OsRR24/LEPTO1, a type-B response regulator, plays a significant role in establishing leptotene chromosomes after entry into meiosis in rice.
Abstract
Response regulators play significant roles in controlling various biological processes; however, their roles in plant meiosis remain unclear. Here, we report the identification of OsRR24/LEPTOTENE1 (LEPTO1), a rice (Oryza sativa) type-B response regulator that participates in the establishment of key molecular and morphological features of chromosomes in leptotene, an early stage of prophase I in meiosis. Although meiosis initiates normally, as indicated by staining of the centromere-specific histone CENH3, the meiotic chromosomes in lepto1 mutant pollen mother cells fail to form the thin thread-like structures that are typical of leptotene chromosomes in wild-type pollen mother cells. Furthermore, lepto1 mutants fail to form chromosomal double-strand breaks, do not recruit meiosis-specific proteins to the meiotic chromosomes, and show disrupted callose deposition. LEPTO1 also is essential for programmed cell death in tapetal cells. LEPTO1 contains a conserved signal receiver domain (DDK) and a myb-like DNA binding domain at the N terminus. LEPTO1 interacts with two authentic histidine phosphotransfer (AHP) proteins, OsAHP1 and OsAHP2, via the DDK domain, and a phosphomimetic mutation of the DDK domain relieves its repression of LEPTO1 transactivation activity. Collectively, our results show that OsRR24/LEPTO1 plays a significant role in the leptotene phase of meiotic prophase I.
INTRODUCTION
In sexually reproducing eukaryotes, the successful completion of meiosis to generate haploid cells is essential for the propagation of species. The entry into and progression of meiosis preserve the faithful segregation of homologous chromosomes and contribute to evolution. During the meiotic cell cycle, a single round of DNA replication is followed by two successive rounds of chromosome segregation. The critical decision to enter meiosis is conserved in eukaryotes and is thought to be made before the premeiotic S phase (Pawlowski et al., 2007; Nonomura et al., 2011). It is widely accepted that a specifically organized premeiotic interphase is required for normal progression of the meiotic process. In Arabidopsis (Arabidopsis thaliana), a model eudicot species, reloading of the centromere histone H3 variant (CENH3) takes place during the G2 stage, and the premeiotic stage involves specialized loading of CENH3 (Lermontova et al., 2006; Ravi et al., 2011). Also, observations in the monocot rice (Oryza sativa) demonstrate that prophase I meiocytes have larger CENH3 foci than those in the somatic cells, suggesting that a greater amount of CENH3 loading is related to the specialized meiotic process (Ren et al., 2018).
Following DNA replication, sister chromatids are held together by a cohesin complex. Cohesin complexes involved in mitosis and meiosis share three of the same protein subunits: STRUCTURAL MAINTENANCE OF CHROMOSOMES1, STRUCTURAL MAINTENANCE OF CHROMOSOMES3, and SISTER-CHROMATID COHESION PROTEIN3. The meiotic cohesin complex contains the meiosis-specific REC8 protein, which is recruited in place of the mitotic subunit SISTER-CHROMATID COHESION PROTEIN1 (Chelysheva et al., 2005). During the early stage of prophase I, known as leptotene, the gradual loading of cohesion facilitates the assembly of the dispersed chromosomes into thin, thread-like individualized units.
A series of highly coordinated meiotic events take place at leptotene. SPO11-induced DNA double-strand breaks (DSBs) start to form at leptotene and are essential for the onset of meiotic recombination (Mahadevaiah et al., 2001). The SPO11 protein is homologous to the A subunit of archaeal topoisomerase. Other than SPO11, recent studies have reported that the B-like topoisomerase subunit also is involved in generating an active topoisomerase VI complex for meiotic DSB formation in plants and animals (Robert et al., 2016; Vrielynck et al., 2016). Meiotic DSB formation relies on the assembly of chromosomes into a “loop-axis” structure through the loading of a proteinaceous axial element (AE) (Borde and Lichten, 2014). In plants, rice PAIR3, Arabidopsis ASY3, and maize (Zea mays) DSY2 are homologous proteins that exhibit conserved loading behavior on the AE. In maize, DSY2 becomes discontinuous, but dense foci form along the chromosome at leptotene, which are important for facilitating the recruitment of DSB-forming proteins and maintaining chromosome integrity (Lee et al., 2015). Moreover, the location of proteins participating in DSB processing and repair is obvious in leptotene nuclei, suggesting that DSB repair is initiated immediately following DSB formation (Ji et al., 2012; Tang et al., 2014). These critical events involved in leptotene progression are the basis for the subsequent pairing, recombination, and faithful separation of homologous chromosomes.
Response regulators (RR) are important elements in two-component signal transduction systems and participate in many cellular processes (Galperin, 2010). Based on the similarity of their sequences and domain structures, RRs in plants are divided mainly into three groups: type A, type B, and type C (Tsai et al., 2012). The type-B RRs have a DDK domain and a MYB domain at the N terminus, followed by a long C-terminal extension, and may act as transcriptional regulators (Hosoda et al., 2002; Mason et al., 2005). Previous studies demonstrated that Arabidopsis type-B RRs play significant roles in growth and development, organ regeneration, and stress responses (Argyros et al., 2008; Nguyen et al., 2016; Meng et al., 2017; Zhang et al., 2017). In rice, Early heading date1, a type-B RR, promotes flowering by regulating the expression of FLOWERING LOCUS T-like genes (Doi et al., 2004). The function of type-B RRs has been widely studied; nevertheless, the roles of type-B RRs in meiosis are still unclear.
In this study, we identified OsRR24/LEPTOTENE1 (LEPTO1), which encodes a type-B RR that is essential for establishing meiotic leptotene chromosomes in rice pollen mother cells (PMC). Mutation of LEPTO1 resulted in abnormal megasporogenesis. LEPTO1 can interact with two rice authentic histidine phosphotransfer proteins, OsAHP1 and OsAHP2, via its DDK domain. Moreover, the phosphorylation of the second conserved Asp of the DDK domain can relieve its repression on the transcriptional activation ability of LEPTO1.
RESULTS
Identification of the OsRR24/LEPTO1 Gene
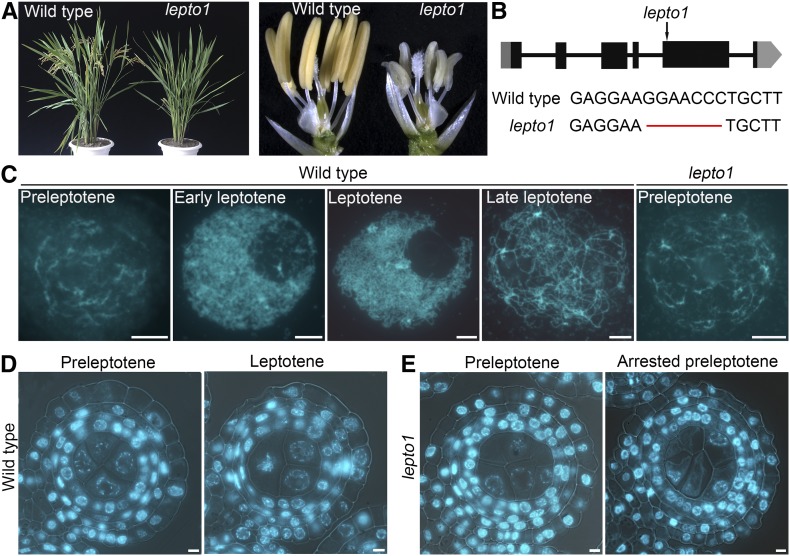
To identify genes that are essential in meiosis, we performed a genetic screen and identified a completely sterile mutant, which we designated lepto1. The progeny of plants heterozygous for lepto1 showed a 3:1 (fertile:sterile) segregation ratio, suggesting that a single recessive mutation is responsible for the sterile phenotype. The lepto1 mutant showed no detectable differences from the wild type during vegetative growth (Figure 1A). However, lepto1 anthers were small and white and contained no pollen grains, as revealed by I2-KI staining (Figure 1A; Supplemental Figure 1). The mutant set no seeds when pollinated with mature pollen from wild-type plants, suggesting that the mutant is both male and female sterile.
Figure 1.
PMCs in lepto1 Are Arrested at Preleptotene.
(A) Morphology of the rice sterile mutant lepto1. The left panel shows plant morphology after heading. The right panel shows opened spikelets of the wild type and lepto1.
(B) Gene structure of LEPTO1 and the mutation site of lepto1. Black boxes represent exons, black lines represent introns, and gray boxes represent the untranslated regions.
(C) Chromosome morphology of meiocytes at preleptotene, early leptotene, leptotene, and late leptotene in the wild type and at preleptotene in lepto1.
(D) Transverse sections of wild-type anthers stained with DAPI at preleptotene and leptotene.
(E) Transverse sections of lepto1 anthers stained with DAPI at preleptotene and arrested preleptotene.
Bars = 5 μm.
Using a map-based cloning strategy, the mutated gene was delimited to an approximately 120-kb region on the short arm of chromosome 2 (Supplemental Figure 1A). DNA sequencing allowed us to identify a 7-bp deletion in the fifth exon of LOC_Os02g08500 that caused a predicted reading frame shift, resulting in mistranslation at the C terminus of the putative protein (Figure 1B). To verify that the sterile phenotype indeed resulted from the mutation of LOC_Os02g08500, we conducted a complementation test by transformation of plasmid containing the coding sequence of LOC_Os02g08500, which rescued the fertility of lepto1 (Supplemental Figure 1E). In addition, we generated a CRISPR-Cas9-derived transgenic line, lepto1-cas9, which has a 1-bp insertion in the first exon (Supplemental Figure 2A). The lepto1-cas9 mutant also exhibited the sterile phenotype. These results demonstrate that the mutations in LOC_Os02g08500 are indeed responsible for the observed sterile phenotype, and subsequently, LOC_Os02g08500 was designated as LEPTO1. This locus was identified previously as OsRR24 (Tsai et al., 2012).
We next performed RACE PCR to obtain the full-length cDNA of LEPTO1. Analysis of the cDNA sequence indicated that LEPTO1 had six exons, five introns, and a 1878-bp open reading frame. In addition, LEPTO1 had a 350-bp 5′-untranslated region and a 255-bp 3′-untranslated region (Supplemental Figure 3).
LEPTO1 Is Essential for Leptotene Chromosome Organization
To explore the cause of sterility in the lepto1 mutant, we examined the cytological morphology and chromosome behavior of wild-type and lepto1 anthers at different developmental stages by observing transverse sections or squashed tissues stained with 4′,6-diamidino-phenylindole (DAPI). The first meiotic stage that can be distinguished is preleptotene, during which chromosomes in PMCs were discrete and partially contracted (Figure 1D). As observed in the PMCs of squashed anthers, chromosomes at preleptotene exhibited a hairy appearance with blurred outlines. Subsequently, chromosomes in PMCs formed thin threads at early leptotene. The thin threads became more distinct at late leptotene (Figures 1C and 1D). Chromosomes then underwent two successive rounds of segregation, giving rise to tetrads (Supplemental Figure 2D).
The development of lepto1 anthers showed no obvious difference from wild-type anthers before the four-layer stage. At the four-layer stage, the morphology of chromosomes in lepto1 germ cells resembled that of preleptotene chromosomes in the wild type. Nevertheless, we observed few chromosomes with the typical thin thread-like morphology in lepto1 anthers in the subsequent development stages (Figures 1C and 1E). Moreover, the germ cells showed a similarly arrested phenotype in lepto1-cas9 anthers (Supplemental Figure 2I). These results indicate that the germ cells in the lepto1 mutant failed to establish the characteristic leptotene chromosome morphology and likely arrested at preleptotene. Moreover, the arrested chromosomes in lepto1 PMCs eventually underwent degeneration as the anthers expanded (Supplemental Figures 2E to 2G).
Meiotic Fate Acquisition Is Not Affected in the lepto1 Mutant
Meiosis involves a specific CENH3 loading pattern that is different from that in mitosis, and the CENH3 foci in preleptotene meiocytes become brighter than those in somatic cells (Ren et al., 2018). Therefore, we detected the immunofluorescence signal intensity of CENH3 foci in transverse sections of anthers to investigate whether meiosis initiates in the germ cells of the lepto1 mutant. CENH3 foci showed no obvious differences in the germ cells of wild-type and lepto1 anthers, all of which exhibited a larger and brighter signal than that in the peripheral somatic cells (Figures 2A and 2B). These results suggest that the initiation of meiosis is not affected in lepto1 anthers and support the speculation that the meiotic cycle is able to proceed into preleptotene.
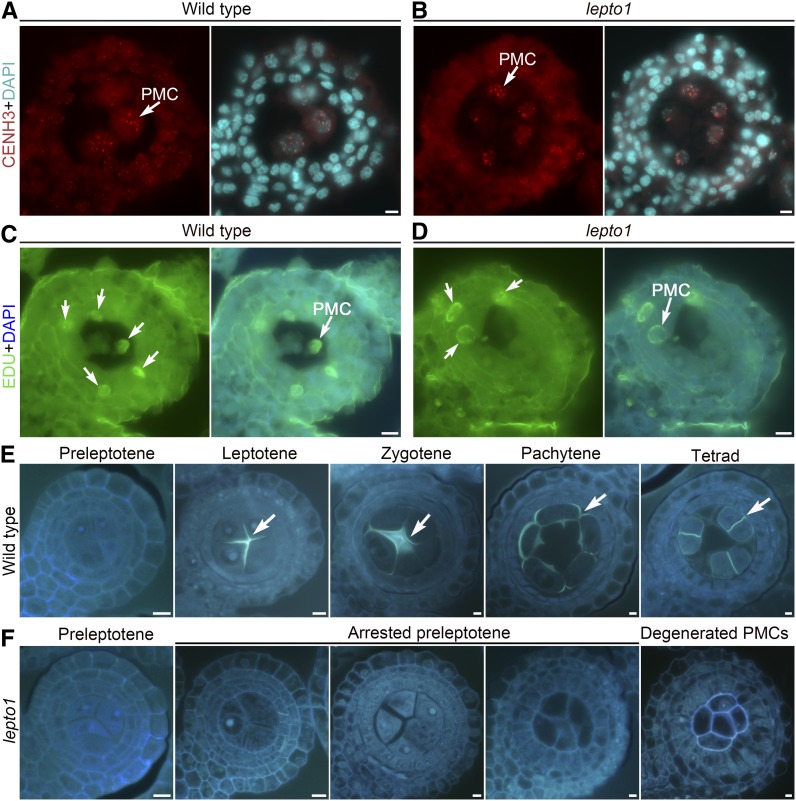
Figure 2.
The lepto1 Mutant Shows Normal Acquisition of Meiotic Fate but Disrupted Deposition of Meiotic Callose.
(A) and (B) Immunodetection of CENH3 (red) in PMCs of the wild type (A) and lepto1 (B). Chromosomes were stained with DAPI (light blue). Arrows indicate CENH3 signals in PMCs.
(C) and (D) Meiotic DNA replication occurred in lepto1 PMCs. Green fluorescence indicates EDU signals, and blue fluorescence indicates DAPI signals. Anthers containing four layers of somatic cells were selected for observation. Arrows indicate EDU signals, and EDU signals were detected in both wild-type and lepto1 PMCs.
(E) Transverse sections of wild-type anthers at various developmental stages stained with aniline blue. Arrows indicate aniline blue fluorescence signals.
(F) Transverse sections of lepto1 anthers at various developmental stages stained with aniline blue.
Bars = 5 μm.
As LEPTO1 is required for the organization of leptotene chromosomes, we further detected DNA replication in lepto1 PMCs. We conducted a 5-ethynyl-2′-deoxyuridine (EDU) incorporation experiment to detect whether meiotic DNA replication occurred in lepto1. Anthers containing four layers of somatic cells were selected for observation using transverse sections after treatment with EDU for 4 h. The EDU signals were detected in both wild-type and lepto1 PMCs, suggesting that meiotic DNA replication indeed occurred in lepto1 (Figures 2C and 2D).
In addition, rice sporocyteless (Osspl) mutants show abnormal development of sporogenous cells with failed acquisition of meiotic fate (Supplemental Figure 4A). We then constructed a double mutant of lepto1 and Osspl. Examination of transverse sections showed that the generation of preleptotene meiocytes was blocked completely in the Osspl lepto1 double mutant, suggesting that the function of LEPTO1 in preleptotene progression depends on OsSPL-mediated meiosis initiation (Supplemental Figure 4B).
Normal Callose Deposition Is Disrupted in lepto1 PMCs
The deposition of callose during meiosis helps the PMCs separate from each other (Chen et al., 2007). Therefore, we next asked whether this process was normal in lepto1 mutants. We investigated callose deposition by observing aniline blue-stained transverse sections of wild-type and lepto1 anthers. The callose deposition started to become visible at leptotene in wild-type PMCs. At zygotene, aniline blue signals around the PMCs were enhanced further. A circle of callose on the periphery of each individual PMC was then visible at pachytene. At the tetrad stage, the callose plate developed a cross-wall between the four daughter cells, which may promote the separation of the daughter cells from one another (Figure 2E). However, callose deposition was not observed in lepto1 PMCs, which exhibited a status similar to that of wild-type preleptotene PMCs (Figure 2F). In addition, PMCs were arranged into a long bunch with cell walls in the center of lepto1 anthers, while the similarly sized wild-type spikelet had already generated isolated microspores (Supplemental Figures 5A to 5C).
Rice UDP-Glucose Pyrophosphorylase1 (UGP1) and Glucan Synthase-Like5 (GSL5) encode two important enzymes involved in callose deposition during male gametophyte development. UGP1 is predominately expressed in PMCs during meiosis, and loss of UGP1 leads to abnormal callose deposition in PMCs (Chen et al., 2007). GSL5 is highly expressed in meiotic anthers, and the gsl5 mutant shows defects in male fertility and callose metabolism (Shi et al., 2015). RT-qPCR assays showed that the relative expression levels of UGP1 and GSL5 in lepto1 mutant anthers were downregulated by 45 and 89%, respectively, which was consistent with the results of aniline blue staining (Supplemental Figures 5D and 5E).
Loss of LEPTO1 Causes Failure of DSB Formation and Lack of Recruitment of Meiosis-Specific Proteins
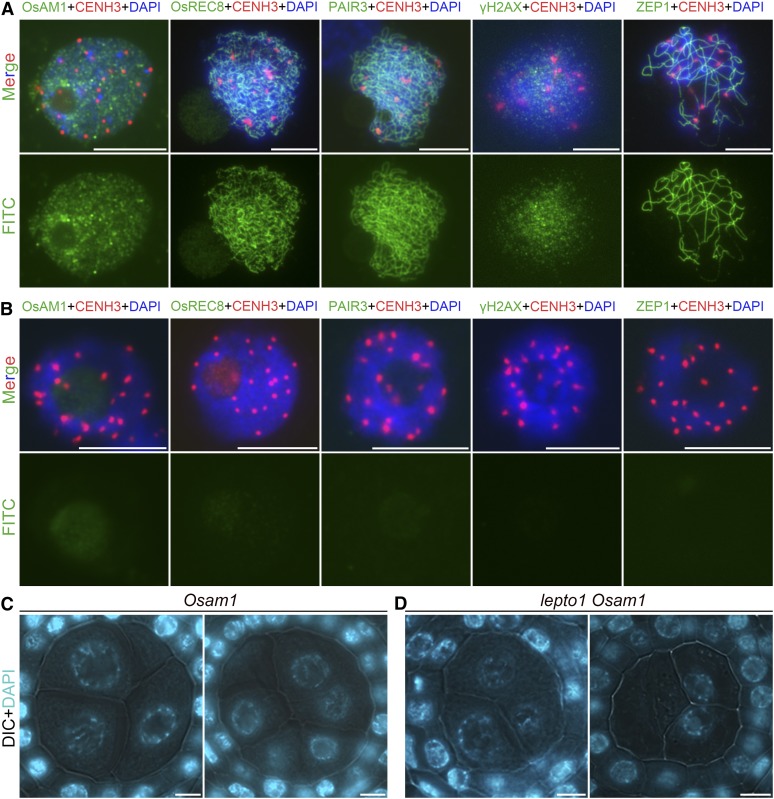
The cytological observations described above suggest that LEPTO1 might be required for the establishment of characteristic leptotene chromosomes in PMCs and imply that mutations in LEPTO1 may affect the loading of proteins that participate in prophase I progression. To test this, we performed dual immunolocalization experiments, in which a bright CENH3 signal indicated the PMCs. Rice AMEIOTIC1 (OsAM1) is a meiosis-specific coiled-coil protein that is recruited to meiotic chromosomes at leptotene (Che et al., 2011). No obvious OsAM1 signals could be detected in lepto1 PMCs (Figures 3A and 3B). In addition, PMCs of the lepto1 Osam1 double mutant showed similar defects to the lepto1 single mutant, supporting the hypothesis that LEPTO1 functions upstream of OsAM1 (Figures 3C and 3D).
Figure 3.
The lepto1 Mutant Has Defects in DSB Formation and Meiosis-Specific Protein Loading.
(A) Dual immunolocalization of meiotic proteins (green) and CENH3 (red) in PMCs of the wild type. The top row shows merged signals. The bottom row shows immunosignals of different proteins. Chromosomes were stained with DAPI (blue). FITC, fluorescein isothiocyanate.
(B) Dual immunolocalization of meiotic proteins (green) and CENH3 (red) in PMCs of lepto1. The top row shows merged signals. The bottom row shows immunosignals of different proteins. Chromosomes were stained with DAPI (blue).
(C) and (D) Transverse sections of Osam1 and lepto1 Osam1 anthers stained with DAPI. DIC, differential interference contrast.
Bars = 5 μm.
Moreover, the meiosis-specific cohesion subunit OsREC8 and the AE component PAIR3 failed to be installed onto chromosomes in lepto1 PMCs (Figures 3A and 3B). In the lepto1 mutant, aborted loading of γH2AX, a phosphorylated form of the histone variant H2AX that marks the sites of meiotic DSBs, was observed, indicating that meiotic DSB formation did not take place (Figures 3A and 3B). ZEP1 is the central element of the synaptonemal complex in rice meiosis. As expected, we did not detect any ZEP1 signals in lepto1 PMCs, suggesting that there is a defect in homologous recombination and synaptonemal complex formation (Figures 3A and 3B). Therefore, we suspect that the key events involved in prophase I progression are not initiated due to the loss of LEPTO1.
The lepto1 Mutant Has Defects in the Development of Anther Somatic Cells
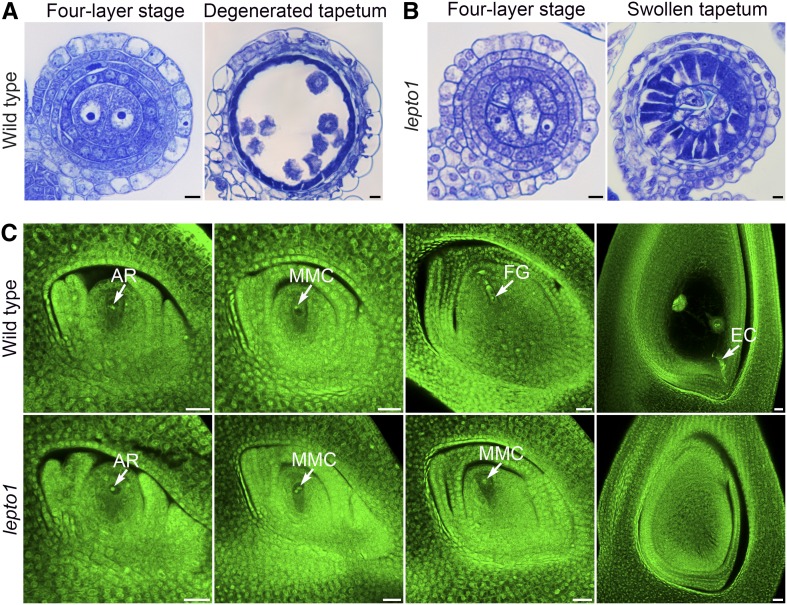
To determine whether the development of somatic layers in lepto1 anthers was normal, transverse sections of wild-type and lepto1 anthers were stained with toluidine blue O (TBO) for further examination. We found that there was no cytological difference between wild-type and lepto1 anthers up to the four-layer stage. Somatic layers with normal morphology were found in both wild-type and lepto1 anthers, which included the epidermis, endothecium, middle layer, and tapetal layer (Figures 4A and 4B). In the wild type, the three hypodermal somatic layers then underwent degeneration upon the completion of meiosis (Figure 4A). However, these somatic layers did not degrade in lepto1 anthers, and lepto1 tapetum swelled as the anther volume increased (Figure 4B). lepto1-cas9 also showed a similar swollen tapetum, and the inward growth of the tapetum in lepto1 was quite different from the weakly expanded tapetum observed in the meiotic mutant Osspo11-1 and the meiotic arrest mutant Osam1 (Supplemental Figures 6B, 6C, and 6E). Moreover, the tapetal defects in the lepto1 Osam1 double mutant were similar to those observed in lepto1 (Supplemental Figures 6C and 6D), indicating a role of LEPTO1 in regulating the development of anther somatic layers.
Figure 4.
LEPTO1 Is Required for the Development of Anther Somatic Cells and Megasporogenesis.
(A) Transverse sections of wild-type anthers stained with TBO. Bars = 5 μm.
(B) Transverse sections of lepto1 anthers stained with TBO. Bars = 5 μm.
(C) The examination of ovule development in the wild type (archesporial cell [AR], megaspore mother cell [MMC], functional megaspore [FG], and mature embryo sac) and lepto1. No functional megaspore was observed in lepto1. EC, egg cell. Bars = 20 μm.
In addition, we performed the terminal deoxynucleotidyl transferase-mediated dUTP nick-end labeling assay to explore whether the lepto1 mutant has defects in the programmed cell death (PCD) of somatic cells. In the wild type, PCD signals were detectable at the microspore stage (Supplemental Figures 6F and 6G). However, no obvious PCD signals were detected in lepto1 somatic cells even when the tapetum swelled (Supplemental Figures 6H and 6I). Consistent with this, TAPETUM DEGENERATION RETARDATION (TDR) and ETERNAL TAPETUM 1 (EAT1), which encode key regulators of tapetal PCD, were downregulated by 47 and 35%, respectively, in the lepto1 mutant (Supplemental Figures 6J and 6K). Moreover, we detected downregulated expression of OsCP1, a Cys protease-encoding gene regulated by TDR (Supplemental Figure 6L). Also, two direct targets of EAT1, OsAP25 and OsAP37, which encode aspartic proteases, showed decreased levels of expression (Supplemental Figures 6M and 6N). Therefore, we conclude that lepto1 anthers also may have defects in tapetal PCD.
LEPTO1 Is Required for Megasporogenesis
The lepto1 mutant failed to set any seeds when pollinated with wild-type pollen, indicating that LEPTO1 also plays a role in female reproductive development. Therefore, we performed whole-mount stain-clearing laser scanning confocal microscopy (WCLSM) to examine the developmental defects in megasporogenesis in the lepto1 mutant. We observed that, in lepto1 ovules, archesporial cells were able to differentiate into megaspore mother cells (MMC), and the MMCs showed no morphological differences compared with those of the wild type (Figure 4C). In the wild type, MMCs further underwent meiosis to form four megaspores, among which a single surviving megaspore located at the chalazal end underwent three rounds of mitosis and gave rise to a mature embryo sac. By contrast, meiotic processes were not observed in lepto1 MMCs. Consistent with this, the mutant failed to produce normal embryo sacs (Figure 4C). These results from WCLSM observation show that meiotic events also are affected in lepto1 ovules.
LEPTO1 Is Expressed Preferentially in Early-Meiosis PMCs
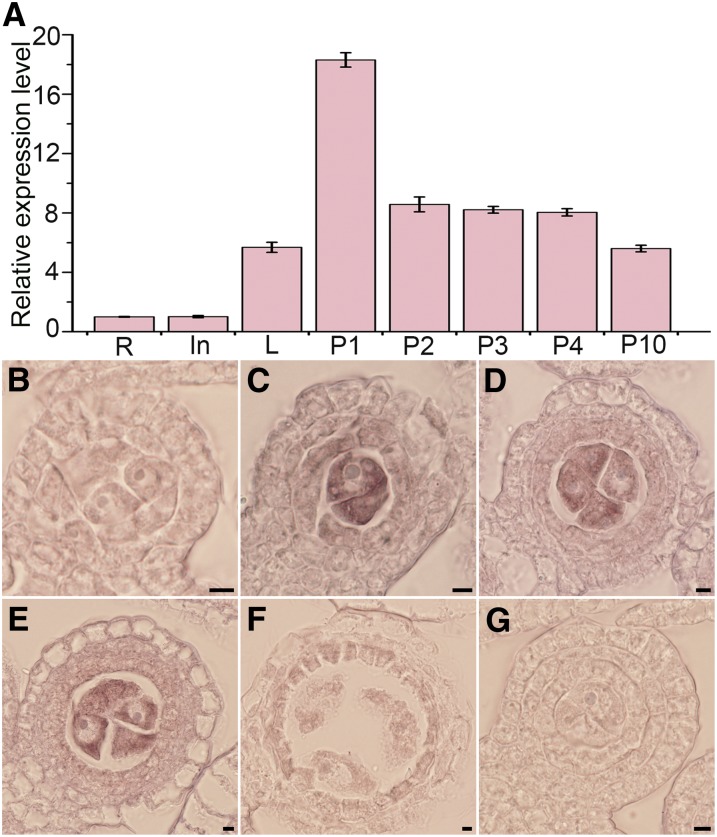
We conducted RT-qPCR analysis and RNA in situ hybridization to examine the spatiotemporal expression pattern of LEPTO1. Based on the results of RT-qPCR, LEPTO1 was expressed weakly in roots and internodes and at a low level in leaves. LEPTO1 mRNA accumulated to high levels in the 1-cm-long panicle but decreased dramatically as the panicles developed (Figure 5A). Furthermore, RNA in situ hybridization showed that LEPTO1 transcripts were barely observable in wild-type anthers at the two-layer stage (Figure 5B). LEPTO1 transcripts peaked later at the three-layer stage, and LEPTO1 was found to be most highly expressed in germ cells. Weak LEPTO1 mRNA signals also were observed in hypodermal somatic layers (Figure 5C). Following the formation of a four-layer pattern, LEPTO1 transcripts remained at a relatively high level and accumulated primarily in PMCs (Figures 5D and 5E). Subsequently, the relative expression of LEPTO1 decreased and became almost undetectable thereafter (Figure 5F). By contrast, no signal was detected in the anther section with the sense probe (Figure 5G). Collectively, the results of expression analysis show that LEPTO1 is expressed mainly in early-meiosis PMCs, supporting the potential roles of LEPTO1 in leptotene chromosome organization and the development of somatic cell layers.
Figure 5.
Expression Profiles of LEPTO1.
(A) RT-qPCR analysis of LEPTO1 expression. In, internode; L, leaf; P1, 1-cm-long panicle; P2, 2-cm-long panicle; P3, 3-cm-long panicle; P4, 4-cm-long panicle; P10, 10-cm-long panicle; R, root. Expression values based on RT-qPCR are means ± sd of three biological replicates.
(B) to (G) Expression of LEPTO1 in wild-type anthers determined by RNA in situ hybridization: two-layer stage (B), three-layer stage (C), early four-layer stage ([D] and [E]), pachytene (F), and negative control using sense-strand probe (G). Bars = 5 μm.
LEPTO1 Encodes a Type-B RR in Rice
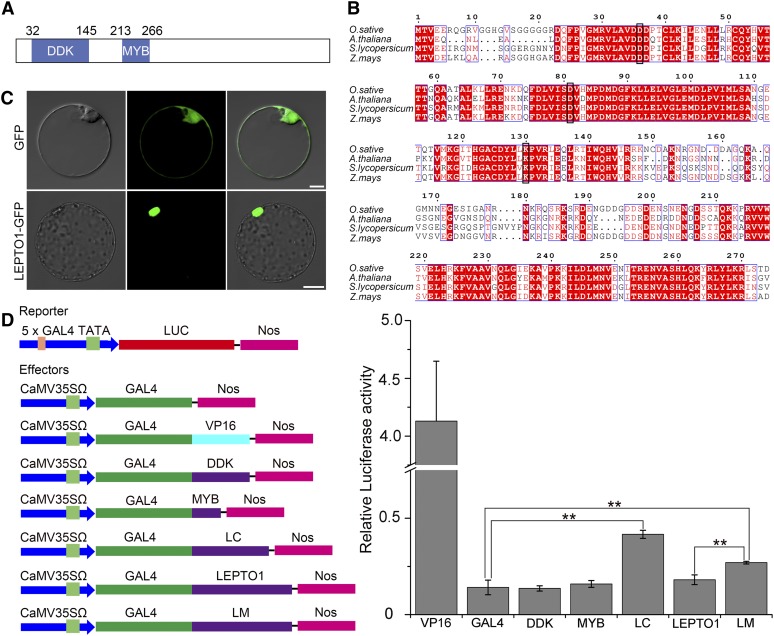
LEPTO1 encodes a predicted 626-amino acid protein belonging to the type-B RR family, and LEPTO1 contains a signal receiver domain (DDK; amino acids 32–145) and a myb-like DNA binding domain (MYB; amino acids 213–266) at the N terminus (Figure 6A). A multiple alignment of the predicted full-length protein sequences of LEPTO1 and its homologs from other plants revealed that the DDK and MYB domains were highly conserved in plants (Figure 6B). In a previous study, LEPTO1 was named rice OsRR24 and distributed in the B-I subfamily (Tsai et al., 2012). As shown in Supplemental Figure 7, LEPTO1 was most closely related to ZmRR9 in maize by phylogenetic analysis.
Figure 6.
LEPTO1 Encodes a Nuclear Type-B RR.
(A) Schematic representation of LEPTO1.
(B) Multiple sequence alignment of LEPTO1 with its homologs from Arabidopsis (AtARR12, NP_180090.6), tomato (SlRR9, XP_004242565.1), and maize (ZmRR9, NP_001104863.1). Black boxes show the conserved Asp and Lys of the DDK domain.
(C) Subcellular localization of LEPTO1. The LEPTO1:GFP fusion and the GFP control were expressed separately in rice protoplasts. Bars = 10 μm.
(D) Relative luciferase activity detected in the Arabidopsis transient assay. DDK, the DDK domain of LEPTO1; GAL4, negative control; LC, the C-terminal region of LEPTO1 (amino acids 301–626); LEPTO1, the full-length amino acid sequence of LEPTO1; LM; the mutated LEPTO1 in which Asp-81 was substituted with Glu; MYB, the MYB domain of LEPTO1; VP16, positive control. Ratios of firefly luciferase (Luc) to Renilla luciferase activity in Arabidopsis protoplasts are means ± sd of three biological replicates. ** P < 0.01, two-tailed Student’s t test.
Studies in Arabidopsis have shown that exogenous cytokinin treatment affects the expression of type-A RR genes, but the expression levels of type-B RR genes are not regulated by cytokinin (D’Agostino et al., 2000). Therefore, we examined the transcription change of LEPTO1 in response to exogenous cytokinin treatment in rice seedlings and panicles. The expression level of LEPTO1 in the aboveground parts of 2-week-old wild-type seedlings treated with 6-benzylaminopurine for 6 h showed no significant differences from the control, which was different from OsRR1, a cytokinin-induced type-A RR gene (Supplemental Figures 8A and 8B). In addition, the expression of LEPTO1 in panicles also was not induced after treatment with 50 ppm of a synthetic cytokinin (N-2-chloro-4-pyridyl benzene-N′-phenyl urea) for 12 h (Supplemental Figures 8C and 8D). These results suggest that the expression of LEPTO1 is not induced by cytokinin treatment, which is similar to the expression characteristics of type-B RRs in Arabidopsis.
LEPTO1 Is a Nuclear Protein with Potential Transactivation Ability
To examine the subcellular localization of LEPTO1, we fused LEPTO1 with a green fluorescent protein (GFP) tag and introduced the construct into rice protoplasts. The GFP signal was located in the nucleus, suggesting that LEPTO1 is a nuclear protein (Figure 6C). Consistent with this, GFP signals in the spikelets of Pactin:LEPTO1-GFP transgenic lines were observed in the nuclei (Supplemental Figure 9). We also examined the transcriptional activity of LEPTO1 using the dual luciferase reporter assay system in Arabidopsis protoplasts. Various portions of LEPTO1 were fused with the yeast GAL4 DNA binding domain (GAL4) as effectors, and the reporter was the firefly luciferase gene preceded by the GAL4 target sequence. The results showed that transactivation occurred when the effector was the C terminus of LEPTO1 (LEPTO1C; amino acids 301–626). This result also was supported by the fact that LEPTO1C exhibited self-activation in a yeast two-hybrid (Y2H) system (Figure 7A). However, the DDK domain, the MYB domain, and the full-length LEPTO1 protein did not show transactivation ability (Figure 6D). Therefore, the results indicate that LEPTO1C possesses the ability to promote transcription, but the N-terminal amino acid sequence of LEPTO1 may repress this activity.
Figure 7.
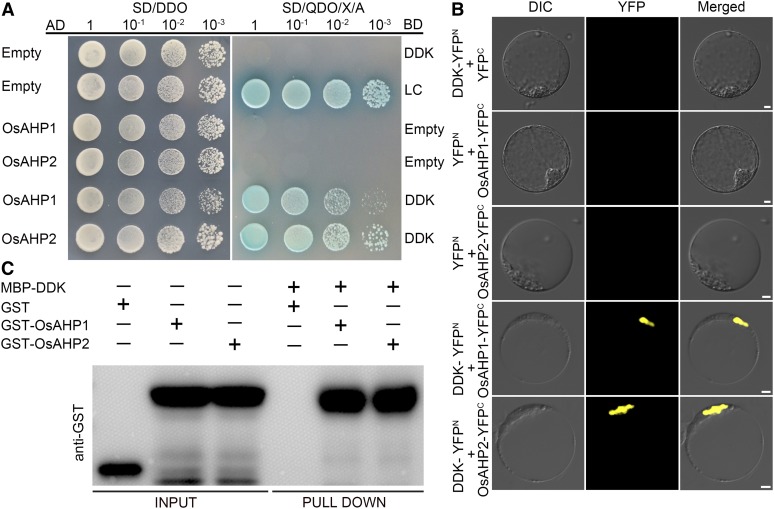
The DDK Domain of LEPTO1 Interacts with OsAHP1 and OsAHP2.
(A) The DDK domain of LEPTO1 interacts with OsAHP1 and OsAHP2 in Y2H assays. The transformation efficiency test was performed on SD-Leu-Trp medium (SD/DDO). The interactions were detected on SD/QDO/X/A. AD, prey vector; BD, bait vector.
(B) The DDK domain of LEPTO1 interacts with OsAHP1 and OsAHP2 in BiFC assays. Various vector pair combinations were cotransformed into rice protoplasts. DIC, differential interference contrast. Bars = 5 μm.
(C) The DDK domain of LEPTO1 interacts with OsAHP1 and OsAHP2 as determined by pull-down assays. Glutathione S-transferase (GST)-OsAHP1 and GST-OsAHP2 were pulled down by the MBP-DDK protein using immunoblotting analysis. The GST tag alone was used as the negative control.
Studies in Arabidopsis have indicated that phosphorylation of the second conserved Asp of the DDK domain of ARR1, a type-B RR, can relieve its repression of the transcriptional activation ability of ARR1 (Wang et al., 2017). Therefore, we substituted the second conserved Asp of the DDK domain (Asp-81) with Glu (LEPTO1D81E) to mimic the phosphorylated state (To et al., 2007). Compared with wild-type LEPTO1, this phosphorylated mimic form of LEPTO1 significantly improved relative luciferase activity (Figure 6D). Taking these results together, we propose that LEPTO1, as a type-B RR in rice, very likely acquires transactivation ability with the phosphorylation of the DDK domain at the second conserved Asp.
The DDK Domain of LEPTO1 Interacts with OsAHP1 and OsAHP2
We performed Y2H screening to identify LEPTO1-interacting proteins using a cDNA library from anthers of 1.5- to 2.5-mm spikelets as the prey. As LEPTO1C has self-activation ability and the MYB domain is thought to be involved in DNA binding, we selected the DDK domain of LEPTO1 as the bait. We screened ∼2 × 106 yeast transformants and obtained 12 positive clones. Among these 12 positive clones, 8 were found to encode OsAHP1 and 4 were found to encode OsAHP2; both proteins belong to the HP family and have been reported to be involved in the cytokinin signaling pathway in rice (Sun et al., 2014).
To verify these interactions, the full-length coding sequences of OsAHP1 and OsAHP2 were cloned separately into pGADT7, and the DDK domain coding sequence of LEPTO1 was cloned into pGBKT7. Yeast strains cotransformed with OsAHP1-AD or OsAHP2-AD and DDK-BD grew normally on SD/-Ade/-His/-Leu/-Trp medium containing X-α-Gal and aureobasidin A (SD/QDO/X/A; Figure 7A). We further conducted bimolecular fluorescence complementation (BiFC) assays to confirm these interactions. Yellow fluorescent protein (YFP) signals were detected in the nuclei of protoplasts coexpressing DDK-YFPN and either OsAHP1-YFPC or OsAHP2-YFPC (Figure 7B). Furthermore, the interactions between the DDK domain of LEPTO1 and OsAHP1 and OsAHP2 were validated using in vitro pull-down assays (Figure 7C). Moreover, when the full-length LEPTO1 sequence was inserted into pGADT7, the protein also could interact with OsAHP1-BD and OsAHP2-BD in Y2H assays (Supplemental Figure 10). These results suggest that the DDK domain of LEPTO1 mediates the interactions between LEPTO1 and the two OsAHP proteins.
LEPTO1 Regulates the Expression of Several Cyclin Genes
Because LEPTO1 may function as a transcriptional activator, we investigated whether the failure of loading of meiotic elements described above was caused by the downregulation of their expression in lepto1. OsAM1 and OsREC8 are thought to be installed at the beginning of meiosis, and both are required for the normal localization of most meiotic proteins (Che et al., 2011; Shao et al., 2011). Nevertheless, the results of RNA in situ hybridization showed that mRNA signals of both OsAM1 and OsREC8 were not affected in the lepto1 mutants. Moreover, the expression levels of PAIR3, OsDMC1, and OsSPO11-1 also were not affected in lepto1. However, the ZEP1 mRNA signal was not detectable in wild-type or lepto1 PMCs during the preleptotene stage (Supplemental Figure 11). Therefore, we speculate that the loading defects were more likely due to an inappropriate chromosome state in lepto1 meiocytes.
In Arabidopsis, multiple cyclins have potential roles in meiotic cycle progression (Bulankova et al., 2013). And in rice, plants carrying a mutation in a meiosis-specific cyclin, OsSDS, have defects in meiotic DSB formation (Wu et al., 2015). Interestingly, we found that the expression of several cyclin genes was downregulated significantly in lepto1. As shown in Supplemental Figure 12, an A-type cyclin, CYCA1-1, a B-type cyclin gene, CYCB1-3, and OsSDS were highly expressed in wild-type panicles undergoing meiosis. However, these genes showed reduced expression in lepto1 anthers (Supplemental Figure 13), indicating that cyclin-dependent meiotic processes may rely on the normal function of LEPTO1.
DISCUSSION
Meiosis is a specific and highly coordinated process that requires precise regulation and produces observable changes in the structure of chromosomes. Previous studies have uncovered the existence of a preleptotene stage that occurs around the time that meiosis starts (Zickler and Kleckner, 1998). Preleptotene chromosome contraction has been shown to participate in the establishment of leptotene chromosomes, and this chromosome behavior may be conserved in both plants and animals (Burns, 1972; Bennett and Stern, 1975; Hartung and Stahl, 1977; Luciani et al., 1977; Zickler and Kleckner, 1998). However, the regulatory mechanisms involved in this important process have not yet been studied in plants. In this study, we characterized a sterile rice mutant with defects in this meiotic event and identified the OsRR24/LEPTO1 gene using map-based cloning.
Combining previous studies and our results suggests that the appearance of chromosomes at preleptotene resembles that of prometaphase chromosomes, but the edges of preleptotene chromosomes appear to be more irregular (Bennett and Stern, 1975; Hartung and Stahl, 1977). Preleptotene meiocytes can be observed easily in early developed rice anthers. PMCs possess a specialized pattern for CENH3 loading and show distinguishable immunofluorescence signals of CENH3 at preleptotene (Ravi et al., 2011; Ren et al., 2018). In the lepto1 mutant, PMCs were normal in appearance, with a large nucleus and nucleolus, and preleptotene chromosomes were observed in lepto1 PMCs with bright CENH3 signals. However, further chromosome progression was abolished in lepto1 PMCs, and the thin thread-like structures did not form. Based on these cytological observations, we suspect that LEPTO1 is probably required for the establishment of the characteristic leptotene chromosome structure.
The molecular events involved in leptotene meiocytes have been well described. During early leptotene, the appropriate relationship is established between sister chromatids, which includes the assembly of the cohesin complex and the formation of the loop-axis chromosome structure (Borde and Lichten, 2014). However, two important meiotic proteinaceous elements, OsREC8 and PAIR3, are not able to load onto lepto1 chromosomes, suggesting failures in the formation of meiotic axial chromosome structure. Moreover, meiotic DSB formation is absent in lepto1 meiocytes, and DSB-dependent meiotic recombination is abolished correspondingly. The AM1 protein is involved in many crucial events in the early stage of meiosis and is required for the establishment of sister chromatid cohesion in both Arabidopsis and rice. Loss of function in AM1 in Arabidopsis and rice leads to different degrees of defects in chromosome condensation (Mercier et al., 2001; Che et al., 2011). Osam1 mutant PMCs show arrested condensation of chromosomes at leptotene. The absence of OsAM1 loading on lepto1 chromosomes supports the hypothesis that OsAM1 may function downstream of LEPTO1 in leptotene chromosome organization. Because the expression levels of OsREC8, OsAM1, PAIR3, and OsSPO11-1 in PMCs showed no obvious differences between the wild type and lepto1, we suspect that the initiation of these critical events at leptotene may require the normal function of LEPTO1 to generate an appropriate chromosome state, providing further evidence for the important role of LEPTO1 in leptotene chromosome establishment.
Rice LEPTO1 encodes the protein OsRR24, which belongs to the nuclear type-B RR family. We found that the DDK domain of LEPTO1 can interact with OsAHP1 and OsAHP2, two putative functional HPs in rice (Sun et al., 2014). Also, the protein sequence alignment shows that LEPTO1 contains a conserved Asp (Asp-81) in the DDK domain that may serve as a potential phosphorylation site. This suggests that LEPTO1 may be able to receive a phosphate group through direct interactions with OsAHPs. Experiments using the dual luciferase reporter assay system and Y2H assays demonstrated that the C terminus of LEPTO1 has transactivation ability. Furthermore, this ability may be repressed by the N-terminal amino acids of LEPTO1. The phosphorylation mimic LEPTO1D81E relieved the inhibition of the DDK domain on its transcriptional activity. Therefore, we speculate that the activity of LEPTO1 may be regulated by HPs in rice. Moreover, these results indicate a potential role of cytokinin in the meiotic process, which needs to be investigated further.
The progression of the cell cycle requires the elevated expression of cyclin genes. In Arabidopsis, most cyclins are thought to have a role in the cell cycle (Schaller et al., 2014). For example, the D-type cyclin CYCD3 forms a CDKA/CYCD complex to promote the G1/S transition (Scofield et al., 2013). Studies in model organisms indicate that some cyclins play significant roles in the meiotic cell cycle. In mouse, loss of function in cyclin A1 blocks meiosis at diplotene (Liu et al., 1998). In budding yeast, two type-B cyclins, Clb5 and Clb6, are required for premeiotic DNA replication and activation of the S/M checkpoint (Stuart and Wittenberg, 1998). Moreover, COSA-1, a cyclin-related protein, participates in the conversion of DSBs into crossovers in Caenorhabditis elegans (Yokoo et al., 2012). Massive expansion has occurred in the cyclin gene family in angiosperms (Bulankova et al., 2013). Arabidopsis SDS is essential for homologous chromosome paring, and CYCA1-2 is required for the transition from meiosis I to meiosis II (Azumi et al., 2002; d’Erfurth et al., 2010). The meiosis-specific cyclin OsSDS also is required for the meiotic process but plays a role in DSB formation in rice (Wu et al., 2015). In this study, we showed that three cyclin genes expressed preferentially in meiotic panicles are significantly downregulated in lepto1. The disrupted function of OsSDS may contribute to the impaired DSB formation in lepto1 meiocytes. Nevertheless, unlike most mutants with meiotic defects, including Ossds, the lepto1 mutant exhibits arrested meiosis at preleptotene. This stage may represent a meiosis-specific G2/M transition phase or the early stage of leptotene (Hartung and Stahl, 1977). In plants, A-type and B-type cyclins have been implicated in the G2/M transition in mitosis (Francis, 2011). The expression of A/B-type cyclin genes is regulated by conserved MYB transcription factors in both plants and animals (Ito et al., 2001; Araki et al., 2004; Nakata et al., 2007). Unlike animals and yeast, there is no cyclin mutant showing arrested prophase I in plants, which may be due to a functional redundancy. Considering that LEPTO1 contains a conserved MYB domain, we suspect that the mutation in LEPTO1 may prevent preleptotene chromosomes from proceeding into thin thread-like structures by affecting the expression of A/B-type cyclin genes and a variety of other coexpressed genes (Schaller et al., 2014).
In addition, lepto1 also has defects in the development of anther somatic cells, such as the abnormally swollen tapetum. Our terminal deoxynucleotidyl transferase-mediated dUTP nick-end labeling analysis further showed that the PCD process is retarded in the swollen tapetal cells. The basic helix-loop-helix members TDR and EAT1, and their downstream target genes OsCP1, OsAP25, and OsAP37, are believed to be responsible for the activation of tapetal PCD (Niu et al., 2013). Decreased expression of these PCD-related genes may be the main reason for the abnormal development of somatic cells in lepto1. However, mutations in TDR and EAT1 do not affect the completion of the meiotic process (Li et al., 2006; Niu et al., 2013). Given that the accumulation of LEPTO1 transcripts was detected in both PMCs and hypodermal somatic layers, we propose that LEPTO1 is likely to regulate meiosis progression and also the development of tapetum. Moreover, previous studies have suggested a non-cell-autonomous regulation between tapetum and PMCs (Ono et al., 2018). There exists a possibility that mutations in LEPTO1 may disturb tapetum-meiocyte communication and lead to more serious defects in meiosis progression.
To conclude, we have shown that OsRR24/LEPTO1 encodes a type-B RR involved in the establishment of meiotic leptotene chromosome and the degradation of tapetum in rice. We have provided evidence that LEPTO1 can interact with OsAHP1 and OsAHP2 via its DDK domain and that LEPTO1 has potential transcriptional activity. Therefore, we propose that LEPTO1 may act as a transcription factor to regulate the expression of related genes to affect the development of meiocytes and nurse cells.
METHODS
Plant Material and Growth Conditions
The rice (Oryza sativa) lepto1 mutant was obtained from the progeny of the indica rice variety, Guangluai 4, which had been treated with 60Co γ-radiation. The lepto1-cas9 mutant was produced by CRISPR-Cas9 gene editing in the japonica rice variety Yandao 8. The Osspl and Osam1 mutants used in this study were described previously (Che et al., 2011; Ren et al., 2018). Guangluai 4 was used as the wild type. All rice plants were cultivated in paddy fields under natural conditions in Beijing. Arabidopsis (Arabidopsis thaliana) ecotype Columbia used for transcriptional activity assays was grown under ∼100 μmol m−2 s−1 white light with an 8-h photoperiod in a growth room at 22°C.
Map-Based Cloning of LEPTO1
To create mapping populations, we crossed plants heterozygous for lepto1 with the japonica rice variety Wuyunjing 8. For linkage analysis and fine mapping, 210 sterile F2 progeny and 450 sterile F3 progeny plants were used, respectively. The primer sequences used for map-based cloning are given in Supplemental Table 1.
Phenotypic Characterization
Fresh panicles representing different developmental stages in both the wild type and lepto1 were fixed in Carnoy’s solution (ethanol:glacial acetic acid, 3:1). To examine anther development, the samples were dehydrated in an ethanol series, embedded in Technovit 7100 resin (Hereaus Kulzer), and polymerized at 37°C for 4 to 5 d. The spikelets were transversely sectioned into 4-μm-thick slices using a Leica microtome. The samples were stained with DAPI or 0.25% (w/v) TBO and observed with a light microscope.
Immunofluorescence
For immunofluorescence assays, fresh young panicles of both the wild type and the lepto1 mutant were fixed in 4% (w/v) paraformaldehyde for 30 min at room temperature. The spikelets were then soaked in phosphate-buffered saline (PBS) and squashed on glass slides. The slides were frozen in liquid nitrogen and dehydrated using an ethanol series (70, 90, and 100%). The slides were then incubated with different antibodies diluted 1:500 in TNB buffer (0.1 M Tris-HCl, pH 7.5, 0.15 M NaCl, and 0.5% blocking reagent) for 4 h in a humid chamber at 37°C. Texas red-conjugated goat anti-rabbit antibody and fluorescein isothiocyanate-conjugated sheep anti-mouse antibody (1:1000) were added to the slides after three rounds of washing in PBS. Finally, the slides were incubated in a humid chamber for 45 min at 37°C and counterstained with DAPI after washing in PBS three times (Hu et al., 2017a). Fluorescence signals were observed using a Zeiss A2 fluorescence microscope.
WCLSM
For the detection of female gametophytes by WCLSM, florets from both the wild type and the lepto1 mutant were first fixed in Carnoy’s solution. Dissected ovaries were then sequentially hydrated in 50% ethanol, 30% ethanol, and distilled water, each step for 30 min. Following pretreatment in 2% aluminum potassium sulfate for 20 min, the ovaries were stained with 10 mg/L eosin B for 10 to 12 h at room temperature. Following this, the samples were dehydrated in a graded ethanol series (30, 50, 70, and 100%) and pretreated in a mixture of ethanol and methyl salicylate (1:1) for 1 h. The ovaries were finally cleared in pure methylsalicylate and observed with a laser scanning confocal microscope (Leica TCS SP5).
Real-Time PCR and RACE
For spatiotemporal expression pattern analysis of LEPTO1, total RNA from various tissues of the wild type (root, internode, leaf, 1-cm-long panicle, 2-cm-long panicle, 3-cm-long panicle, 4-cm-long panicle, and 10 cm-long panicle) was extracted using TRIzol reagent. For expression analysis of other genes, meiotic anthers of both the wild type and the lepto1 mutant were collected for RNA extraction. After digestion to remove contaminating genomic DNA and cDNA synthesis (Invitrogen), real-time PCR assays were conducted on a Bio-Rad CFX96 instrument using SsoFast EvaGreen. Rapid amplification of cDNA ends (RACE) was conducted to isolate full-length cDNA of LEPTO1 using the SMARTer RACE 5′/3′ Kit (Takara) according to the manufacturer’s instructions. Primer sequences used for real-time PCR and RACE assays are given in Supplemental Table 2.
RNA in Situ Hybridization
LEPTO1 riboprobes were transcribed with T7 RNA polymerase, and the transcripts were labeled using the digoxigenin RNA labeling kit (Roche). Sample fixation and RNA in situ hybridization were performed according to a previous study (Wang et al., 2014).
Phylogenetic Analysis
The full-length LEPTO1 sequence was used as a query to conduct PSI-BLAST searches. The conserved N-terminal amino acid sequences (DDK and MYB domains) of LEPTO1 and other type-B subfamily I RRs from four plant species (rice, Arabidopsis, tomato [Solanum lycopersicum], and maize [Zea mays]) were used for phylogenetic analysis. A subfamily B-III RR in Arabidopsis, AtARR20, was used as the outgroup gene. The protein sequences were aligned with ClustalW, and a neighbor-joining phylogenetic tree was constructed using MEGA 5.2 software. The number of bootstrap replications was set as 1000. The resulting phylogenetic tree and the alignments used are shown in Supplemental Figure 7 and Supplemental File.
Multiple Sequence Alignments
Multiple alignments were performed using MAFFT (https://toolkit.tuebingen.mpg.de/mafft) and processed with ESPRIPT3 (http://espript.ibcp.fr/ESPript/ESPript/).
Subcellular Localization of LEPTO1
We cloned the coding sequence of LEPTO1, and the PCR product was inserted into the pJIT163-GFP vector. Both the LEPTO1-GFP fusion construct and the empty GFP vector were introduced into rice protoplasts. Following incubation in the dark at 28°C for 20 h, protoplasts with GFP signals were observed with a laser scanning confocal microscope (Leica TCS SP5).
Transcriptional Activity Analysis
Transcriptional activity analysis was performed in Arabidopsis protoplasts, and the relative level of firefly luciferase activity was detected using the dual-luciferase reporter assay system (Promega). The DDK domain, the MYB domain, LEPTO1C, LEPTO1, or LEPTO1D81E were fused with the DNA binding domain of yeast GAL4 and used as effectors. The point mutation that converts Asp-81 to Glu was introduced into the DDK domain using a site-directed mutagenesis system (Thermo Fisher Scientific). The reporter was the firefly luciferase gene preceded by the GAL4 target sequence and the TATA box, the core element of the CaMV 35S promoter. Renilla luciferase driven by the 35S promoter was used as the internal control. Each effector was cotransformed with the reporter and internal control into Arabidopsis protoplasts. After transformation, the protoplasts were incubated in the dark at 23°C for 16 to 20 h. The relative firefly luciferase activity, representing the firefly luciferase:Renilla luciferase ratio, was determined using a GloMax 20-20 Luminometer (Promega). Each cotransformation was repeated at least three times.
CRISPR-Cas9 Targeting of LEPTO1
For CRISPR-Cas9 targeting of LEPTO1, one specific target sequence was selected using an online toolkit (https://www.genome.arizona.edu/crispr/). The target sequence was ligated with the intermediate vector SK-gRNA, which had been digested with AarI. The DNA fragment was released by AarI digestion and then inserted into the AarI-linearized CRISPR-Cas9 binary vector pC1300-Cas9s (Hu et al., 2017b; Hua et al., 2017). The construct was introduced into the japonica rice variety Yandao 8 using Agrobacterium tumefaciens-mediated transformation.
Library Screening and Y2H Assays
For library screening, the DDK domain of LEPTO1 was introduced into the pGBKT7 vector (Clontech) as the bait. Wild-type anthers from 1.5- to 2.5-mm spikelets were used to construct the prey library according to the Mate and Plate Library System (Clontech). For Y2H assays, the full-length coding sequences of OsAHP1 and OsAHP2 were amplified and cloned into pGADT7 (Clontech). Yeast Y2H Gold strain cells were cotransformed with pGBKT7-DDK and pGADT7-OsAHP1 or pGADT7-OsAHP2 using the Matchmaker Gold Yeast Two-Hybrid system (Clontech). Both the assays for the self-activating effect and protein interactions were conducted on SD/QDO/X/A plates. Primer sequences used for plasmid vector construction are given in Supplemental Table 3.
BiFC Assays
For BiFC assays, the DDK domain of LEPTO1 was fused with the pSCYNE vector. The full-length coding sequences of OsAHP1 and OsAHP2 were cloned into the pSCYCE vector. Rice protoplasts were cotransformed with various vector combinations. After incubation in the dark at 28°C for 16 to 20 h, YFP signals were observed using a laser scanning confocal microscope (Leica TCS SP5).
Pull-Down Assays
The DDK domain coding sequence of LEPTO1 was inserted into the pMAL-c5X vector (New England Biolabs). The full-length coding sequences of OsAHP1 and OsAHP2 were cloned into the pGEX-4T-2 vector (GE Healthcare). The DDK domain fused with the MBP and OsAHP1 and OsAHP2 fused with glutathione S-transferase were expressed in Escherichia coli BL21 (DE3) cells at 37°C for 5 h and induced by 1 mM IPTG. Pull-down assays were performed as described previously (Bai et al., 2012).
Accession Numbers
Sequence data used in this article can be found in the National Center for Biotechnology Information databases under the following accession numbers: AtARR14, NP_178285.1; AtARR11, NP_176938.1; AtARR1, NP_566561.2; AtARR2, NP_193346.5; AtARR18, NP_200616.4; AtARR10, NP_194920.1; AtARR12, NP_180090.6; AtARR20, NP_001319821.1; SlRR14, XP_010319384.1; SlRR26, XP_004239497.1; SlRR1, XP_004239797.1; SlRR2, XP_004229281.1; SlRR24, XP_004251765.1; SlRR9, XP_004242565.1; OsRR21, XP_015630577.1; OsRR22, XP_015640894.1; OsRR23, XP_015625496.1; LEPTO1, XP_015626716.1; OsRR25, XP_025882103.1; OsRR26, XP_015622017.1; ZmRR8, NP_001104861.2; ZmRR9, NP_001104863.1; ZmRR22, XP_008657992.1; ZmRR23, PWZ23280.1; ZmRR25, XP_008659833.1; ZmRR26, XP_008656969.1.
Supplemental Data
Supplemental Figure 1. Map-based cloning of LEPTO1 and characterization of the rice sterile mutant lepto1.
Supplemental Figure 2. Cytological behavior of lepto1 and lepto1-cas9.
Supplemental Figure 3. The cDNA sequence of LEPTO1.
Supplemental Figure 4. Cytological behavior of the lepto1 osspl double mutant.
Supplemental Figure 5. lepto1 PMCs are difficult to separate from each other.
Supplemental Figure 6. lepto1 has defects in the development and PCD of somatic cell layers.
Supplemental Figure 7. Phylogenetic tree of LEPTO1 and its orthologs from three other angiosperms.
Supplemental Figure 8. Relative gene expression levels of OsRR1 and LEPTO1 in response to exogenous cytokinin treatment.
Supplemental Figure 9. LEPTO1 is located in the nuclei.
Supplemental Figure 10. The full-length LEPTO1 interacts with OsAHP1 and OsAHP2.
Supplemental Figure 11. In situ expression analysis of meiotic genes in wild-type and lepto1 anthers.
Supplemental Figure 12. The expression pattern of cyclin genes in the wild type.
Supplemental Figure 13. Relative expression levels of cyclin genes in wild-type and lepto1 anthers.
Supplemental Table 1. Primers designed for map-based cloning of LEPTO1.
Supplemental Table 2. Primers for RACE and real-time PCR.
Supplemental Table 3. Primers for plasmid vector construction.
Supplemental File. Text file of the alignment used to generate the phylogenetic tree in Supplemental Figure 7.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
This work was supported by grants from the National Key Research and Development Program of China (2016YFD0100901), the National Natural Science Foundation of China (31670313 and 31872859), and the Key Project of Jiangsu Education Department (15KJA180010).
AUTHOR CONTRIBUTIONS
Z.C. and T.Z. conceived the original screening and research plans; T.Z., L.R., X.C., and C.L. performed most of the experiments; Y.S., D.T., H.Y., Y.L., G.D., W.S., and B.M. designed the experiments and analyzed the data; T.Z. and L.R. wrote the article with contributions of all the authors; Z.C. supervised and complemented the writing.
Footnotes
Articles can be viewed without a subscription.
References
- Araki S., Ito M., Soyano T., Nishihama R., Machida Y. (2004). Mitotic cyclins stimulate the activity of c-Myb-like factors for transactivation of G2/M phase-specific genes in tobacco. J. Biol. Chem. 279: 32979–32988. [DOI] [PubMed] [Google Scholar]
- Argyros R.D., Mathews D.E., Chiang Y.H., Palmer C.M., Thibault D.M., Etheridge N., Argyros D.A., Mason M.G., Kieber J.J., Schaller G.E. (2008). Type B response regulators of Arabidopsis play key roles in cytokinin signaling and plant development. Plant Cell 20: 2102–2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azumi Y., Liu D., Zhao D., Li W., Wang G., Hu Y., Ma H. (2002). Homolog interaction during meiotic prophase I in Arabidopsis requires the SOLO DANCERS gene encoding a novel cyclin-like protein. EMBO J. 21: 3081–3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai M.Y., Shang J.X., Oh E., Fan M., Bai Y., Zentella R., Sun T.P., Wang Z.Y. (2012). Brassinosteroid, gibberellin and phytochrome impinge on a common transcription module in Arabidopsis. Nat. Cell Biol. 14: 810–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett M.D., Stern H. (1975). The time and duration of preleptotene chromosome condensation stage in Lilium hybrid cv. Black Beauty. Proc. R. Soc. Lond. B Biol. Sci. 188: 477–493. [DOI] [PubMed] [Google Scholar]
- Borde V., Lichten M. (2014). A timeless but timely connection between replication and recombination. Cell 158: 697–698. [DOI] [PubMed] [Google Scholar]
- Bulankova P., Akimcheva S., Fellner N., Riha K. (2013). Identification of Arabidopsis meiotic cyclins reveals functional diversification among plant cyclin genes. PLoS Genet. 9: e1003508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns J.A. (1972). Preleptotene chromosome contraction in Nicotiana species. J. Hered. 63: 175–178. [Google Scholar]
- Che L., Tang D., Wang K., Wang M., Zhu K., Yu H., Gu M., Cheng Z. (2011). OsAM1 is required for leptotene-zygotene transition in rice. Cell Res. 21: 654–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chelysheva L., et al. (2005). AtREC8 and AtSCC3 are essential to the monopolar orientation of the kinetochores during meiosis. J. Cell Sci. 118: 4621–4632. [DOI] [PubMed] [Google Scholar]
- Chen R., Zhao X., Shao Z., Wei Z., Wang Y., Zhu L., Zhao J., Sun M., He R., He G. (2007). Rice UDP-glucose pyrophosphorylase1 is essential for pollen callose deposition and its cosuppression results in a new type of thermosensitive genic male sterility. Plant Cell 19: 847–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Agostino I.B., Deruère J., Kieber J.J. (2000). Characterization of the response of the Arabidopsis response regulator gene family to cytokinin. Plant Physiol. 124: 1706–1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- d’Erfurth I., Cromer L., Jolivet S., Girard C., Horlow C., Sun Y., To J.P., Berchowitz L.E., Copenhaver G.P., Mercier R. (2010). The cyclin-A CYCA1;2/TAM is required for the meiosis I to meiosis II transition and cooperates with OSD1 for the prophase to first meiotic division transition. PLoS Genet. 6: e1000989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doi K., Izawa T., Fuse T., Yamanouchi U., Kubo T., Shimatani Z., Yano M., Yoshimura A. (2004). Ehd1, a B-type response regulator in rice, confers short-day promotion of flowering and controls FT-like gene expression independently of Hd1. Genes Dev. 18: 926–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis D. (2011). A commentary on the G2/M transition of the plant cell cycle. Ann. Bot. 107: 1065–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galperin M.Y. (2010). Diversity of structure and function of response regulator output domains. Curr. Opin. Microbiol. 13: 150–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartung M., Stahl A. (1977). Preleptotene chromosome condensation in mouse oogenesis. Cytogenet. Cell Genet. 18: 309–319. [DOI] [PubMed] [Google Scholar]
- Hosoda K., Imamura A., Katoh E., Hatta T., Tachiki M., Yamada H., Mizuno T., Yamazaki T. (2002). Molecular structure of the GARP family of plant Myb-related DNA binding motifs of the Arabidopsis response regulators. Plant Cell 14: 2015–2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Q., Li Y., Wang H., Shen Y., Zhang C., Du G., Tang D., Cheng Z. (2017a). Meiotic Chromosome Association 1 interacts with TOP3α and regulates meiotic recombination in rice. Plant Cell 29: 1697–1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X., Wang C., Liu Q., Fu Y., Wang K. (2017b). Targeted mutagenesis in rice using CRISPR-Cpf1 system. J. Genet. Genomics 44: 71–73. [DOI] [PubMed] [Google Scholar]
- Hua Y., Wang C., Huang J., Wang K. (2017). A simple and efficient method for CRISPR/Cas9-induced mutant screening. J. Genet. Genomics 44: 207–213. [DOI] [PubMed] [Google Scholar]
- Ito M., Araki S., Matsunaga S., Itoh T., Nishihama R., Machida Y., Doonan J.H., Watanabe A. (2001). G2/M-phase-specific transcription during the plant cell cycle is mediated by c-Myb-like transcription factors. Plant Cell 13: 1891–1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji J., Tang D., Wang K., Wang M., Che L., Li M., Cheng Z. (2012). The role of OsCOM1 in homologous chromosome synapsis and recombination in rice meiosis. Plant J. 72: 18–30. [DOI] [PubMed] [Google Scholar]
- Lee D.H., Kao Y.H., Ku J.C., Lin C.Y., Meeley R., Jan Y.S., Wang C.J.R. (2015). The axial element protein DESYNAPTIC2 mediates meiotic double-strand break formation and synaptonemal complex assembly in maize. Plant Cell 27: 2516–2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lermontova I., Schubert V., Fuchs J., Klatte S., Macas J., Schubert I. (2006). Loading of Arabidopsis centromeric histone CENH3 occurs mainly during G2 and requires the presence of the histone fold domain. Plant Cell 18: 2443–2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N., et al. (2006). The rice tapetum degeneration retardation gene is required for tapetum degradation and anther development. Plant Cell 18: 2999–3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D., Matzuk M.M., Sung W.K., Guo Q., Wang P., Wolgemuth D.J. (1998). Cyclin A1 is required for meiosis in the male mouse. Nat. Genet. 20: 377–380. [DOI] [PubMed] [Google Scholar]
- Luciani J.M., Devictor M., Stahl A. (1977). Preleptotene chromosome condensation stage in human foetal and neonatal testes. J. Embryol. Exp. Morphol. 38: 175–185. [PubMed] [Google Scholar]
- Mahadevaiah S.K., Turner J.M.A., Baudat F., Rogakou E.P., de Boer P., Blanco-Rodríguez J., Jasin M., Keeney S., Bonner W.M., Burgoyne P.S. (2001). Recombinational DNA double-strand breaks in mice precede synapsis. Nat. Genet. 27: 271–276. [DOI] [PubMed] [Google Scholar]
- Mason M.G., Mathews D.E., Argyros D.A., Maxwell B.B., Kieber J.J., Alonso J.M., Ecker J.R., Schaller G.E. (2005). Multiple type-B response regulators mediate cytokinin signal transduction in Arabidopsis. Plant Cell 17: 3007–3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng W.J., Cheng Z.J., Sang Y.L., Zhang M.M., Rong X.F., Wang Z.W., Tang Y.Y., Zhang X.S. (2017). Type-B ARABIDOPSIS RESPONSE REGULATORs specify the shoot stem cell niche by dual regulation of WUSCHEL. Plant Cell 29: 1357–1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercier R., Vezon D., Bullier E., Motamayor J.C., Sellier A., Lefèvre F., Pelletier G., Horlow C. (2001). SWITCH1 (SWI1): A novel protein required for the establishment of sister chromatid cohesion and for bivalent formation at meiosis. Genes Dev. 15: 1859–1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakata Y., Shetzline S., Sakashita C., Kalota A., Rallapalli R., Rudnick S.I., Zhang Y., Emerson S.G., Gewirtz A.M. (2007). c-Myb contributes to G2/M cell cycle transition in human hematopoietic cells by direct regulation of cyclin B1 expression. Mol. Cell. Biol. 27: 2048–2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen K.H., et al. (2016). Arabidopsis type B cytokinin response regulators ARR1, ARR10, and ARR12 negatively regulate plant responses to drought. Proc. Natl. Acad. Sci. USA 113: 3090–3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu N., Liang W., Yang X., Jin W., Wilson Z.A., Hu J., Zhang D. (2013). EAT1 promotes tapetal cell death by regulating aspartic proteases during male reproductive development in rice. Nat. Commun. 4: 1445. [DOI] [PubMed] [Google Scholar]
- Nonomura K., Eiguchi M., Nakano M., Takashima K., Komeda N., Fukuchi S., Miyazaki S., Miyao A., Hirochika H., Kurata N. (2011). A novel RNA-recognition-motif protein is required for premeiotic G1/S-phase transition in rice (Oryza sativa L.). PLoS Genet. 7: e1001265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono S., Liu H., Tsuda K., Fukai E., Tanaka K., Sasaki T., Nonomura K.I. (2018). EAT1 transcription factor, a non-cell-autonomous regulator of pollen production, activates meiotic small RNA biogenesis in rice anther tapetum. PLoS Genet. 14: e1007238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawlowski W.P., Sheehan M.J., Ronceret A. (2007). In the beginning: The initiation of meiosis. BioEssays 29: 511–514. [DOI] [PubMed] [Google Scholar]
- Ravi M., Shibata F., Ramahi J.S., Nagaki K., Chen C., Murata M., Chan S.W.L. (2011). Meiosis-specific loading of the centromere-specific histone CENH3 in Arabidopsis thaliana. PLoS Genet. 7: e1002121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren L., Tang D., Zhao T., Zhang F., Liu C., Xue Z., Shi W., Du G., Shen Y., Li Y., Cheng Z. (2018). OsSPL regulates meiotic fate acquisition in rice. New Phytol. 218: 789–803. [DOI] [PubMed] [Google Scholar]
- Robert T., Nore A., Brun C., Maffre C., Crimi B., Bourbon H.M., de Massy B. (2016). The TopoVIB-Like protein family is required for meiotic DNA double-strand break formation. Science 351: 943–949. [DOI] [PubMed] [Google Scholar]
- Schaller G.E., Street I.H., Kieber J.J. (2014). Cytokinin and the cell cycle. Curr. Opin. Plant Biol. 21: 7–15. [DOI] [PubMed] [Google Scholar]
- Scofield S., Dewitte W., Nieuwland J., Murray J.A.H. (2013). The Arabidopsis homeobox gene SHOOT MERISTEMLESS has cellular and meristem-organisational roles with differential requirements for cytokinin and CYCD3 activity. Plant J. 75: 53–66. [DOI] [PubMed] [Google Scholar]
- Shao T., Tang D., Wang K., Wang M., Che L., Qin B., Yu H., Li M., Gu M., Cheng Z. (2011). OsREC8 is essential for chromatid cohesion and metaphase I monopolar orientation in rice meiosis. Plant Physiol. 156: 1386–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X., Sun X., Zhang Z., Feng D., Zhang Q., Han L., Wu J., Lu T. (2015). GLUCAN SYNTHASE-LIKE 5 (GSL5) plays an essential role in male fertility by regulating callose metabolism during microsporogenesis in rice. Plant Cell Physiol. 56: 497–509. [DOI] [PubMed] [Google Scholar]
- Stuart D., Wittenberg C. (1998). CLB5 and CLB6 are required for premeiotic DNA replication and activation of the meiotic S/M checkpoint. Genes Dev. 12: 2698–2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L., Zhang Q., Wu J., Zhang L., Jiao X., Zhang S., Zhang Z., Sun D., Lu T., Sun Y. (2014). Two rice authentic histidine phosphotransfer proteins, OsAHP1 and OsAHP2, mediate cytokinin signaling and stress responses in rice. Plant Physiol. 165: 335–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang D., Miao C., Li Y., Wang H., Liu X., Yu H., Cheng Z. (2014). OsRAD51C is essential for double-strand break repair in rice meiosis. Front. Plant Sci. 5: 167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- To J.P., Deruère J., Maxwell B.B., Morris V.F., Hutchison C.E., Ferreira F.J., Schaller G.E., Kieber J.J. (2007). Cytokinin regulates type-A Arabidopsis Response Regulator activity and protein stability via two-component phosphorelay. Plant Cell 19: 3901–3914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai Y.C., Weir N.R., Hill K., Zhang W., Kim H.J., Shiu S.H., Schaller G.E., Kieber J.J. (2012). Characterization of genes involved in cytokinin signaling and metabolism from rice. Plant Physiol. 158: 1666–1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrielynck N., Chambon A., Vezon D., Pereira L., Chelysheva L., De Muyt A., Mézard C., Mayer C., Grelon M. (2016). A DNA topoisomerase VI-like complex initiates meiotic recombination. Science 351: 939–943. [DOI] [PubMed] [Google Scholar]
- Wang J., Tian C., Zhang C., Shi B., Cao X., Zhang T.Q., Zhao Z., Wang J.W., Jiao Y. (2017). Cytokinin signaling activates WUSCHEL expression during axillary meristem initiation. Plant Cell 29: 1373–1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Wang J., Shi B., Yu T., Qi J., Meyerowitz E.M., Jiao Y. (2014). The stem cell niche in leaf axils is established by auxin and cytokinin in Arabidopsis. Plant Cell 26: 2055–2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z., Ji J., Tang D., Wang H., Shen Y., Shi W., Li Y., Tan X., Cheng Z., Luo Q. (2015). OsSDS is essential for DSB formation in rice meiosis. Front. Plant Sci. 6: 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoo R., Zawadzki K.A., Nabeshima K., Drake M., Arur S., Villeneuve A.M. (2012). COSA-1 reveals robust homeostasis and separable licensing and reinforcement steps governing meiotic crossovers. Cell 149: 75–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T.Q., Lian H., Zhou C.M., Xu L., Jiao Y., Wang J.W. (2017). A two-step model for de novo activation of WUSCHEL during plant shoot regeneration. Plant Cell 29: 1073–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zickler D., Kleckner N. (1998). The leptotene-zygotene transition of meiosis. Annu. Rev. Genet. 32: 619–697. [DOI] [PubMed] [Google Scholar]