CRISPR/Cas9-mediated knockout of S2-SLFI of Petunia inflata established the essential role of SLF proteins in self-compatibility and reveals their complex interactions with S-RNases.
Abstract
Self-incompatibility (SI) in Petunia is regulated by a polymorphic S-locus. For each S-haplotype, the S-locus contains a pistil-specific S-RNase gene and multiple pollen-specific S-locus F-box (SLF) genes. Both gain-of-function and loss-of-function experiments have shown that S-RNase alone regulates pistil specificity in SI. Gain-of-function experiments on SLF genes suggest that the entire suite of encoded proteins constitute the pollen specificity determinant. However, clear-cut loss-of-function experiments must be performed to determine if SLF proteins are essential for SI of pollen. Here, we used CRISPR/Cas9 to generate two frame-shift indel alleles of S2-SLF1 (SLF1 of S2-haplotype) in S2S3 plants of P. inflata and examined the effect on the SI behavior of S2 pollen. In the absence of a functional S2-SLF1, S2 pollen was either rejected by or remained compatible with pistils carrying one of eight normally compatible S-haplotypes. All results are consistent with interaction relationships between the 17 SLF proteins of S2-haplotype and these eight S-RNases that had been determined by gain-of-function experiments performed previously or in this work. Our loss-of-function results provide definitive evidence that SLF proteins are solely responsible for SI of pollen, and they reveal their diverse and complex interaction relationships with S-RNases to maintain SI while ensuring cross-compatibility.
INTRODUCTION
Self-incompatibility (SI) is a reproductive strategy widely used by flowering plants producing bisexual flowers to circumvent the tendency to self-fertilize, thereby promoting outcrossing to generate genetic variability (de Nettancourt, 2001). For the Solanaceae, SI is regulated by a polymorphic locus named the S-locus. If the S-haplotype of pollen matches either S-haplotype of the pistil, growth of the pollen tube is inhibited. In Petunia, the S-locus of each haplotype houses a single pistil-specific S-RNase gene (Lee et al., 1994) and a suite of pollen-specific S-locus F-box (SLF) genes (Sijacic et al., 2004; Kubo et al., 2010, 2015; Williams et al., 2014a, 2014b). The polymorphic S-RNase gene is solely responsible for pistil specificity in SI, as has been demonstrated by gain-of-function and loss-of-function experiments (Lee et al., 1994; Murfett et al., 1994). For example, in P. inflata, a wild parent of garden petunia (P. hybrida; Bombarely et al., 2016), expression of S3-RNase (S-RNase of S3-haplotype) in the pistils of S1S2 transgenic plants resulted in the pistil’s gaining the ability to reject S3 pollen. Conversely, expression of an antisense S3-RNase gene in the pistils of S2S3 transgenic plants abolished their ability to reject S3 pollen but did not affect their ability to reject S2 pollen (Lee et al., 1994). Based on pollen transcriptome analysis, both S2-haplotype and S3-haplotype of P. inflata possess the same set of 17 SLF genes, named SLF1 to SLF17 (Williams et al., 2014a). These 17 SLF genes, plus one other type, have been found in eight additional S-haplotypes of P. hybrida, with the number of SLF genes in each of these S-haplotypes ranging from 16 to 18 (Kubo et al., 2015).
S-RNases may act as a cytotoxin to degrade pollen tube RNAs, as their ribonuclease activity is essential for their function in SI (Huang et al., 1994). During initial pollen tube growth in the pistil, S-RNases are taken up by the pollen tube (Luu et al., 2000; Goldraij et al., 2006); however, only self S-RNase (having an S-haplotype matching that of pollen) can inhibit further tube growth to the ovary. A model, named collaborative non-self recognition, was proposed to explain why self S-RNase, but not any non-self S-RNase, inhibits pollen tube growth (Kubo et al., 2010). The model predicts that, for a given S-haplotype, each SLF functions as the F-box protein subunit of an SCF (Skp1–Cullin1– F-box) type E3 ubiquitin ligase to mediate ubiquitination and degradation of the non-self S-RNase(s) with which the SLF interacts. Indeed, all 17 SLF proteins of S2-haplotype and S3-haplotype of P. inflata have been shown to assemble into similar SCF complexes (Li et al., 2014, 2016), with both the Skp1-like and Cullin1 components being pollen specific (named PiSSK1 and PiCUL1-P, respectively). Moreover, S-RNases expressed in Escherichia coli or isolated from pistils have been shown to be ubiquitinated and degraded in pollen extracts in a 26S proteasome-dependent manner (Hua and Kao, 2006; Entani et al., 2014).
The collaborative non-self recognition model further predicts that a complete suite of SLF proteins is required to detoxify all non-self S-RNases to allow cross-compatible pollination, and that none of the SLF proteins can interact with their self S-RNase, allowing it to degrade pollen tube RNAs to result in self-incompatible pollination. The role of SLF genes in Petunia has been examined using an in vivo gain-of-function approach (Sijacic et al., 2004; Hua et al., 2007; Kubo et al., 2010, 2015; Sun and Kao, 2013; Williams et al., 2014b). For example, a pollen-specific promoter of tomato (Solanum lycopersicum), LAT52 (Twell et al., 1990), was used to express green fluorescent protein (GFP)-fused S2-SLF1 in pollen of S2S3 transgenic plants, with the result that S3 transgenic pollen expressing S2-SLF1 was able to successfully pollinate S3-carrying pistils, whereas S2 transgenic pollen expressing S2-SLF1 remained incompatible with S2-carrying pistils (Hua et al., 2007). Thus, expression of S2-SLF1 in S3 pollen allows the transgenic pollen tube to gain the ability to detoxify S3-RNase, suggesting that S2-SLF1 interacts with S3-RNase to mediate its ubiquitination and degradation. We previously used this assay to determine a total of 40 pairwise interaction relationships (indicated by brackets in Table 1) between six SLF proteins of S2-haplotype (S2-SLF1, -SLF3, -SLF4, -SLF5, -SLF6, -SLF8) and eight S-RNases (S2-, S3-, S5-, S6a-, S7-, S11-, S12-, S13-RNase).
Table 1. Summary of Genetic Interaction Relationships among 17 SLF Proteins Produced by S2 Pollen and 9 S-RNases of Petunia inflata.
| S2- RNase | S3- RNase | S5- RNase | S6a- RNase | S7- RNase | S11- RNase | S12- RNase | S13- RNase | S16- RNase | |
|---|---|---|---|---|---|---|---|---|---|
| S2-SLF1 | [—] | [+] | [—] | — | [+] | [—] | + | [+] | — |
| S2-SLF2 | — | — | + | — | |||||
| S2-SLF3 | [—] | [—] | — | — | — | — | — | — | — |
| S2-SLF4 | [—] | [—] | [+] | [—] | [—] | [—] | [—] | [—] | — |
| S2-SLF5 | [—] | [—] | [—] | [—] | [—] | [—] | [+] | [—] | — |
| S2-SLF6 | [—] | [—] | [—] | [—] | [—] | [—] | [—] | [—] | — |
| S2-SLF7 | — | — | — | — | |||||
| S2-SLF8 | [—] | [—] | [—] | [+] | [—] | [—] | [—] | [—] | — |
| S2-SLF9 | — | — | — | — | — | — | — | — | — |
| S2-SLF10 | — | — | — | — | — | — | — | — | — |
| S2-SLF11 | — | — | — | ||||||
| S2-SLF12 | — | — | — | ||||||
| S2-SLF13 | — | — | — | — | |||||
| S2-SLF14 | — | — | — | ||||||
| S2-SLF15 | — | — | — | — | — | — | — | — | — |
| S2-SLF16 | — | — | — | ||||||
| S2-SLF17 | — | — | — |
+ indicates positive interaction, determined in this study; — indicates no interaction, determined in this study; [+] indicates positive interaction, determined in previous studies; [—] indicates no interaction, determined in previous studies; blank indicates relationship not yet determined.
To definitely establish the role of SLF genes in SI, it is imperative that their function also be examined by loss-of-function experiments. For example, if SLF1 of P. inflata is essential for SI of pollen, and if S2-SLF1 is the only SLF of the 17 produced by S2 pollen that can detoxify S3-RNase, then in the absence of a functional S2-SLF1, we would expect S2 pollen to be rejected by normally compatible S3-carrying pistils. We previously used the approach of artificial microRNA (amiRNA) to knock down the expression of S2-SLF1 in S2 pollen and found that the S2 transgenic pollen remained compatible with S3-carrying pistils (Sun and Kao, 2013). These results could be interpreted to mean that SLF1 is not required for SI of pollen. However, we cannot rule out the possibility that a small amount of residual S2-SLF1 produced in S2 transgenic pollen, due to incomplete suppression of the transcript of S2-SLF1, might be responsible for the normal SI phenotype. The results could also be interpreted to mean that at least one of the 11 SLF proteins whose interaction relationship with S3-RNase had not been determined by the gain-of-function assay at that time might also interact with S3-RNase. Thus, to date, definitive evidence for the role of SLF proteins in SI remains lacking.
We had succeeded inusing the polycistronic tRNA-gRNA (PTG)-based CRISPR/Cas9 genome-editing system (Xie et al., 2015) to knock out PiSSK1 (Sun and Kao, 2018). This attainment guided our decision to use this more definitive loss-of-function approach to examine the role of the SLF genes of P. inflata in SI. In the present study, we chose S2-SLF1 as the target of CRISPR/Cas9, as we previously found that, among the 40 interaction relationships established between six SLF proteins and eight S-RNases, only S2-SLF1 interacted with multiple S-RNases: S3-, S7-, and S13-RNase (Sun and Kao, 2013). Thus, knocking out S2-SLF1 would allow us to study the effect on the compatibility of S2 pollen with pistils of different S-haplotypes. We observed that S2* pollen (denoting S2 pollen with an indel allele of S2-SLF1) carrying one of the two frame-shift indel alleles identified was rejected by S3S3 and S13S13 pistils, but remained compatible with S5S5, S6aS6a, S7S7, S11S11, S12S12, and S16S16 pistils. We then used gain-of-function experiments to identify 68 additional interaction relationships between the 17 SLF proteins of S2-haplotype and nine S-RNases (the eight previously studied plus S16-RNase). Based on a total of 108 interaction relationships, we showed that the SI behavior of S2* pollen lacking a functional S2-SLF1 with pistils carrying different S-haplotypes is entirely consistent with whether S2 pollen employs S2-SLF1 as the only SLF in detoxifying a particular S-RNase, whether S2 pollen employs S2-SLF1 and at least one other SLF protein in detoxifying a particular S-RNase, and whether S2 pollen employs SLF protein(s) other than S2-SLF1 in detoxifying a particular S-RNase. Thus, the results of CRISPR/Cas9-mediated knockout of S2-SLF1, coupled with analysis of the SLF-S-RNase interaction relationships, provide definitive evidence that SLF proteins are solely responsible for SI of pollen and reveal the complexity and diversity of the interactions between SLF proteins and S-RNases.
RESULTS
Four Different Indel Alleles of S2-SLF1 Generated by CRISPR/Cas9-Mediated Genome Editing
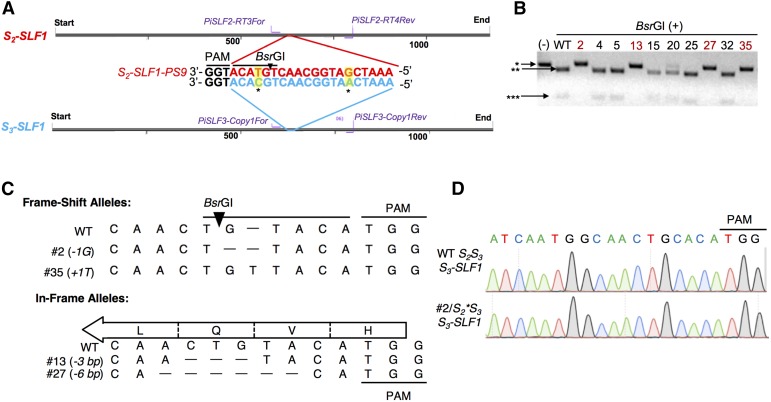
To edit the coding sequence of S2-SLF1 in S2S3 plants without affecting S3-SLF1 (with which it shares 94% nucleotide sequence identity), we designed a guide RNA (gRNA) to target a 20-bp protospacer sequence (624 to 643 bp, counting from the start codon) of the antisense strand of S2-SLF1; this protospacer (named S2-SLF1-PS9) is followed by the protospacer adjacent motif (PAM), TGG (Figure 1A). The corresponding 20-bp sequence in S3-SLF1 contains two nucleotide differences (Figure 1A) and was not expected to be the target of the gRNA. No other potential off-target sites were found in any of the other 16 SLF genes of S2-haplotype and S3-haplotype, as their sequences differed from the protospacer sequence by more than two nucleotides (Supplemental Figure 1). The PTG fragment-containing Ti-plasmid construct (Supplemental Figure 2A) was used in Agrobacterium-mediated transformation of S2S3 plants.
Figure 1.
Generation of S2-SLF1 Indel Alleles by CRISPR/Cas9-Mediated Genome Editing.
(A) Design of a gRNA specifically targeting S2-SLF1. A 20-bp sequence (named S2-SLF1-PS9) of the antisense strand of S2-SLF1 followed by the PAM motif (TGG) was chosen as the protospacer for CRISPR/Cas9; two mismatches (highlighted in yellow and indicated with asterisks) are found in the corresponding 20-bp region in S3-SLF1 (highlighted in blue). “Start” indicates the start codon (ATG) on the sense strand (5′ to 3′) of these two genes, and “End” indicates the stop codon (TAG). The positions of the PCR primers specific to S2-SLF1 (PiSLF2-RT-3For/PiSLF2-RT-4Rev) and those specific to S3-SLF1 (PiSLF3-Copy1For/PiSLF3-Copy1Rev) are indicated by purple lines. Black triangle indicates the cleavage site of BsrGI in the wild-type S2-SLF1 sequence.
(B) PCR-restriction enzyme digestion screen for edited S2-SLF1 alleles in 10 transgenic plants. (-): PCR product amplified from genomic DNA of one of the transgenic plants by the S2-SLF1 specific primers, without digestion by BsrGI. BsrGI (+): BsrGI digestion of the PCR products amplified from genomic DNA of a wild-type S2S3 plant and the 10 transgenic plants. Asterisk (*) indicates the ∼220-bp PCR product resistant to, or not subjected to, BsrGI digestion; double asterisks (**) indicate the ∼180-bp BsrGI fragment; triple asterisks (***) indicates the ∼40-bp BsrGI fragment. The plant numbers of those T0 plants carrying mutant S2-SLF1 alleles resistant to BsrGI digestion are highlighted in red.
(C) Sequences of four indel alleles in the edited region of S2-SLF1. The sequences shown are those of the antisense strand of S2-SLF1 (5′ to 3′ from left to right). The black triangle indicates the cleavage site of BsrGI in the wild-type S2-SLF1 sequence. The open arrow indicates the direction of translation, and the encoded amino acids in the wild-type S2-SLF1 are shown. The 3-bp in-frame deletion in plant #13 abolishes the codon 5′-CAG-3′ for Gln-210. The 6-bp in-frame deletion in plant #27 abolishes the codon for Gln-210 and disrupts the codon 5′-GTA-3′ for Val-209 and the codon 5′-TTG-3′ for Leu-211. However, as the Val codon is restored as 5′-GTG-3′, only Gln-210 and Leu-211 are deleted from the encoded protein.
(D) Sequencing chromatograms of PCR amplicons of S3-SLF1 from T0 plant #2/S2*S3 and from a wild-type S2S3 plant. The sequences are those of the antisense strand from 5′ to 3′ (left to right).
PCR analysis of 36 plants regenerated from transformation showed that 10 carried the 35S:Cas9 transgene (Supplemental Figure 2B; sequences of all primers used in this work are listed in Supplemental Table 1). The wild-type sequence of S2-SLF1-PS9 contains a cleavage site of restriction enzyme BsrGI close to the PAM. The PCR products amplified with a pair of primers specific to S2-SLF1 from T0 plants #2, #13, # 27, and #35 were resistant to digestion, suggesting a loss of this BsrGI cleavage site caused by genome editing (Figure 1B). Sequencing of the PCR products of these four T0 plants revealed the exact sequence of each indel allele of S2-SLF1 in the targeted region (Figure 1C). Genome editing in #2 and #35 resulted in a 1-bp deletion (denoted -1G) and a 1-bp insertion (denoted +1T), respectively, causing frame-shift after the codon for Val-209 (Supplemental Figure 3). Editing in #13 and #27 resulted in 3-bp and 6-bp in-frame deletions, respectively, yielding mutated forms of S2-SLF1 with Gln-210 (Q210) deleted, and with both Q210 and Leu-211 (L211) deleted, respectively (Supplemental Figure 3). To confirm that no off-target editing occurred in S3-SLF1, we sequenced the PCR products (217 bp) amplified from leaf genomic DNA of these four T0 plants and a wild-type S2S3 plant using a pair of primers specific to S3-SLF1 (Figure 1A). The sequences of the four T0 plants were completely identical to the wild-type sequence (Figure 1D).
Analysis of Self-Incompatibility Behavior of S2 Pollen Carrying One of the Two Frame-Shift Indel Alleles of S2-SLF1
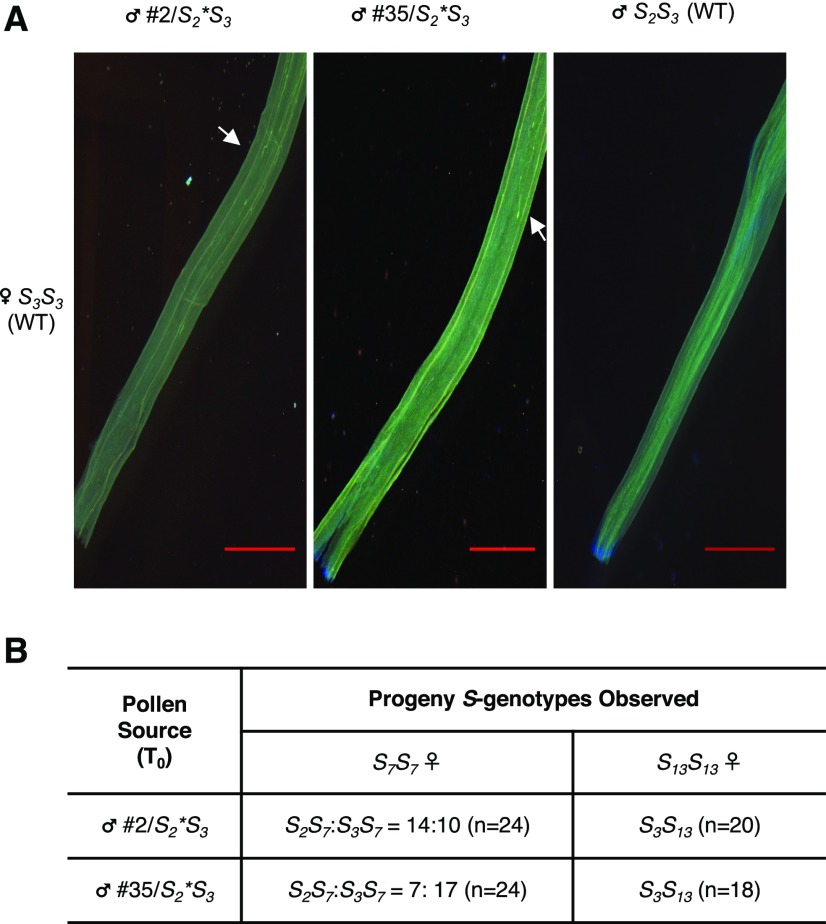
To examine the effect of the two frame-shift indel mutations in S2-SLF1 on the SI behavior of S2* pollen, we first used pollen from T0 plants #2/S2*S3 and #35/S2*S3 to separately pollinate the wild-type S3S3 plants. No fruits were set from these pollinations, and aniline blue staining of pollen tubes inside S3S3 pistils showed few pollen tubes in the bottom segment of the style (Figure 2A). These results suggest that S2* pollen lacking a functional S2-SLF1 cannot detoxify S3-RNase; and that, if SLF proteins are required for SI of pollen, no other SLF proteins produced by S2 pollen can interact with S3-RNase.
Figure 2.
Analysis of SI Behavior of T0 Plants Carrying Either a 1-bp Deletion or a 1-bp Insertion Frame-Shift Allele of S2-SLF1.
(A) Aniline blue staining of pollen tubes in the bottom segment of the style after a wild-type S3S3 plant was separately pollinated by pollen from T0 plants, #2/S2*S3 and #35/S2*S3, and a wild-type S2S3 plant. White arrows indicate where growth of most pollen tubes stopped, in the case of incompatible pollinations. Scale bar = 1 mm.
(B) Progeny analysis of crosses using pollen from #2/S2*S3 or #35/S2*S3 to separately pollinate the wild-type S7S7 and S13S13 pistils. n indicates the number of plants in each progeny analyzed.
We next used pollen from #2/S2*S3 and #35/S2*S3 to separately pollinate the wild-type S7S7 and S13S13 plants. S3 pollen produced by these two transgenic plants should be compatible with both S7S7 and S13S13 pistils to yield S3S7 and S3S13 progeny plants, respectively. As expected, all these pollinations set fruits, and we used PCR to determine the S-genotypes of at least 18 randomly selected plants in each progeny. If S2* pollen remained compatible with S7S7 and S13S13 pistils, we would expect to obtain S2*S7 and S2*S13 progeny plants, respectively. We identified both S2*S7 and S3S7 genotypes in the progeny from crosses with S7S7 pistils (Figure 2B, Supplemental Figure 4A), but we found only S3S13 genotype in the progeny from crosses with S13S13 pistils (Figure 2B, Supplemental Figure 4B). Thus, S2* pollen produced by both #2/S2*S3 and #35/S2*S3 remained compatible with S7-carrying pistils, but it was incompatible with normally compatible S13-carrying pistils.
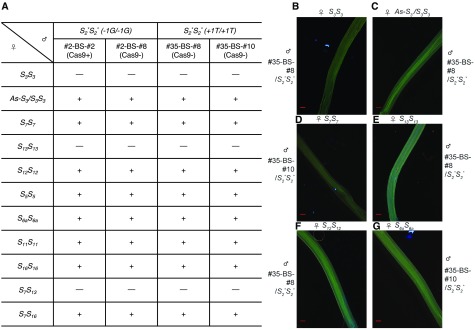
To further examine the effects of the two frame-shift indel mutations of S2-SLF1 on the SI behavior of S2* pollen, we performed bud-selfing (BS, self-pollination of immature flower buds) on #2/S2*S3 and #35/S2*S3 to obtain S2*S2*, plants each carrying one of the indel alleles. BS circumvents SI, because immature buds produce very low levels of S-RNases that are insufficient to inhibit growth of self-pollen tubes (Lee et al., 1994; Sun and Kao, 2013). Two S2*S2* plants in each progeny (#2-BS-#2 and -#8 in the progeny of #2/S2*S3, and #35-BS-#8 and -#10 in the progeny of #35/S2*S3) were identified (Supplemental Figures 5A to 5C). Using a primer pair specific to the Cas9 transgene, we found that only #2-BS-#8 inherited the Cas9 transgene (Supplemental Figure 5D). We used all four BS plants for subsequent analysis to assess the SI behavior of S2* pollen in the absence of S3 pollen. By using the three transgene-free BS plants, we could eliminate any possible complications that might be caused by the presence of the Cas9-containing transgene.
We used pollen from those four BS plants to pollinate the wild-type plants of 10 different S-genotypes: S3S3, S5S5, S6aS6a, S7S7, S11S11, S12S12, S13S13, S16S16, S7S13, and S7S16. For each S-genotype, the same results were obtained for all four BS plants (described below and summarized in Figure 3A). We used aniline blue to stain and visualize pollen tubes in the pollinated pistils of all crosses. Representative results are shown in Figures 3B to 3G.
Figure 3.
Analysis of SI Behavior of S2S2 Plants Homozygous for Either Frame-Shift Indel Allele of S2-SLF1.
(A) Results of pollination using pollen from two bud-selfed (BS) progeny plants of #2/S2*S3 (#2-BS-#2 and #2-BS-#8), and two BS progeny plant of #35/S2*S3 (#35-BS-#8 and #35-BS-#10) to separately pollinate pistils of various S-genotypes. S2*S2* indicates that all four BS progeny plants were S2S2 and homozygous for the indel allele of S2-SLF1 inherited from their respective T0 plants. (Cas9+) and (Cas9-) indicate presence and absence of the Cas9 transgene, respectively, in the BS plants. —: incompatible pollination (no fruit set); +: compatible pollination (fruit set). As-S3/S3S3: a self-compatible transgenic plant not producing any S3-RNase in the pistil due to expression of an antisense S3-RNase gene.
(B) to (G) Aniline blue staining of pollen tubes in the bottom segment of the style of the pistil from each of the wild-type plants of five different S-genotypes, as indicated, and from a transgenic plant As-S3/S3S3. These plants were separately pollinated with pollen from #35-BS-#8 or #35-BS-#10, as indicated. Scale bar = 0.25 mm.
Pollinations with S3S3 pistils were incompatible (Figure 3B), consistent with the results obtained for T0 plants #2/S2*S3 and #35/S2*S3 (Figure 2A). To determine whether the incompatibility of S2*pollen with normally compatible S3S3 pistils was due to the inability of S2* pollen to detoxify S3-RNase, we also used pollen of these four BS plants to pollinate a self-compatible transgenic plant, As-S3/S3S3, whose production of S3-RNase in the pistil is completely suppressed by an antisense S3-RNase transgene (Lee et al., 1994; Sun and Kao, 2013). Pollinations with pistils of As-S3/S3S3 were fully compatible (Figure 3C), setting normal size fruits. Therefore, S2* pollen, which lacks a functional S2-SLF1, was incompatible with the wild-type S3S3 pistils, but it was compatible with As-S3/S3S3 pistils. These results suggest the necessary presence of S2-SLF1 for S2 pollen to detoxify S3-RNase.
Pollinations with S7S7 pistils were compatible (Figure 3D), but pollinations with S13S13 pistils were incompatible (Figure 3E), consistent with progeny analysis of the crosses using pollen from T0 plants #2/S2*S3 and #35/S2*S3 to pollinate the wild-type S7S7 and S13S13 plants (Figure 2B, Supplemental Figure 4). These results together suggest that for S2 pollen, S2-SLF1 is not the only SLF that interacts with S7-RNase, but that S2-SLF1 is the only SLF that interacts with S13-RNase.
All pollinations with S5S5, S6aS6a, S11S11, and S12S12 pistils were compatible (Figures 3A, 3F, and 3G), consistent with our previous findings that (a) S2 pollen did not use S2-SLF1 to interact with S5-RNase or S11-RNase; (b) S2 pollen used S2-SLF4 to interact with S5-RNase, S2-SLF8 to interact with S6a-RNase, and S2-SLF5 to interact with S12-RNase (Table 1; Sun and Kao, 2013; Williams et al., 2014b). Pollinations with S16S16 pistils were also compatible (Figure 3A), suggesting that some other SLF protein(s) produced by S2 pollen is (are) responsible for detoxifying S16-RNase. All these results also suggest that the gRNA used for editing S2-SLF1 does not affect the other SLF genes, or genes involved in pollen development or fertilization.
Consistent with the results described above, pollinations with S7S13 pistils were incompatible, because S2* pollen without a functional S2-SLF1 cannot detoxify S13-RNase. Pollinations with S7S16 pistils were compatible, suggesting that S2* pollen can still use some other SLF protein(s) to detoxify S7-RNase and S16-RNase.
Interaction Relationships between 17 SLF Proteins of S2-Haplotype and Various S-RNases
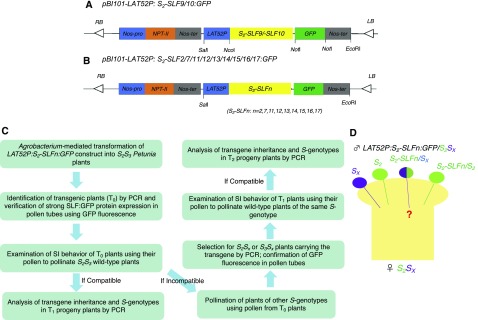
To further examine the results obtained from loss-of-function of S2-SLF1, we used the in vivo gain-of-function assay to determine 68 additional interaction relationships (denoted + or — in Table 1) between the 17 SLF proteins of S2-haplotype and nine S-RNases. These, together with the 40 previously determined (denoted [+] or [—] in Table 1), bring the total of interaction relationships to 108. To perform this assay, we first made transgene constructs for the 11 SLF genes that had not been previously studied, each fused with the coding sequence for GFP (Figures 4A and 4B). We used each construct to generate S2S3 transgenic plants. All the T0 lines of S2S3 used in the gain-of-function assay are listed in Supplemental Table 2, and the workflow of this assay is outlined in Figure 4C. Pollen from at least three transgenic plants found to express a particular GFP-tagged SLF at high levels, based on intensity of GFP fluorescence in the pollen tubes (Supplemental Figure 6), was used to pollinate the wild-type S2S3 plants to examine the interaction relationship between this SLF and S3-RNase. None of the pollinations involving pollen expressing one of the 11 SLF proteins set fruits (Supplemental Table 2). These results (illustrated in Figure 4D) suggest that none of these 11 SLF proteins interact with S3-RNase (denoted — in the “S3-RNase” column of Table 1). Of the six SLF proteins we previously examined, five did not interact with S3-RNase (denoted [—] in the “S3-RNase” column of Table 1; Hua et al., 2007; Williams et al., 2014b). Thus, among the 17 SLF proteins produced in S2 pollen, only S2-SLF1 interacts with S3-RNase, consistent with the finding that S2* pollen carrying one of the frame-shift indel alleles of S2-SLF1 was rejected by S3-carrying pistils. These results also confirmed that none of the 17 SLF proteins of S2-haplotype interact with their self S2-RNase (denoted — or [—] in the “S2-RNase” column of Table 1). These resultswere consistent with the predictionby the collaborative, non-self recognition model, that none of the SLF proteins of a given S-haplotype interact with their self S-RNase (Kubo et al., 2010).
Figure 4.
The in Vivo Gain-of-Function Assay Used for Determining Interaction Relationships between SLF Proteins and S-RNases.
(A) Schematic of the transgene constructs for S2-SLF9 and S2-SLF10.
(B) Schematic of nine additional transgene constructs, each containing one of the SLF genes indicated (denoted S2-SLFn). All constructs shown in (A) and (B) were made using Ti plasmid pBI101 as the backbone. RB, right border of the T-DNA; Nos-pro, promoter of the gene encoding nopaline synthase; NPT-II, gene encoding neomycin phosphotransferase II (conferring resistance to kanamycin); Nos-ter, transcription terminator of the gene encoding nopaline synthase; LAT52P: promoter of the pollen-specific LAT52 gene from tomato; GFP, gene encoding green fluorescent protein; LB, left border of the T-DNA.
(C) Workflow of the in vivo gain-of-function assay.
(D) Graphic illustration of the genetic basis for determining interaction relationships between SLF proteins and S-RNases. The transgene construct for an SLF gene of S2-haplotype, denoted S2-SLFn, is introduced into S2Sx plants, with Sx being an S-haplotype different from S2. Pollen from the LAT52P:S2-SLFn:GFP/S2Sx transgenic plant is used to pollinate a wild-type S2Sx plant. Among the four genotypes of pollen produced by the transgenic plant, S2 and Sx should be rejected by the S2Sx pistil, and S2 carrying the transgene is expected be rejected, as the S2-SLFn transgene is from the same S-haplotype as pollen. Thus, whether or not this pollination is compatible is determined solely by the SI behavior of Sx pollen carrying the transgene. If S2-SLFn interacts with Sx-RNase to mediate its ubiquitination and degradation in the LAT52P:S2-SLFn:GFP/Sx pollen tube, then the pollination should be compatible, and all the progeny will inherit the transgene and carry Sx-haplotype. If S2-SLFn does not interact with Sx-RNase, then the LAT52P:S2-SLFn:GFP/Sx pollen tube should be rejected in the style and the pollination should be incompatible.
We then examined the other 46 of the 68 additional interaction relationships, including those between the 17 SLF proteins of S2-haplotype and seven non-self S-RNases (S5-, S6a-, S7-, S11-, S12-, S13-, and S16-RNase). We used pollen from the S2S3 transgenic plants expressing one of these 17 SLF proteins (denoted S2-SLFn in Figures 4C and 4D) to pollinate plants of appropriate S-genotypes to obtain S2Sx or S3Sx transgenic plants (Sx being S5, S6a, S7, S11, S12, S13, or S16). Subsequently we used pollen produced by these transgenic plants to pollinate the wild-type plants of the same S-genotype to test the interaction relationship between each SLF protein and Sx-RNase. The transgenic lines generated and their SI behavior are summarized in Supplemental Table 2.
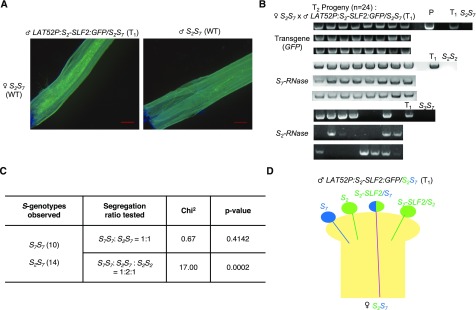
Among the pollinations performed, the only ones that set fruits were (a) pollinations of the wild-type S2S7 plants by pollen of the S2S7 transgenic plants expressing S2-SLF2:GFP (Figure 5A), and (b) pollinations of the wild-type S2S12 plants by pollen of the S2S12 transgenic plants expressing S2-SLF1:GFP (Supplemental Table 2). We then randomly selected 24 progeny plants raised from compatible pollinations involving the wild-type S2S7 and wild-type S2S12 plants and used PCR to analyze segregation of the S-haplotype and SLF transgene in each progeny. Among the progeny from pollinations of the wild-type S2S7 plants by pollen of the S2S7 transgenic plants expressing S2-SLF2:GFP, all carried S7-haplotype and the S2-SLF2:GFP transgene (Figure 5B). The chi-square test supported the 1:1 ratio of S2S7:S7S7, and the 1:2:1 ratio of S2S2:S2S7:S7S7 was rejected with a P-value < 0.05 (Figure 5C). These results suggest that expression of S2-SLF2:GFP causes breakdown of SI in S7 transgenic pollen (Figure 5D). In the progeny from pollinations of the wild-type S2S12 plants by pollen of the S2S12 transgenic plants expressing S2-SLF1:GFP, all carried S12-haplotype and the S2-SLF1:GFP transgene (Figure 6A). The chi-square test supported the 1:1 ratio of S2S12:S12S12, and the 1:2:1 ratio of S2S2:S2S12:S12S12 was rejected with a P-value <0.05 (Figure 6B). These results suggest that expression of S2-SLF1:GFP causes breakdown of SI in S12 transgenic pollen. Thus, in addition to S2-SLF1 and S2-SLF5, which we previously found to interact with S7-RNase and S12-RNase, respectively, S2-SLF2 also interacts with S7-RNase, and S2-SLF1 also interacts with S12-RNase (Table 1). The functional redundancy employed by S2 pollen in detoxifying S7-RNase and S12-RNase is consistent with the finding that S2* pollen lacking a functional S2-SLF1 remained compatible with S7S7 and S12S12 pistils.
Figure 5.
Assessment of Interaction between S2-SLF2 and S7-RNase by in Vivo Gain-of-Function Assay.
(A) Aniline blue staining of pollen tubes in the bottom segment of the style after a wild-type S2S7 plant was self-pollinated (right) and pollinated with pollen from transgenic plant LAT52P:S2-SLF2:GFP/S2S7 (left). This transgenic plant (a T1 plant) was obtained by pollinating a wild-type S7S7 plant with pollen from a T0 transgenic plant LAT52P:S2-SLF2:GFP/S2S3. Scale bar = 1 mm.
(B) Analysis of 24 T2 plants resulting from the cross, S2S7 x LAT52P:S2-SLF2:GFP/S2S7, shown in (A). T1 indicates genomic DNA from the T1 plant LAT52P:S2-SLF2:GFP/S2S7; P indicates plasmid DNA of pBI101-LAT52P:S2-SLF2:GFP (as positive control for the PCR amplification of the GFP transgene). S2S7 indicates genomic DNA from a wild-type S2S7 plant (as negative control for the PCR amplification of the GFP transgene). S2S2 indicates genomic DNA from a wild-type S2S2 plant (as negative control for the PCR amplification of the S7-RNase gene). S3S7 indicates genomic DNA from a wild-type S3S7 plant (as negative control for the PCR amplification of the S2-RNase gene).
(C) Chi-square analysis of the S-haplotype inheritance in the 24 T2 plants analyzed in (B). Chi-square analysis was used to test the null hypothesis of S-haplotype inheritance, 1:1 ratio of S2S7:S7S7 versus 1:2:1 ratio of S2S2:S2S7:S7S7.
(D) Graphic illustration of interpretation of the results of progeny analysis from S2S7 x LAT52P:S2-SLF2:GFP/S2S7 shown in (B). The observation that all progeny plants inherited the GFP transgene and none were S2S2 indicates that only the transgenic S7 pollen carrying the LAT52P:S2-SLF2:GFP transgene can effect fertilization. This result suggests that S2-SLF2 produced in the transgenic S7 pollen interacts with and detoxifies S7-RNase to render the transgenic S7 pollen compatible with the S2S7 pistil.
Figure 6.
Assessment of Interaction between S2-SLF1 and S12-RNase by in Vivo Gain-of-Function Assay.
(A) Analysis of 24 T2 plants resulting from the cross, S2S12 x LAT52P:S2-SLF1:GFP/S2S12. T1 indicates genomic DNA from the T1 plant LAT52P:S2-SLF1:GFP/S2S12; S2S3 indicates genomic DNA from a wild-type S2S3 plant (as negative control for the PCR amplification of the GFP transgene and S12-RNase gene); S3S12 indicates genomic DNA from a wild-type S3S12 plant (as negative control for the PCR amplification of the S2-RNase gene).
(B) Chi-square analysis of the S-haplotype inheritance in the 24 T2 plants analyzed in (A). Chi-square was used to test the null hypothesis of S-haplotype inheritance, 1:1 ratio of S2S12:S12S12 versus 1:2:1 ratio of S2S2:S2S12:S12S12.
As none of the other pollinations set fruits, these results, combined with the previously established 40 interaction relationships, led us to these four conclusions: (a) among the nine SLF proteins examined for their interaction relationships with S5-RNase, S2-SLF4 is the only one that interacts; (b) among the 10 SLF proteins examined for their interaction relationships with S6a-RNase, S2-SLF8 is the only one that interacts; (c) none of the nine SLF proteins examined for their interaction relationships with S11-RNase interact with this S-RNase, and none of the 11 SLF proteins examined for their interaction relationships with S16-RNase interact with this S-RNase; (d) S2-SLF1 is the only one among the nine SLF proteins examined that interacts with S13-RNase (Table 1; Sun and Kao, 2013; Williams et al., 2014b). The findings that S2-SLF1 is not responsible for interacting with S5-, S6a-, S11-, or S16-RNase are consistent with the findings that S2* pollen lacking a functional S2-SLF1 remained compatible with S5-, S6a-, S11-, and S16-carrying pistils. For S11-RNase and S16-RNase, we would predict that at least one of the eight (for S11-RNase) and at least one of the six (for S16-RNase) SLF proteins yet to be examined interact with S11- and S16-RNase, respectively. Moreover, based on the finding that S2* pollen was incompatible with S13-carrying pistils, we would predict that none of the eight SLF proteins yet to be examined interact with S13-RNase.
Analysis of Self-Incompatibility Behavior of S2 Pollen Carrying One of the Two In-Frame Indel Alleles of S2-SLF1
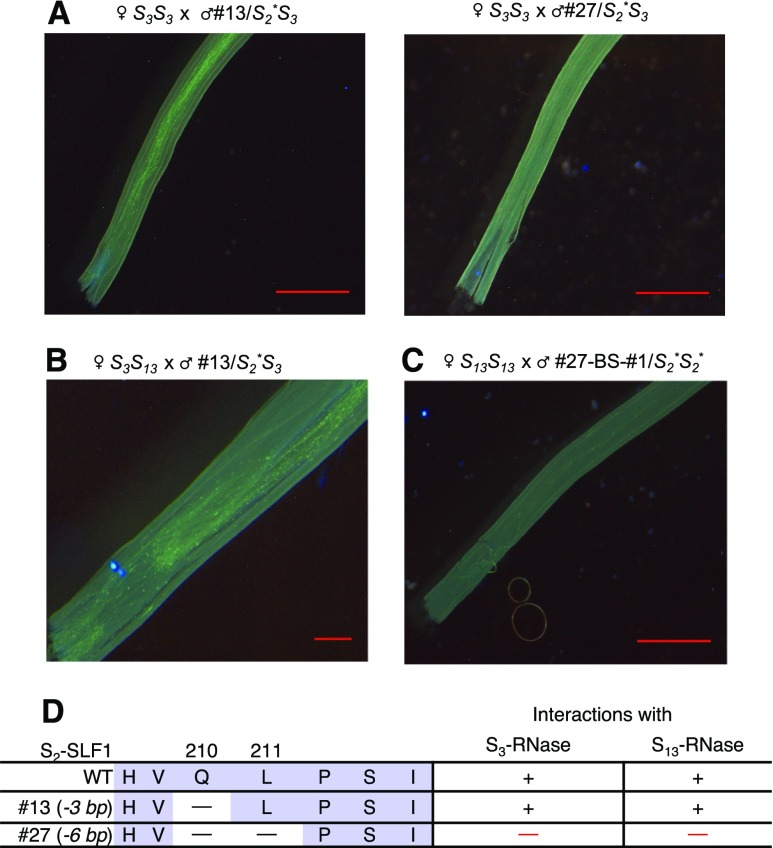
We also examined the in-frame indel alleles of S2-SLF1 identified in T0 plants #13/S2*S3 and #27/S2*S3 (Figure 1C) for their effects on the SI behavior of S2* pollen. We used pollen from both transgenic plants to pollinate the wild-type S3S3 plants. Pollinations by pollen of #13/S2*S3 were compatible (left panel of Figure 7A), whereas pollinations by pollen of #27/S2*S3 were incompatible (right panel of Figure 7A). Pollen produced by both transgenic plants was either S3 or S2*, and S3 pollen should be rejected by S3S3 pistils. Thus, the finding of compatible pollinations between pollen of #13/S2*S3 and S3-carrying pistils suggests that deletion of Q210 of S2-SLF1 does not affect its ability to detoxify S3-RNase, and the finding of incompatible pollinations between pollen of #27/S2*S3 and S3-carrying pistils suggests that deletion of Q210 and L211 of S2-SLF1 abolishes its ability to detoxify S3-RNase.
Figure 7.
Analysis of SI Behavior of S2* Pollen Carrying a 3-bp or a 6-bp In-Frame Deletion Allele of S2-SLF1.
(A) Pollen tube growth in the style after a wild-type S3S3 plant was separately pollinated with pollen from T0 plants #13/S2*S3 (carrying a 3-bp deletion allele) and #27/S2*S3 (carrying a 6-bp deletion allele).
(B) Pollen tube growth in the style after a wild-type S3S13 plant was pollinated with pollen from T0 plant #13/S2*S3.
(C) Pollen tube growth in the style after a wild-type S13S13 plant was pollinated with pollen from a progeny plant, #27-BS-#1/S2*S2*, obtained by bud-selfing T0 plant #27/S2*S3. Scale bar = 1 mm in all microscopy images of aniline blue staining in (A), (B), and (C).
(D) Effect of a single amino-acid deletion (Q210) and a two-amino acid deletion (Q210 and L211) of S2-SLF1 on its ability to detoxify S3-RNase and S13-RNase. Seven amino acids of the wild-type S2-SLF1 in the region where deletions occur are shown for comparison. + indicates ability to detoxify S3-RNase and S13-RNase, and — indicates inability to detoxify these two S-RNases.
We then used pollen of #13/S2*S3 and #27/S2*S3 to pollinate pistils of the wild-type S3S13 and S13S13 plants, respectively, to examine whether the two mutated forms of S2-SLF1 could still detoxify S13-RNase. Pollinations of S3S13 pistils by pollen of #13/S2*S3 were compatible (Figure 7B), suggesting that deletion of Q210 of S2-SLF1 does not affect its ability to interact with and detoxify S13-RNase. Pollinations of S13S13 pistils by #27/S2*S3 were compatible, as pollen of S3-haplotype should be compatible with S13S13 pistil. We then used PCR to determine the S-genotypes of 24 randomly selected progeny plants and found all of them to be S3S13 (Supplemental Figure 7A); the absence of S2S13 plants in the progeny suggests that S2* pollen from #27/S2*S3 was incompatible with S13-carrying pistils. Thus, deletion of both Q210 and L211 also abolishes the ability of S2-SLF1 to detoxify S13-RNase. To further confirm the results obtained from the crosses involving #27/S2*S3, we bud-selfed this plant and identified two Cas9 transgene-free S2*S2* progeny plants, #27-BS-#1 and -#2 (Supplemental Figures 7B to 7E). We then used pollen of each plant to separately pollinate S3S3, S13S13, and As-S3/S3S3 pistils. Pollinations of S3S3 and S13S13 pistils were incompatible (Figure 7C, Supplemental Figure 7F), consistent with the results obtained for #27/S2*S3; but pollinations of As-S3/S3S3 pistils, which did not produce a detectable level of S3-RNase, were compatible (Supplemental Figure 7F). Taking together all these results, we conclude that S2-SLF1 with both Q210 and L211 deleted fails to detoxify S3-RNase and S13-RNase, while S2-SLF1 with Q210 alone deleted can still interact with and detoxify both S-RNases (summary in Figure 7D).
Q210 and L211 are conserved between S2-SLF1 and S3-SLF1 (Supplemental Figure 3), suggesting that they are not involved in differential interactions between these two SLF proteins and S-RNases (e.g., S2-SLF1, but not S3-SLF1, interacts with S3-, S7- and S13-RNases; Hua et al., 2007; Kubo et al., 2010; Wu et al., 2018). We previously used computational modeling and molecular docking to predict the interaction surface between S2-SLF1 and S3-RNase (Supplemental Figure 8A; Wu et al., 2018), and here we used the same approach to predict the interaction surface between S2-SLF1 and S13-RNase (Supplemental Figure 8B). The docking results show that Q210 and L211 are not located at the predicted interface between S2-SLF1 and S3-RNase or S13-RNase. Thus, the inability of S2-SLF1 (with both Q210 and L211 deleted) to detoxify S3-RNase and S13-RNase is unlikelycaused by direct disruption of the interaction surface between S2-SLF1 and S3-RNase or S13-RNase, and the inability may be caused by conformational changes to S2-SLF1, which indirectly affect the interaction.
DISCUSSION
The molecular and biochemical basis of three different SI systems have been studied extensively. Both the Brassicaceae and Papaveraceae systems involve highly specific “one-to-one” self-recognition between pollen and pistil S-specificity determinants, with each determinant encoded by a single polymorphic gene (Kachroo et al., 2001; Takayama et al., 2001; Wheeler et al., 2009, 2010; Iwano and Takayama, 2012; Fujii et al., 2016). In contrast, the Solanaceae system involves complex non-self recognition between pollen and pistil specificity determinants. A single polymorphic S-RNase gene encodes the pistil specificity determinant, as has been demonstrated by both gain-of-function and loss-of-function experiments (Lee et al., 1994), whereas multiple polymorphic SLF genes collectively encode the pollen specificity determinant (Kubo et al., 2010, 2015; Williams et al., 2014b). According to the collaborative non-self recognition model (Kubo et al., 2010), each SLF protein produced by pollen of a given S-haplotype is only capable of interacting with a subset of its non-self S-RNases. Thus, a complete suite of SLF proteins is required to detoxify all their non-self S-RNases, but not self S-RNase, to result in cross-compatible but self-incompatible pollination (Kubo et al., 2010).
The evidence for the involvement of SLF proteins in pollen specificity was obtained by the gain-of-function assay developed based on the phenomenon of “competitive interaction,” which refers to the breakdown of SI in diploid pollen carrying two different S-haplotypes (Stout and Chandler, 1941, 1942). For example, SI breaks down in diploid SmSn pollen produced by a tetraploid SmSmSnSn plant, derived from a self-incompatible SmSn plant. The collaborative non-self recognition model can explain this interesting observation. Because SmSn pollen produces all SLF proteins of Sm-haplotype and all SLF proteins of Sn-haplotype, it could use (a) at least one of the SLF proteins of Sm-haplotype to interact with and detoxify Sn-RNase (a non-self S-RNase for Sm pollen), and (b) at least one of the SLF proteins of Sn-haplotype to interact with and detoxify Sm-RNase (a non-self S-RNase for Sn pollen). As a result, SmSn pollen tubes can detoxify both Sm-RNase and Sn-RNase, and they are thus compatible with pistils of both tetraploid SmSmSnSn and diploid SmSn plants. Using the gain-of-function assay to identify which of the 17 SLF proteins of S2-haplotype interact(s) with S3-RNase, we previously found that expressing S2-SLF1 alone in S3 pollen was sufficient to render S3 transgenic pollen compatible with S3-carrying pistils (Sijacic et al., 2004; Hua et al., 2007). This finding suggests that S2-SLF1 interacts with and detoxifies S3-RNase in the S3 transgenic pollen tube. It would seem counter-intuitive that gain-of-function of S2-SLF1 in the recipient S3 pollen actually results in loss of the SI function in S3 pollen. We further used the gain-of-function assay to show that S2-SLF1 also interacted with S7-RNase and S13-RNase (Table 1; Sun and Kao, 2013).
In this work, we have used a loss-of-function approach, CRISPR/Cas9-mediated genome editing, to definitively establish that SLF proteins are solely responsible for SI of pollen. Most notably, we found that S2* pollen lacking a functional S2-SLF1 was incompatible with normally compatible S3-carrying pistils, but that it remained compatible with S7- and S12-carrying pistils. Moreover, we found that S2* pollen was compatible with a transgenic plant (As-S3/S3S3) whose production of S3-RNase in the pistil was non-detectable, and was also compatible with pistils of immature S2S3 pistils that produce very low levels of S3-RNase insufficient to inhibit S3 pollen (Lee et al., 1994; Sun and Kao, 2013). We reasoned that if the SLF proteins are required for SI of pollen, then these results would suggest that S2-SLF1 is the only SLF of the 17 produced by S2 pollen that interacts with S3-RNase. Furthermore, we reasoned that there are additional SLF protein(s) that interact with S7-RNase and S12-RNase. For S3-RNase and S7-RNase, we have used the gain-of-function assay to completely determine their interaction relationships with all 17 SLF proteins of S2-haplotype. We found that none of the other 16 SLF proteins interact with S3-RNase (Table 1), whereas one of them, S2-SLF2, also interacts with S7-RNase (Figure 5). For S12-RNase, we have so far determined their interaction relationships with nine SLF proteins (Table 1) and found that both S2-SLF1 and S2-SLF5 interact with S12-RNase (Figure 6). These interaction relationships established by the gain-of-function experiments are entirely consistent with the SI behavior of S2* pollen with S3-, S7-, and S12-carrying pistils. That is, in the absence of S2-SLF1, S2* pollen cannot use any other SLF proteins to detoxify S3-RNase, but S2* pollen can still use S2-SLF2 to detoxify S7-RNase and at least S2-SLF5 to detoxify S12-RNase.
The 108 pairwise interaction relationships between the 17 SLF proteins of S2-haplotype and nine S-RNases that we have determined so far (Table 1) also reveal the complexity and diversity involving both “one-to-one” interactions (one SLF protein recognizing a particular S-RNase) and redundant interactions (at least two SLF proteins interacting with the same S-RNase). Functional redundancy of recognition molecules has been observed in other non-self recognition systems. For example, in plant-bacteria interactions, the effector proteins AvrPto and AvrPtoB produced by Pseudomonas syringae pv tomato DC300 function redundantly to block the pathogen-associated molecular pattern (PAMP) bacterial flagellin protein fliC produced by host plants (Kvitko et al., 2009). Functional redundancy enhances the robustness of a biological system by making it more “fail-safe” (Kitano, 2004). In the Petunia SI system, redundancy in the use of SLF proteins to detoxify a given non-self S-RNase could be advantageous, as this fail-safe mechanism would minimize the possibility of losing cross-compatibility in situations where mutations abolish the recognition function of certain SLF proteins (Sun and Kao, 2013). It has also been proposed that SLF proteins with overlapping specificities could gain new interaction specificity at the same time, which makes evolution of S-RNases with new specificities possible (Fujii et al., 2016). However, evolving and maintaining SLF proteins with redundant, or overlapping, interaction specificity with S-RNases may also be evolutionarily costly and therefore may not always be favored by natural selection (Kubo et al., 2015). In cases in which a single SLF is responsible for detoxifying a particular S-RNase, the deleterious effect of loss-of-function mutations could be alleviated by (a) a decrease in the frequency of the plants whose pistils produce this S-RNase in the population, and/or (b) transmission of the mutated SLF gene through the female. The complexity of the interaction network between S-RNases and SLF proteins, therefore, is likely to be shaped by the evolutionary history of the S-RNase gene and the SLF gene repertoire at the S-locus.
We have thus far found that the largest number of SLF proteins produced by S2-haplotype that recognize the same non-self S-RNase is two: S2-SLF1 and S2-SLF2 for S7-RNase, and S2-SLF1 and S2-SLF5 for S12-RNase (Sun and Kao, 2013; Williams et al., 2014b; this study). This is also the case for the interactions between SLF proteins and S-RNases of P. hybrida: PhS5-SLF1 and PhS5-SLF2 for PhS9-RNase; PhS7-SLF1 and PhS7-SLF2 for PhS9-RNase; and PhS7-SLF2 and PhS7-SLF9A for PhS19-RNase (Kubo et al., 2010, 2015). It would be interesting to determine, for a given S-haplotype, what is the maximum number of SLF proteins that can interact with the same non-self S-RNase. The upper limit of different SLFs with overlapping specificities may be restricted by the time required for the evolution of different interaction specificity between SLF proteins and S-RNases and/or by the biochemical properties of the interaction surface of SLF proteins and S-RNases. Determining more interaction relationships between SLF proteins and S-RNases, and theoretical modeling of their interactions, will likely shed light on the question of why certain S-RNases only interact with one SLF, while others interact with more than one.
It is important to note that our laboratory previously used artificial microRNA (amiRNA) targeting S2-SLF1 to knock down S2-SLF1 in pollen and reported that S2 pollen in which the S2-SLF1 transcript level was significantly reduced remained compatible with S3-carrying and S13-carrying pistils (Sun and Kao, 2013). Those results are different from our finding in this work that CRISPR/Cas9-mediated knockout of S2-SLF1 rendered S2 pollen incompatible with S3-carrying and S13-carrying pistils. Accurate assessment of the level of suppression of a pollen-expressed gene is often difficult, because if a single copy of the transgene is integrated into the genome of a transgenic plant, only half of the pollen produced carries the transgene. Thus, even if an amiRNA completely suppresses the transcript level of its target gene in the half of the pollen that carries the transgene, the other half of the pollen will still produce the wild-type levels of the transcript. In this case, when equal amounts of total pollen RNA were examined, the total transcript level of the target gene will be ∼50%—not ∼0%, that of pollen produced by the wild-type plants. Also, in the amiRNA-mediated knockdown experiments of S2-SLF1, the native (and weak) promoter of S2-SLF1 was used to drive the transcription of amiRNA in the generative nucleus of the pollen where the SLF genes are expressed (Sun and Kao, 2013). Therefore, the phenotypic difference may be due to detoxification of S3-RNase and S13-RNase by the residual S2-SLF1 in S2 transgenic pollen. This explanation also suggests that SLF proteins are efficient in detoxifying S-RNases in vivo, and thus it is essential to use the knockout approach to address the function of any SLF.
The generation of two in-frame deletion alleles of S2-SLF1 has allowed us to also address the effect of a small number of amino acid deletions on the function of S2-SLF1 in interactions with S3-RNase and S13-RNase. Interestingly, deletion of one amino acid, Q210, near the middle of the protein does not affect the ability of S2-SLF1 to interact with and detoxify these two S-RNases. However, deletion of Q210 and its adjacent amino acid, L211, results in the loss of the ability of S2-SLF1 to detoxify both S-RNases (Figure 7D). The results of molecular docking show that Q210 and L211 are not located at the interface of S2-SLF1 and S3-RNase or S13-RNase (Supplemental Figure 8; Wu et al., 2018), suggesting that their deletion most likely indirectly affects the interaction interface of S2-SLF1 with these two S-RNases. Q210 and L211 are conserved in S3-SLF1, which, unlike S2-SLF1, does not interact with S3-RNase or S13-RNase (Hua et al., 2007; Kubo et al., 2010; Wu et al., 2018). These results suggest that conserved amino acids outside the interface between SLF proteins and S-RNases can also contribute to the establishment of interactions, even though they do not directly contribute to the specificity of interactions. Thus, interactions between SLF proteins and S-RNases are under strict and intricate constraints. Further structural studies of SLF-S-RNase complexes will help shed light on the amino acid residues of SLF proteins that are critical for the establishment of their interactions with S-RNases.
In summary, in this work we have extensively characterized the effect of loss-of-function of S2-SLF1 on the SI behavior of S2 pollen, using two frame-shift indel alleles of S2-SLF1 generated by CRISPR/Cas9-mediated genome editing. The results, coupled with the comprehensive analysis of interaction relationships between the 17 SLF proteins of S2-haplotype and nine S-RNases determined via the in vivo gain-of-function assay, provide definitive evidence for the essential role of SLF proteins in the SI of pollen and lend strong support for the validity of the collaborative non-self recognition model (Kubo et al., 2010). The interaction relationships also reveal that the Petunia SI system has evolved complex and diverse interaction patterns between SLF proteins and S-RNases, with both “fail-safe” and “one-to-one” interactions. The results from this work and determination of more SLF-S-RNase interaction relationships will provide valuable insights for investigations into the biochemical and structural basis of differential interactions between SLF proteins and S-RNases, as well as for studies of the evolutionary dynamics of SLF repertoires and S-RNases during the long evolutionary history of this SI system.
METHODS
Plant Materials and Growth Conditions
All the S-haplotypes of Petunia inflata used in this work (S2, S3, S5, S6a, S7, S11, S12, S13, and S16) were from our laboratory’s genetic stock (Ai et al., 1990; Wang et al., 2001; Sun and Kao, 2013). The As-S3/S3S3 plants were obtained by bud-selfing the previously generated As-S3/S2S3 plants (Lee et al., 1994; Sun and Kao, 2013). Petunia seedlings were grown at 30°C with a light cycle of 16 h (2600 lumens cool white light, Philips 40-Watt Cool White Linear Fluorescent Light Bulbs). Mature plants (over 30-cm tall) in individual pots were maintained in the greenhouse at Pennsylvania State University. The temperature in the greenhouse was kept at 25°C, with a light cycle of 16 h under a high-pressure sodium (HPS) light system (1080-watt PL 2000, P.L. Light Systems).
Generation and Characterization of CRISPR/Cas9-Mediated Knockout Mutants of S2-SLF1
To generate the CRISPR/Cas9 construct targeting S2-SLF1, the pre-tRNA and the gRNA scaffold fragments were ligated to the S2-SLF1-Protospacer-9 (S2-SLF1-PS9) fragment, and the resulting DNA fragment was cloned into pKSE401 plasmid (Xing et al., 2014; a gift from Qi-Jun Chen; Addgene plasmid # 62202) following the protocol described previously (Xie et al., 2015; Sun and Kao, 2018). The pGTR plasmid (obtained from Yinong Yang laboratory, Pennsylvania State University) was used as a template to synthesize the tRNA-gRNA fragment (PTG gene) in two separate PCRs using Phusion DNA polymerase (Thermo Scientific). S2-SLF1-PS9-F/L3AD5-R and S2-SLF1-PS9-R/L5AD51-F were used to synthesize two halves of the PTG gene, and they were ligated together using the Golden Gate Assembly method. The assembled product was further amplified using S51AD5-F and S3AD5-R primers, and it was digested with FokI (New England BioLabs). The FokI-digested PTG fragment was ligated to BsaI-digested pKSE401 to create the PTG-S2-SLF1-PS9-pKSE401 construct. The binary plasmid PTG-S2-SLF1-PS9-pKSE401 was transformed into Agrobacterium tumefaciens (LBA4404) by electroporation. Transformation of P. inflata of S2S3 genotype was performed as described previously (Meng et al., 2011). Genomic DNA from regenerated plants was extracted as described previously (Meng et al., 2011). A pair of primers was used for the identification of transgenic plants. The pair of primers (35S-gt-F/Cas9-gt-R) were flanking a 580-bp fragment, from the 3′-end of the CaMV 35S promoter to the 5′-end of Cas9 on the PTG-S2-SLF1-PS9-pKSE401 Ti plasmid.
The target region in S2-SLF1 was amplified with PiSLF2-RT-3For/4Rev primers using Phusion DNA polymerase. The PCR products were subjected to BsrGI-HF (New England BioLabs) digestion overnight, and the digested PCR products were electrophoresed on 2% agarose gels. The PCR products that were not digested by BsrGI were sequenced to determine the nature of indel mutations. The region in S3-SLF1 corresponding to the target region in S2-SLF1 was amplified with PiSLF3-Copy1For/Rev primers using Phusion DNA polymerase, and the PCR products were sequenced. The S-genotype of BS progeny plants were determined by PCR using primers PiSLF3-Copy1For/Rev (for S3-SLF1).
Generation of Transgenic Plants Used for Gain-of-Function Assays
A total of 11 Ti plasmid constructs were made in this work for the in vivo gain-of-function assay, all of which were in the pBI101 backbone. Each construct contained the LAT52 promoter (LAT52P; Twell et al., 1990) driving the expression of one SLF fused at its last codon with the coding sequence of GFP. These 11 constructs are schematically detailed in Figures 4A and 4B. Generation of the pBI101-LAT52P:S2-SLF9/S2-SLF10 constructs (Figure 4A) was as described by Hua et al. (2007). pBI101-LAT52P:S2-SLF2/-SLF7/S2-SLF11/-SLF12/-SLF13/-SLF14/-SLF15/-SLF16/-SLF17 constructs (Figure 4B) were generated using the In-fusion HD Cloning Kit from Clontech (Williams et al., 2014b). Primers used for making all 11 constructs are listed in Supplemental Table 1. All Ti plasmid constructs were electroporated into Agrobacterium tumefaciens (LBA4404) competent cells, and subsequently they were transformed into P. inflata plants of S2S3 or S2S7, as described previously (Meng et al., 2011). Transgenic lines were identified using a primer pair specific to the GFP transgene (GFP-001For and GFP-500Rev).
Visualization of GFP Fluorescence of Pollen Tubes
Mature pollen was collected and germinated in pollen germination medium for two hours and visualized with a Nikon Eclipse 90i epifluorescence microscope, as described previously (Meng et al., 2011; Williams et al., 2014b).
Pollination Assay
Stigmas of emasculated mature flowers were manually pollinated with pollen from mature anthers. For aniline blue staining of pollen tubes, the pollinated pistils were collected 20 h after manual pollination, fixed with a mix of acetic acid with 95% ethanol (1:3), macerated with 8 M KOH, and then stained with 0.1 mg/ml aniline blue dye diluted with 1 mM KH2PO4 (1:20). The stained pistils were visualized under the DAPI-filtered UV light of a Nikon Eclipse 90i epifluorescence microscope.
Computational Modeling of Protein Tertiary Structures and Protein-Protein Docking Analysis
Wu et al. (2018) reported the predicted tertiary structures of S2-SLF1 (without the first 95 amino acids that contain the F-box domain) and S3-RNase, as well as their molecular docking. The tertiary structure of S13-RNase was modeled using I-TASSER (Zhang, 2008; Yang et al., 2015), and structure refinement was similarly performed as previously described by Wu et al. (2018). Molecular docking of S13-RNase (as the ligand) with S2-SLF1 (without the first 95 amino acids, as the receptor) was similarly performed using ClusPro, as previously described (Comeau et al., 2004; Kozakov et al., 2017; Wu et al., 2018). All structures were visualized using the PyMOL molecular visualization package (The PyMOL Molecular Graphics System, Version 1.8 Schrödinger, LLC).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: S2-SLF1 (AAS79485), S3-SLF1 (AAS79486), S2-SLF2 (KJ670474), S2-SLF3 (EF614187), S2-SLF4 (KF524351), S2-SLF5 (KF524352), S2-SLF6 (KF524353), S2-SLF7 (EF614189), S2-SLF8 (EF614188), S2-SLF9 (AY363971), S2-SLF10 (AY363974), S2-SLF11(KJ670428), S2-SLF12 (KJ670433), S2-SLF13 (KJ670438), S2-SLF14 (KJ670443), S2-SLF15 (KJ670448), S2-SLF16 (KJ670453), S2-SLF17 (KJ670458), S2-RNase (AAG21384), S3-RNase (AAA33727), S6a-RNase (AF301167), S7-RNase (AF301168), S11-RNase (AF301172), S12-RNase (AF301173), S13-RNase (AF301174), S16-RNase (AF301176), S5-SLF1(KC590092), S7-SLF1 (KC590093), S11-SLF1(KC590094), and S13-SLF1 (KC590095). All genes are from Petunia inflata.
Supplemental Data
Supplemental Figure 1. Alignment of nucleotide sequences of the protospacer region of S2-SLF1 and the corresponding regions of all SLF genes of S2-Haplotype and S3-Haplotype.
Supplemental Figure 2. Generation of the PTG-CRISPR/Cas9 construct targeting S2-SLF1, and analysis of regenerated plants for the presence of the transgene.
Supplemental Figure 3. Alignment of deduced amino acid sequences of S2-SLF1, S3-SLF1, and four indel alleles of S2-SLF1 in the region targeted by CRISPR/Cas9.
Supplemental Figure 4. PCR analysis of S-Genotypes of plants in each progeny obtained from pollination of S7S7 and S13S13 plants by pollen from T0 plants #2/S2*S3 and #35/S2*S3.
Supplemental Figure 5. Identification of bud-selfed (BS) progeny plants homozygous for S2-Haplotype and for the indel alleles inherited from their respective T0 plants #2/S2*S3 and #35/S2*S3.
Supplemental Figure 6. GFP fluorescence of pollen tubes germinated in vitro from pollen of a representative T0 plant of each of the 12 transgenic lines generated in this study.
Supplemental Figure 7. Analysis of SI behavior of plants carrying a 6-bp In-frame deletion allele of S2-SLF1.
Supplemental Figure 8. Computational modeling of interactions of S2-SLF1 with S3-RNase and S13-RNase.
Supplemental Table 1. Primers used in this study
Supplemental Table 2. Summary of T0 and T1 plants used in the in vivo gain-of-function assay and their SI behavior
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank Y.-N. Yang and B. Minkenberg for providing the pGTR plasmid and technical advice on CRISPR/Cas9-mediated genome editing; S. Burghard for greenhouse management; and H. Hao for general laboratory help. This work was supported by the National Science Foundation (IOS-1645557 to T.-h.K.).
AUTHOR CONTRIBUTIONS
L.S., J.S.W., S.L., and T.-h.K. designed the experiments. L.S., J.S.W., S.L., L.W., W.A.K., P.G.S., and M.D.K. performed the experiments and analysis. L.S. and T.-h.K. wrote the article.
References
- Ai Y., Singh A., Coleman C.E., Ioerger T.R., Kheyr-Pour A., Kao T.H. (1990). Self-incompatibility in Petunia inflata: Isolation and characterization of cDNAs encoding three S-allele-associated proteins. Sex. Plant Reprod. 3: 130–138. [Google Scholar]
- Bombarely A., et al. (2016). Insight into the evolution of the Solanaceae from the parental genomes of Petunia hybrida. Nat. Plants 2: 16074. [DOI] [PubMed] [Google Scholar]
- Comeau S.R., Gatchell D.W., Vajda S., Camacho C.J. (2004). ClusPro: A fully automated algorithm for protein-protein docking. Nucleic Acids Res. 32: W96–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Nettancourt D. (2001). Incompatibility and Incongruity in Wild and Cultivated Plants. (Berlin, NY: Springer; ). [Google Scholar]
- Entani T., Kubo K., Isogai S., Fukao Y., Shirakawa M., Isogai A., Takayama S. (2014). Ubiquitin-proteasome-mediated degradation of S-RNase in a solanaceous cross-compatibility reaction. Plant J. 78: 1014–1021. [DOI] [PubMed] [Google Scholar]
- Fujii S., Kubo K., Takayama S. (2016). Non-self- and self-recognition models in plant self-incompatibility. Nat. Plants 2: 16130. [DOI] [PubMed] [Google Scholar]
- Goldraij A., Kondo K., Lee C.B., Hancock C.N., Sivaguru M., Vazquez-Santana S., Kim S., Phillips T.E., Cruz-Garcia F., McClure B. (2006). Compartmentalization of S-RNase and HT-B degradation in self-incompatible Nicotiana. Nature 439: 805–810. [DOI] [PubMed] [Google Scholar]
- Hua Z., Kao T.H. (2006). Identification and characterization of components of a putative petunia S-locus F-box-containing E3 ligase complex involved in S-RNase-based self-incompatibility. Plant Cell 18: 2531–2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua Z., Meng X., Kao T.H. (2007). Comparison of Petunia inflata S-Locus F-box protein (Pi SLF) with Pi SLF like proteins reveals its unique function in S-RNase based self-incompatibility. Plant Cell 19: 3593–3609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S., Lee H.S., Karunanandaa B., Kao T.H. (1994). Ribonuclease activity of Petunia inflata S proteins is essential for rejection of self-pollen. Plant Cell 6: 1021–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwano M., Takayama S. (2012). Self/non-self discrimination in angiosperm self-incompatibility. Curr. Opin. Plant Biol. 15: 78–83. [DOI] [PubMed] [Google Scholar]
- Kachroo A., Schopfer C.R., Nasrallah M.E., Nasrallah J.B. (2001). Allele-specific receptor-ligand interactions in Brassica self-incompatibility. Science 293: 1824–1826. [DOI] [PubMed] [Google Scholar]
- Kitano H. (2004). Biological robustness. Nat. Rev. Genet. 5: 826–837. [DOI] [PubMed] [Google Scholar]
- Kozakov D., Hall D.R., Xia B., Porter K.A., Padhorny D., Yueh C., Beglov D., Vajda S. (2017). The ClusPro web server for protein-protein docking. Nat. Protoc. 12: 255–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo K., Entani T., Takara A., Wang N., Fields A.M., Hua Z., Toyoda M., Kawashima S., Ando T., Isogai A., Kao T.H., Takayama S. (2010). Collaborative non-self recognition system in S-RNase-based self-incompatibility. Science 330: 796–799. [DOI] [PubMed] [Google Scholar]
- Kubo K., Paape T., Hatakeyama M., Entani T., Takara A., Kajihara K., Tsukahara M., Shimizu-Inatsugi R., Shimizu K.K., Takayama S. (2015). Gene duplication and genetic exchange drive the evolution of S-RNase-based self-incompatibility in Petunia. Nat. Plants 1: 14005. [DOI] [PubMed] [Google Scholar]
- Kvitko B.H., Park D.H., Velásquez A.C., Wei C.F., Russell A.B., Martin G.B., Schneider D.J., Collmer A. (2009). Deletions in the repertoire of Pseudomonas syringae pv. tomato DC3000 type III secretion effector genes reveal functional overlap among effectors. PLoS Pathog. 5: e1000388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H.S., Huang S., Kao T.H. (1994). S proteins control rejection of incompatible pollen in Petunia inflata. Nature 367: 560–563. [DOI] [PubMed] [Google Scholar]
- Li S., Sun P., Williams J.S., Kao T.H. (2014). Identification of the self-incompatibility locus F-box protein-containing complex in Petunia inflata. Plant Reprod. 27: 31–45. [DOI] [PubMed] [Google Scholar]
- Li S., Williams J.S., Sun P., Kao T.H. (2016). All 17 S-locus F-box proteins of the S2- and S3-haplotypes of Petunia inflata are assembled into similar SCF complexes with a specific function in self-incompatibility. Plant J. 87: 606–616. [DOI] [PubMed] [Google Scholar]
- Luu D.T., Qin X., Morse D., Cappadocia M. (2000). S-RNase uptake by compatible pollen tubes in gametophytic self-incompatibility. Nature 407: 649–651. [DOI] [PubMed] [Google Scholar]
- Meng X., Hua Z., Sun P., Kao T.H. (2011). The amino terminal F-box domain of Petunia inflata S-locus F-box protein is involved in the S-RNase-based self-incompatibility mechanism. AoB Plants 2011: plr016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murfett J., Atherton T.L., Mou B., Gasser C.S., McClure B.A. (1994). S-RNase expressed in transgenic Nicotiana causes S-allele-specific pollen rejection. Nature 367: 563–566. [DOI] [PubMed] [Google Scholar]
- Sijacic P., Wang X., Skirpan A.L., Wang Y., Dowd P.E., McCubbin A.G., Huang S., Kao T.H. (2004). Identification of the pollen determinant of S-RNase-mediated self-incompatibility. Nature 429: 302–305. [DOI] [PubMed] [Google Scholar]
- Stout A.B., Chandler C. (1941). Change from self-incompatibility to self-compatibility accompanying change from diploidy to tetraploidy. Science 94: 118. [DOI] [PubMed] [Google Scholar]
- Stout A.B., Chandler C. (1942). Hereditary transmission of induced tetraploidy and compatibility in fertilization. Science 96: 257–258. [DOI] [PubMed] [Google Scholar]
- Sun L., Kao T.H. (2018). CRISPR/Cas9-mediated knockout of PiSSK1 reveals essential role of S-locus F-box protein-containing SCF complexes in recognition of non-self S-RNases during cross-compatible pollination in self-incompatible Petunia inflata. Plant Reprod. 31: 129–143. [DOI] [PubMed] [Google Scholar]
- Sun P., Kao T.H. (2013). Self-incompatibility in Petunia inflata: The relationship between a self-incompatibility locus F-box protein and its non-self S-RNases. Plant Cell 25: 470–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takayama S., Shimosato H., Shiba H., Funato M., Che F.S., Watanabe M., Iwano M., Isogai A. (2001). Direct ligand-receptor complex interaction controls Brassica self-incompatibility. Nature 413: 534–538. [DOI] [PubMed] [Google Scholar]
- Twell D., Yamaguchi J., McCormick S. (1990). Pollen-specific gene expression in transgenic plants: Coordinate regulation of two different tomato gene promoters during microsporogenesis. Development 109: 705–713. [DOI] [PubMed] [Google Scholar]
- Wang X., Hughes A.L., Tsukamoto T., Ando T., Kao T.H. (2001). Evidence that intragenic recombination contributes to allelic diversity of the S-RNase gene at the self-incompatibility (S) locus in Petunia inflata. Plant Physiol. 125: 1012–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler M.J., de Graaf B.H., Hadjiosif N., Perry R.M., Poulter N.S., Osman K., Vatovec S., Harper A., Franklin F.C.H., Franklin-Tong V.E. (2009). Identification of the pollen self-incompatibility determinant in Papaver rhoeas. Nature 459: 992–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler M.J., Vatovec S.,Franklin-Tong V.E. (2010). The pollen S-determinant in Papaver: Comparisons with known plant receptors and protein ligand partners. J. Exp. Bot. 61: 2015–2025. [DOI] [PubMed] [Google Scholar]
- Williams J.S., Der J.P., dePamphilis C.W., Kao T.H. (2014a). Transcriptome analysis reveals the same 17 S-locus F-box genes in two haplotypes of the self-incompatibility locus of Petunia inflata. Plant Cell 26: 2873–2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams J.S., Natale C.A., Wang N., Li S., Brubaker T.R., Sun P., Kao T.H. (2014b). Four previously identified Petunia inflata S-locus F-box genes are involved in pollen specificity in self-incompatibility. Mol. Plant 7: 567–569. [DOI] [PubMed] [Google Scholar]
- Wu L., Williams J.S., Wang N., Khatri W.A., San Román D., Kao T.H. (2018). Use of domain-swapping to identify candidate amino acids involved in differential interactions between two allelic variants of Type-1 S-locus F-box protein and S3-RNase in Petunia inflata. Plant Cell Physiol. 59: 234–247. [DOI] [PubMed] [Google Scholar]
- Xie K., Minkenberg B., Yang Y. (2015). Boosting CRISPR/Cas9 multiplex editing capability with the endogenous tRNA-processing system. Proc. Natl. Acad. Sci. USA 112: 3570–3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing H.L., Dong L., Wang Z.P., Zhang H.Y., Han C.Y., Liu B., Wang X.C., Chen Q.J. (2014). A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant Biol. 14: 327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Yan R., Roy A., Xu D., Poisson J.,Zhang Y. (2015). The I-TASSER Suite: Protein structure and function prediction. Nat. Methods 12: 7–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. (2008). I-TASSER server for protein 3D structure prediction. BMC Bioinformatics 9: 40. [DOI] [PMC free article] [PubMed] [Google Scholar]