Abstract
Background
Pseudomonas syringae is a γ-proteobacterium causing economically relevant diseases in practically all cultivated plants. Most isolates of this pathogen contain native plasmids collectively carrying many pathogenicity and virulence genes. However, P. syringae is generally an opportunistic pathogen primarily inhabiting environmental reservoirs, which could exert a low selective pressure for virulence plasmids. Additionally, these plasmids usually contain a large proportion of repeated sequences, which could compromise plasmid integrity. Therefore, the identification of plasmid stability determinants and mechanisms to preserve virulence genes is essential to understand the evolution of this pathogen and its adaptability to agroecosystems.
Results
The three virulence plasmids of P. syringae pv. savastanoi NCPPB 3335 contain from one to seven functional stability determinants, including three highly active toxin-antitoxin systems (TA) in both pPsv48A and pPsv48C. The TA systems reduced loss frequency of pPsv48A by two orders of magnitude, whereas one of the two replicons of pPsv48C likely confers stable inheritance by itself. Notably, inactivation of the TA systems from pPsv48C exposed the plasmid to high-frequency deletions promoted by mobile genetic elements. Thus, recombination between two copies of MITEPsy2 caused the deletion of an 8.3 kb fragment, with a frequency of 3.8 ± 0.3 × 10− 3. Likewise, one-ended transposition of IS801 generated plasmids containing deletions of variable size, with a frequency of 5.5 ± 2.1 × 10− 4, of which 80% had lost virulence gene idi. These deletion derivatives were stably maintained in the population by replication mediated by repJ, which is adjacent to IS801. IS801 also promoted deletions in plasmid pPsv48A, either by recombination or one-ended transposition. In all cases, functional TA systems contributed significantly to reduce the occurrence of plasmid deletions in vivo.
Conclusions
Virulence plasmids from P. syringae harbour a diverse array of stability determinants with a variable contribution to plasmid persistence. Importantly, we showed that multiple plasmid-borne TA systems have a prominent role in preserving plasmid integrity and ensuring the maintenance of virulence genes in free-living conditions. This strategy is likely widespread amongst native plasmids of P. syringae and other bacteria.
Electronic supplementary material
The online version of this article (10.1186/s13100-019-0149-4) contains supplementary material, which is available to authorized users.
Keywords: Postsegregational killing, Native plasmid evolution, IS801, Replicative transposition, IS91 family, Olive knot disease, Pathogenicity, MITEs, One-ended transposition, Pseudomonas savastanoi
Background
Plasmids are dispensable extrachromosomal elements widely distributed in bacteria, facilitating their survival and the colonization of eukaryotic hosts [1–4]. The plasticity and transmissibility of plasmids contribute to a rapid dissemination of resistance and virulence genes, thus promoting the emergence of uncontrollable bacterial diseases, both in clinical and agricultural settings [5–8]. However, plasmids are usually large and exist in several copies per cell, potentially imposing a significant metabolic burden to the cell, which might facilitate the emergence of plasmid-free derivatives in the absence of selection for plasmid-borne characters [7, 9]. This metabolic cost can be lowered by diverse plasmid-host adaptations, such as deletions, mutations in the plasmid replication machinery, or chromosomal mutations [7, 9]. Additionally, plasmids can increase their stability by conjugal transfer and/or by carrying a battery of specifically dedicated genetic determinants, classified into three main categories [9–11]. Partition determinants, in the first category, direct the active segregation of plasmid molecules during cell division. All low-copy plasmids appear to contain a partition system, which usually consists of an operon of two genes plus a specific DNA sequence for recognition. Multimer resolution systems comprise the second category and include recombinases that resolve plasmid cointegrates and maximize the number of plasmid copies available at cell division. The third category, postsegregational killing systems, include toxin-antitoxin (TA) systems and, less prominently, restriction modification loci; these systems ensure plasmid maintenance by inhibiting cell growth.
The Pseudomonas syringae complex is considered the most important bacterial plant pathogen in the world [12]. Most strains contain plasmids with an array of adaptive genes that increase aggressiveness, expand their host range, and confer resistance to antibacterials or to UV light [1, 6, 13–15]. Most of these plasmids belong to the so-called pPT23A-family plasmids (PFP) group, characterized by sharing the highly conserved RepA-PFP replicon. These replicons are highly plastic and adaptable, and strains often contain two or more stably co-existing PFP plasmids [6, 16–18]. Insertion sequences, transposons and miniature inverted-repeat transposable elements (MITEs) can account for at least a third of a PFP plasmid, actively participating in the acquisition and exchange of adaptive characters [17–21]. Insertion sequence IS801 (1.5 kb), and its isoforms, is particularly significant because of its relatively high transposition frequency, its common association with virulence genes and its ability to undergo one-ended transposition, whereby the element can mobilize adjacent DNA [19, 21, 22]. Additionally, plasmids of P. syringae have a mosaic structure and often share extensive regions of similarity, suggesting their evolution through the acquisition and loss of large DNA regions in a multistep process [14–17, 20, 23]. Despite this, plasmid profiles of individual strains appear to be characteristic and stable, although certain plasmids can be lost with high frequency under certain culture conditions [1, 24–27]. Agricultural settings exert a strong selection pressure on P. syringae populations, generally towards highly virulent clones adapted to single hosts, which can be accomplished both by gain and loss of certain virulence genes [23, 28]. However, P. syringae is an opportunistic pathogen whose life cycle primarily occurs in a variety of outside-host environments, including living on the surface of plants without causing disease [29]. It is not clear what mechanisms are driving the maintenance of virulence genes in free-living populations, where selection pressure for pathogenicity should be predictably low. Although diverse potential stability determinants were identified among PFP plasmids [15–18, 30–32], it is not yet clear whether or not they are functional and what their role in the bacterial life cycle is.
P. syringae pv. savastanoi NCPPB 3335 causes tumours in olive (Olea europaea) and is a prominent model for the study of the molecular basis of pathogenicity on woody hosts [33, 34]. This strain contains three PFP virulence plasmids pPsv48A (80 kb), pPsv48B (45 kb) and pPsv48C (42 kb) [18]. Plasmid pPsv48A carries the virulence gene ptz, involved in the biosynthesis of cytokinins, and the Type III effector gene hopAF1; pPsv48B carries the Type III effector gene hopAO1 and, in turn, plasmid pPsv48C carries the virulence gene idi, potentially involved in cytokinin biosynthesis. Both pPsv48A and pPsv48C are essential for the production of tumours in olive plants [18, 35], whereas pPsv48B contributes to fitness and virulence in planta [36]. Although pPsv48A and pPsv48B can be cured, pPsv48C is remarkably stable and could not be evicted from strain NCPPB 3335 [18], perhaps because it carries two different replicons [37]. We were interested in the identification and characterization of the stability determinants of the plasmid complement of strain NCPPB 3335, to gain insights into the mechanisms allowing the long-term maintenance of PFP plasmids and the dynamics of virulence genes.
Here, we determined that the three virulence plasmids from P. syringae pv. savastanoi NCPPB 3335 carry from one to seven functional stability determinants of different types, including three highly active TA systems in both pPsv48A and pPsv48C, although the two replicons in pPsv48C are likely sufficient for full stability. We serendipitously discovered that the mobile genetic elements IS801 and MITEPsy2 promote plasmid deletions and reorganizations with very high frequency. These derivatives are, however, efficiently excluded from the bacterial populations thanks to multiple plasmidic TA systems, which simultaneously favour the maintenance of virulence genes ptz and idi when outside the plant.
Results
Identification of putative stability determinants in the three native plasmids
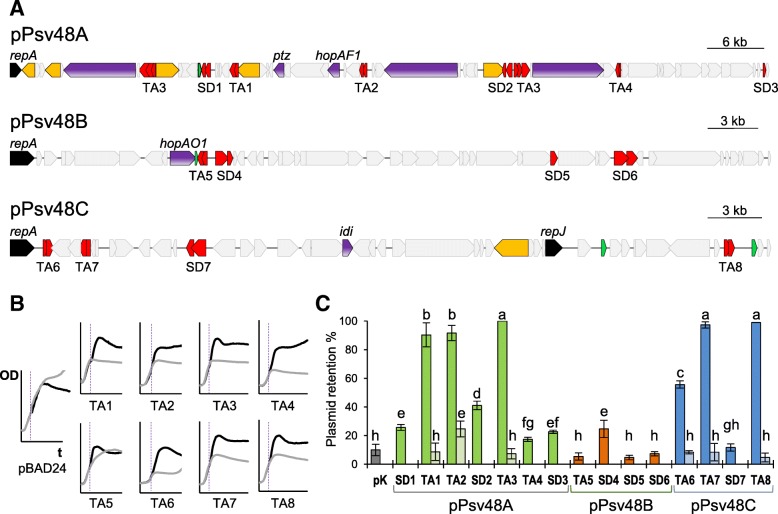
We identified a total of 15 putative stability determinants, each consisting of one to three coding sequences (CDSs), from the complete sequence of pPsv48A, pPsv48B and pPsv48C (Table 1 and Fig. 1a; see Materials and Methods). These were annotated as four partition systems (SD1, SD4, SD6 and SD7), a multimer resolution system (SD2), a CopG plasmid copy-number regulator (SD3), a plasmid killer protein (SD5), and eight TA systems (TA1 to TA8).
Table 1.
Putative stability determinants identified in the three native plasmids of P. syringae pv. savastanoi NCPPB 3335
| Plasmid and determinanta | Locus Tagb | Deduced product (InterPro family or signature matches) |
|---|---|---|
| pPsv48A | ||
| SD1 | PSPSV_A0016 | Putative partition protein A (IPR027417, IPR025669) |
| PSPSV_A0015 | Ribbon-helix-helix protein, CopG family (none predicted) | |
| TA1 | PSPSV_A0020 | Putative addiction module antitoxin, RelB/DinJ family protein (IPR007337) |
| PSPSV_A0019 | Putative toxin of the YafQ-DinJ toxin-antitoxin system (IPR004386), addiction module toxin, RelE/StbE family (IPR007712) | |
| TA2 | PSPSV_A0032 | Predicted transcriptional regulator, ribbon-helix-helix protein, CopG (IPR010985) |
| PSPSV_A0031 | Putative plasmid stabilization system protein; RelE/ParE toxin family (IPR007712) | |
| SD2c | PSPSV_A0042 | StbA, putative stability/partitioning determinant, resolvase (IPR036162) |
| PSPSV_A0041 | MvaT-like transcriptional regulator (IPR035616) | |
| TA3 | PSPSV_A0043d | StbC, Arc-type ribbon-helix-helix protein, putative antitoxin (IPR013321) |
| PSPSV_A0044d | StbB, putative ribonuclease of the VapC family (IPR022907) | |
| PSPSV_A0045d | StbA, resolvase (IPR036162) | |
| TA4 | PSPSV_A0051 | Putative RelB/DinJ family addiction module antitoxin (none predicted) |
| PSPSV_A0050 | Putative RelE/StbE family addiction module toxin (IPR007712) | |
| SD3 | PSPSV_A0067 | Putative transcriptional regulator; CopG/Arc/MetJ DNA-binding domain-containing protein, possibly responsible for the regulation of plasmid copy number (IPR002145) |
| pPsv48B | ||
| TA5 | PSPSV_B0012 | Hypothetical protein, putative plasmid maintenance component (IPR021558, DUF3018) |
| PSPSV_B0011 | Putative Mazf-like toxin, ccdB family (IPR003477) | |
| SD4 | PSPSV_B0013 | ParA/YafB type stability/partitioning protein, cobyrinic acid ac-diamide synthase (IPR027417) |
| PSPSV_B0014 | Stability/partitioning protein (none predicted) | |
| SD5 | PSPSV_B0038 | IncN plasmid killer protein (IPR009989) |
| SD6 | PSPSV_B0042 | Putative stability/partitioning determinant (IPR027417) |
| PSPSV_B0043 | Hypothetical protein (none predicted) | |
| pPsv48C | ||
| TA6 | PSPSV_C0003 | Putative RelE/StbE family antitoxin, stability determinant (none predicted) |
| PSPSV_C0004 | Putative RelE/StbE family toxin, stability determinant (IPR007712) | |
| TA7 | PSPSV_C0008 | Putative CopG family transcriptional regulator (IPR010985; IPR013321) |
| PSPSV_C0007 | Putative addiction module toxin, plasmid stabilization protein (IPR007712) | |
| SD7e | PSPSV_C0017 | Putative ParA family protein (IPR027417) |
| not annotated | Hypothetical protein (none predicted) | |
| TA8 | PSPSV_C0050 | Putative antitoxin (none predicted) |
| PSPSV_C0051 | Putative addiction module toxin, RelE/StbE family (IPR007712) | |
aTA toxin-antitoxin system, SD generic stability determinant
bThe listing order indicates direction of transcription
cGene PSPSV_A0042 shows 90% nt identity to gene PSPSV_A0045 in determinant TA3
dGenes PSPSV_A0043/44/45 are 100% identical to genes PSPSV_A0007/8/9, respectively
eSD7 was cloned containing an unannotated CDS 3′ of PSPSV_C0017 (fragment containing positions 9861–11,121 of FR820587), which could be part of a par operon
Fig. 1.
Functional analysis of putative stability determinants from the three native plasmids of P. syringae pv. savastanoi NCPPB 3335. a Maps of the native plasmids showing the relative position of the stability determinants analysed (red; Table 1), replication initiator protein genes (black), copies of the IS801 isoform CRR1 (orange), MITEs (green) and virulence genes (purple). b Growth patterns of E. coli NEB10β containing the toxin gene from the indicated TA systems cloned behind a PBAD promoter, or the empty vector (pBAD24). The vertical dashed line indicates the time when cultures received glucose (black lines), which repressed expression, or arabinose (grey lines), which induced expression. Values of OD600 (OD) versus time (t) are the average of three replicates; graphs are representative of at least 4 independent clones. c Bars indicate the percentage (mean ± sd) of P. syringae pv. syringae B728a cells retaining pKMAG-C alone (pK) or the cloned stability determinants tested in this study (panel a; Table 1). For TA systems leading to > 50% of plasmid retention, we show to their right retention values given by their corresponding antitoxins cloned alone. Experiments were repeated three times, each with three replicates. Means with different letters are significantly different (one-way ANOVA and Duncan’s multiple range test; p < 0.05)
The deduced products of the putative TA systems had typical protein signatures (Table 1), except the antitoxins of systems TA4, TA6 and TA8. Moreover, the eight respective toxin genes, except that from TA5, lead to cell growth arrest when highly expressed in E. coli NEB10β (Fig. 1b). Together, these results indicate that systems TA1-TA8 are indeed toxin-antitoxin systems, although TA5 might be non-functional or E. coli NEB10β might be resistant to the TA5 toxin.
Plasmids pPsv48A, pPsv48B and pPsv48C contain diverse functional stability determinants
The 15 putative stability determinants from plasmids pPsv48A, pPsv48B and pPsv48C were cloned into pKMAG-C, and the stability conferred to the vector was assayed in the plasmidless strain P. syringae pv. syringae B728a (Fig. 1c). pKMAG-C is able to replicate in both E. coli and pseudomonads [37], and is highly unstable in P. syringae.
All seven determinants tested from pPsv48A (Table 1), significantly increased stability of pKMAG-C to varying degrees (Fig. 1c). Four are TA systems, although only three of them conferred very high levels of stability. As expected, these TA systems were functional only when cloned completely, but not when the putative antitoxin was cloned by itself (Fig. 1c), although the antitoxin from system TA2 on its own conferred moderate levels of stability. System TA3 is widespread in pseudomonads e.g. [32, 38, 39] and it is an operon of the TA genes stbCB plus the putative resolvase stbA (Table 1). Constructs containing either stbCBA or only genes stbCB conferred equal high levels of stability (not shown); therefore, we evaluated the possible contribution of stbA to stability by cloning it separately. stbA is the last CDS in the stbCBA operon and predictably lacks a promoter; thus, we tested functionality of the stbA allele PSPSV_A0042, which is the first CDS of another putative operon (SD2 in Fig. 1) and shows 90% nt identity to the allele in operon stbCBA. Operon SD2 also significantly increased stability of pKMAG-C, likely through resolution of plasmid multimers by the StbA resolvase [11], suggesting that operon stbCBA might contribute to stability through different mechanisms.
Only one of the four determinants from pPsv48B evaluated here (Table 1) appeared to contribute, albeit modestly, to plasmid stability (Fig. 1c). This was unexpected because low-copy number plasmids usually carry diverse maintenance determinants [40]. The four determinants from pPsv48B showed similar retention values in UPN912 than in strain B728a (not shown), suggesting that lack of activity of three of them (TA5, SD5 and SD6) is not strain-related. Nevertheless, it is possible that pPsv48B contains stability genes that were not included or whose activity was not detected in our assays, and/or that its stability is increased by conjugation [10].
Three TA systems, out of the four determinants tested from pPsv48C, contributed to plasmid stability (Table 1); again, the putative antitoxins did not confer any stability by themselves (Fig. 1c). Remarkably, the eight different TA systems showed distinct behaviours in our assays (Fig. 1c), which varied from no apparent contribution to stability (TA5) to conferring moderate (TA4) to very high stability levels (e.g. TA3 or TA8).
The two replicons from pPsv48C confer distinct stability
To explore the basis of the very high stability of pPsv48C, we evaluated the contribution of the RepA-PFP and RepJ replicons to its maintenance. Therefore, we cloned them into the E. coli vector pKMAG and, as before, evaluated stability in the plasmidless strain P. syringae pv. syringae B728a (Fig. 2). However, plasmid replicons are often adapted to increase their persistence in their bacterial host e.g. [41, 42]. Therefore, we also tested stability in the plasmidless strain P. syringae pv. savastanoi UPN912 (Fig. 2), which derives from the original host strain NCPPB 3335 (Table 2).
Fig. 2.

Stability of constructs containing the native RepA-PFP and RepJ replicons from pPsv48C, and their chimeras. a Fragments of the RepA-PFP (black) or RepJ (white) replicons, and their chimeras, were cloned at the indicated positions into pKMAG; small and large arrows represent the putative leader peptide and the replication initiator genes, respectively. TT, T4 transcription terminator; MCS, multiple cloning site; kan, kanamycin resistance gene. b Percentage (mean ± sd) of P. syringae pv. syringae B728a cells (dark grey) or of P. syringae pv. savastanoi UPN912 cells (light grey) retaining each of the constructs of panel a means with different letters are significantly different (two-way ANOVA and Duncan’s multiple range test; p < 0.05). Experiments were repeated three times, each with three replicates
Table 2.
Bacterial strains and plasmids used in this study
| Strain/plasmid | Main featuresa | Source or reference |
|---|---|---|
| Escherichia coli | ||
| NEB10β | Δ(mrr-hsdRMS-mcrB) deoR recA1 endA1 araD139 Δ(ara, leu)7697 galU galK λ− rpsL nupG | New England Biolabs |
| S17–1 | Strain used to transfer pDR1 plasmid | [43] |
| P. syringae pv. savastanoi | ||
| NCPPB 3335 | Pathotype strain, isolated from a diseased olive tree; contains three native plasmids (pPsv48A, pPsv48B and pPsv48C) | [78] |
| Psv48ΔAB | NCPPB 3335 derivative, cured of pPsv48A and pPsv48B | [18] |
| UPN25.1 | UPN827 derivative containing pPsv48CΔ25, a 5.5 kb spontaneous deletion derivative of pPsv48C that spans the RepJ replicon, with no TA systems. | [37] |
| UPN508 | Derivative of NCPPB 3335 containing pPsv48A::Tn5-GDYN1 | [18] |
| UPN827 | Derivative of Psv48ΔAB containing plasmid pPsv48C::Tn5-GDYN1 | [18] |
| UPN864 | UPN827 derivative, which contains plasmid pPsv48CΔ1 | This work |
| UPN912 | Plasmidless derivative of strain NCPPB3335; was obtained by curing pPsv48C::sacB from strain UPN1007 | [35] |
| UPN1007 | Derivative of Psv48ΔAB containing plasmid pPsv48C::sacB | This work |
| P. syringae pv. syringae | ||
| B728a | Plasmidless bean pathogen CuR, RifR, SmR | [79] |
| Plasmids | ||
| pBlueScript II SK | E. coli cloning vector; 2.96 kb, AmpR | Stratagene |
| pBAD24 | E. coli expression vector, contains a PBAD promoter inducible with arabinose; 4.5 kb, AmpR | [76] |
| pDR1 | Delivery vector for Tn5-GDYN1, based on pSUP2021; KmR, GmR, SmR, SpR; confers sucrose-dependent lethality | [43] |
| pGEM-T Easy | E. coli cloning vector; 3 kb, AmpR | Promega |
| pJET1.2 | E. coli cloning vector 2.9 kb, AmpR | Thermo Fisher Scientific |
| pK18mobsacB | Mobilizable cloning vector, confers sucrose-dependent lethality; KmR, SucS | [71] |
| pKMAG | E. coli vector derived from pK184, devoid of the Plac promoter and containing the transcriptional terminator and polylinker from pME6041; 2.6 kb, KmR; accession no. KX714576 | [37] |
| pKMAG-C | pKMAG containing the minimal RepA-PFP replicon from pPsv48C, replicates in E. coli and in Pseudomonas; 4.3 kb, KmR; accession no. KX714577 | [37] |
| pME6041 | Broad host range cloning vector; 5.6 kb, KmR | [80] |
| pPsv48A | Virulence plasmid of NCPPB 3335 (accession n° FR820585); with RepA-PFP replicon; 80.1 kb | [18] |
| pPsv48A::Tn5-GDYN1 | Plasmid pPsv48A tagged with Tn5-GDYN1 at position 1469 | [18] |
| pPsv48B | Native plasmid of NCPPB 3335 (accession no. FR820586); with RepA-PFP replicon; 45.2 kb | [18] |
| pPsv48C | Virulence plasmid of NCPPB 3335 (accession no. FR820587); with RepA-PFP and RepJ replicons; 42.1 kb | [18] |
| pPsv48C::sacB | pPsv48C derivative containing the KmR-sacB cassette from pK18mobsacB inserted at position 26,916; KmR, SucS | This work |
| pPsv48C::Tn5-GDYN1 | pPsv48C containing Tn5-GDYN1 inserted at position 37,036; KmR, GmR, SucS | This work |
| pPsv48CΔ1 | Derives from pPsv48C::Tn5GDYN1 by the spontaneous deletion of 8.3 kb (positions 32,807–41,121 of FR820587) mediated by recombination between two copies of MITEPsy2 | This work |
| pPsv48CΔ25 | Spontaneous sucrose-resistant deletion derivative from pPsv48C::sacB, generated by a one-ended transposition of IS801; this plasmid is 5.5 kb long, spanning positions 27,019–32,557 of FR820587 | [37] |
| pRK3A | pRK415 derivative containing genes PSPSV_A0043, PSPSV_A0032 and PSPSV_A0020 cloned in tandem in this order, each with their own promoter and under the control of the Plac promoter from the vector; TcR | This work |
| pRK3C | pRK415 derivative containing genes PSPSV_C0050, PSPSV_C0008 and PSPSV_C0003 cloned in tandem in this order, each with their own promoter and under the control of the Plac promoter from the vector; TcR | This work |
| pRK415 | Broad host range cloning vector; 10.5 kb, TcR | [81] |
aAbbreviations: Amp ampicillin, Cu copper, Km kanamycin, Gm gentamicin, Rif rifampicin, Sm streptomycin, Sp spectinomycin, Suc sucrose, Tc tetracyclin. Superscripts R and S denote resistance or susceptibility, respectively
Construct pKMAG-C, containing the RepA-PFP replicon cloned outside the polylinker of the vector, was highly unstable and was nearly completely lost after only one night of growth (Figs. 1c and 2). This was probably due to a destabilization of the replication control system from an increase in transcription by read-through from the constitutive kanamycin promoter, a phenomenon previously described for replicon RepJ [37]. In fact, its cloning after the transcription terminator of pKMAG significantly increased stability (2 in Fig. 2). Gene ssb, which is frequently found downstream of the repA gene [17, 18, 31] only showed a marginal contribution to stability (compare 2 and 3, Fig. 2). In turn, the RepJ replicon conferred a significantly higher stability than the RepA-PFP replicon (compare 2 and 4, Fig. 2). Noticeably, all the constructs were significantly more stable in strain UPN912 than in B728a (Fig. 2), suggesting that these replicons are adapted to the bacterial host in which they occur naturally, to maximize their survival.
The RepA-PFP and RepJ replicons consist of two separable functional fragments: a control region, containing the promoter, a putative antisense RNA and a leader peptide, and a replication region, containing the replication initiator protein (rep) gene [37]. The approx. 0.3 kb control region determines the transcription rate of the rep gene. The RepA-PFP and RepJ replicons share very similar, but not identical control regions preceding the rep gene [37], and we hypothesized that this could potentially influence replicon stability. We therefore evaluated the stability of constructs containing chimeric replicons, with the replication control region (Rex-C module) reciprocally swapped [37]. The highest stability in UPN912, but not in strain B728a, was reached with the chimera RepA-PFP:RepJ (control:replication modules; construct 5, Fig. 2), indicating that replicon stability is mostly dependent on the activity of the replication module, but it can be modulated by the control module (Fig. 2).
The significant values of plasmid loss observed for RepJ (Fig. 2) conflicted with the high stability observed for pPsv48C deletion derivatives (not shown), suggesting that we did not clone all the replicon sequences needed for stable replication. We therefore tested the stability of a spontaneous 5.5 kb deletion derivative of pPsv48C (clone pPsv48CΔ25; Table 2), containing the minimal RepJ replicon [37] plus additional DNA that did not include any other potential plasmid maintenance genes. Plasmid pPsv48CΔ25 was maintained in 100% of the cells obtained from starting cultures and after seven sequential culture transfers (1622 and 2804 colonies tested, respectively). In contrast, the RepJ construct in pKMAG (construct 4 in Fig. 2) was retained by 94 ± 2% of UPN912 cells from starting cultures and by only 63 ± 2% of the cells after seven transfers (2366 and 2666 colonies tested, respectively). These results indicate that the native RepJ replicon is larger than the minimal replicon [37] and underscore its high stability in its genetic context.
A toxin-antitoxin system prevents a deletion in pPsv48C mediated by MITEs
We sought to obtain derivatives of NCPPB 3335 cured of plasmid pPsv48C, and to evaluate the contribution of its three TA systems to stability. We thus constructed strain UPN827, containing a transposon carrying the sacB gene (Tn5-GDYN1) inserted into pPsv48C (Fig. 3a; Table 2); this allowed us to easily select for plasmid loss by growth in the presence of sucrose [43]. To inactivate functionally the TA systems [44] and facilitate plasmid loss, we constructed pRK3C, containing the three antitoxin genes from pPsv48C cloned in pRK415 (Table 2), and introduced it into UPN827 to neutralise the three corresponding toxins.
Fig. 3.

Recombination between two directly repeated copies of MITEPsy2 causes a deletion on pPsv48C. a Partial map of pPsv48C::Tn5-GDYN1 (pC::Tn5) showing the relative positions of its only copy of the IS801 isoform, its two replication initiation protein genes (repJ and repA), and toxin-antitoxin system 8 (TA8). Green block arrows, MITEPsy2; inverted black triangle, Tn5-GDYN1 (Tn). pCΔ1 is pPsv48CΔ1, containing an 8.3 kb deletion resulting from MITEPsy2 recombination. b Electrophoresed uncut plasmid preparations from: (1) P. syringae pv. savastanoi NCPPB 3335; (2) Psv48ΔAB; (3) UPN827, and (4) UPN864. pA, pPsv48A; pB; pPsv48B; pC, pPsv48C; pCΔ1, pPsv48CΔ1; clp, chromosomal DNA and linearized plasmids. Lanes were loaded with equivalent amounts of cell lysates; results are representative of at least 20 independent plasmid preparations
We routinely obtained 50 times more sucrose-resistant (sucR) colonies with strain UPN827(pRK3C) (38 ± 3 × 10− 4 sucR colonies) than with its parental strain UPN827(pRK415) (0.8 ± 0.4 × 10− 4 sucR colonies), and this difference was statistically significant. All sucR colonies examined contained an 8.3 kb deletion in pPsv48C caused by the recombination of two direct copies of MITEPsy2, as assessed by sequencing, eliminating the sacB transposon Tn5-GDYN1 (Fig. 3a). One of these plasmids was retained and designated pPsv48CΔ1 (Fig. 3a). These results indicate that, despite its small size (228 nt), MITEPsy2 is a hot spot for recombination.
In plasmid profile gels of the wild type strain NCPPB 3335, pPsv48C routinely appears with lower intensity than the other two native plasmid bands (Fig. 3b) [18]. Remarkably, bands of plasmid pPsv48CΔ1 were repetitively more intense than those of the wild type plasmid or of pPsv48C::Tn5-GDYN1 (Fig. 3b), suggesting that the 8.3 kb deletion caused a higher copy number. We estimated a moderate copy number for plasmids pPsv48A (8.0 ± 1.0), pPsv48B (8.6 ± 1.6) and pPsv48C (6.6 ± 1.2), with no significant differences among them. These are as expected for medium-size native plasmids [45] and similar to the five copies reported for the native plasmid pFKN from P. syringae pv. maculicola [20]. Unexpectedly, the estimated copy number of pPsv48CΔ1 (6.9 ± 0.8) was not significantly different from that of pPsv48C. These results indicate that each of the three native plasmids from strain NCPPB 3335 exist in 6–9 copies per cell, and that the 8.3 kb fragment from pPsv48C does not carry any determinant involved in copy number control. This also suggests that structural differences among plasmids could differentially impact their purification by alkaline lysis and questions the use of agarose gel electrophoresis to estimate relative plasmid DNA quantities.
Toxin-antitoxin systems from pPsv48C prevent accumulation of plasmid deletions mediated by IS801
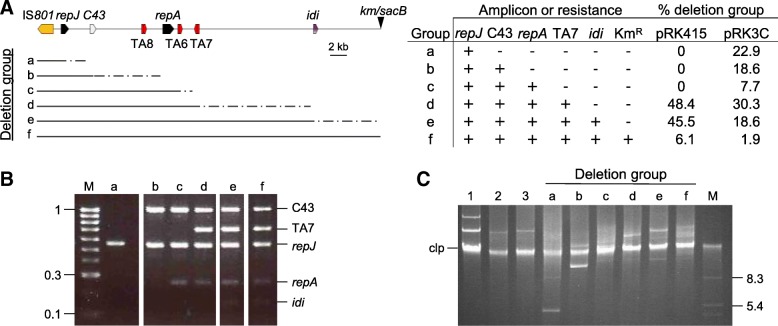
Our preliminary experiments soon indicated that the inactivation of the three TA systems of pPsv48C did not facilitate the isolation of plasmid-cured strains but, instead, led to the recovery of deletion derivatives generated by one-ended transposition of the IS801 isoform CRR1 (Fig. 4) [18]; for clarity, we will henceforth refer to this isoform as IS801. Therefore, strain UPN1007 was used to better estimate the causes and frequency of the different deletions. This strain carries plasmid pPsv48C::sacB, containing a KmR-sacB cassette immediately adjacent to the only IS801 copy of pPsv48C (Fig. 5); thus, the selection of sucR colonies would allow for the identification and quantification of all types of deletions mediated by one-ended transposition of IS801.
Fig. 4.
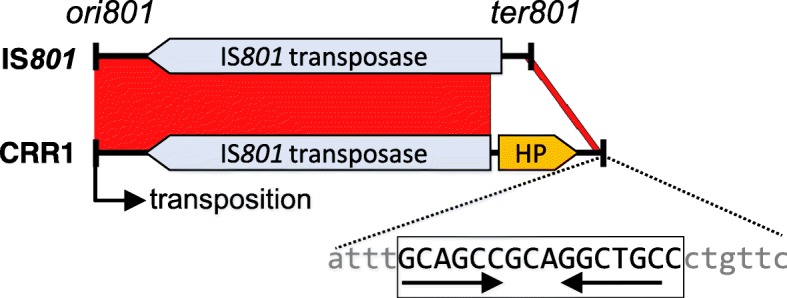
Comparison of the wild type IS801 with its isoform CRR1. Blastn alignment of IS801 (X57269; 1512 nt) and CRR1 (from FR820587; 1765 nt); the red bands connecting the two elements indicate collinear regions of identity. CRR1 contains an insertion of 365 nt, causing a deletion of 112 nt that removes the predicted transposase start codon and trims the ter801 terminus to the endmost 26 nt (expanded sequence). This 26 nt region contains a conserved motif (capital letters) with an inverted repeat sequence (horizontal arrows), probably involved in recognition and interaction with the transposase [46]. HP, hypothetical protein
Fig. 5.
Types of deletions of pPsv48C::sacB as influenced by functional toxin-antitoxin systems. a Left: Map of pPsv48C::sacB; TA6, TA7 and TA8, toxin-antitoxin systems; C43, locus PSPSV_C0043; inverted triangle, KmR-sacB cassette cloned 0.1 kb 3′ of the IS801 isoform. Lines under the map indicate the minimum (black line) and maximum (dotted line) extent of DNA transposed by IS801 on each group of sucR plasmids. Right: Presence (+) or absence (−) of specific amplicons for each of the genes shown, or of resistance (+) and sensitivity (−) to kanamycin. Last two columns indicate the percentage of sucR colonies containing each plasmid group in UPN1007 containing the empty vector pRK415 (310 colonies analysed) or pRK3C, leading to functional inactivation of the TA systems (323 colonies analysed). Gels showing typical patterns of multiplex PCR amplifications (panel b) and uncut plasmids (panel c) of example clones from each plasmid group. M, molecular weight markers, in kb; clp, chromosomal DNA and linearized plasmids. Lanes: (1) P. syringae pv. savastanoi NCPPB 3335; (2) Psv48ΔAB, containing only pPsv48C; and (3) UPN864, containing only pPsv48C::sacB
The frequency of sucR colonies was 1.8 ± 0.7 × 10− 4 for UPN1007 containing the empty vector but significantly higher (5.5 ± 2.1 × 10− 4) for strain UPN1007(pRK3C), in which the three TA systems are functionally inactivated (Fig. 5). The plasmid profile and PCR analyses of > 700 independent clones, plus sequencing of 13 of them, indicated that none had lost pPsv48C but showed a plasmid band of ca. 4 to 42 kb resulting from deletions of variable size in this plasmid. All deletion derivatives contained IS801 and repJ (Fig. 5), and sequencing showed that all had a common left border corresponding to the 3′ end of IS801 (position 27,019 of pPsv48C; Fig. 5a), containing the ori801 where transposition of this element initiates [46]. The right border of the different plasmid derivatives was GAAC (5 clones) or CAAG (8 clones), which were described as consensus tetramers immediately adjacent to insertions of IS801 and places for one-ended transposition events to finish [19, 47].
The extent and frequency of deletions generated in pPsv48C, both in UPN1007(pRK415) and in UPN1007(pRK3C), was evaluated in clones growing in SNA by a multiplex PCR analysis (Fig. 5b). Additionally, loss of kanamycin resistance indicated the loss of the KmR-sacB cassette in the largest deletion derivatives (notice that transpositions ending closer from IS801 result in the deletion of larger DNA fragments from pPsv48C). The 310 sucR clones examined from strain UPN1007(pRK415) retained plasmids of at least 22 kb, all spanning the three TA operons (TA6–8; Fig. 5a). This was expected because the three TA systems are functional in UPN1007 and their loss would predictably result in growth inhibition. However, around half of the clones had lost gene idi, indicating the spontaneous loss of this gene in routine culture conditions with a frequency of 0.9 ± 0.3 × 10− 4. The types of deletions were more varied in the 323 sucR clones of UPN1007(pRK3C), containing functionally inactivated TA systems, with nearly half of the clones losing the RepA-PFP replicon and around 80% (4.4 ± 1.9 × 10− 4) of them lacking gene idi (Fig. 5). Notably, IS801 was able to transpose the complete length of pPsv48C in both strains (plasmid group f in Fig. 5), although at a low frequency of around 10− 5, suggesting that IS801 is capable of mobilizing more than 40 kb of adjacent DNA. Incidentally, the generation of circular deletion variants of pPsv48C mediated by IS801 also indicates that, as predicted [47], this element transposes by a rolling circle mechanism.
Toxin-antitoxin systems also contribute to the maintenance of plasmid pPsv48A and to reducing the occurrence of deletions
Because IS801 is pervasive in P. syringae genomes, we wanted to know if deletions mediated by this element also occurred in other plasmids, and whether or not TA systems are contributing to decrease their frequency. For this, we used strain UPN508, a derivative of strain NCPPB 3335 containing plasmid pPsv48A with an insertion of Tn5-GDYN1 located at 1.9 kb 3′ of gene repA (Fig. 6) [18]. pPsv48A contains only one replicon and Tn5-GDYN1 is inserted between two of the five copies of IS801 in the plasmid, limiting the types and size of deletions that we can detect, although the experimental setting still allowed us to evaluate the possible occurrence of deletions.
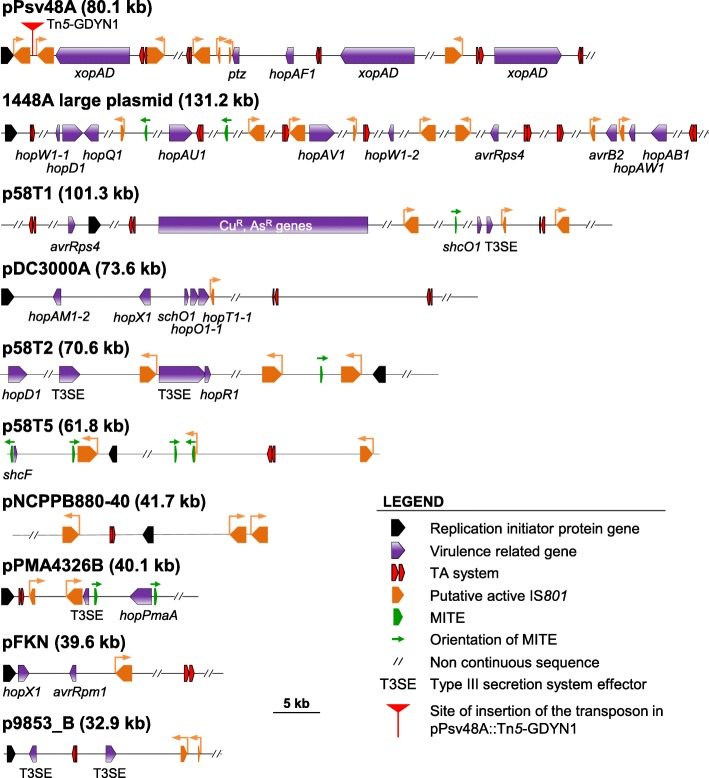
Fig. 6.
Schematic representation of relevant features found in closed plasmid sequences of Pseudomonas syringae. The diagram shows the replication initiator protein genes, virulence genes, TA systems, putative active IS801 elements and MITEs found in closed plasmid sequences of the P. syringae complex. Features are drawn to scale but, for clarity, only pertinent plasmid fragments are shown. The direction of transposition of IS801 fragments and isoforms is indicated with orange arrows. Harbouring organism and accession numbers for the plasmids are P. syringae pv. savastanoi NCPPB 3335, NC_019265 (pPsv48A); P. syringae pv. phaseolicola 1448A, NC_007274 (p1448A); P. syringae pv. tomato DC3000, NC_004633 (pDC3000A); P. cerasi 58T, NZ_LT222313 (p58T1), NZ_LT222314 (p58T2), NZ_LT222317 (p58T5); P. syringae pv. tomato NCPPB 880, NC_019341 (pNCPPB880–40); P. cannabina pv. alisalensis ES4326, NC_005919 (pPMA4326B); P. syringae pv. maculicola M6, NC_002759 (pFKN); P. syringae pv. actinidiae ICMP 9853, NZ_CP018204 (p9853_B)
Strain UPN508(pRK415) generated sucR clones with a frequency of 1.1 ± 0.8 × 10− 4. From 282 of these sucR clones, plasmid pPsv48A::Tn5-GDYN1 was lost in two clones, it contained spontaneous mutations inactivating sacB in nine clones, and was reorganized or contained deletions in the remaining ones (Table 3). The majority of the sucR clones, around 90% of the total, contained derivatives of ca. 76 kb; sequencing of three of these clones suggests that they resulted from recombination between the two isoforms of IS801 flanking the insertion point of Tn5-GDYN1 (Table 3), causing its deletion.
Table 3.
Type and proportion of sucrose-resistant derivatives of pPsv48A::Tn5-GDYN1 in the presence or absence of functional toxin-antitoxin systems
| Number (%) of sucR clonesb | ||||
|---|---|---|---|---|
| Plasmid sizea | UPN508(pRK415) | UPN508(pRK3A) | ptz c | Type of event d |
| - (cured) |
2 (0.7) | 110 (41.4) | – | |
| 57 | 1 (0.4) | 38 (14.3) | + | Recombinatione |
| 70 | 16 (5.7) | 4 (1.5) | + | Reorganizationsf |
| 76 | 251 (89.0) | 111 (41.7) | + | Recombination?g |
| 89 | 9 (3.2) | 2 (0.8) | + | Spontaneous mutation in sacB |
| > 90 | 3 (1.1) | 1 (0.4) | + | Reorganizationsh |
| Total | 282 | 266 | ||
aApproximate size (kb) of the deletion derivative. Total size of pPsv48A::Tn5-GDYN1 is around 89 kb
bStrain UPN508 contains pPsv48A::Tn5-GDYN1, with functional TA systems. The three TA systems of this plasmid are functionally inactivated in the presence of pRK3A, containing the three cloned antitoxins from pPsv48A
cPresence (+) or absence (−) of the virulence gene ptz, for cytokinin biosynthesis, in the resulting deletion derivatives
dAll events resulted in deletion of Tn5-GDYN1, except the spontaneous mutation in sacB
eRecombination between IS801–1 and IS801–4
fDiverse group of clones with different, uncharacterized intramolecular reorganizations
gThe sequence of five clones confirmed that they resulted from recombination between IS801–1 and IS801–2, but we cannot discard the possibility that some or all of the remaining clones resulted from a transposition of IS801–2
hThese plasmids appeared to result from a transposition of IS801–2, terminating precisely at the end of IS801–1 as determined by sequencing, that eliminate Tn5-GDYN1. According to their relative size on plasmid profile gels, these plasmids must contain an uncharacterized insertion
Functional inactivation of the three TA systems, in strain UPN508(pRK3A), lead to only a modest, but significant increase of the frequency of sucR clones to 3.6 ± 1.5 × 10− 4, and to a dramatic change on the plasmid content of these clones (Table 3). The first major difference was that the frequency of loss of pPsv48A was around 1.5 ± 0.2 × 10− 4, two orders of magnitude higher than that in UPN508(pRK415) (Table 3). The second major difference was that deletion derivatives of approx. 57 kb, all of which had lost system TA1, appeared around 40 times more frequently than in strain UPN508(pRK415) (Table 3). The frequency of occurrence of the other reorganizations (Table 3) varied no more than four times between both strains. Noticeably, and contrasting with pPsv48C, most of the deletions affecting pPsv48A are likely due to recombination between IS801 elements instead to one-ended transpositions of IS801. This indicates that IS801 promotes plasmid deletions with high frequency by diverse mechanisms.
Are multiple toxin-antitoxin systems commonly safeguarding virulence plasmids of P. syringae?
Many plasmids of P. syringae contain virulence genes and a large amount of mobile genetic elements [1, 2, 6, 17, 18], of which MITEs and IS801 transpose most frequently [19]. Here we showed that these mobile elements also mediate frequent deletions and reorganizations in two virulence plasmids of P. syringae pv. savastanoi NCPPB 3335, and that their carriage of multiple toxin-antitoxin systems allows avoiding these deletions and maintain plasmid integrity. We therefore questioned if this could be a common strategy among virulence plasmids of P. syringae.
We found sequences homologous to IS801 in 53 out of the 78 available closed plasmid sequences from strains of the P. syringae group (including P. cerasi; December, 2018), with around two thirds of them containing at least one complete or truncated copy of CRR1. This indicates a frequent occurrence of this mobile element in the P. syringae pangenome. The sequence of nine of these plasmids, chosen as examples, contained one to eight copies of ori801 potentially capable of initiating one-ended transposition (Fig. 6); four of them also contained one to four copies of MITEPsy1. Likewise, eight of the nine plasmids harboured at least one putative TA system; an extreme case is p1448A-A (131.2 kb), containing eight ori801 and seven putative TA systems (Fig. 6). These TA systems are also likely limiting the occurrence of deletions, which could potentially eliminate one or more of the virulence genes included in these plasmids (Fig. 6).
Discussion
Native plasmids of P. syringae and other phytopathogenic bacteria often carry genes contributing to virulence and resistance to bactericides, sometimes being essential for pathogenicity [2, 6, 14, 15, 17, 18, 48]. Although they are generally considered moderately to highly stable in the few tested P. syringae strains [18, 27], there is a general lack of knowledge of the molecular mechanisms involved in long-term plasmid survival. Here we show that the virulence plasmids from P. syringae pv. savastanoi NCPPB 3335 use diverse mechanisms to persist in the cell and maintain their physical integrity.
We identified 11 functional stability determinants among the 15 determinants examined from the three native plasmids of strain NCPPB 3335. These included seven TA systems, two putative partition systems, one putative multimer resolution system and one putative CopG-type copy number control regulator. The four remaining determinants evaluated (TA5, SD5, SD6 and SD7) appeared to be non-functional. It is nevertheless possible that the high instability of the vector used for testing, pKMAG-C, did not allow us to detect their activity as stability determinants, although TA5 is probably non-functional since it did not show activity in P. syringae strains B728a and UPN912, and in E. coli. We showed that the TA systems are a major stability determinant only for plasmid pPsv48A, increasing its stability by two orders of magnitude. The TA systems do not appear to contribute to the stability of pPsv48C because this plasmid carries two replicons [37] conferring a very high stability level by themselves. In particular, RepJ can be maintained with no apparent plasmid loss for seven sequential culture transfers in the absence of any identifiable maintenance determinants. Notably, these two replicons appear to be adapted to its native host to maximize their stability (Fig. 2). The carriage of several strong stability determinants clearly favours the maintenance of virulence genes but also likely the acquisition of new plasmids and adaptive characters. Virulence genes are frequently found on PFP plasmids [1, 6], which are often exchanged horizontally [2, 49, 50]. This, however, appears to not disturb the previous plasmid complements, because strains of P. syringae usually harbour two to six different PFP plasmids [16]. Thus, strong stability determinants likely contribute to the retention of newly acquired PFP plasmids until they accumulate changes allowing their full compatibility with other resident plasmids. Indeed, we have shown that as little as five nt changes in the replication control region are sufficient to overcome incompatibility between PFP plasmid replicons [37].
The virulence plasmids pPsv48A and pPsv48C are structurally very fragile, experiencing high frequency intramolecular deletions and reorganizations promoted by the mobile genetic elements MITEPsy2 and IS801. The TA systems carried by these plasmids, however, significantly reduce the accumulation of structural variants by selectively excluding them from the bacterial population. TA systems are bicistronic operons coding for a stable toxin and an unstable antitoxin that neutralises the activity of the toxin [51]. If the operon is lost, for instance due to a deletion, the antitoxin is rapidly degraded and bacterial growth is arrested due to the action of the stable toxin; thus, only cells that did not suffer the deletion and still contain the TA system can grow.
Our functional inactivation of the TA systems significantly increased the frequency of the pPsv48C deletions mediated by MITEPsy2 by 50 times and by three times those mediated by IS801. This would indicate that the TA systems might be only moderately successful in preventing deletions mediated by IS801. However, we should consider that inactivation of the TA systems lead to a fivefold increase in the loss rate of gene idi, which is essential for tumour formation in the plant host [35]. Noticeably, it appears that the loss of gene idi was reduced even in those cases where deletion of this gene would not determine loss of any TA system (Fig. 5a). This could be a general feature, because a TA system from a virulence plasmid of Shigella spp. favoured the retention of nearby sequences, maintaining plasmid integrity [52].
Likewise, the occurrence of intramolecular deletions and reorganizations of pPsv48A increased three times upon functional inactivation of its TA systems (Table 3). This phenomenon has been termed post-recombinational killing [52], whereby the occurrence of insertion sequence-mediated rearrangements involving the deletion of TA systems lead to bacterial growth arrest and the consequent exclusion of the reorganized variants from the bacterial population. The modest protection offered by TA systems of pPsv48A is predictably an underestimate because of the limited number and types of events that we could detect with the pPsv48A::Tn5-GDYN1 construct used. Nevertheless, the TA systems of pPsv48A are contributing to the maintenance of virulence gene ptz (Table 3), which is essential for the induction of full-size tumours and the development of mature xylem vessels within them [18]. The occurrence of multiple, apparently redundant, TA systems in plasmids is intriguing. However, plasmids are highly dynamic entities undergoing a continuous trade of genetic material [2, 4]; as such it is feasible that multiple TA systems are selected to ensure the survival of different plasmid fragments. This is clearly exemplified by the 8.3 kb fragment that is “protected” by TA8 (Fig. 3).
In this work, we concentrated on examining the plasmids of strain NCPPB 3335. However, we would expect that the structural fragility of native plasmids and the protective role of TA systems are common phenomena in the P. syringae complex, and likely in other plant pathogens, for three main reasons. First, repetitive mobile genetic elements, and particularly IS801, are widespread in the P. syringae complex, can represent at least one third of diverse native plasmids, and are often associated to virulence genes [18, 19, 22, 27, 53]. IS801 is remarkable, because it can efficiently transpose with a transposase provided in trans and because it follows a rolling circle replicative mechanism, leading to permanent insertions [19, 46, 47]. This implies that any fragment of IS801 containing ori801 is potentially mobilizable, that every transposition generates a potentially recombining site, and that one-ended transposition events can immediately lead to the generation of small to very large plasmid deletions. Additionally, other highly repetitive genes, such as the rulAB operon for resistance to UV light and many other DNA repair genes, are also commonly associated to virulence and other adaptive genes in P. syringae and many other bacteria [54–56]. All these repetitive genetic elements favour the mobility of virulence genes, promoting the high plasticity and adaptability of native plasmids [6, 16–18]; however, at the same time, represent recombination hotspots that can mediate deletion of key virulence genes [57], as highlighted by our results, and of many other adaptive genes. Second, the frequencies of recombination between MITEs and of transposition of IS801 were very high, suggesting that they could be very active in promoting genomic changes. Third, and although largely ignored, TA systems are increasingly being found associated to native plasmids in many diverse plant pathogens, including P. syringae (see also Fig. 6) [17, 58, 59]. It is also noteworthy that most of these plasmids possess several TA systems, as occurs with plasmids from other bacteria [4, 57, 58].
Conclusions
Here we show that TA systems are frequently found in plasmids of P. syringae and that they significantly contribute to plasmid stability, to preserve plasmid integrity and to maintain virulence genes in free living conditions. TA systems have been involved in a disparity of functions including, among others, the stabilization of plasmids and other mobile genetic elements, biofilm formation, modulation of bacterial persistence, resistance to antibacterial compounds, and prevention of large scale deletions in the chromosome, plasmids and episomes [51, 52, 60–62]. Our results show that genes found in plasmids of the plant pathogen P. syringae can be eliminated with high frequency because of plasmid loss and rearrangements mediated by mobile genetic elements. The occurrence of multiple toxin-antitoxin systems in plasmids effectively increase the survival of virulence genes and virulence plasmids in bacterial populations, facilitating their preservation in a diversity of environments lacking the strong selective pressure exerted by the plant host.
Methods
Bacterial strains, plasmids and growth conditions
Table 2 summarizes strains, native plasmids and constructions used in this study. LB medium [63] was routinely used for growing both E. coli (at 37 °C) and Pseudomonas strains (at 25 °C). Counter selection of cells carrying the sacB gene, which confers lethality in the presence of sucrose, was carried out in nutrient agar medium (Oxoid, Basingstoke, UK) supplemented with 5% sucrose (medium SNA). When necessary, media were supplemented with (final concentrations, in μg ml− 1): ampicillin, 100; gentamicin, 12.5; kanamycin, 7 for P. syringae and 50 for E. coli; tetracycline, 12.5.
General molecular procedures and bioinformatics
DNA was amplified using a high fidelity enzyme (PrimeStar HS, Takara Bio Inc., Japan), or a standard enzyme (BIOTaq, Bioline, UK), and primers detailed in Additional file 1 Table S1. Amplicons were cloned using the CloneJET PCR Cloning Kit (Thermo Scientific) or the pGEM-T Easy Vector System (Promega). Purification of plasmids from E. coli was carried out following a boiling method [64] or using a commercial kit (Illustra plasmidPrep Mini Spin Kit, GE Healthcare). For plasmid profile gels, DNA was purified by alkaline lysis and separated by electrophoresis in 0.8% agarose gels with 1xTAE as described [25]. Plasmids were transferred to P. syringae by electroporation [65].
DNA sequences were compared and aligned using the BLAST algorithms [66], as well as the on-line MULTALIN [67] and EMBL-EBI server tools (http://www.ebi.ac.uk/Tools/msa/). The InterPro interface [68] (http://www.ebi.ac.uk/interpro/) was used to search for protein motifs. Nucleotide sequence visualization and manipulation was performed using the Artemis genome browser and ACT [69]. Primers were designed using the Primer3plus software [70].
Manipulation of native plasmids of P. syringae pv. savastanoi
Native plasmids of P. syringae pv. savastanoi were tagged with Tn5-GDYN1 by conjugation using E. coli S17.1 as a donor; this transposon carries the levansucrase gene sacB, which allows for the identification of derivatives cured of plasmids by selection in medium with sucrose [18, 43]. Sites of Tn5-GDYN1 insertion were determined by sequencing of cloned EcoRI fragments containing the GmR end of the transposon and the adjacent sequences using primer IS50_F (Additional file 1 Table S1).
We constructed a derivative of pPsv48C containing a KmR-sacB cassette, immediately 5′ of the IS801 isoform (100 nt upstream), as a tool to analyse the diverse deletions generated by the activity of this mobile element. The KmR-sacB cassette was amplified from pK18mobsacB [71] by PCR with specific primers (Additional file 1 Table S1), and introduced into an EcoRV site of pPsv48C (position 26,919 in accession no. FR820587) by allelic exchange recombination.
Estimation of plasmid copy number
Plasmid copy number was estimated by quantitative PCR (qPCR) using as template total DNA purified with the JET flex Genomic DNA Purification Kit (Genomed, Germany). qPCR was performed using the CX96™ Real-Time System and analysed using CFX Manager software version 3.0 (BioRad, CA, USA), essentially as described [72] . A ten-fold serial dilution series of DNA was used to construct the standard curve for the single-copy chromosomal gene gyrA, used as reference [72], and the plasmids genes ptz (PSPSV_A0024; pPsv48A), hopAO1 (PSPSV_B0010, pPsv48B) and idi (PSPSV_C0024, pPsv48C), using the primers indicated in Additional file 1 Table S1. Plasmid copy numbers were estimated using the ΔΔCt method [73, 74].
Identification of putative plasmid stability determinants
For identification of putative stability determinants from plasmids pPsv48A (FR820585), pPsv48B (FR820586) and pPsv48C (FR820587), we manually inspected the annotation of the three plasmids and searched for those CDSs containing terms (stability, partition and related forms), or whose products contained typical domains associated to plasmid maintenance. Additionally, we selected putative toxin-antitoxin operons with a significant score (higher than 70) in the web tool RASTA-bacteria [75]. The complete set of loci identified and tested is summarized in Table 1.
The functionality of toxin genes from the putative TA systems was tested using the expression vector pBAD24 [76]. Toxin genes were amplified by high-fidelity PCR using primers with adapters for KpnI and PstI (Additional file 1 Table S1), cloned in the same sites of pBAD24, generating translational fusions with the first or second codon of the toxin gene, and transformed into E. coli NEB10β. Single colonies of appropriate clones grown overnight on LB + Amp were resuspended in LB, and two wells per clone of a microtiter plate were inoculated with 5 μl of the bacterial suspension and 150 μl of LB + Amp. Plates were incubated in a BioTek Gen5 (BioTek Instruments, VT, USA) microplate reader at 37 °C with 3 min of shaking every 15 min; after 3–4 h, one of the wells for each clone received 0.5% arabinose (final concentration, to induce the PBAD promoter) and the other well received 0.2% glucose (final concentration, to further repress the PBAD promoter). The OD600 of each well was recorded every 15 min, for a total of 20 h. The fidelity of clones was confirmed by sequencing, and at least four independent clones were tested for each toxin gene.
Replication and stability assays
For functional analyses, the putative stability determinants from the three native plasmids of NCPPB 3335 (Table 1) were amplified by PCR with their own promoters, using specific primers, and cloned as BamHI fragments into the polylinker of vector pKMAG-C (construct 1 in Fig. 2). pKMAG-C replicates in E. coli through a p15a replicon and in pseudomonads through the cloned RepA-PFP replicon from pPsv48C [37]. The stability of these constructions, as well as that of the RepA-PFP and RepJ replicons from the pPsv48C plasmid and previously constructed chimeras [37], was tested after transformation into the plasmidless strain P. syringae pv. syringae B728a, essentially as described [77]. Briefly, transformants were grown overnight on LB plates with kanamycin, and twenty colonies per clone were collected and resuspended together in 500 μl of Ringer’s solution (1/4 strength; Oxoid, Basingstoke, UK). Serial dilutions were then plated on LB agar to get isolated colonies and, once developed, 100 colonies were picked to LB plates with and without kanamycin to determine the percentage of plasmid-containing colonies (KmR). The same procedure was followed to test these constructs in strain UPN912. The unstable cloning vector pKMAG-C was also included in the analyses as the baseline reference.
The stability of the minimal RepJ replicon [37], cloned into pKMAG (construct 4 in Fig. 2), was compared to that of plasmid pPsv48CΔ25, a naturally occurring 5.5 kb deletion derivate of pPsv48C that contains the RepJ replicon plus around 2 kb of downstream DNA, but no other maintenance systems. Both plasmids were maintained in strains derived from NCPPB 3335 and with no other native plasmids. Short-term stability was evaluated as stated above for strain B728a. For long-term stability, three independent LB cultures of each strain were started from single colonies and incubated at 25 °C with shaking and, after overnight growth, 10 μl of each culture were transferred to 3 ml of LB and incubated in the same conditions. We obtained LB plates containing 200–300 colonies both from the starting culture, immediately after single-colony inoculation, and after seven serial transfers in LB. These colonies were transferred to nylon membranes and analysed by colony hybridization [63], using an internal probe for repJ. The number of hybridizing colonies out of the total was scored to assess the prevalence of the RepJ replicon in both populations.
Inactivation of TA systems
To evaluate the role of TA systems on plasmid maintenance, we proceeded to their functional inactivation, by supplying in trans the cognate antitoxins cloned in the broad-host range vector pRK415; resulting in the neutralization of the toxin by the cloned antitoxin, as described [44]. Antitoxin genes PSPSV_A0043, PSPSV_A0032 and PSPSV_A0020 from pPsv48A were amplified by PCR with their own promoters, cloned into pGEM-T Easy, excised as BamHI or NcoI-SacI (for PSPSV_A0032) fragments, and sequentially cloned into the BamHI, NcoI-SacI and BglII sites of vector pME6041, respectively. Primers A1_R and TA3_F were used to amplify these three elements as a single fragment, which was cloned into pJET 2.1 (CloneJET PCR Cloning Kit, Thermo Scientific), excised as a BglII fragment and cloned into the BamHI site of pRK415, downstream of the constitutive Plac promoter in the vector, resulting in pRK3A. Essentially the same procedure was followed to clone in tandem and in this order, using primers A6_R and TA8_F, antitoxin genes PSPSV_C0050, PSPSV_C0008 and PSPSV_C0003 from pPsv48C into the vector pRK415, resulting in pRK3C. The integrity and fidelity of all clones was confirmed by nucleotide sequencing.
Statistical procedures
All data are given as the mean ± standard deviation (sd). Each experiment was repeated from three to six times, with three technical replicates for each of the conditions tested. Means were compared using an analysis of variance (ANOVA) followed, when needed, by Duncan’s multiple range test (p < 0.05). We used software R Project 3.3.3 (R Core Team (2017); Vienna, Austria) to perform the statistics.
Additional file
Table S1. List and application of primers used in this work. (PDF 186 kb)
Acknowledgements
We are indebted to Theresa Osinga for her help with the English language.
Funding
This work was funded by the Spanish Plan Nacional I + D + I grants AGL2014–53242-C2–1-R, AGL2014–53242-C2–2-R, AGL2017-82492-C2-1-R, and AGL2017-82492-C2-2-R from the Ministerio de Economía y Competitividad (MINECO), co-financed by the Fondo Europeo de Desarrollo Regional (FEDER). The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Availability of data and materials
All the data supporting the findings are presented in the manuscript and its supplementary information file. For raw data, please contact authors for data requests.
Abbreviations
- MITE
Miniature inverted-repeat transposable element
- PFP
pPT23A-family plasmids
- SD
Stability determinant
- sd
Standard deviation
- sucR
Resistant to 5% sucrose
- TA
Toxin-antitoxin system
Author’s contributions
LB and JM conceived the study and designed the experiments; LB, MA and ME performed the experiments; LB, CR, and JM analysed the data and interpreted the results; LB and JM drafted the manuscript with contributions from MA and CR; all authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Leire Bardaji, Email: leire.bardaji@unavarra.es.
Maite Añorga, Email: maite.anorga@unavarra.es.
Myriam Echeverría, Email: myriam.echeverria@unavarra.es.
Cayo Ramos, Email: crr@uma.es.
Jesús Murillo, Email: jesus.murillo@unavarra.es.
References
- 1.Vivian A, Murillo J, Jackson RW. The role of plasmids in phytopathogenic bacteria: mobile arsenals? Microbiology. 2001;147:763–780. doi: 10.1099/00221287-147-4-763. [DOI] [PubMed] [Google Scholar]
- 2.Sundin GW, Murillo J. Gene traders: characteristics of native plasmids from plant pathogenic bacteria. In: Jackson RW, editor. Plant pathogenic bacteria: genomics and molecular biology. Cambs: Caister: Academic Press; 2009. p. 295–310.
- 3.Norman A, Hansen LH, Sørensen SJ. Conjugative plasmids: vessels of the communal gene pool. Phil Trans R Soc B. 2009;364:2275–2289. doi: 10.1098/rstb.2009.0037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pilla G, Tang CM. Going around in circles: virulence plasmids in enteric pathogens. Nat Rev Microbiol. 2018;16:484–495. doi: 10.1038/s41579-018-0031-2. [DOI] [PubMed] [Google Scholar]
- 5.Kelly BG, Vespermann A, Bolton DJ. The role of horizontal gene transfer in the evolution of selected foodborne bacterial pathogens. Food Chem Toxicol. 2009;47:951–968. doi: 10.1016/j.fct.2008.02.006. [DOI] [PubMed] [Google Scholar]
- 6.Sundin GW. Genomic insights into the contribution of phytopathogenic bacterial plasmids to the evolutionary history of their hosts. Annu Rev Phytopathol. 2007;45:129–151. doi: 10.1146/annurev.phyto.45.062806.094317. [DOI] [PubMed] [Google Scholar]
- 7.Porse A, Schønning K, Munck C, Sommer MOA. Survival and evolution of a large multidrug resistance plasmid in new clinical bacterial hosts. Mol Biol Evol. 2016;33:2860–2873. doi: 10.1093/molbev/msw163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carattoli A. Plasmids and the spread of resistance. Int J Med Microbiol. 2013;303:298–304. doi: 10.1016/j.ijmm.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 9.San Millan A, MacLean RC. Fitness costs of plasmids: a limit to plasmid transmission. Microbiol Spectr. 2017;5:MTBP-0016-2017. doi: 10.1128/microbiolspec.MTBP-0016-2017. [DOI] [PubMed] [Google Scholar]
- 10.De Gelder L, Ponciano JM, Joyce P, Top EM. Stability of a promiscuous plasmid in different hosts: no guarantee for a long-term relationship. Microbiology. 2007;153:452–463. doi: 10.1099/mic.0.2006/001784-0. [DOI] [PubMed] [Google Scholar]
- 11.Sengupta M, Austin S. Prevalence and significance of plasmid maintenance functions in the virulence plasmids of pathogenic bacteria. Infect Immun. 2011;79:2502–2509. doi: 10.1128/IAI.00127-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mansfield J, Genin S, Magori S, Citovsky V, Sriariyanum M, Ronald P, et al. Top 10 plant pathogenic bacteria in molecular plant pathology. Mol Plant Pathol. 2012;13:614–629. doi: 10.1111/j.1364-3703.2012.00804.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sundin GW, Murillo J. Functional analysis of the Pseudomonas syringae rulAB determinant in tolerance to ultraviolet B (290-320 nm) radiation and distribution of rulAB among P. syringae pathovars. Environ Microbiol. 1999;1:75–88. doi: 10.1046/j.1462-2920.1999.00008.x. [DOI] [PubMed] [Google Scholar]
- 14.Jackson RW, Athanassopoulos E, Tsiamis G, Mansfield JW, Sesma A, Arnold DL, et al. Identification of a pathogenicity island, which contains genes for virulence and avirulence, on a large native plasmid in the bean pathogen Pseudomonas syringae pathovar phaseolicola. Proc Natl Acad Sci U S A. 1999;96:10875–10880. doi: 10.1073/pnas.96.19.10875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gutiérrez-Barranquero JA, Cazorla FM, de Vicente A, Sundin GW. Complete sequence and comparative genomic analysis of eight native Pseudomonas syringae plasmids belonging to the pPT23A family. BMC Genomics. 2017;18:365. doi: 10.1186/s12864-017-3763-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Murillo J, Keen NT. Two native plasmids of Pseudomonas syringae pathovar tomato strain PT23 share a large amount of repeated DNA, including replication sequences. Mol Microbiol. 1994;12:941–950. doi: 10.1111/j.1365-2958.1994.tb01082.x. [DOI] [PubMed] [Google Scholar]
- 17.Stavrinides J, Guttman D. Nucleotide sequence and evolution of the five-plasmid complement of the phytopathogen Pseudomonas syringae pv. maculicola ES4326. J Bacteriol. 2004;186:5101–15. [DOI] [PMC free article] [PubMed]
- 18.Bardaji L, Pérez-Martínez I, Rodríguez-Moreno L, Rodríguez-Palenzuela P, Sundin GW, Ramos C, et al. Sequence and role in virulence of the three plasmid complement of the model tumor-inducing bacterium Pseudomonas savastanoi pv. savastanoi NCPPB 3335. PLoS One. 2011;6:e25705. [DOI] [PMC free article] [PubMed]
- 19.Bardaji L, Añorga M, Jackson RW, Martínez-Bilbao A, Yanguas N, Murillo J. Miniature transposable sequences are frequently mobilized in the bacterial plant pathogen Pseudomonas syringae pv. phaseolicola. PLoS One. 2011;6:e25773. [DOI] [PMC free article] [PubMed]
- 20.Rohmer L, Kjemtrup S, Marchesini P, Dangl JL. Nucleotide sequence, functional characterization and evolution of pFKN, a virulence plasmid in Pseudomonas syringae pathovar maculicola. Mol Microbiol. 2003;47:1545–1562. doi: 10.1046/j.1365-2958.2003.03402.x. [DOI] [PubMed] [Google Scholar]
- 21.Alarcón-Chaidez FJ, Peñaloza-Vázquez A, Ullrich M, Bender CL. Characterization of plasmids encoding the phytotoxin coronatine in Pseudomonas syringae. Plasmid. 1999;42:210–220. doi: 10.1006/plas.1999.1424. [DOI] [PubMed] [Google Scholar]
- 22.Garcillán-Barcia MP, de la Cruz F. Distribution of IS91 family insertion sequences in bacterial genomes: evolutionary implications. FEMS Microbiol Ecol. 2002;42:303–313. doi: 10.1111/j.1574-6941.2002.tb01020.x. [DOI] [PubMed] [Google Scholar]
- 23.O'Brien HE, Thakur S, Guttman DS. Evolution of plant pathogenesis in Pseudomonas syringae: a genomics perspective. Annu Rev Phytopathol. 2011;49:269–289. doi: 10.1146/annurev-phyto-072910-095242. [DOI] [PubMed] [Google Scholar]
- 24.Pérez-Martínez I, Zhao Y, Murillo J, Sundin GW, Ramos C. Global genomic analysis of Pseudomonas savastanoi pv. savastanoi plasmids. J Bacteriol. 2008;190:625–635. doi: 10.1128/JB.01067-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Murillo J, Shen H, Gerhold D, Sharma AK, Cooksey DA, Keen NT. Characterization of pPT23B, the plasmid involved in syringolide production by Pseudomonas syringae pv. tomato PT23. Plasmid. 1994;31:275–287. doi: 10.1006/plas.1994.1029. [DOI] [PubMed] [Google Scholar]
- 26.Sundin GW, Demezas DH, Bender CL. Genetic and plasmid diversity within natural populations of Pseudomonas syringae with various exposures to copper and streptomycin bactericides. Appl Environ Microbiol. 1994;60:4421–4431. doi: 10.1128/aem.60.12.4421-4431.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sundin GW, Bender CL. Relative fitness in vitro and in planta of Pseudomonas syringae strains containing copper and streptomycin resistance plasmids. Can J Microbiol. 1994;40:279–285. [Google Scholar]
- 28.Vinatzer BA, Monteil CL, Clarke CR. Harnessing population genomics to understand how bacterial pathogens emerge, adapt to crop hosts, and disseminate. Annu Rev Phytopathol. 2014;52:19–43. doi: 10.1146/annurev-phyto-102313-045907. [DOI] [PubMed] [Google Scholar]
- 29.Morris CE, Monteil CL, Berge O. The life history of Pseudomonas syringae: linking agriculture to earth system processes. Annu Rev Phytopathol. 2013;51:85–104. doi: 10.1146/annurev-phyto-082712-102402. [DOI] [PubMed] [Google Scholar]
- 30.Gibbon MJ, Sesma A, Canal A, Wood JR, Hidalgo E, Brown J, et al. Replication regions from plant-pathogenic Pseudomonas syringae plasmids are similar to ColE2-related replicons. Microbiology. 1999;145:325–334. doi: 10.1099/13500872-145-2-325. [DOI] [PubMed] [Google Scholar]
- 31.Sundin GW, Mayfield CT, Zhao Y, Gunasekera TS, Foster GL, Ullrich MS. Complete nucleotide sequence and analysis of pPSR1 (72,601 bp), a pPT23A-family plasmid from Pseudomonas syringae pv. syringae A2. Mol Gen Genomics. 2004;270:462–76. [DOI] [PubMed]
- 32.Joardar V, Lindeberg M, Jackson RW, Selengut J, Dodson R, Brinkac LM, et al. Whole-genome sequence analysis of Pseudomonas syringae pv. phaseolicola 1448A reveals divergence among pathovars in genes involved in virulence and transposition. J Bacteriol. 2005;187:6488–98. [DOI] [PMC free article] [PubMed]
- 33.Ramos C, Matas IM, Bardaji L, Aragón IM, Murillo J. Pseudomonas savastanoi pv. savastanoi: some like it knot. Mol Plant Pathol. 2012;13:998–1009. doi: 10.1111/j.1364-3703.2012.00816.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Caballo-Ponce E, Murillo J, Martínez-Gil M, Moreno-Pérez A, Pintado A, Ramos C. Knots untie: molecular determinants involved in knot formation induced by Pseudomonas savastanoi in woody hosts. Front Plant Sci. 2017;8:1089. doi: 10.3389/fpls.2017.01089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Añorga M. Biología de plásmidos y dinámica génica en el complejo Pseudomonas syringae [Ph. D. Thesis] Pamplona: Universidad Pública de Navarra; 2017. [Google Scholar]
- 36.Castañeda-Ojeda MP, Moreno-Pérez A, Ramos C, López-Solanilla E. Suppression of plant immune responses by the Pseudomonas savastanoi pv. savastanoi NCPPB 3335 type III effector tyrosine phosphatases HopAO1 and HopAO2. Front Plant Sci. 2017;8:680. [DOI] [PMC free article] [PubMed]
- 37.Bardaji L, Añorga M, Ruiz-Masó JA, del Solar G, Murillo J. Plasmid replicons from Pseudomonas are natural chimeras of functional, exchangeable modules. Front Microbiol. 2017;8:190. doi: 10.3389/fmicb.2017.00190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hanekamp T, Kobayashi D, Hayes S, Stayton MM. Avirulence gene D of Pseudomonas syringae pv. tomato may have undergone horizontal gene transfer. FEBS Lett. 1997;415:40–44. doi: 10.1016/s0014-5793(97)01089-2. [DOI] [PubMed] [Google Scholar]
- 39.Buell CR, Joardar V, Lindeberg M, Selengut J, Paulsen IT, Gwinn ML, et al. The complete genome sequence of the Arabidopsis and tomato pathogen Pseudomonas syringae pv. tomato DC3000. Proc Natl Acad Sci U S A. 2003;100:10181–10186. doi: 10.1073/pnas.1731982100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nordström K, Austin SJ. Mechanisms that contribute to the stable segregation of plasmids. Annu Rev Genet. 1989;23:37–69. doi: 10.1146/annurev.ge.23.120189.000345. [DOI] [PubMed] [Google Scholar]
- 41.Stalder T, Rogers LM, Renfrow C, Yano H, Smith Z, Top EM. Emerging patterns of plasmid-host coevolution that stabilize antibiotic resistance. Sci Rep. 2017;7:4853. [DOI] [PMC free article] [PubMed]
- 42.Yano H, Wegrzyn K, Loftie-Eaton W, Johnson J, Deckert GE, Rogers LM, et al. Evolved plasmid-host interactions reduce plasmid interference cost. Mol Microbiol. 2016;101:743-56. [DOI] [PMC free article] [PubMed]
- 43.Flores M, Brom S, Stepkowski T, Girard ML, Dávila G, Romero D, et al. Gene amplification in Rhizobium: identification and in vivo cloning of discrete amplifiable DNA regions (amplicons) from Rhizobium leguminosarum biovar phaseoli. Proc Natl Acad Sci U S A. 1993;90:4932–4936. doi: 10.1073/pnas.90.11.4932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kopfmann S, Roesch S, Hess W. Type II toxin–antitoxin systems in the unicellular cyanobacterium Synechocystis sp. PCC 6803. Toxins. 2016;8:228. doi: 10.3390/toxins8070228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Summers DK. The biology of plasmids. Osney, Oxford OX; Cambridge, Mass.: Blackwell Science; 1996. ix, 157 p. p.
- 46.Garcillán-Barcia MP, Bernales I, Mendiola MV, de la Cruz F. IS91 rolling-circle transposition. In: Craig NL, Craigie R, Gellert M, Lambowitz AM, editors. Mobile DNA II. Washington, D.C.: ASM Press; 2002. p. 891–904.
- 47.Richter GY, Björklöf K, Romantschuk M, Mills D. Insertion specificity and trans-activation of IS801. Mol Gen Genet. 1998;260:381–387. doi: 10.1007/s004380050907. [DOI] [PubMed] [Google Scholar]
- 48.Sesma A, Aizpún MT, Ortiz A, Arnold D, Vivian A, Murillo J. Virulence determinants other than coronatine in Pseudomonas syringae pv. tomato PT23 are plasmid-encoded. Physiol Mol Plant Pathol. 2001;58:83–93. [Google Scholar]
- 49.Sesma A, Sundin GW, Murillo J. Phylogeny of the replication regions of pPT23A-like plasmids from Pseudomonas syringae. Microbiology. 2000;146:2375–2384. doi: 10.1099/00221287-146-10-2375. [DOI] [PubMed] [Google Scholar]
- 50.Ma Z, Smith JJ, Zhao Y, Jackson RW, Arnold DL, Murillo J, et al. Phylogenetic analysis of the pPT23A plasmid family of Pseudomonas syringae. Appl Environ Microbiol. 2007;73:1287–1295. doi: 10.1128/AEM.01923-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Díaz-Orejas R, Espinosa M, Yeo CC. The importance of the expendable: toxin–antitoxin genes in plasmids and chromosomes. Front Microbiol. 2017;8:1479. doi: 10.3389/fmicb.2017.01479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pilla G, McVicker G, Tang CM. Genetic plasticity of the Shigella virulence plasmid is mediated by intra- and inter-molecular events between insertion sequences. PLoS Genet. 2017;13:e1007014. doi: 10.1371/journal.pgen.1007014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim JF, Charkowski AO, Alfano JR, Collmer A, Beer SV. Sequences related to transposable elements and bacteriophages flank avirulence genes of Pseudomonas syringae. Mol Plant-Microbe Interact. 1998;11:1247–1252. [Google Scholar]
- 54.Arnold DL, Jackson RW, Fillingham AJ, Goss SC, Taylor JD, Mansfield JW, et al. Highly conserved sequences flank avirulence genes: isolation of novel avirulence genes from Pseudomonas syringae pv. pisi. Microbiology. 2001;147:1171–82. [DOI] [PubMed]
- 55.Jackson RW, Vinatzer B, Arnold DL, Dorus S, Murillo J. The influence of the accessory genome on bacterial pathogen evolution. Mob Genet Elements. 2011;1:55–65. doi: 10.4161/mge.1.1.16432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rhodes G, Bosma H, Studholme D, Arnold DL, Jackson RW, Pickup RW. The rulB gene of plasmid pWW0 is a hotspot for the site-specific insertion of integron-like elements found in the chromosomes of environmental Pseudomonas fluorescens group bacteria. Environ Microbiol. 2014;16:2374–2388. doi: 10.1111/1462-2920.12345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hülter N, Ilhan J, Wein T, Kadibalban AS, Hammerschmidt K, Dagan T. An evolutionary perspective on plasmid lifestyle modes. Curr Opin Microbiol. 2017;38:74–80. doi: 10.1016/j.mib.2017.05.001. [DOI] [PubMed] [Google Scholar]
- 58.Shidore T, Triplett LR. Toxin-antitoxin systems: implications for plant disease. Annu Rev Phytopathol. 2017;55:161–179. doi: 10.1146/annurev-phyto-080516-035559. [DOI] [PubMed] [Google Scholar]
- 59.McCann HC, Rikkerink EHA, Bertels F, Fiers M, Lu A, Rees-George J, et al. Genomic analysis of the kiwifruit pathogen Pseudomonas syringae pv. actinidiae provides insight into the origins of an emergent plant disease. PLoS Pathog. 2013;9:e1003503. [DOI] [PMC free article] [PubMed]
- 60.Szekeres S, Dauti M, Wilde C, Mazel D, Rowe-Magnus DA. Chromosomal toxin–antitoxin loci can diminish large-scale genome reductions in the absence of selection. Mol Microbiol. 2007;63:1588–1605. doi: 10.1111/j.1365-2958.2007.05613.x. [DOI] [PubMed] [Google Scholar]
- 61.Martins PMM, Merfa MV, Takita MA, De Souza AA. Persistence in phytopathogenic bacteria: do we know enough? Front Microbiol. 2018;9:1099. doi: 10.3389/fmicb.2018.01099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Slayden RA, Dawson CC, Cummings JE. Toxin–antitoxin systems and regulatory mechanisms in Mycobacterium tuberculosis. Pathog Dis. 2018;76:fty039. [DOI] [PubMed]
- 63.Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. cold Spring Harbor, NY: cold Spring Harbor. Laboratory. 1989.
- 64.Holmes DS, Quigley M. A rapid boiling method for the preparation of bacterial plasmids. Anal Biochem. 1981;114:193–197. doi: 10.1016/0003-2697(81)90473-5. [DOI] [PubMed] [Google Scholar]
- 65.Choi KH, Kumar A, Schweizer HP. A 10-min method for preparation of highly electrocompetent Pseudomonas aeruginosa cells: application for DNA fragment transfer between chromosomes and plasmid transformation. J Microbiol Meth. 2006;64:391–397. doi: 10.1016/j.mimet.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 66.Hubbard TJ, Aken BL, Ayling S, Ballester B, Beal K, Bragin E, et al. Ensembl 2009. Nucleic Acids Res. 2008;37:D690–D6D7. doi: 10.1093/nar/gkn828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Corpet F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988;16:10881–10890. doi: 10.1093/nar/16.22.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mitchell A, Chang H-Y, Daugherty L, Fraser M, Hunter S, Lopez R, et al. The InterPro protein families database: the classification resource after 15 years. Nucleic Acids Res. 2015;43:D213–DD21. doi: 10.1093/nar/gku1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Carver T, Berriman M, Tivey A, Patel C, Bohme U, Barrell BG, et al. Artemis and ACT: viewing, annotating and comparing sequences stored in a relational database. Bioinformatics. 2008;24:2672–2676. doi: 10.1093/bioinformatics/btn529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, et al. Primer3—new capabilities and interfaces. Nucleic Acids Res. 2012;40:e115. doi: 10.1093/nar/gks596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schäfer A, Tauch A, Jager W, Kalinowski J, Thierbach G, Pühler A. Small mobilizable multipurpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene. 1994;145:69–73. doi: 10.1016/0378-1119(94)90324-7. [DOI] [PubMed] [Google Scholar]
- 72.Matas IM, Pilar Castaneda-Ojeda M, Aragon IM, Antunez-Lamas M, Murillo J, Rodriguez-Palenzuela P, et al. Translocation and functional analysis of Pseudomonas savastanoi pv. savastanoi NCPPB 3335 type III secretion system effectors reveals two novel effector families of the Pseudomonas syringae complex. Mol Plant-Microbe Interact. 2014;27:424–36. [DOI] [PubMed]
- 73.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 74.Tao L, Jackson RE, Cheng Q. Directed evolution of copy number of a broad host range plasmid for metabolic engineering. Metab Eng. 2005;7:10–17. doi: 10.1016/j.ymben.2004.05.006. [DOI] [PubMed] [Google Scholar]
- 75.Sevin EW, Barloy-Hubler F. RASTA-Bacteria: a web-based tool for identifying toxin-antitoxin loci in prokaryotes. Genome Biol. 2007;8:R155. doi: 10.1186/gb-2007-8-8-r155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Guzman L-M, Belin D, Carson MJ, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol. 1995;177:4121-30. [DOI] [PMC free article] [PubMed]
- 77.López-Villarejo J, Diago-Navarro E, Hernández-Arriaga AM, Díaz-Orejas R. Kis antitoxin couples plasmid R1 replication and parD (kis,kid) maintenance modules. Plasmid. 2012;67:118–127. doi: 10.1016/j.plasmid.2011.12.015. [DOI] [PubMed] [Google Scholar]
- 78.Rodríguez-Palenzuela P, Matas I, Murillo J, López-Solanilla E, Bardaji L, Pérez-Martínez I, et al. Annotation and overview of the Pseudomonas savastanoi pv. savastanoi NCPPB 3335 draft genome reveals the virulence gene complement of a tumour-inducing pathogen of woody hosts. Environ Microbiol. 2010;12:1604–20. [DOI] [PubMed]
- 79.Feil H, Feil WS, Chain P, Larimer F, DiBartolo G, Copeland A, et al. Comparison of the complete genome sequences of Pseudomonas syringae pv. syringae B728a and pv. tomato DC3000. Proc Natl Acad Sci. 2005;102:11064–9. [DOI] [PMC free article] [PubMed]
- 80.Heeb S, Itoh Y, Nishijyo T, Schnider U, Keel C, Wade J, et al. Small, stable shuttle vectors based on the minimal pVS1 replicon for use in gram-negative, plant-associated bacteria. Mol Plant-Microbe Interact. 2000;13:232–237. doi: 10.1094/MPMI.2000.13.2.232. [DOI] [PubMed] [Google Scholar]
- 81.Keen NT, Tamaki S, Kobayashi D, Trollinger D. Improved broad-host-range plasmids for DNA cloning in gram-negative bacteria. Gene. 1988;70:191–197. doi: 10.1016/0378-1119(88)90117-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. List and application of primers used in this work. (PDF 186 kb)
Data Availability Statement
All the data supporting the findings are presented in the manuscript and its supplementary information file. For raw data, please contact authors for data requests.