Summary
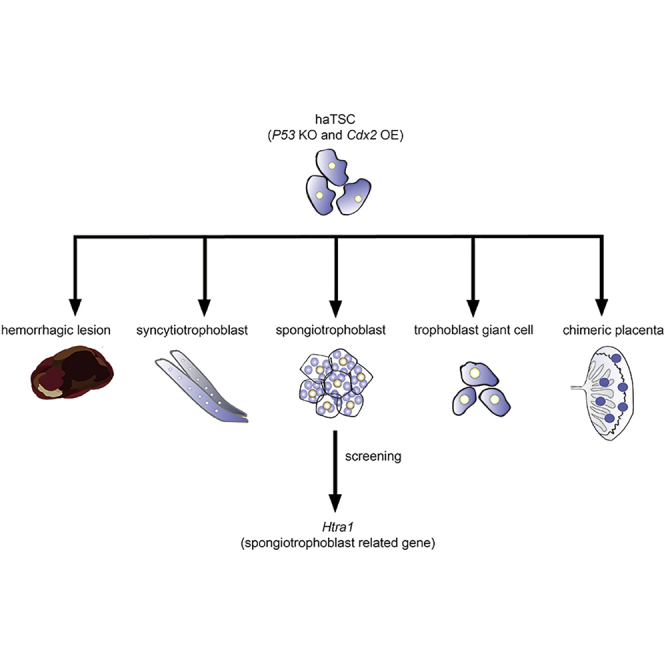
Owing to their single genome, haploid cells are powerful to uncover unknown genes by performing genetic screening in mammals. However, no haploid cell line from an extraembryonic lineage has been achieved yet, which limits the application of haploid cells in placental genetic screening. Here, we show that overexpression of Cdx2 can convert haploid embryonic stem cells to trophoblast stem cells (TSCs). p53 deletion reduces diploidization during the conversion and guarantees the generation of haploid-induced TSCs (haiTSCs). haiTSCs not only share the same molecular characterization with trophoderm-derived TSCs but also possess multipotency to placental lineages in various procedures. In addition, haiTSCs can maintain haploidy in the long term, assisted by periodic sorting and with reliance on FGF4 and heparin. Finally, we perform piggyBac-mediated high-throughput mutation in haiTSCs and use them in trophoblast lineage genetic screening. Deep sequencing analysis and validation experiments prove that Htra1 is a blocker for spongiotrophoblast specification.
Subject Areas: Cell Biology, Stem Cells Research, Genomics
Graphical Abstract
Highlights
-
•
A haploid cell line of extraembryonic lineages with self-renewal ability
-
•
haiTSCs have multipotency to functional trophoblast lineages both in vitro and in vivo
-
•
High-throughput screening of spongiotrophoblast specification-related genes in haiTSCs
-
•
Htra1 is a blocker for spongiotrophoblast-specific differentiation
Cell Biology; Stem Cells Research; Genomics
Introduction
Haploid cells serve as a powerful tool in forward and reverse genetic screening owing to their single-set chromosome feature (Shuai and Zhou, 2014). To date, haploid embryonic stem cells (haESCs) have been achieved in many species assisted by Hoechst 33342 staining and fluorescence-activated cell sorting (FACS) (Leeb and Wutz, 2011, Sagi et al., 2016), which are important for recessive gene discovery (Elling et al., 2011, Leeb et al., 2014). Recent derivation of haploid somatic cell lines has facilitated lineage-specific genetic screening (He et al., 2017, Gao et al., 2018). Nevertheless, all haploid cell cultures prefer to double back to diploids, the mechanism of which is still unclear. According to previous reports, the addition of cell cycle inhibitors (Takahashi et al., 2014, He et al., 2017) or editing of specific genes (Olbrich et al., 2017, He et al., 2018) can stabilize the haploid genome and reduce self-diploidization to some degree. However, no haploid cell line has been reported for an extraembryonic lineage. Trophoblast stem cells (TSCs) are one type of placental progenitor cell and are derived from blastocysts or the extraembryonic ectoderm of implantation embryos (Tanaka et al., 1998). They can self-renew by relying on FGF4 and heparin (F4H in vitro and retain the potential to contribute exclusively to the placenta (Oda et al., 2006). Therefore the derivation of TSCs provides considerable insight into the mechanisms that regulate extraembryonic lineage specification and placental development.
Previously, haploid cells were shown to be detectable in preimplantation blastocysts (Liu et al., 2002) and implantation epiblast-stage embryos (Shuai et al., 2015), which indicated that haploid cells were reasonable in trophectoderm (TE) lineages. By regulating the expression of Oct4 and Cdx2, embryonic stem cells (ESCs) and TSCs could switch from one to the other easily (Niwa et al., 2005, Wu et al., 2011). In addition, transcriptional induction reprograms somatic cells to functional TSCs (Kubaczka et al., 2015, Benchetrit et al., 2015), suggesting that overexpression of TSC-specific transcription factors can commit cell fate to TE lineages. Hence, haESCs may have the potential to be converted to haploid-induced TSCs (haiTSCs) via overexpression of Cdx2.
Here, we overexpressed Cdx2 in haESCs by using a Tet-On inducible system to alternate cell fate. We demonstrated that haiTSCs were generated from p53-deleted haESCs in vitro under defined conditions. haiTSCs maintained haploidy and contributed to the placenta in a chimeric experiment, proving that they potentially differentiated into functional trophoblast terminal cells. Then we performed a proof-of-concept screening in haiTSCs to identify key genes regulating spongiotrophoblast specification.
Results
Overexpression of Cdx2 Converts haESCs to TSCs
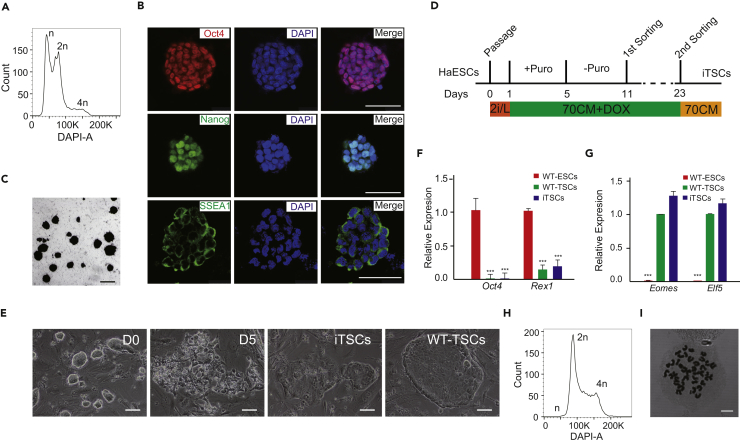
To generate haiTSCs from haESCs by conversion in vitro, we adopted an inducible overexpression strategy. Parthenogenetic haESC lines were established from 129Sv/Jae background chemical-activated oocytes, and one line with a high percentage of haploid cells was chosen to perform subsequent experiments (Figure 1A). We then designed two piggyBac (PB) vectors to introduce Tet-On inducible Cdx2 overexpression into haESCs: vector 1 had the rtTA (Tet-On Advanced transactivator) and neomycin selection genes, driven separately by an SV40 promoter, and vector 2 had the Cdx2 and puromycin resistance genes, driven by a tetracycline response element with a minimal cytomegalovirus promoter (Figure S1A). We transfected these two PB vectors and a PBase vector into haESCs by electroporation. Transfected cells were selected in 2i/L (inhibitor PD0325901, inhibitor CHIR99021, and mLif) medium (Ying et al., 2008), supplemented with G418 (250 μg/mL) for 6 days. To evaluate the pluripotency of the transfected haESCs (which we termed OE-Cdx2 haESCs), we performed immunofluorescent staining of pluripotent markers and alkaline phosphatase (AP) staining. The results showed that OE-Cdx2 haESCs were positive not only for Oct4, Nanog, and SSEA1 (Figure 1B) but also for AP (Figure 1C), which demonstrated that vector insertion did not jeopardize haESC pluripotency. Thereafter we cultured newly sorted haploid OE-Cdx2 haESCs in standard TSC culture medium supplemented with doxycycline (Dox) and puromycin to induce Cdx2 overexpression (Figure 1D). Obvious morphological change was observed 5 days after Dox induction, and many cells died during puromycin selection. Approximately 11 days after induction, typical TSC-like colonies were formed (Figure 1E) and expanded with trypsin, which meant that an inducible TSC (iTSC) line was established. Immunofluorescence results revealed that iTSCs expressed the TSC-specific markers Cdx2 and Eomes, rather than the ESC marker Oct4 (Figure S2D). Quantitative PCR (qPCR) results further confirmed that iTSCs did not express pluripotent genes (Oct4 and Rex1) relative to wild-type (WT) ESCs (Figure 1F) and showed similar expression levels of TSC markers (Eomes and Elf5) relative to WT-TSCs (Figure 1G). Therefore iTSCs were derived by conversion of haESCs in vitro through Cdx2 overexpression.
Figure 1.
Overexpression of Cdx2 Converts haESCs to TSCs
(A) DNA content analysis of haESCs. The percentage of 1n (G0/G1) peak was 50.2%.
(B) Immunofluorescence staining of pluripotent markers (Oct4, Tetramethylrhodamine [TRITC] channel; Nanog and SSEA1, fluorescein isothiocyanate channel) in haESCs. DNA is stained with DAPI. Scale bar, 50 μm.
(C) Alkaline phosphatase-stained haESCs cultured on mouse embryonic fibroblasts. Scale bar, 100 μm.
(D) Schematic overview of iTSC derivation from haESCs via Cdx2 overexpression.
(E) The morphological changes of colonies during the conversion process. WT-TSCs are used as control. Scale bar, 100 μm.
(F) The expression levels of pluripotent marker genes (Oct4 and Rex1) in iTSCs, WT-TSCs, and WT-ESCs by qPCR. t test, ***p < 0.001. Data are represented as mean ± SEM.
(G) The expression levels of TSC marker genes (Eomes and Elf5) in iTSCs, WT-TSCs and WT-ESCs. t test, ***p < 0.001. Data are represented as mean ± SEM.
(H) DNA content analysis of iTSCs derived from haESCs. The results indicated that there were no haploid cells in the iTSCs.
(I) Chromosome spreads of iTSCs. Each single cell spread had 40 chromosomes. Scale bar, 7.5 μm.
See also Figures S1 and S2.
We utilized Hoechst 33342 staining and FACS to isolate haploid cells during conversion. After optimization, we first sorted the haploid cells on day 11 post conversion; the 1n peak (haploid cells) was 4.38% and was further expanded with good viability (Figure S1B). There was no haploid cell left among the iTSCs according to the second round of sorting by FACS (Figures 1H and S2C) and chromosome spread analysis (Figure 1I). We reasoned that different PB insertions might affect haploid maintenance ability; thus we randomly picked six subclones from among OE-Cdx2 haESCs. We genotyped the subclones, and the results suggested that all subclones carried Cdx2 and rtTA (Figure S1C). Among the six subclones, #1 and #2 were stable in terms of haploid maintenance and were further assessed (Figure S2A). Although #1 and #2 carried a few insertions (Figure S1D) and could be converted to typical iTSCs easily (Figure S2B), neither of them could generate haiTSC lines (Figure S2C). We reasoned that the nature of diploidization in haESCs during conversion hindered the derivation of haiTSCs.
Deletion of p53 Facilitates Derivation of HaiTSCs
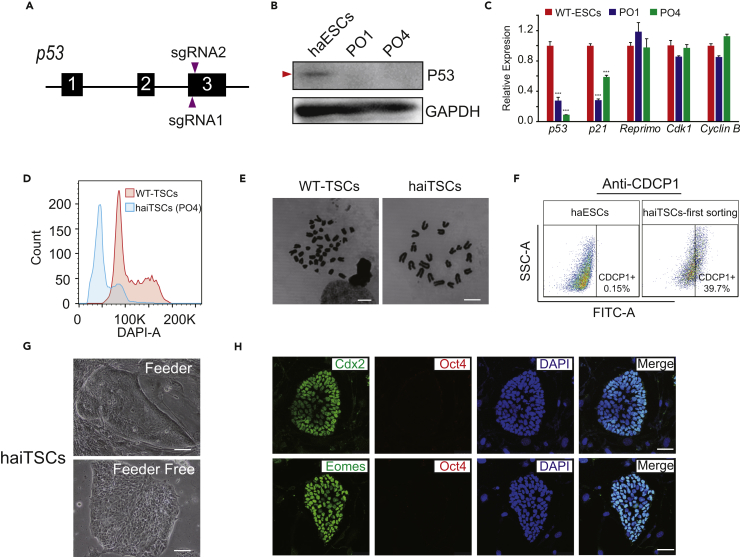
A previous study showed that p53 gene deficiency could stabilize haploidy in mouse haESCs by promoting the viability and proliferation of haploid cells in daily culture (Olbrich et al., 2017). To achieve haiTSCs in our OE-Cdx2 system, we knocked out p53 in OE-Cdx2 haESCs through CRISPR/Cas9-mediated non-homologous end joining. We transfected plasmids carrying Cas9-GFP and two single guide RNAs (sgRNAs, targeting the third exon of p53) (Figure S3B) into OE-Cdx2 haESCsubclone #1, which had a high percentage of haploid cells (Figure 2A). Approximately 36 hr after transfection, haploid cells expressing Cas9-GFP were enriched by FACS (Figure S3A). To address whether p53 deletion occurred, we randomly picked four subclones (we termed them PO1, PO2, PO3, and PO4) and performed T7 endonuclease I (T7ENI) cleavage analysis. The results showed that all subclones underwent gene editing (Figure S3C). Further sequencing results confirmed that all four subclones carried mutations with small deletions at the target sites (Figure S3D). We also detected p53 at the protein level in PO1 and PO4 by western blot and found that the p53 protein was absent in these subclones (Figure 2B). Furthermore, we analyzed the expression levels of p53-related genes in PO1 and PO4 by qPCR relative to a WT-ESC line. p53 and P21 in PO1 and PO4 were downregulated, whereas other cell cycle genes, including Reprimo, Cdk1, and Cyclin B, exhibited no significant differences (Figure 2C). Taken together, the results indicated that p53 deletion was successfully realized in OE-Cdx2 haESCs.
Figure 2.
Deletion of p53 Facilitates Derivation of haiTSCs
(A) Schematic diagram of the strategy to knock out p53 via the CRISPR/Cas9 system. The two sgRNAs are designed to target exon 3 of p53.
(B) Western blot to detect p53 in PO1, PO4, and WT-haESCs. GAPDH is used as a loading control.
(C) The expression levels of p53-related genes and cell-cycle-related genes (p53, P21, Reprimo, Cdk1, and CyclinB) in PO1, PO4, and WT-ESCs by qPCR. t test, ***p < 0.001. Data are represented as mean ± SEM.
(D) DNA content analysis of haiTSCs derived from the cell line PO4. The percentage of the 1n (G0/G1) peak was 70.4%. Diploid WT-TSCs are used as a control.
(E) Chromosome spreads of haiTSCs and WT-TSCs. haiTSCs have a 20-chromosome set, whereas WT-TSCs show 40 chromosomes in a single cell. Scale bar, 7.5 μm.
(F) TSC-specific CDCP1 antibody analysis of derived haiTSCs at first sorting. The percentage of CDCP1-positive cells is 39.7%.
(G) Images of haiTSCs colonies on feeder cells and on Matrigel. Scale bar, 100 μm.
(H) Immunofluorescence staining of TSC markers (Cdx2 and Eomes, fluorescein isothiocyanate channel) and pluripotent markers (Oct4, Tetramethylrhodamine [TRITC] channel) in haiTSCs. DNA is stained with DAPI. Scale bar, 50 μm.
See also Figure S3.
To determine whether p53 mutation could stabilize haploidy in OE-Cdx2 haESCs, we used a serum ESC medium without 2i (Elling et al., 2011) to trigger severe diploidization. We cultured PO1, PO2, PO3, and PO4 separately in this serum ESC medium for five passages without sorting and found that the PO4 cell line maintained haploidy at a very high percentage relative to the other subclones (Figure S3E). In another repeat trial, we cultured PO4 and #1 in 2i/L medium; 40.2% of PO4 cells remained in the 1n peak (haESCs at G0/G1 phase), whereas the 1n peak percentage in #1 was reduced to 16% during the same period (Figure S3F). Given that PO4 with p53 deletion maintained haploidy in a steady manner, we performed conversion with PO4 to derive haiTSCs with Dox and puromycin monitoring. After conversion for approximately 11 days and three subsequent rounds of haploidy purification by FACS (Figure S3G), a haploid cell line was established and showed mostly one set of chromosomes by FACS (Figure 2D) and karyotype analysis (Figure 2E). To enrich authentic haiTSCs from these haploid derivatives, we purified them with the TSC-specific antibody CDCP1 (Rugg-Gunn et al., 2012). The FACS results showed that approximately 39.7% of cells were already CDCP1 positive (Figure 2F); these cells were harvested and further cultured in either feeder or feeder-free culture systems with typical TSC colony morphology (Figure 2G). Immunofluorescence staining results revealed that haiTSCs expressed the TSC-specific markers Cdx2 and Eomes instead of the ESC marker Oct4 (Figure 2H).
Molecular Characterization and Differentiation Potentials of haiTSCs
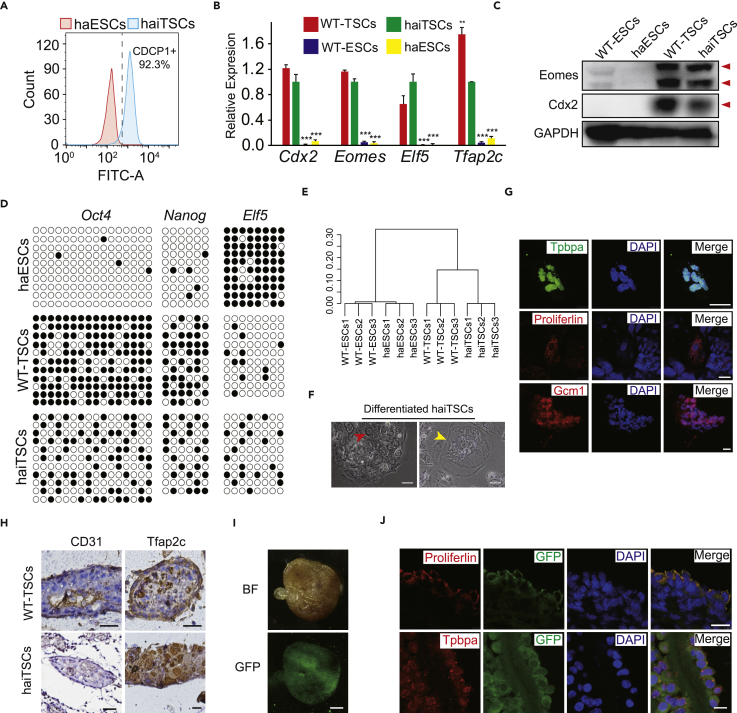
To assess the purity of haiTSCs after CDCP1-positive cell sorting, we passaged them several times and reanalyzed them with the CDCP1 antibody by FACS. CDCP1-positive cells remained at a high percentage (92.3%) in haiTSCs, which meant that haiTSCs could sustain TE identity during self-renewal (Figure 3A). The CDCP1-antibody-purified haiTSCs were expanded several times (three passages), and 15.3% of haploid cells remained (Figure S4A), indicating that haiTSCs could maintain haploidy in a TSC-specific manner. qPCR further confirmed that the expression levels of TSC-specific genes (Cdx2, Eomes, Elf5, and Tfap2c) were high in haiTSCs and WT-TSCs (Figure 3B), whereas expression levels of pluripotent genes (Oct4 and Nanog) in haiTSCs and WT-TSCs were much lower than those in haESCs and WT-ESCs (Figure S4B). Western blot results also suggested that haiTSCs and WT-TSCs were positive for Cdx2 and Eomes relative to ESCs (Figure 3C). DNA-methylation-mediated gene regulation is crucial in the determination of specific transcription factors, which are extensively utilized to judge cell identities (Wu et al., 2011). We analyzed the DNA methylation status of the Oct4, Nanog, and Elf5 promoters in haiTSCs by performing bisulfite sequencing, with haESCs and WT-TSCs as controls. Results revealed that the Elf5 promoter in haiTSCs was hypomethylated, whereas the Oct4 and Nanog promoters in haiTSCs were hypermethylated (Figure 3D), which were consequent with previous report (Ng et al., 2008). To elucidate the properties of haiTSCs on the transcriptome scale, we analyzed the global RNA levels of haiTSCs by performing RNA sequencing (RNA-seq). Cluster analysis revealed that haiTSCs resembled WT-TSCs but were distinct from either haESCs or WT-ESCs (Figures 3E and S4C). As female WT-TSCs exhibit an inactive X chromosome in a single cell, we assessed the state of the sole X chromosome in haiTSCs by costaining for histone 3 lysine 27 trimethylation (H3K27me3) and Cdx2, with female and male WT-TSCs as controls. Only H3K27me3 in female WT-TSCs (XX) accumulated in the nuclei, visualized as bright spots; however, H3K27me3 in both haiTSCs and male WT-TSCs (XY) was located all over the nuclei (Figure S4D), which indicated that the X chromosome in haiTSCs was active.
Figure 3.
Identification of haiTSCs Properties and Differentiation Potential
(A) FACS analysis of CDCP1-positive cells among established haiTSCs. haESCs are used as negative control.
(B) The expression levels of TSC-specific marker genes (Cdx2, Eomes, Elf5, and Tfap2c) in haiTSCs, WT-TSCs, haESCs, and WT-ESCs. t test, **p < 0.01, ***p < 0.001. Data are represented as mean ± SEM.
(C) Western blot analysis of Eomes and Cdx2 in WT-ESCs, haESCs, WT-TSCs, and haiTSCs. GAPDH is used as a loading control.
(D) DNA methylation status in the promoter regions of Oct4, Nanog, and Elf5. An haESCs line and a WT-TSCs line were used as controls.
(E) Global gene expression cluster analysis of transcripts in WT-ESCs, haESCs, WT-TSCs, and haiTSCs.
(F) Images of differentiated haiTSCs. Red arrow indicates syncytiotrophoblast cells, and yellow arrow indicates trophoblast giant cells. Scale bar, 100 μm.
(G) Immunofluorescence staining of three trophoblast-lineage-specific markers, Tpbpa (fluorescein isothiocyanate channel), proliferin (Tetramethylrhodamine [TRITC] channel), and GCM1 (TRITC channel), in cells differentiated from haiTSCs in vitro. DNA is stained with DAPI. Scale bar, 25 μm.
(H) Immunohistochemical (IHC) analysis of hemorrhagic lesions derived from haiTSCs. Fixed sample is IHC stained against the endothelial marker CD31 and the trophoblast marker Tfap2c.
(I) Images of BF (top) and GFP (bottom) of a chimeric placenta following blastocyst injection of haiTSCs-eGFP cells. Scale bar, 2 mm.
(J) Immunofluorescence staining of chimeric placenta following sectioning, with trophoblast-lineage-specific antibodies against proliferin and Tpbpa. Scale bar, 25 μm.
See also Figures S4 and S5.
To test the differentiation potential of haiTSCs, we cultured the haiTSCs by withdrawing FGF4, heparin, and mouse embryonic fibroblasts in vitro. Obvious polyploidy trophoblast giant cells (TGCs) and syncytiotrophoblast (SyT) cells were observed (Figure 3F) and increased gradually (Figure S5A). Immunofluorescence staining also revealed that the spongiotrophoblast-cell-specific marker Tpbpa, the TGC-specific marker proliferin, and the labyrinth-progenitor-specific marker Gcm1 were observed in differentiated cells from haiTSCs (Figure 3G). We analyzed trophoblast-lineage-specific gene expression levels of differentiated cells (4 days) from haiTSCs, iTSCs (diploid), and WT-TSCs by qPCR. Accordingly, differentiated cells from haiTSCs expressed all lineage-specific genes; in particular, Ctsq, Prl2d1, and Tpbpa increased significantly (Figure S5B). Taken together, the results indicated that haiTSCs could differentiate into diverse trophoblast lineage cells in vitro by random differentiation. To investigate the in vivo differentiation potential of haiTSCs, we transplanted approximately 1 × 106 haiTSCs into the testis of ICR mice, with WT-TSCs and ESCs as parallel controls. After 3 weeks, hemorrhagic lesions formed in the haiTSC and WT-TSC groups, whereas teratomas formed in the ESC group (Figure S5C). Blood vessels with TGC invasion were confirmed in haiTSC-derived hemorrhagic lesions by immunohistochemical staining of Tfap2c and CD31 (Figure 3H), indicating that haiTSCs could mimic placental development by undergoing differentiation in vivo. Next, we microinjected GFP-labeled haiTSCs into blastocysts to test whether they could contribute to functional placenta. In the reconstructed blastocysts, the GFP-positive haiTSCs integrated into the TE instead of the inner cell mass (ICM), showing their trophoderm nature (Figure S5D). To evaluate further development, the reconstructed embryos were transferred to the uteruses of pseudopregnant mice. On embryonic day 10.5 (E10.5), placentas and embryos were dissected from the pseudopregnant mice. We found GFP-positive cells contributing to the placentas (Figure 3I), and these cells were positive for proliferin and Tpbpa antibody staining according to fluorescence histology analysis (Figure 3J). Our findings revealed that haiTSCs could not only mimic placental development in vivo but also contributed to functional placenta, which are typical features of trophoblast progenitor cells.
piggyBac Transposon-Mediated High-Throughput Gene Trapping in haiTSCs
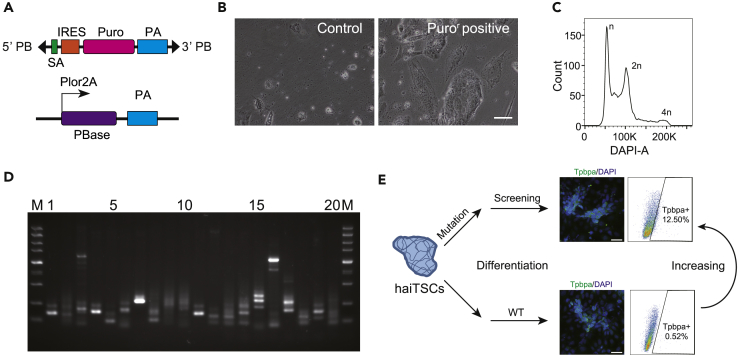
Haploid cells are easy to use for generating genome-wide homozygous mutant libraries via PB integration or virus infection (Li and Shuai, 2017). To determine the feasibility of using haiTSCs in this process, approximately 1 × 107 haiTSCs were transfected with PB-based gene trap vectors (Leeb and Wutz, 2011) carrying a puromycin resistance (Puror) gene (Figure 4A). After 4 days of puromycin selection, integrated haiTSCs with the Puror gene survived, and the control group mostly died (Figure 4B). We analyzed the mutant haiTSCs by FACS and found that 38.1% of haploid cells remained, suggesting high efficiency for obtaining homozygous mutant haiTSCs (Figure 4C). We harvested mutant haiTSCs and amplified the integration sites by performing splinkerette PCR. Obvious strands demonstrating integration were visualized (Figure 4D). We linked the products into plasmids and sequenced them by Sanger sequencing. In total, 50 different sites were addressed, of which 24 were located inside the gene body (Figures S6A and S6B). These data indicate that haiTSCs could undergo gene manipulation and generate homozygous mutant libraries.
Figure 4.
High-throughput Mutaion and Genetic screening in Derived haiTSCs
(A) Schematic diagram of piggyBac-based gene trapping vectors. The PB vector contains: 5′ PB and 3′ PB, the inverted terminal repeats (ITRs); SA, splice acceptor; IRES, internal ribozyme entry site; Puro, the coding region of puromycin resistance gene; and PA, the poly(A) sequence; the PBase vector includes Plor2A, the promoter of pbase; the Pbase, the coding sequence region of Pbase; and PA.
(B) haiTSCs transfected with piggyBac-based gene trapping vectors are selected by puromycin for 4 days. haiTSCs without transfection are used as a control. Scale bar, 100 μm. After selection, only transfected haiTSCs can survive.
(C) DNA content analysis of haiTSCs after transfection and puromycin selection. The percentage of the 1n (G0/G1) peak was 38.1%.
(D) Splinkerette PCR analysis of the transposition sites in the mutant haiTSCs. Each lane corresponds to one subclone; each strand corresponds to one PB integration.
(E) Schematic diagram of screening for relevant genes in Tpbpa-positive cell. Random differentiation was performed with mutated and nonmutated haiTSCs cells independently for 3 days. Immunofluorescence staining (Tpbpa, left panel), FACS analysis, and sorting of Tpbpa-positive cells (right panel).
See also Figure S6.
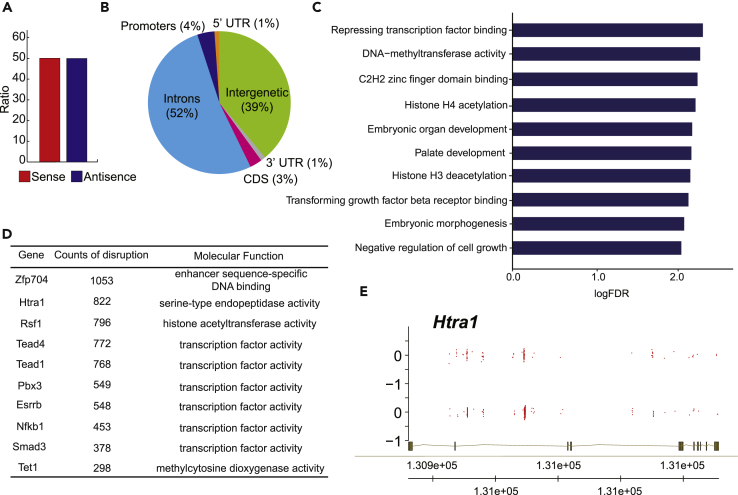
To apply haiTSCs in trophoblast lineage-specific genetic screening, we focused on key genes regulating spongiotrophoblast differentiation and utilized a specific antibody against Tpbpa (Latos and Hemberger, 2016) as indicated. Briefly, we performed random differentiation in mutated haiTSCs, as previously described, and nonmutated haiTSCs separately for 3 days. Tpbpa-positive cells demonstrating spongiotrophoblast features were analyzed by immunostaining and FACS (Figure 4E). If the percentage of Tpbpa-positive cells increased significantly in the mutated haiTSCs group relative to the nonmutated haiTSCs group, the mutated Tpbpa-positive cells were harvested with a FACS-assisted antibody for further bioinformatics analysis. We performed this experiment several times and deep-sequenced two repeats with the Tpbpa-positive cells increasing group (Figures 4E, S6C, and S6D). According to deep sequencing, approximately 4 million independent insertions across more than 20,000 genes were identified, of which 49.8% were derived from the sense orientation (Figure 5A). In addition, approximately 57% of the insertions were located in intragenetic regions (coding regions + intron + 5′/3′ UTR), whereas 39% of insertions landed in intergenetic regions (Figure 5B). Enrichment analysis with gene ontology databases showed that insertions preferred genes carrying specific functions for epigenetic modifications, such as repressing transcription factor binding and DNA methyltransferase activity (Figure 5C). Ten genes, including Zfp704, Htra1, and Rsf1, were identified (Figure 5D) due to both frequent insertions determined by PB screening and higher transcription activity measured from RNA-seq (Figures 5E and S7). Of the top candidates, Htra1 was chosen for further validation experiments in WT-TSCs (Figure 5E).
Figure 5.
Bioinformatics Analysis of Integrations in haiTSCs
(A) Proportion of insertional orientation (sense/antisense) after piggyBac integration. A total of 49.8% insertions were derived from the sense orientation.
(B) Proportion of integration sites across various genomic regions: promoters (1 kb upstream of the transcription starting sites), 5′ UTR, exons, introns, 3′ UTR, and intergenic regions. Genome-wide PB transposon integration site analysis. Promoter region is identified as the 5-kb region upstream of a known gene.
(C) Enriched gene ontologies of the top 100 genes with the most frequent insertions.
(D) List of the top 10 genes with the most insertion sites.
(E) Strand-specific coverage tracks of the gene Htra1 for the selected library (red). Gene model and chromosome coordinates are shown at the bottom.
See also Figure S7.
Htra1 As a Blocker for Spongiotrophoblast Differentiation
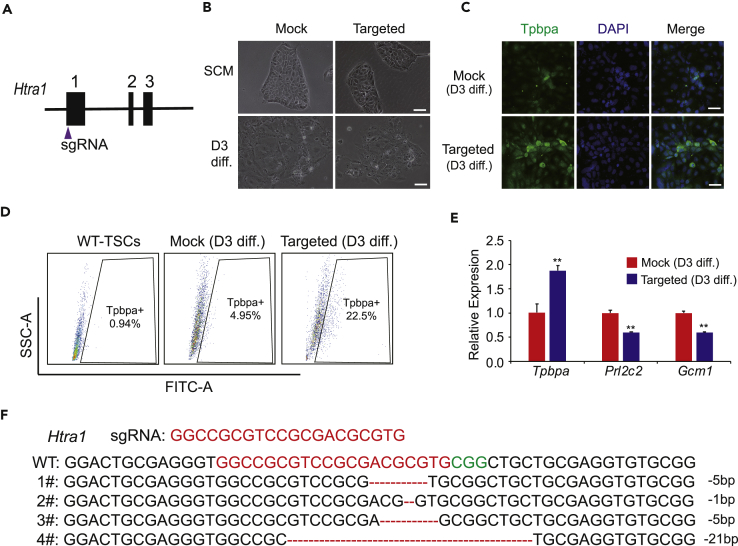
To testify whether Htra1 is an essential block against spongiotrophoblast differentiation in TSCs, we designed specific sgRNAs to knock out Htra1 in WT-TSCs with the CRISPR/Cas9 system (Figure 6A). We transfected TSCs with a Cas9-sgRNAs vector (targeted), and control TSCs were transfected with an empty vector (mock). We performed random differentiation independently with the targeted TSCs (gene edited) and mock TSCs (non-gene edited) for 3 days. Spongiotrophoblast cells were observed by morphology (Figure 6B) analysis and Tpbpa antibody immunostaining (Figure 6C). According to a FACS analysis, the percentage of Tpbpa-positive cells in the targeted group significantly increased relative to the mock TSCs group (Figure 6D). In a parallel experiment, we analyzed the expression levels of trophoblast-lineage-specific markers in differentiated cells from the targeted and mock groups and found that the expression level of the spongiotrophoblast-specific marker Tpbpa in the targeted group increased significantly relative to the mock group, whereas the other two lineage markers in the targeted group decreased (Figure 6E). We tested the genotypes of targeted TSCs, and the results indicated that the cells carried a gene mutation in Htra1 (Figure 6F). Thus Htra1 plays a very important role in spongiotrophoblast specification.
Figure 6.
Validation of Candidate Genes in a WT-TSC Differentiation Assay
(A) Schematic diagram of the strategy to knock out Htra1 in WT-TSCs using the CRISPR/Cas9 system.
(B) Bright-field images of TSCs and differentiated cells from the non-gene-edited group (mock) and the gene edited group (targeted). Scale bar, 100 μm.
(C) Immunofluorescence staining for Tpbpa (fluorescein isothiocyanate channel) in differentiated cells of the mock group and targeted group. DNA is stained with DAPI. Scale bar, 50 μm.
(D) FACS analysis of Tpbpa-positive cells in WT-TSCs, the mock group (differentiation for 3 days), and the targeted group (differentiation for 3 days). The percentage of the targeted group is 22.5%, which is higher than that of the mock group (4.95%).
(E) The expression levels of trophoblast-lineage-specific genes (Tpbpa, Prl2c2, and Gcm1) in differentiated cells (day 3) from the mock group and targeted group. t test, **p < 0.01. Data are represented as mean ± SEM.
(F) Htra1-deleted genotypes in the targeted group.
Discussion
In this study, we demonstrate that haiTSCs can be generated by the conversion of haESCs in vitro and maintain haploidy well, with the potential to differentiate into placental cells. We utilized a Dox-inducible Tet-On system to introduce Cdx2 overexpression in haESCs, which was reported to be a key regulator of TE development (Strumpf et al., 2005). Although a severe diploidization phenomenon was observed during ESC-TSC conversion, we adopted two strategies to establish haiTSCs: (1) knock out p53 in the initial OE-Cdx2 haESCs to stabilize the haploid genome and (2) modify the TSC-specific culture system to promote cell fate alternation (including specific growth factors and inhibitors, TSC-specific antibody sorting, etc.). With these modifications, our haiTSCs maintained haploidy stably through p53 gene deletion and guaranteed their potential in a subsequent gene trapping procedure. The results proved that the p53 gene was a vital regulator of diploidization, which was consistent with a previous report (Olbrich et al., 2017). However, the exact mechanism by which p53 triggers diploidization remains unknown. In addition to gene modification, we optimized the TSC culture medium with a serum-free system, named TX medium (Kubaczka et al., 2014). Haploidy was better sustained in TX medium than in traditional TSC (70CM) medium (Tanaka et al., 1998), potentially due to some unknown ingredients in serum (data not shown). In another parallel experiment, we attempted to derive haploid TSCs from haploid blastocysts in TX medium. However, no haploid cell line was established, mainly because embryonic TSCs tended to diploidize more severely. This leaves an open question: can haploid TSCs be established directly from haploid embryos and what is the mechanism underlying the thorough diploidization of haploid TE? We can compare the transcriptome differences between haiTSCs and haploid TEs from blastocysts in a future study.
As our haiTSCs were generated from haESCs in vitro, their purity and TSC properties needed to be confirmed. CDCP1 is a TSC-specific marker that has been widely used in research related to trophoblast cell fate determination (Rugg-Gunn et al., 2012, Nosi et al., 2017). Although our haiTSCs were established from an inducible system, a high percentage of CDCP1-positive cells was obtained (Figure 3A) after two rounds of FACS, and these cells maintained TSC properties. haiTSCs were maintained in a haploid state assisted by Hoechst 33342 purification and possessed differentiation potential to mimic placental development (Figures S5C and 3G), even contributing to functional placenta according to a chimeric experiment (Figures 3H and 3I). These features make haiTSCs perfect tools to evaluate the function of recessive genes related to placental development. Recently, the derivation of human TSCs was reported, raising extensive concerns regarding the study of placental abnormality in diseases associated with abortion (Okae et al., 2018). In this study, haiTSCs served as a platform to screen essential placental development genes through reverse genetic screening. Abundant mutations were introduced into haiTSCs by the PB transposon (Figures 4D, S6A and S6B), consistent with a previous report (Wang et al., 2018). Stable maintenance of haploidy by haiTSCs facilitated the production of homozygous mutant libraries (Figure 4C), which are quite valuable for genetic screening. Therefore, in this study, we used haiTSCs in high-throughput screening to identify Htra1 as a crucial gene regulating trophoblast specification. Early cell fate determination induces blastomeres to divide into the TE and ICM (Vogel, 2005, Takaoka and Hamada, 2012); however, the molecular mechanism remains unclear. haiTSCs can also serve as a tool for the discovery of key genes regulating TE and ICM interactions.
In summary, we converted haESCs to haiTSCs by controlling the expression level of Cdx2 with a Dox-inducible system. Although diploidization accompanied the conversion process, deletion of the p53 gene reduced diploidization to some degree and facilitated the derivation of haiTSCs. haiTSCs not only showed standard TSC colonies and expressed TSC-specific genes but also held the potential to differentiate into placental cells. Therefore these haploid extraembryonic progenitor cells show great advantages for trophoblast lineage genetic screening.
Limitations of the Study
In this experiment, we adapted p53 knockout strategy to establish authentic haploid extraembryonic cell line in a haploid nature. Whether the p53 knockout method could also benefit derivation of haploid TSCs from haploid blastocysts remains unknown. Meanwhile, p53 is an essential gene related to many important biological processes, including mitosis (Cross et al., 1995) and cancer (Li et al., 2010). Strikingly, p53 knockout rescued the viability of tetraploid ESCs and enabled the generation of late-stage mouse tetraploid embryos (Horii et al., 2015). Whether p53 knockout genotype would affect some unknown function of haiTSCs warrants more investigations.
Methods
All methods can be found in the accompanying Transparent Methods supplemental file.
Acknowledgments
We acknowledge Dr. Xudong Wu from Tianjin Medical University for cell sorting. This work was supported by the National Key Research and Development Program of China (2018YFC1004101 to L.S.), the National Natural Science Foundation of China (31501186, 31671538, and 31872841 to L.S.), the Natural Science Foundation of Tianjin (15JCZDJC65300 to L.S.), and the Fundamental Research Funds for the Central Universities.
Author Contributions
L.S. designed and supervised this project. K.P., X.L., Y.W., J.Z., Q.G., W.Z., and Q.Z. performed the experiments. J.Y. and C.W. analyzed the bioinformatics. L.S.,Y.F., and Y.Y. wrote the manuscript with contribution by all authors.
Declaration of Interests
The authors declare no competing interests.
Published: January 8, 2019
Footnotes
Supplemental Information includes Transparent Methods, seven figures, and one table and can be found with this article online at https://doi.org/10.1016/j.isci.2018.12.014.
Supplemental Information
References
- Benchetrit H., Herman S., van Wietmarschen N., Wu T., Makedonski K., Maoz N., Yom Tov N., Stave D., Lasry R., Zayat V. Extensive nuclear reprogramming underlies lineage conversion into functional trophoblast stem-like cells. Cell Stem Cell. 2015;17:543–556. doi: 10.1016/j.stem.2015.08.006. [DOI] [PubMed] [Google Scholar]
- Cross S.M., Sanchez C.A., Morgan C.A., Schimke M.K., Ramel S., Idzerda R.L., Raskind W.H., Reid B.J. A p53-dependent mouse spindle checkpoint. Science. 1995;267:1353–1356. doi: 10.1126/science.7871434. [DOI] [PubMed] [Google Scholar]
- Elling U., Taubenschmid J., Wirnsberger G., O'Malley R., Demers S.P., Vanhaelen Q., Shukalyuk A.I., Schmauss G., Schramek D., Schnuetgen F., von Melchner H. Forward and reverse genetics through derivation of haploid mouse embryonic stem cells. Cell Stem Cell. 2011;9:563–574. doi: 10.1016/j.stem.2011.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Q., Zhang W., Ma L., Li X., Wang H., Li Y., Freimann R., Yu Y., Shuai L., Wutz A. Derivation of haploid neural stem cell lines by selection for a Pax6-GFP reporter. Stem Cells Dev. 2018;27:479–487. doi: 10.1089/scd.2017.0193. [DOI] [PubMed] [Google Scholar]
- He W., Zhang X., Zhang Y., Zheng W., Xiong Z., Hu X., Wang M., Zhang L., Zhao K., Qiao Z., Lai W. Reduced self-diploidization and improved survival of semi-cloned mice produced from androgenetic haploid embryonic stem cells through overexpression of Dnmt3b. Stem Cell Reports. 2018;10:477–493. doi: 10.1016/j.stemcr.2017.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Z.Q., Xia B.L., Wang Y.K., Li J., Feng G.H., Zhang L.L., Li Y.H., Wan H.F., Li T.D., Xu K., Yuan X.W. Generation of mouse haploid somatic cells by small molecules for genome-wide genetic screening. Cell Rep. 2017;20:2227–2237. doi: 10.1016/j.celrep.2017.07.081. [DOI] [PubMed] [Google Scholar]
- Horii T., Yamamoto M., Morita S., Kimura M., Nagao Y., Hatada I. p53 suppresses tetraploid development in mice. Sci. Rep. 2015;5:8907. doi: 10.1038/srep08907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubaczka C., Senner C., Arauzo-Bravo M.J., Sharma N., Kuckenberg P., Becker A., Zimmer A., Brustle O., Peitz M., Hemberger M., Schorle H. Derivation and maintenance of murine trophoblast stem cells under defined conditions. Stem Cell Reports. 2014;2:232–242. doi: 10.1016/j.stemcr.2013.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubaczka C., Senner C.E., Cierlitza M., Arauzo-Bravo M.J., Kuckenberg P., Peitz M., Hemberger M., Schorle H. Direct induction of trophoblast stem cells from murine fibroblasts. Cell Stem Cell. 2015;17:557–568. doi: 10.1016/j.stem.2015.08.005. [DOI] [PubMed] [Google Scholar]
- Latos P.A., Hemberger M. From the stem of the placental tree: trophoblast stem cells and their progeny. Development. 2016;143:3650–3660. doi: 10.1242/dev.133462. [DOI] [PubMed] [Google Scholar]
- Leeb M., Dietmann S., Paramor M., Niwa H., Smith A. Genetic exploration of the exit from self-renewal using haploid embryonic stem cells. Cell Stem Cell. 2014;14:385–393. doi: 10.1016/j.stem.2013.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leeb M., Wutz A. Derivation of haploid embryonic stem cells from mouse embryos. Nature. 2011;479:131–134. doi: 10.1038/nature10448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Fang X., Baker D.J., Guo L., Gao X., Wei Z., Han S., van Deursen J.M., Zhang P. The ATM-p53 pathway suppresses aneuploidy-induced tumorigenesis. Proc. Natl. Acad. Sci. U S A. 2010;107:14188–14193. doi: 10.1073/pnas.1005960107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Shuai L. A versatile genetic tool: haploid cells. Stem Cell Res. Ther. 2017;8:197. doi: 10.1186/s13287-017-0657-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Trimarchi J.R., Keefe D.L. Haploidy but not parthenogenetic activation leads to increased incidence of apoptosis in mouse embryos. Biol. Reprod. 2002;66:204–210. doi: 10.1095/biolreprod66.1.204. [DOI] [PubMed] [Google Scholar]
- Ng R.K., Dean W., Dawson C., Lucifero D., Madeja Z., Reik W., Hemberger M. Epigenetic restriction of embryonic cell lineage fate by methylation of Elf5. Nat. Cell Biol. 2008;10:1280–1290. doi: 10.1038/ncb1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa H., Toyooka Y., Shimosato D., Strumpf D., Takahashi K., Yagi R., Rossant J. Interaction between Oct3/4 and Cdx2 determines trophectoderm differentiation. Cell. 2005;123:917–929. doi: 10.1016/j.cell.2005.08.040. [DOI] [PubMed] [Google Scholar]
- Nosi U., Lanner F., Huang T., Cox B. Overexpression of trophoblast stem cell-enriched MicroRNAs promotestrophoblast fate in embryonic stem cells. Cell Rep. 2017;19:1101–1109. doi: 10.1016/j.celrep.2017.04.040. [DOI] [PubMed] [Google Scholar]
- Oda M., Shiota K., Tanaka S. Trophoblast stem cells. Methods Enzymol. 2006;419:387–400. doi: 10.1016/S0076-6879(06)19015-1. [DOI] [PubMed] [Google Scholar]
- Okae H., Toh H., Sato T., Hiura H., Takahashi S., Shirane K., Kabayama Y., Suyama M., Sasaki H., Arima T. Derivation of human trophoblast stem cells. Cell Stem Cell. 2018;22:50–63.e6. doi: 10.1016/j.stem.2017.11.004. [DOI] [PubMed] [Google Scholar]
- Olbrich T., Mayor-Ruiz C., Vega-Sendino M., Gomez C., Ortega S., Ruiz S., Fernandez-Capetillo O. A p53-dependent response limits the viability of mammalian haploid cells. Proc. Natl. Acad. Sci. U S A. 2017;114:9367–9372. doi: 10.1073/pnas.1705133114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rugg-Gunn P.J., Cox B.J., Lanner F., Sharma P., Ignatchenko V., McDonald A.C., Garner J., Gramolini A.O., Rossant J., Kislinger T. Cell-surface proteomics identifies lineage-specific markers of embryo-derived stem cells. Dev. Cell. 2012;22:887–901. doi: 10.1016/j.devcel.2012.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagi I., Chia G., Golan-Lev T., Peretz M., Weissbein U., Sui L., Sauer M.V., Yanuka O., Egli D., Benvenisty N. Derivation and differentiation of haploid human embryonic stem cells. Nature. 2016;532:107–111. doi: 10.1038/nature17408. [DOI] [PubMed] [Google Scholar]
- Shuai L., Wang Y., Dong M., Wang X., Sang L., Wang M., Wan H., Luo G., Gu T., Yuan Y. Durable pluripotency and haploidy in epiblast stem cells derived from haploid embryonic stem cells in vitro. J. Mol. Cell Biol. 2015;7:326–337. doi: 10.1093/jmcb/mjv044. [DOI] [PubMed] [Google Scholar]
- Shuai L., Zhou Q. Haploid embryonic stem cells serve as a new tool for mammalian genetic study. Stem Cell Res. Ther. 2014;5:20. doi: 10.1186/scrt409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strumpf D., Mao C.A., Yamanaka Y., Ralston A., Chawengsaksophak K., Beck F., Rossant J. Cdx2 is required for correct cell fate specification and differentiation of trophectoderm in the mouse blastocyst. Development. 2005;132:2093–2102. doi: 10.1242/dev.01801. [DOI] [PubMed] [Google Scholar]
- Takahashi S., Lee J., Kohda T., Matsuzawa A., Kawasumi M., Kanai-Azuma M., Kaneko-Ishino T., Ishino F. Induction of the G2/M transition stabilizes haploid embryonic stem cells. Development. 2014;141:3842–3847. doi: 10.1242/dev.110726. [DOI] [PubMed] [Google Scholar]
- Takaoka K., Hamada H. Cell fate decisions and axis determination in the early mouse embryo. Development. 2012;139:3–14. doi: 10.1242/dev.060095. [DOI] [PubMed] [Google Scholar]
- Tanaka S., Kunath T., Hadjantonakis A.K., Nagy A., Rossant J. Promotion of trophoblast stem cell proliferation by FGF4. Science. 1998;282:2072–2075. doi: 10.1126/science.282.5396.2072. [DOI] [PubMed] [Google Scholar]
- Vogel G. Embryology. Embryologists polarized over early cell fate determination. Science. 2005;308:782–783. doi: 10.1126/science.308.5723.782. [DOI] [PubMed] [Google Scholar]
- Wang H., Zhang W., Yu J., Wu C., Gao Q., Li X., Li Y., Zhang J., Tian Y., Tan T. Genetic screening and multipotency in rhesus monkey haploid neural progenitor cells. Development. 2018;145 doi: 10.1242/dev.160531. [DOI] [PubMed] [Google Scholar]
- Wu T., Wang H., He J., Kang L., Jiang Y., Liu J., Zhang Y., Kou Z., Liu L., Zhang X., Gao S. Reprogramming of trophoblast stem cells into pluripotent stem cells by Oct4. Stem Cells. 2011;29:755–763. doi: 10.1002/stem.617. [DOI] [PubMed] [Google Scholar]
- Ying Q.L., Wray J., Nichols J., Batlle-Morera L., Doble B., Woodgett J., Cohen P., Smith A. The ground state of embryonic stem cell self-renewal. Nature. 2008;453:519–523. doi: 10.1038/nature06968. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.