Human induced pluripotent stem cells (iPSC) have been used to generate intestinal organoids that mimic key intestinal properties without the requirement for invasive procedures to obtain human tissues. The main protocols that have been described result in gut organoids that contain both intestinal epithelium as well as mesenchymal cells.1, 2 We have previously reported on human iPSC-derived intestinal organoids (iPSCo) that can be propagated in long-term culture that contain solely epithelial cells.3, 4, 5, 6, 7 A pure epithelial model offers unique opportunities to study epithelial cell intrinsic and cell-type–specific mechanisms. Among these cellular processes are epigenetic mechanisms such as DNA methylation, which acts as a key regulator of intestinal epithelial development and regional identity.1, 7 The purpose of this study was to characterize iPSCo by comparing these cultures with primary purified intestinal epithelial cells (IECs).
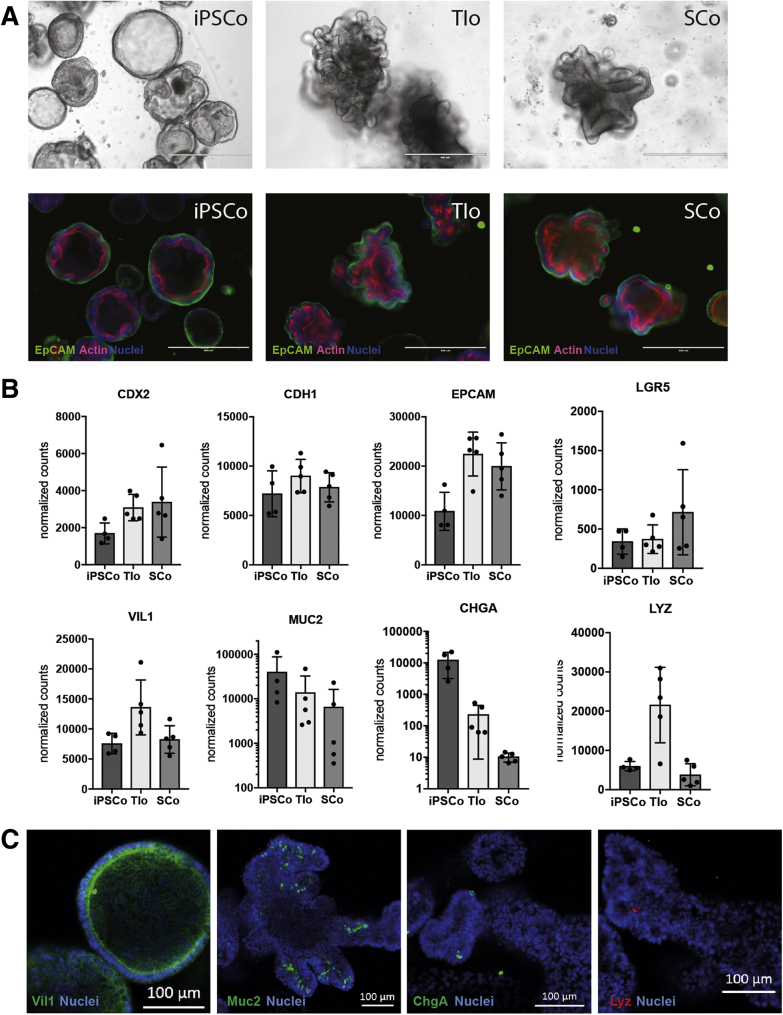
Intestinal epithelial organoid cultures were derived from at least 3 different lines of iPSCs according to the described protocol3, 4, 5 (see also Supplementary Figure 1), and showed a morphology comparable with mucosal biopsy-derived intestinal epithelial organoids. This was highlighted by the presence of epithelial cell adhesion molecules and epithelial polarization (Figure 1A). Furthermore, iPSCos were found to express several epithelial cell markers including E-cadherin (CDH1) and the intestinal stem cell marker LGR5 (Figure 1B). Immunofluorescent staining showed ubiquitous positivity for the enterocyte marker villin, with specific subsets of cells expressing mucin 2, chromogranin A, and lysozyme, markers associated with the epithelial cell subsets Goblet, enteroendocrine, and Paneth cells, respectively (Figure 1B and C).
Figure 1.
Characterization of iPSC-derived intestinal epithelial organoids. (A) Intestinal epithelial organoids derived from PSCs (iPSCo), terminal ileum organoids (TIo), or SC organoids (SCo) in microscopic brightfield view (upper panel), or immunofluorescent staining for epithelial cell adhesion molecule (epithelial cell adhesion molecule [EpCAM], green), actin (red), and nuclei (blue). Scale bars: 400 μm. (B) Expression of IEC-type marker as normalized counts from RNA sequencing data of the respective organoids. Bar shows means ± SD, n = 4–5 per group. (C) Immunofluorescent staining for enterocyte marker villin (VIL1, green), goblet cell marker mucin 2 (MUC2, green), enteroendocrine marker chromogranin A (CHGA, green), Paneth cell marker lysozyme (LYZ, red), and nuclear counterstain (blue), Scale bars: 100 um.
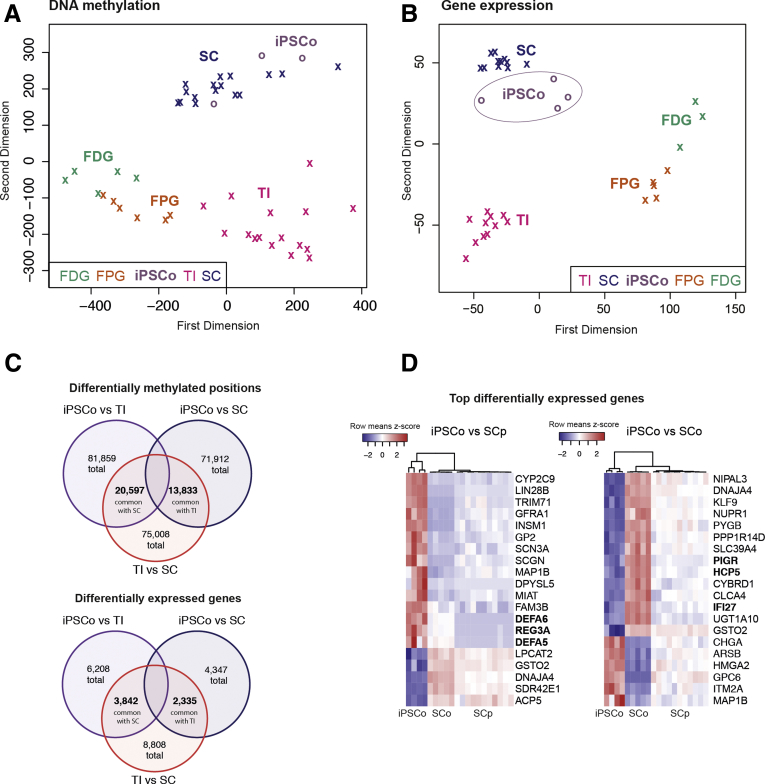
To further evaluate regional identity and the degree of developmental maturation of iPSCos, we performed DNA methylation and transcriptomic profiling using Illumina (Illumina, Cambridge, UK) Infinium bead arrays and RNA sequencing, respectively. We compared these genome-wide profiles with data sets we previously derived from purified IECs from mature terminal ileum and sigmoid colon (SC), as well as human fetal proximal gut and fetal distal gut (Supplementary Tables 1 and 2).6
Relative sample similarity based on multidimensional scaling indicated that iPSCos clustered more closely to mature colonic epithelium, both on an epigenetic (ie, DNA methylation) and transcriptomic level (Figure 2A and B). This was confirmed further by performing unsupervised, hierarchical clustering (Supplementary Figure 2), as well as differential DNA methylation and gene expression analyses. The latter showed that iPSCos had a greater number of significantly differentially methylated positions (adjusted P < .01) and differentially expressed genes (adjusted P < .01) in common with the SC than with the terminal ileum (Figure 2C, Supplementary Table 3).
Figure 2.
Genome-wide profiling of human iPSCo and purified IECs. (A) Multidimensional scaling plot showing sample similarity of genome-wide DNA methylation measured by an Illumina 450K array of iPSCo, in the context of IECs derived from terminal ileum (TI), SC, fetal proximal gut (FGP), and fetal distal gut (FDG).1 Each symbol represents a sample, x = IEC, o = organoid. (B) MDS plot of RNA-sequencing profiles showing the same sample groups as in panel A. Sample distance in the plot is based on regularized-logarithmic transformed read counts of all expressed genes. (C) Venn diagram indicating the number of differentially methylated CpG positions (top) or differentially expressed genes (bottom) (adjusted P < .01) between the different groups. (D) Heatmap of expression values (regularized-logarithmic normalized counts) of the top 20 most significantly differentially expressed genes comparing iPSCo and SC purified epithelium (SCp) (left), and iPSCo and SCo (right) for all 3 sample groups. The scale indicates the row mean Z-score (means, 0). The dendrogram above the heatmap clusters samples based on the expression similarity of those genes.
Given the overall similarity with pediatric sigmoid colon epithelium, we examined this relationship in more detail using mucosa-derived SC organoids (SCos) as an additional reference (Supplementary Figure 3A and B). We focused on the most significant differentially expressed genes (adjusted P < .01) between iPSCo and either purified IECs (SC purified epithelium) or organoids (SCo), respectively. Interestingly, some key small intestinal markers for Paneth cells (eg, α-defensins DEFA5 and DEFA6) were abundant in iPSCo, while several genes involved in innate defense (eg, IFI27, HCP5, PIGR) showed markedly lower expression levels (Figure 2D). This indicated that iPSCos may not reach the full level of differentiation into small or large intestinal epithelium as found in vivo. Thus, iPSCo models may require further optimization of the differentiation protocol to permit further maturation and regionalization. Studying the signaling pathways that could drive this maturation could offer unique insight into the processes involved in intestinal development.
Together, our findings indicate that iPSC-derived human intestinal epithelial cell organoids more closely resemble mature colonic epithelium, which is in keeping with reports from other groups using alternative protocols to achieve distalization and maturation.8, 9, 10 However, using a genome-wide approach to validate the iPSCo model also uncovered distinct differences and incomplete regionalization of iPSC-derived organoids compared with human primary cells. These observations provide an ideal starting point to further investigate the factors required to model complete epithelial development in vitro.
Acknowledgments
The authors would like to thank the members of the Zilbauer group who contributed to the original cohort, with special thanks to Komal M. Nayak, Kate J. Howell, Marco Gasparetto, Claire Lee, and Felicity Payne. The authors would like to express their gratitude to their collaborators Dr Bon-Kyoung Koo (Institute of Molecular Biotechnology, Vienna), Dr Oliver Stegle (European Bioinformatics Institute), Philip Rosenstiel (Christian-Albrechts University of Kiel), and Roger Barker (University of Cambridge). Furthermore, the authors are grateful to the clinical team of Paediatric Gastroenterology, Hepatology and Nutrition at Cambridge University Hospitals under Dr Robert Heuschkel, and all patients participating in this study. The authors would also like to thank the Wellcome Trust Sanger Institute Core Scientific Operations team for conducting transcriptome sequencing.
Profiling data of iPSC-derived organoids is available at ArrayExpress under accession numbers E-MTAB-7289 (DNA methylation) and E-MTAB-7306 (RNA sequencing). Previously published data are accessible for DNA methylation under ArrayExpress accession number E-MTAB-4957; for RNA sequencing under ArrayExpress accession number E-MTAB-5015, or in the European Nucleotide Archive Study PRJEB15114 1.1
Footnotes
Conflicts of interest This author discloses the following: Ludovic Vallier is a funder of, stockholder of, and consultant for DefiniGEN. The remaining authors disclose no conflicts.
Funding Supported by Crohn’s and Colitis UK and Crohn’s in Childhood Research association (J.K.); by core support from the Wellcome Trust (G.D. and J.F.); RNA sequencing was funded by the Wellcome Trust (206194); also supported by the European Research Council advanced grant 741707, Cambridge University Hospitals National Institute for Health Research Biomedical Research Center, core support from the Wellcome Trust, and Medical Research Council to the Cambridge Stem Cell Institute (PSAG028) and the European Union grant Intestinal Tissue ENgineering Solution (INTENS) (LV); and supported by the Wellcome Trust Interdisciplinary Programme in Translational Medicine and Therapeutics (100138/B/12/Z to A.R.).
Supplementary Material
References
- 1.Spence J.R. Nature. 2011;470:105–109. doi: 10.1038/nature09691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Forster R. Stem Cell Reports. 2014;2:838–852. doi: 10.1016/j.stemcr.2014.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Forbester J.L. Methods Mol Biol. 2016 doi: 10.1007/7651_2016_7. [DOI] [Google Scholar]
- 4.Forbester J.L. Infect Immun. 2015;83:2926–2934. doi: 10.1128/IAI.00161-15. (2015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hannan N.R.F. Stem Cell Reports. 2013;1:293–306. doi: 10.1016/j.stemcr.2013.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kraiczy J. Gut. 2019;68:49–61. doi: 10.1136/gutjnl-2017-314817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kraiczy J. Mucosal Immunol. 2016;9:647–658. doi: 10.1038/mi.2015.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Múnera J.O. Cell Stem Cell. 2017;21:51–64.e6. doi: 10.1016/j.stem.2017.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crespo M. Nat Med. 2017;23:878–884. doi: 10.1038/nm.4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Watson C.L. Nat Med. 2014;20:1310–1314. doi: 10.1038/nm.3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
References
- 1.Kraiczy J. Gut. 2017 Epub ahead of print. [Google Scholar]
- 2.Sato T. Gastroenterology. 2011;141:1762–1772. doi: 10.1053/j.gastro.2011.07.050. [DOI] [PubMed] [Google Scholar]
- 3.Forbester J.L. Methods Mol Biol. 2016 Epub ahead of print. [Google Scholar]
- 4.Hannan N.R.F. Stem Cell Rep. 2013;1:293–306. doi: 10.1016/j.stemcr.2013.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Forbester J.L. Infect Immun. 2015;83:2926–2934. doi: 10.1128/IAI.00161-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yusa K. Nature. 2011;478:391–394. doi: 10.1038/nature10424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kraiczy J. Mucosal Immunol. 2016;9:647–658. doi: 10.1038/mi.2015.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jenke A.C. PLoS One. 2013;8:e55636. doi: 10.1371/journal.pone.0055636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aryee M.J. Bioinformatics. 2014;30:1363–1369. doi: 10.1093/bioinformatics/btu049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Du P. BMC Bioinformatics. 2010;11:587. doi: 10.1186/1471-2105-11-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leek JT, et al. R Package version 3180.
- 12.GK http://bioconductor.org/packages/release/bioc/html/limma.html Available at: Accessed: November 7, 2014.
- 13.Martin M. EMBnet J. 2011;17:10. [Google Scholar]
- 14.Kim D. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Langmead B. Curr Protoc Bioinformatics. 2010 doi: 10.1002/0471250953.bi1107s32. Chapter 11:Unit 11.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li H. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Anders S. Bioinformatics. 2014;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Risso D. Nat Biotechnol. 2014;32:896–902. doi: 10.1038/nbt.2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Love M.I. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao S. https://cran.r-project.org/web/packages/heatmap3/index.html Available at:
- 21.Kim S.K. Science. 2001;293:2087–2092. doi: 10.1126/science.1061603. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.