin biosynthesis (Ji et al., 2014; Shen et al., 2016); in Oryza sativa (rice) for diterpenoid phytoalexin biosynthetic gene expression (Yamamura et al., 2015); in Salvia miltiorrhiza (red sage) for tanshinone biosynthesis (Zhou et al., 2016); and in Aquilaria sinensis (incense tree) for agarwood sesquiterpene biosynthesis (Xu et al., 2017). Two bHLHs from Betula platyphylla Suk (Japanese white birch) were recently shown to be involved in regulating triterpenoid biosynthesis (Yin et al., 2017). Interestingly, some bHLH transcription factors appear to act as repressors of terpenoid biosynthesis, including the biosynthesis of the diterpene paclitaxel in Taxus cuspidata (Japanese yew; Lenka et al., 2015) and the monoterpenoid indole alkaloid biosynthetic pathway in C. roseus (Patra et al., 2018).
The objective of this study was to explore the roles of SlMYC1 in the regulation of volatile terpene biosynthesis in tomato trichomes and trichome development via gene knockdown, knockout, and overexpression. Our results shed light on the roles of SlMYC1 in these two very different processes.
RESULTS
SlMYC1 Knockdown Affects the Expression of Mono- and Sesquiterpene Synthase Genes
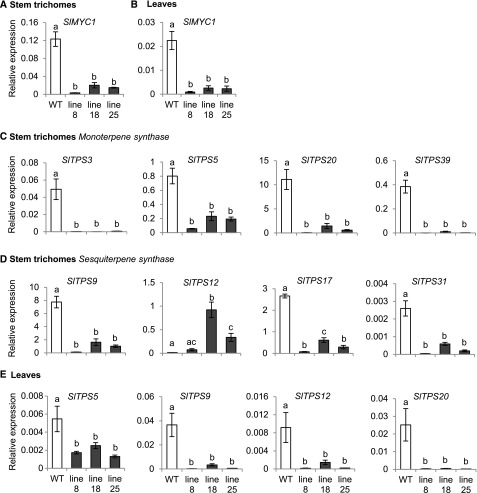
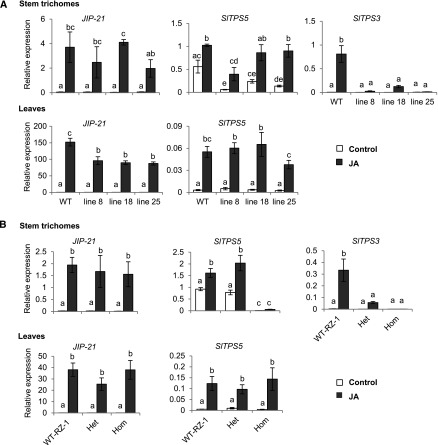
To investigate whether SlMYC1 participates in the regulation of volatile terpene biosynthesis, we used an RNA interference strategy to downregulate SlMYC1 expression in transgenic tomato plants. An SlMYC1-inverted-repeat (ir) construct driven by the Cauliflower mosaic virus 35S promoter was introduced into S. lycopersicum cultivar (cv) Moneymaker. We obtained many independent transgenic lines and selected three lines (ir-SlMYC1 lines 8, 18, and 25) for further analysis. All three lines had significantly reduced SlMYC1 transcript levels (by 80 to 95%) in stem trichomes and leaves compared with wild type (Figures 1A and 1B; Supplemental Data Set). Additionally, the transcript levels of monoterpene synthase genes SlTPS3, SlTPS5, SlTPS20, and SlTPS39 and sesquiterpene synthase genes SlTPS9, SlTPS17, and SlTPS31 were reduced in stem trichomes (Figures 1C and 1D). By contrast, SlTPS12, encoding a β-caryophyllene/α-humulene sesquiterpene synthase, had significantly higher transcript levels in stem trichomes of ir-SlMYC1 lines 18 and 25 compared with wild type (Figure 1D). In leaves, the downregulation of SlMYC1 led to significantly reduced transcript levels of SlTPS12 and other TPS genes (Figure 1E).
Figure 1.
Effect of SlMYC1 Downregulation on the Expression of Terpene Synthase Genes in Stem Trichomes and Leaves.
Relative transcript levels of SlMYC1 in (A) stem trichomes and (B) leaves from wild-type (WT) Moneymaker and ir-SlMYC1 plants. Relative transcript levels of (C) several monoterpene and (D) sesquiterpene synthase genes in stem trichomes from wild-type (WT) Moneymaker and ir-SlMYC1 plants. (E) Relative transcript levels of terpene synthase genes in leaves from wild-type (WT) Moneymaker and ir-SlMYC1 plants. Lines 8, 18, and 25 are three independent transgenic ir-SlMYC1 lines. All transcript levels were determined by qRT-PCR and normalized to Actin transcript levels. Bars represent the mean values (±se) of three to four biological replicates, each consisting of multiple stem pieces or leaflets from different plants. Bars annotated with different letters were significantly different according to Fisher’s LSD test (P ≤ 0.05) after ANOVA, see Supplemental Data Set for details (Brady et al., 2015).
SlMYC1 Knockdown Differentially Affects Mono- and Sesquiterpene Levels in Stem and Leaf Trichomes
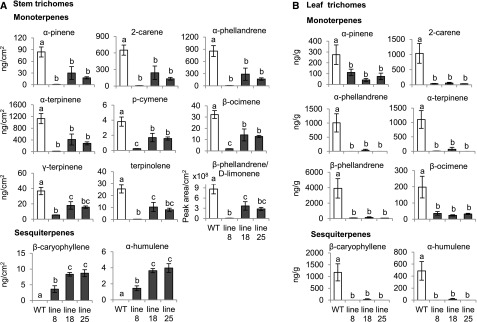
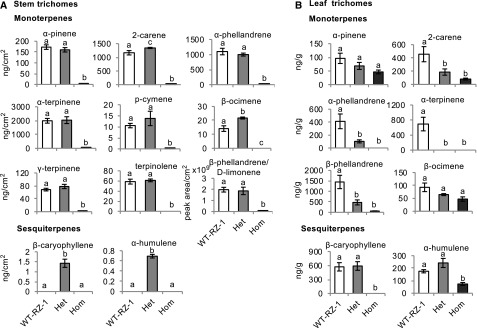
Quantification of the volatile terpenes in stem trichomes using gas chromatography-mass spectrometry revealed that the levels of ten monoterpenes identified as α-pinene, 2-carene, α-phellandrene, α-terpinene, p-cymene, β-ocimene, γ-terpinene, terpinolene, β-phellandrene, and D-limonene were significantly reduced in the ir-SlMYC1 lines (Figure 2A). The levels of all of these monoterpenes were lowest in line 8, which had the lowest SlMYC1 transcript level. Volatile sesquiterpenes were not detected in wild-type Moneymaker stem trichomes. However, considerable amounts of β-caryophyllene and α-humulene were present in the stem trichomes of the ir-SlMYC1 lines (Figure 2A), which is consistent with the elevated SlTPS12 transcript levels (Figure 1D). β-caryophyllene and α-humulene are normally produced in leaf trichomes rather than stem trichomes in cultivated tomato plants (Schilmiller et al., 2010a). In leaf trichomes, the ir-SlMYC1 plants produced significantly less β-caryophyllene and α-humulene than the wild type, and the levels of all monoterpenes were also reduced (Figure 2B). These results indicate that SlMYC1 plays different roles in the regulation of β-caryophyllene and α-humulene biosynthesis in tomato leaf and stem trichomes.
Figure 2.
Effect of SlMYC1 Downregulation on Volatile Terpene Levels in Stem and Leaf Trichomes of Tomato.
Levels of volatile terpenes in (A) stem and (B) leaf trichomes from wild-type (WT) and ir-SlMYC1 line 8, 18, and 25 plants, as quantified by gas chromatography-mass spectrometry (GC-MS). The terpene levels in stem trichomes were normalized by stem surface area and in leaf trichomes by leaf fresh weight. Bars represent the mean values (±se) of three to four biological replicates, each consisting of multiple stem pieces or leaflets from different plants. Bars annotated with different letters were significantly different according to Fisher’s LSD test (P ≤ 0.05) after ANOVA.
SlMYC1 Is Involved in Type VI Glandular Trichome Development
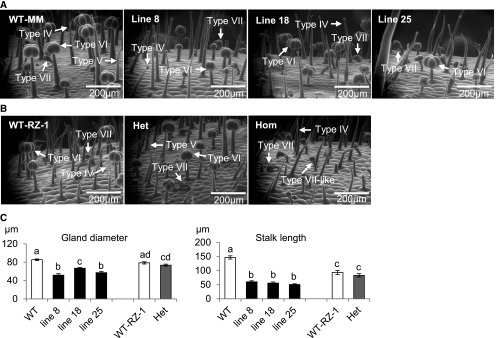
Type VI trichomes contain four glandular cells on top of an intermediate cell and a stalk, which form the head, where metabolites are stored in a cavity under a waxy cuticle (McDowell et al., 2011; Bergau et al., 2015). Since SlMYC1 regulates terpene biosynthesis in tomato trichomes, we also examined type VI glandular trichome morphology by environmental scanning electron microscopy (ESEM). As shown in Figure 3A, type VI glandular trichomes on the stems of 4-week-old ir-SlMYC1 plants had shorter stalks and smaller glandular heads compared with wild-type Moneymaker (WT-MM). The gland diameters and stalk lengths of type VI trichomes on ir-SlMYC1 stems were 60 to 80% and 30 to 45% of those on wild-type stems, respectively (Figure 3C). The glandular cells of type VI trichomes in ir-SlMYC1 plants occasionally appeared to have undergone an irregular cell division, leading to the formation of five cells (Supplemental Figure 1B). The morphology of type IV, V, and VII trichomes was unaltered in the ir-SlMYC1 plants (Figure 3A).
Figure 3.
Morphology and Size of Type VI Glandular Trichomes on Stems.
(A) Stem surfaces of wild-type Moneymaker (WT-MM) and ir-SlMYC1 line 8, 18, and 25 plants.
(B) Stem surfaces of wild-type RZ (WT-RZ-1), heterozygous MYC1/myc1 (Het), and homozygous myc1 (Hom) plants. Arrows indicate different types of trichomes.
(C) Gland diameter and stalk length of type VI trichomes on the stem surfaces of wild-type (WT) Moneymaker and ir-SlMYC1 line 8, 18, and 25 plants and wild-type RZ (WT-RZ-1) and heterozygous MYC1/myc1 (Het) plants. All plants were 4 weeks old. The bars represent the mean values (±se), calculated from three to four scanning-electron micrographs of stems from different plants. Bars annotated with different letters were significantly different according to Fisher’s LSD test (P ≤ 0.05) after ANOVA.
To further explore the role of SlMYC1 in the formation of type VI glandular trichomes, we identified a myc1 mutant from an ethyl methane sulfonate (EMS)-treated S. lycopersicum population. The myc1 mutant has a single nucleotide change of G to T at position 1477 in the coding sequence of SlMYC1, resulting in an early stop codon at amino acid position 493. This truncated SlMYC1 was unable to activate the promoter of SlTPS5 in N. benthamiana leaves (Supplemental Figure 2), indicating that this mutant allele does not provide an active form of SlMYC1. Therefore, we examined the trichomes in homozygous myc1 and heterozygous MYC1/myc1 plants. Figure 3B shows that type VI glandular trichomes were absent from the stems of homozygous myc1 plants. By contrast, the type VI trichomes on the stem surfaces of heterozygous MYC1/myc1 plants were similar to those of wild-type plants (Figures 3B and 3C). We also occasionally observed some irregular divisions of type VI glandular cells in MYC1/myc1 plants (Supplemental Figure 1B) similar to those observed in ir-SlMYC1 plants. The type IV, V, and VII trichomes of MYC1/myc1 and myc1 plants were similar to those of wild-type plants (Figure 3B). We also observed some type VII-like glandular trichomes in addition to normal type VII glandular trichomes in homozygous myc1 plants (Figure 3B). These peculiar type VII-like trichomes contained a single stalk cell with a wide intermediate cell and a berry-shaped glandular head composed of multiple cells, which is similar to the morphology of type VII glandular trichomes. However, the unusual trichomes stood straight up, unlike type VII trichomes, which bent downward at an angle of ∼45° (Supplemental Figure 1A). Due to the presence of a wide intermediate cell, we designated these trichomes type VII-like trichomes, but theoretically, they could also have been malformed type VI trichomes. To further confirm the crucial role of SlMYC1 in type VI glandular trichome formation, we constructed SlMYC1 knockout lines via CRISPR-Cas9 genome editing (Barrangou et al., 2007; Mali et al., 2013). Two independent transgenic lines with different nucleotide deletions in SlMYC1 (Supplemental Figure 3) lacked type VI glandular trichomes on the stem surface (Supplemental Figure 4A). Like homozygous myc1 plants, these two CRISPR-Cas9_myc1 lines also had type VII-like glandular trichomes (Supplemental Figure 4B).
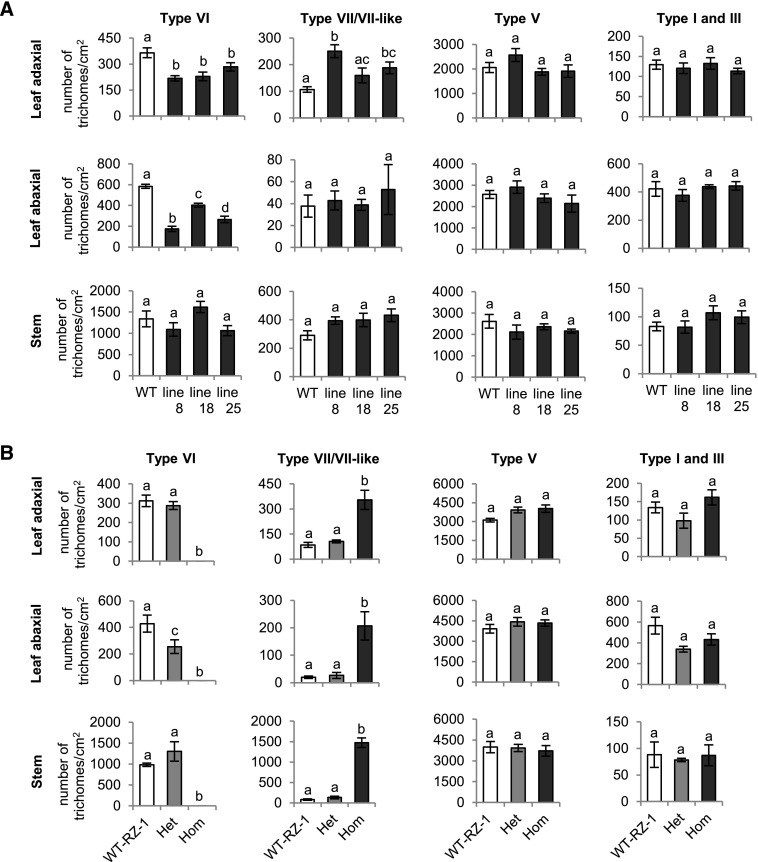
The effect of reduced SlMYC1 expression on type VI trichome morphology prompted us to ask whether trichome density was also affected in these plants. In 7-week-old ir-SlMYC1 plants, the densities of type VI glandular trichomes were lower on both the adaxial and abaxial leaf surfaces but were unchanged on stems compared with wild type (Figure 4A). The type VI trichome densities on the adaxial leaf surface and stem were similar in MYC1/myc1 and wild-type plants but were reduced on the abaxial leaf surface in MYC1/myc1 plants (Figure 4B). The densities of type I, III, and V trichomes on leaves and stems were not affected in any of the lines (Figures 4A and 4B). We also counted type VII-like trichomes together with type VII trichomes, as it was difficult to distinguish between the two. The myc1 mutant had higher densities of type VII/VII-like trichomes than wild type (Figure 4B), as did ir-SlMYC1 line 8 and 25 on the adaxial leaf surface (Figure 4A). In these 7-week-old plants, type IV glandular trichomes were absent from the leaf surfaces and occasionally present on stems. However, these trichomes were present on the stems of 4-week-old plants and, interestingly, the densities of these type IV trichomes were higher in ir-SlMYC1 line 8 and 25 than in wild-type plants (Supplemental Figure 5). These results indicate that the downregulation or absence of SlMYC1 led to an increase in the numbers of other trichomes.
Figure 4.
Trichome Density on the Leaf and Stem Surfaces of Tomato.
Trichome density on the adaxial and abaxial leaf and stem surfaces of (A) wild-type (WT) Moneymaker and ir-SlMYC1 line 8, 18, and 25 plants and (B) wild-type RZ (WT-RZ-1), heterozygous MYC1/myc1 (Het) and homozygous myc1 (Hom) plants. All plants were 7 weeks old. Bars represent the mean values (±se) of three to four leaf or stem samples from different plants. Bars annotated with different letters were significantly different according to Fisher’s LSD test (P ≤ 0.05) after ANOVA.
Regulation of Terpene Biosynthesis by SlMYC1 Occurs in Type VI Glandular Trichomes
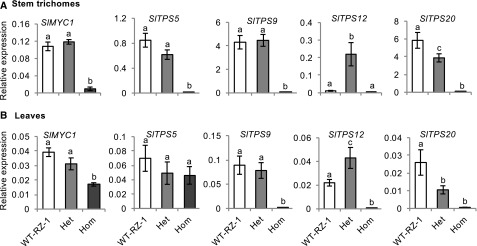
In cultivated tomato plants, volatile mono- and sesquiterpenes are mainly synthesized by the glandular cells of type VI trichomes (Schilmiller et al., 2009; Kang et al., 2010a; Widhalm et al., 2015). As predicted, homozygous myc1 plants produced very few monoterpenes and sesquiterpenes in the remaining stem trichomes. The heterozygous MYC1/myc1 plants contained similar amounts of most monoterpenes to wild-type plants (Figure 5A) but interestingly, the levels of β-caryophyllene and α-humulene were higher in these plants than in wild type (Figure 5A), which is similar to what we observed in ir-SlMYC1 plants (Figure 2A). The terpene levels coincided well with the transcript levels of TPS genes. In stem trichomes, MYC1/myc1 plants had significantly higher transcript levels of β-caryophyllene and α-humulene synthase (SlTPS12) genes than wild type, whereas the myc1 plants had extremely low transcript levels of all TPS genes (Figure 6A). The leaf trichomes of the myc1 plants had lower levels of most terpenes (Figure 5B), which is consistent with the low expression levels of TPS genes (Figure 6B). SlTPS5 was still expressed in myc1 leaf trichomes but not stem trichomes, and these leaf trichomes produced tiny amounts of terpenes. MYC1/myc1 plants had reduced levels of monoterpenes and similar levels of sesquiterpenes in their leaf trichomes compared with wild-type plants (Figure 5B), although SlTPS12 expression levels were higher (Figure 6B).
Figure 5.
Volatile Terpene Levels in Trichomes of Hetero- and Homozygous myc1 Mutants.
Volatile terpene levels in (A) stem and (B) leaf trichomes from wild-type RZ (WT-RZ-1), heterozygous MYC1/myc1 (Het), and homozygous myc1 (Hom) plants, as quantified by gas chromatography-mass spectrometry (GC-MS). Terpene levels in stem trichomes were normalized by stem surface area and in leaf trichomes by leaf fresh weight. Bars represent the mean values (±se) of three to four biological replicates, each consisting of multiple stem pieces or leaflets from different plants. Bars annotated with different letters were significantly different according to Fisher’s LSD test (P ≤ 0.05) after ANOVA.
Figure 6.
Expression of Terpene Synthase Genes in Hetero- and Homozygous myc1 Mutants.
Relative transcript levels in (A) stem trichomes and (B) leaves from wild-type RZ (WT-RZ-1), heterozygous MYC1/myc1 (Het), and homozygous myc1 (Hom) plants. All transcript levels were determined by qRT-PCR and normalized to Actin transcript levels. Bars represent the mean values (±se) of three to four biological replicates, each consisting of multiple stem pieces or leaflets from different plants. Bars annotated with different letters were significantly different according to Fisher’s LSD test (P ≤ 0.05) after ANOVA.
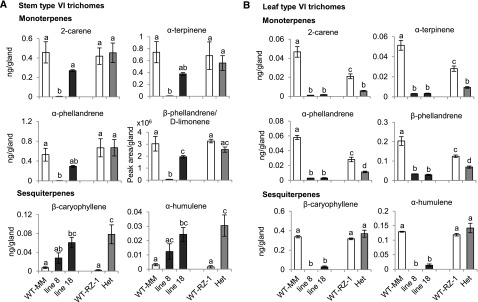
To further explore the role of SlMYC1 in regulating terpene levels, we examined the terpene composition of isolated type VI glandular cells. For each type of analysis, we collected 200 individual type VI glands from the stems or leaves of each plant with a stretched glass pipette. We could not collect type VI trichomes from homozygous myc1 plants, as these were absent. As shown in Figure 7A, the monoterpene levels were the most strongly reduced in line 8, in which the expression level of SlMYC1 was lowest (Figure 1A). The MYC1/myc1 plants had the same monoterpene levels as wild-type plants in type VI stem trichomes (Figure 7A). Conversely, the MYC1/myc1 and ir-SlMYC1 plants exhibited elevated levels of β-caryophyllene and α-humulene in these trichomes (Figure 7A). Among type VI leaf trichomes, monoterpene levels were reduced in ir-SlMYC1 and MYC1/myc1 plants compared with wild type (Figure 7B). Also, β-caryophyllene and α-humulene levels were reduced in type VI trichomes on the leaves of ir-SlMYC1 plants but were similar to wild type in MYC1/myc1 (Figure 7B). These results indicate that SlMYC1 differentially regulates of mono- and sesquiterpene biosynthesis in type VI glandular trichomes.
Figure 7.
Effect of SlMYC1 Downregulation on Volatile Terpene Levels in Type VI Glandular Trichomes.
Volatile terpene levels in (A) stem and (B) leaf type VI glandular trichomes from wild-type Moneymaker (WT-MM) and ir-SlMYC1 line 8 and 18 plants and wild-type RZ (WT-RZ-1) and heterozygous MYC1/myc1 (Het) plants. For each sample, 200 type VI glandular trichomes were collected with a glass capillary to quantify volatile terpene levels by gas chromatography-mass spectrometry (GC-MS). Bars represent the mean values (±se) of three to four biological replicates, each consisting of 200 trichomes collected from multiple leaves from different plants. Bars annotated with different letters were significantly different according to Fisher’s LSD test (P ≤ 0.05) after ANOVA.
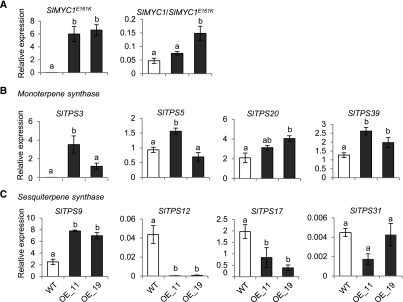
SlMYC1E161K Expression Leads to the Downregulation of SlTPS12 in Stem Trichomes
The Arabidopsis myc2-322B mutant is a gain-of-function AtMYC2 mutant in which a glutamate to lysine (E165K) substitution in the transcriptional activation domain causes enhanced transcriptional activity and the partial release of AtMYC2 from repression by jasmonate ZIM-domain proteins (Gasperini et al., 2015). We investigated whether the corresponding E161K mutation in SlMYC1 would affect the expression of TPS genes. SlMYC1E161K driven by the glandular trichome-specific SlTPS5 promoter (Spyropoulou et al., 2014b) was transformed into S. lycopersicum cv Moneymaker. Accordingly, SlMYC1E161K was expressed in the stem trichomes of two independent SlTPS5p:SlMYC1E161K lines (Figure 8A). The total SlMYC1 transcript levels (SlMYC1/SlMYC1E161K) differed between the two lines (Figure 8A). SlMYC1E161K expression led to the upregulation of most monoterpene synthase genes (Figure 8B). SlMYC1E161K expression had several effects on sesquiterpene synthase genes: (1) SlTPS12 expression was almost abolished in SlTPS5p:SlMYC1E161K plants; (2) SlTPS17 expression was reduced; (3) SlTPS31 expression did not change; and (4) SlTPS9 expression increased (Figure 8C). These experiments using a “stabilized” SlMYC1E161K help confirm the finding that SlMYC1 is involved in regulating the expression of TPS genes. Additionally, we examined type VI trichome morphology in SlTPS5p:SlMYC1E161K plants. The type VI stem trichomes in these plants were similar to those of wild-type plants (Supplemental Figures 4A and 4B).
Figure 8.
Transcript Levels of SlMYC1 and Terpene Synthase Genes in the Stem Trichomes of SlTPS5p:SlMYC1E161K Plants.
Relative transcript levels of (A) SlMYC1E161K and SlMYC1 (SlMYC1/SlMYC1E161K) and (B) several monoterpene and (C) sesquiterpene synthase genes in stem trichomes from wild-type (WT) Moneymaker and SlTPS5p:SlMYC1E161K plants determined by qRT-PCR. Line OE_11 and OE_19 are two independent transgenic SlTPS5p:SlMYC1E161K lines of the T1 generation. Bars represent the mean values (±se) of three to four biological replicates, each consisting of multiple stem pieces from different plants normalized to Actin transcript levels. Bars annotated with different letters were significantly different according to Fisher’s LSD test (P ≤ 0.05) after ANOVA.
JA Application Fails to Restore the Phenotype of Type VI Trichomes in SlMYC1 Knockdown Plants
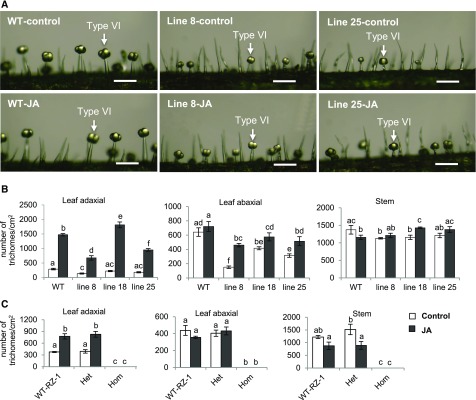
Consecutive applications of methyl jasmonate increase the density of type VI glandular trichomes on newly formed tomato leaves (Boughton et al., 2005). Therefore, we treated ir-SlMYC1 and myc1 plants with JA to determine whether type VI trichome morphology could be restored. After multiple applications of JA, the gland size and stalk length of type VI trichomes on newly expanded leaves were similar in untreated versus treated plants. As shown in Figure 9, the stalks of type VI trichomes on newly developed stems were shorter in both untreated and JA-treated ir-SlMYC1 plants compared with wild type. Therefore, consecutive treatments with JA failed to restore the changes in type VI trichome morphology caused by the reduced expression of SlMYC1.
Figure 9.
Type VI Glandular Trichome Morphology and Density after JA Treatment.
(A) Morphology of type VI trichomes on the stem surfaces of control and JA-sprayed wild-type (WT) Moneymaker, and ir-SlMYC1 line 8 and 25 plants (scale bars, 200 μm), 28 days after JA treatment.
(B) and (C) Type VI trichome density in 7-week-old control plants and plants sprayed with jasmonic acid (JA) on newly formed stems and leaves 28 days after treatment; (B) wild-type Moneymaker (WT) and ir-SlMYC1 line 8, 18, and 25 plants; (C) wild-type RZ (WT-RZ-1), heterozygous MYC1/myc1 (Het), and homozygous myc1 (Hom) plants. Bars annotated with different letters were significantly different according to Fisher’s LSD test (P ≤ 0.05) after ANOVA. Mean values (±se) of three to four biological replicates are shown.
We also investigated type VI trichome density after exogenous JA application. JA treatment did not rescue the loss of type VI trichomes in homozygous myc1 plants (Figure 9C). Treatment with JA resulted in an increase in type VI trichome density on the adaxial leaf surface in wild type, ir-SlMYC1, and MYC1/myc1 plants (Figures 9B and 9C). The increase was significantly higher in wild-type plants and ir-SlMYC1 line 18 than in the two other ir-SlMYC1 lines (Figure 9B). The increases in type VI trichome density on the adaxial leaf surfaces of JA-treated MYC1/myc1 and wild-type plants were similar (Figure 9C). On the abaxial leaf surface, however, JA treatment led to significantly increased type VI trichome density only in ir-SlMYC1 plants (Figures 9B and 9C). These results indicate that the reduced expression of SlMYC1 did not impair the induction of type VI trichome formation by JA. Interestingly, JA application led to a reduction in trichome density on the stems of wild type (Figure 9B) and MYC1/myc1 plants (Figure 9C).
SlMYC1 Is Required for the Induction of SlTPS3 by JA
JA induces the expression of several TPS genes in S. lycopersicum (Falara et al., 2011). For instance, SlTPS3 and SlTPS5 are induced by JA treatment (Falara et al., 2011), and both of their promoters can be transactivated by SlMYC1 (Spyropoulou et al., 2014a). Thus, we investigated whether SlMYC1 also controls the JA-induced expression of TPS genes. After 24 h of JA treatment, the JA-responsive marker gene Jasmonate-inducible protein-21 was significantly induced by JA treatment in the stem trichomes and leaves of all genotypes (Figures 10A and 10B). In stem trichomes of the myc1 mutant, SlTPS5 and SlTPS3 were not induced by JA (Figures 10A and 10B), but SlTPS5 was induced in ir-SlMYC1 and heterozygous MYC1/myc1 plants, in contrast to SlTPS3 (Figures 10A and 10B). SlTPS3 was not expressed in leaves, but interestingly, JA still induced SlTPS5 expression in the leaves of all genotypes (Figures 10A and 10B). SlMYC1 itself was not induced by JA in stem trichomes (Supplemental Figure 6A), as was shown previously (Spyropoulou et al., 2014a), but it was induced by JA in leaves, albeit not in ir-SlMYC1 plants (Supplemental Figure 6B). These results demonstrate that SlMYC1 is essential for the induction of SlTPS3, but not of SlTPS5, in response to JA.
Figure 10.
Effect of SlMYC1 Downregulation on JA-Induced Expression of Terpene Synthase Genes.
Relative transcript levels of Jasmonate-inducible protein-21 (JIP-21) and terpene synthase genes in stem trichomes and leaves of (A) wild-type (WT) Moneymaker and ir-SlMYC1 line 8, 18, and 25 and (B) wild-type RZ (WT-RZ-1), heterozygous MYC1/myc1 (Het), and homozygous myc1 (Hom) plants. The plants were sprayed with JA solution (1 mM JA and 0.05% SilwetL-77 in tap water) or control solution (0.05% SilwetL-77 in tap water) and samples were collected 24 h later. Transcript levels were determined by qRT-PCR. Bars represent mean values (±se) of three to four biological replicates normalized to Actin transcript levels. Bars annotated with different letters were significantly different according to Fisher’s LSD test (P ≤ 0.05) after ANOVA.
SlMYC2 Is Not Involved in Regulating Type VI Glandular Trichome Development or Volatile Terpene Biosynthesis in Trichomes
SlMYC1 and SlMYC2 are homologous proteins belonging to the bHLH IIIe subclade in S. lycopersicum (Goossens et al., 2017). The key role of SlMYC2 in various JA-mediated processes (Du et al., 2017) prompted us to investigate whether this protein also participates in the regulation of volatile terpene biosynthesis and type VI glandular trichome development. We used an RNA interference strategy to downregulate SlMYC2 in transgenic tomato plants. We obtained three independent transgenic lines with a 90% reduction in SlMYC2 transcript levels but no consistent effect on SlMYC1 expression (Supplemental Figures 7A and 7B). However, downregulation of SlMYC2 did not lead to significant changes in the transcript levels of the TPS genes, which are regulated by SlMYC1 (Supplemental Figures 7C and 7D), nor were type VI glandular trichomes affected in these lines (Supplemental Figures 8A and 8B). We also investigated whether SlMYC2 is required for the induction of TPS genes by JA. In wild-type tomato plants, SlMYC2 was induced by JA in both stem trichomes and leaves (Supplemental Figures 7E and 7F). Although SlMYC2 was not induced by JA in 35S:ir-SlMYC2 plants, the JA-regulated induction of SlTPS3 and SlTPS5 was not affected in these plants (Supplemental Figures 7E and 7F). These results indicate that unlike SlMYC1, SlMYC2 is not involved in regulating TPS genes or the formation of type VI trichomes.
DISCUSSION
Our study demonstrates that the bHLH transcription factor SlMYC1 is involved in the formation of type VI glandular trichomes and positively regulates monoterpene biosynthesis in tomato leaf and stem trichomes. In addition, SlMYC1 plays differential roles in regulating sesquiterpene biosynthesis, as it is necessary for β-caryophyllene and α-humulene biosynthesis in leaf trichomes, whereas it inhibits their biosynthesis in stem trichomes.
SlMYC1 Regulates Glandular Trichome Formation
The current model for trichome differentiation is based the development of nonglandular trichomes in Arabidopsis, in which an R2R3 repeat MYB protein (GLABRA1), a bHLH IIIf subfamily protein (GL3 or ENHANCER OF GLABRA3), and a WD40 repeat protein (TRANSPARENT TESTA GLABRA1) constitute an active MYB/bHLH/WD40 complex that triggers the expression of GLABRA2, a class IV HD-ZIP (HD-ZIP IV) transcription factor to induce trichome formation (Rerie et al., 1994; Schiefelbein, 2003; Pesch and Hülskamp, 2004, 2009; Serna and Martin, 2006; Ishida et al., 2008). In addition, several R3 MYBs (e.g., AtTRY) move from early trichome cells to neighboring cells, where they inhibit trichome initiation by competing with R2R3-repeat MYB proteins for interaction with bHLH proteins (Esch et al., 2004). Whether the initiation and differentiation of glandular trichomes involves analogous MYB/bHLH/WD40 complexes and downstream HD-ZIP and R3 MYB transcription factors has been unclear. MYB and HD-ZIP IV transcription factors have been implicated in the development of glandular trichomes in some species (Glover et al., 1998; Yang et al., 2011; Matías-Hernández et al., 2017; Yan et al., 2017; Shi et al., 2018).
Here, we show that one partner of this potential complex, the bHLH transcription factor SlMYC1, functions in glandular trichome development in tomato. In addition, tomato appears to contain a functional ortholog of the R3 MYB gene AtTRY, as heterologous expression of tomato SlTRY in Arabidopsis inhibits trichome formation (Tominaga-Wada et al., 2013). Furthermore, tomato Wo, which encodes an HD-ZIP IV transcription factor, specifically controls the initiation and development of type I trichomes (Yang et al., 2011). Together, these data suggest that MYB/bHLH/WD40 complexes and downstream HD-ZIP IV and TRY transcription factors may indeed function in glandular trichome development.
We found that knocking down SlMYC1 (in the ir-SlMYC1 lines) led to the production of fewer type VI trichomes on both sides of the leaf but also to more type IV glandular trichomes on stems (Supplemental Figure 5), likely due to reduced inhibition of the formation of other types of trichomes. In addition, an aberrant type of glandular trichome formed in these knockdown lines and in the homozygous myc1 mutant. Since the intermediate cell in this type of trichome was wider than the stalk, as is the case for type VII glandular trichomes, we considered these to be type VII-like glandular trichomes (Supplemental Figure 1A). However, since these stalks did not bend but were instead straight, like type VI glandular trichomes, one could argue that they were type VI-like glandular trichomes, a notion that requires further investigation. The total numbers of glandular trichomes on the abaxial leaf surfaces of these knockdown and knockout lines were lower than in wild-type plants (Figure 4), indicating that SlMYC1 plays a role in trichome initiation. In addition, the observation that trichomes of the ir-SlMYC1 lines had shorter stalks and smaller glandular heads than the wild type indicates that SlMYC1 is involved in glandular trichome development. We thus conclude that SlMYC1 is involved in both the initiation and maturation of type VI glandular trichomes.
The Role of SlMYC1 in Regulating Terpene Biosynthesis
The downregulation of SlMYC1 resulted in reduced terpene levels as well as altered trichome densities. The changes in the overall terpene profiles of total trichomes (Figures 2 and 5) were similar to those of individual type VI glands (Figure 7), demonstrating that this was not due to the presence of fewer type VI glandular trichomes. Stem trichomes of the knockdown plants and MYC1/myc1 plants produced more sesquiterpenes than the wild type, while the leaf trichomes produced reduced amounts of monoterpenes. The sizes of type VI trichomes in MYC1/myc1 plants were similar to those of the wild type, indicating that the altered terpene production was not simply due to altered type VI trichome development. Our findings thus indicate that SlMYC1 itself is a regulator of TPS genes. Consistent with this conclusion, the stem trichomes of transgenic tomato plants expressing SlMYC1E161K, encoding a (putatively) more stable SlMYC1 (Gasperini et al., 2015), under the control of the type VI-specific SlTPS5 promoter (Spyropoulou et al., 2014b) had higher SlTPS3 and lower SlTPS12 transcript levels than the wild type, indicating that SlMYC1 regulates these genes.
One glandular head of a type VI stem trichome produced more than 10 times the amount of monoterpenes found in the leaf but almost no sesquiterpenes (Figure 7). In cultivated tomato, most sesquiterpenes are synthesized from precursors provided by the mevalonate pathway, which functions in the cytosol, endoplasmic reticulum, and peroxisomes, whereas the plastidial methylerythritol-4-phosphate pathway is mainly responsible for producing precursors for monoterpene biosynthesis (Simkin et al., 2011; Pulido et al., 2012). Although they are physically separated, metabolic crosstalk occurs between these pathways (Gutensohn et al., 2013; Henry et al., 2018), and we thought that the decrease in monoterpene biosynthesis could have caused an increase in sesquiterpene biosynthesis. However, in the stem trichomes of MYC1/myc1 plants, monoterpene levels were not reduced, although sesquiterpene levels were elevated. This observation indicates that SlMYC1, and not metabolic crosstalk, modulates the expression of mono- and sesquiterpene synthase genes in tomato leaf and stem trichomes.
Although bHLH transcription factors are involved in regulating terpenoid biosynthesis in other species, most function as positive regulators of biosynthesis genes. In Arabidopsis, for instance, AtMYC2 forms homo- or heterodimers with bHLH transcription factors AtMYC3 and AtMYC4 to regulate the expression of target genes (Fernández-Calvo et al., 2011). Perhaps the differential regulation of sesquiterpene biosynthesis in tomato trichomes could be explained by a model in which another bHLH transcription factor expressed in stem but not leaf trichomes regulates SlTPS12 expression. Such a process would lead to the formation of a bHLH complex in which SlMYC1 inhibits the expression of terpene biosynthesis genes in stem trichomes, and this inhibition would be alleviated under reduced SlMYC1 levels.
The Role of SlMYC1 in JA Responses
In tomato, the leaves of both JA receptor and JA biosynthesis mutants have reduced numbers of type VI glandular trichomes (Li et al., 2004; Boughton et al., 2005; Yan et al., 2013; Bosch et al., 2014). In addition, reduced levels of JA lead to lower levels of volatile terpenes in individual type VI trichomes (Yan et al., 2013). The components that act downstream of JA remain to be identified. Our data show that SlMYC1 is an essential downstream component, as JA failed to restore the formation of type VI trichomes in the myc1 mutant and failed to increase the size of the smaller type VI trichomes in the ir-SlMYC1 plants. Interestingly, our data also demonstrate that the induction of TPS genes by JA is not entirely dependent on SlMYC1, and thus, other transcription factors, such as SlEOT1, might be involved in this process (Spyropoulou et al., 2014b). Further research using the tools that we have developed should allow these regulators to be identified. A challenge for future studies will be to identify other molecular players involved in type VI trichome development in tomato that might act in a network with the bHLH transcription factor SlMYC1.
METHODS
Plant Growth Conditions and Sampling
Tomato (Solanum lycopersicum cv Moneymaker) and Nicotiana benthamiana plants were grown in soil in a greenhouse with day/night temperatures of 23/18°C and a 16/8 h light/dark regime with supplemental light when necessary (150 µE m−2s−1; Philips Master Green Power; www.usa.lighting.philips.com). Four-week-old tomato plants and 4- to 5-week-old N. benthamiana plants were used for all experiments unless otherwise indicated. For all experiments, multiple stem pieces or leaflets were harvested from multiple plants for one biological replicate.
Constructs and Generation of Transgenic Plants
To create the 35S:ir-SlMYC1 and 35S:ir-SlMYC2 constructs, Gateway technology (Invitrogen; www.thermofisher.com) was used. Primer pairs ir-MYC1_attB1 and ir-MYC1_attB2 (targeting bases 1953 to 2174 of SlMYC1 mRNA) and ir-MYC2_attB1 and ir-MYC2_attB2 (targeting bases 2071 to 2361 of SlMYC2 mRNA), containing attB sites, were used to amplify the SlMYC1 and SlMYC2 fragments, respectively (Supplemental Table). The fragments were cloned into the pDONR207 vector (Invitrogen) and subsequently recombined in destination vector pK7GWIWG2(1) (Karimi et al., 2002). To create the SlTPS5p:SlMYC1E161K construct, primer pairs MYC1_F and MYC1E161K_R and MYC1E161K_F and MYC1_R were used to amplify two halves of the fragments (nucleotides 1 to 491 and 471 to 1833) for SlMYC1E161K with a single nucleotide mutation introduced into position 481 of the coding sequence. The open reading frame of SlMYC1E161K was amplified with a mixture of the two fragments using primer pair MYC1_F and MYC1_R. The SlMYC1E161K sequence was verified and cloned into the pDONR207 vector with primer pair MYC1_attB1 and MYC1_attB2, creating SlMYC1E161K-pENTR. The SlTPS5 promoter (comprising 1254 bp of the genomic sequence upstream of the start codon of SlTPS5) was amplified with primer pair SlTPS5p_attB4 and SlTPS5p_attB1r and cloned into the pGEM BOX1-P4P1R vector, creating SlTPS5p-pENTR. SlTPS5p-pENTR and SlMYC1E161K-pENTR were recombined in destination vector pK7m24GW(3) (Karimi et al., 2002). To create the truncated SlMYC1 construct 35S:SlMYC1tru493, primer pair MYC1_attB1 and MYC1tru493_attB2 with a single nucleotide mutation introduced (from G to T at position 1477 of the SlMYC1 coding sequence, resulting in an early stop codon at amino acid position 493) was used to amplify the fragment. The fragment was cloned into pDONR207 and subsequently recombined in destination vector pK2GW7(0) (Karimi et al., 2002). To create the CRISPR-Cas9_myc1 construct, three guide RNAs (gRNAs), designed to target the open reading frame of SlMYC1 (583 to 603 bp, 1825 to 1844 bp, and 1092 1111 bp) were synthesized (www.eurofinsgenomics) and cloned into vector pYPQ131, pYPQ132, and pYPQ133, respectively (Addgene; www.addgene.org; Lowder et al., 2015), creating pYPQ131-gRNA1, pYPQ132-gRNA2, and pYPQ133-gRNA3. The three guide RNAs were then assembled into pYPQ143 using Golden Gate technology to create the gRNA entry vector pYPQ143-gRNAs. The Cas9 entry vector pYPQ150 (Addgene) and pYPQ143-gRNAs were recombined in destination vector pK2GW7(0) using Gateway technology. All constructs were verified by sequencing and introduced into Agrobacterium tumefaciens strain GV3101 (pMP90). Stably transformed transgenic tomato plants were created using explants derived from the cotyledons of sterile S. lycopersicum cv Moneymaker seedlings as previously described (Cortina and Culiáñez-Macià, 2004). The kanamycin-resistant transformants were screened for the presence of the nptII gene.
Identification of the myc1 Mutant
Seeds of S. lycopersicum breeding line Rijk Zwaan (RZ)-1 were treated with EMS by submerging ∼10,000 seeds in an aerated solution of 0.5% (w/v) EMS for 24 h at room temperature. The treated seeds were germinated, and the resulting plants were grown in a greenhouse to produce M2 seeds. M2 seeds were harvested from mature plants and bulked in one pool. The resulting pool of M2 seeds was used as starting material to screen 8000 M2 plants to identify individual M2 plants that showed an aberrant type VI glandular trichome phenotype under a stereomicroscope. Using quantitative trait loci-mapping of an F2 population of the mutant and the parent, the recessive trait was localized to chromosome 8. The mutant had a single nucleotide change, G to T at position 1477, in the coding sequence of SlMYC1, resulting in an early stop codon at amino acid position 493. This mutation was predictive for the phenotype in an F4 population.
RNA Isolation and Quantitative RT-PCR
For RNA isolation, stems from whole plants and the second pair of leaflets from the fourth leaf were collected and frozen in liquid nitrogen. Trichomes were isolated from the frozen stems by shaking in liquid nitrogen in 50-mL tubes with a vortex mixer. For RNA extraction, total RNA was isolated using TRIzol (Invitrogen) and treated with TURBO DNase (Ambion; www.thermofisher.com) to remove contaminating DNA. RNA quantity was determined with a NanoDrop spectrophotometer (Thermo Fisher; www.thermofisher.com), and complementary DNA was synthesized from 1.5 μg RNA using RevertAid M-MuLV Reverse Transcriptase (Fermentas; www.thermofisher.com). For quantitative RT-PCR (qRT-PCR), cDNA equivalent to 5 ng total RNA was used as a template, and PCR was performed with SYBR Green qPCR SuperMix (Solis Biodyne; www.sbd.ee) and 300 nM of each primer (Supplemental Table) in a total volume of 10 μL in an ABI 7500 Real-Time PCR System (Applied Biosystems; www.appliedbiosystems.com) with the following cycling program: 2 min 50°C, 15 min 95°C, 45 cycles of 15 s at 95°C, and 1 min at 60°C. Primer pair efficiencies were calculated by analyzing amplification curves from a standard cDNA dilution range. Expression levels were normalized to the transcript level of ACTIN. To analyze the SlTPS5p:SlMYC1E161K transgenic lines, SlMYC1E161K-QF (bases 1791 to 1810 of SlMYC1 mRNA) and SlMYC1E161K-QR (16 to 36 bp of primer MYC1_attB2) were used to check SlMYC1E161K expression, and SlMYC1-QF (731 to 750 bp of SlMYC1 mRNA) and SlMYC1-QR (877 to 858 bp of SlMYC1 mRNA) were used to measure total SlMYC1 transcript levels (SlMYC1/SlMYC1E161K).
Analysis of Volatile Terpenes
To measure volatile terpenes, stem pieces from the third internodes of plants were collected and frozen in liquid nitrogen. Trichomes were isolated from the frozen stems by shaking in liquid nitrogen in 15-mL tubes with a vortex mixer. Terpene extraction was performed using 1 mL of hexane plus 0.5 ng µL−1 benzyl acetate (Sigma-Aldrich; www.sigmaaldrich.com) as an internal standard. Na2CO3 (Sigma-Aldrich) was used to remove any water from the hexane. For leaf trichomes, the second pair of leaflets from the fourth leaf was washed with 1 mL of hexane plus 0.5 ng µL−1 benzyl acetate. For terpenes in type VI glandular trichomes, 200 individual type VI glands were collected from stems or leaves with a pulled Pasteur pipette and dissolved in 100 μL of the same solvent. The volatiles were analyzed using an Agilent (www.agilent.com) 7890A gas chromatograph, coupled to an Agilent 7200 accurate-mass quadrupole time-of-flight mass spectrometer operating in electron-impact mode. Liquid injection of a 1-μL sample was performed at 50°C, with the injector port directly heated to 275°C at a rate of 240°C min−1. The oven temperature was maintained at 40°C for 3 min and increased by 5°C per min until it reached 140°C. The temperature was then increased by 10°C per min until it reached 250°C and maintained at 250°C for 5 min. Compounds were separated on a capillary HP-5ms column (30 m × 250 µm, 0.25 µm film thickness; Agilent) with helium as the carrier gas at a flow rate of 1 mL min−1. Terpene standards were used for compound identification and quantification.
Trichome Density and Morphology
Scanning electron micrographs of fresh, unfixed plant material were produced with an environmental scanning electron microscope (ESEM) type XL-30 FEG (FEI/Philips; www.fei.com) operating in wet mode. The gas pressure in the ESEM chamber (1.5 mBar) was regulated by introducing water vapor. A gaseous secondary electron detector was used for imaging. Stem trichomes from the third fully grown internode of each plant were scanned to determine their size. An average of four to five images were taken per line. Light microscopy images of trichomes were obtained under a Leica MZFLIII microscope (www.leica-microsystems.com), which was also used to count to the trichomes. Images were captured with a Nikon DS-Fi2 5-megapixel CCD camera (www.nikon.com) using NIS-Elements (version F4.30.00). To measure trichome density on leaves, the second pair of leaflets of the seventh leaf of each 7-week-old tomato plant was used. To measure trichome density on stems, stem pieces from the third internodes of 4-week-old plants and the sixth internodes of 7-week-old plants were used. The morphology of stem trichomes submerged in water was also examined under an EVOSfl (www.thermofisher.com) inverted microscope.
JA Treatment
For JA treatment, 4-week-old tomato plants were sprayed with 1 mM JA (Duchefa; www.duchefa-biochemie.com) in tap water plus 0.05% SilwetL-77; the control plants were sprayed with tap water plus 0.05% SilwetL-77. Stem pieces and leaflets were collected for RNA isolation and volatile terpene measurements 24 h later. To induce trichome formation on newly formed leaves, the plants were sprayed weekly with JA or control solution. On day 28 after the first treatment, newly developed stems and leaflets were used for trichome counting and to determine the morphology of type VI glandular trichomes.
Transient Transactivation Assay in N. benthamiana Leaves
The β-glucuronidase (GUS) gene driven by the SlTPS5 promoter (Spyropoulou et al., 2014b) was used as a reporter to verify the transactivation of the effectors. The 35S:SlMYC1 and 35S:RFP effector constructs were used as positive and negative controls, respectively (Spyropoulou et al., 2014a). A. tumefaciens cultures were grown overnight from a single colony until an OD600 between 1.0 and 1.5 was reached. After centrifugation, the bacterial pellets were resuspended in infiltration buffer (50 mM MES pH 5.8, 0.5% glucose, 2 mM Na2HPO4, 100 μM acetosyringone; Sigma-Aldrich) to OD600 of 0.3 and incubated for 1 h at room temperature. A construct containing the luciferase (LUC) gene driven by the Cauliflower mosaic virus 35S promoter (Spyropoulou et al., 2014b) was used to normalize for transformation efficiency and protein extraction efficiency. Leaves from 4- to 5-week-old N. benthamiana plants were infiltrated with A. tumefaciens cultures carrying various MYC1 or RFP constructs, the SlTPS5p:GUS reporter construct, and a 35S:LUC construct at a 5:5:2 ratio. Two leaves each from three plants were infiltrated for each combination. Two days later, leaf disks from the infiltrated areas were collected, frozen in liquid nitrogen, and used to prepare crude extracts in extraction buffer containing 25 mM TRIS phosphate pH 7.8, 2 mM dithiothreitol, 2 mM trans-1,2-diaminocyclohexane-N,N,N',N'-tetraacetic acid pH 7.8, 10% glycerol, and 1% Triton X-100 (Sigma-Aldrich). 4-methylumbelliferyl-β-d-glucuronid and LUC activities were measured using a Fluorocount SYNERGY H1 microplate reader (BioTek; www.biotek.com). Enzymatic GUS activity was determined by normalizing 4-methylumbelliferyl-β-d-glucuronide activity with the LUC activity of each sample.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL/Solgenomics databases under the following accession numbers: SlMYC1, KF430611; SlMYC2, Solyc08g076930; SlTPS3, Solyc01g105870; SlTPS5, Solyc01g105890; SlTPS9, Solyc06g059930; SlTPS12, Solyc06g059910; SlTPS17, Solyc12g006570; SlTPS20, Solyc08g005670; SlTPS31, Solyc01g101170; SlTPS39, Solyc10g005390; Jasmonate-inducible protein-21, Solyc03g098790; ACT, Solyc03g078400.
Supplemental Data
Supplemental Figure 1. Environmental scanning electron micrographs of stem trichomes.
Supplemental Figure 2. Truncated SlMYC1 does not transactivate the SlTPS5 promoter in Nicotiana benthamiana leaves.
Supplemental Figure 3. Nucleotide sequences of SlMYC1 in the CRISPR-Cas9 knockout lines.
Supplemental Figure 4. Morphology of type VI glandular trichomes on stems.
Supplemental Figure 5. Type IV glandular trichome density on the tomato stem surface.
Supplemental Figure 6. SlMYC1 expression in tomato stem trichomes and leaves treated with JA.
Supplemental Figure 7. Effect of SlMYC2 downregulation on the expression of TPS genes in stem trichomes and leaves.
Supplemental Figure 8. Morphology of type VI glandular trichomes on the stem surface in ir-MYC2 plants.
Supplemental Table. List of primers used in this study.
Supplemental Data Set. ANOVA tables.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank Frank Syrowatka (Martin-Luther-Universität Halle-Wittenberg) for taking the ESEM images of the trichomes; Alain Tissier (Leibniz Institute of Plant Biochemistry) for growing the plants; Carlos Galvan and Christa Testerink for the pGEM P4-P1r BOX1 vector; Eleni Spyropoulou and Michel de Vries for technical assistance; Yanaika Sylvana Hok-a-Hin for performing the trichome density assays; Ludek Tikovsky, Harold Lemereis, and Thijs Hendrix for taking care of the plants; and Plant Editors (planteditors.com) for assisting with editing the manuscript. J. Xu was supported by the Chinese Scholarship Council (CSC).
AUTHOR CONTRIBUTIONS
J.X. performed most of the experiments and analyzed the data; R.C.S. designed the study; R.C.S. and M.A.H. supervised the research; Z.O.v.H. and D.B.B. generated the myc1 mutant; C.S. created the CRISPR-Cas9_myc1 construct; J.X. and R.C.S. wrote the manuscript; and M.A.H. revised the manuscript.
References
- Barrangou R., Fremaux C., Deveau H., Richards M., Boyaval P., Moineau S., Romero D.A., Horvath P. (2007). CRISPR provides acquired resistance against viruses in prokaryotes. Science 315: 1709–1712. [DOI] [PubMed] [Google Scholar]
- Bergau N., Bennewitz S., Syrowatka F., Hause G., Tissier A. (2015). The development of type VI glandular trichomes in the cultivated tomato Solanum lycopersicum and a related wild species S. habrochaites. BMC Plant Biol. 15: 289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besser K., Harper A., Welsby N., Schauvinhold I., Slocombe S., Li Y., Dixon R.A., Broun P. (2009). Divergent regulation of terpenoid metabolism in the trichomes of wild and cultivated tomato species. Plant Physiol. 149: 499–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleeker P.M., Spyropoulou E.A., Diergaarde P.J., Volpin H., De Both M.T., Zerbe P., Bohlmann J., Falara V., Matsuba Y., Pichersky E., Haring M.A., Schuurink R.C. (2011). RNA-seq discovery, functional characterization, and comparison of sesquiterpene synthases from Solanum lycopersicum and Solanum habrochaites trichomes. Plant Mol. Biol. 77: 323–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch M., Wright L.P., Gershenzon J., Wasternack C., Hause B., Schaller A., Stintzi A. (2014). Jasmonic acid and its precursor 12-oxophytodienoic acid control different aspects of constitutive and induced herbivore defenses in tomato. Plant Physiol. 166: 396–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boughton A.J., Hoover K., Felton G.W. (2005). Methyl jasmonate application induces increased densities of glandular trichomes on tomato, Lycopersicon esculentum. J. Chem. Ecol. 31: 2211–2216. [DOI] [PubMed] [Google Scholar]
- Brady S.M., Burow M., Busch W., Carlborg Ö., Denby K.J., Glazebrook J., Hamilton E.S., Harmer S.L., Haswell E.S., Maloof J.N., Springer N.M., Kliebenstein D.J. (2015). Reassess the t Test: Interact with all your data via ANOVA. Plant Cell 27: 2088–2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang J., Yu T., Gao S., Xiong C., Xie Q., Li H., Ye Z., Yang C. (2016). Fine mapping of the dialytic gene that controls multicellular trichome formation and stamen development in tomato. Theor. Appl. Genet. 129: 1531–1539. [DOI] [PubMed] [Google Scholar]
- Chang J., Yu T., Yang Q., Li C., Xiong C., Gao S., Xie Q., Zheng F., Li H., Tian Z., Yang C., Ye Z. (2018). Hair, encoding a single C2H2 zinc-finger protein, regulates multicellular trichome formation in tomato. Plant J. 96: 90–102. [DOI] [PubMed] [Google Scholar]
- Cortina C., Culiáñez-Macià F.A. (2004). Tomato transformation and transgenic plant production. Plant Cell Tissue Organ Cult. 76: 269–275. [Google Scholar]
- Du M., Zhao J., Tzeng D.T.W., Liu Y., Deng L., Yang T., Zhai Q., Wu F., Huang Z., Zhou M., Wang Q., Chen Q., et al. (2017). MYC2 orchestrates a hierarchical transcriptional cascade that regulates jasmonate-mediated plant immunity in tomato. Plant Cell 29: 1883–1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esch J.J., Chen M.A., Hillestad M., Marks M.D. (2004). Comparison of TRY and the closely related At1g01380 gene in controlling Arabidopsis trichome patterning. Plant J. 40: 860–869. [DOI] [PubMed] [Google Scholar]
- Ewas M., Gao Y., Wang S., Liu X., Zhang H., Nishawy E.M.E., Ali F., Shahzad R., Ziaf K., Subthain H., Martin C., Luo J. (2016). Manipulation of SlMXl for enhanced carotenoids accumulation and drought resistance in tomato. Sci. Bull. (Beijing) 61: 1413–1418. [Google Scholar]
- Ewas M., Gao Y.Q., Ali F., Nishawy E.M., Shahzad R., Subthain H., Amar M., Martin C., Luo J. (2017). RNA-seq reveals mechanisms of SlMX1 for enhanced carotenoids and terpenoids accumulation along with stress resistance in tomato. Sci. Bull. (Beijing) 62: 476–485. [DOI] [PubMed] [Google Scholar]
- Falara V., Akhtar T.A., Nguyen T.T., Spyropoulou E.A., Bleeker P.M., Schauvinhold I., Matsuba Y., Bonini M.E., Schilmiller A.L., Last R.L., Schuurink R.C., Pichersky E. (2011). The tomato terpene synthase gene family. Plant Physiol. 157: 770–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falara V., Alba J.M., Kant M.R., Schuurink R.C., Pichersky E. (2014). Geranyllinalool synthases in solanaceae and other angiosperms constitute an ancient branch of diterpene synthases involved in the synthesis of defensive compounds. Plant Physiol. 166: 428–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Calvo P., et al. (2011). The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. Plant Cell 23: 701–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridman E., Wang J., Iijima Y., Froehlich J.E., Gang D.R., Ohlrogge J., Pichersky E. (2005). Metabolic, genomic, and biochemical analyses of glandular trichomes from the wild tomato species Lycopersicon hirsutum identify a key enzyme in the biosynthesis of methylketones. Plant Cell 17: 1252–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao S., Gao Y., Xiong C., Yu G., Chang J., Yang Q., Yang C., Ye Z. (2017). The tomato B-type cyclin gene, SlCycB2, plays key roles in reproductive organ development, trichome initiation, terpenoids biosynthesis and Prodenia litura defense. Plant Sci. 262: 103–114. [DOI] [PubMed] [Google Scholar]
- Gasperini D., Chételat A., Acosta I.F., Goossens J., Pauwels L., Goossens A., Dreos R., Alfonso E., Farmer E.E. (2015). Multilayered organization of jasmonate signalling in the regulation of root growth. PLoS Genet. 11: e1005300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gershenzon J., Dudareva N. (2007). The function of terpene natural products in the natural world. Nat. Chem. Biol. 3: 408–414. [DOI] [PubMed] [Google Scholar]
- Glas J.J., Schimmel B.C., Alba J.M., Escobar-Bravo R., Schuurink R.C., Kant M.R. (2012). Plant glandular trichomes as targets for breeding or engineering of resistance to herbivores. Int. J. Mol. Sci. 13: 17077–17103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover B.J., Perez-Rodriguez M., Martin C. (1998). Development of several epidermal cell types can be specified by the same MYB-related plant transcription factor. Development 125: 3497–3508. [DOI] [PubMed] [Google Scholar]
- Goossens J., Mertens J., Goossens A. (2017). Role and functioning of bHLH transcription factors in jasmonate signalling. J. Exp. Bot. 68: 1333–1347. [DOI] [PubMed] [Google Scholar]
- Gutensohn M., Orlova I., Nguyen T.T., Davidovich-Rikanati R., Ferruzzi M.G., Sitrit Y., Lewinsohn E., Pichersky E., Dudareva N. (2013). Cytosolic monoterpene biosynthesis is supported by plastid-generated geranyl diphosphate substrate in transgenic tomato fruits. Plant J. 75: 351–363. [DOI] [PubMed] [Google Scholar]
- Henry L.K., Thomas S.T., Widhalm J.R., Lynch J.H., Davis T.C., Kessler S.A., Bohlmann J., Noel J.P., Dudareva N. (2018). Contribution of isopentenyl phosphate to plant terpenoid metabolism. Nat. Plants 4: 721–729. [DOI] [PubMed] [Google Scholar]
- Hong G.J., Xue X.Y., Mao Y.B., Wang L.J., Chen X.Y. (2012). Arabidopsis MYC2 interacts with DELLA proteins in regulating sesquiterpene synthase gene expression. Plant Cell 24: 2635–2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida T., Kurata T., Okada K., Wada T. (2008). A genetic regulatory network in the development of trichomes and root hairs. Annu. Rev. Plant Biol. 59: 365–386. [DOI] [PubMed] [Google Scholar]
- Ji Y., Xiao J., Shen Y., Ma D., Li Z., Pu G., Li X., Huang L., Liu B., Ye H., Wang H. (2014). Cloning and characterization of AabHLH1, a bHLH transcription factor that positively regulates artemisinin biosynthesis in Artemisia annua. Plant Cell Physiol. 55: 1592–1604. [DOI] [PubMed] [Google Scholar]
- Kang J.H., Liu G., Shi F., Jones A.D., Beaudry R.M., Howe G.A. (2010b). The tomato odorless-2 mutant is defective in trichome-based production of diverse specialized metabolites and broad-spectrum resistance to insect herbivores. Plant Physiol. 154: 262–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J.H., Shi F., Jones A.D., Marks M.D., Howe G.A. (2010a). Distortion of trichome morphology by the hairless mutation of tomato affects leaf surface chemistry. J. Exp. Bot. 61: 1053–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J.H., McRoberts J., Shi F., Moreno J.E., Jones A.D., Howe G.A. (2014). The flavonoid biosynthetic enzyme chalcone isomerase modulates terpenoid production in glandular trichomes of tomato. Plant Physiol. 164: 1161–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J.H., Campos M.L., Zemelis-Durfee S., Al-Haddad J.M., Jones A.D., Telewski F.W., Brandizzi F., Howe G.A. (2016). Molecular cloning of the tomato Hairless gene implicates actin dynamics in trichome-mediated defense and mechanical properties of stem tissue. J. Exp. Bot. 67: 5313–5324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M., Inzé D., Depicker A. (2002). GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7: 193–195. [DOI] [PubMed] [Google Scholar]
- Kortbeek R.W., Xu J., Ramirez A., Spyropoulou E., Diergaarde P., Otten-Bruggeman I., de Both M., Nagel R., Schmidt A., Schuurink R.C., Bleeker P.M. (2016). Engineering of tomato glandular trichomes for the production of specialized metabolites. Methods Enzymol. 576: 305–331. [DOI] [PubMed] [Google Scholar]
- Lenka S.K., Nims N.E., Vongpaseuth K., Boshar R.A., Roberts S.C., Walker E.L. (2015). Jasmonate-responsive expression of paclitaxel biosynthesis genes in Taxus cuspidata cultured cells is negatively regulated by the bHLH transcription factors TcJAMYC1, TcJAMYC2, and TcJAMYC4. Front. Plant Sci. 6: 115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Zhao Y., McCaig B.C., Wingerd B.A., Wang J., Whalon M.E., Pichersky E., Howe G.A. (2004). The tomato homolog of CORONATINE-INSENSITIVE1 is required for the maternal control of seed maturation, jasmonate-signaled defense responses, and glandular trichome development. Plant Cell 16: 126–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowder L.G., Zhang D., Baltes N.J., Paul J.W. III, Tang X., Zheng X., Voytas D.F., Hsieh T.F., Zhang Y., Qi Y. (2015). A CRISPR/Cas9 toolbox for multiplexed plant genome editing and transcriptional regulation. Plant Physiol. 169: 971–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P., Yang L., Esvelt K.M., Aach J., Guell M., DiCarlo J.E., Norville J.E., Church G.M. (2013). RNA-guided human genome engineering via Cas9. Science 339: 823–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matías-Hernández L., Jiang W., Yang K., Tang K., Brodelius P.E., Pelaz S. (2017). AaMYB1 and its orthologue AtMYB61 affect terpene metabolism and trichome development in Artemisia annua and Arabidopsis thaliana. Plant J. 90: 520–534. [DOI] [PubMed] [Google Scholar]
- McDowell E.T., et al. (2011). Comparative functional genomic analysis of Solanum glandular trichome types. Plant Physiol. 155: 524–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertens J., Pollier J., Vanden Bossche R., Lopez-Vidriero I., Franco-Zorrilla J.M., Goossens A. (2016). The bHLH transcription factors TSAR1 and TSAR2 regulate triterpene saponin biosynthesis in Medicago truncatula. Plant Physiol. 170: 194–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patra B., Pattanaik S., Schluttenhofer C., Yuan L. (2018). A network of jasmonate-responsive bHLH factors modulate monoterpenoid indole alkaloid biosynthesis in Catharanthus roseus. New Phytol. 217: 1566–1581. [DOI] [PubMed] [Google Scholar]
- Pesch M., Hülskamp M. (2004). Creating a two-dimensional pattern de novo during Arabidopsis trichome and root hair initiation. Curr. Opin. Genet. Dev. 14: 422–427. [DOI] [PubMed] [Google Scholar]
- Pesch M., Hülskamp M. (2009). One, two, three...models for trichome patterning in Arabidopsis? Curr. Opin. Plant Biol. 12: 587–592. [DOI] [PubMed] [Google Scholar]
- Pulido P., Perello C., Rodriguez-Concepcion M. (2012). New insights into plant isoprenoid metabolism. Mol. Plant 5: 964–967. [DOI] [PubMed] [Google Scholar]
- Rerie W.G., Feldmann K.A., Marks M.D. (1994). The GLABRA2 gene encodes a homeo domain protein required for normal trichome development in Arabidopsis. Genes Dev. 8: 1388–1399. [DOI] [PubMed] [Google Scholar]
- Sallaud C., Rontein D., Onillon S., Jabès F., Duffé P., Giacalone C., Thoraval S., Escoffier C., Herbette G., Leonhardt N., Causse M., Tissier A. (2009). A novel pathway for sesquiterpene biosynthesis from Z,Z-farnesyl pyrophosphate in the wild tomato Solanum habrochaites. Plant Cell 21: 301–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiefelbein J. (2003). Cell-fate specification in the epidermis: A common patterning mechanism in the root and shoot. Curr. Opin. Plant Biol. 6: 74–78. [DOI] [PubMed] [Google Scholar]
- Schilmiller A.L., Last R.L., Pichersky E. (2008). Harnessing plant trichome biochemistry for the production of useful compounds. Plant J. 54: 702–711. [DOI] [PubMed] [Google Scholar]
- Schilmiller A.L., Schauvinhold I., Larson M., Xu R., Charbonneau A.L., Schmidt A., Wilkerson C., Last R.L., Pichersky E. (2009). Monoterpenes in the glandular trichomes of tomato are synthesized from a neryl diphosphate precursor rather than geranyl diphosphate. Proc. Natl. Acad. Sci. USA 106: 10865–10870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilmiller A.L., Miner D.P., Larson M., McDowell E., Gang D.R., Wilkerson C., Last R.L. (2010a). Studies of a biochemical factory: Tomato trichome deep expressed sequence tag sequencing and proteomics. Plant Physiol. 153: 1212–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilmiller A., Shi F., Kim J., Charbonneau A.L., Holmes D., Daniel Jones A., Last R.L. (2010b). Mass spectrometry screening reveals widespread diversity in trichome specialized metabolites of tomato chromosomal substitution lines. Plant J. 62: 391–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt A., Li C., Shi F., Jones A.D., Pichersky E. (2011). Polymethylated myricetin in trichomes of the wild tomato species Solanum habrochaites and characterization of trichome-specific 3′/5′- and 7/4′-myricetin O-methyltransferases. Plant Physiol. 155: 1999–2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serna L., Martin C. (2006). Trichomes: Different regulatory networks lead to convergent structures. Trends Plant Sci. 11: 274–280. [DOI] [PubMed] [Google Scholar]
- Shen Q., Lu X., Yan T., Fu X., Lv Z., Zhang F., Pan Q., Wang G., Sun X., Tang K. (2016). The jasmonate-responsive AaMYC2 transcription factor positively regulates artemisinin biosynthesis in Artemisia annua. New Phytol. 210: 1269–1281. [DOI] [PubMed] [Google Scholar]
- Shi P., et al. (2018). The roles of AaMIXTA1 in regulating the initiation of glandular trichomes and cuticle biosynthesis in Artemisia annua. New Phytol. 217: 261–276. [DOI] [PubMed] [Google Scholar]
- Simkin A.J., Guirimand G., Papon N., Courdavault V., Thabet I., Ginis O., Bouzid S., Giglioli-Guivarc’h N., Clastre M. (2011). Peroxisomal localisation of the final steps of the mevalonic acid pathway in planta. Planta 234: 903–914. [DOI] [PubMed] [Google Scholar]
- Simmons A.T., Gurr G.M. (2005). Trichomes of Lycopersicon species and their hybrids: effects on pests and natural enemies. Agric. For. Entomol. 7: 265–276. [Google Scholar]
- Spyropoulou E.A., Haring M.A., Schuurink R.C. (2014a). RNA sequencing on Solanum lycopersicum trichomes identifies transcription factors that activate terpene synthase promoters. BMC Genomics 15: 402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spyropoulou E.A., Haring M.A., Schuurink R.C. (2014b). Expression of Terpenoids 1, a glandular trichome-specific transcription factor from tomato that activates the terpene synthase 5 promoter. Plant Mol. Biol. 84: 345–357. [DOI] [PubMed] [Google Scholar]
- Tholl D. (2015). Biosynthesis and biological functions of terpenoids in plants. Adv. Biochem. Eng. Biotechnol. 148: 63–106. [DOI] [PubMed] [Google Scholar]
- Tominaga-Wada R., Nukumizu Y., Sato S., Wada T. (2013). Control of plant trichome and root-hair development by a tomato (Solanum lycopersicum) R3 MYB transcription factor. PLoS One 8: e54019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Moerkercke A., et al. (2015). The bHLH transcription factor BIS1 controls the iridoid branch of the monoterpenoid indole alkaloid pathway in Catharanthus roseus. Proc. Natl. Acad. Sci. USA 112: 8130–8135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Moerkercke A., Steensma P., Gariboldi I., Espoz J., Purnama P.C., Schweizer F., Miettinen K., Vanden Bossche R., De Clercq R., Memelink J., Goossens A. (2016). The basic helix-loop-helix transcription factor BIS2 is essential for monoterpenoid indole alkaloid production in the medicinal plant Catharanthus roseus. Plant J. 88: 3–12. [DOI] [PubMed] [Google Scholar]
- van Schie C.C., Haring M.A., Schuurink R.C. (2007). Tomato linalool synthase is induced in trichomes by jasmonic acid. Plant Mol. Biol. 64: 251–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vendemiatti E., Zsögön A., Silva G.F.F.E., de Jesus F.A., Cutri L., Figueiredo C.R.F., Tanaka F.A.O., Nogueira F.T.S., Peres L.E.P. (2017). Loss of type-IV glandular trichomes is a heterochronic trait in tomato and can be reverted by promoting juvenility. Plant Sci. 259: 35–47. [DOI] [PubMed] [Google Scholar]
- Widhalm J.R., Jaini R., Morgan J.A., Dudareva N. (2015). Rethinking how volatiles are released from plant cells. Trends Plant Sci. 20: 545–550. [DOI] [PubMed] [Google Scholar]
- Williams W.G., Kennedy G.G., Yamamoto R.T., Thacker J.D., Bordner J. (1980). 2-Tridecanone: A naturally occurring insecticide from the wild tomato Lycopersicon hirsutum f.glabratum. Science 207: 888–889. [DOI] [PubMed] [Google Scholar]
- Xu Y.H., Liao Y.C., Lv F.F., Zhang Z., Sun P.W., Gao Z.H., Hu K.P., Sui C., Jin Y., Wei J.H. (2017). Transcription factor AsMYC2 controls the jasmonate-responsive expression of ASS1 regulating sesquiterpene biosynthesis in Aquilaria sinensis (Lour.) Gilg. Plant Cell Physiol. 58: 1924–1933. [DOI] [PubMed] [Google Scholar]
- Yamamura C., Mizutani E., Okada K., Nakagawa H., Fukushima S., Tanaka A., Maeda S., Kamakura T., Yamane H., Takatsuji H., Mori M. (2015). Diterpenoid phytoalexin factor, a bHLH transcription factor, plays a central role in the biosynthesis of diterpenoid phytoalexins in rice. Plant J. 84: 1100–1113. [DOI] [PubMed] [Google Scholar]
- Yan L., Zhai Q., Wei J., Li S., Wang B., Huang T., Du M., Sun J., Kang L., Li C.B., Li C. (2013). Role of tomato lipoxygenase D in wound-induced jasmonate biosynthesis and plant immunity to insect herbivores. PLoS Genet. 9: e1003964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan T., Chen M., Shen Q., Li L., Fu X., Pan Q., Tang Y., Shi P., Lv Z., Jiang W., Ma Y.N., Hao X., et al. (2017). HOMEODOMAIN PROTEIN 1 is required for jasmonate-mediated glandular trichome initiation in Artemisia annua. New Phytol. 213: 1145–1155. [DOI] [PubMed] [Google Scholar]
- Yang C., Li H., Zhang J., Luo Z., Gong P., Zhang C., Li J., Wang T., Zhang Y., Lu Y., Ye Z. (2011). A regulatory gene induces trichome formation and embryo lethality in tomato. Proc. Natl. Acad. Sci. USA 108: 11836–11841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin J., Li X., Zhan Y., Li Y., Qu Z., Sun L., Wang S., Yang J., Xiao J. (2017). Cloning and expression of BpMYC4 and BpbHLH9 genes and the role of BpbHLH9 in triterpenoid synthesis in birch. BMC Plant Biol. 17: 214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Hedhili S., Montiel G., Zhang Y., Chatel G., Pré M., Gantet P., Memelink J. (2011). The basic helix-loop-helix transcription factor CrMYC2 controls the jasmonate-responsive expression of the ORCA genes that regulate alkaloid biosynthesis in Catharanthus roseus. Plant J. 67: 61–71. [DOI] [PubMed] [Google Scholar]
- Zhou Y., Sun W., Chen J., Tan H., Xiao Y., Li Q., Ji Q., Gao S., Chen L., Chen S., Zhang L., Chen W. (2016). SmMYC2a and SmMYC2b played similar but irreplaceable roles in regulating the biosynthesis of tanshinones and phenolic acids in Salvia miltiorrhiza. Sci. Rep. 6: 22852. [DOI] [PMC free article] [PubMed] [Google Scholar]