Abstract
Background
Glucose-6-phosphate dehydrogenase (G6PDH or G6PD) functions in supply of NADPH, which is required for plant defense responses to stresses. However, whether G6PD functions in the abscisic acid (ABA) signaling pathway remains to be elucidated. In this study, we investigated the involvement of the cytosolic G6PD5 in the ABA signaling pathway in Arabidopsis.
Results
We characterized the Arabidopsis single null mutant g6pd5. Phenotypic analysis showed that the mutant is more sensitive to ABA during seed germination and root growth, whereas G6PD5-overexpressing plants are less sensitive to ABA compared to wild type (WT). Furthermore, ABA induces excessive accumulation of reactive oxygen species (ROS) in mutant seeds and seedlings. G6PD5 participates in the reduction of H2O2 to H2O in the ascorbate-glutathione cycle. In addition, we found that G6PD5 suppressed the expression of Abscisic Acid Insensitive 5 (ABI5), the major ABA signaling component in dormancy control. When G6PD5 was overexpressed, the ABA signaling pathway was inactivated. Consistently, G6PD5 negatively modulates ABA-blocked primary root growth in the meristem and elongation zones. Of note, the suppression of root elongation by ABA is triggered by the cell cycle B-type cyclin CYCB1.
Conclusions
This study showed that G6PD5 is involved in the ABA-mediated seed germination and root growth by suppressing ABI5.
Electronic supplementary material
The online version of this article (10.1186/s12870-019-1647-8) contains supplementary material, which is available to authorized users.
Keywords: Abscisic acid, Germination, Glucose-6-phosphate dehydrogenase 5, NADPH oxidases, Reactive oxygen species, Root system architecture
Background
The oxidative pentose phosphate pathway (OPPP) is the main pathway of NADPH production, which is used for biosynthesis [1–4] and redox balance in plant cells [5, 6]. The main regulatory step of OPPP is catalysed by glucose-6-phosphate dehydrogenase (G6PDH or G6PD). Many experiments have proven that G6PD is induced by adverse biotic and abiotic stresses, including salinity, drought and ABA [7–11]. Enhanced G6PD activity is linked to the promotion of plant survival and tolerance [9, 11, 12]. Arabidopsis genome-wide analysis indicates the presence of two cytosolic (Cy-G6PD) and four plastidial (Pla-G6PD) isoforms of G6PD [13]. The Cy-G6PD includes G6PD5 and G6PD6. Based on the difference in amino acid sequence, the Pla-G6PD is divided into P1, P2 and P0 type: P1 mainly exists in the chloroplast (G6PD1); P2 mainly exists in plastids and some non-oxygen cells (G6PD2, G6PD3), while P0 is a non-functional gene (G6PD4) [13]. Extensive studies indicate that cytosolic and plastidic G6PD play different roles in plant survival and tolerance [9, 11, 12]. For example, Pla-G6PD is crucial in regulating biochemical responses of heavy metals [14], while Cy-G6PD is involved in aluminum toxicity of soybean under high aluminum concentration [15]. In Arabidopsis, G6PD6 constitutes an immune signaling module downstream of pattern recognition receptors, linking protein phosphorylation cascades to metabolic regulation [16].
Many stresses cause water deficit and ion imbalance, which leads to inhibition of essential enzymes, destabilization of cell membranes, decrease in nutrient supply, and overproduction of reactive oxygen species (ROS) [12, 17]. ROS serve as signaling molecules to regulate many biological processes including seed germination and root growth in plants [12, 17–19]. It has been documented that ROS are produced through both enzymatic and non-enzymatic reactions in plants [20, 21]. In Arabidopsis, ROS are directly originated from AtrbohD and AtrbohF, two ROS-generating NADPH oxidases, impairing stress inhibition of primary root elongation [18, 22]. Recent studies showed that G6PD plays a primary role in stress responses, favoring ROS-scavenging functions [23]. In fact, during drought stress, plant cells increase their needs for reducing power in order to sustain the antioxidant defense system and counteract ROS accumulation and consequent damages [23, 24].
Abscisic acid (ABA) synthesis is significantly induced by stresses and the ABA signaling has an important function in abiotic stress responses, such as seed maturation and dormancy, stomatal closure, and root growth and developmental regulation [19, 25]. ABA-mediated gene regulation occurs through the conserved ABA-responsive elements (ABREs) in gene promoters [26]. ABREs contain ACGT as the core nucleotide sequence, which acts as a binding site for bZIP transcription factors [2, 26, 27]. In Arabidopsis, Abscisic Acid-Insensitive 5 (ABI5), a bZIP transcription factor, plays a vital role in mediating ABA signaling during seed maturation [28]. ABA-enhanced stress tolerance is associated with the induction of ROS scavenging systems [29–32]. Furthermore, recent studies showed that ABA affects the activity and expression of barley plastidial G6PD [2]. ABA mediates drought-induced increase of the cytosolic G6PD activity, and the enhanced cytosolic G6PD activity maintains cellular redox homeostasis by regulating the AsA-GSH cycle in soybean roots [11].
With regard to the cytosolic isoforms, the Arabidopsis cy-G6PD mutants produce seeds with higher oil content, suggesting that cy-G6PD is essential for the fatty acid metabolism in developing seeds [11, 13]. Interestingly, when G6PD6 knockout plants were tested for their stress sensitivity, the germination rate of mutant seeds was significantly reduced under salinity conditions and the root growth was strongly affected by NaCl [12]. However, little is known about the expression and function of G6PD5. In this work, we used genetic and molecular approaches to study the function of G6PD5. We characterized the function of G6PD5 in seed germination and root growth. In addition, our results demonstrate that G6PD5 functions antagonistically with ABI5 to maintain the ABA signaling level necessary for seed germination and subsequent seedling establishment. We uncovered a novel interplay between ROS, ABA, and G6PD5.
Methods
Plant materials and growth conditions
Arabidopsis thaliana Col-0 was used as the wild-type. T-DNA insertion mutants g6pd5 (CS804669) and g6pd6 (SALK_016157C) were obtained from the Arabidopsis Biological Resource Center (http://www.arabidopsis.org/). The T-DNA in the g6pd5 mutant is inserted in the coding region of At3g2300, and in the g6pd6 mutant, T-DNA is inserted in the coding region of At5g40760. The G6PD5 overexpressing plants (OE#1, OE#9) was obtained by transforming the G6PD5-containing constructs into Col-0. The following mutants or transgenic lines were also used in this study: aba2–1, aba2–3, abi4–102. The NADPH oxidase gene single mutants atrbohD1 (CS9555) and atrbohF1 (CS9557) and the double mutant atrbohD1/F1 were obtained from the Arabidopsis Biological Resource Center. ABI5::GUS was friendly given by Zuhua He (Chinese Academy of Sciences). Seeds of abi3 and abi5–2 were provided in courtesy from Yinggao Liu (Shandong Agricultural University, China). The transgenic line CYCB1;1::GUS was kindly provided by Guangqin Guo (Lanzhou University, China). All of them are in the Col-0 background. Seeds were sterilized with 1.5% NaClO for 15 min, washed with sterile water for three times, placed at 4 °C for 3 d. Cold-treated seeds were germinated on the half-strength Murashige and Skoog (1/2 MS) medium (pH 5.8) containing 1% sucrose and 0.8% agar in a growth room at 23 °C under 100–120 μmol photons·m− 2·s− 1 with a 16 h/8 h light/dark photoperiod.
Phenotypic analysis
In germination assays, WT, g6pd5, OE#1, and OE#9 seeds (approximately 50 seeds for each replicate) were surface-sterilized. The seeds were sown on 1/2 MS medium with or without different concentrations of ABA and then incubated at 23 °C with a 16-h light/8-h dark photoperiod. The number of planted and germinated seeds was recorded 5 d after planting. Radicle emergence of > 1 mm indicated seed germination. Three replicate plates were used for each treatment.
In root elongation measurements, Arabidopsis seeds were sown on 1/2 MS medium as above, stratified for 3 d, and then germinated at 23 °C for 5 d. For root elongation measurements, 15 seedlings were used per replicate, and three replicates were made for each treatment. Five-day-old seedlings with roots 1–1.5 cm long were transferred from 1/2 MS agar plates onto a new agar medium supplemented with different concentrations of ABA. Increases in root length were measured after 3 d of treatment [33, 34]. The length of primary roots was measured with the NIH Image software (Image J, version 1.43). β-glucuronidase (GUS)-staining sites were counted using an anatomical microscope.
Generation of plants with different genotypes
To generate g6pd5 plants harboring CYCB1;1::GUS (g6pd5/CYCB1;1 plants) or ABI5::GUS (g6pd5/ABI5 plants), the emasculated flowers of g6pd5 plants were crossed with pollen from Col-0 plants harboring CYCB1;1::GUS (Col/CYCB1;1 plants) or ABI5::GUS (Col/ABI5 plants). The resulting homozygous g6pd5/CYCB1;1 and g6pd5/ABI5 plants were identified by PCR using G6PD5-specific primers (left genomic primer, 5΄-CACCATGGGTTCTGGTCAATGGC-3΄; right genomic primer, 5΄-CAATGTAGGAGGGATCTAAATGTAG-3΄) and a T-DNA left-border primer LBb1. Identification of the CYCB1;1::GUS and ABI5::GUS background was performed by GUS staining; lines in which all seedlings stained blue were used for the experiments.
Confocal microscopy
For analysis of fluorescence from the probe propidium iodide (PI), seedling roots were stained with PI (Molecular Probes, Eugene, OR, USA) according to the method described by Mei et al. [35]. Seedlings were incubated in the dark with 10 μg/ml of PI for 5–10 min at 25 °C and then washed three times with ddH2O. The roots were then imaged under a confocal microscope (Olympus FV 1000; excitation 488 nm, emission 570–650 nm). The length of the elongation zone and the meristem zone in roots, the epidermal cell size, and the cell number in the root elongation region in WT and mutant seedlings were determined after staining with PI under a confocal microscope.
Histochemical staining and assay of reactive oxygen species (ROS)
The hydrogen peroxide (H2O2) accumulation was determined using 2,7-dichlorodihydrofluorescein diacetate (H2DCF, Molecular Probes). Seeds of 2-day-old or roots of 5-day-old seedlings were treated with 20 μM H2DCF for 10–15 min, and the fluorescence intensity was monitored under a fluorescence microscope (Olympus FV 1000, excitation 488 nm and emission 500–550 nm). For the determination of H2O2 or O2.- in roots, histochemical staining in root tips was performed using 0.01% (w/v) 3,3-diaminobenzidine (DAB, pH 5.0, for H2O2 detection) or 0.005% (w/v) nitrotetrazolium blue chloride (NBT, pH 7.4, for O2.- detection), respectively. In all cases, stained materials were photographed using a digital camera (Canon, PC1146). H2O2 and O2.- content were visually detected according to Wang et al. [15].
Activity determination of antioxidant enzymes, NADPH oxidase and G6PD
Ten-day-old seedlings were soaked in solutions with or without 10 μM ABA for 12 h. After treatment, the activities of antioxidant enzymes, NADPH oxidase and G6PD were evaluated according to the methods of Liu et al. [36] and Wang et al. [37].
GUS staining
For GUS staining, seedlings were incubated in GUS staining buffer (1 mM 5-bromo-4-chloro-3-indolyl-β-D-glucuronic acid (X-Gluc), 100 mM sodium phosphate (pH 7.5), 0.5 mM K3[Fe (CN)6], 0.5 mM K4[Fe (CN)6], 10 mM EDTA, and 0.1% Triton X-100) for 6–18 h at 37 °C and mounted in solution for microscopy analysis [38]. Quantitative GUS activity assay was performed as described by Nan et al. [33].
Quantitative real-time PCR analysis
Trizol (TaKaRa) was used for the extraction of total RNA from seedlings. After total RNA was digested with the RNase-free DNase I (Promega) for 45 min at 37 °C, it was used for the cDNA synthesis with PrimeScript II 1st Strand cDNA Synthesis Kit (Takara). The diluted cDNA was used for qRT-PCR analysis with SYBR Premix Ex Taq™ II (Takara). Primer sequences used in the study were shown in Additional file 1: Table S1. The cycle threshold 2(−ΔΔC(T))-based method was used for relative quantitation of gene expression. Expression levels were normalized to Actin 2.
Arabidopsis Genome Initiative locus identifiers for the genes mentioned in this article are as follows: ACTIN2 (AT3G18780), G6PD5 (AT3G27300), G6PD6 (AT5G40760), AtrbohD (AT5G47910), AtrbohF (AT1G64060), APX1 (At1G07890), GR2 (At3G54660), NCED6 (AT3G24220), NCED9 (AT1G78390), CYP707A3 (AT5G45340), CYP707A4 (AT3G19270), ABI3 (AT3G24650), ABI4 (AT2G40220), ABI5 (AT2G36270), CYCB1;1 (AT4G37490), PLT1 (AT3G20840), PLT2 (AT1G51190).
Generation of G6PD5-overexpressing (G6PD5-OE) lines
Full-length Arabidopsis G6PD5 cDNA was obtained using reverse transcription PCR, cloned into the pENTR-TOPO cloning vector (Invitrogen) and sequenced. After the LR reaction, G6PD5 cDNA was inserted into the pGWB2 vector; this construct was named pGWB2-G6PD5. Transformed plants were selected on hygromycin-containing medium. Plants of the second generation after transformation were used for the experiments. The empty pGWB5 vector (the ccdb gene was substituted by a non-sense segment with a termination codon) was also transferred into Col-0 and used as control plants.
Statistical analysis
Each experiment was repeated at least three times. Data were analyzed by one-way variance analysis (ANOVA, P < 0.05), and expressed as mean ± SE.
Results
Responsiveness of g6pd mutants and G6PD5-overexpressing lines to ABA treatment during seed germination and root growth
ABA is significantly induced by stresses and its signaling has an important function in abiotic stress responses. Therefore, we tested whether G6PD5 is involved in the regulation of ABA signaling. To study the underlying role of G6PD5 in Arabidopsis, we obtained a T-DNA insertion mutant from the Arabidopsis Biological Resource Center. The g6pd5 mutant did not display any visible phenotype at the germination and seedling stages compared to WT under normal growth conditions (Fig. 1a-c). However, the g6pd5 mutant exhibited severely reduced seed germination rate with increased ABA concentrations compared to WT (Figs. 1, 2a-c). In addition, the primary root growth of g6pd5 also showed an ABA-sensitive phenotype (Fig. 2d, e). In contrast, seeds of the g6pd6 mutant exhibited inconspicuous seed germination compared to WT with ABA treatment (Fig. 1a-c). Moreover, the expression of G6PD5 was induced by ABA treatment, whereas G6PD6 was inhibited (Fig. 1). Therefore, we mainly focused on G6PD5 in the following experiments.
Fig. 1.
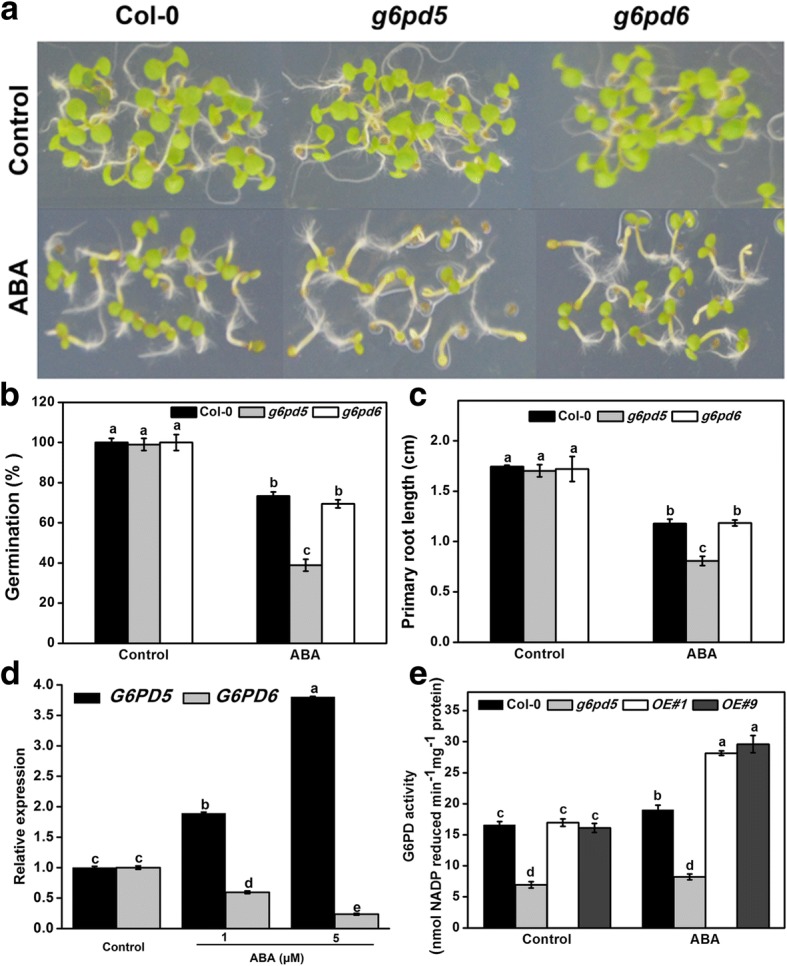
Seed germination and root growth of wild type (WT), g6pd5, and g6pd6 mutant in response to ABA. a and b Seeds were germinated on 1/2 MS agar plates with 1 μM ABA. Photographs were taken 4 d after treatment. c 5-day-old seedlings were grown vertically on 1/2 MS agar plates supplemented with 10 μM ABA. Root growth was monitored and analyzed using ImageJ software. d Relative transcript levels of G6PD5 and G6PD6 in wild-type (Col-0) with different concentrations of ABA treatment. The transcript levels were normalized to Actin2. e The activities of G6PD in Arabidopsis WT and mutants exposed to ABA treatment. One-way Duncan’s test was performed, and statistically significant differences are indicated by different lower case letters (P < 0.05). Bar, 1 cm. The experiments were repeated at least three times with similar results, and data from one representative experiment are presented
Fig. 2.
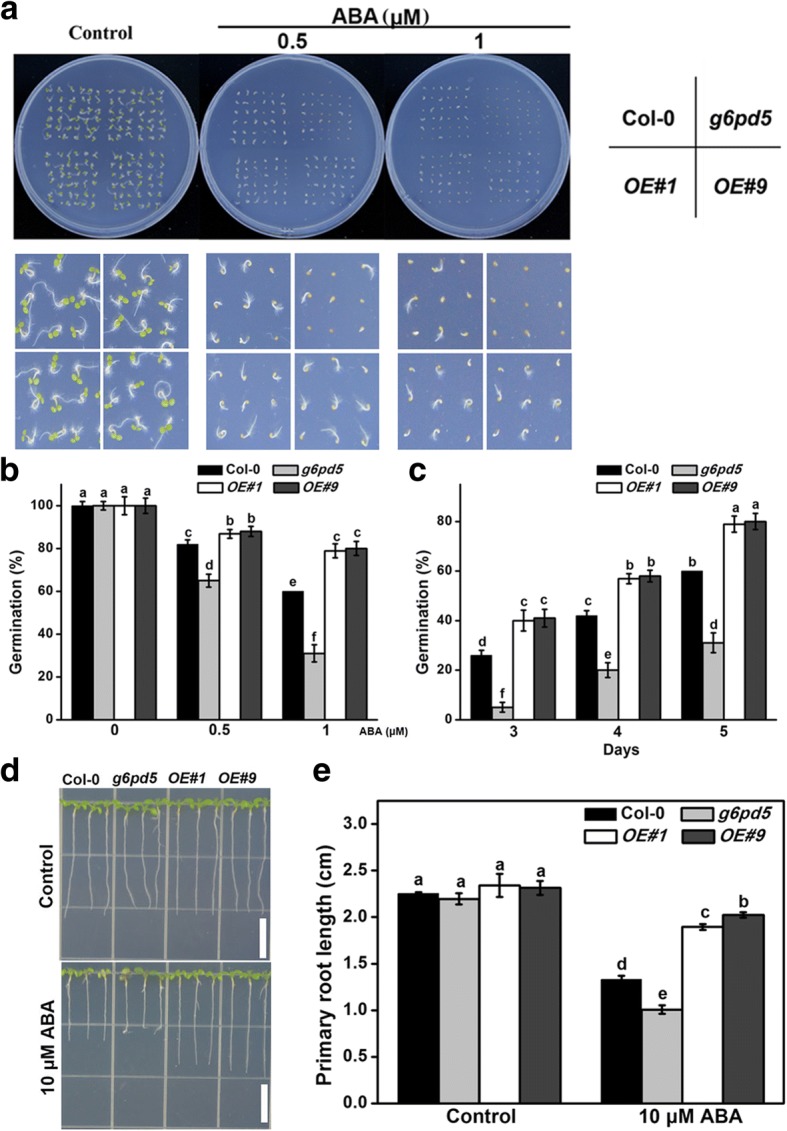
Seed germination and root growth of wild type (WT), g6pd5 mutant, and G6PD5-OE lines in response to ABA. a Seeds were germinated on 1/2 MS agar plates with or without ABA. Photographs were taken 4 d after ABA treatment. b Percentage of seed germination with or without ABA treatment for 4 d. c Percentage of seed germination with 1 μM ABA for 3–5 d. d and e 5-day-old seedlings were grown vertically on 1/2 MS agar plates supplemented with indicated concentrations of ABA for 3 d and the length of newly grown roots was measured. Root growth was monitored and analyzed using the ImageJ software
Under normal growth conditions, no significant difference in the germination rate and primary root growth was observed between WT and the G6PD5 overexpression (OE) lines (Fig. 2). However, with ABA treatment, the OE lines exhibited a significantly higher seed germination rate than WT (Fig. 2a, b) and the primary root growth of OE plants was also hyposensitive to ABA treatment (Fig. 2c, d). These data indicate that overexpression of G6PD5 increases ABA tolerance in Arabidopsis.
Oxidative damage in G6PD5-overexpressing and g6pd5 mutant plants
Previous studies showed that ROS play a key regulatory role in the germination program under abiotic stress [39, 40]. ROS can modulate root elongation by loosening cell walls under favorable conditions [22, 41]. ABA causes oxidative damage and ROS production, so we evaluated the ROS levels in seeds and seedlings of g6pd5 and G6PD5-overexpressing (OE) lines treated with ABA. The H2O2 content in both seeds and seedlings of the g6pd5 mutant and WT increased in response to ABA treatment (Fig. 3a, Additional file 1: Figure S1, Additional file 1: Figure S2a). It is noteworthy that the O2.- content was significantly enhanced in the g6pd5 mutant but attenuated in OE lines (Fig. 3b, Additional file 1: Figure S2b). To further dissect the role of G6PD5 involvement in ROS signaling, exogenous H2O2 was supplied to the medium. The g6pd5 mutant showed increased sensitivity to oxidative stress, as manifested by delayed germination and root elongation relative to WT (data not shown). These results suggest that the oxidative level in g6pd5 is higher than that in WT.
Fig. 3.
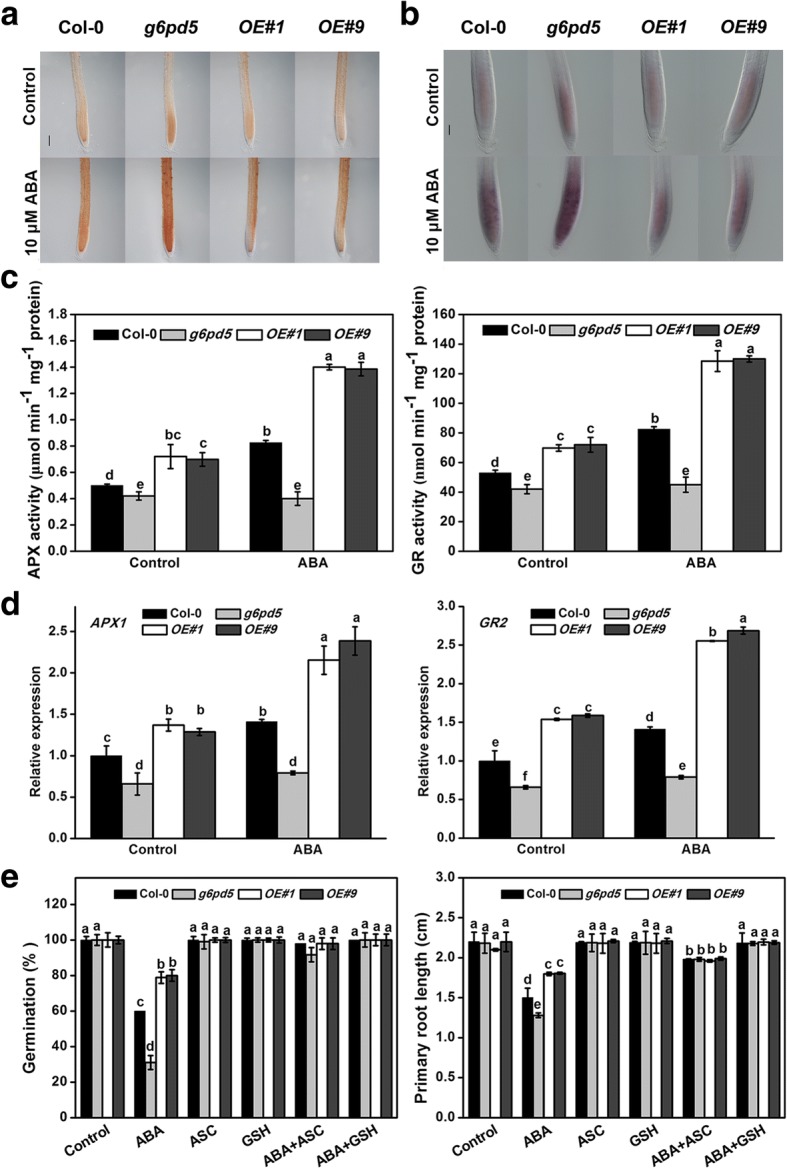
G6PD5 affects the ROS levels with ABA treatment and the activities and transcript levels of antioxidant enzymes in Arabidopsis seedlings. 5-day-old seedlings were grown vertically on 1/2 MS agar plates supplemented with the 10 μM ABA for 6 h. a Staining of roots with DAB (brown color displaying H2O2) after ABA treatment. Bar = 100 μm. b Staining of roots with NBT (blue color displaying O2.-) after ABA treatment. Bar = 50 μm. c and d The activities of antioxidant enzymes and transcript levels of antioxidant enzyme responsive genes. The transcript levels were normalized to Actin2. e The seeds and seedlings are incubated with 0.25 μM ASC or 5 μM GSH
G6PD5 influences NADPH oxidases AtrbohD and AtrbohF
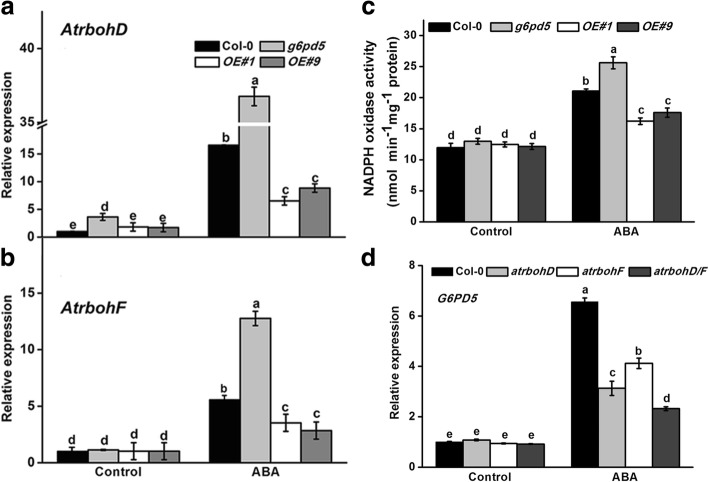
In Arabidopsis, ROS could be generated by peroxidases, amino oxidases and oxygen photoreduction. However, the main source of ROS production is from the NADPH oxidases AtrbohD and AtrbohF, which play an important role in stress-inhibited primary root growth [22]. To determine whether the function of G6PD5 in response to ABA is achieved through the NADPH oxidase signaling pathway, we analyzed the expression of NADPH oxidase genes in WT and g6pd5 with or without ABA treatment. As shown in Fig. 4, the expression of the NADPH oxidases AtrbohD and AtrbohF was markedly increased by ABA treatment in all materials, especially in the g6pd5 mutant (Fig. 4a, b). These results suggest that G6PD5 influences the expression of NADPH oxidases. Expectedly, the activity of the NADPH oxidase was higher in g6pd5 than in WT under ABA treatment (Fig. 4c). These results suggest that cy-G6PD is involved in RBOH-dependent ROS production in ABA-treated seedlings. To prove the hypothesis, we used the NADPH oxidase single mutants, atrbohD1 (CS9555) and atrbohF1 (CS9557), and the double mutant atrbohD1/F1. In these mutants, the expression of G6PD5 was lower than that in WT plants (Fig. 4d).
Fig. 4.
Response of G6PD5 to ABA through NADPH oxidase signaling pathway. a and b Relative transcript levels of NADPH oxidase genes AtrbohD and AtrbohF in Arabidopsis seedlings with or without 20 μM ABA treatment. c Activity of the NADPH oxidase in WT and mutants exposed to ABA treatment. d Relative transcript levels of G6PD5 in WT (Col-0) and NADPH oxidase mutant seeds (atrbohD1, atrbohF1, atrbohD1/F1) with or without 20 μM ABA treatment. The transcript levels were normalized to Actin2. Results are averages ± SE (n = 3). All experiments were repeated at least three times with similar results
G6PD5 enhances the expression of antioxidant enzymes
Antioxidant enzymes respond to stresses to remove extra ROS to maintain the equilibrium between ROS production and scavenging [19]. To investigate the effects of G6PD5 on the transcript and activity of antioxidant enzymes, we determined the activities and expression levels of antioxidant enzymes, including APX and GR. Results showed that the ABA-induced activity and expression levels of APX and GR (Fig. 3c) in the g6pd5 mutant were significantly lower than that in WT plants. In contrast, the expression levels of APX and GR in OE lines were higher than that in WT (Fig. 3d). These results suggest that G6PD5 enhances the capacity of plants to scavenge excessive ROS under ABA treatment to maintain the balance between ROS production and scavenging. The enhanced G6PD5 activity provides more NADPH for the antioxidant system to remove excess ROS.
Further analysis showed that exogenous application of ascorbic acid (ASC) or glutathione (GSH) partially or fully rescued the seed germination and root growth defects in the g6pd5 mutant (Fig. 3e). It is noteworthy that GSH was more effective than ASC (Fig. 3f). Therefore, G6PD5 participates in the reduction of H2O2 to H2O in both the glutathione peroxidase cycle and the ascorbate-glutathione cycle.
G6PD5 affects genes in ABA biosynthesis and catabolism
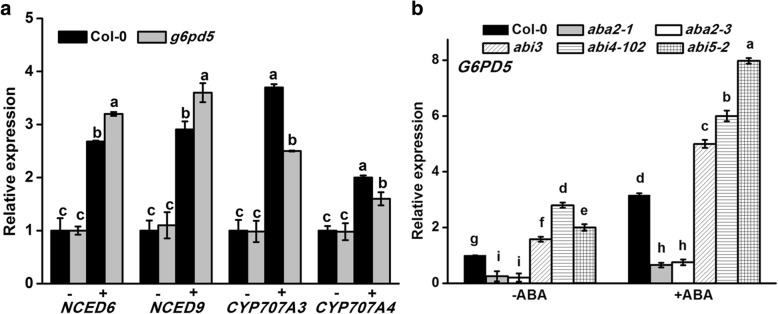
As a stress phytohormone, ABA has a pivotal regulatory function in numerous stress responses [19, 42]. To determine the function of G6PD5 in ABA metabolism, we analyzed the expression of genes encoding enzymes in ABA biosynthesis and catabolism in WT and g6pd5 plants. qRT-PCR results showed that ABA biosynthesis genes (NCED6 and NCED9) had higher expression levels in the g6pd5 mutant than in WT under ABA treatment (Fig. 6a). In contrast, the catabolic genes CYP707A3 and CYP707A4 were expressed at an obviously lower level in the g6pd5 mutant than in WT (Fig. 6a). This result indicates that G6PD5 is involved in the expression regulation of genes in ABA metabolism.
Fig. 6.
Gene expression. a Expression of ABA-biosynthesis genes NCED6 and NCED9 and catabolic genes CYP707A3 and CYP707A4 in WT and g6pd5 was analyzed before or after ABA treatment. b The expression of G6PD5 in ABA mutants. Relative transcript levels of G6PD5 in wild-type (Col-0) and ABA mutant seeds (aba2–1, aba2–3, abi4–102) with or without ABA. In all experiments, the expression of Actin2 was used as the control. Three replicates were made for each treatment with similar results. Values are mean ± SE of three different experiments. Means denoted by the same letter do not significantly differ at P < 0.05 according to Duncan’s multiple range test
G6PD5 suppresses the ABA-signaling pathway
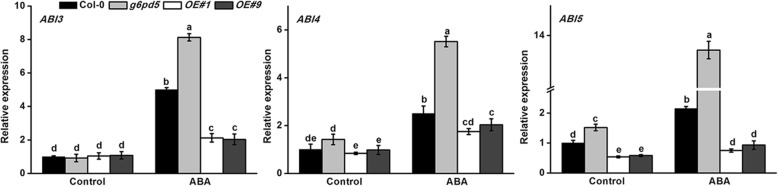
ABA signal transduction genes are involved in the regulation of seed germination and root growth [43]. The germination defects in g6pd5 seeds strongly pointed to the involvement of ABA. We investigated whether differences in the expression of ABA signaling-related genes account for the hypersensitivity of g6pd5 to ABA (Fig. 5). We examined the expression of ABA signaling genes ABI3, ABI4, and ABI5. qRT-PCR results showed that significant differences in the expression of these ABA-regulated genes were observed between WT and g6pd5 mutant plants (Fig. 5). The transcript level of ABI5 was significantly induced in the mutant plant (an increase of 6 fold relative to WT), indicating that G6PD5 probably acts upstream of these genes in the ABA signaling pathway. We also examined the expression level of G6PD5 in ABA-deficient mutants (aba2–1, aba2–3) and ABA-responsive mutant (abi3, abi4–102, abi5–2). Results showed that the G6PD5 expression level in ABA-deficient mutants (aba2–1, aba2–3) was markedly lower than that in WT, whereas in ABA-responsive mutants the expression level of G6PD5 was significantly higher than that in WT (Fig. 6b).
Fig. 5.
G6PD5 attenuates the expression of several ABA responsive genes. Relative expression levels of ABA-signaling genes ABI3, ABI4, ABI5 in plants. Two-week-old seedlings were incubated in liquid MS medium with or without 10 μM ABA for 12 h. The transcript levels were determined by qRT-PCR analysis. The transcript levels were normalized to Actin2. Results are averages ± SE (n = 3). All experiments were repeated at least three times with similar results
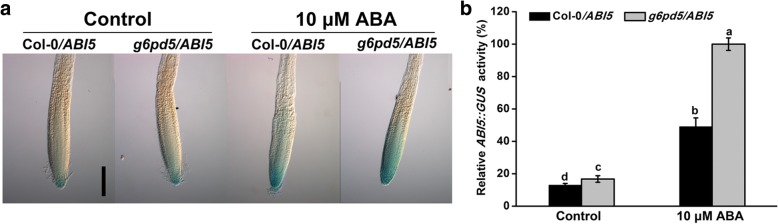
To confirm the ABI5 expression level in the g6pd5 mutant, we crossed the g6pd5 mutant with Col-0 plants harboring ABI5::GUS (Col/ABI5 plants). We obtained homozygous g6pd5 plants harboring ABI5::GUS (g6pd5/ABI5 plants) and compared the ABI5 expression in Col/ABI5 and g6pd5/ABI5 seedlings (Fig. 7). Five-day-old g6pd5/ABI5 seedlings showed a strong blue coloration in the QC of root tips relative to Col/ABI5 with or without ABA treatment (Fig. 7). These results confirmed a higher ABI5 expression level in g6pd5 seedlings, suggesting that G6PD5 is likely involved in the regulation of ABA signaling.
Fig. 7.
G6PD5 knockout on the expression of ABI5 genes in Arabidopsis. a Expression of ABI5::GUS in Col-0/ABI5 and g6pd5/ABI5 seedling roots grown for 7 d then treated with or without 20 μM ABA for 12 h using GUS staining. b Quantification of the GUS activity in Col-0/ABI5 and g6pd5/ABI5 seedlings. The GUS activity of Col-0/ABI5 root was adjusted to 100%. Mean values and SE were calculated from three independent experiments (n = 20). Within each set of experiments, bars with different letters were significantly different at the p < 0.05 level
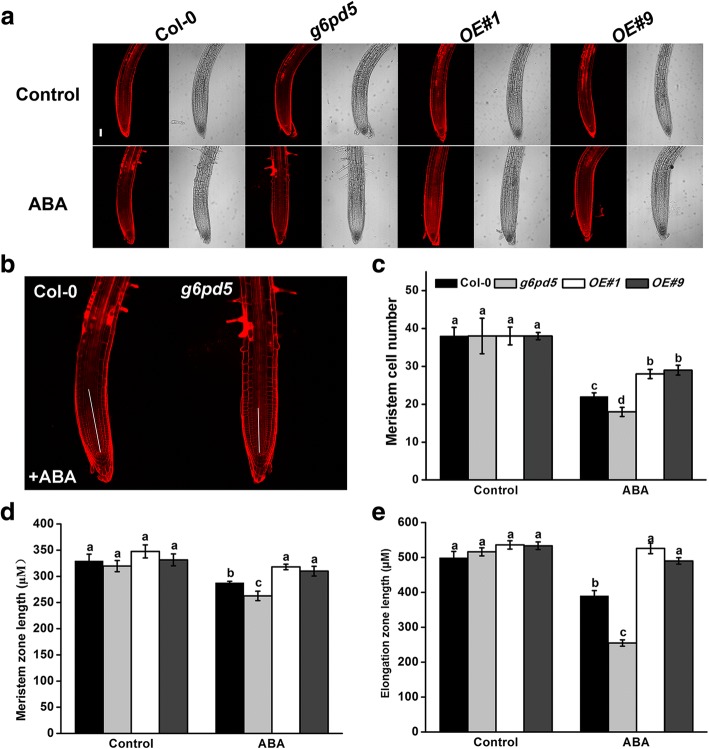
G6PD5 is involved in ABA-inhibited cell elongation in root meristem and elongation zones
To further dissect the mechanisms of G6PD5 function in ABA-repressed root growth in Arabidopsis, we measured the primary root length in WT and g6pd5 mutants grown on 1/2 MS media supplied with ABA. The root growth of WT and mutants was similar on ABA-free medium (Fig. 8a). However, when grown on ABA-containing medium for 12 h, g6pd5 mutant plants exhibited dramatically shortened root elongation zone compared to WT (Fig. 8a, e). These results indicate that ABA suppresses cell elongation in the elongation zone of g6pd5 roots. In addition to cell elongation in the elongation zone, cell division in the root meristem zone also contributes to root growth. Therefore, we examined the size of root apical meristem. The length and cell number of the meristem zone in g6pd mutant plants were less than that in WT in the presence of ABA (Fig. 8a-d), implying that G6PD5 is required for cell division in the root meristem.
Fig. 8.
G6PD5 regulates root meristem and elongation zone. a and b Root meristems of propidium iodide (PI)-stained images in Arabidopsis WT seedlings. Bars = 100 μm. c Meristem cell number. d Root meristem zone size. e Root elongation zone size. The 5-day-old seedlings were treated with 10 μM ABA for 12 h. Except in a and b, mean values and SE were calculated from three independent experiments (n = 20). Within each set of experiments, bars with different letters were significantly different at the p < 0.05 level
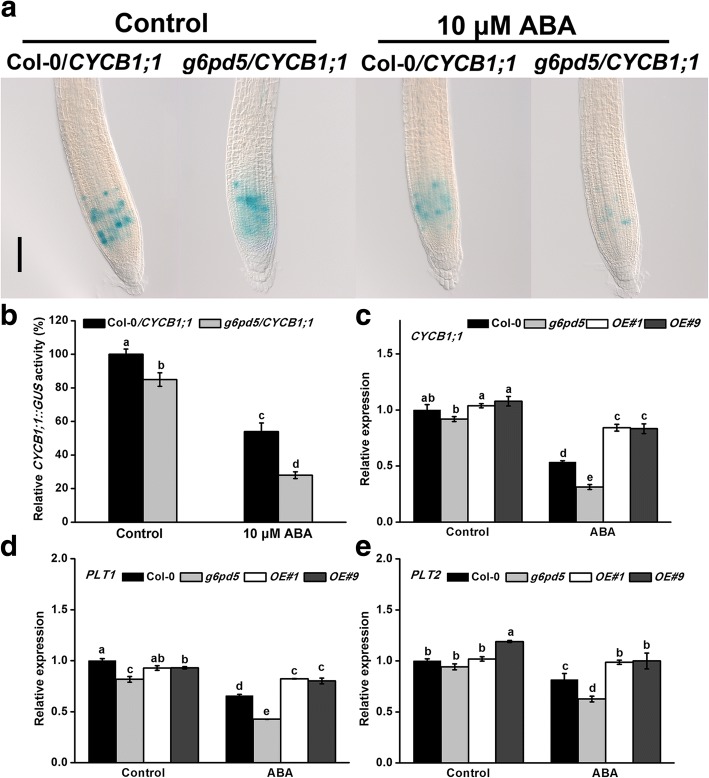
Furthermore, we found that the cell cycle B-type cyclin CYCB1 accounts for the reduction in primary root growth in g6pd5 mutants under ABA treatment (Fig. 9). To further investigate the protective mechanism of G6PD5 under ABA condition, we first examined the expression of CYCB1 gene in the g6pd5 mutant and WT. Results showed that the expression of CYCB1 was decreased under ABA treatment in seedlings, especially in g6pd5 mutant (Fig. 9c). Next, we used the cell cycle marker line CYCB1;1::GUS and observed that homozygous g6pd5 plants harboring CYCB1;1::GUS (g6pd5/CYCB1;1 plants) had weaker blue coloration than Col-0/CYCB1;1 in the root meristem under ABA treatment (Fig. 9a, b). Based on the above results, we concluded that G6PD5 is involved in the transcriptional regulation of the CYCB1 gene, and in the process, G6PD5 plays a key role by participating in CYCB accumulation under ABA treatment in primary roots.
Fig. 9.
G6PD5 is involved in cell division in the absence or presence of ABA. a Expression of CYCB1;1::GUS in Col-0/CYCB1;1 and g6pd5/CYCB1;1 seedlings grown for 5 d with or without ABA treatment for 12 h. Scale bars, 200 μm. b Quantification of the GUS activity in CYCB1;1::GUS seedlings. The GUS activity in Col-0/CYCB1;1 roots was adjusted to 100%. c-e Relative transcript levels of CYCB1, PLT1 and PLT2 in WT (Col-0), g6pd5 mutant and OE lines plants with or without ABA treatment. Data are presented as mean values ± SD of three independent experiments. One-way Duncan’s test was performed, and statistically significant differences are indicated by different lower case letters (P < 0.05). The experiments were repeated at least three times with similar results, and data from one representative experiment are presented
The activity of the root meristem is regulated by root meristem-specific genes, among which the PLETHORA (PLTs) genes are master regulators of root development [44]. PLT1 and PLT2 are predominantly localized in the QC and redundantly regulate the root stem cell niche in root apical meristem (RAM) [44]. We investigate the expression of RAM-related genes in WT and g6pd5. Results showed that the expression levels of PLT1 and PLT2 were lowered in the g6pd5 mutant compared to WT with ABA treatment (Fig. 9d, e). These findings indicate that G6PD5 is crucial for RAM maintenance in the presence of ABA.
Discussion
To date, several studies have reported the expression patterns of G6PDs during plant development and in response to stresses [11, 37]. Cy-G6PD acts as the main isoforms in the regulatory step of OPPP and is involved in regulating redox balance in cells during salt tolerance [11, 37]. Therefore, Cy-G6PD plays positive roles in enhancing plant tolerance to environmental stresses in several species [23, 45]. In this study, we investigated the involvement of G6PD5 in the response of Arabidopsis to ABA during seed germination and root development. We showed that G6PD5 is a central component in seed germination and seedling development under ABA treatment. Seed germination rate of the g6pd5 mutant under ABA treatment was reduced by approximately 50% compared to WT, implying that G6PD5 has critical functions in plant development and ABA responses (Fig. 2a, b). In contrast, g6pd6 mutant seeds exhibited inconspicuous germination compared to WT under ABA treatment (Fig. 1a-c). These results suggest that G6PD5 plays a more important role than G6PD6 in Arabidopsis seed germination. The g6pd5 mutant exhibited severely reduced seed germination rate and shortened primary roots with increased ABA concentrations compared to WT. Why is G6PD5 responsive to ABA treatment? We found ABREs in the promoter region of G6PD5 but not in G6PD6 (Additional file 1: Table S2).
Under favorable conditions, ROS, such as H2O2, superoxide anion and hydroxyl radical, are produced at low concentrations in plasma membrane, chloroplasts, mitochondria and peroxisomes [46]. ROS are not only the ever-present danger due to their physicochemical toxicity, but also important signaling molecules that accumulate under many stress conditions [11, 19]. Previous reports showed that H2O2 plays a role in drought-induced increase of total G6PD activity [11]. Thus, we examined the effect of H2O2 on G6PD5 under ABA treatment. We confirmed that the oxidative level is higher in g6pd5 than in WT. ROS originated from the NADPH oxidases AtrbohD and AtrbohF play an important role in stress-inhibited primary root growth in Arabidopsis [22]. Further investigation demonstrated that the ABA-induced H2O2 generation results from the enhanced NADPH oxidase activity. To consolidate this observation, we examined the antioxidant enzyme activities and transcription levels in g6pd5 and WT plants (Fig. 3c, d). These results suggest that G6PD5 is enhanced to scavenge the excessive ROS under ABA treatment in order to maintain the balance of ROS production and scavenging. Results also indicate that the enhanced G6PD5 activity provides more NADPH for the antioxidant system to remove excessive ROS. Reduction of H2O2 to H2O can then be achieved through either the glutathione peroxidase cycle or the ascorbate-glutathione cycle.
ABA plays a central role in plant development and adaptation to numerous stress responses [19, 47, 48]. ABI3 has long been recognized as the major regulator of seed dormancy and ABA inhibition of seed germination [39, 49]. ABI5 functions downstream of ABI3 and promotes dormancy in concert with ABI3 [39, 43, 50]. Furthermore, ABI5 plays a role in regulating ROS homeostasis by activating CATALASE 1 transcription during seed germination [28]. CATALASE 1 is a key positive regulator of ABA signaling involved in mediating seed germination and subsequent primary root establishment [51]. Mechanistic investigations revealed other important modulators that interact with ABI5 to fine-tune seed germination [51]. ABI4 is involved in redox regulation and oxidative challenges in Arabidopsis leaves [39, 43]. Interestingly, the transcript levels of ABI3, ABI4, and ABI5 were increased in the g6pd5 mutant (Fig. 5). This indicates that G6PD5 probably acts upstream of these genes in the ABA signaling pathway. The g6pd5 mutant showed less sensitivity to ABA compared with WT. Taken together, our results suggest that the germination defects in the g6pd5 mutant is mediated by ABI genes, especially by ABI5.
In addition, ABA inhibition of primary root growth is mediated by the ABA-induced regulation of CYCB1 expression at the G2/M checkpoint [22, 52]. One mechanism in plant response to primary root growth is to regulate the expression of cell cycle-related genes. We found that the expression of CYCB1 was decreased under ABA treatment in g6pd5 mutant seedlings (Fig. 9). Therefore, we propose that G6PD5 is involved in the transcriptional regulation of CYCB1 gene under ABA treatment. Certainly, G6PD5 is crucial for RAM maintenance in the presence of ABA by regulating the expression levels of PLT1 and PLT2 (Fig. 9d, e).
Conclusions
Our results showed that ABA, H2O2, APX, GR and ABI are required for ABA-induced G6PD5 gene function. Our findings point to a different node of this crosstalk that is activated by an increase in cytosolic G6PD5 and that is involved in dormancy and germination control. Based on these results, as well as those reported previously, we proposed a hypothetical model shown in Fig. 10. In this model, ABA induces G6PD5, which subsequently suppresses H2O2 generation by activating the NADPH oxidase in seeds and roots under ABA treatment. The enhanced G6PD5 is involved in regulating key enzymes (APX and GR) in the ASC-GSH cycle by utilizing NADPH. The enhanced antioxidant ability can facilitate to maintain a steady-state level of H2O2 in cells, thus avoiding ROS damages to plant cells. G6PD5 is crucial for RAM maintenance in the presence of ABA by regulating the expression levels of CYCB1, PLT1 and PLT2. Moreover, G6PD5 affects genes in ABA biosynthesis and catabolism after ABA treatment and G6PD5 probably acts upstream of ABI genes in the ABA signaling pathway.
Fig. 10.
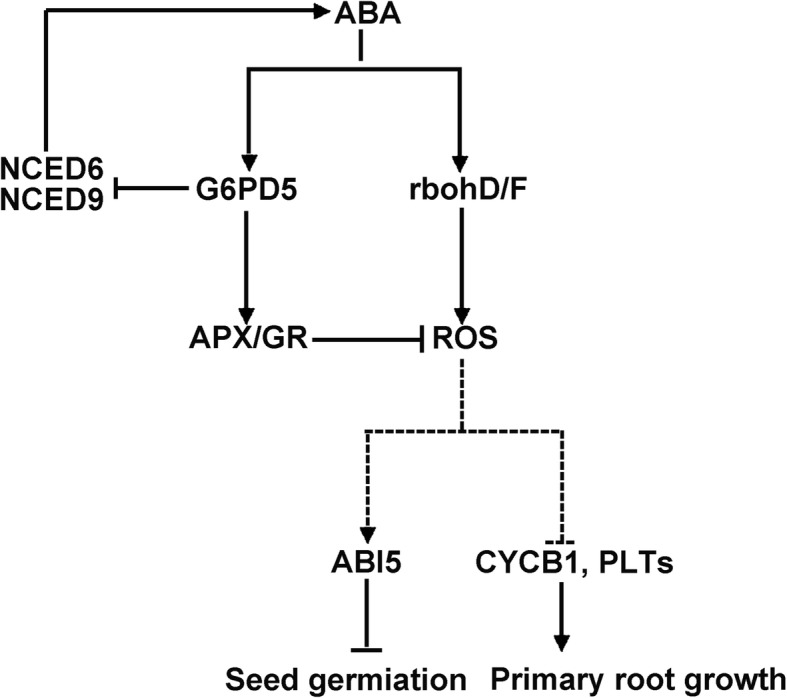
Schematic illustration of a proposed model during Arabidopsis seed germination and root growth. In this model, arrows indicate positive regulation and bars indicate negative regulation. Dotted arrows indicate results from the literature
Additional file
Table S1. Primer sequences used in the study. Figure S1. ROS levels in g6pd5 and OE lines. 1-day-old seeds were grown vertically on 1/2 MS agar plates supplemented with 10 μM ABA for 6 h. Quantification of the H2DCF-DA fluorescence in Arabidopsis seeds with ABA treatment. Figure S2. H2O2 and O2− levels in g6pd5 and OE lines. 5-day-old seedlings were grown vertically on 1/2 MS agar plates supplemented with the 10 μM ABA for 6 h. Table S2. Cis-acting regulatory elements identified in the promoter region of G6PD5 and G6PD6. The online search tool PlantCARE was used to detect putative cis-acting regulatory elements. (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/). (DOC 610 kb)
Acknowledgements
None.
Funding
This work was supported by the National Natural Science Foundation of China (31671595; 31670244), the Agricultural Biotechnology Research and Application Development Program of Gansu Province (GNSW-2016-23), the Fundamental Research Funds for the Central Universities (lzujbky-2017-kb05), the Foundation of Science and Technology Program of Lanzhou City (2015-3-53), the Project of Qinghai Science & Technology Department (2016-ZJ-Y01), and the Open Project of State Key Laboratory of Plateau Ecology and Agriculture, Qinghai University (201 -KF-05). The funding bodies did not play any role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Availability of data and materials
The datasets generated and analyzed during the current study are available from the corresponding author upon reasonable request.
Abbreviations
- ABA
Abscisic acid
- ABI
Abscisic acid insensitive
- APX
Ascorbate peroxidase
- AsA
Ascorbate acid
- Cy-G6PD
Cytosolic glucose-6-phosphate dehydrogenase
- CYP707A
Aabscisic acid 8′-hydroxylase
- DAB
3, 3′-diaminobenzidine
- G6PD
Glucose-6-phosphate dehydrogenase
- GR
Glutathione reductase
- GSH
Glutathione
- H2DCF-DA
Dichlorodihydrofluorescein diacetate
- H2O2
Hydrogen peroxide
- NBT
Nitroblue tetrazolium
- NCED
9-cis-epoxycarotenoid dioxygenase
- PPP
Pentose phosphate pathway
- ROS
Reactive oxygen species
Authors’ contributions
Conceived and designed the experiments: LY, YB, XW. Performed the experiments: LY, SW, MR, LS, SL, RH, WZ and CL Analyzed the data: LY Wrote the paper: LY. All authors reviewed and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Lei Yang, Email: yangleisllyyn@163.com.
Shengwang Wang, Email: wangshw16@lzu.edu.cn.
Lili Sun, Email: sunlilimeng@163.com.
Mengjiao Ruan, Email: ruanmj16@lzu.edu.cn.
Sufang Li, Email: lisf17@lzu.edu.cn.
Rui He, Email: her2016@lzu.edu.cn.
Wenya Zhang, Email: zhangwy16@lzu.edu.cn.
Cuifang Liang, Email: 15002517278@163.com.
Xiaomin Wang, Email: wangxiaomin@lzu.edu.cn.
Yurong Bi, Phone: +86-931-8911781, Email: yrbi@lzu.edu.cn.
References
- 1.Bowsher C, Hucklesby D, Emes M. Nitrite reduction and carbohydrate metabolism in plastids purified from roots of Pisum sativum. Planta. 1989;177:359–366. doi: 10.1007/BF00403594. [DOI] [PubMed] [Google Scholar]
- 2.Cardi M, Chibani K, Cafasso D, Rouhier N, Jacquot JP, Esposito S. Abscisic acid effects on activity and expression of barley (Hordeum vulgare) plastidial glucose-6-phosphate dehydrogenase. J Exp Bot. 2011;62(11):4013–4023. doi: 10.1093/jxb/err100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Esposito S, Massaro G, Vona V, Di Martino Rigano V, Carfagna S. Glutamate synthesis in barley roots: the role of the plastidic glucose-6-phosphate dehydrogenase. Planta. 2003;216(4):639–647. doi: 10.1007/s00425-002-0892-4. [DOI] [PubMed] [Google Scholar]
- 4.Hutchings D, Rawsthorne S, Emes MJ. Fatty acid synthesis and the oxidative pentose phosphate pathway in developing embryos of oilseed rape (Brassica napus L.) J Exp Bot. 2005;56(412):577–585. doi: 10.1093/jxb/eri046. [DOI] [PubMed] [Google Scholar]
- 5.Kruger NJ, von Schaewen A. The oxidative pentose phosphate pathway: structure and organisation. Curr Opin Plant Biol. 2003;6(3):236–246. doi: 10.1016/S1369-5266(03)00039-6. [DOI] [PubMed] [Google Scholar]
- 6.Scheibe R. Malate valves to balance cellular energy supply. Physiol Plant. 2004;120:21–26. doi: 10.1111/j.0031-9317.2004.0222.x. [DOI] [PubMed] [Google Scholar]
- 7.Cardi M, Castiglia D, Ferrara M, Guerriero G, Chiurazzi M, Esposito S. The effects of salt stress cause a diversion of basal metabolism in barley roots: possible different roles for glucose-6-phosphate dehydrogenase isoforms. Plant Physiol Bioch. 2015;86:44–54. doi: 10.1016/j.plaphy.2014.11.001. [DOI] [PubMed] [Google Scholar]
- 8.Liu J, Wang X, Hu Y, Hu W, Bi Y. Glucose-6-phosphate dehydrogenase plays a pivotal role in tolerance to drought stress in soybean roots. Plant Cell Rep. 2013;32(3):415–429. doi: 10.1007/s00299-012-1374-1. [DOI] [PubMed] [Google Scholar]
- 9.Liu X, Wei W, Zhu W, Su L, Xiong Z, Zhou M, Zheng Y, Zhou DX. Histone deacetylase AtSRT1 regulates metabolic flux and stress response in Arabidopsis. Mol Plant. 2017;10(12):1510–1522. doi: 10.1016/j.molp.2017.10.010. [DOI] [PubMed] [Google Scholar]
- 10.Meyer T, Holscher C, Schwoppe C, von Schaewen A. Alternative targeting of Arabidopsis plastidic glucose-6-phosphate dehydrogenase G6PD1 involves cysteine-dependent interaction with G6PD4 in the cytosol. Plant J. 2011;66(5):745–758. doi: 10.1111/j.1365-313X.2011.04535.x. [DOI] [PubMed] [Google Scholar]
- 11.Wang H, Yang L, Li Y, Hou J, Huang J, Liang W. Involvement of ABA- and H2O2-dependent cytosolic glucose-6-phosphate dehydrogenase in maintaining redox homeostasis in soybean roots under drought stress. Plant Physiol Bioch. 2016;107:126–136. doi: 10.1016/j.plaphy.2016.05.040. [DOI] [PubMed] [Google Scholar]
- 12.Dal Santo S, Stampfl H, Krasensky J, Kempa S, Gibon Y, Petutschnig E, Rozhon W, Heuck A, Clausen T, Jonak C. Stress-induced GSK3 regulates the redox stress response by phosphorylating glucose-6-phosphate dehydrogenase in Arabidopsis. Plant Cell. 2012;24(8):3380–3392. doi: 10.1105/tpc.112.101279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wakao S, Benning C. Genome-wide analysis of glucose-6-phosphate dehydrogenases in Arabidopsis. Plant J. 2005;41(2):243–256. doi: 10.1111/j.1365-313X.2004.02293.x. [DOI] [PubMed] [Google Scholar]
- 14.De Lillo A, Cardi M, Landi S, Esposito S. Mechanism(s) of action of heavy metals to investigate the regulation of plastidic glucose-6-phosphate dehydrogenase. Sci Rep. 2018;8(1):13481. doi: 10.1038/s41598-018-31348-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang H, Hou J, Li Y, Zhang Y, Huang J, Liang W. Nitric oxide-mediated cytosolic glucose-6-phosphate dehydrogenase is involved in aluminum toxicity of soybean under high aluminum concentration. Plant Soil. 2017;416(1–2):39–52. doi: 10.1007/s11104-017-3197-x. [DOI] [Google Scholar]
- 16.Stampfl H, Fritz M, Dal Santo S, Jonak C. The GSK3/shaggy-like kinase ASKa contributes to pattern-triggered immunity. Plant Physiol. 2016;171(2):1366–1377. doi: 10.1104/pp.15.01741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hasegawa P, Bressan R, Zhu J, Bohnert H. Plant cellular and molecular responses to high salinity. Annu Rev Plant Physiol Plant Mol Biol. 2000;51:463–499. doi: 10.1146/annurev.arplant.51.1.463. [DOI] [PubMed] [Google Scholar]
- 18.Kwak JM, Nguyen V, Schroeder JI. The role of reactive oxygen species in hormonal responses. Plant Physiol. 2006;141(2):323–329. doi: 10.1104/pp.106.079004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu R, Liu Y, Ye N, Zhu G, Chen M, Jia L, Xia Y, Shi L, Jia W, Zhang J. AtDsPTP1 acts as a negative regulator in osmotic stress signalling during Arabidopsis seed germination and seedling establishment. J Exp Bot. 2015;66(5):1339–1353. doi: 10.1093/jxb/eru484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Apel K, Hirt H. Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu Rev Plant Biol. 2004;55:373–399. doi: 10.1146/annurev.arplant.55.031903.141701. [DOI] [PubMed] [Google Scholar]
- 21.Ma L, Zhang H, Sun L, Jiao Y, Zhang G, Miao C, Hao F. NADPH oxidase AtrbohD and AtrbohF function in ROS-dependent regulation of Na+/K+ homeostasis in Arabidopsis under salt stress. J Exp Bot. 2012;63(1):305–317. doi: 10.1093/jxb/err280. [DOI] [PubMed] [Google Scholar]
- 22.Jiao Y, Sun L, Song Y, Wang L, Liu L, Zhang L, Liu B, Li N, Miao C, Hao F. AtrbohD and AtrbohF positively regulate abscisic acid-inhibited primary root growth by affecting Ca2+ signalling and auxin response of roots in Arabidopsis. J Exp Bot. 2013;64(14):4183–4192. doi: 10.1093/jxb/ert228. [DOI] [PubMed] [Google Scholar]
- 23.Landi S, Nurcato R, De Lillo A, Lentini M, Grillo S, Esposito S. Glucose-6-phosphate dehydrogenase plays a central role in the response of tomato (Solanum lycopersicum) plants to short and long-term drought. Plant Physiol Bioch. 2016;105:79–89. doi: 10.1016/j.plaphy.2016.04.013. [DOI] [PubMed] [Google Scholar]
- 24.Gill SS, Tuteja N. Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol Bioch. 2010;48(12):909–930. doi: 10.1016/j.plaphy.2010.08.016. [DOI] [PubMed] [Google Scholar]
- 25.Fujita Y, Fujita M, Shinozaki K, Yamaguchi-Shinozaki K. ABA-mediated transcriptional regulation in response to osmotic stress in plants. J Plant Res. 2011;124(4):509–525. doi: 10.1007/s10265-011-0412-3. [DOI] [PubMed] [Google Scholar]
- 26.Hattori T, Totsuka M, Hobo T, Kagaya Y, Yamamoto-Toyoda A. Experimentally determined sequence requirement of ACGT-containing abscisic acid response element. Plant Cell Physiol. 2002;43(1):136–140. doi: 10.1093/pcp/pcf014. [DOI] [PubMed] [Google Scholar]
- 27.Kashiwakura Y, Kobayashi D, Jikumaru Y, Takebayashi Y, Nambara E, Seo M, Kamiya Y, Kushiro T, Kawakami N. Highly sprouting-tolerant wheat grain exhibits extreme dormancy and cold imbibition-resistant accumulation of abscisic acid. Plant Cell Physiol. 2016;57(4):715–732. doi: 10.1093/pcp/pcw051. [DOI] [PubMed] [Google Scholar]
- 28.Bi C, Ma Y, Wu Z, Yu YT, Liang S, Lu K, Wang XF. Arabidopsis ABI5 plays a role in regulating ROS homeostasis by activating CATALASE 1 transcription in seed germination. Plant Mol Biol. 2017;94(1–2):197–213. doi: 10.1007/s11103-017-0603-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shi H, Chen Y, Qian Y, Chan Z. Low temperature-induced 30 (LTI30) positively regulates drought stress resistance in Arabidopsis: effect on abscisic acid sensitivity and hydrogen peroxide accumulation. Front Plant Sci. 2015;6:893. doi: 10.3389/fpls.2015.00893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang T, Zhang L, Hao H, Zhang P, Zhu H, Cheng W, Wang Y, Wang X, Wang C. Nuclear-localized AtHSPR links abscisic acid-dependent salt tolerance and antioxidant defense in Arabidopsis. Plant J. 2015;84(6):1274–1294. doi: 10.1111/tpj.13080. [DOI] [PubMed] [Google Scholar]
- 31.Zhao ZG, Chen GC, Zhang CL. Interaction between reactive oxygen species and nitric oxide in drought-induced abscisic acid synthesis in root tips of wheat seedlings. Aust J Plant Physiol. 2001;28(10):1055–1061. [Google Scholar]
- 32.Xia Z, Huo Y, Wei Y, Chen Q, Xu Z, Zhang W. The Arabidopsis LYST INTERACTING PROTEIN 5 acts in regulating abscisic acid signaling and drought response. Front Plant Sci. 2016;7:758. doi: 10.3389/fpls.2016.00758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nan W, Wang X, Yang L, Hu Y, Wei Y, Liang X, Mao L, Bi Y. Cyclic GMP is involved in auxin signalling during Arabidopsis root growth and development. J Exp Bot. 2014;65(6):1571–1583. doi: 10.1093/jxb/eru019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rosado A, Amaya I, Valpuesta V, Cuartero J, Botella MA, Borsani O. ABA- and ethylene-mediated responses in osmotically stressed tomato are regulated by the TSS2 and TOS1 loci. J Exp Bot. 2006;57(12):3327–3335. doi: 10.1093/jxb/erl094. [DOI] [PubMed] [Google Scholar]
- 35.Mei Y, Jia WJ, Chu YJ, Xue HW. Arabidopsis phosphatidylinositol monophosphate 5-kinase 2 is involved in root gravitropism through regulation of polar auxin transport by affecting the cycling of PIN proteins. Cell Res. 2012;22(3):581–597. doi: 10.1038/cr.2011.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu Y, Wu R, Wan Q, Xie G, Bi Y. Glucose-6-phosphate dehydrogenase plays a pivotal role in nitric oxide-involved defense against oxidative stress under salt stress in red kidney bean roots. Plant Cell Physiol. 2007;48(3):511–522. doi: 10.1093/pcp/pcm020. [DOI] [PubMed] [Google Scholar]
- 37.Wang X, Ma Y, Huang C, Wan Q, Li N, Bi Y. Glucose-6-phosphate dehydrogenase plays a central role in modulating reduced glutathione levels in reed callus under salt stress. Planta. 2008;227(3):611–623. doi: 10.1007/s00425-007-0643-7. [DOI] [PubMed] [Google Scholar]
- 38.Pelagio-Flores R, Ortiz-Castro R, Mendez-Bravo A, Macias-Rodriguez L, Lopez-Bucio J. Serotonin, a tryptophan-derived signal conserved in plants and animals, regulates root system architecture probably acting as a natural auxin inhibitor in Arabidopsis thaliana. Plant Cell Physiol. 2011;52(3):490–508. doi: 10.1093/pcp/pcr006. [DOI] [PubMed] [Google Scholar]
- 39.Chen C, Letnik I, Hacham Y, Dobrev P, Ben-Daniel BH, Vankova R, Amir R, Miller G. ASCORBATE PEROXIDASE6 protects Arabidopsis desiccating and germinating seeds from stress and mediates cross talk between reactive oxygen species, abscisic acid, and auxin. Plant Physiol. 2014;166(1):370–383. doi: 10.1104/pp.114.245324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sarath G, Hou G, Baird LM, Mitchell RB. Reactive oxygen species, ABA and nitric oxide interactions on the germination of warm-season C4-grasses. Planta. 2007;226(3):697–708. doi: 10.1007/s00425-007-0517-z. [DOI] [PubMed] [Google Scholar]
- 41.Liszkay A, van der Zalm E, Schopfer P. Production of reactive oxygen intermediates O2.-, H2O2, and .OH by maize roots and their role in wall loosening and elongation growth. Plant Physiol. 2004;136(2):3114–3123. doi: 10.1104/pp.104.044784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cutler SR, Rodriguez PL, Finkelstein RR, Abrams SR. Abscisic acid: emergence of a core signaling network. Annu Rev Plant Biol. 2010;61:651–679. doi: 10.1146/annurev-arplant-042809-112122. [DOI] [PubMed] [Google Scholar]
- 43.Lopez-Molina L, Mongrand S, McLachlin DT, Chait BT, Chua NH. ABI5 acts downstream of ABI3 to execute an ABA-dependent growth arrest during germination. Plant J. 2002;32(3):317–328. doi: 10.1046/j.1365-313X.2002.01430.x. [DOI] [PubMed] [Google Scholar]
- 44.Galinha C, Hofhuis H, Luijten M, Willemsen V, Blilou I, Heidstra R, Scheres B. PLETHORA proteins as dose-dependent master regulators of Arabidopsis root development. Nature. 2007;449(7165):1053–1057. doi: 10.1038/nature06206. [DOI] [PubMed] [Google Scholar]
- 45.Scharte J, Schon H, Tjaden Z, Weis E, von Schaewen A. Isoenzyme replacement of glucose-6-phosphate dehydrogenase in the cytosol improves stress tolerance in plants. Proc Natl Acad Sci U S A. 2009;106(19):8061–8066. doi: 10.1073/pnas.0812902106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhou W, Zhou T, Li MX, Zhao CL, Jia N, Wang XX, Sun YZ, Li GL, Xu M, Zhou RG, et al. The Arabidopsis J-protein AtDjB1 facilitates thermotolerance by protecting cells against heat-induced oxidative damage. New Phytol. 2012;194(2):364–378. doi: 10.1111/j.1469-8137.2012.04070.x. [DOI] [PubMed] [Google Scholar]
- 47.Boudsocq M, Lauriere C. Osmotic signaling in plants: multiple pathways mediated by emerging kinase families. Plant Physiol. 2005;138(3):1185–1194. doi: 10.1104/pp.105.061275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhong C, Xu H, Ye S, Wang S, Li L, Zhang S, Wang X. Gibberellic acid-stimulated Arabidopsis6 serves as an integrator of gibberellin, abscisic acid, and glucose signaling during seed germination in Arabidopsis. Plant Physiol. 2015;169(3):2288–2303. doi: 10.1104/pp.15.00858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bentsink L, Koornneef M. Seed dormancy and germination. Arabidopsis Book. 2008;6:e0119. doi: 10.1199/tab.0119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gampala SS, Finkelstein RR, Sun SS, Rock CD. ABI5 interacts with abscisic acid signaling effectors in rice protoplasts. J Biol Chem. 2002;277(3):1689–1694. doi: 10.1074/jbc.M109980200. [DOI] [PubMed] [Google Scholar]
- 51.Pan J, Wang H, Hu Y, Yu D. Arabidopsis VQ18 and VQ26 proteins interact with ABI5 transcription factor to negatively modulate ABA response during seed germination. Plant J. 2018;95(3):529–544. doi: 10.1111/tpj.13969. [DOI] [PubMed] [Google Scholar]
- 52.West G, Inze D, Beemster GT. Cell cycle modulation in the response of the primary root of Arabidopsis to salt stress. Plant Physiol. 2004;135(2):1050–1058. doi: 10.1104/pp.104.040022. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Primer sequences used in the study. Figure S1. ROS levels in g6pd5 and OE lines. 1-day-old seeds were grown vertically on 1/2 MS agar plates supplemented with 10 μM ABA for 6 h. Quantification of the H2DCF-DA fluorescence in Arabidopsis seeds with ABA treatment. Figure S2. H2O2 and O2− levels in g6pd5 and OE lines. 5-day-old seedlings were grown vertically on 1/2 MS agar plates supplemented with the 10 μM ABA for 6 h. Table S2. Cis-acting regulatory elements identified in the promoter region of G6PD5 and G6PD6. The online search tool PlantCARE was used to detect putative cis-acting regulatory elements. (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/). (DOC 610 kb)
Data Availability Statement
The datasets generated and analyzed during the current study are available from the corresponding author upon reasonable request.