Abstract
Background
The relationship between the gut microbiome and the human host is dynamic and we may expect adjustments in microbiome function if host physiology changes. Metatranscriptomic approaches should be key in unraveling how such adjustments occur.
Methods
We employ metatranscriptomic sequencing analyses to study gene expression in the gut microbiota of infants through their first year of life, and of their mothers days before delivery and one year afterwards.
Findings
In infants, hallmarks of aerobic metabolism disappear from the microbial metatranscriptome as development proceeds, while the expression of functions related to carbohydrate transport and metabolism increases and diversifies, approaching that observed in non-pregnant women. Butyrate synthesis enzymes are overexpressed at three months of age, even though most butyrate-producing organisms are still rare. In late pregnancy, the microbiota readjusts the expression of carbohydrate-related functions in a manner consistent with a high availability of glucose.
Interpretation
Our findings suggest that butyrate production may be ensured in the gut of young infants before the typical butyrate synthesizers of the adult gut become abundant. The late pregnancy gut microbiota may be able to access the high levels of blood glucose characteristic of this period. Moreover, late pregnancy gut bacteria may reach stationary phase, which may affect their likelihood of translocating across the intestinal epithelium.
Funds
This work was supported by grants CSD2009-00006 (CONSOLIDER Program) and SAF2009-13032-C02-02 from MICINN (Ministry of Science and Innovation, Spain), and by grant SAF2012-31187 from MINECO (Ministry of Economics and Competitiveness, Spain).
Keywords: Metatranscriptomics, Microbiota, Gut, Metabolism, Infant, Pregnancy
Research in context.
Evidence before this study
Previous studies have analyzed how the bacterial community that inhabits the human gut develops during infancy and how it is altered during pregnancy. However, these studies have not investigated gene expression patterns in the gut microbiota during such critical periods.
Added value of this study
Our metatranscriptomic analyses provide an overall picture of how gene expression in the gut microbiota evolves during the first year of life and of how it adapts to the physiological conditions of late pregnancy. As such, they reveal new insights into the global metabolism of this community and its plasticity.
Implications of all the available evidence
The new insights inform us about the pace at which the microbiota deploys new capacities to process more complex carbohydrates as infants acquire more varied diets. Our results also tell us when the bacterial metabolic pathways that synthesize important molecules for gut health, such as short chain fatty acids, start to be expressed. In the case of mothers, our analyses highlight significant metabolic alterations in the gut microbiota during late pregnancy that are likely caused by the hyperglycemic conditions characteristic of this period, and suggest that the capacity of these bacteria to translocate across the intestinal epithelium may also be affected.
Alt-text: Unlabelled Box
1. Introduction
The microbiota of the gut develops mainly during infancy, in close interaction with immune and metabolic development, and with extensive variability across individuals. Recent works have significatively advanced our understanding of microbial succession in the infant's gut by investigating this process through metagenomic sequencing [4,17,20,22,36,37]. These studies have detected some overarching patterns in microbiota development, such as a strong directionality of change throughout infancy towards the taxonomic and functional composition of the adult microbiota, with a key role for the introduction of solid foods and the cessation of breastfeeding in directing the colonization process. However, to date, no metatranscriptomic studies of this process had been performed, therefore limiting our understanding of the functional maturation of the gut microbiota during this critical period.
Similarly, although 16S rRNA and metagenomic studies have revealed changes in gut microbiota composition during pregnancy [19,37], no metatranscriptomic analyses have investigated whether there are significant adjustments at the gene expression level that might adapt the microbiota to the important immune and metabolic modifications that characterize this period. In fact, metagenomic analyses have suggested that the gene repertoire of the gut microbiota remains relatively stable during pregnancy in spite of taxonomic composition changes. However, metatranscriptomic studies are indispensable for understanding the actual functioning of a microbial community and can reveal substantial aspects of its biology that remain obscured when gene expression is not taken into account ([12,13,14,27]; Franzosa et al., 2014).
We have previously produced a detailed metagenomic analysis of intestinal microbial succession in a birth cohort of Spanish infants [37]. Here, we analyze a subset of samples from this cohort with a metatrancriptomic approach, in order to capture the temporal development of gene expression patterns in the gut microbiota community throughout the first year of life. In addition, we analyze the gut microbiota in the mothers of the infants at two points in time, i. e. days before delivery and one year after delivery. These analyses allow us to establish stage-specific patterns of expression of gut microbiome functions. These patterns bear the hallmarks of adaptation to the varying physiological conditions associated with late pregnancy and with the different developmental phases of the first year of an infant's life.
2. Materials and methods
2.1.1. Human subjects
We recruited to this study women having healthy pregnancies and stating their intention to exclusively breastfeed their infants during at least three months [37]. Women were asked to fill a questionnaire regarding their medical and reproductive history. Here, we analyze the microbial metatranscriptome in fecal samples for seven of these women and their infants (Mother Infant Pair, MIP), whose main characteristics are described in Table 1, Table 2. None of the seven women had any history of diabetes, inflammatory bowel disease, asthma or allergies and none developed gestational diabetes during pregnancy. Their Body Mass Index before pregnancy implied normal weight or slight overweight, and most gained adequate weight during pregnancy (with the exception of MIP01 who had an elevated weight gain). At the moment of delivery, these women were between 29 and 42 years of age and had not taken antibiotics in at least three months before the onset of labor. Three women (MIP06, MIP07 and MIP16) received antibiotics during delivery. All infants were born at term (>37 weeks of gestation), five of them by vaginal delivery and two by C-section (MIP06 and MIP07). Six of the infants were exclusively breastfed during at least three months, while one was partially breastfed during the first month and formula-fed thereafter (MIP09).
Table 1.
Mothers' information obtained from self-administered questionnaires.
| MIP | Age at delivery | Pre-pregnancy BMI | Pregnancy weight gain | Antibiotics at delivery | Antibiotics during the year after delivery |
|---|---|---|---|---|---|
| MIP01 | 29 years | 19.6 | 23 Kg | – | – |
| MIP03 | 30 years | 20.0 | 14 Kg | – | Amoxicillin, Cefuroxime |
| MIP06 | 42 years | 26.7 | 9 Kg | Amoxicillin | – |
| MIP07 | 31 years | 21.2 | 13 Kg | Amoxicillin | – |
| MIP08 | 30 years | 25.4 | 15 Kg | – | – |
| MIP09 | 30 years | 26.2 | 12 Kg | – | Amoxicillin |
| MIP16 | 39 years | 20.3 | 15 Kg | Amoxicillin | – |
MIP, Mother Infant Pair.
Table 2.
Infants' information obtained from questionnaires answered by parents.
| Sample | Age | Sex | Delivery | Antibiotics | Dieta |
|---|---|---|---|---|---|
| MIP01-I1 | 1 week | M | Vaginal | – | Breast Milk |
| MIP01-I3 | 3 months | – | – | – | Breast Milk |
| MIP01-I4 | 7 months | – | – | – | Solids (5 months) |
| MIP01-I5 | 1 year | – | – | – | Solids |
| MIP03-I1 | 1 week | F | Vaginal | Oftalmowellb | Breast Milk |
| MIP03-I3 | 3 months | – | – | – | Breast Milk |
| MIP03-I4 | 7 months | – | – | – | Solids (5 months 7 days) |
| MIP03-I5 | 1 year | – | – | Cefuroxime | Solids |
| MIP06-I1 | 1 week | F | C-section | – | Breast Milk |
| MIP06-I3c | 3 months | – | – | – | Breast Milk |
| MIP06-I4 | 7 months | – | – | – | Solids (5 months 12 days) |
| MIP06-I5 | 1 year | – | – | Amoxicillin | Solids |
| MIP07-I1 | 1 week | M | C-section | – | Breast Milk |
| MIP07-I3 | 3 months | – | – | – | Breast Milk |
| MIP07-I4 | 7 months | – | – | – | Solids (5 months 23 days) |
| MIP07-I5 | 1 year | – | – | – | Solids |
| MIP08-I1 | 1 week | F | Vaginal | – | Breast Milk |
| MIP08-I3 | 3 months | – | – | – | Breast Milk |
| MIP08-I4 | 7 months | – | – | – | Solids (5 moths 5 days) |
| MIP08-I5 | 1 year | – | – | – | Solids |
| MIP09-I1 | 1 week | M | Vaginal | – | Mixed |
| MIP09-I3 | 3 months | – | – | – | Formula |
| MIP09-I4 | 7 months | – | – | – | Solids (4 months) |
| MIP09-I5 | 1 year | – | – | – | Solids |
| MIP16-I1 | 1 week | M | Vaginal | – | Breast Milk |
| MIP16-I3 | 3 months | – | – | – | Breast Milk |
| MIP16-I4 | 7 months | – | – | – | Solids (6 months) |
| MIP16-I5 | 1 year | – | – | – | Solids |
MIP, Mother Infant Pair. Infant samples collected at one week (I1), three months (I3, before introduction of solid foods), seven months (I4, after introduction of solid foods) and one year after birth (I5).
The time of solid food introduction for each infant is reported in parentheses in the I4 row.
Oftalmowell is an eye drops solution containing a combination of gramicidin, neomycin and polymyxin B.
This sample was not included in the analyses as sufficient sequencing reads could not be obtained.
In addition to fecal samples, throughout the 12-months sampling period we obtained information regarding the infants' diet, general health and intake of antibiotics and other drugs, by means of specifically designed questionnaires that were given to the parents. This information allowed us to establish that all infants remained healthy throughout most of the sampling period and that solid foods were introduced into their diets between 4 and 6 months after birth (Table 2), following patterns typical of Spanish Mediterranean infant diets [6].
This study was approved by the Ethics Committee of the Center for Public Health Research (CSISP), Valencia, Spain. All women participating in the study read and signed forms of informed consent specifically approved for this project by the Ethics Committee.
2.1.2. Sample collection
Infant samples were collected at one week (I1), three months (I3, before introduction of solid foods), seven months (I4, after introduction of solid foods) and one year after birth (I5), and maternal samples were collected within one week prior to delivery (MA) and one year after (MB). Mothers were called on the day before a sample had to be collected and overall compliance with the sampling schedule was high. All fecal samples were collected by the mothers and stored in home freezers until brought to the laboratory, where they were stored at −80 °C until processing [37].
2.1.3. RNA purification and sequencing
The fecal samples were defrosted and resuspended in a 50% RNAlater (Invitrogene)/phosphate-buffered saline (PBS) (containing, per liter, 8 g of NaCl, 0.2 g of KCl, 1.44 g of Na2HPO4, and 0.24 g of KH2PO4 [pH 7.2]). Then, they were centrifuged at 2000 rpm at 4 °C for 2 min to remove fecal debris. The supernatant was centrifuged at 13,000 rpm for 5 min to pellet bacterial cells.
Total RNA was extracted using the RiboPure™ Bacteria kit (Ambion) and then treated with Baseline-ZERO™ DNAse (Illumina). The rRNA removal was performed using ScriptSeq™ Complete Kit (Illumina). Then, the rRNA-depleted RNA was processed using the above kit (ScriptSeq™ Complete Kit) to generate the double stranded cDNA and the sequencing libraries according to the manufacturer's instructions. The metatranscriptomes were sequenced using the Kit v3 (2 × 230 cycles) in a MiSeq platform (Illumina) at FISABIO-Salut Pública.
2.1.4. Sequence processing, assembly and annotation
Raw paired-end reads were trimmed to remove adapters and poly A/T tails using the cutadapt software (v1.9.1) [24]. Then, the PRINSEQ tool (v0.20.4) [32] was used to trim low-quality bases and to filter low-quality sequences. Next, each read was joined with its correspoding pair on overlapping ends by means of the fastq-join application [2].
The reads derived from the 16S rRNA gene and from the human genome were identified by aligning all reads with megaBLAST (v2.2.26) [1] and with BWA [21] against the SILVA database (release 122) [30] and the human genome (GRCh38.p1), respectively, and they were subsequently discarded.
NEWBLER (v2.9) [25] was used to assemble the paired-end sequences in order to obtain contigs. Next, these contigs (as well as the sequences not assembled into the contigs) were translated into amino acids at the six possible open reading frames using in-house R scripts (v3.1) [31]. The resulting dataset was aligned via HMMER (v3.1b2) [10] against the TIGRFAM database of prokaryotic protein family models (v9.0) [34]. After obtaining the functional annotation and the alignment coordinates for each obtained match, these coordinates were used to identify putatitive genes within the contigs. Finally, we aligned the filtered sequencing reads to the putative annotated genes in the contigs via megaBLAST and we quantified the abundace of each gene by counting the aligned reads, using in-house R scripts. We quantified the abundance of each TIGRFAM model in a sample by adding the abundance of all genes annotated with the corresponding TIGRFAM annotation.
2.1.5. Analyses of metagenome and metatranscriptome variation
We used the metagenome dataset from Vallès et al. [37] to compare variation at the metagenome and metatranscriptome levels. TIGRFAM relative abundance values in both data sets were arcsine square root-transformed and quantile normalized to variance-stabilize the data and to better approximate normality. Also, a smoothing step was applied to avoid divisions by zero and other numerical irregularities that can occur when too many zeros are present in contingency tables. The smoothing attempts to compensate for the possibility that uneven sequencing coverage might have affected the detection of lowly abundant genes in some samples. The procedure is based on random imputation, taking values falling into an interval defined for each biologically homogeneous group of samples and going from zero up to the smallest non zero observed abundance.
To statistically assess the effect of life-stage on the composition of the metatranscriptomes and metagenomes, we used ANOSIM to test whether there is a significant difference between two or more groups of samples by comparing Bray-Curtis distances between sample groups to those within groups. In addition, Canonical Correspondence Analysis (CCA), Principal Coordinates Analysis (PCoA), clustering and heatmaps were generated with in-house R scripts (v3.1).
Self-Organizing Maps (SOM) were constructed using the R function ‘som’ from the ‘som’ library. These maps are artificial neural networks that use a neighborhood function to separate a complex, high-dimensional input space into a reduced number of discrete groups with unique behaviors through time. In order to get reliable SOM-based clusters we used the bootstrap method. Firstly, we built 200 different sets of resampled temporal profiles for each feature (function) by resampling entire profiles of randomly selected individuals. Then, we carried out a SOM-based clustering over this 200-fold-sized data set. We retrieved only those features whose profiles were classified into the same cluster in at least 90% of the resampling sets.
2.1.6. Biomarker detection
The linear discriminant analysis (LDA) effect size (LEfSe) algorithm [35] was applied to identify biomarkers of the metatranscriptomes from the different life-stages analyzed, by combining Kruskal–Wallis and pairwise Wilcoxon rank-sum tests for statistical significance and feature selection. Default parameters were used for significance (p-value<0.05) and linear discriminant analysis threshold (>2.0).
2.1.7. Gene expression analysis
To detect over- or underexpressed TIGRFAM families and subroles, we compared their relative abundance in the metagenome and metatranscriptome datasets at each timepoint by means of paired Wilcoxon signed-rank tests and Student's t-tests. To control the false discovery rate, we validated the statistical tests by adjusting all p-values using the Benjamini-Hochberg correction (R library “stats”, function “p.adjust”) (BH adjusted p-value <0.05). A fold-change value was also computed for every TIGRFAM family and subrole by dividing the mean relative abundance in the metatranscriptome by the mean relative abundance in the metagenome; fold-changes >2 or <0.5 were considered significant.
2.1.8. Data availability
All sequences have been deposited in the European Nucleotide Archive server under accession number PRJEB25404. Gene expression comparisons are available upon request.
3. Results
3.1.1. Functional composition of the metatranscriptome in infants and mothers
We analyzed the microbial metatranscriptome for fecal samples of seven MIPs. Metatranscriptome analysis was performed for infant samples collected at one week (I1), three months (I3), seven months (I4) and one year after birth (I5), and for maternal samples collected within one week prior to delivery (MA) and one year after (MB). Sample MIP09-MB was not included in the analyses as sufficient amounts of total RNA could not be extracted. After several sequencing attempts, the number of sequencing reads that could be obtained for two other samples (MIP06-I3 and MIP16-MB) was significantly lower than those obtained for the rest, so these two samples were excluded from further analyses, which then proceeded with an overall total of 39 samples.
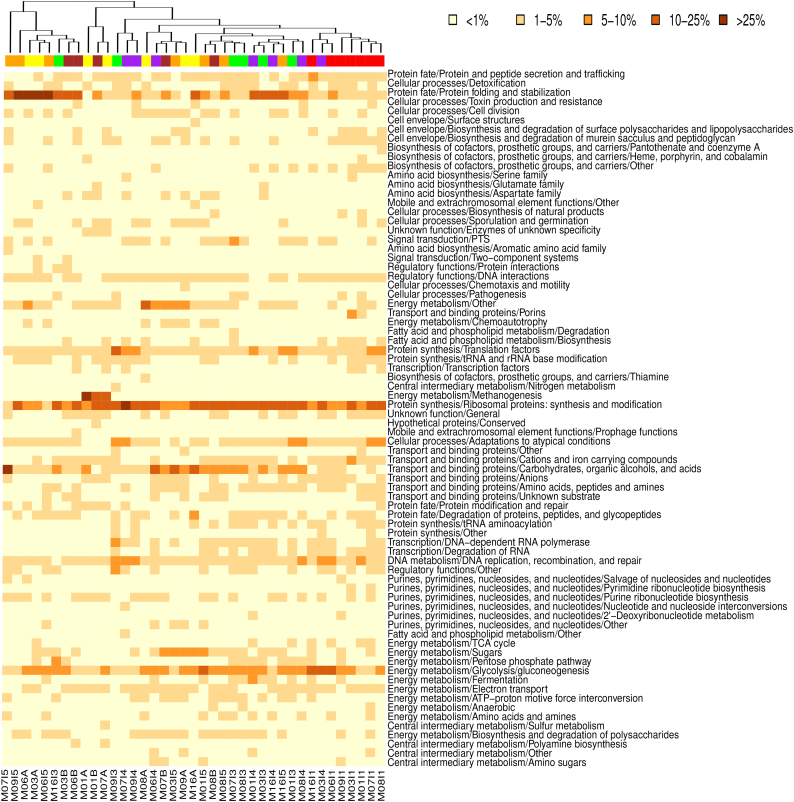
Fig. 1 presents the functional composition of the metatranscriptome of all 39 infant and maternal samples with basis on the TIGRFAM protein family classification, at subrole level (TIGRFAM is a hierarchical classification entailing main roles, the highest functional levels, and subroles, which represent more specific functions within each main role). As is the case in the metagenome [37], functions related to the main role “protein synthesis” predominate across most samples, followed by those related to “transport and binding proteins” and “energy metabolism”, with “ribosomal proteins synthesis and modification”, “carbohydrates, organic alcohols and acids” and “glycolysis and gluconeogenesis” as the most abundant subroles within the respective main roles. In addition, the “protein fate” subrole “protein folding and stabilization” is highly represented in the metatranscriptome of a large subset of the samples, although this was not the case at the metagenome level.
Fig. 1.
Heatmap and clustering of all samples based on functional composition of the metatranscriptome. Functional composition was based on TIGRFAM mainroles and subroles. Clustering was based on Bray-Curtis distances. Colors on top of the heatmap represent the timepoints to which samples belong: red, infant samples collected at one week (I1); green, at three months (I3); purple, at seven months (I4); orange, at one year (I5); yellow, maternal samples collected within one week prior to delivery (MA); brown, maternal samples collected one year after delivery (MB). Heatmap colors depict the percentage ranges of sequences assigned to the functional subroles (abundance >1% in at least one sample).
Overall, the clustering pattern does not separate the different samples according to timepoint; however, all I1 samples do group together, although the cluster also contains a single I4 sample (MIP03). The fact that the I1 cluster includes the two infants born by C-section (MIP06 and MIP07) and the infant who received formula supplementation (MIP09) indicates that the metatranscriptome differences between one-week-olds and older infants are larger than those among one-week-olds, irrespectively of their mode of birth or feeding regime. The rest of the samples from infants and mothers are distributed across different clusters without assorting according to age. This sample clustering pattern is very different from that observed for metagenomic data, where I1 samples are distributed among several clusters while all maternal samples group closely together [37].
3.1.2. Defined directionality of change in the metatranscriptome along infant development
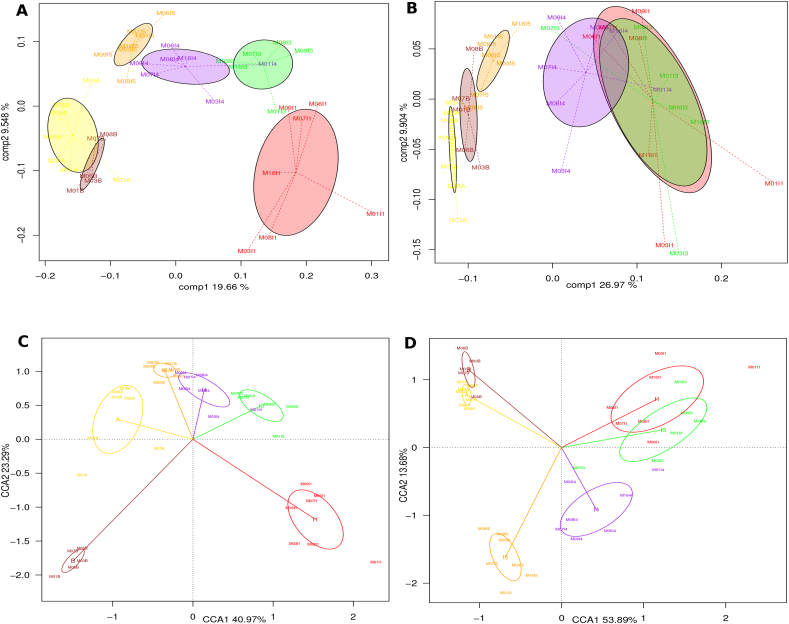
To analyze the variation existing among samples at the metatranscriptome level, we employed analysis of similarities (ANOSIM) and ordination techniques and compared the results to those obtained for the same samples with metagenomic data (using the corresponding subset of data from [37]). ANOSIM indicates that there is significant variation among the different infant and mother timepoints at both the metagenome and metatranscriptome levels (p = 0.001; Fig. S1). In order to further visualize and compare the distribution of metagenomic and metatranscriptomic variation, we performed Principal Coordinates Analysis (PCoA). Fig. 2A clearly shows a time-ordered displacement of the positions of the infants' metatranscriptomes towards those of the mothers along the first PCoA axis. The I4 samples occupy a central space, with some of them lying on either side of the axis. The colored ellipses in the figure indicate standard deviations around each timepoint centroid, showing that the different timepoints are well separated on the ordination graph. This indicates that the metatranscriptome differences among infants within a timepoint (including those that may result from variation in mode of birth, feeding regime or antibiotic use) are smaller than those present among infants of different ages. Therefore, the main source of variation in the metatranscriptome is driven by the age-related maturation of the gut microbiota. For the metagenomes, the progressive change in the gene repertoire of the infants' samples in the direction of the mothers is less evident; in this case, samples are more clearly divided into two groups, with I1, I3 and I4 on the right and I5, MA and MB on the left of the first axis, and the metagenomes of I1 and I3 remain very similarly distributed (Fig. 2B). When the time variable is directly incorporated into a Canonical Correspondence Analysis (CCA), a progressive change from timepoint to timepoint with clear directionality towards the adult state can be seen in both the metatranscriptome and the metagenome (Fig. 2C, D), in accordance with the analyses of Vallès et al. [37]. Again, in both cases the first axis of the CCA graph separates the majority of infant samples from the one-year-old and maternal samples (I5, MB and MA), indicating that progressive functional change throughout the first year has resulted in a bacterial gene repertoire that is more similar in both composition and expression to that of the mothers. As in the PCoA, in the case of the metatranscriptome some of the I4 samples lie on the left side of the axis along with the I5 and maternal samples.
Fig. 2.
Directionality of change in the metatranscriptome and metagenome along infant development. Principal Coordinates Analyses (PcoA) for functional data obtained from metatranscriptome (A) and from metagenome (B). Canonical Correspondence Analysis (CCA) of metatranscriptomic (C) and metagenomic (D) functional data. The analyses were performed at the level of TIGRFAM protein families. See also Fig. S1.
In addition, both ordination techniques and ANOSIM (Fig. S1) indicate that I1 displays the largest functional heterogeneity among coetaneous samples of infants, at both the RNA and the DNA level. At the metagenome level, I3 and I4 still have large heterogeneities; however, at the metatranscriptome level, these timepoints are already much less heterogeneous than I1. This suggests that different infants at these stages selectively express similar functions in spite of the existing interindividual differences in gene repertoires. By I5, both gene repertoire and expression have low heterogeneity levels, similar to those observed for MB. To ensure that this decrease of functional heterogeneity is not due to a reduced effect of mode of birth on the variability among individual infants as time progresses, we repeated the analyses including only the 5 infants born by vaginal delivery. Fig. S1 shows that the patterns of functional heterogeneity through time in this subset of infants are identical to those in the entire data set, reinforcing the notion that the main cause of the decrease in heterogeneity is the age-related maturation of the gut microbiota.
Regarding the maternal samples, both ordination techniques and ANOSIM indicate that, for the metatranscriptome, the heterogeneity of the perinatal samples (MA) is much larger than that of the samples taken one year later (MB), whereas for the metagenome the heterogeneity of the MA and MB samples is similar. This may suggest a decrease in the host's capacity to regulate microbiota function during late pregnancy, perhaps due to the numerous physiological changes that take place at this time.
3.1.3. Temporal dynamics of specific functions in the metatranscriptome
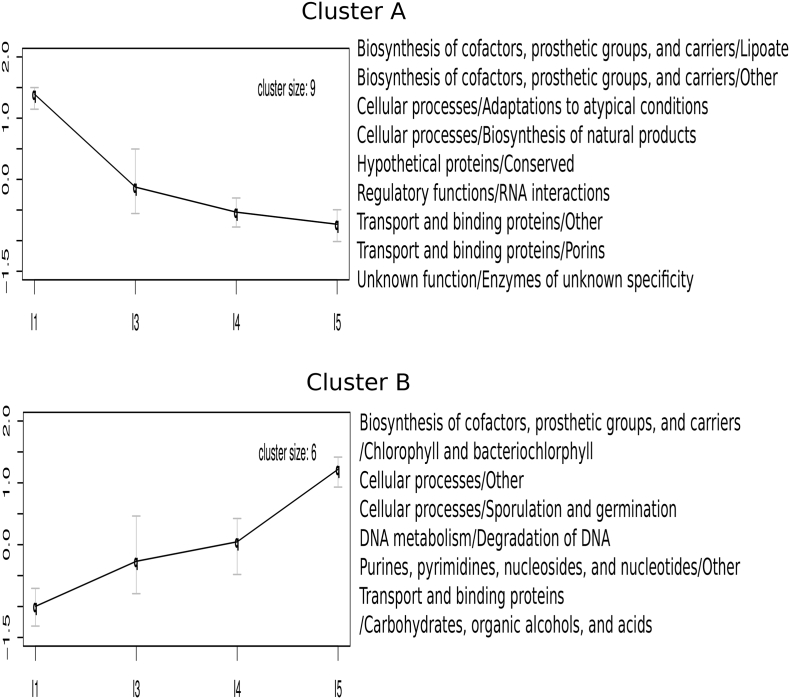
We analyzed the dynamics of specific functions in the metatranscriptome by means of a Self-Organizing Map approach (SOM). This approach identifies functions with similar abundance profiles across individual infants along development and classifies them into groups that follow the same pattern. Fig. 3 shows the two clusters of TIGRFAM subroles identified as having distinct temporal profiles with 90% support in a bootstrapped SOM procedure. Cluster A regroups nine subroles that decrease from I1 to I5, with the sharpest decrease between I1 and I3. These include several biosynthetic functions, such as “biosynthesis of natural products” and the subroles “lipoate” (an essential cofactor of aerobic metabolism) and “other” of the main role “biosynthesis of cofactors, prosthetic groups, and carriers”. A similar decreasing trend was detected at the metagenome level for the functions related to biosynthesis of natural products and biosynthesis of lipoate, as well as for several other functions related to aerobic metabolism [37]. Two subroles of the main role “transport and binding proteins” also decrease, i. e. the subroles “porins” and “other“. Other decreasing subroles include “RNA interactions” and “adaptations to atypical conditions”, as well as the uncharacterized functions “enzymes of unknown specificity” and “hypothetical proteins/conserved”.
Fig. 3.
Temporal dynamics of metatranscriptome functions. The clusters of functional subroles are based on their expression profile throughout infant development, analyzed by a Self-Organizing Map (SOM) approach. The subroles included in each of the clusters are indicated. For each cluster, average values at each timepoint are shown along with their corresponding 90% confidence intervals, in a scale centered at the mean of all samples and scaled by the standard deviation.
Cluster B regroups six subroles that increase throughout the first year, with a sharp increase between I4 and I5. These include nucleic acid related functions (“purines, pyrimidines, nucleosides and nucleotides/other”; “degradation of DNA”) as well as functions related to “sporulation and germination”, biosynthesis of “chlorophyll and bacteriochlorophyll”, and “transport and binding proteins/carbohydrates, alcohols and acids”.
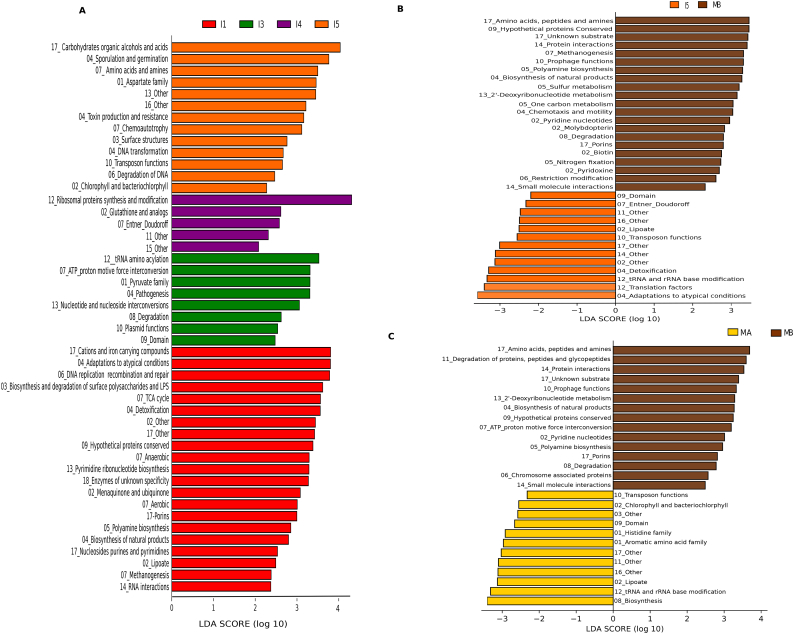
Comparisons using the linear discriminant analysis (LDA) effect size (LEfSe) method confirm that I1 and I5 are the times that display the highest numbers of characteristic functions, i. e. functional subroles that are more frequent at one of these timepoints than at any other (21 and 13, respectively). LEfSe also confirms that all of the subroles in Cluster A are significantly more frequent in the metatranscriptome of I1 than at all other timepoints in the infants, while, except for “cellular processes/other”, all subroles in Cluster B are significantly more frequent at I5 (Fig. 4A). This approach also identifies 12 other subroles that are more frequent in I1, such as additional transport and binding proteins and biosynthetic functions, as well as several subroles related to detoxification and energy metabolism, including aerobic metabolism functions. At I5, LEfSe identifies 8 subroles that are more frequent at this stage but were not identified by SOM, such as additional nucleic acid related functions (transcription, DNA transformation) as well as functions related to amino acid metabolism, chemoautotrophy, sporulation and germination, and toxin production and resistance. In addition, LEfSe identifies some functional subroles that are more frequent at timepoints I3 or I4. At I3, these involve functions related to protein synthesis, energy metabolism, metabolism of aminoacids, lipids, bases and nucleotides, plasmids, and pathogenesis. Only 5 subroles are more frequent at I4 than at any other time, and these involve functions related to protein synthesis and fate, glutathione biosynthesis, energy metabolism and signal transduction.
Fig. 4.
Changes in metabolic functions using LEfSe biomarker discovery tool at the different life-stages. (A) LEfSe analysis of subroles between infant timepoint samples. Positive LDA scores indicate enriched functions in each group. (B) LEfSe analysis of subroles between I5 and MB samples. LDA scores (log 10) for most discriminative functions in I5 are represented on the positive scale, whereas negative LDA scores indicate enriched functions in MB. (C) LEfSe analysis of subroles between MA and MB samples. LDA scores (log 10) for most discriminative functions in MB are represented on the positive scale, whereas negative LDA scores indicate enriched functions in MA. The main roles to which the subroles belong are indicated by a number: 1, amino acid biosynthesis; 2, biosynthesis of cofactors, prosthetic groups, and carriers; 3, cell envelope; 4, cellular processes; 5, central intermediary metabolism; 6, DNA metabolism; 7, energy metabolism; 8, fatty acid and phospholipid metabolism; 9, hypothetical proteins; 10, mobile and extrachromosomal element functions; 11, protein fate; 12, protein synthesis; 13, purines, pyrimidines, nucleosides, and nucleotides; 14, regulatory functions; 15, signal transduction; 16, transcription; 17, transport and binding proteins; 18, unknown function.
LEfSe also identifies that some functions that decrease during the first year are further reduced in MB (Fig. 4B); these include the biosynthesis of lipoate, as well as functions related to detoxification and adaptation to atypical conditions. In addition, several functions related to transcription, protein synthesis and fate are also less frequent in the mothers than in one-year-olds, although they had not been detected to decrease in the course of the first year. On the other hand, LEfSe identifies many subroles that are more frequent in the mothers than in one-year-olds, and, in this case, none of them had been detected to increase in the course of the first year. These include subroles related to central intermediary metabolism (polyamine biosynthesis, sulfur metabolism, one-carbon metabolism and nitrogen fixation); biosynthesis of several cofactors, prosthetic groups or carriers; different groups of transport and binding proteins; protein and small molecule interactions; chemotaxis and motility; degradation of fatty acids and phospholipids; methanogenesis; nucleotide metabolism; restriction-modification; and prophage functions. Interestingly, many of these functions that are more frequent in the metatranscriptome of MB mothers than in one-year-olds are also more frequent in MB as compared to the maternal samples taken perinatally (Fig. 4C). In contrast, MA metatranscriptomes are enriched in several biosynthetic functions, including the biosynthesis of histidine, aromatic amino acids, fatty acids and phospholipids, and lipoate.
3.1.4. Over- and underexpressed functions in the gut metatranscriptome
We used the metagenome dataset from Vallès et al. [37] to analyze gene expression level. To detect over- or underexpressed TIGRFAM families, we compared their relative abundance in the metagenome and metatranscriptome datasets at each timepoint. Taking a significance threshold of p < 0.05 (BH-adjusted Wilcoxon test) and fold-change >2 or <0.5, we identified, out of the 2261 TIGRFAM families detected in these datasets, 1530 (68%) that are differentially expressed in at least one timepoint. The specific TIGRFAM families that are differentially expressed, as well as the percentage of the total families that they represent, differs across timepoints, with a large increase at MB: 450 (21%), 445 (21%), 528 (23%), 616 (27%), 430 (19%) and 742 (40%) families are differentially expressed at I1, I3, I4, I5, MA and MB, respectively. Of these differentially expressed families, the vast majority is underexpressed (72% to 83% depending on the timepoint).
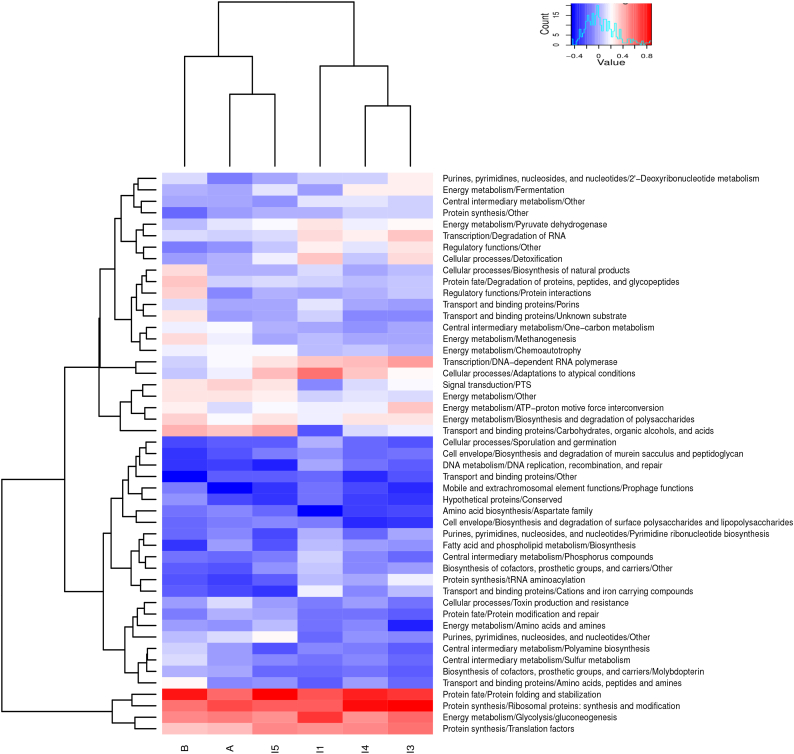
Fig. 5 shows all functional subroles that are globally over- or underexpressed at one or more timepoints. There are only four functional subroles that are consistently overexpressed across timepoints: “glycolysis/gluconeogenesis”, “synthesis and modification of ribosomal proteins”, “translation factors”, and “protein folding and stabilization”. On the other hand, numerous functional subroles are underexpressed across most or all timepoints, including those related to the transport and degradation of amino acids and peptides, as well as many of those involved in biosynthetic processes. Other generally underexpressed subroles include “sporulation/germination”, “toxin production and resistance” and “prophage functions”, as well as the information-related subroles “DNA replication, recombination, and repair”, “tRNA aminoacylation” and “protein modification and repair”.
Fig. 5.
Over- and underexpressed functions in the gut metatranscriptome.
Heatmap and hierarchical clustering of relative gene expression at subrole level, expressed as log (RNA/DNA). The represented values are means for all individuals at a given timepoint. Only subroles having an adjusted p < 0.05 in the Wilcoxon paired test for at least one timepoint are represented.
There is also a substantial fraction of functional roles that are either over- or underexpressed depending on the timepoint. The profiles of the different timepoints clearly show a separation between two main clusters, formed by I1-I3-I4, on one hand, and I5-MA-MB, on the other. Within these two main clusters, I3 and I4 are more similar to each other than they are to I1, and, remarkably, I5 and MA are more similar to each other than they are to MB.
3.1.5. Stage-specific gene expression during the first year of life
We identify numerous functions that are differentially expressed along the first year of the infants' lifes.
At I1, many functions are more strongly overexpressed than at other timepoints (Fig. 5). In particular, the subrole “adaptations to atypical conditions” is strongly overexpressed, whereas it is moderately so at other infant timepoints and it is underexpressed in mothers. Moreover, within the subrole, the most overexpressed function at I1 is TIGR03140 (Table 3), which encodes an alkyl hydroperoxide reductase subunit involved in protection from oxydative stress. “Pyruvate dehydrogenase” and “detoxification” are also most overexpressed at I1; these two subroles remain overexpressed at I3, but are underexpressed at all other timepoints. This pattern of overexpression of functions related to aerobic metabolism agrees with the higher concentration of oxygen in the gut environment at one week of birth.
Table 3.
Over- (O) or under- (U) expressed status of selected TIGRFAMs discussed in the text. The main roles to which the subroles belong are indicated by a number: 1, amino acid biosynthesis; 2, biosynthesis of cofactors, prosthetic groups, and carriers; 3, cell envelope; 4, cellular processes; 5, central intermediary metabolism; 6, DNA metabolism; 7, energy metabolism; 8, fatty acid and phospholipid metabolism; 9, hypothetical proteins; 10, mobile and extrachromosomal element functions; 11, protein fate; 12, protein synthesis; 13, purines, pyrimidines, nucleosides, and nucleotides; 14, regulatory functions; 15, signal transduction; 16, transcription; 17, transport and binding proteins; 18, unknown function.
| TIGRFAM # | Function | Main role/Subrole | I1 | I3 | I4 | I5 | MA | MB |
|---|---|---|---|---|---|---|---|---|
| TIGR03140 | alkyl hydroperoxide reductase subunit F | 4/Adaptations to atypical conditions | O | |||||
| TIGR03796 | NHLM bacteriocin system ABC transporter | – | O | O | ||||
| TIGR02706 | phosphate butyryltransferase | 7/Fermentation | O | |||||
| TIGR02707 | butyrate kinase | 7/Fermentation | O | |||||
| TIGR01216 | ATP synthase F1, ε subunit | 7/ATP-proton motive force interconversion | O | O | O | |||
| TIGR02487 | anaerobic ribonucleoside-triphosphate reductase | 13/2′-Deoxyribonucleotide metabolism | O | O | O | O | ||
| TIGR01394 | GTP-binding protein TypA/BipA | 4/Adaptations to atypical conditions | O | O | ||||
| TIGR01232 | tagatose 1,6-diphosphate aldolase | 7/Biosynthesis and degradation of polysaccharides | O | O | ||||
| TIGR01252 | alpha-acetolactate decarboxylase | 7/Fermentation | O | O | O | |||
| TIGR01501 | methylaspartate mutase, S subunit | 7/Amino acids and amines | O | O | ||||
| TIGR00822 | PTS, mannose-fructose-sorbose, IIC component | 17/Carbohydrates, organic alcohols, and acids | O | |||||
| TIGR00824 | PTS, mannose-fructose-sorbose, IIA component | 17/Carbohydrates, organic alcohols, and acids | O | |||||
| TIGR00854 | PTS, mannose-fructose-sorbose, IIB component | 17/Carbohydrates, organic alcohols, and acids | O | O | ||||
| TIGR00849 | PTS, glucitol-sorbitol-specific IIA component | 17/Carbohydrates, organic alcohols, and acids | O | O | ||||
| TIGR00792 | glycoside-pentoside-hexuronide transporter | 17/Carbohydrates, organic alcohols, and acids | O | U | O | |||
| TIGR02633 | D-xylose ABC transporter, ATP-binding protein | 17/Carbohydrates, organic alcohols, and acids | O | |||||
| TIGR02634 | D-xylose ABC transporter, D-xylose-binding protein | 17/Carbohydrates, organic alcohols, and acids | O | O | ||||
| TIGR02392 | alternative sigma factor RpoH | 4/Adaptations to atypical conditions | O | U | ||||
| TIGR01118 | galactose-6-phosphate isomerase, LacA subunit | 7/Biosynthesis and degradation of polysaccharides | O | O | ||||
| TIGR01322 | sucrose-6-phosphate hydrolase | 7/Biosynthesis and degradation of polysaccharides | O | |||||
| TIGR00410 | PTS, lactose-cellobiose family IIC component | 17/Carbohydrates, organic alcohols, and acids | O | |||||
| TIGR00776 | RhaT L-rhamnose-proton symporter family protein | 17/Carbohydrates, organic alcohols, and acids | O | |||||
| TIGR01039 | ATP synthase F1, β subunit | 7/ATP-proton motive force interconversion | O | |||||
| TIGR01114 | methyltransferase, subunit H | 7/Methanogenesis | O | |||||
| TIGR01149 | S-methyltransferase, subunit G | 7/Other | O | |||||
| TIGR03277 | putative methanogenesis marker domain 9 | – | O | |||||
| TIGR00333 | nrdI protein | 13/2′-Deoxyribonucleotide metabolism | O | |||||
| TIGR03284 | thymidylate synthase | 13/2′-Deoxyribonucleotide metabolism | U | U | O | |||
| TIGR03006 | polysaccharide deacetylase family protein | 3/Biosynthesis and degradation of surface polysaccharides | O | |||||
| TIGR02038 | periplasmic serine peptidase DegS | 11/Degradation of proteins, peptides, and glycopeptides | O | |||||
| TIGR01932 | HflC protein | 11/Degradation of proteins, peptides, and glycopeptides | U | O | ||||
| TIGR02091 | glucose-1-phosphate adenylyltransferase | 7/Biosynthesis and degradation of polysaccharides | O | |||||
| TIGR00828 | PTS, mannose-fructose-sorbose, IID component | 17/Carbohydrates, organic alcohols, and acids | O | |||||
| TIGR02975 | phage shock protein G | 4/Adaptations to atypical conditions | U | U | ||||
| TIGR00087 | 5′/3′-nucleotidase SurE | 4/Adaptations to atypical conditions | U | U | U | |||
| TIGR02474 | pectate lyase | 7/Biosynthesis and degradation of polysaccharides | U | U | U |
The subrole “glycolysis/gluconeogenesis”, which is one of the most abundant subroles in the metatranscriptome and one of the few that are overexpressed at all timepoints, is also most strongly upregulated at I1. At the same time, numerous functions are less strongly underexpressed at I1 than at other timepoints, including biosynthetic functions related to “fatty acid and phospholipid metabolism”, “biosynthesis of cofactors, prosthetic groups, and carriers” and “pyrimidine ribonucleotide biosynthesis”, as well as functions related to “DNA replication, recombination, and repair”, “central intermediary metabolism”, “transport and binding proteins/cations and iron carrying compounds” and “sporulation and germination”. In contrast, there is one functional subrole that is underexpressed at I1 but overexpressed at all later infant timepoints (i. e. “biosynthesis and degradation of polysaccharides”), as well as two subroles that are strongly underexpressed at I1, moderately underexpressed at I3 and I4 and overexpressed at I5 and in mothers (i. e. “PTS” and “transport and binding proteins/carbohydrates, organic alcohols, and acids”). Overall, in comparison to later stages, these specific patterns of gene expression at I1 suggest an active biosynthetic metabolism that is less dependent on complex carbohydrates. Also, the fact that glycolysis is more strongly overexpressed at I1, and that numerous functions related to central intermediary metabolism, and even sporulation and germination, are less strongly underexpressed, suggests that at least some of the microbiota populations have a very active metabolic status at this stage.
In turn, I3 is also characterized by several functions that are more strongly overexpressed at this timepoint than at any other (Fig. 5). These include the subroles “2′-deoxyribonucleotide metabolism”, which is only overexpressed at I3, and “ATP-proton motive force interconversion”, which is overexpressed at I3 and, weakly, at MB. There are also functions that were already overexpressed at I1 and become more strongly so at I3, including various subroles related to transcription and translation, such as “DNA−dependent RNA polymerase”, “degradation of RNA” and “translation factors”. Similarly, “tRNA-aminoacylation” is less underexpressed at I3 than at any other timepoint. This suggests a high protein synthesis rate at this stage, probably related to fast growth. Another main translation-related subrole, “ribosomal protein synthesis and modification”, which is one of the most abundant subroles in the metatranscriptome, is overexpressed at I3 beyond the levels observed at other timepoints, with the exception of I4. The “fermentation” subrole is also similarly overexpressed at I3 and I4, whereas it is underexpressed at all other timepoints. However, the specific fermentation-related functions overexpressed at the two timepoints differ. Of particular interest, TIGR02706 and TIGR02707, involved in butyrate synthesis, are only overexpressed at I3 (Table 3).
At I4, there are no functional subroles that are the most or the least strongly overexpressed in comparison to all other timepoints. All of the subroles that are overexpressed at I4 were already overexpressed at least as strongly at I3 or at I1, although there are specific protein families that become overexpressed for the first time at this point, including TIGR01394, involved in stress responses, TIGR01232 involved in lactose catabolism by the tagatose-6-phosphate pathway, and TIGR01252 and TIGR01501, related to fermentation of pyruvate and glutamate, respectively (Table 3). There are, however, subroles that had been overexpressed at I1 and I3 and become underexpressed at I4, such as “pyruvate dehydrogenase”, “regulatory functions/other” and “detoxification”.
It is at I5 that several new functional subroles become overexpressed (Fig. 5). These subroles are also overexpressed in mothers and include “PTS”, “transport and binding proteins/carbohydrates, organic alcohols, and acids”, and “energy metabolism/other”. In relation to carbohydrates, I5 displays the overexpression of numerous functions enabling the uptake of a variety of different compounds (Table 3), including, among others, the PTS Man family (TIGR00822, TIGR00824, TIGR00854), which is unique in having a broad specificity for a range of different sugars, the glucitol/sorbitol PTS (TIGR00849), the D-xylose ABC transporter (TIGR02633, TIGR02634), and the GPH:cation symporter family (TIGR00792), which includes transporters for melibiose, lactose, raffinose, glucuronides, pentosides and isoprimeverose.
In the other direction, the subrole “degradation of RNA” that had been overexpressed in younger infants is underexpressed at I5, and several subroles that were already underexpressed become more strongly so at this time, including “tRNA aminoacylation”. At the same time, “DNA-dependent RNA polymerase” remains overexpressed but at a level lower than that detected in younger infants. Together, these changes may indicate a slowing down of the growth rate.
3.1.6. Expression differences remaining at one year of life in comparison to mothers
In order to assess whether gut microbiota gene expression patterns at one year have reproduced those of adults, or whether the remaining differences in taxonomic richness, diversity and complexity of interactions among taxa [37] are reflected at this level, we compared gene expression between one-year-olds and mothers. The gene expression pattern at I5 retains several differences with those observed in mothers (Fig. 5). The subrole “DNA−dependent RNA polymerase” is no longer overexpressed in mothers, and is actually underexpressed at MB. Also, the subrole “translation factors” is less strongly overexpressed in the mothers than at I5 and the younger infants. This suggests that the growth rate of the gut microbial populations is lower in the mothers than in the one-year-olds. Similarly, the subrole “adaptations to atypical conditions” is overexpressed in infants up to I5 but underexpressed in mothers, particularly at MB. Specific functions in this subrole that are overexpressed at I5 (Table 3) but underexpressed at MB (Table 3) relate to heat shock (TIGR02392) and other stress responses (TIGR01394).
At the same time, a variety of functional subroles that are not overexpressed at I5, or at any other point in infants, are overexpressed in mothers, but, remarkably, only in the samples taken one year after delivery, not in those of the perinatal period. These subroles include “methanogenesis”, “protein interactions”, “degradation of proteins, peptides, and glycopeptides”, “biosynthesis of natural products”, and “transport and binding proteins/unknown substrates”. Another functional subrole is also overexpressed in MB but not in I5 or MA, “ATP−proton motive force interconversion”, although this subrole had already been overexpressed at I3.
3.1.7. Altered expression of specific bacterial functions during late pregnancy
We identify numerous differences between the pattern of gene expression detected in the maternal samples collected in the last week of pregnancy and that detected in the samples collected one year after delivery (Fig. 5). In particular, MB displays several overexpressed functional subroles that are not overexpressed in MA, whereas no functional subrole is overexpressed uniquely in MA. One of the functional subroles overexpressed in MB but not in MA is “biosynthesis and degradation of polysaccharides”, specifically due to the overexpression in MB of lactose catabolism enzymes of the tagatose-6-phosphate pathway (TIGR01118, TIGR01232) and of sucrose-6-phosphate hydrolase (TIGR01322), and to the underexpression of pectate lyase (TIGR02474) in MA (Table 3). Remarkably, the only function within the “biosynthesis and degradation of polysaccharides” subrole that is overexpressed in MA but not significantly so in MB is glucose-1-phosphate adenylyltransferase (TIGR02091), involved in glycogen synthesis. Furthermore, MA and MB also differ in the expression of several PTS and other carbohydrate transport systems. MB overexpresses components of PTS families specific for lactose/cellobiose (TIGR00410) and glucitol/sorbitol (TIGR00849), as well as transporters or binding proteins for L-rhamnose (TIGR00776), D-xylose (TIGR02634) and glycosides (TIGR00792). On the other hand, MA only overexpresses components of the broad specificity PTS Man family (TIGR00828, TIGR00854), which can transport glucose and other sugars. Overall, this differential pattern of expression suggests that the MB microbiota relies on a wide variety of carbohydrates, while the MA microbiota is less dependent on carbohydrates other than glucose and has an increased capacity for the storage of glucose in the form of glycogen.
Two other functional subroles related to energy metabolism are also overexpressed in MB but not in MA, i. e. the “ATP-proton motive force interconversion” and “methanogenesis” subroles. These differences are driven by the overexpression in MB of subunits of the respiratory F0F1-ATP synthase (TIGR01039, TIGR01216), and, on the other hand, of several functions that are considered markers of archaeal methanogenesis (TIGR01114, TIGR01149, TIGR03277). TIGR01216 encodes the F1 epsilon subunit, which selectively inhibits ATP hydrolysis, suggesting that the activity of the F0F1-ATP synthase in MB is mainly in the synthetic direction.
Also overexpressed in MB but not in MA are functions involved in the synthesis of deoxyribonucleotides such as ribonucleotide reductases (TIGR00333, TIGR02487) and thymidylate synthase (TIGR03284), an enzyme that produces dTMP from dUMP-, the export of proteins and bacteriocins (TIGR03006, TIGR03796), and the regulation of protein interactions, including various regulatory proteases and regulators of proteolysis (TIGR02038, TIGR01932).
On the other hand, although no functional subrole is overexpressed uniquely in MA, some subroles are less strongly underexpressed than they are in MB. This is the case of the subroles “DNA−dependent RNA polymerase” and “adaptations to atypical conditions”, with activities related to stationary phase survival (TIGR00087), heat shock (TIGR02392) and phage shock (TIGR02975) being underexpressed in MB but not in MA.
4. Discussion
4.1.1. Adaptation of microbiota gene expression to late pregnancy conditions
During pregnancy, metabolic adaptation takes place in the mother to ensure an adequate supply of nutrients to the fetus. From early pregnancy onwards, maternal food intake and fat deposition increase. In mid-to-late gestation, a series of metabolic changes promote insulin resistance and facilitate lipolysis and the utilization of the resulting glycerol molecules for gluconeogenesis [5,8]. This diabetogenic state maintains hyperglycaemia and provides a continuous supply of glucose to the growing fetus.
The observed metatranscriptomic pattern indicates that the late pregnancy microbiota adjusts gene expression to take advantage of a hyperglycaemic condition. The overexpression of numerous functions related to the binding, transport and degradation of a variety of carbohydrates observed in MB, as well as in samples from male subjects [12], is absent in MA, and is replaced by the overexpression of a single PTS transporter capable of internalizing glucose, and of enzymes of the glycogen synthesis pathway. This suggests that at least part of the gut microbiota is exposed to an increased glucose concentration in the environment. This is consistent with the fact that the low-grade inflammation and increased intestinal permeability that are characteristic of late pregnancy could facilitate an increased diffusion of glucose from the gut epithelium towards the lumen. The presence of glucose in the environment would then result in the repression of enzymes involved in the transport and metabolism of alternate carbohydrates, which are tightly regulated in response to substrate availability by carbon catabolite repression [11]. At the same time, in many bacterial species, the synthesis of glycogen will be stimulated when glucose is abundant in the medium [28].
Furthermore, energy metabolism functions other than glycolysis that are overexpressed in MB, such as “ATP-proton motive force interconversion” and “methanogenesis”, are globally under-expressed in MA. Functions related to methanogenesis, as well as some subunits of the respiratory F0F1-ATP synthase, are also overexpressed in samples from male subjects [12]. This suggests that the late pregnancy microbiota relies more heavily on glucose fermentation, rather than exploiting the variety of energy-providing strategies present in males and non-pregnant women.
The detected differences in gene expression could also suggest that the late pregnancy microbiota may be growing less actively and that some of its populations may have reached stationary phase. This is supported by the observation that some functions that are important during stationary phase growth are underexpressed in MB but not in MA. Such functions include a phage shock operon protein (TIGR02975) that down-regulates motility, thereby adjusting the use of energy and proton motive force [15]. Furthermore, under high glucose concentration, synthesis of glycogen is often stimulated when limiting growth conditions or stationary phase are reached. Finally, the fact that several biosynthetic and regulatory activities, most notably the synthesis of DNA replication precursors, are overexpressed in MB but not in MA also suggests that the late pregnancy microbiota may be growing at a slower pace. This may have important physiological consequences, as growth phase and motility could affect the likelihood of bacterial translocation, a phenomenon that is facilitated during late pregnancy as a result of the elevated intestinal permeability [7,9,29].
4.1.2. Adaptation of microbiota gene expression during the first year of life
The pattern of gene expression changes observed throughout the first year of life also indicates several lines of adaptation to the specific conditions at each stage. Gene expression at one week of age is in accordance with the presence of oxygen in the gut environment, involving an over-representation of a variety of aerobic metabolism functions as well as detoxification functions related to protection from oxydative stress. Such over-expression reinforces the elevated abundance of these functions in the metagenome at this stage, linked to the high prevalence of aerobic organisms such as the enteric bacteria in these infants during their first weeks [37]. At the same time, the energy metabolism pattern observed at I1 is unique, in that glycolysis is more upregulated than at any other point, while the biosynthesis and degradation of polysaccharides is downregulated. Interestingly, this subrole is upregulated at all other timepoints except in the mothers during the perinatal period. This indicates that, as seems to be the case in late pregnancy, the microbiota at I1 is not strongly dependent on complex carbohydrates. But, unlike what occurs in MA, other characteristics of I1 gene expression suggest that the microbiota at one week has a very active metabolic status, with an elevated biosynthetic activity.
Many of the biosynthetic and aerobiosis-related functions that are strongly overexpressed at I1 decrease by I3, while functions related to carbohydrate transport and metabolism gain in relevance. This likely indicates that by three months oxygen tensions have been largely reduced in the infant gut and that the use of carbohydrates has diversified, possibly in relation to the assimilation of the varied repertoire of human milk oligosaccharides, which are mostly utilized by the Bifidobacterium and Bacteroides species that dominate the milk-adapted microbiota of these infants [23,33,37]. Interestingly, we detect that I3 is the only timepoint where enzymes involved in butyrate synthesis are overexpressed, especifically those of the less common of the two pathways for butyrate production from butyryl-CoA, the phosphotransbutyrylase pathway. This is surprising because most butyrate-producing organisms are not yet abundant at I3 and because breastfed infants have a metabolic profile with very low proportions of butyrate, compared with adults [16]. Bifidobacterium and Bacteroides do not encode any pathway for butyrate synthesis, but Clostridium species can be abundant in some of the infants at I3 and often encode only the phosphotransbutyrylase pathway [38]. Therefore, Clostridium species may be responsible for the detected overexpression of butyrate synthesis at this stage. Although it has been assumed that butyrate is not as important in infants and that infants' enterocytes may use an alternative energy source [26], the overexpression of the phosphotransbutyrylase pathway at I3 suggests that the biosynthesis of this short chain fatty acid may be ensured even if most butyrate producers are rare. On the other hand, the higher expression at I3 of functions related to transcription and translation suggests a high protein synthesis rate, probably related to fast growth in at least some of the microbiota populations.
In spite of the fact that they are separated by the introduction of solid food to the infants' diets, timepoints I3 and I4 display relatively similar gene expression patterns, at least at the subrole level. Gene expression at I4 shows few unique characteristics, may be in correspondence to the fact that this is a transitional state in terms of diet. It is nevertheless remarkable that, in spite of the addition to the diet of cereals, vegetables and fruits, which were consumed by >80% of infants at this point [37], carbohydrate utilization subroles such as the phosphotransferase system and other transport and binding proteins remain under-regulated and expressed at levels comparable to those observed at I3. Moreover, this is despite the fact that microbiota composition at I4 has responded to the dietary changes with an increase in Bacteroides and in a diverse array of Firmicutes that are proficient at the utilization of a variety of complex carbohydrates [37]. Nevertheless, Bifidobacterium is still preponderant in many of the 7-months infants, which may contribute to maintain the overall similarity in the expression profile. The main change observed in I4 with respect to the previous timepoints is the decrease in aerobiosis-related functions, in agreement with the substantial decrease in enteric bacteria observed in the gut of these infants [37].
In contrast, the expression profile at I5 changes importantly as many new energy metabolism and carbohydrate transport functions that are expressed in adults become upregulated. This may reflect the further diversification of the diet in these infants, that by one-year of age were consuming a variety of products of plant and animal origin. This diet diversification was accompanied by a substantial increase in many Firmicutes genera, particularly of the main butyrate producers in the gut, and a decrease in Bifidobacterium, eventhough 70% of the infants were still being partially breastfed at this point. As a result of these changes, the ranking of taxon abundances observed in the one-year-old infants becomes remarkably similar to that of the mothers, with Bacteroides, Faecalibacterium, Clostridium and Ruminococcus present among the five top genera in I5, MA and MB [37]. Nevertheless, gene expression patterns at I5 retain several differences with those observed in mothers. On one hand, the decrease in the expression of transcription and translation functions seen at I5 is deepened in the mothers, suggesting a further slowing down of the growth rate, possibly linked to a saturation of the environment and a stabilization of the microbial community. On the other hand, MB, but not MA, presents a more diversified metabolism, notably including a higher expression of archaeal methanogenesis functions, and of functions related to the degradation of proteins and peptides or to the biosynthesis of natural products, likely mediated by the concomitant upregulation of regulatory protein interactions. These differences in gene expression between one-year-olds and mothers reinforce the notion that the gut microbiota assembly process is far from completed at this age, in agreement with the fact that the levels of taxonomic richness, diversity and complexity of interactions among taxa are still far from those observed in adults [37]. It is also noteworthy that many of the functions that are not yet seen to be upregulated at I5 also fail to be upregulated in the maternal microbiota at times of altered conditions such as late pregnancy. This may indicate that these latecoming functions are less central to the functioning of the gut ecosystem and can be downgraded as the microbiota adapts to specific physiological situations.
The following are the supplementary data related to this article.
ANOSIM analyses for metagenome and metatranscriptome at protein family level among (A) all samples (metagenome: R = 0.624, P = 0.001; metatranscriptome: R = 0.688, P = 0.001), and (B) samples for vaginally delivered infants only (metagenome: R = 0.647, P = 0.001; metatranscriptome: R = 0.656, P = 0.001). The length of the bows indicates the level of heterogeneity and the width the number of compared samples.
Acknowledgments
Acknowledgments
The authors thank all the mothers and babies who participated in the study for their generous collaboration.
This work was supported by grants CSD2009-00006 (CONSOLIDER Program) and SAF2009-13032-C02-02 from MICINN (Ministry of Science and Innovation, Spain), and by grant SAF2012-31187 from MINECO (Ministry of Economics and Competitiveness, Spain). The funding sources did not have any role in study design, data collection, data analysis, interpretation, or writing of the report.
Author contributions
YV, MJG and MPF designed the experiments. MJG and YV supervised the work. JC, SM and NJH performed the experiments. XP, AA, MJG and MPF analyzed the results. MJG and MPF wrote the paper.
Declaration of interests
The authors have no competing interests to declare.
References
- 1.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Aronesty E. Ea-utils: Command-line tools for processing biological sequencing data. 2011. https://github.com/ExpressionAnalysis/ea-utils
- 4.Bäckhed F., Roswall J., Peng Y., Feng Q., Jia H., Kovatcheva-Datchary P. Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe. 2015;17:690–703. doi: 10.1016/j.chom.2015.04.004. [DOI] [PubMed] [Google Scholar]
- 5.Barbour L.A., McCurdy C.E., Hernandez T.L., Kirwan J.P., Catalano P.M., Friedman J.E. Cellular mechanisms for insulin resistance in normal pregnancy and gestational diabetes. Diabetes Care. 2007;30(Suppl. 2) doi: 10.2337/dc07-s202. [DOI] [PubMed] [Google Scholar]
- 6.Capdevila F., Vizmanos B., Martí-Henneberg C. Implications of the weaning pattern on macronutrient intake, food volume and energy density in non-breastfed infants during the first year of life. J Am Coll Nutr. 1998;17:256–262. doi: 10.1080/07315724.1998.10718756. [DOI] [PubMed] [Google Scholar]
- 7.de Andrés J., Jiménez E., Chico-Calero I., Fresno M., Fernández L., Rodríguez J.M. Physiological translocation of lactic acid bacteria during pregnancy contributes to the composition of the milk microbiota in mice. Nutrients. 2018;10(14) doi: 10.3390/nu10010014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Diderholm B., Stridsberg M., Ewald U., Lindeberg-Nordén S., Gustafsson J. Increased lipolysis in non-obese pregnant women studied in the third trimester. BJOG. 2005;112:713–718. doi: 10.1111/j.1471-0528.2004.00534.x. [DOI] [PubMed] [Google Scholar]
- 9.Donnet-Hughes A., Perez P.F., Dore J., Leclerc M., Levenez F., Benyacoub J. Potential role of the intestinal microbiota of the mother in neonatal immune education. Proc Nutr Soc. 2010;69:407–415. doi: 10.1017/S0029665110001898. [DOI] [PubMed] [Google Scholar]
- 10.Durbin R., Eddy S.R., Krogh A., Mitchison G.J. 1998. Biological sequence analysis: probabilistic models of proteins and nucleic acids (Cambridge University Press) [Google Scholar]
- 11.Flint H.J., Scott K.P., Duncan S.H., Louis P., Forano E. Microbial degradation of complex carbohydrates in the gut. Gut Microbes. 2012;3:289–306. doi: 10.4161/gmic.19897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Franzosa E.A., Morgan X.C., Segata N., Waldron L., Reyes J., Earl A.M., Giannoukos G., Boylan M.R., Ciulla D., Gevers D. Relating the metatranscriptome and metagenome of the human gut. Proc Natl Acad Sci. 2014;111:E2329–E2338. doi: 10.1073/pnas.1319284111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frias-Lopez J., Shi Y., Tyson G.W., Coleman M.L., Schuster S.C., Chisholm S.W. Microbial community gene expression in ocean surface waters. Proc Natl Acad Sci. 2008;105:3805–3810. doi: 10.1073/pnas.0708897105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gosalbes M.J., Durbán A., Pignatelli M., Abellan J.J., Jiménez-Hernández N., Pérez-Cobas A.E. Metatranscriptomic approach to analyze the functional human gut microbiota. PLoS One. 2011;6 doi: 10.1371/journal.pone.0017447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kanematsu T., Yasunaga A., Mizoguchi Y., Kuratani A., Kittler J.T., Jovanovic J.N. Modulation of GABAA receptor phosphorylation and membrane trafficking by phospholipase C-related inactive protein/protein phosphatase 1 and 2A signaling complex underlying brain-derived neurotrophic factor-dependent regulation of GABAergic inhibition. J Biol Chem. 2006;281:22,180–22,189. doi: 10.1074/jbc.M603118200. [DOI] [PubMed] [Google Scholar]
- 16.Knol J., Scholtens P., Kafka C., Steenbakkers J., Groß S., Helm K. Colon microflora in infants fed formula with galacto- and fructo-oligosaccharides: More like breast-fed infants. J Pediatr Gastroenterol Nutr. 2005;40:36–42. doi: 10.1097/00005176-200501000-00007. [DOI] [PubMed] [Google Scholar]
- 17.Koenig J.E., Spor A., Scalfone N., Fricker A.D., Stombaugh J., Knight R. Succession of microbial consortia in the developing infant gut microbiome. Proc Natl Acad Sci. 2011;108:4578–4585. doi: 10.1073/pnas.1000081107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koren O., Goodrich J.K., Cullender T.C., Spor A., Laitinen K., Kling Bäckhed H. Host remodeling of the gut microbiome and metabolic changes during pregnancy. Cell. 2012;150:470–480. doi: 10.1016/j.cell.2012.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kostic A.D., Gevers D., Siljander H., Vatanen T., Hyötyläinen T., Hämäläinen A.M. The dynamics of the human infant gut microbiome in development and in progression towards type 1 diabetes. Cell Host Microbe. 2015;17:260–273. doi: 10.1016/j.chom.2015.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lim E.S., Zhou Y., Zhao G., Bauer I.K., Droit L., Ndao I.M. Early life dynamics of the human gut virome and bacterial microbiome in infants. Nat Med. 2015;21:1228–1234. doi: 10.1038/nm.3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marcobal A., Barboza M., Sonnenburg E.D., Pudlo N., Martens E.C., Desai P. Bacteroides in the infant gut consume milk oligosaccharides via mucus-utilization pathways. Cell Host Microbe. 2011;10:507–514. doi: 10.1016/j.chom.2011.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011;17:10–12. [Google Scholar]
- 25.Miller J.R., Delcher A.L., Koren S., Venter E., Walenz B.P., Brownley A. Aggressive assembly of pyrosequencing reads with mates. Bioinformatics. 2008;24:2818–2824. doi: 10.1093/bioinformatics/btn548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Parrett A.M., Edwards C.A. In vitro fermentation of carbohydrate by breast fed and formula fed infants. Arch Dis Child. 1997;76:249–253. doi: 10.1136/adc.76.3.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pérez-Cobas A.E., Gosalbes M.J., Friedrichs A., Knecht H., Artacho A., Eismann K. Gut microbiota disturbance during antibiotic therapy: a multi-omic approach. Gut. 2013;62:1591–1601. doi: 10.1136/gutjnl-2012-303184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Preiss J., Romeo A. Vol. 30. 1989. Physiology, biochemistry and genetics of bacterial glycogen synthesis. Advances in microbial physiology; pp. 183–233. [DOI] [PubMed] [Google Scholar]
- 29.Perez P.F., Dore J., Leclerc M., Levenez F., Benyacoub J., Serrant P. Bacterial imprinting of the neonatal immune system: lessons from maternal cells? Pediatrics. 2007;119:e724–e732. doi: 10.1542/peds.2006-1649. [DOI] [PubMed] [Google Scholar]
- 30.Quast C., Pruesse E., Yilmaz P., Gerken J., Schweer T., Yarza P. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Opens external link in new window. Nucleic Acids Res. 2013;4:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Development Core Team R. 2011. R: A language and environment for statistical computing (Vienna, R Foundation for Statistical Computing) [Google Scholar]
- 32.Schmieder R., Edwards R. Quality control and preprocessing of metagenomic datasets. Bioinformatics. 2011;27:863–864. doi: 10.1093/bioinformatics/btr026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sela D.A., Mills D.A. Nursing our microbiota: molecular linkages between bifidobacteria and milk oligosaccharides. Trends Microbiol. 2011;18:298–307. doi: 10.1016/j.tim.2010.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Selengut J.D., Haft D.H., Davidsen T., Ganapathy A., Gwinn-Giglio M., Nelson W.C. TIGRFAMs and Genome Properties: tools for the assignment of molecular function and biological process in prokaryotic genomes. Nucleic Acids Res. 2007;35(Database issue):D260–D264. doi: 10.1093/nar/gkl1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Segata N., Izard J., Waldron L., Gevers D., Miropolsky L., Garrett W.S. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12:R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vallès Y., Gosalbes M.J., de Vries L.E., Abellán J.J., Francino M.P. Metagenomics and development of the gut microbiota in infants. Clin Microbiol Infect. 2012;18:21–26. doi: 10.1111/j.1469-0691.2012.03876.x. [DOI] [PubMed] [Google Scholar]
- 37.Vallès Y., Artacho A., Pascual-García A., Ferrús M.L., Gosalbes M.J., Abellán J.J. Microbial succession in the gut: Directional trends of taxonomic and functional change in a birth cohort of spanish infants. PLoS Genet. 2014;10:e1004406. doi: 10.1371/journal.pgen.1004406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vital M., Howe A.C., Tiedje J.M. Revealing the bacterial butyrate synthesis pathways by analyzing (meta)genomic data. MBio. 2014;5 doi: 10.1128/mBio.00889-14. e00889–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
ANOSIM analyses for metagenome and metatranscriptome at protein family level among (A) all samples (metagenome: R = 0.624, P = 0.001; metatranscriptome: R = 0.688, P = 0.001), and (B) samples for vaginally delivered infants only (metagenome: R = 0.647, P = 0.001; metatranscriptome: R = 0.656, P = 0.001). The length of the bows indicates the level of heterogeneity and the width the number of compared samples.
Data Availability Statement
All sequences have been deposited in the European Nucleotide Archive server under accession number PRJEB25404. Gene expression comparisons are available upon request.