Abstract
Background
Malignant pleural mesothelioma (MPM) is a tumour arising from pleural cavities with poor prognosis. Multimodality treatment with pemetrexed combined with cisplatin shows unsatisfying response-rates of 40%. The reasons for the rather poor efficacy of chemotherapeutic treatment are largely unknown. However, it is conceivable that DNA repair mechanisms lead to an impaired therapy response. We hypothesize a major role of homologous recombination (HR) for genome stability and survival of this tumour. Therefore, we analysed genes compiled under the term “BRCAness”. An inhibition of this pathway with olaparib might abrogate this effect and induce apoptosis.
Methods
We investigated the response of three MPM cell lines and lung fibroblasts serving as a control to treatment with pemetrexed, cisplatin and olaparib. Furthermore, we aimed to find possible correlations between response and gene expression patterns associated with BRCAness phenotype. Therefore, 91 clinical MPM samples were digitally screened for gene expression patterns of HR members.
Results
A BRCAness-dependent increase of apoptosis and senescence during olaparib-based treatment of BRCA-associated-protein 1 (BAP1)-mutated cell lines was observed. The gene expression pattern identified could be found in approx. 10% of patient samples. Against this background, patients could be grouped according to their defects in the HR system. Gene expression levels of Aurora Kinase A (AURKA), RAD50 as well as DNA damage-binding protein 2 (DDB2) could be identified as prognostic markers in MPM.
Conclusions
Defects in HR compiled under the term BRCAness are a common event in MPM. The present data can lead to a better understanding of the underlaying cellular mechanisms and leave the door wide open for new therapeutic approaches for this severe disease with infaust prognosis. Response to Poly (ADP-ribose)-Polymerase (PARP)-Inhibition could be demonstrated in the BAP1-mutated NCI-H2452 cells, especially when combined with cisplatin. Thus, this combination therapy might be effective for up to 2/3 of patients, promising to enhance patients’ clinical management and outcome.
Electronic supplementary material
The online version of this article (10.1186/s12885-019-5314-0) contains supplementary material, which is available to authorized users.
Keywords: Malignant pleural mesothelioma - overall survival, PARP1, BRCAness - BAP1, Olaparib
Background
Malignant pleural mesothelioma (MPM) is a highly aggressive tumour arising from the pleural cavities [1, 2]. Despite treatment, MPM patients have poor prognosis with a median survival of approximately 12 months [3–5]. The state-of-the-art systemic treatment of unresectable and advanced MPM is chemotherapy with a combination of cisplatin and pemetrexed [6, 7]. However, even with aggressive treatment approaches, recurrence or progression occurs in most cases as a result of chemotherapy resistance [6, 8, 9].
The term “BRCAness” is defined as a defect in double-strand break repair (DSBR) of DNA by the homologous recombination repair (HRR) pathway [2, 10].
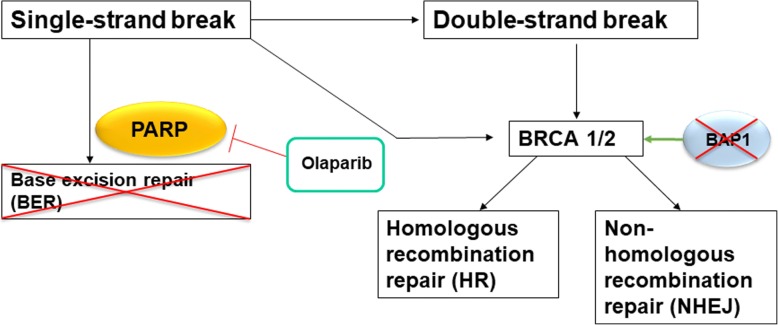
HRR is involved in the repair of DNA lesions blocking the replication fork or inducing double-strand breaks (DSBs) [10]. Alterations in various genes, associated to BRCAness phenotype, were assessed in several tumours. Interestingly, BAP1 loss-of-function mutation has been found in 26–64% of MPMs [11]. BRCAness leads to genomic instability and therefore could make the tumour more susceptible to different chemotherapeutics targeting these features [12, 13]. Furthermore, alternative repair mechanisms could overcome the lack of HRR and tumour cells could evade apoptosis. Poly (ADP-ribose)-Polymerase (PARP) is essential for base excision repair (BER) and non-homologous end joining (NHEJ) and may be a target to inhibit alternative repair mechanisms in case of BRCAness [14] (Fig. 1).
Fig. 1.
Enhanced base excision repair, due to defective HRR that is caused by BRCAness phenotype, increases the reliance on PARP1. It is suggested that loss-of-function mutation of BAP1 also results in BRCAness phenotype. Inhibition of PARP1 prevents the alternative repair pathway and thus could lead to apoptosis of the cell
PARP-inhibitors are already FDA approved drugs for treatment of several cancers with Breast Cancer 1 (BRCA1) or BRCA2 mutations [15]. We hypothesize, that not BRCA1/2 mutations are exclusively necessary for efficient PARP-inhibition therapy. We hypothesize, that PARP-inhibitors might also be effective in cancers with alterations in related genes of the whole homologous recombination repair pathway. In addition, a combination of platinum-based chemotherapy with PARP-inhibitors is assumed to be more effective than PARP-inhibitors alone [12].
The present study was designed to
Find predictive gene expression patterns to olaparib treatment, based on HRR key players. Analyse different human MPM cell lines for the presence of defects in HRR pathway. Furthermore, we investigated if MPM cell lines shows sensitivity against PARP-inhibition. In this case, we also aimed to find gene expression patterns predictive for treatment with PARP-inhibitors.
Define patients with altered HRR based on the results of the MPM cell lines analysed in 1) and validate the potential predictive gene expression pattern associated with response to PARP-inhibitors in clinical specimens.
Investigate associations between patients’ survival prognosis and response to cisplatin, respectively, in association with HRR key players. Based on this study, a preselection of patients with a beneficial response to olaparib treatment might be done.
Methods
Study design
Finding predictive gene expression patterns, based on HRR key players
We investigated the response of MPM cell lines and lung fibroblasts to treatment with pemetrexed, cisplatin and olaparib. The control lung fibroblast cell line (MRC-5) and MPM cell lines: MSTO-211H, NCI-H2052 and BAP1-mutant NCI-H2452 were used for the cell culture experiments. Either single agent olaparib, cisplatin and pemetrexed, or cisplatin in combination with either olaparib or pemetrexed, was added to the cells. Response to treatment was assessed by using three luminescent-based assays detecting apoptosis, necrosis and senescence of cell lines. In addition, we aimed to find possible correlations between response and gene expression patterns associated with BRCAness phenotype.
Finding patients with apparent predictive expression patterns
For determination of BRCAness phenotypes, RNA of the cell lines and 90 formalin-fixed, paraffin-embedded (FFPE) patient samples, was isolated and used for digital gene expression analysis of genes listed in Table 1. Correlation tests were used to determine response associated patterns in cell lines. These results were compared with the gene expression pattern of the patient cohort.
Table 1.
List of genes for gene express ion analysis. The short name and full name of the genes as well as their function are listed [40]
| Gene | Official Full Name | Function (40) |
|---|---|---|
| BAP1 | BRCA1 Associated Protein 1 | Promotes DSB repair |
| BRCA1 | Breast Cancer 1 | Cell cycle checkpoints activation, resection of 5′ ends of the DSB, necessary for RAD51 function |
| BRCA2 | Breast Cancer 2 | Localizes RAD51 to the DSB |
| PARP1 | Poly (ADP-Ribose) Polymerase 1 | Initiates SSB repair |
| RAD51 | RAD51 Recombinase | RAD 51 activity allows DNA to invade homologous double helix serving as template |
| RAD50 | RAD50 double strand break repair protein | Forming of the MRN complex |
| ATM | ATM serine/threonine kinase | Detection of DSB |
| ATR | ATR serine/threonine kinase | Detection of DSB |
| PALB2 | Partner and localizer of BRCA2 | Localizes BRCA2 to the DSB |
| BARD1 | BRCA1 associated RING domain 1 | Binding partner of BAP1 |
| EMSY | BRCA2 interacting transcriptional repressor | Transcription regulator that interacts with BRCA2 |
| DDB2 | Damage specific DNA binding protein 2 | Required for DNA binding in DNA damage repair |
| BACH1 | BTB domain and CNC homolog 1 | Transcriptional regulator that acts as repressor or activator |
| MRE11 | MRE11 homolog, double strand break repair nuclease | Forming of the MRN complex |
| NBN | nibrin | Forming of the MRN complex |
| AURKA | Aurora kinase A | Kinase in cell-cycle, is involved in microtubule formation and stabilization at spindle pole during chromosome segregation |
| FANCD2 | Fanconi anaemia complementation group D2 | Involved in HRR, is monoubiquinated in response to DNA damage |
| BRIP1 | BRCA1 interacting protein C-terminal helicase 1 | Involved in HHR by interaction with BRCA1 |
| CHEK2 | Checkpoint kinase 2 | Cell cycle checkpoint regulator and putative tumour suppressor |
| RPA1 | Replication protein A1 | Activates ATR |
| GUSB | Glucuronidase beta | Normalization for digital expression analysis |
| POLR1B | RNA polymerase I subunit B | Normalization for digital expression analysis |
| TUBB | Tubulin beta class I | Normalization for digital expression analysis |
| PGK1 | Phosphoglycerate kinase 1 | Normalization for digital expression analysis |
Investigation of associations between survival prognosis of patients and response to cisplatin
We aimed to find associations between defects in HRR and response to cisplatin using survival prognosis with the help of the results of the digital gene expression analysis (Table 1).
Patient cohort
Specimens
90 FFPE specimens from MPM patients were collected from the Department of Pathology, Helios hospital Emil von Behring (Berlin, Germany) (46 specimens) and of the Institute of Pathology, university hospital Essen (Germany) (44 specimens)
Tumour classification
Tumour classification is based on the WHO classification of tumours guidelines (2015) [16]. TNM-staging is based on the Union internationale contre le cancer (UICC) classification of malignant tumours [17]. All samples were confirmed by two experienced pathologists (JW, TM).
Eligibility criteria
The study included MPM patients, treated at the West German Cancer Centre or the West German Lung Centre (Essen) between 2006 and 2009 and the Helios Hospital Emil von Behring (Berlin) between 2002 and 2009. Inclusion criteria were the availability of sufficient tumour material and the case to be listed in the clinical registry for tumour response and survival with a complete set of data concerning follow-up and treatment. Each patient underwent first-line chemotherapy regimen consisting of cisplatin and pemetrexed.
The study was conducted retrospectively to identify gene expression-based biomarkers. It was approved by the institutional ethics review board (Ethics Committee of the Medical Faculty of the University Duisburg-Essen, identifier: 14–5775-BO). The investigations conform to the principles of the declaration of Helsinki.
Clinical and pathological data
Response to chemotherapy was determined radiologically according to modRECIST [18]. Response was classified as complete response (CR), partial response (PR), stable disease (SD) or progressive disease (PD). Remission was classified as CR or PR vs. SD or PD. Progression was classified as CR or PR or SD vs. PD. Progression-free survival was calculated from start of treatment until first radiological progression (modRECIST). Overall survival was determined from initial diagnosis until death or loss of follow up. Collection of all specimens was performed prior to systemic treatment. Surveillance for this study was stopped on August 31, 2014. General patient data are summarized in Table 2.
Table 2.
Summary of general statistical patient data
| Number of Patients | |
|---|---|
| Histology | |
| Biphasic | 7 |
| Epithelioid | 73 |
| Sarcomatoid | 6 |
| Unknown | 5 |
| Age [Years] | |
| Minimum | 34.57 |
| Median | 64.86 |
| Mean | 64.26 |
| Maximum | 81.76 |
| Unknown | 4 |
| Time to death [month] | |
| Minimum | 0.77 |
| Median | 17.33 |
| Mean | 21.69 |
| Maximum | 81.73 |
| Unknown | 4 |
| Clinical outcome | |
| Alive | 9 |
| Dead | 78 |
| Unknown | 4 |
Cell culture
MPM cell lines MSTO-211H (biphasic subtype, pemetrexed-sensitive) and NCI-H2052 (epithelioid subtype, cisplatin-sensitive) as well as the cell line NCI-H2452 (BAP1-mutant, sarcomatoid subtype) were cultured in Roswell Park Memorial Institute (RPMI) -1640 medium (Thermo Fisher Scientific, Massachusetts), USA. The human lung-fibroblast cell line MRC-5 was used as control cell line. MRC-5 cells were cultured in Minimal Essential medium (Thermo Fisher Scientific). All culture media were supplemented with 10% foetal calf serum and 1% penicillin and streptomycin (Thermo Fisher Scientific).
Treatment of MPM cell lines with cytostatic agents cisplatin and Pemetrexed or with PARP-inhibitor Olaparib
For the treatment of cells, 5000 cells/well were used. The concentrations of the agents were 0.25 μM for pemetrexed (Selleckchem, Houston, USA) and 10 μM for cisplatin (Selleckchem). To evaluate the most efficient concentration for olaparib (Selleckchem), a dilution series comprising 0.1, 0.5, 1, 5, and 10 μM was applied. In addition to single-agent-treatment, cisplatin was combined with pemetrexed as well as 0.5 μM, 1 μM and 5 μM of olaparib to identify synergistic effects.
Cell state analysis
5000 cells per reaction were applied to detect apoptosis, senescence and necrosis. All reactions were measured using a luminometer (Glo Max Multi + Detection System; Promega).
Senescence was analyzed using the CellTiter-Glo® Luminescent Cell Viability Assay kit (Promega, Wisconsin, USA). 10 μl of Digitonin (30 μg/ml), added to the cells in a separate well 15 min before cell lysis, served as positive control to measure a decrease of cellular viability of 100%.
Necrosis was analysed using the CytoTox-Glo® Assay kit (Promega). Ionomycin (Selleckchem) was used for positive control. Two hours before measurement, 50 μl of Ionomycin (100 μM), was added to the cells in a separate well. After adding 50 μl of the AAF-Glo® reagent to each well, cells were incubated for 15 min at room temperature, protected from light.
The apoptotic potential of the cells was analysed using the Caspase-Glo® 3/7 Assay (G8093, Promega). 100 μl of required cells/well were placed into a white 96-well plate. Staurosporine (10 μM, Selleckchem) served as positive control and was given to the cells in a separate well 4 h before measurement. After adding 100 μl of Caspase-Glo® reagent to each well, cells were incubated for 30 min at room temperature.
Changes in cell state were calculated as percentage of signal gained by the positive control normalized to the baseline (untreated cells).
RNA isolation from eukaryotic cells and FFPE patient samples using the automated Maxwell system
RNA-isolation of 1 × 106 cells per sample was performed by using the Maxwell purification platform with appertaining reagents (Maxwell RSC simplyRNA Cells Kit, Promega).
RNA-purification of FFPE specimens was performed by using the Maxwell RSC RNA FFPE Kit (Promega).
The concentration of RNA was determined via fluorometric quantification (Qubit, Thermo Scientific) using the RNA Broad range assay kit according to the manufacturer’s instructions. 1 μl of each isolated RNA sample was applied for measurement.
Gene expression analysis using the NanoString PlexSet assay
The multiplexed digital gene expression assay (PlexSet, NanoString, Seattle, USA) was used for the investigation of BRCAness phenotype gene expression patterns.
PlexSet assay was performed according to the manufacturer’s instructions. 150 ng FFPE derived RNA from patients suffering from MPM and 70 ng of RNA freshly isolated from cell lines MRC-5, MSTO-211H, NCI-H2052 and NCI-H2452 were applied. Hybridization reaction was executed for 18 h. nCounter Prep-Station processing was performed using the high-sensitivity protocol. Cartridges were scanned on the Digital Analyzer (NanoString) with maximal sensitivity (555 fields of view (FOV)). Investigated genes in samples with counts < 100 are considered as not expressed.
Statistical analysis
For statistical and graphical analyses, the R statistical programming environment (v3.2.3) was used.
Nanostring data processing has been performed as described previously [19, 20]. In detail, Nanostring counts for each gene underwent technical normalization, based on positive controls included in a code set. Subsequently, biological normalization has been performed by calculating a normalization factor for each sample out of the geometric mean of the included mRNA reference genes.
Additionally, all counts with p > 0.05 after one-sided t-test versus negative controls plus 2x standard deviations were interpreted as not expressed to overcome basal noise.
Statistical analysis has been performed as described elsewhere [21]. For exploratory data analysis of dichotomous variables either the Wilcoxon Mann-Whitney rank sum test (non-parametric) or two-sided students t-test (parametric) was applied. For ordinal variables with more than two groups, either the Kruskal-Wallis test (non-parametric) or analysis of variance (ANOVA) (parametric) was used to detect group differences.
Double dichotomous contingency tables were analysed using Fisher’s Exact test. For more than two groups, the dependency of ranked parameters was calculated by using the Pearson’s Chi-squared test. Correlations between metric variables were tested by using the Spearman’s rank correlation test as well as the Pearson’s product moment correlation coefficient for linear modelling.
To further specify the different candidate pattern, each unsupervised and supervised clustering to overcome commonalities as well as principal component analysis to overcome differences were performed.
For the assessment of associations between gene expression and progression-free survival (PFS) or overall survival (OS), Kaplan-Meier analysis was performed. Significant differences in PFS or OS between tested groups were determined by using the COXPH-model. Therefore, the Wald-test, likelihood-ratio test and the Score (log rank) test were used.
P-values were adjusted by using the false discovery rate (FDR) with a subsequently defined level of statistical significance of p ≤ 0.05.
Results
Treatment of MPM cell lines with cytostatic agents
The human lung-fibroblast cell line MRC-5, BAP1wt/wt MPM cell lines MSTO-211H and NCI-H2052, as well as the BAP1 mutant MPM cell line NCI-H2452 were analysed for apoptosis, senescence and necrosis during treatment with pemetrexed, cisplatin and olaparib.
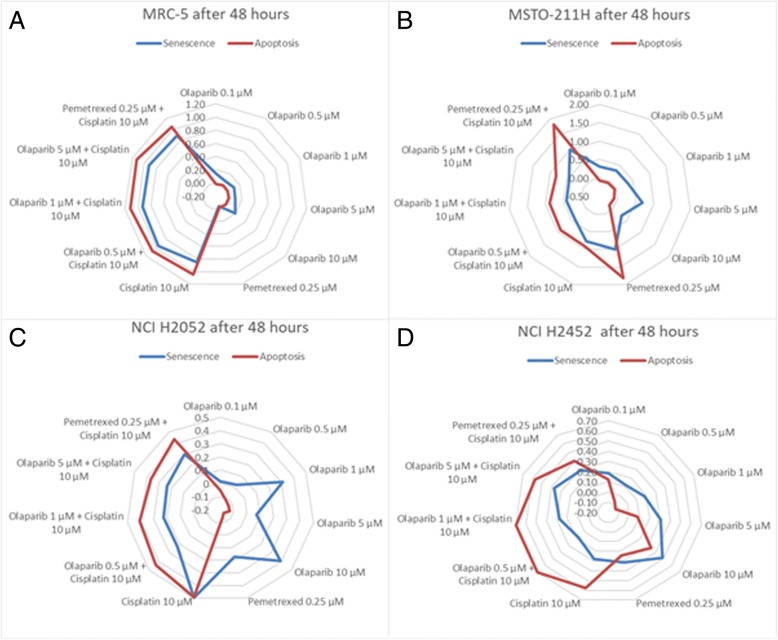
While cell lines MRC-5 and MSTO-211H showed a strong induction of apoptosis and senescence by treatment with cisplatin or pemetrexed, olaparib did not have any apoptotic effect on these cells. NCI-H2052 and NCI-H2452 cells showed a notably lower induction of apoptosis than MRC-5 or MSTO-211H cells. The treatment of NCI-H2052 cells with 1 μM and 10 μM single-agent olaparib showed 30–50% induction of senescence. In BAP1-mutant NCI-H2452, 0.5 μM and 1 μM olaparib combined with 10 μM cisplatin induced the highest apoptotic effect with low induction of senescence (Fig. 2).
Fig. 2.
Senescence and apoptosis rate of cell lines after 48 h of incubation. a: The effect of both senescence and apoptosis is comparative in MRC-5. b: In MSTO-211H, the sharp increase of apoptotic effect of 181% of pemetrexed alone and in combination with cisplatin is illustrated, while senescence showed an effect of 100%. The well treated with 10 μM olaparib showed a senescence effect of 80%, while no apoptotic effect was detected. c: NCI H2052 cells showed apoptotic effects only in wells treated with cisplatin or in combination with pemetrexed or olaparib and senescence of 30–50%. Wells treated with 1 μM and 10 μM olaparib showed 40% of senescence, while no apoptotic effect was detected. d: NCI H2452 cells showed 70% of apoptosis and only 20% of senescence in wells treated with 0.5 μM, 1 μM or 10 μM olaparib combined with cisplatin. 10 μM Olaparib alone showed 15% higher senescence than apoptosis
No induction of necrosis could be observed with neither of the used agents, except for the MSTO-211H cells (data not shown). Especially, wells treated with pemetrexed as single agent or in combination showed a necrotic effect.
BRCAness mRNA marker profiling
Differences in gene expression patterns
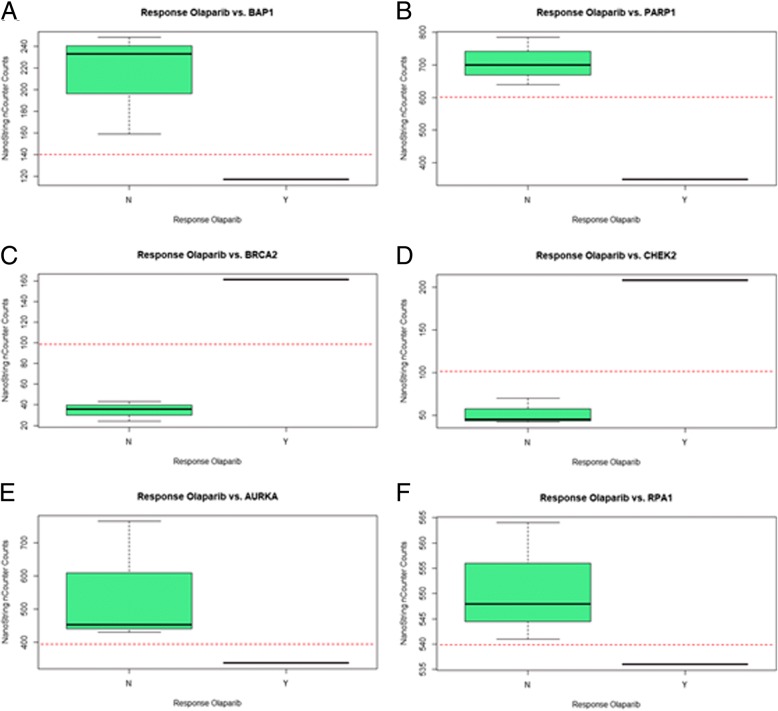
Differences in gene expression patterns of each cell line with respect to response during olaparib-treatment were observed. The only cell line showing response to olaparib was the BAP1-mutant NCI-H2452. AURKA, replication protein A1 (RPA1), BAP1 and PARP1 were significantly lower or not expressed in NCI-H2452, compared to other cell lines (Fig. 3 a, b, e and f). BRCA2 and checkpoint kinase2 (CHEK2) were expressed in NCI-H2452, while other cell lines showed no expression with counts < 100 (Fig. 3 c and d).
Fig. 3.
Comparison of significant differences in gene expression level between cell lines with respect to response to treatment with olaparib. Red dotted lines were placed and may represent thresholds between gene expression patterns leading to response to olaparib treatment or not. a: BAP1 was rarely expressed in the cell line that showed response to olaparib treatment (NCI H2452), while it was expressed in other cell lines that showed no response (160 to 250 counts). Threshold was set at 140 counts. b: PARP1 was expressed in NCI H2452 with 350 counts, but other cell lines showed significantly higher expression patterns with 640–785 counts. Threshold was set at 600 counts. c/d: BRCA2 and CHEK2 are expressed in the cell line that showed response to olaparib (NCI H2452), while no expression was detected in cell lines that showed no response. Thresholds were set at 100 counts for BRCA2 and CHEK2. e/f: AURKA and RPA1 are more expressed in cell lines that showed no response, than in NCI H2452. Thresholds were set at 400 counts for AURKA and 540 counts for RPA1
The expression pattern of these genes in BAP1-mutant NCI-H2452 cells could be found in approximately 10% of patient samples (Additional file 1: Table S1).
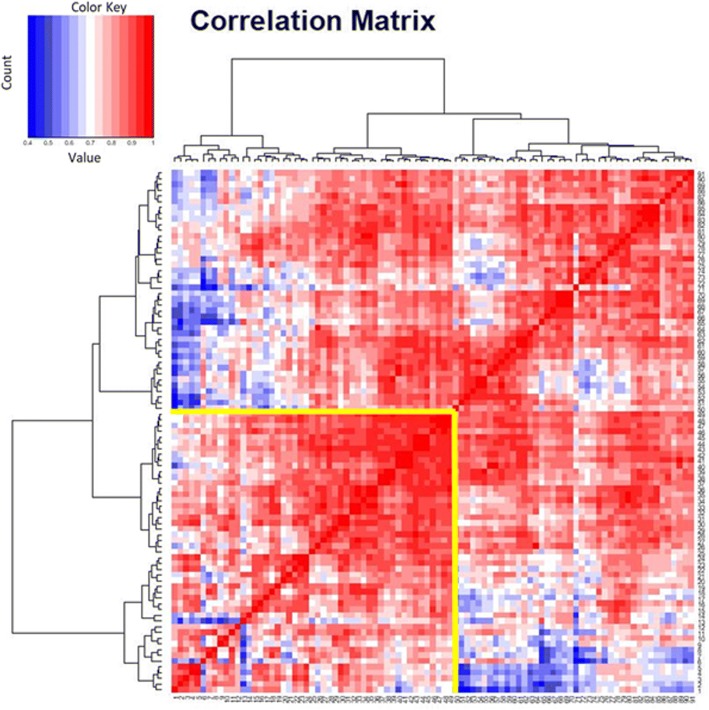
Correlation between samples
Overall correlation pattern between different MPM samples is based on Pearson product moment correlation. Unsupervised clustering revealed two distinct groups, each comprising about half of all patients (Fig. 4). One group includes all samples with altered HRR (yellow box), including olaparib-responsive NCI-H2052 as well as BAP1-mutated NCI-H2452 cells. Of note, these 49 samples showed similar gene expression pattern of key enzymes involved in formation of BRCAness-like cell state, overall indicating two different phenotypes of MPM with respect to HRR based repair.
Fig. 4.
Overall correlation pattern between samples. Unsupervised clustering revealed two distinct groups. The group in the yellow box includes all samples with altered HRR. This box also includes expression patterns of olaparib-responsive NCI-H2052 as well as BAP1-mutated NCI-H2452 cells
The histomorphology of patient tumours revealed no significant differences in gene expression patterns (Additional file 2: Figure S1).
Association with therapy response and survival
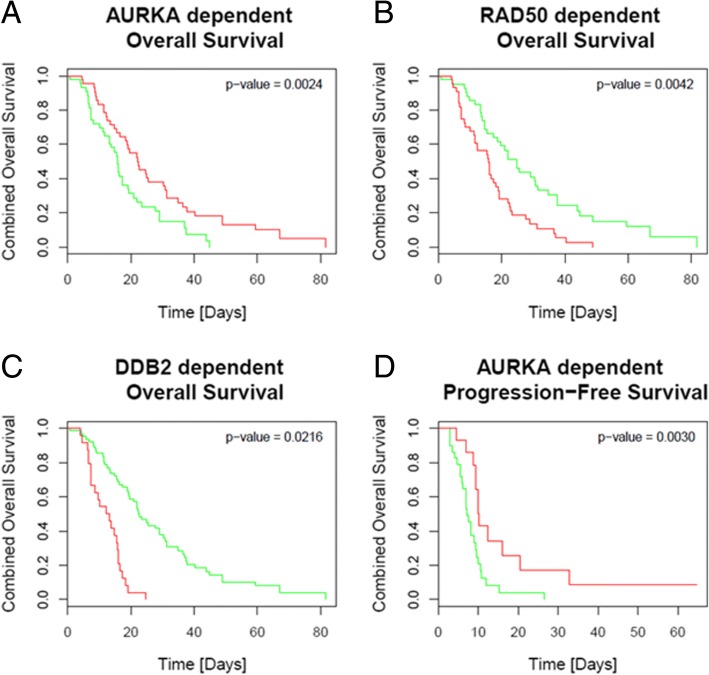
By determining associations between gene expression and therapy response due to survival, AURKA, RAD50 and damage specific DNA-damage-binding-protein-2 (DDB2) were significantly associated to overall survival with FDR-adjusted p-values ≤0.02 (Additional file 3: Table S2). Patients with low expression of AURKA have a 2.4-fold higher chance of prolonged overall survival, while high expression of RAD50 and resulted in 2.3-fold higher risk of dying from their disease. High expression of DDB2 in patients showed a 4.373-fold chance of prolonged survival. In addition, low expression of AURKA showed also a 2.3-fold higher chance of prolonged progression-free survival (Fig. 5).
Fig. 5.
Overall and progression-free survival dependent on gene expression of AURKA, RAD50 and DDB2. Low expression of AURKA and high expression of RAD50 and DDB2 resulted in prolonged overall survival (A-C). Low expression of AURKA (D) also resulted in prolonged progression-free survival with p < 0.0030. Low expression is highlighted in red, high expression is highlighted in green. The median was used to set a cut-off between high and low gene expression
Discussion
This study was designed to evaluate gene expression of BRCAness related genes in MPM and their impact on susceptibility to olaparib.
BRCAness is a common event in MPM
Defects in individual genes that modulate HRR, compiled under the term “BRCAness”, has been found in various tumours [10]. Studies revealed, that impaired HRR results in enhanced use of NHEJ for DSB repair [10, 22–24]. NHEJ repair is more error-prone compared to HRR, often leading to DNA mutations, especially by deletions [10, 25]. This genomic instability predisposes cancer susceptibility caused by BRCAness [26].
In the MPM patients investigated, ten of the 24 tested genes necessary for intact HRR were not significantly expressed (Table 3). Additionally, to this lack of expression, a loss-of-function of three of those genes could already be determined by Betti et al., who also found deleterious mutations in BRCA1, BRCA2 and PALB2 gene loci [27]. The authors described BRCA1 and PALB2 exhibiting a deletion, leading to a nonsense mutational effect, while BRCA2 showed a frameshift due to deletion [27].
Table 3.
Tested genes that showed no or very low expression (counts < 100) in MPM patients. FANCD2 showed partially basal expression with a maximum of 214 counts
| AURKA | BRCA1 | CHEK2 | BRIP1 | FANCD2 |
| BARD1 | BRCA2 | EMSY | PALB2 | RAD51 |
The loss-of-expression of RAD51 is not surprising, as it is commonly deleted in malignant mesothelioma due to frequent losses from 15q11.1–22 [28]. Nevertheless, as central element of HR by mediating the invasion into the homologous double helix of the sister chromatid and thereby directly regulated binding partner of BRCA2, RAD51 plays an important role for the shaping of BRCAness.
Beside these main key-players of BRCAness phenotype development, genes of the Fanconi Anaemia (FA) pathway play an important role within this context of DSB repair. Of note, Fanconi anaemia complementation group D2 (FANCD2) as a key member within the FA, shows only basal gene expression levels in all analysed patient samples, indicating an impaired function of the FA pathway. It directly interacts with BRCA1 and BRCA2 but also ataxia telangiectasia mutated (ATM), thereby promoting DNA repair by HR [26, 29, 30]. In contrast, Røe et al. found an overexpression of FANCD2 in microarray data of malignant pleural mesothelioma patients [31], but the analysed sample size of five mesothelioma tissues is quite small. Therefore, these contradictory findings should be examined more detailed.
The correlation matrix of gene expression patterns indicates a cluster of patient samples showing features similar to the olaparib-sensitive NCI-H2452 as well as NCI-H2052 cell lines. It could be assumed, that these patients could benefit from treatment with PARP-inhibitors. Assuming a favourable chance of response.
BRCAness is a strong prognostic factor in MPM
In association with clinicopathological data, high expression of AURKA was associated with shortened overall survival and poor prognosis. The high AURKA expression pattern possibly explains the high mitotic and proliferative activity, rather found in more aggressive tumours with higher risk of metastatic spread.
In contrast, high expression of DDB2 and RAD50 is significantly associated with prolonged survival. DDB2 is involved in DNA damage repair and is modulated by BRCA1 and/or p53 [32]. It might be suggested, that DDB2 together with CHEK1, activates p53 and thereby triggers TP53-induced apoptosis and senescence in response to DNA damage [20]. Barakat et al. showed, that overexpression of DDB2 enhances cisplatin-sensitivity in ovarian cancer cells [33]. The present data indicate that these observations in ovarian cancer cells could also enhance cisplatin-sensitivity in MPM, resulting in 4.373-fold higher chance of prolonged overall survival.
Zhang et al. demonstrated likewise, that loss of RAD50 is a key marker of BRCAness in ovarian cancers (OvCa) [34]. Cell culture analysis demonstrated, that loss of RAD50 augmented OvCa cell’s response to cisplatin and PARP-inhibitors [34].
Olaparib induces superior apoptotic response in BAP1-mutant tumours in vitro. It is shown that BAP1 is commonly mutated in MPM and thereby might cause defects in HRR as well as a BRCAness phenotype of affected cells [1, 35–37]. We hypothesized, that the PARP-inhibitor olaparib could have a therapeutic effect on BAP1-mutant MPM and, furthermore, that a combination with cisplatin enhances the inhibitory effect of the former one.
A distinct expression pattern of investigated genes could be found in NCI-H2452 cells. In this cell line, induction of apoptosis could be proven during olaparib-based treatment. Furthermore, we could confirm enhanced therapeutic effects using olaparib in combination with cisplatin. Response of treated NCI-H2452 cells to olaparib has also been shown by Srinivasan et al. [11].
Necrosis was not detected in treated cells, which is preferable, because necrosis leads, compared to apoptosis, to adverse complications in therapy.
The benign human fibroblast cell line MRC-5 was used as benign control. They derive from the same cotyledon (mesoderm), like malignant pleural mesothelioma cells. Therefore, it was preferred, as the e.g. control cell line MET-5a was SV40-immortalized and therefore shows altered culture performance [38]. The TERT1-immortalized cell line LP-9 would be another option for using as control cell line [39]. However, the use of this cell line as control has to be investigated and validated.
Response of Olaparib on MPM patients might be predicted by similar expression patterns compared to responsive NCI-H2452 cells
Gene expression pattern of NCI-H2452 showed weak correlations to other cell lines, suggesting that gene expression pattern of BAP1 is associated with enhanced response to PARP-inhibitor olaparib. Furthermore, AURKA, RPA1, PARP1, BRCA2 and CHEK2 showed significantly different gene expression pattern compared to other cell lines. Transferred to patient samples, approximately 10% of patients have the same gene expression pattern as NCI-H2452 cells, making them suspicious for susceptibility to olaparib based treatment approaches. The BAP1 mutational status in combination with certain gene expression patterns seem to contribute significantly to cellular response to olaparib. Therefore, this may be a promising therapeutic approach for a substantial portion of MPM patients, especially when combined with platin-based agents.
Conclusions
In conclusion, response could be demonstrated during treatment of BAP1-mutant NCI-H2452 cells with olaparib in combination with cisplatin. Thus, this combined therapy might be effective for up to 2/3 of patients suffering from MPM.
Investigation of BRCAness related genes in MPM patients showed similarities in gene expression patterns compared to MPM cell lines (BACH1, FANCD2 and RAD51), particularly BAP1-mutant NCI-H2452 (BAP1, PARP1, BRCA2, CHEK2, AURKA and RPA1). These gene expression patterns represent a novel and promising tool for the prediction of response to the PARP-inhibitor olaparib.
Additional files
Table S1. Normalized counts of genes measured by the Digital Analyzer. (PDF 343 kb)
Figure S1. Expression of tested genes is independent of histomorphology of MPM. The boxplot shows no significant differences between gene expression patterns due to biphasic (B), epithelioid (E), or sarcomatoid (S) MPM. (PDF 59 kb)
Table S2. AURKA, RAD50, and DDB2 showed statistically significant dependencies on overall and progression-free survival. (PDF 33 kb)
Acknowledgements
Not applicable.
Funding
The study was financed by the Department of Pathology, University Hospital Essen. No additional funding support was used.
Availability of data and materials
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- °C
Degree Celsius
- AAF
Alanin-Alanin-Phenylalanin-Aminoluciferin
- ADP
Adenosine diphosphate
- Anova
Analysis of variance
- ATM
Ataxia telangiectasia mutated
- ATR
ATM and RAD3-related
- BAP1
BRCA1-associated protein 1
- BARD1
BRCA1 associated RING domain 1
- BER
Base excision repair
- BRCA1/2
Breast Cancer 1/2
- BRIP1
BRCA1 interacting protein C-terminal helicase 1
- CART
Classification and regression tree algorithm
- CHEK2
Checkpoint kinase 2
- CR
Complete response
- CTree
Conditional interference tree
- DMSO
Dimethyl sulfoxide
- DNA
Deoxyribonucleic acid
- DPBS
Dulbecco’s Phosphate Buffered Saline
- DSB
Double-strand break
- DSBR
Double-strand break repair
- FANCD2
Fanconi anaemia complementation group D2
- FDA
Food and Drug Administration
- FDR
False discovery rate
- FFPE
Formalin-fixed paraffin-embedded
- FOV
Fields of view
- h
Hours
- HCF-1
Host cell factor-1
- HRR
Homologous recombination repair
- min
minutes
- MPM
Malignant pleural mesothelioma
- MRE11A
Meiotic recombination 11 homologue A
- NBS1
Nijmegen breakage syndrome 1
- NER
Nucleotide excision repair
- Ng
Nanogram
- NHEJ
Non-homologous end joining
- OS
Overall survival
- PALB2
Partner localizer of BRCA2
- PARP1
Poly (ADP-ribose) polymerase 1
- PD
Progressive disease
- PFS
Progression-free survival
- PR
Partial response
- RLU
Relative light units
- RNA
Ribonucleic acid
- ROC
Receiver operating characteristic
- RPMI
Roswell park memorial institute
- SD
Stable disease
- SV40
Simian virus 40
- UICC
Union international contre le cancer
- μl
Microliter
- μM
Micro molar
Authors’ contributions
Sample acquisition and histomorphology assessment were performed by JW and TM. Study design was outlined by KS, FM and TP. Experiments were performed by SB. Data analysis was mainly performed by FM, MW, TH1 and SB. Covariate acquisition was performed by WE, EM, CA, JK and JS. Clinical aspects of data analysis and interpretation were highlighted by WE, CA, JK, DC and TP. Pathological aspects of study was done by TH1, TM, JW and KS. For biological and cell-biological data interpretation, main contributions were made by SB, MW, JS, EM, TH2, RW and FM. In general, data interpretation as well as raising important points or drawing conclusions for discussion of the results was performed by all authors (SB, MW, JS, EM, JK, TH1, TM, TH2, DC, RW, WE, TP, JW, CA, KS and FM). Manuscript preparation was mainly performed by FM, KS and SB. All authors (SB, MW, JS, EM, JK, TH1, TM, TH2, DC, RW, WE, TP, JW, CA, KS and FM) reviewed and revised the manuscript critically. FM and RW and DC coordinated the project. Ethics committees’ approval was obtained by CA, JW, JK and TM. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The study conforms to the principles outlined in the declaration of Helsinki and was approved by the institutional review board (Ethics Committee of the Medical Faculty of the University Duisburg-Essen), protocol no.: 14–5775-BO. Due to the ethics approval, a patients’ informed consent has not been obtained.
Consent for publication
Not applicable.
Competing interests
All authors have read and approved the manuscript and are aware of the submission. The authors declare no conflict of interest.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Sabrina Borchert, Phone: +49 (0) 201 / 723 - 83035, Email: sabrina.borchert@uk-essen.de.
Michael Wessolly, Email: michael.wessolly@uk-essen.de.
Jan Schmeller, Email: jan.schmeller@uk-essen.de.
Elena Mairinger, Email: elena.mairinger@uk-essen.de.
Jens Kollmeier, Email: jens.kollmeier@helios-gesundheit.de.
Thomas Hager, Email: thomas.hager@uk-essen.de.
Thomas Mairinger, Email: thomas.mairinger@helios-gesundheit.de.
Thomas Herold, Email: thomas.herold@uk-essen.de.
Daniel C. Christoph, Email: d.christoph@kliniken-essen-mitte.de
Robert F. H. Walter, Email: robert.walter@uk-essen.de
Wilfried E. E. Eberhardt, Email: Wilfried.Eberhardt@uk-essen.de
Till Plönes, Email: till.ploenes@ruhrlandklinik.uk-essen.de.
Jeremias Wohlschlaeger, Email: jeremias.wohlschlaeger@uk-essen.de.
Clemens Aigner, Email: clemens.aigner@ruhrlandklinik.uk-essen.de.
Kurt Werner Schmid, Email: KW.Schmid@uk-essen.de.
Fabian D. Mairinger, Email: fabian.mairinger@uk-essen.de
References
- 1.Carbone M, Yang H. Mesothelioma: recent highlights. Ann Transl Med. 2017;5(11):238. doi: 10.21037/atm.2017.04.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mairinger FD, Werner R, Flom E, Schmeller J, Borchert S, Wessolly M, et al. miRNA regulation is important for DNA damage repair and recognition in malignant pleural mesothelioma. Virchows Arch. 2017;470(6):627–637. doi: 10.1007/s00428-017-2133-z. [DOI] [PubMed] [Google Scholar]
- 3.Turkcu G, AlabalIk U, Keles AN, Ibiloglu I, Kucukoner M, Sen HS, et al. Comparison of SKIP expression in malignant pleural mesotheliomas with Ki-67 proliferation index and prognostic parameters. Pol J Pathol. 2016;67(2):108–113. doi: 10.5114/pjp.2016.61445. [DOI] [PubMed] [Google Scholar]
- 4.Montanaro F, Rosato R, Gangemi M, Roberti S, Ricceri F, Merler E, et al. Survival of pleural malignant mesothelioma in Italy: a population-based study. Int J Cancer. 2009;124(1):201–207. doi: 10.1002/ijc.23874. [DOI] [PubMed] [Google Scholar]
- 5.Borasio P, Berruti A, Bille A, Lausi P, Levra MG, Giardino R, et al. Malignant pleural mesothelioma: clinicopathologic and survival characteristics in a consecutive series of 394 patients. Eur J Cardiothorac Surg. 2008;33(2):307–313. doi: 10.1016/j.ejcts.2007.09.044. [DOI] [PubMed] [Google Scholar]
- 6.Christoph DC, Eberhardt WE. Systemic treatment of malignant pleural mesothelioma: new agents in clinical trials raise hope of relevant improvements. Curr Opin Oncol. 2014;26(2):171–181. doi: 10.1097/CCO.0000000000000053. [DOI] [PubMed] [Google Scholar]
- 7.Boons CC, MW VANT, Burgers JA, Beckeringh JJ, Wagner C, Hugtenburg JG. The value of pemetrexed for the treatment of malignant pleural mesothelioma: a comprehensive review. Anticancer Res. 2013;33(9):3553–3561. [PubMed] [Google Scholar]
- 8.Christoph DC, Asuncion BR, Mascaux C, Tran C, Lu X, Wynes MW, et al. Folylpoly-glutamate synthetase expression is associated with tumor response and outcome from pemetrexed-based chemotherapy in malignant pleural mesothelioma. J Thorac Oncol. 2012;7(9):1440–1448. doi: 10.1097/JTO.0b013e318260deaa. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Walter RF. Molecular biological analysis of malignant pleural mesothelioma [dissertation] Essen: Ruhrlandklinik, west German lung Centre, At the university hospital Essen – university hospital: university Duisburg-Essen; 2014. [Google Scholar]
- 10.Lord CJ, Ashworth A. BRCAness revisited. Nat Rev Cancer. 2016;16(2):110–120. doi: 10.1038/nrc.2015.21. [DOI] [PubMed] [Google Scholar]
- 11.Srinivasan G, Sidhu GS, Williamson EA, Jaiswal AS, Najmunnisa N, Wilcoxen K, et al. Synthetic lethality in malignant pleural mesothelioma with PARP1 inhibition. Cancer Chemother Pharmacol. 2017;80(4):861–867. doi: 10.1007/s00280-017-3401-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rottenberg S, Jaspers JE, Kersbergen A, van der Burg E, Nygren AO, Zander SA, et al. High sensitivity of BRCA1-deficient mammary tumors to the PARP inhibitor AZD2281 alone and in combination with platinum drugs. Proc Natl Acad Sci U S A. 2008;105(44):17079–17084. doi: 10.1073/pnas.0806092105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Menear KA, Adcock C, Boulter R, Cockcroft XL, Copsey L, Cranston A, et al. 4-[3-(4-cyclopropanecarbonylpiperazine-1-carbonyl)-4-fluorobenzyl]-2H-phthalazin- 1-one: a novel bioavailable inhibitor of poly (ADP-ribose) polymerase-1. J Med Chem. 2008;51(20):6581–6591. doi: 10.1021/jm8001263. [DOI] [PubMed] [Google Scholar]
- 14.Parrotta R, Okonska A, Ronner M, Weder W, Stahel R, Penengo L, et al. A novel BRCA1-associated Protein-1 isoform affects response of mesothelioma cells to drugs impairing BRCA1-mediated DNA repair. J Thorac Oncol. 2017;12(8):1309–1319. doi: 10.1016/j.jtho.2017.03.023. [DOI] [PubMed] [Google Scholar]
- 15.Fong PC, Boss DS, Yap TA, Tutt A, Wu P, Mergui-Roelvink M, et al. Inhibition of poly (ADP-ribose) polymerase in tumors from BRCA mutation carriers. N Engl J Med. 2009;361(2):123–134. doi: 10.1056/NEJMoa0900212. [DOI] [PubMed] [Google Scholar]
- 16.Galateau-Salle F, Churg A, Roggli V, Travis WD. World Health Organization Committee for tumors of the P. The 2015 World Health Organization classification of tumors of the pleura: advances since the 2004 classification. J Thorac Oncol. 2016;11(2):142–154. doi: 10.1016/j.jtho.2015.11.005. [DOI] [PubMed] [Google Scholar]
- 17.Patriarca S, Ferretti S, Zanetti R. TNM classification of malignant tumours - eighth edition: which news? Epidemiol Prev. 2017;41(2):140–143. doi: 10.19191/EP17.2.P140.034. [DOI] [PubMed] [Google Scholar]
- 18.Byrne MJ, Nowak AK. Modified RECIST criteria for assessment of response in malignant pleural mesothelioma. Ann Oncol. 2004;15(2):257–260. doi: 10.1093/annonc/mdh059. [DOI] [PubMed] [Google Scholar]
- 19.Mairinger FD, Schmeller J, Borchert S, Wessolly M, Mairinger E, Kollmeier J, et al. Immunohistochemically detectable metallothionein expression in malignant pleural mesotheliomas is strongly associated with early failure to platin-based chemotherapy. Oncotarget. 2018;9(32):22254–22268. doi: 10.18632/oncotarget.24962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Walter RF, Vollbrecht C, Werner R, Mairinger T, Schmeller J, Flom E, et al. Screening of pleural mesotheliomas for DNA-damage repair players by digital gene expression analysis can enhance clinical Management of Patients Receiving Platin-Based Chemotherapy. J Cancer. 2016;7(13):1915–1925. doi: 10.7150/jca.16390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wessolly M, Walter RFH, Vollbrecht C, Werner R, Borchert S, Schmeller J, et al. Processing escape mechanisms through altered proteasomal cleavage of epitopes affect immune response in pulmonary neuroendocrine tumors. Technol Cancer Res Treat. 2018;17:1533033818818418. doi: 10.1177/1533033818818418. [DOI] [Google Scholar]
- 22.Tutt A, Bertwistle D, Valentine J, Gabriel A, Swift S, Ross G, et al. Mutation in Brca2 stimulates error-prone homology-directed repair of DNA double-strand breaks occurring between repeated sequences. EMBO J. 2001;20(17):4704–4716. doi: 10.1093/emboj/20.17.4704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xia F, Taghian DG, DeFrank JS, Zeng ZC, Willers H, Iliakis G, et al. Deficiency of human BRCA2 leads to impaired homologous recombination but maintains normal nonhomologous end joining. Proc Natl Acad Sci U S A. 2001;98(15):8644–8649. doi: 10.1073/pnas.151253498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moynahan ME, Pierce AJ, Jasin M. BRCA2 is required for homology-directed repair of chromosomal breaks. Mol Cell. 2001;7(2):263–272. doi: 10.1016/S1097-2765(01)00174-5. [DOI] [PubMed] [Google Scholar]
- 25.Lord CJ, Ashworth A. The DNA damage response and cancer therapy. Nature. 2012;481(7381):287–294. doi: 10.1038/nature10760. [DOI] [PubMed] [Google Scholar]
- 26.Turner N, Tutt A, Ashworth A. Hallmarks of ‘BRCAness’ in sporadic cancers. Nat Rev Cancer. 2004;4(10):814–819. doi: 10.1038/nrc1457. [DOI] [PubMed] [Google Scholar]
- 27.Betti M, Casalone E, Ferrante D, Aspesi A, Morleo G, Biasi A, et al. Germline mutations in DNA repair genes predispose asbestos-exposed patients to malignant pleural mesothelioma. Cancer Lett. 2017;405:38–45. doi: 10.1016/j.canlet.2017.06.028. [DOI] [PubMed] [Google Scholar]
- 28.De Rienzo A, Balsara BR, Apostolou S, Jhanwar SC, Testa JR. Loss of heterozygosity analysis defines a 3-cM region of 15q commonly deleted in human malignant mesothelioma. Oncogene. 2001;20(43):6245–6249. doi: 10.1038/sj.onc.1204828. [DOI] [PubMed] [Google Scholar]
- 29.Patel KJ, Joenje H. Fanconi anemia and DNA replication repair. DNA Repair (Amst) 2007;6(7):885–890. doi: 10.1016/j.dnarep.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 30.D'Andrea AD, Grompe M. The Fanconi anaemia/BRCA pathway. Nat Rev Cancer. 2003;3(1):23–34. doi: 10.1038/nrc970. [DOI] [PubMed] [Google Scholar]
- 31.Roe OD, Anderssen E, Sandeck H, Christensen T, Larsson E, Lundgren S. Malignant pleural mesothelioma: genome-wide expression patterns reflecting general resistance mechanisms and a proposal of novel targets. Lung Cancer. 2010;67(1):57–68. doi: 10.1016/j.lungcan.2009.03.016. [DOI] [PubMed] [Google Scholar]
- 32.De Luca P, Moiola CP, Zalazar F, Gardner K, Vazquez ES, De Siervi A. BRCA1 and p53 regulate critical prostate cancer pathways. Prostate Cancer Prostatic Dis. 2013;16(3):233–238. doi: 10.1038/pcan.2013.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barakat BM, Wang QE, Han C, Milum K, Yin DT, Zhao Q, et al. Overexpression of DDB2 enhances the sensitivity of human ovarian cancer cells to cisplatin by augmenting cellular apoptosis. Int J Cancer. 2010;127(4):977–988. doi: 10.1002/ijc.25112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang M, Liu G, Xue F, Edwards R, Sood AK, Zhang W, et al. Copy number deletion of RAD50 as predictive marker of BRCAness and PARP inhibitor response in BRCA wild type ovarian cancer. Gynecol Oncol. 2016;141(1):57–64. doi: 10.1016/j.ygyno.2016.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bott M, Brevet M, Taylor BS, Shimizu S, Ito T, Wang L, et al. The nuclear deubiquitinase BAP1 is commonly inactivated by somatic mutations and 3p21.1 losses in malignant pleural mesothelioma. Nat Genet. 2011;43(7):668–672. doi: 10.1038/ng.855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Testa JR, Cheung M, Pei J, Below JE, Tan Y, Sementino E, et al. Germline BAP1 mutations predispose to malignant mesothelioma. Nat Genet. 2011;43(10):1022–1025. doi: 10.1038/ng.912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Emri SA. The Cappadocia mesothelioma epidemic: its influence in Turkey and abroad. Ann Transl Med. 2017;5(11):239. doi: 10.21037/atm.2017.04.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Singh A, Pruett N, Hoang CD. In vitro experimental models of mesothelioma revisited. Transl Lung Cancer Res. 2017;6(3):248–258. doi: 10.21037/tlcr.2017.04.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shukla A, Barrett TF, MacPherson MB, Hillegass JM, Fukagawa NK, Swain WA, et al. An extracellular signal-regulated kinase 2 survival pathway mediates resistance of human mesothelioma cells to asbestos-induced injury. Am J Respir Cell Mol Biol. 2011;45(5):906–914. doi: 10.1165/rcmb.2010-0282OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Database GHG. www.genecards.org. Accessed 20 June 2018.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Normalized counts of genes measured by the Digital Analyzer. (PDF 343 kb)
Figure S1. Expression of tested genes is independent of histomorphology of MPM. The boxplot shows no significant differences between gene expression patterns due to biphasic (B), epithelioid (E), or sarcomatoid (S) MPM. (PDF 59 kb)
Table S2. AURKA, RAD50, and DDB2 showed statistically significant dependencies on overall and progression-free survival. (PDF 33 kb)
Data Availability Statement
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.