Abstract
Plant voltage-gated K+ channels have been referred to as “plant Shakers” in reference to animal Shaker channels, the first K+ channels identified. Recent advances in our knowledge of K+ channel evolution and structure have significantly deepened the divide between these plant and animal K+ channels, suggesting that it is time to completely retire the “plant Shaker” designation. Evolutionary genomics reveals that plant voltage-gated K+ channels and metazoan Shakers derive from distinct prokaryotic ancestors. The plant channels belong to a lineage that includes cyclic nucleotide-gated channels and metazoan ether-à-go-go and hyperpolarization-activated, cyclic nucleotide-gated channels. We refer to this lineage as the CNBD channel superfamily, because all these channels share a cytoplasmic gating domain homologous to cyclic nucleotide binding domains. The first structures of CNBD superfamily channels reveal marked differences in coupling between the voltage sensor and ion-conducting pore relative to metazoan Shaker channels. Viewing plant voltage-gated K+ channel function through the lens of CNBD superfamily structures should lead to insights into how these channels are regulated.
INTRODUCTION
Organisms in all domains of life possess ion channels that mediate rapid, energetically downhill flux of ions across cell membranes. At a basic level, ion channels are characterized by two properties. The first property is conductance, defined by the ion selectivity of the channel and the rate at which ions pass through the pore. For example, some ion channels are highly selective for a specific ionic species, while others are selective simply for cations or anions. The second property is channel gating, which refers to the biophysical mechanisms that alter the probability of the channel residing in an open or closed state. Some channels are gated by the voltage difference across the membrane in which they reside, and thus they are referred to as voltage-gated channels. Other channels are gated by sensory stimuli such as light or pressure, or are gated or regulated by interactions with proteins, small molecules, nonpermeant ionic species, or by posttranslational modifications, e.g., by kinases. The rich variety of gating mechanisms provides pathways for channels to modulate membrane voltage and ion flux on the time scale of milliseconds to minutes.
Examples of all of these types of channels and regulatory mechanisms are present in the Plantae. This review focuses on a particular category of K+-selective voltage-gated plant ion channels (Table 1), which informally have been referred to as “plant Shaker,” “plant Shaker-like,” or “plant Shaker-type” channels. Among the important processes that these channels mediate are K+ uptake from the soil solution, K+-dependent changes in guard cell volume that cause stomatal opening and closure, K+ release from xylem parenchyma cells to the transpiration stream, and K+ fluxes during pollen tube growth. New awareness of their evolutionary history and probable structure now greatly strengthens some of the initial observations concerning distinctions between animal Shaker channels and these plant channels. In this review, we highlight these distinctions and describe these channels within the larger context of cation channel diversity in the Plantae and in all three domains of life across the great span of evolution.
Table 1. The Arabidopsis Members of the Plant Voltage-Gated K+ Channel Family.
| Channel | Refseq ID | TAIR Locus ID | Voltage Activation | CNBD | Ankyrin Domain |
|---|---|---|---|---|---|
| KAT1 | NM_123993 | AT5g46240 | Hyperpolarization | Present | Absent |
| KAT2 | NM_117939 | AT4g18290 | Hyperpolarization | Present | Absent |
| KAT3 | NM_119417 | AT4g32650 | Hyperpolarization | Present | Absent |
| AKT1 | NM_128222 | AT2g26650 | Hyperpolarization | Present | Present |
| AKT2 | NM_118342 | AT4g22200 | Hyperpolarization | Present | Present |
| AKT5 | NM_119402 | AT4g32500 | Hyperpolarization | Present | Present |
| AKT6 | NM_128118 | AT2g25600 | Hyperpolarization | Present | Present |
| GORK | NM_123109 | AT5g37500 | Depolarization | Present | Present |
| SKOR | NM_111153 | AT3g02850 | Depolarization | Present | Present |
PLANT VOLTAGE-GATED K+ CHANNELS AND METAZOAN SHAKER CHANNELS HAVE DISTINCT DOMAIN STRUCTURES
A major historical reason why plant voltage-gated (VG) channels such as the plasma membrane K+ channels KAT1 and SKOR (Table 1) picked up the Shaker moniker is that they were among the earliest plant K+ channels cloned, having been discovered in the early 1990s at a time when the only other commonly known VG K+ channels were the Shaker family channels which had been discovered in Drosophila (Kamb et al., 1987; Papazian et al., 1987) and later found in vertebrates (Wei et al., 1990). Shaker channels derive their name from the shaker mutant phenotype, characterized by leg shaking of fruit flies under ether anesthesia (Kaplan and Trout, 1969; Salkoff and Tanouye, 1986). Plant VG K+ channel subunits and metazoan Shakers do share a homologous transmembrane core consisting of a voltage sensor domain (VSD) and a pore domain (PD) that constitutes the conduction pathway in the assembled tetrameric channel (Figure 1). The VSD consists of four transmembrane domains commonly known as S1-S4 (Long et al., 2005a). In Shaker channels, basic gating charges lining one face of the S4 helix sense the electric field and move the helix outward in response to depolarization (Liman et al., 1991; Aggarwal and MacKinnon, 1996; Long et al., 2005a, 2005b). Acidic countercharges in the other VSD helices stabilize the activated and resting states of the VSD by forming charge clusters with the S4 gating charges that approach them in each state (Seoh et al., 1996; Silverman et al., 2003; Long et al., 2005b). The PD of each subunit consists of two transmembrane domains, S5 and S6, bridged by a selectivity filter motif that forms ion binding sites within the extracellular side of the pore (Doyle et al., 1998; Long et al., 2005a). The S6 domain lines the pore and forms the activation gate at the intracellular vestibule of the pore, while the S5 domain forms the outer surface of the PD facing the VSD (Long et al., 2005a). Biophysical (MacKinnon, 1991; MacKinnon et al., 1993) and later structural (Long et al., 2005a) analyses of metazoan Shaker channels unequivocally demonstrate that Shaker channels function as tetramers with a single PD formed by the S5 and S6 domains from each subunit, surrounded by four independent VSDs (Figure 1).
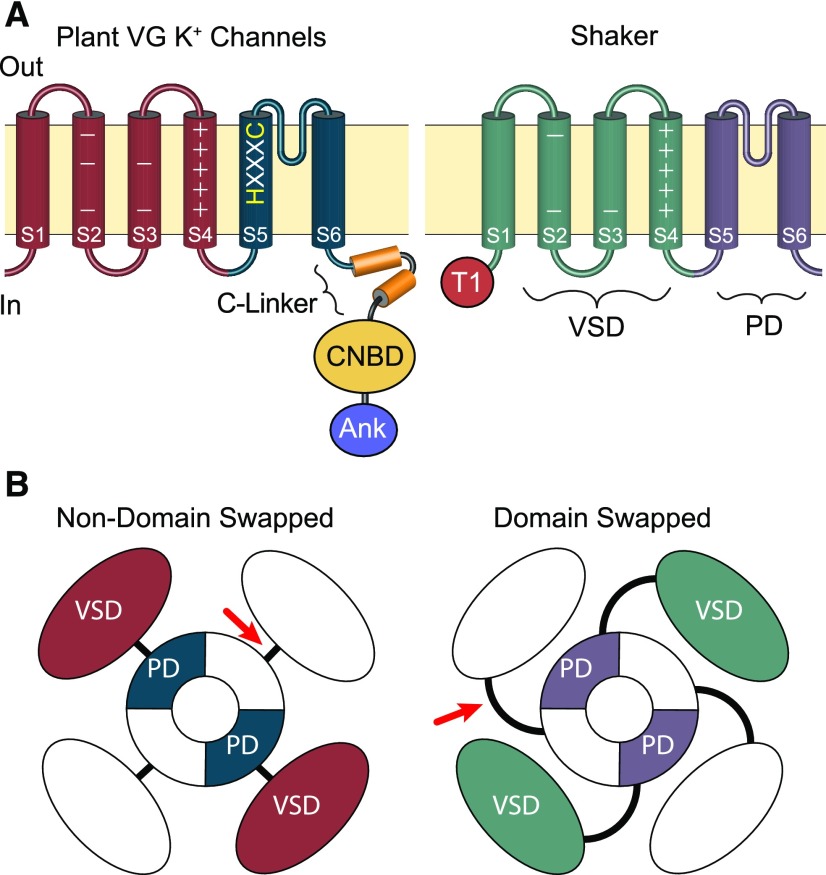
Figure 1.
Comparison of the Domain Architecture of Plant VG K+ Channels and Metazoan Shaker Channels.
(A) Side view schematic drawings of channel subunits. The plasma membrane is represented by the tan horizontal boxes; extracellular and intracellular sides are marked “Out” and “In”; and transmembrane domains within the subunits are depicted with cylinders. In both channels, the S1-S4 transmembrane segments comprise the voltage sensor domain (VSD, marked on Shaker); the S5-S6 transmembrane segments form the channel PD (marked on Shaker); and the extracellular loop between S5 and S6 forms the pore’s selectivity filter. Basic voltage-sensing gating charges reside in S4 and are indicated by + signs. The S5 of plant voltage-gated K+ channels includes an HXXXC amino acid motif that is highly conserved in CNBD superfamily channels but is absent from Shaker channels. Plant VG K+ channels and Shakers also differ in the composition of their cytoplasmic domains. The C terminus of plant VG K+ channels has a CNBD connected to the PD through a conserved helical linker (C-linker). In contrast, Shaker channels have neither a CNBD nor a C-linker and contain a distinctive N-terminal tetramerization domain (T1), which plays a role in subunit assembly. Many plant VG K+ channels also contain a series of Ankyrin repeats (Ank) in the C terminus distal to the CNBD, which are also absent from Shaker channels.
(B) Aerial view—from the extracellular membrane face—of channel tetramers showing the relative positions of VSDs within the channel tetramers. Structural analysis of multiple metazoan CNBD superfamily channels shows the VSD is positioned directly adjacent to the PD from the same subunit and connected by a short S4-S5 linker (red arrow); this arrangement is therefore likely to be conserved in plant VG K+ channels. For Shaker lineage channels, the VSD is domain-swapped and sits nearest the neighboring subunit’s PD, although it still gates the PD from the same subunit through an extended S4-S5 linker (red arrow).
Assumptions of a Shaker-type channel structure were not unreasonable, because plant VG K+ channel subunits share this same transmembrane core, recognizable even with the vast evolutionary distance between metazoans and plants. Furthermore, electrophysiological analyses support their tetramerization (Dreyer et al., 1997; Duby et al., 2008; Lebaudy et al., 2008). However, from the very beginning it was also noted that the plant channels differ from Shakers in other important functional domains. In particular, the plant channels have a cytoplasmic domain in the C terminus homologous to the cyclic nucleotide binding domain (CNBD) (Figure 1) (Sentenac et al., 1992; Talke et al., 2003). The CNBD is appended to a broad functional diversity of eukaryotic cation channels that also share the VSD-PD subunit core, and it has long been recognized that plant VG K+ channels share higher sequence identity with animal CNBD-containing channels than with animal Shakers (Anderson et al., 1992; Hoshi, 1995). Eukaryotic CNBD-containing or “CNBD superfamily” channels differ in their ion selectivity, cyclic nucleotide-sensitivity and voltage-gating (Figure 2); but in all of them, the CNBD interfaces with the transmembrane channel core through a conserved domain known as the C-linker, which forms a flexible gating ring positioned underneath the VSD and PDs (Figure 1). It is now known that the C-linker comprises a major intersubunit interface (Zagotta et al., 2003; Whicher and MacKinnon, 2016), based on crystal and Cryo-EM structures of metazoan HCN and K+ channel CNBD/C-linker complexes, solved decades after the initial observation of CNBD sequence homology in AKT1 (Sentenac et al., 1992).The C-linker plays a key role in transducing conformational changes in the CNBD to the PD to modulate channel gating (Craven and Zagotta, 2006).
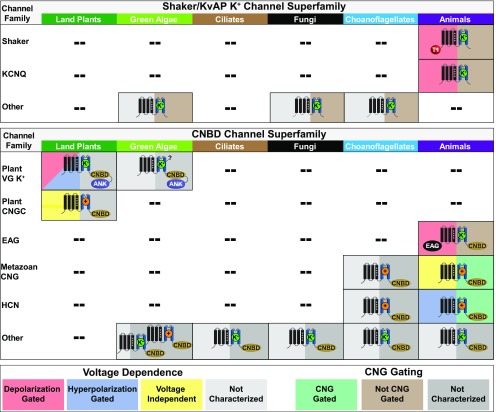
Figure 2.
Major Families of Eukaryotic Shaker/KvAP Superfamily and CNBD Superfamily Channels Listed by Phylogenetic Group.
Characteristic cytoplasmic domains are depicted (Shaker tetramerization domain, T1; CNBD; ankyrin repeat domain, ANK; eag domain, EAG). Selectivity is indicated by the superimposed ion (K+-selective, green, K+; non-selective cation channel, orange, +). Protozoan, fungal and algal channels that have not been functionally expressed are listed as K+-selective or non-selective based on the presence or absence of a canonical K+-selectivity filter sequence. The split color background for each icon indicates voltage dependence on the left (red = depolarization gated; blue = hyperpolarization gated; yellow = voltage-insensitive) and CNBD-mediated cyclic nucleotide gating on the right (green = cyclic nucleotide-gated, brown = not cyclic nucleotide gated). Uncertain voltage- and cyclic nucleotide-gating phenotypes due to lack of conclusive data or lack of functional expression are indicated with gray backgrounds. Note that the plant VG K+ channel family includes both hyperpolarization- and depolarization-gated channels and that direct cyclic nucleotide binding for plant CNGCs is considered an open question. CNBD and Shaker/KvAP superfamily channels that are not members of well-characterized gene families are listed as “Other.” Green algae Shaker/KvAP and CNBD “Other” channels are described in this review in Figures 4 and 5. Remaining “Other” channels are described in the following references: 1) Shaker/KvAP superfamily: choanoflagellates (Li et al., 2015c) and fungi (Prole and Taylor, 2012); 2) CNBD superfamily: choanoflagellates and animals (Fechner et al., 2015), fungi (Avelar et al., 2014), and ciliates (Jegla and Salkoff, 1994, 1995).
In land plants, the CNBD and C-linker are found in all plant VG K+ channels (Nieves-Cordones et al., 2014; Nieves-Cordones and Gaillard, 2014) and are also found in all plant cyclic nucleotide-gated channels (plant CNGCs, a distinct gene family within the CNBD superfamily; Figure 2) (Kohler et al., 1999), which share homology with the plant VG K+ channels but are not K+-selective (Leng et al., 1999; Hua et al., 2003; Ali et al., 2006). In contrast, these domains are not found in true metazoan Shakers (Figures 1 and 2). Thus, despite the fact that evolutionarily distant channels within the CNBD superfamily do not show close conservation from the perspective of biophysical properties and physiological roles, their shared CNBD and C-linker structures suggests a common origin, one that is independent of metazoan Shakers. The next two sections reveal how new whole genome sequences of multiple prokaryotes, particularly within the Archaea, inform on the distinct evolutionary origins of Shaker channels versus CNBD superfamily channels. We then delve deeper into the widening biophysical distinctions between metazoan Shakers and CNBD superfamily channels, as illuminated by recent structural analyses.
ARCHAEAN ORIGIN OF SHAKER CHANNELS AND THE LOSS OF THE SHAKER/KvAP LINEAGE IN LAND PLANTS
Metazoan Shaker channels likely arose from a lineage of prokaryotic VG K+ channels found in the Archaea, the direct ancestors of Eukaryotes (Figure 3). These channels are typified by KvAP (from Archaea, Aeropyrum pernix), the first VG K+ channel structurally characterized (Jiang et al., 2003; Lee et al., 2005); and they contain the canonical VSD and PD, but no other conserved domains. Even in a phylogeny built from the common VSD/PD channel core that is broadly shared, KvAP and metazoan Shakers group together and separately from CNBD superfamily channels (Figure 4). These results suggest that KvAP and true Shakers form a channel lineage that is separate from CNBD superfamily channels and traceable to Archaea. The Archaea channels lack an N-terminal cytoplasmic domain (T1) that aids Shaker channel assembly (Shen and Pfaffinger, 1995; Kreusch et al., 1998) and is a diagnostic feature for the metazoan Shaker family (Li et al., 2015c). The T1 domain appears to have been appended to a VG K+ channel ancestor to form the Shaker family only in metazoans (Li et al., 2015c), and thus true Shakers are limited to metazoans (Figures 2 and 3), even though Shaker lineage channels without the T1 domain have previously been described in choanoflagellates and fungi (Prole and Taylor, 2012; Li et al., 2015c). Therefore, we refer to this lineage as the Shaker/KvAP lineage or superfamily.
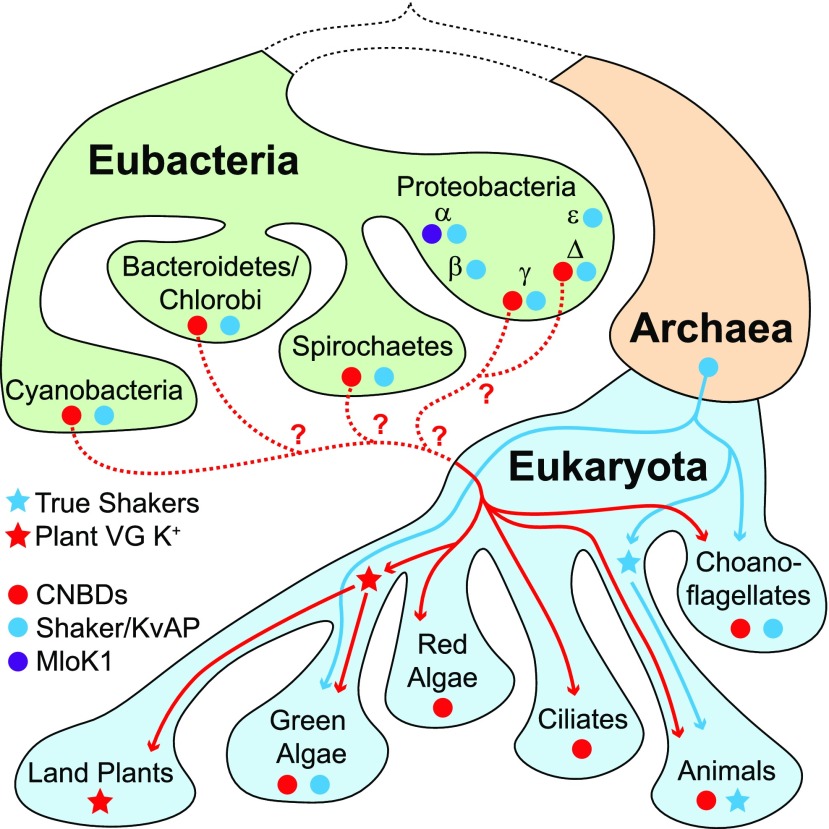
Figure 3.
Proposed Paths for Evolutionary Transfer of CNBD Superfamily and Shaker/KvAP Superfamily Channels from Eubacteria into Extant Eukaryotes Based on Phylogenetic Distribution.
Only the most relevant prokaryotic and eukaryotic clades are depicted, for simplicity. The presence of CNBD and Shaker/KvAP superfamily channels is indicated with red and blue circles, respectively. The origin and distribution of the plant VG K+ channels (red stars) and metazoan Shakers (blue stars) are marked. Red and blue lines indicate probable paths for evolutionary inheritance of CNBD and Shaker/KvAP channels, respectively. We propose that Shaker/KvAP channels were inherited directly from the Archaean ancestor of eukaryotes, while CNBD superfamily channels, which are absent from Archaea, must have been acquired through lateral gene transfer from the Eubacteria, although the specific group of origin is unclear (dotted red lines). MLoK1-like K+ channels (purple circle), which include a CNBD but not the C-linker of CNBD superfamily channels, are restricted to α-proteobacteria and do not appear to have been transferred to eukaryotes.
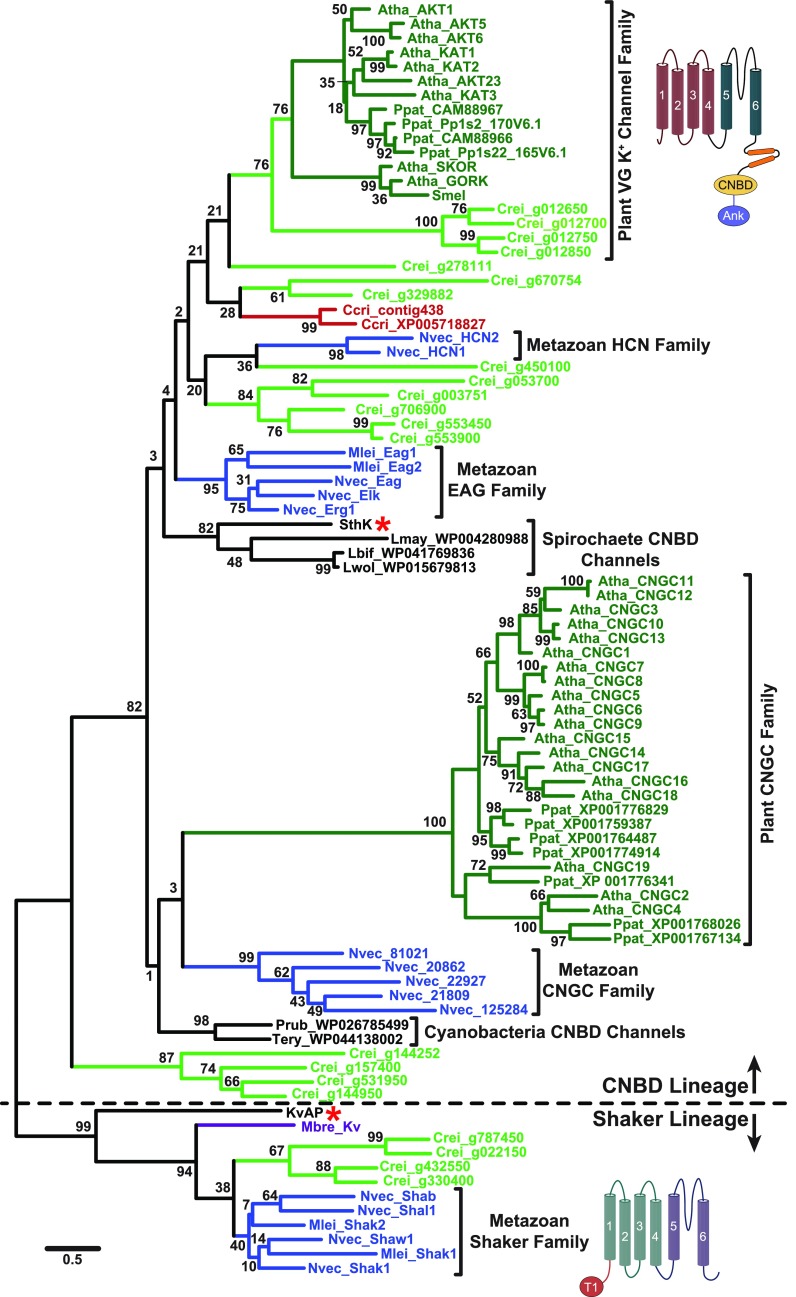
Figure 4.
Maximum Likelihood Phylogeny Supports Separate Prokaryotic Origins for Both Plant VG K+ Channels and Metazoan Shakers.
The phylogeny is based on an alignment of the common VSD-PD transmembrane core shared by channels in the CNBD and Shaker/KvAP superfamilies. It contains algal and other plant channels that have a C-terminal C-linker/CNBD (all group in the CNBD lineage) or no identifiable C-terminal domains (all group in the Shaker lineage; algae only). Structural cartoons next to the plant VG K+ channel clade and the metazoan Shaker clade highlight their distinct domain structure outside this core region. The phylogeny is unrooted but is shown with a root between the CNBD and Shaker/KvAP lineages (separated by dotted line) for display only. The scale bar indicates branch length in substitutions/site, and numbers at nodes indicate % support for the clade in 1000 bootstrap replications. Gene family clades supported by bootstrap analysis are indicated with colored terminal branches (dark green, land plants, Atha, Arabidopsis thaliana, Ppat, Physcomitrella patens, Smel, Selaginella moellendorffii; light green, green algae, Crei, Chlamydomonas reinhardtii; blue, metazoans, MLei, Mnemiopsis leidyi, Nvec, Nematostella vectensis; purple, choanoflagellates, Mbre, Monosiga brevicollis; red, red algae, Ccri, Chondrus crispus; black, eubacteria, Lbif, Leptospira biflexa, Lmay, Leptospira mayottensis, Lwol, Leptospira wolbachii, Prub, Planktothrix rubescens, SthK, Spirochaeta thermophila, Tery, Trichodesmium erythraeum). Gene families discussed in this review are labeled with brackets. Red asterisks mark SthK and KvAP, the prototypical prokaryotic CNBD and Shaker/KvAP lineage channels. Note that most connections between channels from distinct phylogenetic groups within the CNBD superfamily lineage are not supported by bootstrap analysis. Sequences used in the phylogeny were aligned using MUSCLE as implemented in MEGA7 (Kumar et al., 2016) and adjusted by hand as necessary; aligned sequences are presented in the Supplemental Data Set with links to original database sources. The phylogeny was constructed in MEGA7 (Kumar et al., 2016) using Maximum Likelihood methods with an LG substitution matrix (Le and Gascuel, 2008) and a discrete Gamma distribution to model evolutionary rate differences among sites (5 categories, +G = 4.4246); positions with less than 75% sequence coverage were eliminated from the analysis. The phylogeny includes 195 sites, and the tree with the highest log likelihood (-26965.57) is shown.
If plant VG K+ channels are not Shakers, what happened to the Shaker/KvAP lineage during plant evolution? Early eukaryotic ancestors of plants presumably inherited Shaker/KvAP-like channels from Archaea and phylogenetic analysis suggests that the lineage persisted into the green algae. Four channels in the green alga Chlamydomonas that contain the VSD/PD core but lack the C-linker/CNBD fall into the Shaker/KvAP lineage with metazoan Shakers in our phylogeny (Figure 4). So far, no similar channels have been found in land plants, indicating that the Shaker/KvAP lineage was probably lost prior to the radiation of extant land plants. Although we did not include them in our analysis, green and red algae also contain a third lineage of VSD/PD-containing K+ channels that have C-terminal cytoplasmic Regulate Conductance K+ (RCK) channel domains (Gomez-Porras et al., 2012). RCK-containing K+ channels are present in prokaryotes (Jiang et al., 2001, 2002), although phylogenetic data currently available cannot distinguish whether the algal RCK-containing K+ channels arose from Archaea, Prokaryotes, or both clades. In eukaryotes, RCK-containing K+ channels include metazoan large-conductance calcium-activated K+ channels (Jiang et al., 2001). RCK-containing K+ channels are also found in mosses, but these channels seem to have been lost in vascular plants (Gomez-Porras et al., 2012). It has been proposed (Gomez-Porras et al., 2012) that voltage-gated channel diversity went through an evolutionary bottleneck in the transition to land plants in which only a few of the channel types present in algae survived, with the loss at least partially compensated by subsequent functional diversification of the plant VG K+ channel and plant CNGC families within the plant CNBD superfamily. The tonoplast channel typified by Arabidopsis TPC1, which is a cation channel that contains a tandem repeat of the VSD/PD motif (Guo et al., 2016; Kintzer and Stroud, 2016), is not a close relative of voltage-gated K+ channels and thus is not a subject of this review. It instead falls in a separate eukaryotic family of voltage-gated cation channels containing tandem VSD/PD motifs (Furuichi et al., 2001; Guo et al., 2016; Kintzer and Stroud, 2016) that are closely related to metazoan and fungal voltage-gated Na+ and Ca2+ channels (Liebeskind et al., 2013; Rahman et al., 2014).
PROKARYOTIC ORIGIN OF THE CNBD SUPERFAMILY AND ALGAL ORIGIN OF THE LAND PLANT VOLTAGE-GATED K+ FAMILY
The recent discovery of the Lokiarchaeota, a new group of Archaea with strong affinities to eukaryotes, appears to place the origin of the Eukaryota firmly within the Archaea (Spang et al., 2015). One of the most interesting aspects of CNBD channel evolution is that, despite the ubiquity of these channels among eukaryotes, CNBD superfamily channels were absent from the first Archaea genomes to be sequenced (Kuo et al., 2005) and have still not been found in the Archaea despite a deluge of new genome sequences. CNBD-containing K+ channels typified by MLoK1 were found in α-proteobacteria over a decade ago, but they lack the C-linker (Nimigean et al., 2004). They also lack the signature sequence (HXXXC) found in the S5 of most eukaryotic CNBD superfamily channels, including those of land plants (Figure 5B), making their relationship to the eukaryotic CNBD family unclear (Figures 5A and 5B). However, more recently, K+ channels with the C-linker/CNBD and HXXXC have been identified in diverse eubacteria groups, typified by SthK, a cAMP-activated K+ channel cloned from the spirochaete Spirochaeta thermophila (Brams et al., 2014) (Figures 5A and 5B). Eukaryotes therefore probably acquired the CNBD superfamily by lateral gene transfer from eubacteria, although the specific eubacterial group of origin is not clear from phylogenetic analysis (Figures 3 and 4). A detailed analysis of the genome of an extremophile red alga, Galderia sulphuraria, supports the idea that lateral gene transfer from prokaryotes to eukaryotes occurs on a regular basis (Schönknecht et al., 2013). Eukaryotic CNBD gene families originally arose in a single major eukaryotic lineage (Figure 4), indicating that most of the functional diversification of the CNBD superfamily happened after the radiation of extant eukaryotic lineages, including plants and animals (Jegla and Salkoff, 1994, 1995; Zelman et al., 2012; Baker et al., 2015; Li et al., 2015a).
Figure 5.
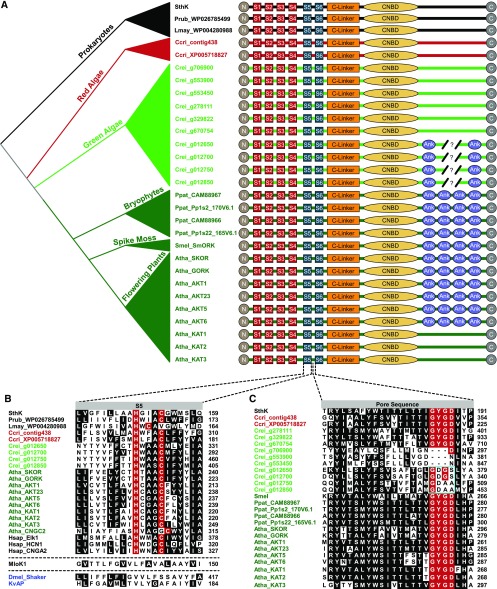
Subunit Structure, S5 Transmembrane Domain and Pore Sequence for Plant VG K+ Channels and Select Algal and Prokaryotic CNBD Channels.
(A) Subunit structures compared among prokaryotic CNBD superfamily channels (black, SthK, Spirochaeta thermophila, Lmay, Leptospira mayottensis, Prub, Planktothrix rubescens); predicted CNBD channel sequences from red algae (red,Ccri, Chondrus crispus) and green algae (light green, Crei, Chlamydomonas reinhardtii); and land plants (dark green, Atha, Arabidopsis thaliana, Ppat, Physcomitrella patens, Smel, Selaginella moellendorffii). Amino acid sequences for the channels can be found in the Supplemental Data Set. Evolutionary relationships between the organisms are indicated in the schematic phylogeny at the left margin. Four VSD transmembrane domains (S1-S4) are depicted with red boxes, two pore helices (S5, S6) with blue boxes, the C-linker with an orange rectangle, the CNBD with a yellow ellipse and ankyrin repeats (Ank) with purple elipses. VG K+ channels in land plants contain 4 or 0 ankyrin repeats, while numbers vary in the four ankyrin repeat-containing gene predictions from green algae (broken line, 4 to 6 repeats) and final numbers have not been confirmed by cloning.
(B) Alignment of the S5 transmembrane domain of the PD from select CNBD family channels from eubacteria, algae, land plants, and metazoans (Hsap_Elk1, NP_653234; Hsap_HCN1, NP_066550; Hsap_CNGA2, NP_005131) illustrates the HXXXC motif characteristic of CNBD channels with MLoK1 (Mesorhizobium loti, WP_010911524), Shaker (Dmel, Drosophila melanogaster, NP_523393), and KvAP provided for comparison. Dotted lines separate MLoK1 and Dmel_Shaker/KvAP to signify they belong to different gene superfamilies. Identical or conservatively substituted residues present in >50% of the sequences are shaded, and the histidine and cysteine of the HXXXC motif are highlighted in red. Note that the cysteine is offset in Arabidopsis CNGC2 and a prokaryotic sequence from Leptospira mayottensis. Sequence positions are listed at the right margin; sequence names (left margin) contain species prefixes and are colored by phylogenetic group as in Figures 4S and 5A.
(C) Amino acid alignment of the pore loop between S5 and S6 with the canonical K+ channel selectivity filter residues (G-Y/F-G-D/N) highlighted in red. Residues conserved or conservatively substituted in >50% of the sequences at other positions are shaded black. Note the lack of conservation in the filter sequence in the four ankyrin repeat-containing green algae orthologs of the plant VG K+ channels (cyan box), suggesting they might not be K+-selective. Sequence names (left) are colored by phylogenetic group as in (A) and (B), and amino acid positions are given at the right margin. Sequences used in the alignments of (B) and (C) can be found in the Supplemental Data Set or have the accession number listed in the figure or legend.
In keeping with the previously noted observations that CNBD family channels diversified separately in the major eukaryotic groups, plant VG K+ channels and plant CNGC channels have no close relatives outside the Plantae (Figure 4). The best clues to understanding the origins of these embryophyte channels can thus be gained by looking at algae. One unique feature of the plant VG K+ channel family is that most members have ankyrin repeats in the cytoplasmic C terminus downstream of the CNBD (Sentenac et al., 1992) (Figures 1 and 5A), but so far no known CNBD family channels outside the plant lineage contain these ankyrin repeats. Four of 16 CNBD family channels found in the green alga Chlamydomonas rheinhardii genome (Merchant et al., 2007) also have the ankyrin repeats (Figure 5A) and also group with embryophyte VG K+ channels in phylogenetic analysis (Figure 4), suggesting that the plant VG K+ channels can be traced back as far as green algae as an independent lineage. As we did not find similar ankyrin repeat-containing CNBD channels in genomes of the red algae Chondrus crispus and Galdieria sulphuraria, further analysis of diverse algal genomes will be needed to determine more precisely when ankyrin repeats were initially appended to a CNBD K+ channel. Interestingly, the absence of the characteristic ankyrin repeats in some embryophyte VG K+ channels (e.g., KAT1, KAT2, and KAT3 in Arabidopsis; Table 1, Figure 5A), seems to represent a recent domain loss—given that the ankyrin repeats are present in the green algae orthologs shown here (Figures 4 and 5A) and in mosses (Gomez-Porras et al., 2012).
One of the most interesting features of many Chlamydomonas CNBD superfamily channels, including the four ankyrin repeat-containing Chlamydomonas channels that group with the plant VG K+ channels (Figures 4 and 5A), is that their pore is missing the canonical K+ selectivity filter sequence (Figure 6). If they are not K+-selective, it could suggest that plant VG K+ channels evolved from non-selective channels, or that K+-selectivity was lost specifically in these extant green algae orthologs after green algae diverged from land plants. We favor the latter explanation because it assumes a K+-selective common ancestor and thus does not invoke de novo evolution of K+ selectivity, which has not yet been observed in any eukaryotic taxa.
Figure 6.
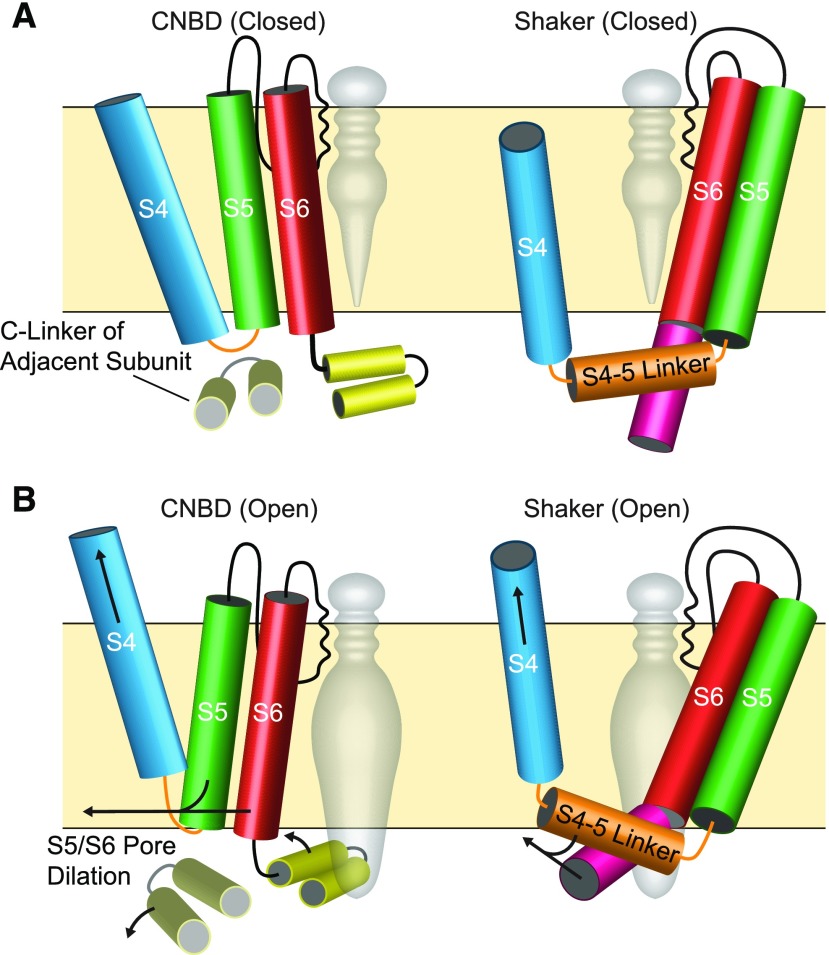
Differences in Voltage-Gating between CNBD and Shaker Superfamily Channels Illustrated with Structural Cartoons.
(A) Diagrams of closed ion channels shown through the plane of the membrane with one of four subunits illustrated and the central pore cavity shown with a gray space fill. Helices depicted as cylinders and subunit features are color-coded as follows: the S4 gating helix (blue), S4-S5 Linker (orange), S5 outer pore helix (green), S6 inner pore helix (red), C-linker (gold, CNBD only), and post-S6 helix (pink, Shaker only). VSD helices S1-S3 have been removed for clarity. The selectivity filter (black line) lines the outer pore cavity, and the pore is closed on the intracellular side.
(B) Open configurations of CNBD and Shaker superfamily channels are shown, with hypothesized gating motions that open the intracellular gate, which are depicted with arrows. In the CNBD superfamily channel (left), outward movement of the S4 allows iris-like rotation of the C-linker away from the central axis of the pore and may also allow outward movement of the S5 and S6. Both movements are predicted to dilate the pore at the intracellular gate. The S4-S5 Linker contacts the C-linker of the adjacent subunit, but the S4 contacts the S5 of the same subunit. Note the CNBD superfamily channel cartoons specifically depict a channel opened by depolarization. In the Shaker superfamily channel (right), outward movement of S4 relieves inward pressure on a tight couple formed by the extended S4-S5 linker helix and post-S6 helix. This allows pore opening by outward flex at an intracellular gating hinge within S6.
VSD/PORE COUPLING DIFFERS BETWEEN SHAKER/KVAP AND CNBD SUPERFAMILY CHANNELS
Until recently, the only structures of voltage-gated K+ channels came from the Shaker/KvAP superfamily: KvAP itself (Lee et al., 2005) and a metazoan Shaker channel chimaera (Long et al., 2005a). The structures are assumed to be in an activated conformation because the PD is open and the S4 gating helix of the VSD is in an outward position consistent with the activated conformation (Aggarwal and MacKinnon, 1996; Larsson et al., 1996; Mannuzzu et al., 1996; Ruta et al., 2005). As in all Shaker/KvAP channels (Figure 1), the VSDs are domain-swapped in these structures, meaning the VSD of one subunit sits next to the PD of a neighboring subunit. It had previously been assumed that CNBD superfamily channels, including plant VG K+ channels, would share the VSD domain swapping and VSD-PD coupling mechanism found in Shaker channels (Grabe et al., 2007). Cryo-EM structures have only recently become available for channels in all three of the metazoan CNBD channel families (EAG, HCN, and CNG; Figure 2). They all share important features in VSD arrangement and VSD-PD coupling that distinguish them from channels in the Shaker/KvAP superfamily (Whicher and MacKinnon, 2016; Lee and MacKinnon, 2017; Li et al., 2017; Wang and MacKinnon, 2017). Note that the S4-S5 linker in these CNBD channels is not a long alpha helix as in Shaker/KvAP channels. It is instead a minimalistic elbow between S4 and S5 (Figure 6). The VSDs and PDs are therefore not domain-swapped and there is no direct helical couple between the S4-S5 linker and the S6 activation gate (Wang and MacKinnon, 2017). Instead, the VSDs sit in close apposition to PDs from the same subunit (Figures 1 and 6). This difference in VSD arrangement explains previous structure-function studies. Those studies showed (based on the gating behavior of mutagenized channels) close proximity between the extracellular ends of S4 from the VSD and S5 from the PD within the same subunit for KAT1 (Lai et al., 2005; Grabe et al., 2007), and for HCN1 from metazoans (Bell et al., 2009). Non-domain-swapped VSDs, a short non-helical S4-S5 linker, and coupling between the VSD and C-linker have now also been observed in a prokaryotic CNBD superfamily member, specifically in the spirochaete Leptospira licerasiae (James et al., 2017). The most parsimonious conclusions are that these features were inherited from the prokaryotic CNBD channels and are present in all eukaryotic CNBD family channels, including the plant VG K+ channels. This difference in VSD arrangement means that CNBD and Shaker/KvAP channels have substantially different interfaces between the VSD and PD within the plane of the membrane.
SUMMARY AND FUTURE DIRECTIONS
In summary, both phylogenetic and structural analyses confirm that it is time to retire the convenient yet incorrect “plant Shaker” designation for the embyrophyte channels listed in Table 1 and their orthologs in other plant species. “Plant VG K+ channels” is a simple, evolutionarily accurate and biophysically correct alternative that we offer and hope the field will adopt, even though it does not directly address the CNBD superfamily lineage of these channels.
Modern structure/function analyses of voltage-gating in plant VG K+ channels with models based on the newly-discovered and unexpected nondomain-swapped structure for CNBD superfamily channels could provide a framework for fresh insights into voltage-gating. A major contrast between CNBD superfamily channels and Shaker KvAP superfamily channels is that while all Shaker/KvAP channels are depolarization-gated, the CNBD superfamily channels (including plant VG K+ channels) can be either hyperpolarization-gated or depolarization-gated, despite an identical direction of voltage-sensor movements in both types of channels (Männikkö et al., 2002; Latorre et al., 2003). Outward rectification of K+ channels appears to have evolved independently in plants and metazoans (Riedelsberger et al., 2015). Structure-function and mutagenesis studies of plant VG K+ channels could therefore address whether multiple mechanisms have evolved in CNBD superfamily channels to establish inward and outward rectification. Mutagenesis of plant VG K+ channels has demonstrated the ability to alter the voltage-gating phenotype of SKOR (Porée et al., 2005; Li et al., 2008; Gajdanowicz et al., 2009; Riedelsberger et al., 2010), but the native mechanism(s) for the reversed voltage-dependence of some CNBD superfamily channels remains to be determined. Renewed analysis of the plant VG K+ channels could play an important role in resolving the issue. For instance, analysis of the HCN1 structure led to a proposal that a longer S4 could underlie reversed polarity voltage gating (Lee and MacKinnon, 2017). However, this hypothesis is unlikely to provide a universal explanation for hyperpolarization-gating in the CNBD superfamily, because the depolarization-gated (GORK and SKOR) and hyperpolarization-gated (KAT1, for example) plant VG K+ channels (Table 1) have identical length S4s and S4-S5 linkers.
The role of the C-linker/CNBD in ligand-modulation of plant VG K+ channel gating also deserves more attention. It is quite interesting that structure/function studies of EAG family K+ channels reveal that the C-linker/CNBD complex retains a central role in regulating voltage gating even in channels that do not bind cyclic nucleotides. EAG family channel gating can be regulated by competition for the CNBD pocket between flavonoids and an intrinsic protein loop (Brelidze et al., 2012, 2013; Carlson et al., 2013a; Dai et al., 2018), and flavonoids also regulate the gating of HCN family channels (Carlson et al., 2013b). The abundance of flavonoids in plants (Winkel-Shirley, 2001; Mouradov and Spangenberg, 2014) raises the interesting possibility that flavonoids could be native ligands other than cyclic nucleotides for the CNBD pocket of plant VG K+ channels or plant CNGCs. The role of phosphoinositides also deserves more attention. For example, PIP2 and PIP3 regulate C-linker/CNBD-dependent gating of the EAG family channel Elk1 (Li et al., 2015b). Furthermore, PIP2 has been found to be essential for the maintenance of KAT1 and SKOR activity in isolated membranes. These findings suggest an important native role of phosphoinositides for plant channels as well (Liu et al., 2005; Wigoda et al., 2010).
There are many other pieces of the plant CNBD channel superfamily puzzle left to assemble. Availability and phylogenetic analysis of additional land plant and algal genomes may help to clarify the evolutionary origins of both the plant VG K+ channels and the embryophyte CNG channels. Functional expression of various CNBD channels from algae could prove a key step in determination of how the characteristic biophysical phenotypes of embryophyte CNBD superfamily channels evolved. Availability of cryoEM or crystal structures of plant VG K+ channels as well as other plant cation channels would greatly advance our knowledge of their gating properties and serve as a guide for structure/function analysis. Structural analysis combined with site-directed mutagenesis and quantitative biochemical and electrophysiological assessment of binding of cyclic nucleotides to both embryophyte CNGC channels and plant VG K+ channelscould definitively answer whether the observed effects of cyclic nucleotides on the gating of these channels (Hoshi, 1995; Gaymard et al., 1996; Talke et al., 2003; Lemtiri-Chlieh and Berkowitz, 2004; Zelman et al., 2012; DeFalco et al., 2016) are mediated directly through the CNBD ligand pocket. Given the low cost of genome sequencing and tools such as cryoEM, the time is right for combined structural, evolutionary, and electrophysiological approaches to yield fresh insights into plant ion channel function.
Supplemental Data
Supplemental Data Set. Source and amino acid sequences for the ankyrins used in phylogenetic analysis.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
This work was supported by the U.S. National Science Foundation IOS (16–21027 to T.J. and S.M.A.).
AUTHOR CONTRIBUTIONS
T.J. and S.M.A. conceived of the review and wrote the text, T.J. analyzed data and contributed figures, and G.B. contributed figures and text.
References
- Aggarwal S.K., MacKinnon R. (1996). Contribution of the S4 segment to gating charge in the Shaker K+ channel. Neuron 16: 1169–1177. [DOI] [PubMed] [Google Scholar]
- Ali R., Zielinski R.E Berkowitz G.A. (2006). Expression of plant cyclic nucleotide-gated cation channels in yeast. J. Exp. Bot. 57: 125–138. [DOI] [PubMed] [Google Scholar]
- Anderson J.A., Huprikar S.S., Kochian L.V., Lucas W.J. Gaber R.F. (1992). Functional expression of a probable Arabidopsis thaliana potassium channel in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 89: 3736–3740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avelar G.M., Schumacher R.I., Zaini P.A., Leonard G., Richards T.A. Gomes S.L. (2014). A rhodopsin-guanylyl cyclase gene fusion functions in visual perception in a fungus. Curr. Biol. 24: 1234–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker E.C., Layden M.J., van Rossum D.B., Kamel B., Medina M., Simpson E. Jegla T. (2015). Functional characterization of cnidarian HCN channels points to an early evolution of Ih. PLoS One 10: e0142730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell D.C., Turbendian H.K., Valley M.T., Zhou L., Riley J.H., Siegelbaum S.A., Tibbs G.R. (2009). Probing S4 and S5 segment proximity in mammalian hyperpolarization-activated HCN channels by disulfide bridging and Cd2+ coordination. Pflugers Arch. 458: 259–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brams M., Kusch J., Spurny R., Benndorf K. Ulens C. (2014). Family of prokaryote cyclic nucleotide-modulated ion channels. Proc. Natl. Acad. Sci. USA 111: 7855–7860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brelidze T.I., Carlson A.E., Sankaran B. Zagotta W.N. (2012). Structure of the carboxy-terminal region of a KCNH channel. Nature 481: 530–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brelidze T.I., Gianulis E.C., DiMaio F., Trudeau M.C. Zagotta W.N. (2013). Structure of the C-terminal region of an ERG channel and functional implications. Proc. Natl. Acad. Sci. USA 110: 11648–11653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson A.E., Brelidze T.I., and Zagotta W.N. (2013a). Flavonoid regulation of EAG1 channels. J. Gen. Physiol. 141: 347–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson A.E., Rosenbaum J.C., Brelidze T.I., Klevit R.E. Zagotta W.N. (2013b). Flavonoid regulation of HCN2 channels. J. Biol. Chem. 288: 33136–33145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craven K.B., Zagotta W.N. (2006). CNG and HCN channels: Two peas, one pod. Annu. Rev. Physiol. 68: 375–401. [DOI] [PubMed] [Google Scholar]
- Dai G., James Z.M. Zagotta W.N. (2018). Dynamic rearrangement of the intrinsic ligand regulates KCNH potassium channels. J. Gen. Physiol. 150: 625–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeFalco T.A., Moeder W. Yoshioka K. (2016). Opening the gates: Insights into cyclic nucleotide-gated channel-mediated signaling. Trends Plant Sci. 21: 903–906. [DOI] [PubMed] [Google Scholar]
- Doyle D.A., Morais Cabral J., Pfuetzner R.A., Kuo A., Gulbis J.M., Cohen S.L., Chait B.T., MacKinnon R. (1998). The structure of the potassium channel: Molecular basis of K+ conduction and selectivity. Science 280: 69–77. [DOI] [PubMed] [Google Scholar]
- Dreyer I., Antunes S., Hoshi T., Müller-Röber B., Palme K., Pongs O., Reintanz B., Hedrich R. (1997). Plant K+ channel alpha-subunits assemble indiscriminately. Biophys. J. 72: 2143–2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duby G., Hosy E., Fizames C., Alcon C., Costa A., Sentenac H., Thibaud J.B. (2008). AtKC1, a conditionally targeted Shaker-type subunit, regulates the activity of plant K+ channels. Plant J. 53: 115–123. [DOI] [PubMed] [Google Scholar]
- Fechner S., Alvarez L., Bönigk W., Müller A., Berger T.K., Pascal R., Trötschel C., Poetsch A., Stölting G., Siegfried K.R., Kremmer E., Seifert R., et al. (2015). A K+-selective CNG channel orchestrates Ca2+ signalling in zebrafish sperm. eLife 4:. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuichi T., Cunningham K.W., Muto S. (2001). A putative two pore channel AtTPC1 mediates Ca2+ flux in Arabidopsis leaf cells. Plant Cell Physiol. 42: 900–905. [DOI] [PubMed] [Google Scholar]
- Gajdanowicz P., Garcia-Mata C., Gonzalez W., Morales-Navarro S.E., Sharma T., González-Nilo F.D., Gutowicz J., Mueller-Roeber B., Blatt M.R., Dreyer I. (2009). Distinct roles of the last transmembrane domain in controlling Arabidopsis K+ channel activity. New Phytol. 182: 380–391. [DOI] [PubMed] [Google Scholar]
- Gaymard F., Cerutti M., Horeau C., Lemaillet G., Urbach S., Ravallec M., Devauchelle G., Sentenac H., Thibaud J.B. (1996). The baculovirus/insect cell system as an alternative to Xenopus oocytes. First characterization of the AKT1 K+ channel from Arabidopsis thaliana. J. Biol. Chem. 271: 22863–22870. [DOI] [PubMed] [Google Scholar]
- Gomez-Porras J.L., Riaño-Pachón D.M., Benito B., Haro R., Sklodowski K., Rodríguez-Navarro A., Dreyer I. (2012). Phylogenetic analysis of K+ transporters in bryophytes, lycophytes, and flowering plants indicates a specialization of vascular plants. Front. Plant Sci. 3:. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabe M., Lai H.C., Jain M., Jan Y.N. Jan L.Y. (2007). Structure prediction for the down state of a potassium channel voltage sensor. Nature 445: 550–553. [DOI] [PubMed] [Google Scholar]
- Guo J., Zeng W., Chen Q., Lee C., Chen L., Yang Y., Cang C., Ren D. Jiang Y. (2016). Structure of the voltage-gated two-pore channel TPC1 from Arabidopsis thaliana. Nature 531: 196–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshi T. (1995). Regulation of voltage dependence of the KAT1 channel by intracellular factors. J. Gen. Physiol. 105: 309–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua B.G., Mercier R.W., Leng Q. Berkowitz G.A. (2003). Plants do it differently. A new basis for potassium/sodium selectivity in the pore of an ion channel. Plant Physiol. 132: 1353–1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James Z.M., Borst A.J., Haitin Y., Frenz B., DiMaio F., Zagotta W.N. Veesler D. (2017). CryoEM structure of a prokaryotic cyclic nucleotide-gated ion channel. Proc. Natl. Acad. Sci. USA 114: 4430–4435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jegla T., Salkoff L. (1994). Molecular evolution of K+ channels in primitive eukaryotes. Soc. Gen. Physiol. Ser. 49: 213–222. [PubMed] [Google Scholar]
- Jegla T., Salkoff L. (1995). A multigene family of novel K+ channels from Paramecium tetraurelia. Receptors Channels 3: 51–60. [PubMed] [Google Scholar]
- Jiang Y., Pico A., Cadene M., Chait B.T., MacKinnon R. (2001). Structure of the RCK domain from the E. coli K+ channel and demonstration of its presence in the human BK channel. Neuron 29: 593–601. [DOI] [PubMed] [Google Scholar]
- Jiang Y., Lee A., Chen J., Cadene M., Chait B.T. MacKinnon R. (2002). Crystal structure and mechanism of a calcium-gated potassium channel. Nature 417: 515–522. [DOI] [PubMed] [Google Scholar]
- Jiang Y., Lee A., Chen J., Ruta V., Cadene M., Chait B.T., MacKinnon R. (2003). X-ray structure of a voltage-dependent K+ channel. Nature 423: 33–41. [DOI] [PubMed] [Google Scholar]
- Kamb A., Iverson L.E. Tanouye M.A. (1987). Molecular characterization of Shaker, a Drosophila gene that encodes a potassium channel. Cell 50: 405–413. [DOI] [PubMed] [Google Scholar]
- Kaplan W.D., Trout W.E., III (1969). The behavior of four neurological mutants of Drosophila. Genetics 61: 399–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kintzer A.F., Stroud R.M. (2016). Structure, inhibition and regulation of two-pore channel TPC1 from Arabidopsis thaliana. Nature 531: 258–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler C., Merkle T. Neuhaus G. (1999). Characterisation of a novel gene family of putative cyclic nucleotide- and calmodulin-regulated ion channels in Arabidopsis thaliana. Plant J. 18: 97–104. [DOI] [PubMed] [Google Scholar]
- Kreusch A., Pfaffinger P.J., Stevens C.F. Choe S. (1998). Crystal structure of the tetramerization domain of the Shaker potassium channel. Nature 392: 945–948. [DOI] [PubMed] [Google Scholar]
- Kumar S., Stecher G. Tamura K. (2016). MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for bigger datasets. Mol. Biol. Evol. 33: 1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo M.M., Haynes W.J., Loukin S.H., Kung C., Saimi Y. (2005). Prokaryotic K+ channels: From crystal structures to diversity. FEMS Microbiol. Rev. 29: 961–985. [DOI] [PubMed] [Google Scholar]
- Lai H.C., Grabe M., Jan Y.N. Jan L.Y. (2005). The S4 voltage sensor packs against the pore domain in the KAT1 voltage-gated potassium channel. Neuron 47: 395–406. [DOI] [PubMed] [Google Scholar]
- Larsson H.P., Baker O.S., Dhillon D.S., Isacoff E.Y. (1996). Transmembrane movement of the shaker K+ channel S4. Neuron 16: 387–397. [DOI] [PubMed] [Google Scholar]
- Latorre R., Olcese R., Basso C., Gonzalez C., Munoz F., Cosmelli D., Alvarez O. (2003). Molecular coupling between voltage sensor and pore opening in the Arabidopsis inward rectifier K+ channel KAT1. J. Gen. Physiol. 122: 459–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le S.Q., Gascuel O. (2008). An improved general amino acid replacement matrix. Mol. Biol. Evol. 25: 1307–1320. [DOI] [PubMed] [Google Scholar]
- Lebaudy A., Hosy E., Simonneau T., Sentenac H., Thibaud J.B., Dreyer I. (2008). Heteromeric K+ channels in plants. Plant J. 54: 1076–1082. [DOI] [PubMed] [Google Scholar]
- Lee C.H., MacKinnon R. (2017). Structures of the human HCN1 hyperpolarization-activated channel. Cell 168: 111–120.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.Y., Lee A., Chen J., MacKinnon R. (2005). Structure of the KvAP voltage-dependent K+ channel and its dependence on the lipid membrane. Proc. Natl. Acad. Sci. USA 102: 15441–15446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemtiri-Chlieh F., Berkowitz G.A. (2004). Cyclic adenosine monophosphate regulates calcium channels in the plasma membrane of Arabidopsis leaf guard and mesophyll cells. J. Biol. Chem. 279: 35306–35312. [DOI] [PubMed] [Google Scholar]
- Leng Q., Mercier R.W., Yao W. Berkowitz G.A. (1999). Cloning and first functional characterization of a plant cyclic nucleotide-gated cation channel. Plant Physiol. 121: 753–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Liu K., Hu Y., Li D., Luan S. (2008). Single mutations convert an outward K+ channel into an inward K+ channel. Proc. Natl. Acad. Sci. USA 105: 2871–2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Zhou X., Wang S., Michailidis I., Gong Y., Su D., Li H., Li X. Yang J. (2017). Structure of a eukaryotic cyclic-nucleotide-gated channel. Nature 542: 60–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Anishkin A., Liu H., van Rossum D.B., Chintapalli S.V., Sassic J.K., Gallegos D., Pivaroff-Ward K., Jegla T. (2015b). Bimodal regulation of an Elk subfamily K+ channel by phosphatidylinositol 4,5-bisphosphate. J. Gen. Physiol. 146: 357–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Liu H., Chu Luo J., Rhodes S.A., Trigg L.M., van Rossum D.B., Anishkin A., Diatta F.H., Sassic J.K., Simmons D.K., Kamel B., Medina M., et al. (2015c). Major diversification of voltage-gated K+ channels occurred in ancestral parahoxozoans. Proc. Natl. Acad. Sci. USA 112: E1010–E1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Martinson A.S., Layden M.J., Diatta F.H., Sberna A.P., Simmons D.K., Martindale M.Q., Jegla T.J. (2015a). Ether-à-go-go family voltage-gated K+ channels evolved in an ancestral metazoan and functionally diversified in a cnidarian-bilaterian ancestor. J. Exp. Biol. 218: 526–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebeskind B.J., Hillis D.M. Zakon H.H. (2013). Independent acquisition of sodium selectivity in bacterial and animal sodium channels. Curr. Biol. 23: R948–R949. [DOI] [PubMed] [Google Scholar]
- Liman E.R., Hess P., Weaver F., Koren G. (1991). Voltage-sensing residues in the S4 region of a mammalian K+ channel. Nature 353: 752–756. [DOI] [PubMed] [Google Scholar]
- Liu K., Li L., Luan S. (2005). An essential function of phosphatidylinositol phosphates in activation of plant shaker-type K+ channels. Plant J. 42: 433–443. [DOI] [PubMed] [Google Scholar]
- Long S.B., Campbell E.B., Mackinnon R. (2005a). Crystal structure of a mammalian voltage-dependent Shaker family K+ channel. Science 309: 897–903. [DOI] [PubMed] [Google Scholar]
- Long S.B., Campbell E.B. Mackinnon R. (2005b). Voltage sensor of Kv1.2: Structural basis of electromechanical coupling. Science 309: 903–908. [DOI] [PubMed] [Google Scholar]
- MacKinnon R. (1991). Determination of the subunit stoichiometry of a voltage-activated potassium channel. Nature 350: 232–235. [DOI] [PubMed] [Google Scholar]
- MacKinnon R., Aldrich R.W. Lee A.W. (1993). Functional stoichiometry of Shaker potassium channel inactivation. Science 262: 757–759. [DOI] [PubMed] [Google Scholar]
- Männikkö R., Elinder F. Larsson H.P. (2002). Voltage-sensing mechanism is conserved among ion channels gated by opposite voltages. Nature 419: 837–841. [DOI] [PubMed] [Google Scholar]
- Mannuzzu L.M., Moronne M.M. Isacoff E.Y. (1996). Direct physical measure of conformational rearrangement underlying potassium channel gating. Science 271: 213–216. [DOI] [PubMed] [Google Scholar]
- Merchant S.S., Prochnik S.E., Vallon O., Harris E.H., Karpowicz S.J., Witman G.B., Terry A., Salamov A., Fritz-Laylin L.K., Maréchal-Drouard L., Marshall W.F. Qu L.H., et al. (2007). The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 318: 245–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouradov A., Spangenberg G. (2014). Flavonoids: A metabolic network mediating plants adaptation to their real estate. Front. Plant Sci. 5(online), [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieves-Cordones M., Gaillard I. (2014). Involvement of the S4-S5 linker and the C-linker domain regions to voltage-gating in plant Shaker channels: Comparison with animal HCN and Kv channels. Plant Signal. Behav. 9: e972892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieves-Cordones M., Chavanieu A., Jeanguenin L., Alcon C., Szponarski W., Estaran S., Chérel I., Zimmermann S., Sentenac H., Gaillard I. (2014). Distinct amino acids in the C-linker domain of the Arabidopsis K+ channel KAT2 determine its subcellular localization and activity at the plasma membrane. Plant Physiol. 164: 1415–1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimigean C.M., Shane T., Miller C. (2004). A cyclic nucleotide modulated prokaryotic K+ channel. J. Gen. Physiol. 124: 203–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papazian D.M., Schwarz T.L., Tempel B.L., Jan Y.N. Jan L.Y. (1987). Cloning of genomic and complementary DNA from Shaker, a putative potassium channel gene from Drosophila. Science 237: 749–753. [DOI] [PubMed] [Google Scholar]
- Porée F., Wulfetange K., Naso A., Carpaneto A., Roller A., Natura G., Bertl A., Sentenac H., Thibaud J.B. Dreyer I. (2005). Plant K(in) and K(out) channels: Approaching the trait of opposite rectification by analyzing more than 250 KAT1-SKOR chimeras. Biochem. Biophys. Res. Commun. 332: 465–473. [DOI] [PubMed] [Google Scholar]
- Prole D.L., Taylor C.W. (2012). Identification and analysis of cation channel homologues in human pathogenic fungi. PLoS One 7: e42404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman T., Cai X., Brailoiu G.C., Abood M.E., Brailoiu E., Patel S. (2014). Two-pore channels provide insight into the evolution of voltage-gated Ca2+ and Na+ channels. Sci. Signal. 7: ra109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedelsberger J., Sharma T., Gonzalez W., Gajdanowicz P., Morales-Navarro S.E., Garcia-Mata C., Mueller-Roeber B., González-Nilo F.D., Blatt M.R. Dreyer I. (2010). Distributed structures underlie gating differences between the kin channel KAT1 and the Kout channel SKOR. Mol. Plant 3: 236–245. [DOI] [PubMed] [Google Scholar]
- Riedelsberger J., Dreyer I., Gonzalez W. (2015). Outward rectification of voltage-gated K+ channels evolved at least twice in life history. PLoS One 10: e0137600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruta V., Chen J., MacKinnon R. (2005). Calibrated measurement of gating-charge arginine displacement in the KvAP voltage-dependent K+ channel. Cell 123: 463–475. [DOI] [PubMed] [Google Scholar]
- Salkoff L.B., Tanouye M.A. (1986). Genetics of ion channels. Physiol. Rev. 66: 301–329. [DOI] [PubMed] [Google Scholar]
- Schönknecht G., Chen W.H., Ternes C.M., Barbier G.G., Shrestha R.P., Stanke M., Bräutigam A., Baker B.J., Banfield J.F., Garavito R.M., Carr K. Wilkerson C., et al. (2013). Gene transfer from bacteria and archaea facilitated evolution of an extremophilic eukaryote. Science 339: 1207–1210. [DOI] [PubMed] [Google Scholar]
- Sentenac H., Bonneaud N., Minet M., Lacroute F., Salmon J.M., Gaymard F. Grignon C. (1992). Cloning and expression in yeast of a plant potassium ion transport system. Science 256: 663–665. [DOI] [PubMed] [Google Scholar]
- Seoh S.A., Sigg D., Papazian D.M., Bezanilla F. (1996). Voltage-sensing residues in the S2 and S4 segments of the Shaker K+ channel. Neuron 16: 1159–1167. [DOI] [PubMed] [Google Scholar]
- Shen N.V., Pfaffinger P.J. (1995). Molecular recognition and assembly sequences involved in the subfamily-specific assembly of voltage-gated K+ channel subunit proteins. Neuron 14: 625–633. [DOI] [PubMed] [Google Scholar]
- Silverman W.R., Roux B., Papazian D.M. (2003). Structural basis of two-stage voltage-dependent activation in K+ channels. Proc. Natl. Acad. Sci. USA 100: 2935–2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spang A., Saw J.H., Jørgensen S.L., Zaremba-Niedzwiedzka K., Martijn J., Lind A.E., van Eijk R., Schleper C., Guy L. Ettema T.J.G. (2015). Complex archaea that bridge the gap between prokaryotes and eukaryotes. Nature 521: 173–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talke I.N., Blaudez D., Maathuis F.J. Sanders D. (2003). CNGCs: Prime targets of plant cyclic nucleotide signalling? Trends Plant Sci. 8: 286–293. [DOI] [PubMed] [Google Scholar]
- Wang W., MacKinnon R. (2017). Cryo-EM structure of the open human ether-à-go-go-related K+ channel hERG. Cell 169: 422–430.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei A., Covarrubias M., Butler A., Baker K., Pak M., Salkoff L. (1990). K+ current diversity is produced by an extended gene family conserved in Drosophila and mouse. Science 248: 599–603. [DOI] [PubMed] [Google Scholar]
- Whicher J.R., MacKinnon R. (2016). Structure of the voltage-gated K+ channel Eag1 reveals an alternative voltage sensing mechanism. Science 353: 664–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigoda N., Ma X., Moran N. (2010). Phosphatidylinositol 4,5-bisphosphate regulates plant K+ channels. Biochem. Soc. Trans. 38: 705–709. [DOI] [PubMed] [Google Scholar]
- Winkel-Shirley B. (2001). Flavonoid biosynthesis. A colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol. 126: 485–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zagotta W.N., Olivier N.B., Black K.D., Young E.C., Olson R. Gouaux E. (2003). Structural basis for modulation and agonist specificity of HCN pacemaker channels. Nature 425: 200–205. [DOI] [PubMed] [Google Scholar]
- Zelman A.K., Dawe A., Gehring C., Berkowitz G.A. (2012). Evolutionary and structural perspectives of plant cyclic nucleotide-gated cation channels. Front. Plant Sci. 3: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]