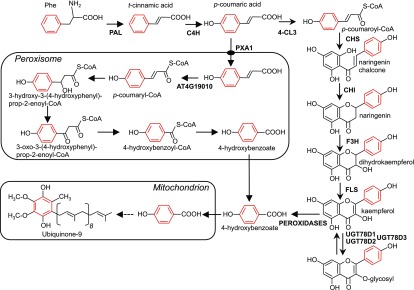
Genetic and biochemical evidence shows that Arabidopsis and tomato can derive the ubiquinone precursor 4-hydroxybenzoate from the B-ring of kaempferol.
Abstract
Land plants possess the unique capacity to derive the benzenoid moiety of the vital respiratory cofactor, ubiquinone (coenzyme Q), from phenylpropanoid metabolism via β-oxidation of p-coumarate to form 4-hydroxybenzoate. Approximately half of the ubiquinone in plants comes from this pathway; the origin of the rest remains enigmatic. In this study, Phe-[Ring-13C6] feeding assays and gene network reconstructions uncovered a connection between the biosynthesis of ubiquinone and that of flavonoids in Arabidopsis (Arabidopsis thaliana). Quantification of ubiquinone in Arabidopsis and tomato (Solanum lycopersicum) mutants in flavonoid biosynthesis pinpointed the corresponding metabolic branch-point as lying between flavanone-3-hydroxylase and flavonoid-3′-hydroxylase. Further isotopic labeling and chemical rescue experiments demonstrated that the B-ring of kaempferol is incorporated into ubiquinone. Moreover, heme-dependent peroxidase activities were shown to be responsible for the cleavage of B-ring of kaempferol to form 4-hydroxybenzoate. By contrast, kaempferol 3-β-d-glucopyranoside, dihydrokaempferol, and naringenin were refractory to peroxidative cleavage. Collectively, these data indicate that kaempferol contributes to the biosynthesis of a vital respiratory cofactor, resulting in an extraordinary metabolic arrangement where a specialized metabolite serves as a precursor for a primary metabolite. Evidence is also provided that the ubiquinone content of tomato fruits can be manipulated via deregulation of flavonoid biosynthesis.
INTRODUCTION
Ubiquinone (coenzyme Q) is a bipartite molecule made up of a redox-active benzoquinone ring and a liposoluble polyprenyl tail. It serves as a vital electron carrier in the prokaryotic and eukaryotic respiratory chains as well as in the photosynthetic reaction center of purple bacteria (Stowell et al., 1997; Lenaz and Genova, 2009; Nowicka and Kruk, 2010). Ubiquinone also doubles as a membrane antioxidant that protects neighboring lipids and proteins from damage by hydroxyl and superoxide radicals (Ohara et al., 2004; Bentinger et al., 2010; Allan et al., 2013). Furthermore, besides its roles as a redox cofactor, recent evidence demonstrates that ubiquinone can serve a purely mechanical function by increasing the thickness of biological membranes in response to osmotic stress (Sévin and Sauer, 2014). The overall architecture of ubiquinone biosynthesis is similar in prokaryotes and eukaryotes: A benzenoid precursor is coupled to a trans-long–chain prenyl diphosphate, and the benzenoid ring is further modified through successive steps of hydroxylation, methylation, decarboxylation, and in some cases deamination (Tran and Clarke, 2007; Pierrel, 2017). In most organisms the benzenoid precursor is 4-hydroxybenzoate; the only exception to date is the yeast Saccharomyces cerevisiae, which can also prenylate and deaminate p-aminobenzoate and then supply the resulting prenylated benzoate to the standard ubiquinone biosynthetic pathway (Marbois et al., 2010; Pierrel et al., 2010).
By contrast, the metabolic origin of 4-hydroxybenzoate is surprisingly disparate across phylogenetic lineages. Eukaryotes, in particular, lack orthologs of the prokaryotic enzyme chorismate lyase, which catalyzes the direct cleavage and aromatization of chorismate into 4-hydroxybenzoate, and instead branch the synthesis of 4-hydroxybenzoate from the metabolism of other aromatic compounds. For instance, vertebrates, S. cerevisiae, and plants are known to be able to derive 4-hydroxybenzoate from Tyr (reviewed in Pierrel, 2017). Yeast can also use the Tyr biosynthetic intermediate 4-hydroxyphenylpyruvate (Payet et al., 2016). Plants are unique in having the ability to synthesize 4-hydroxybenzoate from the β-oxidative metabolism of Phe; this amino acid is actually the preferred precursor for ubiquinone biosynthesis in Arabidopsis (Arabidopsis thaliana; Block et al., 2014). In this pathway, Phe is first deaminated and hydroxylated yielding p-coumarate (Supplemental Figure 1). These reactions, which are both part of the phenylpropanoid pathway and common to all land plants, take place in the cytosol and are catalyzed by Phe ammonia lyase (PAL) and cinnamate-4-hydroxylase (C4H), respectively. The p-coumarate is then imported into peroxisomes and activated by a dedicated ligase (Supplemental Figure 1). The subsequent steps of hydration, oxidation, thiolation, and CoA thioester hydrolysis result in the shortening of the propyl side chain of p-coumaroyl-CoA through loss of one molecule of acetyl-CoA and the formation of 4-hydroxybenzoate (Supplemental Figure 1). The enzymes catalyzing these steps have not been characterized yet, but have been proposed to be identical to those involved in the conversion of cinnamoyl-CoA into benzoate (Block et al., 2014; Widhalm and Dudareva, 2015).
Far more enigmatic, in comparison, are the sporadic reports that plants might be able to synthesize 4-hydroxybenzoate independently of the β-oxidative metabolism of p-coumarate. For instance, CoA-free plant extracts have been shown to catalyze the conversion of p-coumarate into 4-hydroxybenzoate via the formation of 4-hydroxybenzaldehyde (French et al., 1976; Yazaki et al., 1991; Schnitzler et al., 1992). These findings, however, were later attributed to artifacts resulting from the assay conditions (Löscher and Heide, 1994; Wildermuth, 2006). It has also been proposed that similar to the γ-proteobacterium Pseudomonas fluorescens, plants may possess a bifunctional hydratase-lyase capable of catalyzing the retro-aldol cleavage of p-coumaroyl-CoA into p-hydroxybenzaldehyde and acetyl-CoA (Gasson et al., 1998; Mitra et al., 2002). Such activity, however, has, to our knowledge, not been reported in plants.
In this study, we revisited the question of whether the ring of Phe could be incorporated into ubiquinone using a series of Arabidopsis knockouts affecting either the biosynthesis of p-coumarate or its β-oxidative shortening into 4-hydroxybenzoate. Having obtained clues that p-coumarate is incorporated into ubiquinone via functionally redundant routes, we combined gene network modeling, reverse genetics, in vivo isotopic labeling, and in vitro assays to demonstrate that plants have evolved a unique metabolic branch linking the biosynthetic pathway of ubiquinone to that of flavonoids.
RESULTS
Detection of a Functional Connection between Ubiquinone Biosynthesis and Flavonoid Metabolism in Arabidopsis
Three Arabidopsis mutants, peroxisomal abc transporter1 (pxa1), peroxisomal p-coumaroyl-CoA ligase (at4g19010), and reduced epidermal fluorescence3-4 (ref3-4), were selected based on their distinct effects on the use of Phe as a precursor of 4-hydroxybenzoate for ubiquinone biosynthesis. The mutants pxa1 and at4g19010 correspond to knockout alleles of an ABC subfamily D transporter 1 and a p-coumaroyl-CoA ligase, respectively (Supplemental Figure 1). Both proteins are located in peroxisomes and their mutants have been shown to hinder the β-oxidative shortening of the propyl side chain of p-coumarate (Block et al., 2014). By contrast, the ref3-4 mutation, which corresponds to a knockout allele of C4H, results in a complete blockage of the phenylpropanoid pathway upstream of pxa1 and at4g19010 (Supplemental Figure 1).
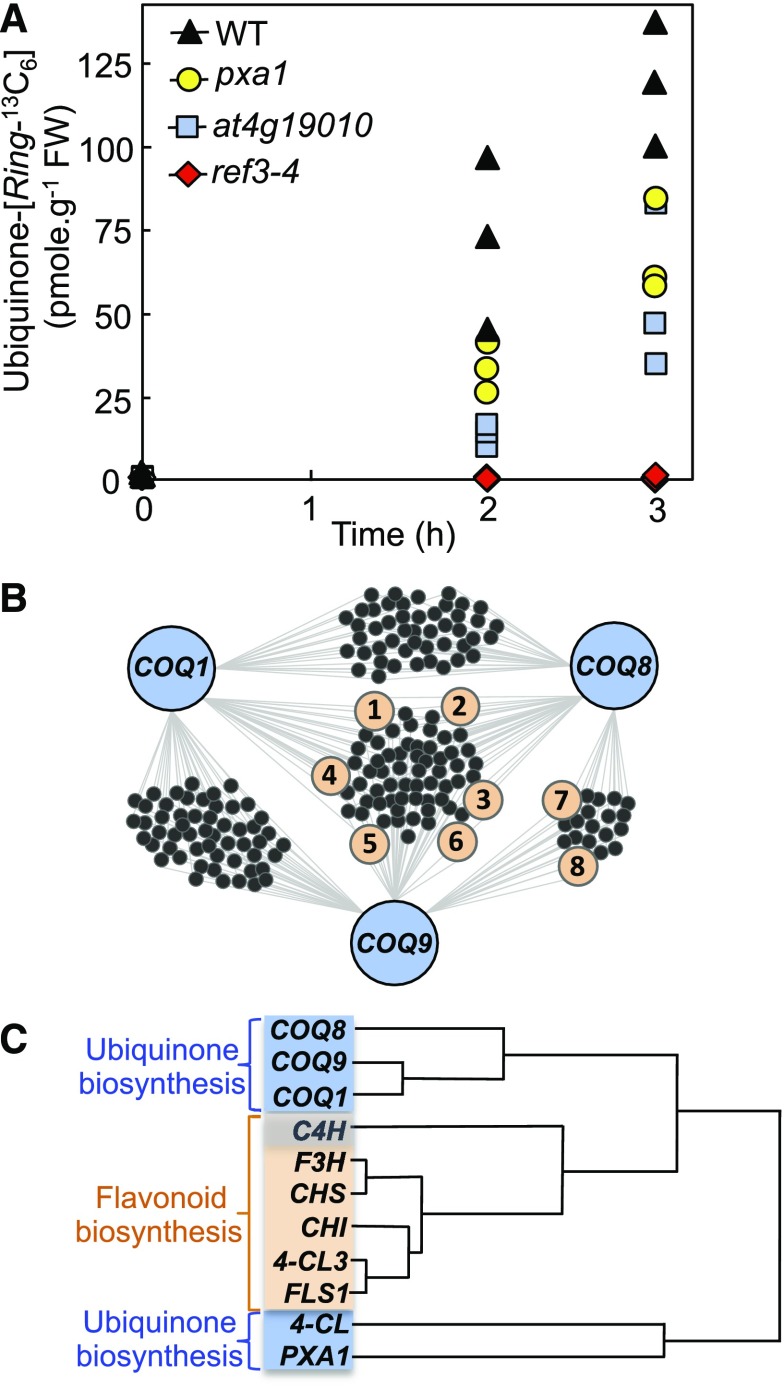
When Phe-[Ring-13C6] was axenically fed for 2 h and 3 h to pxa1, at4g19010, and wild-type plants, quantification of ubiquinone-[Ring-13C6] by liquid chromatography-tandem mass spectrometry indicated that the rates of 13C isotopic enrichment from Phe in the pxa1 and at4g19010 knockouts were approximately half those measured in the wild-type control (Figure 1A). These data coincide with our previous observations that the ubiquinone content of the pxa1 and at4g19010 mutants is decreased by 55% to 65% compared with that of wild-type Arabidopsis (Block et al., 2014). When identical doses of Phe-[Ring-13C6] were given to the ref3-4 knockout, no statistically significant incorporation of Phe into ubiquinone was detected (Figure 1A). It thus emerges from these results that: (1) the phenylpropanoid pathway is the obligatory route for the use of Phe as a precursor for ubiquinone biosynthesis; and (2) Arabidopsis can convert p-coumarate to 4-hydroxybenzoate via some functionally redundant ligases and transporters, and/or via a parallel pathway operating downstream of the C4H-catalyzed reaction.
Figure 1.
Detection of a Functional Connection between the Biosynthesis of Ubiquinone and the Metabolism of Flavonoids.
(A) Ubiquinone-[Ring-13C6] content in the leaves of axenically grown wild-type and ref3-4, pxa1, and at4g19010 knockout Arabidopsis plants fed for 2 h and 3 h with 250 μM doses of Phe-[Ring-13C6]. FW; fresh weight; WT, wild type.
(B) Coexpression network reconstituted from the top 300 elements of the 22,263 expressed loci of the ATTED-II database using Arabidopsis ubiquinone biosynthetic enzymes COQ1 (At2g34630), COQ8 (At4g160660), and COQ9 (At1g19140) as queries. For clarity, the network shows only the coexpressors that intersect with at least two of the query genes, i.e. out of the selected top 300 c-expressors, 47 genes are uncommonly coexpressed between COQ1 and COQ8, 54 are uncommonly coexpressed between COQ1 and COQ9, 19 are uncommonly coexpressed between COQ8 and COQ9, and 63 are uncommonly coexpressed among COQ1, COQ8, and COQ9. Genes involved in flavonoid metabolism are highlighted in orange: (1) F3H; (2) FLS1; (3) UGT78D2; (4) CHI; (5) CHS; (6) UGT89C1; (7) 4-CL3; (8) CHI-L1. The correlation ranks and functional annotations of the associated loci as well as the aggregated gene lists used for network reconstruction are provided in Supplemental Dataset 1.
(C) Hierarchical clustering of the coexpression profiles of ubiquinone (blue) and flavonoid (orange) biosynthetic genes in Arabidopsis. Note that C4H contributes to both the β-oxidative branch of ubiquinone biosynthesis and to flavonoid biosynthesis (Supplemental Figures 1 and 2).
To search for the missing players in p-coumarate metabolism, the top 300 elements of the ATTED-II microarray and RNA-seq database (Obayashi et al., 2018) were mined using Arabidopsis ubiquinone biosynthetic genes (COQ) as queries. These searches identified COQ1 (At2g34630), COQ8 (At4g01660), and COQ9 (At1g19140) as node interactors in the coexpression network of eight genes involved in the metabolism of flavonoids (Figure 1B). Of those genes, six encode for enzymes that synthesize the flavonoid scaffold, namely cytosolic 4-COUMAROYL-COA LIGASE3 (4-CL3), CHALCONE SYNTHASE (CHS), CHALCONE-FLAVANONE ISOMERASE (CHI/CHI-L1), FLAVANONE-3-HYDROXYLASE (F3H), and FLAVONOL SYNTHASE1 (FLS1; Figure 1B; Supplemental Figure 2) and two flavonoid glycosyltransferases: UDP-GLUCOSE:FLAVONOL-3-O-GLUCOSYLTRANSFERASE (UGT78D2) and UDP-RHAMNOSE:FLAVONOL-7-O-RHAMNOSYLTRANSFERASE (UGT89C1; Figure 1B; Supplemental Figure 2). One additional flavonoid biosynthetic gene, FLAVANOID-3′-HYDROXYLASE (F3′H), was identified among the top 300 coexpressors of COQ9 (Supplemental Figure 2).
Remarkably, hierarchical cluster analyses revealed that the overall coexpression profiles of Arabidopsis COQ1, COQ8, and COQ9 correlated more closely with those of flavonoid biosynthetic genes than with those of At4g19010 and the PXA1 transporter (At4g39850), which are known contributors to ubiquinone biosynthesis (Block et al., 2014; Figure 1C). Such marked correlation of expression is not attributable merely to a shared subcellular localization of the cognate pathways, because flavonoid biosynthetic enzymes are cytosolic (Wagner and Hrazdina, 1984), whereas COQ1 is dual targeted to mitochondria and chloroplasts (Ducluzeau et al., 2012), and COQ8 and COQ9 are localized in mitochondria (Cardazzo et al., 1998; Heazlewood et al., 2004). Together, these in silico reconstructions suggest the existence of a functional connection between the biosynthesis of ubiquinone and that of flavonoids in Arabidopsis.
The Biosynthetic Pathways of Flavonoids and Ubiquinone Intersect After the F3H-Catalyzed Reaction in Arabidopsis and Tomato
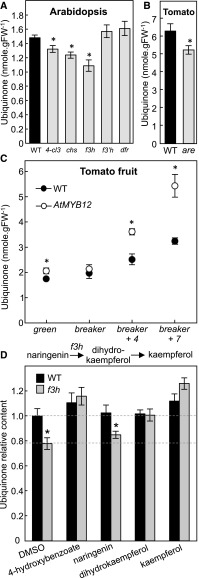
To experimentally verify our in silico reconstructions, ubiquinone levels were quantified in the rosette leaves of wild-type Arabidopsis plants and in a series of previously characterized and verified null alleles (see “Methods”) corresponding to genes encoding enzymes in the central flavonoid biosynthetic pathway. Among these, 4-cl3, f3h, and chs plants displayed a 15% to 25% decrease in ubiquinone content compared with their respective wild-type ecotypes (Figure 2A). By contrast, no statistically significant difference was observed between the ubiquinone content of the f3′h and dfr knockouts and that of wild-type plants (Figure 2A). These results not only confirm that the biosynthetic pathways of flavonoids and ubiquinone are indeed connected, but also indicate that the metabolic node between these branches is located after the F3H-catalyzed reaction and before that catalyzed by F3′H (Supplemental Figure 2). Candidate branch-point metabolites could therefore be dihydrokaempferol and/or kaempferol, but not downstream products of F3′H and DFR, such as quercetin and anthocyanins. It is noteworthy that although a single null allele was selected for each of the flavonoid biosynthetic genes, the combined data obtained with these lines are mutually consistent; that is, all the flavonoid biosynthetic knockouts corresponding to enzymes located upstream of F3H and including the later ones (4-cl3, chs, and f3 h) negatively affect ubiquinone level, whereas those located downstream of this enzyme (f3′h and dfr) do not.
Figure 2.
Ubiquinone Content and Feeding Assays.
(A) Total ubiquinone (reduced + oxidized) levels in 3-week-old rosette leaves of wild-type and 4-cl3, chs, f3 h, f3′h, and dfr Arabidopsis plants. FW; fresh weight; WT, wild type.
(B) Total ubiquinone (reduced + oxidized) levels in the leaves of wild-type (cv VF-36) and are tomato plants.
(C) Total ubiquinone (reduced + oxidized) levels in wild-type (Moneymaker) and AtMYB12-overexpressing tomato fruits. AtMYB12—a transcription factor that functions as a positive regulator of the flavonoid biosynthetic pathway—is under the control of the fruit-specific and ethylene-responsive E8 promoter. Data represent the means of 4–15 biological replicates ±se. Asterisks indicate significant differences from the cognate DMSO control as determined by Fisher’s test (P < α = 0.05) from an analysis of variance.
(D) Relative ubiquinone levels in axenically grown wild-type and f3 h Arabidopsis plants fed for 24 h with 10 μM 4-hydroxybenzoate, naringenin, dihydrokaempferol, or kaempferol. Data are means of 9–14 biological replicates ±se. Asterisks indicate significant differences between wild-type and f3 h knockout plants within each feeding assay as determined by Fisher’s test (P < α = 0.05) from an analysis of variance.
Congruent with these results, a similar decrease in ubiquinone content was observed in the leaves of a tomato (Solanum lycopersicum) anthocyanin reduced (are) mutant, which corresponds to a point mutation in the F3H gene resulting in a partial loss of the cognate activity (Maloney et al., 2014; Zhang et al., 2015; Figure 2B). In a reciprocal experiment, ubiquinone was quantified in the fruits of tomato AtMYB12 transgenics engineered to accumulate high levels of flavonols in their fruits specifically after the breaker ripening stage (Luo et al., 2008; Zhang et al., 2015). Until breaker AtMYB12 fruits displayed either marginal or nonstatistically significant differences in ubiquinone content compared with wild-type fruits (Figure 2C). However, 4 and 7 days after breaker, the ubiquinone content of AtMYB12 fruits was 1.5-fold to 2-fold higher than that of wild-type controls (Figure 2C). All together these results show that the functional linkage between flavonoid and ubiquinone biosynthesis is not specific to Arabidopsis, and that these two pathways also intersect after the F3H-catalyzed reaction in tomato.
To examine the nature of this metabolic branch-point further, axenic cultures of the Arabidopsis f3h knockout were fed with 10 μM doses of naringenin, dihydrokaempferol, or kaempferol. This concentration was determined in pilot experiments to be the lowest amount of 4-hydroxybenzoate (the direct precursor of the aromatic ring of ubiquinone) necessary to restore wild-type level of ubiquinone in the f3h knockout, whereas having no statistically significant effect on ubiquinone biosynthesis in wild-type control cultures (Figure 2D). Dimethyl sulfoxide (DMSO) served as a negative control (Figure 2D). As expected, no rescue was observed with naringenin, the substrate of F3H. Dihydrokaempferol and kaempferol, on the other hand, fully restored ubiquinone biosynthesis in the f3h mutant (Figure 2D). Notably, kaempferol was more efficient than dihydrokaempferol at rescuing the f3h mutant; kaempferol-fed plants displayed a 20% to 30% increase in ubiquinone content as compared with their dihydrokaempferol-fed counterparts (Figure 2D). We will return to this point in the Discussion.
The B-Ring of Kaempferol Is Incorporated into the Benzenoid Moiety of Ubiquinone
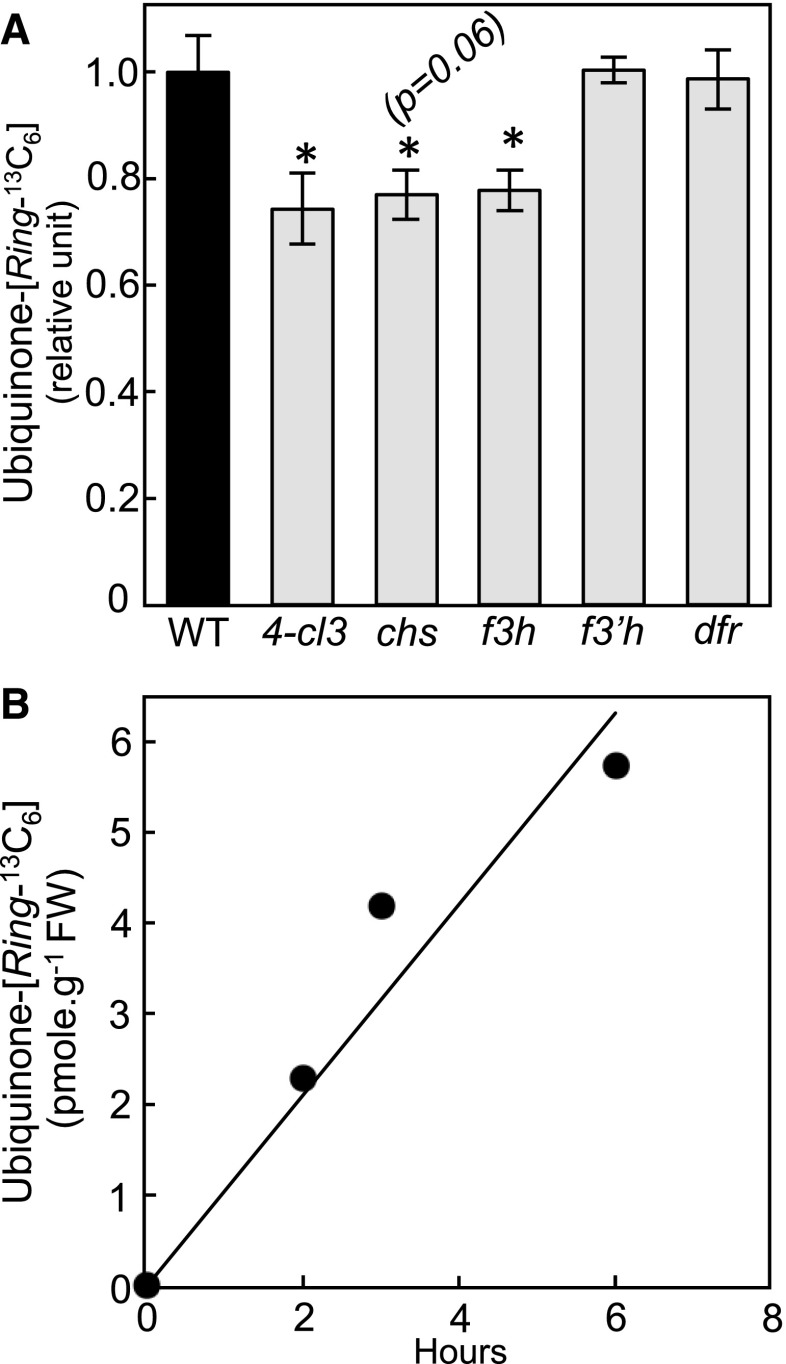
Flavonoids are bipartite molecules made up of chromenone (A-rings and C-rings) and phenyl (B-ring) moieties (Supplemental Figure 2). The A-ring originates from the condensation and subsequent cyclization of malonyl-CoA units, whereas the B-ring comes directly from the aromatic group of Phe via the formation of p-coumarate. After activation by ligation with CoA, the propyl side chain of the latter is conjugated to the A-ring and then cyclized to form the γ-pyrone ring (C-ring) of the chromenone moiety. It results from such an arrangement that the metabolic fate of each ring of the flavonoid scaffold can be traced via the use of labeled precursors. To gain some insight into the molecular mechanism that underlies the metabolic connection between flavonoids and ubiquinone, Phe-[Ring 13C6] was fed to the Arabidopsis 4-cl3, f3h, chs, f3′h, and dfr mutants as well as to their wild-type counterparts. The 13C isotopic enrichment of the benzenoid ring-containing fragment of ubiquinone was decreased by 24% to 33% in the 4-cl3, chs, and f3h knockouts and unchanged in the f3′h and dfr knockouts compared with wild-type controls (Figure 3A). These data recapitulate the measurements of the total ubiquinone content in these mutants (Figure 2A) and are consistent with the B-ring of a flavonol located before the f3′h-catalyzed reaction being incorporated into ubiquinone. To confirm these results, kaempferol-[B-Ring13C6] was high-performance liquid chromatography (HPLC)-purified from Phe-[Ring13C6]-fed Arabidopsis and then added to axenic cultures of the f3h knockout for 2, 3 and 6 h. Quantification of the ion pairs corresponding to the ring of ubiquinone demonstrated the time-dependent transfer of labeling from kaempferol-[B-Ring13C6] to ubiquinone-[Ring13C6] (Figure 3B). No 13C isotope of ubiquinone lighter than that labeled at all ring positions was detected, verifying that the B-ring was not rearranged before its incorporation into ubiquinone.
Figure 3.
De Novo Biosynthesis of Ubiquinone from Phe and Kaempferol in Arabidopsis.
(A) Relative ubiquinone-[Ring-13C6] levels in axenically grown wild-type, 4-cl3, chs, f3 h, f3′h, and dfr Arabidopsis fed for 3 h with 250 μM Phe-[Ring-13C6]. Data are means of four biological replicates ±se. Asterisks indicate significant differences from the wild-type control as determined by Fisher’s test (P < α = 0.05) from an analysis of variance. WT, wild type.
(B) Ubiquinone-[Ring-13C6] levels in f3 h Arabidopsis plant fed for 2, 3, and 6 h with 8.5 μM of [B-Ring13C6]-kaempferol. FW; fresh weight.
Heme-Dependent Peroxidases Are Responsible for the Release of 4-Hydroxybenzoate from the B-ring of Kaempferol
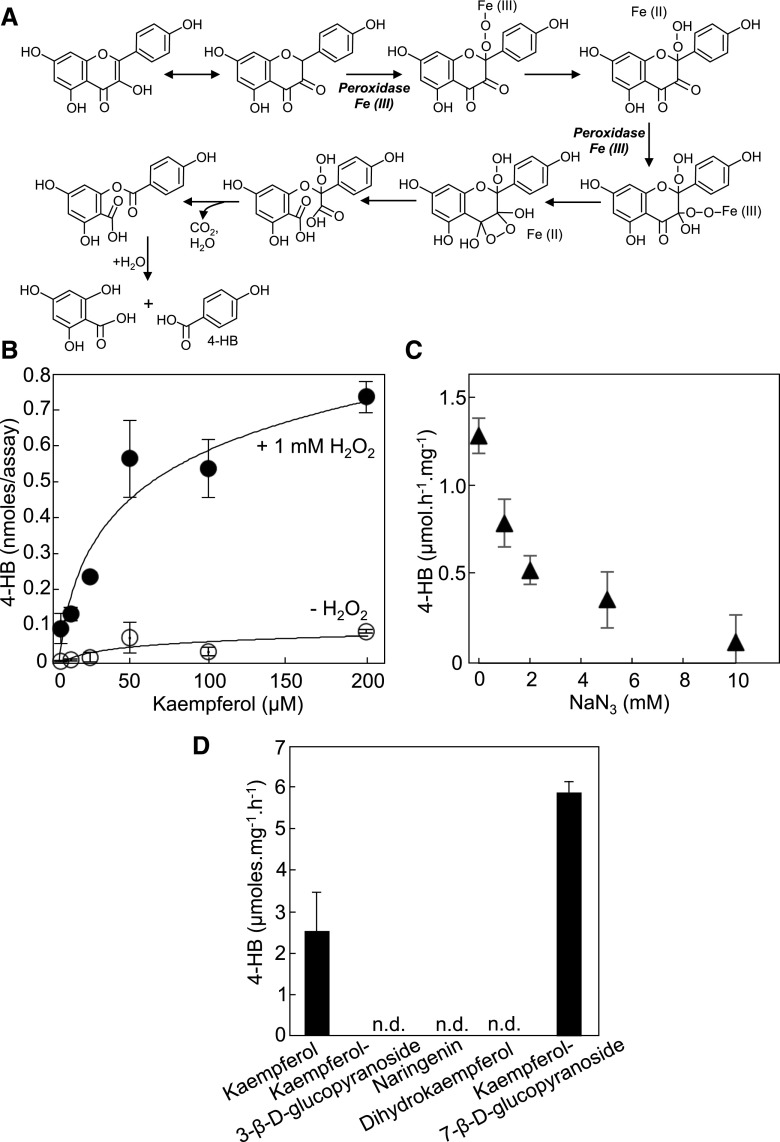
The release of B-ring from the flavonoid scaffold is reminiscent of the oxidative breakdown of quercetin exposed to air (Zenkevich et al., 2007). We hypothesized that in vivo and in the case of kaempferol such an oxidative cleavage would be enzymatic, in particular because plant tissues are notorious for displaying pronounced H2O2-dependent peroxidase activities against a wide range of aromatic compounds including flavonoids (Veitch, 2004; Pourcel et al., 2007). Modeling of the peroxidative cleavage of kaempferol predicts that the double bond between C-2 and C-3 and the hydroxyl group on C-3 are both crucial for the formation of the corresponding keto-tautomer, which is highly sensitive to oxidation and therefore likely to react with the heme group of compound III (Figure 4A).
Figure 4.
The Peroxidative Cleavage of Kaempferol Generates 4-Hydroxybenzoate from B-Ring.
(A) Modeling of the heme-dependent peroxidation of kaempferol and consecutive release of 4-hydroxybenzoate. The heme groups (compound III) of peroxidases react with C-2 and C-3 of the keto-tautomeric form of kaempferol. Electron rearrangements trigger the cleavage of the bond between C-3 and C-4, and loss of C-3 as CO2. In this scenario, the initial formation of the α-diketone tautomer is strictly contingent upon the presence of a double bond between C-2 and C-3 and the presence of a hydroxyl group on C-3. For the purpose of legibility, the prerequisite formation of compound III, which originates from the reaction of the ferric enzyme (ground state) with H2O2 and then reaction of the resulting compound II with H2O2, is not shown here.
(B) Desalted protein extracts (2.5 μg) of Arabidopsis leaves were assayed for 4-hydroxybenzoate formation using kaempferol as a substrate with or without hydrogen peroxide. No formation of 4-hydroxybenzoate was observed when the extract was omitted or when the extract was boiled before the assay. Data are the means of 5–6 replicates ±se.
(C) Inhibition of the peroxidative cleavage of kaempferol by sodium azide. The concentrations of kaempferol and hydrogen peroxide were 50 μM and 1 mM, respectively. Data are the means of three replicates ±se.
(D) Cleavage assays of various flavonoids. Note that all tested flavonoids have the same B-ring and differ only by the saturation level of C-2 and C-3, and/or the substitution of the hydroxyl group on C-3. Assays contained 2.5 μg of protein extract, 50 μM of flavonoid, and 1 mM of hydrogen peroxide. Data are the means of three replicates ±se.
To test this model, desalted protein extracts of Arabidopsis leaves were assayed using kaempferol as a substrate with or without addition of H2O2. The release of 4-hydroxybenzoate was monitored by HPLC with diode array detection. Saturable H2O2-dependent activities of 4-hydroxybenzoate formation were readily detected resulting in an apparent Km of 50 μM (Figure 4B). Only marginal activities were observed when H2O2 was omitted from the assay (Figure 4B). Preincubation of the protein extract with sodium azide, a classic inactivator of prosthetic-heme groups (Ortiz de Montellano et al., 1988), led to pronounced inhibition of 4-hydroxybenzoate formation, confirming that the cognate peroxidase activities were heme-dependent (Figure 4C). No formation of 4-hydroxybenzoate was observed when kaempferol 3-β-D-glucopyranoside, dihydrokaempferol, or naringenin (which lack a free hydroxyl group on C-3 or a C-2 C-3 double bond or both, respectively) were used as substrates, further supporting the proposed reaction mechanism for the peroxidative cleavage of the C-ring (Figure 4D). As predicted, the O-glucosyl ester derivative on the A-ring, kaempferol 7-β-D-glucopyranoside, was cleaved efficiently (Figure 4C).
DISCUSSION
Plants Possess Two CoA-dependent Pathways To Synthesize 4-Hydroxybenzoate from Phe
Here, we provide genetic and biochemical evidence that both Arabidopsis and tomato are able to derive 4-hydroxybenzoate from the B-ring of kaempferol. Figure 5 shows a scheme of the corresponding metabolic pathway that branches from flavonoid biosynthesis and its integration in the overall architecture of ubiquinone biosynthesis. The early steps leading to the formation of p-coumarate in the cytosol are common to both the β-oxidative and the non-β-oxidative pathways. The import of p-coumarate into peroxisomes in effect creates the split between these two branches. Because both pathways entail the activation of p-coumarate with CoA, plants had to evolve two corresponding p-coumaroyl-CoA ligases. In Arabidopsis, these enzymes are represented by cytosolic At1g65060 (4-CL3) and peroxisomal At4g19010, two related members of clade IV and V of the plant superfamily of acyl-activating enzymes, respectively (Shockey and Browse, 2011; Block et al., 2014; Li et al., 2015). The phenyl ring of cytosolic p-coumaroyl-CoA ends up as the B-ring of the flavonoid scaffold; it is then cleaved from kaempferol, without rearrangement with the C-ring, by one or more heme-dependent peroxidases to generate 4-hydroxybenzoate.
Figure 5.
The Flavonoid and β-Oxidative Branches of 4-Hydroxybenzoate Biosynthesis for Ubiquinone Production in Plant Cells.
C4H; 4-CL3; AT4G19010; CHI; CHS; F3H; FLS; PAL; PXA1; UGT78D1; UGT78D2; UGT78D3. Dashed arrows indicate multiple steps. Note that kaempferol glycosylated at the C-3 position is protected from peroxidative cleavage.
The Arabidopsis genome encodes six FLS homologs, two of which have been shown to be catalytically active whereas the respective roles of the four others in flavonol biosynthesis remain enigmatic (Owens et al., 2008; Preuss et al., 2009; Saito et al., 2013). The enzyme leucoanthocyanidin synthase is also known to display side-activity as FLS (Preuss et al., 2009). This leads to a large degree of functional redundancy such that no mutant devoid of FLS activity has been isolated to date (Preuss et al., 2009; Saito et al., 2013). Although the unavailability of flavonol-free plants precludes the examination of how the blockage of the dihydrokaempferol to kaempferol conversion affects 4-hydroxybenzoate production, in vitro assays unequivocally establish that dihydrokaempferol is not a candidate for the peroxidative cleavage of C-ring; the chemistry of this reaction requires indeed the simultaneous presence of a double bond between C-2 and C-3, and a hydroxyl group on C-3. That the exogenous supply of naringenin does not rescue ubiquinone biosynthesis in the f3h knockout is consistent with this mechanism. Similarly, the prerequisite formation of the C-2 C-3 double bond before C-ring cleavage explains why dihydrokaempferol is significantly less efficient than kaempferol at boosting ubiquinone levels in f3h plants. Moreover, if F3H and FLS were to form a metabolon, exogenous dihydrokaempferol would not be as easily converted into kaempferol than its endogenous counterpart. It is noteworthy that the mechanism of kaempferol cleavage in plants contrasts starkly with the catabolism of flavonoids in bacteria. For instance, in Pseudomonas putida, the initial step of quercetin degradation involves its dehydroxylation into naringenin, whereas in Eubacterium ramulus, the C-2 C-3 double bond of flavonols must be reduced before C-ring opening (Schneider and Blaut, 2000; Pillai and Swarup, 2002). Furthermore, in bacteria, opening of the C-ring of quercetin and naringenin generates aliphatic derivatives of the B-ring as intermediates, such as 3,4-dihydroxyphenylacetate, 3,4-dihydroxycinnamate, and 3-(4-hydroxyphenyl) propionate, instead of 3,4-dihydroxybenzoate and 4-hydroxybenzoate, as would be expected in the direct release of the B-ring (Winter et al., 1989; Schneider and Blaut, 2000; Pillai and Swarup, 2002).
3-O-Glycosylation Prevents the Peroxidative Turnover of Kaempferol into 4-Hydroxybenzoate
The chemical modeling of kaempferol oxidation predicts that modification of the hydroxyl group on C-3 blocks the prerequisite formation of the cognate keto-tautomer, and our data confirm that kaempferol 3-β-D-glucopyranoside is resistant to peroxidative cleavage. The observation is important, because in vivo C-3 of the vast majority of kaempferol is locked as O-glucosyl, O-rhamnosyl, or O-arabinosyl ester conjugates (Lepiniec et al., 2006; Yonekura-Sakakibara et al., 2008; Buer et al., 2013). In fact, the pool of aglycone kaempferol is so small that it is undetectable in most Arabidopsis tissues except for some regions of the hypocotyl and root of seedlings (Peer et al., 2001; Buer et al., 2013). However, our finding that 25% to 30% of 4-hydroxybenzoate that is incorporated into ubiquinone originates from the cleavage of kaempferol in Arabidopsis and tomato provides evidence that the C-3 aglycone form does exist in mature leaves.
As one would expect for the biosynthesis of a cofactor, the quantities involved are minute, especially compared with the pool of glycosylated kaempferol in leaf tissues. Thus, it can be calculated from our data and the level of kaempferol in Arabidopsis leaves (∼400 nmol g−1 of fresh weight; Yin et al., 2012), that the amount of 4-hydroxybenzoate of kaempferol origin (0.31 to 0.37 nmol g−1 of fresh weight; Figure 2) represents a mere ∼0.1% of the total kaempferol in leaves. The high kaempferol peroxidase activities further dwarf these values: A cleavage rate equivalent to that measured at 1% of apparent Km (∼15 nmoles min−1 g−1 of fresh weight) would be sufficient to deplete the entire pool of kaempferol, if de-glycosylated on C-3, in <30 min. Although these calculations are only estimates that do not take into account subcellular compartmentation, they suggest strongly that in vivo any kaempferol that escapes glycosylation on C-3 is instantly cleaved. That no aglycone form of kaempferol can be detected in any of the Arabidopsis 3-O-glucosytransferase knockouts (Yin et al., 2012) is consistent with this scenario.
A Paradigm Shift for the Functional and Nutritional Significance of Flavonols in Plant Cells
Flavonoids have long been regarded as archetypal plant “secondary” products: They are conspicuous as flower and leaf pigments (anthocyanins), and also serve as antioxidants, protectants against UV-B and pathogens, signaling molecules, and regulators of auxin transport and metabolism (Peer and Murphy, 2007; Agati et al., 2012; Saito et al., 2013; Maloney et al., 2014; Yin et al., 2014; Tohge et al., 2016). Blurring the classification of flavonoids as strict specialized metabolites, our data show that plants can also use kaempferol as a precursor for the biosynthesis of an essential respiratory cofactor. This finding is consistent with the observation that flavonoid biosynthetic genes are expressed in all tissues and are under strong selective pressure, even in those taxa of the Caryophyllales lineage that lack anthocyanins (e.g. Amaranthaceae, Cactaceae, Phytolaccaceae; Shimada et al., 2005; Brockington et al., 2011). The branch-point between the biosynthesis of flavonoids and that of ubiquinone falls under the evolutionary concept of the “instant new pathway” (Pichersky and Gang, 2000), where an enzyme (here a heme-peroxidase) converts an intermediate from one pathway into an intermediate for another pathway. The atypical feature of this case, however, is that it entails the use of a specialized metabolite as a precursor for a primary metabolite.
The data on ubiquinone level in AtMYB12 tomato transgenics show that the ubiquinone content of fruits can be enhanced via a general increase in flux through the flavonoid biosynthetic pathway. Here again the finding is notable, for ubiquinone supplementation is known to decrease oxidative damage to membrane lipids and to have beneficial effects on mitochondrial metabolism in mammals (Witting et al., 2000; Bello et al., 2005; Sohet et al., 2009; Tarry-Adkins et al., 2013; Fazakerley et al., 2018). Similarly, our findings may reilluminate the understanding of dietary flavonol metabolism in connection to ubiquinone biosynthesis in mammals. Recent evidence indicates indeed that the exogenous supply of kaempferol boosts ubiquinone level in kidney cell cultures, and that those are able to use kaempferol as a ring precursor for ubiquinone biosynthesis (Fernández-Del-Río et al., 2017). Although this study did not identify which part of kaempferol was incorporated into ubiquinone or by which mechanism such an incorporation occurred, it was observed that feeding assays with naringenin and apigenin, which lack a C-2 C-3 double bond and/or a hydroxylated C-3, resulted in either marginal or no effect on ubiquinone content (Fernández-Del-Río et al., 2017). Such a crucial observation suggests that, in vertebrates, dietary kaempferol may contribute to ubiquinone biosynthesis via the same mechanism of peroxidative cleavage as that occurring in plant cells.
METHODS
Chemical and Reagents
Dihydrokaempferol, ubiquinone-9, and ubiquinone-10 were from Sigma-Aldrich. L-Phe-[ring-13C6] was from Cambridge Isotope Laboratories. Unless mentioned otherwise, other reagents were from Thermo Fisher Scientific. Calibration solutions of ubiquinone were prepared in 100% ethanol and quantified using the molar extinction coefficients of 14,700 M−1 cm−1 and 14,600 M−1 cm−1 at 275 nm for ubiquinone-9 and ubiquinone-10, respectively (Dawson et al., 1986). Quinol standards were prepared by reduction of their corresponding quinone forms using sodium borohydride. Calibration solutions of 4-hydroxybenzoate were quantified using the molar extinction coefficient of 13,900 M−1 cm−1 at 255 nm in 0.1 N HCl (Dawson et al., 1986). For the preparation of kaempferol-[B-Ring 13C6], Arabidopsis (Arabidopsis thaliana) f3′h knockout plants were axenically fed for 48 h with 250 µM of Phe-[ring-13C6]. Leaves (100 mg) were homogenized in 900 μL of methanol using a 5 mL Pyrex tissue grinder, and the extract was centrifuged at 18,000g for 10 min. The cleared extract was mixed to an equal volume of 2 N HCl and incubated at 70°C for 40 min. Sample aliquots (200 µL) were then mixed to 100 μL of 100% (v/v) methanol and centrifuged at 18,000g for 15 min. Samples (100 μL) were chromatographed on a Zorbax Eclipse Plus C18 column (4.6 × 100 mm, 3.5 μm; Agilent Technologies) held at 30°C and developed at a flow-rate of 0.8 mL.min−1 using a 25-min linear gradient starting from 10 mM ammonium formate pH 3.5 to 100% (v/v) methanol. The peak of kaempferol (18.7 min) was collected while monitoring the absorbance at 365 nm. Purified kaempferol was evaporated to dryness with gaseous N, resuspended in 100% (v/v) methanol, and quantified using a molar extinction coefficient of 21,242 M−1 cm−1 at 365 nm (calculated from Telange et al., 2014).
Plant Material and Growth Conditions
Arabidopsis mutants of the transparent testa series fully blocked in the formation of anthocyanidins (tt3-1; dfr), anthocyanidins, quercetin, and kaempferol (tt4-1; chs), or quercetin only (tt7-1; f3′h) were those described by Shirley et al. (1995), and were obtained from the Arabidopsis Biological Resource Center at the Ohio State University (Alonso et al., 2003). Knockout mutants corresponding to f3 h (SALK_113321), ref3-4 (GABI_753B06), 4cl (SALK_043310), pxa1-1 (CS3950), and 4-cl3 (Sail_636_b07) were isolated and confirmed as null alleles as described in Zolman et al. (2001), Schilmiller et al. (2009), Block et al. (2014), Masini (2014), and Li et al. (2015). The tomato (Solanum lycopersicum) anthocyanin reduced (are) mutant and its corresponding wild type (cv VF-36) were obtained from Pr. Gloria K. Muday at Wake Forest University. The generation of AtMYB12 tomato transgenics in the Moneymaker background was described in Luo et al. (2008). For the quantification of total ubiquinone in the leaves of the 4-cl3, chs, f3 h, f3′h, and dfr lines and their cognate wild types, plants were grown on potting mix in a growth chamber at 22°C in 16-h days (160 μE m−2 s−1) for 4 weeks. For 13C labeling and chemical rescue experiments, seeds were germinated on Murashige & Skoog (MS) plates containing 1% (w/v) SUC. After 7 d, seedlings were transferred to MS liquid medium containing 1% (w/v) SUC and grown for another week on an orbital shaker at 60 rpm in 10-h days (180 µE m−2 s−1) at 22°C.
Bioinformatics
Gene networks were reconstructed from the ATTED-II (http://atted.jp) database using Arabidopsis genes COQ1 (At2g34630), COQ2 (At4g23660), COQ3 (At2g30920), COQ5 (At5g57300), COQ6 (At3g24200), COQ8 (At4g160660), and COQ9 (At1g19140) as queries. The top 300 coexpressors of each of these genes were then aggregated using jvenn (http://jvenn.toulouse.inra.fr/app/example.html). Genes related to flavonoid metabolism, 4-CL3 (At1g65060) CHS (At5g13930), CHI/CHI-L1 (At3g55120/At5g05270), F3H (At3g51240), FLS1 (At5g08640), UGT78D2 (At5g17050), and UGT89C1 (At1g06000), were identified in the aggregated lists of coexpressors that intersected with at least two of the query genes. Hierarchical clustering of the coexpression profiles of ubiquinone and flavonoid biosynthetic genes was performed using the complete linkage method in Hcluster (http://atted.jp/top_draw.shtml#Hcluster).
Enzyme Assays
Arabidopsis leaves (0.18 g) were ground at 4°C using a 5-mL Pyrex tissue grinder in 3 mL of 50 mM KH2PO4 at pH 7.0, 150 mM KCl, 0.5% (w/v) cross-linked polyvinylpolypyrrolidone. The extract was centrifuged at 18,000g for 10 min, and the supernatant was desalted on a PD-10 column (GE Healthcare) equilibrated in 50 mM KH2PO4 at pH 7.0, 150 mM KCl. Proteins were quantified using the Bradford method (Bradford, 1976) and IgG as a standard. The desalted extract was aliquoted, flash-frozen in liquid N2, and stored at −80°C. Peroxidase assays (100 μL) contained 50 mM KH2PO4 at pH 7.0, 150 mM KCl, 0 μM to 200 μM of flavonols, 0 mM or 1 mM H2O2, and 0 μg to 2.5 μg of protein. For inhibition experiments, the extract was preincubated for 15 min at room temperature with 0 mM to 10 mM NaN3. Reactions were initiated with the addition of H2O2. After 5 min of incubation at 30°C, reactions were stopped with 30 μL of 6 N HCl and the samples were extracted twice with 400 μL of 100% ethyl acetate. Organic phases were evaporated to dryness and the samples were resuspended overnight at 4°C in 100 μL of 10 mM NaH2PO4 at pH 5.5: methanol (90:10, v/v). Samples (10 μL) were analyzed by HPLC with diode array detection (246 nm) on a Zorbax Eclipse Plus C18 (4.6 × 100 mm, 3.5 μm; Agilent Technologies) held at 30°C and developed at a flow-rate of 0.8 mL.min−1 with 10 mM NaH2PO4 at pH 5.5: methanol (95:5, v:v). 4-hydroxybenzoate eluted at 3.4 min and was quantified according to an external calibration standard.
Ubiquinone Analyses
Ubiquinone-9 quantification in Arabidopsis leaf extracts using reverse-phase HPLC coupled to diode array detection was performed as described in Ducluzeau et al. (2012). For LC-electrospray ionization-tandem MS analysis, tissue samples (30 mg to 100 mg fresh weight) were spiked with either 70 pmoles of ubiquinone-10 (Arabidopsis) or 70 pmoles of ubiquinone-9 (tomato) as internal standards and homogenized in 0.4 mL of 95% (v/v) ethanol using a 5-mL Pyrex tissue grinder. The grinder was rinsed twice with 0.3 mL of 95% (v/v) ethanol, and washes were combined to the original extract in a 10-mL Pyrex tube. Water (0.5 mL) was added and the mixture was partitioned twice with 5 mL hexane. Hexane layers were combined, evaporated to dryness with gaseous N, and resuspended in 0.2 mL of methanol:dichloromethane (10:1, v/v). Extracts (5 μL) were chromatographed isocratically on a Zorbax SB-C18 rapid resolution high-throughput column (50 mm × 2.1 mm; 1.8 μm; Agilent Technologies) held at 40°C with 5 mM ammonium formate in 100% methanol at a flow rate of 0.4 mL.min−1. The eluate was electrosprayed in positive mode with a N gas temperature of 300°C at a flow rate of 10 L min−1 into an Agilent 6430 Triple Quadrupole mass spectrometer. Nebulizer pressure was 35 psi and capillary potential voltage was set to 4000V. Reduced and oxidized ubiquinones were analyzed by multiple reaction monitoring using a dwell time of 50 ms and the following ion pairs: ubiquinol-9 (814.6/197) and ubiquinol-9-[13C6] (820.6/203) at 3.94 min; ubiquinol-10 (882.7/197) and ubiquinol-10-[13C6] (888.7/203) at 6.46 min; ubiquinone-9 (812.6/197) and ubiquinone-9-[13C6] (818.6/203) at 7.98 min; and ubiquinone-10 (880.7/197) and ubiquinone-10-[13C6] (886.7/203) at 13.72 min.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: At4g39850 (PXA1), At2g30490 (REF3), At4g19010, At2g34630 (COQ1), At4g23660 (COQ2), At2g30920 (COQ3), At5g57300 (COQ5), At3g24200 (COQ6), At4g01660 (COQ8), At1g19140 (COQ9), At1g65060 (4-CL3), At5g13930 (CHS), At3g55120 (CHI), At5g05270 (CHI-L1), At3g51240 (F3H), At5g07990 (F3′H), At5g08640 (FLS1), At5g42800 (DFR), At5g17050 (UGT78D2), and At1g06000 (UGT89C1).
Supplemental Data
Supplemental Figure 1. Biosynthesis of the ring precursor of ubiquinone, 4-Hydroxybenzoate, via the β-oxidative metabolism of phe in arabidopsis.
Supplemental Figure 2. Scheme of the biosynthetic pathway of flavonoids.
Supplemental Dataset 1. Correlation ranks and functional annotations mined from the ATTED-II databases.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
This work was supported by National Science Foundation grants (MCB-1608088 and MCB-1712608 to G.J.B.), and the USDA-ARS Floriculture and Nursery Research Initiative (to T.A.C.).
The authors thank Pr. Joshua Widhalm at Purdue University for illuminating discussions about benzenoid metabolism, Pr. Clint Chapple at Purdue University for the gift of the ref3-4 and 4-cl3 knockouts, and Pr. Gloria K. Muday at Wake Forest University for the gift of the are mutant.
AUTHOR CONTRIBUTIONS
E.S., T.S.J., and G.J.B. designed and performed the experiments, analyzed the data, and wrote the manuscript; S.L. and A.B. designed and performed the experiments, and analyzed the data; J.K., T.A.C., and M.A.W. analyzed the data and contributed to the preparation of the manuscript; C.M. and E.B. produced and characterized the E8:AtMYB12 tomato plants and characterized them metabolically as well as contributing to the design of the experiments with tomato and the preparation of the manuscript.
Footnotes
Articles can be viewed without a subscription.
References
- Agati G., Azzarello E., Pollastri S., Tattini M. (2012). Flavonoids as antioxidants in plants: location and functional significance. Plant Sci. 196: 67–76. [DOI] [PubMed] [Google Scholar]
- Allan C.M., Hill S., Morvaridi S., Saiki R., Johnson J.S., Liau W.S., Hirano K., Kawashima T., Ji Z., Loo J.A., Shepherd J.N., Clarke C.F. (2013). A conserved START domain coenzyme Q-binding polypeptide is required for efficient Q biosynthesis, respiratory electron transport, and antioxidant function in Saccharomyces cerevisiae. Biochim. Biophys. Acta 1831: 776–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso J.M., Stepanova A.N., Leisse T.J., Kim C.J., Chen H., Shinn P., Stevenson D.K., Zimmerman J., Barajas P., Cheuk R., Gadrinab C., Heller C., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657. [DOI] [PubMed] [Google Scholar]
- Bello R.I., Gómez-Díaz C., Burón M.I., Alcaín F.J., Navas P., Villalba J.M. (2005). Enhanced anti-oxidant protection of liver membranes in long-lived rats fed on a coenzyme Q10-supplemented diet. Exp. Gerontol. 40: 694–706. [DOI] [PubMed] [Google Scholar]
- Bentinger M., Tekle M., Dallner G. (2010). Coenzyme Q—biosynthesis and functions. Biochem. Biophys. Res. Commun. 396: 74–79. [DOI] [PubMed] [Google Scholar]
- Block A., Widhalm J.R., Fatihi A., Cahoon R.E., Wamboldt Y., Elowsky C., Mackenzie S.A., Cahoon E.B., Chapple C., Dudareva N., Basset G.J. (2014). The origin and biosynthesis of the benzenoid moiety of ubiquinone (coenzyme Q) in Arabidopsis. Plant Cell 26: 1938–1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M.M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72: 248–254. [DOI] [PubMed] [Google Scholar]
- Brockington S.F., Walker R.H., Glover B.J., Soltis P.S., Soltis D.E. (2011). Complex pigment evolution in the Caryophyllales. New Phytol. 190: 854–864. [DOI] [PubMed] [Google Scholar]
- Buer C.S., Kordbacheh F., Truong T.T., Hocart C.H., Djordjevic M.A. (2013). Alteration of flavonoid accumulation patterns in transparent testa mutants disturbs auxin transport, gravity responses, and imparts long-term effects on root and shoot architecture. Planta 238: 171–189. [DOI] [PubMed] [Google Scholar]
- Cardazzo B., Hamel P., Sakamoto W., Wintz H., Dujardin G. (1998). Isolation of an Arabidopsis thaliana cDNA by complementation of a yeast abc1 deletion mutant deficient in complex III respiratory activity. Gene 221: 117–125. [DOI] [PubMed] [Google Scholar]
- Dawson R.M.C., Elliot D.C., Elliot W.H., Jones K.M. (1986). Data for Biochemical Research. (New York: Oxford University Press; ). [Google Scholar]
- Ducluzeau A.-L., Wamboldt Y., Elowsky C.G., Mackenzie S.A., Schuurink R.C., Basset G.J.C. (2012). Gene network reconstruction identifies the authentic trans-prenyl diphosphate synthase that makes the solanesyl moiety of ubiquinone-9 in Arabidopsis. Plant J. 69: 366–375. [DOI] [PubMed] [Google Scholar]
- Fazakerley D.J., Chaudhuri R., Yang P., Maghzal G.J., Thomas K.C., Krycer J.R., Humphrey S.J., Parker B.L., Fisher-Wellman K.H., Meoli C.C., Hoffman N.J., Diskin C., et al. (2018). Mitochondrial CoQ deficiency is a common driver of mitochondrial oxidants and insulin resistance. eLife 7: e32111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Del-Río L., Nag A., Gutiérrez Casado E., Ariza J., Awad A.M., Joseph A.I., Kwon O., Verdin E., de Cabo R., Schneider C., Torres J.Z., Burón M.I., et al. (2017). Kaempferol increases levels of coenzyme Q in kidney cells and serves as a biosynthetic ring precursor. Free Radic. Biol. Med. 110: 176–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- French C., Vance C.P., Towers G.H.N. (1976). Conversion of p-coumaric acid to p-hydroxybenzoic acid by cell free extracts of potato tubers and Polyporus hispidus. Phytochemistry 15: 564–566. [Google Scholar]
- Gasson M.J., Kitamura Y., McLauchlan W.R., Narbad A., Parr A.J., Parsons E.L., Payne J., Rhodes M.J., Walton N.J. (1998). Metabolism of ferulic acid to vanillin. A bacterial gene of the enoyl-SCoA hydratase/isomerase superfamily encodes an enzyme for the hydration and cleavage of a hydroxycinnamic acid SCoA thioester. J. Biol. Chem. 273: 4163–4170. [DOI] [PubMed] [Google Scholar]
- Heazlewood J.L., Tonti-Filippini J.S., Gout A.M., Day D.A., Whelan J., Millar A.H. (2004). Experimental analysis of the Arabidopsis mitochondrial proteome highlights signaling and regulatory components, provides assessment of targeting prediction programs, and indicates plant-specific mitochondrial proteins. Plant Cell 16: 241–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenaz G., Genova M.L. (2009). Mobility and function of coenzyme Q (ubiquinone) in the mitochondrial respiratory chain. Biochim. Biophys. Acta 1787: 563–573. [DOI] [PubMed] [Google Scholar]
- Lepiniec L., Debeaujon I., Routaboul J.M., Baudry A., Pourcel L., Nesi N., Caboche M. (2006). Genetics and biochemistry of seed flavonoids. Annu. Rev. Plant Biol. 57: 405–430. [DOI] [PubMed] [Google Scholar]
- Li Y., Kim J.I., Pysh L., Chapple C. (2015). Four isoforms of Arabidopsis 4-coumarate:CoA ligase have overlapping yet distinct roles in phenylpropanoid metabolism. Plant Physiol. 169: 2409–2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Löscher R., Heide L. (1994). Biosynthesis of p-hydroxybenzoate from p-coumarate and p-coumaroyl-CoA in cell-free extracts of Lithospermum erythrorhizon cell cultures. Plant Physiol. 106: 271–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J., Butelli E., Hill L., Parr A., Niggeweg R., Bailey P., Weisshaar B., Martin C. (2008). AtMYB12 regulates caffeoyl quinic acid and flavonol synthesis in tomato: Expression in fruit results in very high levels of both types of polyphenol. Plant J. 56: 316–326. [DOI] [PubMed] [Google Scholar]
- Maloney G.S., DiNapoli K.T., Muday G.K. (2014). The anthocyanin reduced tomato mutant demonstrates the role of flavonols in tomato lateral root and root hair development. Plant Physiol. 166: 614–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marbois B., Xie L.X., Choi S., Hirano K., Hyman K., Clarke C.F. (2010). para-Aminobenzoic acid is a precursor in coenzyme Q6 biosynthesis in Saccharomyces cerevisiae. J. Biol. Chem. 285: 27827–27838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masini L. (2014). Investigation of the molecular basis of PAMP-induced resistance. PhD thesis,University of East Anglia,The Sainsbury Laboratory, Norwich, UK [Google Scholar]
- Mitra A., Mayer M.J., Mellon F.A., Michael A.J., Narbad A., Parr A.J., Waldron K.W., Walton N.J. (2002). 4-Hydroxycinnamoyl-CoA hydratase/lyase, an enzyme of phenylpropanoid cleavage from Pseudomonas, causes formation of C(6)-C(1) acid and alcohol glucose conjugates when expressed in hairy roots of Datura stramonium L. Planta 215: 79–89. [DOI] [PubMed] [Google Scholar]
- Nowicka B., Kruk J. (2010). Occurrence, biosynthesis and function of isoprenoid quinones. Biochim. Biophys. Acta 1797: 1587–1605. [DOI] [PubMed] [Google Scholar]
- Obayashi T., Aoki Y., Tadaka S., Kagaya Y., Kinoshita K. (2018). ATTED-II in 2018: A plant coexpression database based on investigation of the statistical property of the mutual rank index. Plant Cell Physiol. 59: 440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohara K., Kokado Y., Yamamoto H., Sato F., Yazaki K. (2004). Engineering of ubiquinone biosynthesis using the yeast coq2 gene confers oxidative stress tolerance in transgenic tobacco. Plant J. 40: 734–743. [DOI] [PubMed] [Google Scholar]
- Ortiz de Montellano P.R., David S.K., Ator M.A., Tew D. (1988). Mechanism-based inactivation of horseradish peroxidase by sodium azide. Formation of meso-azidoprotoporphyrin IX. Biochemistry 27: 5470–5476. [DOI] [PubMed] [Google Scholar]
- Owens D.K., Alerding A.B., Crosby K.C., Bandara A.B., Westwood J.H., Winkel B.S. (2008). Functional analysis of a predicted flavonol synthase gene family in Arabidopsis. Plant Physiol. 147: 1046–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payet L.A., Leroux M., Willison J.C., Kihara A., Pelosi L., Pierrel F. (2016). Mechanistic details of early steps in coenzyme Q biosynthesis pathway in yeast. Cell Chem. Biol. 23: 1241–1250. [DOI] [PubMed] [Google Scholar]
- Peer W.A., Murphy A.S. (2007). Flavonoids and auxin transport: modulators or regulators? Trends Plant Sci. 12: 556–563. [DOI] [PubMed] [Google Scholar]
- Peer W.A., Brown D.E., Tague B.W., Muday G.K., Taiz L., Murphy A.S. (2001). Flavonoid accumulation patterns of transparent testa mutants of Arabidopsis. Plant Physiol. 126: 536–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichersky E., Gang D.R. (2000). Genetics and biochemistry of secondary metabolites in plants: An evolutionary perspective. Trends in Plant Science 439–445. [DOI] [PubMed] [Google Scholar]
- Pierrel F. (2017). Impact of chemical analogs of 4-hydroxybenzoic acid on coenzyme Q biosynthesis: from inhibition to bypass of coenzyme Q deficiency. Front. Physiol. 8: 436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierrel F., Hamelin O., Douki T., Kieffer-Jaquinod S., Mühlenhoff U., Ozeir M., Lill R., Fontecave M. (2010). Involvement of mitochondrial ferredoxin and para-aminobenzoic acid in yeast coenzyme Q biosynthesis. Chem. Biol. 17: 449–459. [DOI] [PubMed] [Google Scholar]
- Pillai B.V., Swarup S. (2002). Elucidation of the flavonoid catabolism pathway in Pseudomonas putida PML2 by comparative metabolic profiling. Appl. Environ. Microbiol. 68: 143–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pourcel L., Routaboul J.M., Cheynier V., Lepiniec L., Debeaujon I. (2007). Flavonoid oxidation in plants: from biochemical properties to physiological functions. Trends Plant Sci. 12: 29–36. [DOI] [PubMed] [Google Scholar]
- Preuss A., Stracke R., Weisshaar B., Hillebrecht A., Matern U., Martens S. (2009). Arabidopsis thaliana expresses a second functional flavonol synthase. FEBS Lett. 583: 1981–1986. [DOI] [PubMed] [Google Scholar]
- Saito K., Yonekura-Sakakibara K., Nakabayashi R., Higashi Y., Yamazaki M., Tohge T., Fernie A.R. (2013). The flavonoid biosynthetic pathway in Arabidopsis: Structural and genetic diversity. Plant Physiol. Biochem. 72: 21–34. [DOI] [PubMed] [Google Scholar]
- Schilmiller A.L., Stout J., Weng J.K., Humphreys J., Ruegger M.O., Chapple C. (2009). Mutations in the cinnamate 4-hydroxylase gene impact metabolism, growth and development in Arabidopsis. Plant J. 60: 771–782. [DOI] [PubMed] [Google Scholar]
- Schneider H., Blaut M. (2000). Anaerobic degradation of flavonoids by Eubacterium ramulus. Arch. Microbiol. 173: 71–75. [DOI] [PubMed] [Google Scholar]
- Schnitzler J.P., Madlung J., Rose A., Ulrich Seitz H. (1992). Biosynthesis of p-hydroxybenzoic acid in elicitor-treated carrot cell cultures. Planta 188: 594–600. [DOI] [PubMed] [Google Scholar]
- Sévin D.C., Sauer U. (2014). Ubiquinone accumulation improves osmotic-stress tolerance in Escherichia coli. Nat. Chem. Biol. 10: 266–272. [DOI] [PubMed] [Google Scholar]
- Shimada S., Inoue Y.T., Sakuta M. (2005). Anthocyanidin synthase in non-anthocyanin-producing Caryophyllales species. Plant J. 44: 950–959. [DOI] [PubMed] [Google Scholar]
- Shirley B.W., Kubasek W.L., Storz G., Bruggemann E., Koornneef M., Ausubel F.M., Goodman H.M. (1995). Analysis of Arabidopsis mutants deficient in flavonoid biosynthesis. Plant J. 8: 659–671. [DOI] [PubMed] [Google Scholar]
- Shockey J., Browse J. (2011). Genome-level and biochemical diversity of the acyl-activating enzyme superfamily in plants. Plant J. 66: 143–160. [DOI] [PubMed] [Google Scholar]
- Sohet F.M., Neyrinck A.M., Pachikian B.D., de Backer F.C., Bindels L.B., Niklowitz P., Menke T., Cani P.D., Delzenne N.M. (2009). Coenzyme Q10 supplementation lowers hepatic oxidative stress and inflammation associated with diet-induced obesity in mice. Biochem. Pharmacol. 78: 1391–1400. [DOI] [PubMed] [Google Scholar]
- Stowell M.H., McPhillips T.M., Rees D.C., Soltis S.M., Abresch E., Feher G. (1997). Light-induced structural changes in photosynthetic reaction center: implications for mechanism of electron-proton transfer. Science 276: 812–816. [DOI] [PubMed] [Google Scholar]
- Tarry-Adkins J.L., Blackmore H.L., Martin-Gronert M.S., Fernandez-Twinn D.S., McConnell J.M., Hargreaves I.P., Giussani D.A., Ozanne S.E. (2013). Coenzyme Q10 prevents accelerated cardiac aging in a rat model of poor maternal nutrition and accelerated postnatal growth. Mol. Metab. 2: 480–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telange D.R., Patil A.T., Tatode A., Bhoyar B. (2014). Development and validation of UV spectrophotometric method for the estimation of kaempferol in kaempferol: hydrogenated soy phosphatidylcholine (HSPC) complex. Pharm. Methods 5: 34–38. [Google Scholar]
- Tohge T., Wendenburg R., Ishihara H., Nakabayashi R., Watanabe M., Sulpice R., Hoefgen R., Takayama H., Saito K., Stitt M., Fernie A.R. (2016). Characterization of a recently evolved flavonol-phenylacyltransferase gene provides signatures of natural light selection in Brassicaceae. Nat. Commun. 7: 12399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran U.C., Clarke C.F. (2007). Endogenous synthesis of coenzyme Q in eukaryotes. Mitochondrion 7 (Suppl): S62–S71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veitch N.C. (2004). Horseradish peroxidase: a modern view of a classic enzyme. Phytochemistry 65: 249–259. [DOI] [PubMed] [Google Scholar]
- Wagner G.J., Hrazdina G. (1984). Endoplasmic reticulum as a site of phenylpropanoid and flavonoid metabolism in hippeastrum. Plant Physiol. 74: 901–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widhalm J.R., Dudareva N. (2015). A familiar ring to it: Biosynthesis of plant benzoic acids. Mol. Plant 8: 83–97. [DOI] [PubMed] [Google Scholar]
- Wildermuth M.C. (2006). Variations on a theme: Synthesis and modification of plant benzoic acids. Curr. Opin. Plant Biol. 9: 288–296. [DOI] [PubMed] [Google Scholar]
- Winter J., Moore L.H., Dowell V.R. Jr., Bokkenheuser V.D. (1989). C-ring cleavage of flavonoids by human intestinal bacteria. Appl. Environ. Microbiol. 55: 1203–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witting P.K., Pettersson K., Letters J., Stocker R. (2000). Anti-atherogenic effect of coenzyme Q10 in apolipoprotein E gene knockout mice. Free Radic. Biol. Med. 29: 295–305. [DOI] [PubMed] [Google Scholar]
- Yazaki K., Heide L., Tabata M. (1991). Formation of p-hydroxybenzoic acid from p-coumaric acid by cell free extract of Lithospermum erythrorhizon cell cultures. Phytochemistry 30: 2233–2236. [Google Scholar]
- Yin R., Messner B., Faus-Kessler T., Hoffmann T., Schwab W., Hajirezaei M.R., von Saint Paul V., Heller W., Schäffner A.R. (2012). Feedback inhibition of the general phenylpropanoid and flavonol biosynthetic pathways upon a compromised flavonol-3-O-glycosylation. J. Exp. Bot. 63: 2465–2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin R., Han K., Heller W., Albert A., Dobrev P.I., Zažímalová E., Schäffner A.R. (2014). Kaempferol 3-O-rhamnoside-7-O-rhamnoside is an endogenous flavonol inhibitor of polar auxin transport in Arabidopsis shoots. New Phytol. 201: 466–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonekura-Sakakibara K., Tohge T., Matsuda F., Nakabayashi R., Takayama H., Niida R., Watanabe-Takahashi A., Inoue E., Saito K. (2008). Comprehensive flavonol profiling and transcriptome coexpression analysis leading to decoding gene-metabolite correlations in Arabidopsis. Plant Cell 20: 2160–2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenkevich I.G., Eshchenko A.Y., Makarova S.V., Vitenberg A.G., Dobryakov Y.G., Utsal V.A. (2007). Identification of the products of oxidation of quercetin by air oxygen at ambient temperature. Molecules 12: 654–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Butelli E., Alseekh S., Tohge T., Rallapalli G., Luo J., Kawar P.G., Hill L., Santino A., Fernie A.R., Martin C. (2015). Multi-level engineering facilitates the production of phenylpropanoid compounds in tomato. Nat. Commun. 6: 8635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolman B.K., Silva I.D., Bartel B. (2001). The Arabidopsis pxa1 mutant is defective in an ATP-binding cassette transporter-like protein required for peroxisomal fatty acid beta-oxidation. Plant Physiol. 127: 1266–1278. [PMC free article] [PubMed] [Google Scholar]