Abstract
Background
Rhythm abnormalities are crucial for diverse diseases. However, their role in disease progression induced by Helicobacter pylori (H. pylori) remains elusive.
Methods
H. pylori infection was used in in vivo and in vitro experiments to examine its effect on rhythmic genes. The GEO database was used to screen H. pylori affecting rhythm genes, and the effect of rhythm genes on inflammatory factors. Chromatin immunoprecipitation and dual luciferase assays were used to further find out the regulation between molecules. Animal models were used to confirm the relationship between rhythm genes and H. pylori-induced inflammation.
Findings
BMAL1 disorders aggravate inflammation induced by H. pylori. Specifically, H. pylori induce BMAL1 expression in vitro and in vivo through transcriptional activation of LIN28A, breaking the circadian rhythm. Mechanistically, LIN28A binds to the promoter region of BMAL1 and directly activates its transcription under H. pylori infection. BMAL1 in turn functions as a transcription factor and enhances the expression of proinflammatory cytokine TNF-α, thereby promoting inflammation. Of note, BMAL1 dysfunction in the rhythm disorder animal model aggravates inflammatory response induced by H. pylori infection in vivo.
Interpretation
These findings in this study imply the pathogenic relationship between BMAL1 and H. pylori. BMAL1 may serve as a potential diagnostic marker and therapeutic target for the early diagnosis and treatment of diseases related to H. pylori infection.
Fund
National Natural Science Foundation of China.
Keywords: BMAL1, H. pylori, Circadian rhythm, TNF-α, LIN28A
Research in context.
Evidence before this study
Circadian rhythm controls behavior, physiology, and mood, and makes them adapt to 24 h day-night periodicity. Dysfunction of circadian clock genes leads to a variety of disorders, such as metabolic syndrome, obesity, diabetes, autoimmunity, and even cancer.
Gastric cancer (GC) is the fourth most common cancer worldwide and has a high mortality rate. Most gastric adenocarcinomas start from chronic superficial gastritis, and finally evolve to dysplasia and adenocarcinoma. Gastritis paves the way for malignant transformation of gastric mucosa. Helicobacter pylori (H. pylori) was recognized as a type I carcinogen. Several studies uncovered the connection between circadian rhythm, H. pylori, and inflammatory diseases. However, the role of circadian rhythm in disease progression induced by H. pylori remains elusive.
Added value of this study
Through in vivo and in vitro experiments, we for the first time found the pathogenic relationship between H. pylori infection and circadian rhythm gene BMAL1. And we explored one of the specific pathways by which circadian rhythm genes mediated inflammatory responses induced by H. pylori.
Implications of all the available evidence
Therefore, the rhythmic gene BMAL1 may serve as a potential diagnostic marker and therapeutic target for the early diagnosis and treatment of diseases related to H. pylori infection.
Alt-text: Unlabelled Box
1. Introduction
Circadian rhythm controls behavior, physiology, and mood, making them adapt to 24 h day-night periodicity. The external daily stimuli regulate the circadian synchronization. Circadian rhythm is determined by the expression of the circadian clock genes which control approximately 10% of all genes [1]. Circadian clock genes consist of 3 basic helix-loop-helix/PAS domain containing proteins: histone acetyltransferase CLOCK, transcription factors BMAL1 and NPAS2. These proteins can be assembled as transcriptional complex in heterodimer form, CLOCK/BMAL1 or NPAS2/BMAL1. The complex binds to the E-box elements of targeting gene promoters, activating transcription of the period genes (PER1, PER2 and PER3), cryptochrome genes (CRY1 and CRY2), and other core clock genes [2]. Dysfunction of circadian clock genes leads to a variety of disorders, such as metabolic syndrome, obesity, diabetes, autoimmunity, and even cancer [3].
Gastric cancer (GC) is the fourth most common cancer worldwide with a high mortality rate [4]. Most gastric adenocarcinomas start from chronic superficial gastritis, and finally evolve to dysplasia or adenocarcinoma [5]. Gastritis paves the way for malignant transformation of gastric mucosa. There is a bacterial community with hundreds of phylotypes in the stomach [6,7]. However the pH is less than four in the stomach cavity, making it difficult for microbiota to grow excessively under such condition [8]. Helicobacter pylori (H. pylori) infection leads to decreased acid secretion, which may favor different gastric bacteria to grow [9]. In the infected stomach, H. pylori accounts for 93% to 97% of the microbiota, and microbial diversity is significantly reduced [7]. This implies that H. pylori infection triggers a series of abnormal reactions, and contributes most to the gastric diseases. In addition, the global epidemiological distribution of chronic gastritis overlaps that of H. pylori infection despite different causes of the diseases [[10], [11], [12]]. H. pylori has been recognized as a type I carcinogen since 1994, and epidemiological studies have shown its role to increase the risk of GC. Therefore, we focus on the pathogenic effects of H. pylori in this study. Two independent groups showed that individual gastrointestinal cells transcribed and translated circadian clock molecules with circadian periodicity in a cell-autonomous manner, out of phase with the SCN [13], indicating the influence of local factors on circadian rhythm. Interestingly, H. pylori showed effects on circadian gastric acidity which implied the potential relationship between circadian rhythm and H. pylori induced disorders [14]. Several other studies uncovered the connection between circadian rhythm and inflammatory diseases [[15], [16], [17], [18], [19], [20], [21]]. Therefore, we assumed that circadian rhythm may be crucial for the development of inflammatory disease induced by H. pylori.
H. pylori-induced gastric diseases can be activated by inflammatory cytokines, such as IL-1, TNF-α, IL-6, and IL-8 [22,23]. TNF-α, IL-6, and IL-8 are the well-known inflammatory factors among the cytokines secreted by gastric epithelial cells, and they can be induced by H. pylori [24,25]. In addition, dysfunction of CRY1 and CRY2 accelerated the progression of arthritis by promoting the secretion of TNF-α [26]. Therefore, circadian rhythm change, especially the expression of core circadian clock genes may determine the pathogenic effect of H. pylori by regulating the levels of pro-inflammatory cytokines. It is of significance to illustrate the underlying mechanisms.
LIN28 is an RNA binding protein which activates cell transformation by inhibiting let-7 to promote inflammatory responses [27]. LIN28 inhibited the biosynthesis of let-7, and interestingly it was a target of let-7 itself. There was a bidirectional negative feedback regulation loop between them [28]. H. pylori inhibited the expression of most molecules of let-7 family, and some let-7 family molecules may be of value as potential biomarkers for the diagnosis of GC or its precancerous diseases [[29], [30], [31]]. Furthermore, let-7 regulated circadian rhythm and it was controlled by circadian rhythm genes as well [32,33]. The above findings suggested the potential critical role of LIN28/let-7 axis for the circadian rhythm and its involvement in the pathogenic process of H. pylori.
In this study, we demonstrated that H. pylori up-regulated the expression of BMAL1 and disrupted its rhythm through activation of LIN28a. BMAL1 enhanced the transcription of TNF-α, thereby aggravating the inflammatory response caused by H. pylori. Thus, we for the first time found the pathogenic relationship between H. pylori infection and circadian rhythm gene BMAL1. And we explored one of the specific pathways by which circadian rhythm genes mediated inflammatory responses induced by H. pylori. Therefore, the rhythmic gene BMAL1 may be a potential diagnostic marker and therapeutic target for H. pylori induced inflammatory diseases.
2. Methods and materials
2.1. IHC staining
Formalin-fixed, paraffin-embedded (FFPE) sections from mouse or patient samples were subjected to deparaffination and dehydration. After epitope retrieval with citrate buffer and H2O2 treatment, the samples were blocked with goat serum for 30 min, and then incubated with different primary antibodies overnight at 4 °C. And then the sections were incubated with secondary anti-mouse or -rabbit antibodies and the signals were detected by a DAB staining kit (Vector Laboratories, USA). The IHC scores were counted with Imagepro Plus. Primary antibodies used were BMAL1: AB4140 (Millipore), and LIN28A: ab155542 (Abcam).
2.2. RNA extraction, RT-PCR, and qRT-PCR
Total RNA were extracted with Trizol Reagent, and the mRNAs were reverse transcribed with RT reagent Kit gDNA Eraser (TaKaRa) to get cDNAs as the templates for the subsequent qRT-PCR. SYBR-Green (TaKaRa) and the corresponding qRT-PCR protocol were used to detect cDNA expression levels. β-actin was used as internal reference. Primers were shown as follows: hACTIN, Forward (F):5′-AGTTGCGTTACACCCTTTCTTG-3′, Reverse (R):5′-CACCTTCACCGTTCCAGTTTT-3′; mActin, F:5′-GTGACGTTGACATCCGTAAAGA-3′, R:5′-GCCGGACTCATCGTACTCC-3′; hBMAL1, F:5′-ATGTGACCGAGGGAAGATAC-3′, R:5′-GTGCTCCAGAACATAATCGA-3′; mBmal1, F:5′-ACAGTCAGATTGAAAAGAGGCG-3′, R:5′-GCCATCCTTAGCACGGTGAG-3′; hLIN28A, F:5′-CAGGTGCTACAACTGTGGAGGT-3′, R:5′-AGGGCTGTGGATTTCTTCTTCT-3′; hIL-1α, F:5′-TGGTAGTAGCAACCAACGGGA-3′, R:5′-ACTTTGATTGAGGGCGTCATTC-3′; hIL-1β, F:5′-ATGATGGCTTATTACAGTGGCAA-3′, R:5′-GTCGGAGATTCGTAGCTGGA-3′; hTNF-α, F:5′-CTGGGCAGGTCTACTTTGGG-3′, R:5′-CTGGAGGCCCCAGTTTGAAT-3′; hIL-6, F:5′-TCAATATTAGAGTCTCAACCCCCA-3′, R:5′-GCTCCTGGAGGGGAGATAGA-3′; hCXCL8, F:5′-TTTTGCCAAGGAGTGCTAAAGA-3′, R:5′-AACCCTCTGCACCCAGTTTTC-3′; hIL-10, F:5′-GACTTTAAGGGTTACCTGGGTTG-3′, R:5′-TCACATGCGCCTTGATGTCTG-3′; mTnf, F:5′-CAGGCGGTGCCTATGTCTC-3′, R:5′-CGATCACCCCGAAGTTCAGTAG-3′; hCLOCK, F:5′- CTCTTCTCGGAGTTCAAGAAAATCA-3′, R:5′- CATGCCTTGTGGAATTGGTA-3′; hCRY1, F:5′- TCCGATTTGGTTGTTTGTCA-3′, R:5′- GGATTATTTGTTGCTGCTGTAT-3′; hCRY2, F:5′- AACCACGACGAGACCTACGG-3′, R:5′- GGAGGGAGTTGGCGTTCATT-3′; hDBP, F:5′- CTGATCTTGCCCTATCAAGCATT-3′, R:5′- CGATGTCTTCGAGGGTCAAAG-3′; hPER1, F:5′- GCCAACCAGGAATACTACCAGC-3′, R:5′- GTGTGTACTCAGACGTGATGTG-3′; hPER2, F:5′- TCATTGGGAGGCACAAAGTC-3′, R:5′- AGGAGGTCTGGCTCATAAGGT-3′; hPER3, F:5′- CTGTGAGGATTTGAGGAACG-3′, R:5′- CTGATTTAGGTATGGCTGGG-3′; hREV-ERBα, F:5′- GGGAGGTGGTAGAGTTTGCC-3′, R:5′- TGTCTGGTCCTTCACGTTGA-3′; hRORα, F:5′- CACGACGACCTCAGTAACTACA-3′, R:5′- TGGTGAACGAACAGTAGGGAA-3′; hTIMELESS, F:5′- TCTGATCCGCTATTTGAGGCA-3′, R:5′- GGCAGAAGGTCGCTCTGTAG-3′.
2.3. Cell culture and H. pylori infection
BGC-823 cells were cultured in RPMI-1640 containing 10% fetal bovine serum (FBS), and AGS cells were cultured in F12 containing 12% FBS. All cells were supported in a humidified incubator with 37 °C, 5% CO2. H. pylori strains 11,637 and 26,695 were used to challenge gastric epithelial cells with different concentration, and the cells were collected at different time points as shown in the figures. Without special illustration, the gastric epithelial cells were challenged by H. pylori with MOI 1:100, and collected after 8 h.
2.4. Western blot
Total proteins were extracted with RIPA lysis buffer (Beyotime) with phosphatase and protease inhibitors, and the protein concentrations were detected with BCA Protein Assay Kit (Thermo Fisher Scientific). SDS-PAGE was used to separate lysates, and then the proteins were transferred to PVDF membranes. The membranes were blocked with 5% non-fat milk for 2 h at room temperature, and incubated with primary antibody overnight at 4 °C. After being incubated with secondary antibody, the signals were detected with millipore ECL regents. Antibodies used were: β-ACTIN (A1978, Sigma-Aldrich, RRID: AB_476692), BMAL1 (D2L7G,CST, RRID: AB_2728705), CagA (Abcam, RRID:AB_2049729), LIN28A (ab155542, Abcam).
2.5. Resetting of circadian time in gastric epithelial cells
After stimulating the cells with 50% horse serum, the medium was replaced by 2 ml F12 or RPMI-1640 supplemented with 10% or 12% FBS respectively in 6-well plate [34], and then challenged the cells with H. pylori. The cells were collected at 0, 4, 8, 12, 16, 20 h time points after the serum shock.
2.6. Mouse model
80 C57BL/6 mice were separated into two groups randomly: one group was under a 12 h of light-12 h of dark (LD) cycle (N = 40), and another group was under constant darkness (DD, N = 40). They were raised for one week with normal feed to adapt to the environment. Then each group was divided into two subgroups respectively, subjected by intragastric administration (IG) with PBS or H. pylori strain SSI at 12 pm for one month (N = 20). Mice in each group were sacrificed at four time point: 6 am, 12 pm, 6 pm and 12 am (N = 5, for each time point). The method to extract gastric epithelial cells from mice was reported in this study [35]. All animal experiments were approved by Shandong University Research Ethics Committee.
2.7. H. Pylori staining with May-Grunwald-Giemsa
May-Grunwald-Giemsa kit (G1930, Solarbio) was used to stain H. pylori. FFPE sections subjected to deparaffination and dehydration. Stained the sections with May-Grunwald working aliquots for 10 min, and then with Giemsa working aliquots for 20 min. Washed the remaining liquid quickly with anhydrous ethanol, and roasted sections slightly.
2.8. Inflammation score rules
A single pathologist, blind to the treatment, scored the inflammation of the samples. Inflammation was scored according to special inflammatory cells appearing in different parts of stomach, leading to a total score of 12, as previously described [36].
2.9. Patient samples
Patient samples were from Jinan Central Hospital, Shandong, P. R. China. H. pylori infection in patients was detected by 13C urea breath test. The using of patient samples was approved by Shandong University Research Ethics Committee.
2.10. Transfection of plasmids and siRNAs
Lipofectamine 2000 (Invitrogen) was used to transfect BMAL1 and LIN28A siRNAs (Ribobio), the corresponding control siRNAs (Ribobio), mimics and inhibitor of let-7a-3p (Ribobio), and the corresponding negative controls (Ribobio) into gastric epithelial cells according to the protocol provided. For transfection of BMAL1 and LIN28A expression plasmids (Addgene), Roche transfection reagent was used according to the instruction book. Specific experimental steps were from manufacturer's protocols. BMAL1 expression plasmid was kindly donated by Dr. Steven Reppert (Massachusetts General Hospital, Boston, USA).
2.11. Chromatin immunoprecipitation assay (ChIP)
SimpleChIP® Plus Sonication Chromatin IP Kit (#56383, CST) was used to detect the binding of the corresponding proteins to DNA. All experimental procedures were performed according to the instructions. Antibodies used were: LIN28A (Abcam), BMAL1 (CST).
2.12. Dual luciferase reporter gene assay
Human BMAL1 wt-1 and wt-2 promoters and TNF-α promoter fragments were synthesized by company SYE.Biotech. KOD-Plus-Mutagenesis Kit (TOYOBO) was used to mutate or knock out the corresponding sites. Luciferase Assay System (Promega) was used to measure luciferase reporter activity.
2.13. Enzyme linked immunosorbent assay (ELISA)
Human TNF-alpha ELISA Kit (KE00068, proteintech) was used to detect protein secretion of TNF-α. All experimental procedures were performed according to the instructions.
2.14. Statistical analysis
Data of all experimental groups were described as the mean (±) SD. Analysis was evaluated by two-tailed Student's t-tests or Mann-Whitney U test. Statistical analyses were offered by GraphPad PRISM 7. P values <.05 were considered statistically significant.
3. Results
3.1. H. Pylori induced BMAL1 expression and disrupted its circadian rhythm
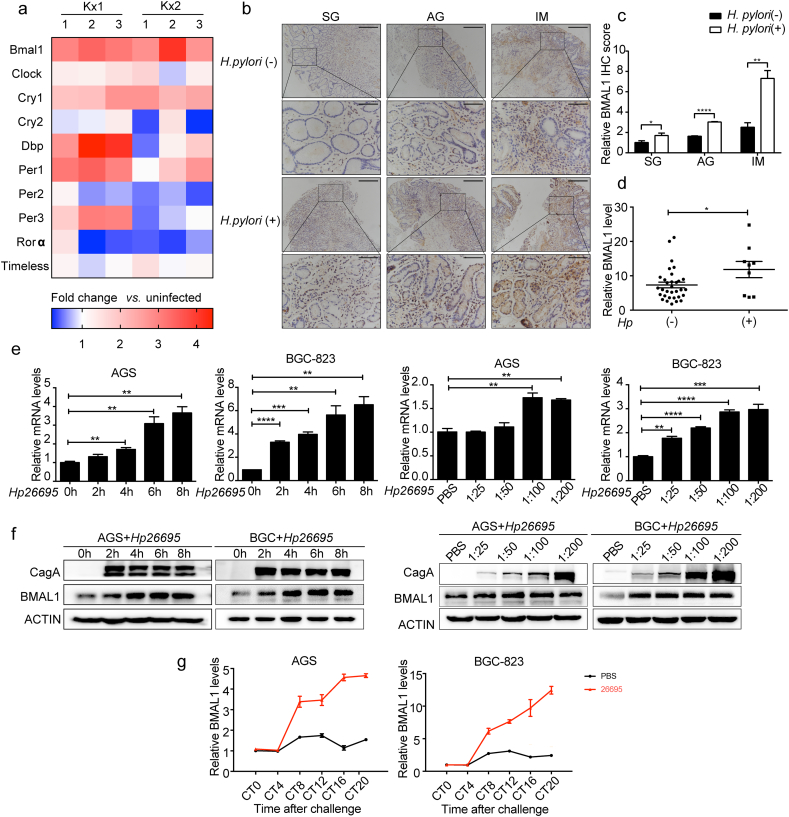
H. pylori infection is a risk factor for gastric diseases [37]. It influenced gastric circadian events, such as gastric acidity, simultaneously [14]. To explore the relationship between H. pylori infection and circadian clock genes in gastric epithelial cells, we analyzed the relative expression of traditional circadian genes in public microarray data from GEO database (GSE10262). In the mouse gastric epithelial progenitor-derived cell line (mGEP), three repeated infection experiments were performed with two H. pylori strains: chronic atrophic gastritis (ChAG)-associated Kx1 and gastric cancer-associated Kx2. Bmal1 expression was significantly increased compared with other genes (Fig. 1a). The analysis of BMAL1 from other public microarray data (Fig. S1a-c) also showed its up-regulation during H. pylori infection. To further verify this, we used diseased 37 patient tissues from superficial gastritis (SG), atrophic gastritis (AG) to Intestinal metaplasia (IM) with or without H. pylori infection. The lamina propria cell composition is complex, and gastric epithelial cells are the frontier cells encountered by H. pylori so that they are closely related to the initiation of gastritis induced by the bacteria. Therefore, we focused on the effect of H. pylori on gastric epithelial cells in this study (Fig. 1b). There is a close relationship between H. pylori infection and the severity of precancerous lesions [38]. CagA-positive H. pylori showed a lower infection density in patients with milder lesions and the density of H. pylori increasesd with the onset of gastritis [39]. There was no obvious elevation of BMAL1 in patients with H. pylori infection in SG patients, which may result from the low density of H. pylori infection. However in AG and IM patients, the expression of BMAL1 in H. pylori infected patients was significantly higher than that in uninfected patients. Therefore H. pylori infection gradually elevated BMAL1 expression with increasing severity of the gastric diseases (Fig. 1c). Correspondingly, in another cohort of specimens, BMAL1 mRNA level was also higher in H. pylori-positive patients than that in H. pylori-negative samples (Fig. 1d). H. pylori infection is continuous and changeable in vivo, and the timing of individual cell infection is in random phase of the circadian curve. To investigate the effect of H. pylori on BMAL1 in random phase in vitro, desynchronized gastric cell lines AGS and BGC-823, which showed nearly constant BMAL1 levels [34], were incubated with two H. pylori strains (11,637 and 26,695). BMAL1 expression was increased both at mRNA and protein levels in cells with H. pylori infection, and the up-regulation was time and concentration dependent (Fig. 1e and f, Fig. S1d and e).
Fig. 1.
H. pylori induced BMAL1 expression and disrupted its circadian rhythm. a, relative expression of circadian clock genes in mouse gastric epithelial progenitor-derived cell line (mGEP) infected with clinically isolated H. pylori strains Kx1 and Kx2, compared with uninfected group. The infection experiments were repeated three times, and represented by the numbers 1, 2, and 3. The data were obtained from the GEO database(GSE10262). b and c, IHC staining for BMAL1 in 37 cases of superficial gastritis(SG), atrophic gastritis(AG) and Intestinal metaplasia(IM) patients respectively with or without H. pylori infection, and the relative values of IHC optical density. Scale bars, 100 μm (insets 50 μm). d, qRT-PCR analysis of BMAL1 in 41 cases of gastritis patients with or without H. pylori infection. e and f, qRT-PCR and Western blot analysis of BMAL1 in AGS and BGC-823 cells infected with H. pylori strain 26,695. The concentration of H. pylori and the cell collection time points were shown. g, AGS and BGC-823 cells were reset circadian time. Subsequently, they were infected with H. pylori strain 26,695. BMAL1 mRNA level was detected at 0, 4, 8, 12, 16, 20 h after the serum shock.
We had confirmed that H. pylori infection up-regulated BMAL1 expression in gastric epithelial cells. Since BMAL1 has stable period and amplitude as a circadian clock gene, it is necessary to identify whether H. pylori infection disturbed BMAL1 circadian rhythm or not. The synchronization model showed the eliciting of circadian gene expression by a serum shock. We reset circadian time of gastric epithelial cells with 50% horse serum before adding H. pylori, to demonstrate circadian oscillators existence in cell lines [34]. The amplitude of BMAL1 expanded dramatically after H. pylori infection. Besides, BMAL1 peak expression and the horizontal baseline were all raised in most phases (Fig. 1g). Therefore, H. pylori infection disrupted the circadian rhythm of BMAL1. In addition, we also examined the effect of H. pylori on the expression of other clock genes in desynchronized and synchronized gastric epithelial cells. H. pylori up-regulated the expression of REV-ERBα and RORα, while showed different effects on other genes. H. pylori infection significantly disturbed the rhythm of CRY1, DBP and REV-ERBα (Fig. S1f and g). This suggested the overall circadian rhythm disorders caused by H. pylori.
3.2. LIN28A directly activated the transcription of BMAL1, independent of let-7a
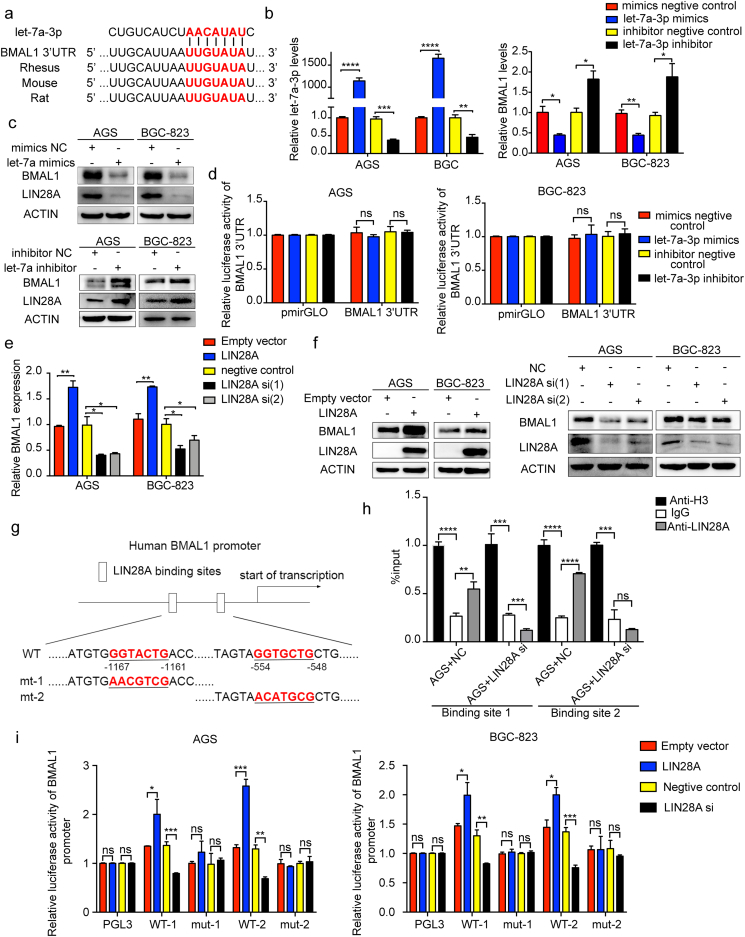
Studies have shown the role of LIN28/let-7 axis in circadian rhythm. Furthermore, BMAL1 contains a binding site of let-7a-3p in the 3′UTR region (Fig. 2a). LIN28 family consists of two molecules, LIN28A and LIN28B. To evaluate the relationship between these two molecules and H. pylori, we analyzed their expression in the GEO database (GSE10262) and speculated the activation of Lin28a by H. pylori (Fig. S2a). As a validation of previous experiments, we demonstrated a negative feedback loop formed by mutual regulation between LIN28A and let-7a-3p (Fig. S2b and c). H. pylori infection down-regulated the expression of let-7a-3p (Fig. S2d). To explore the relationship between let-7a-3p and BMAL1, let-7a-3p mimics and inhibitor were used. We found the inhibition of LIN28A and BMAL1 both at mRNA and protein levels by let-7a-3p (Fig. 2b and c). However, the results of the dual luciferase assay showed the inability of let-7a-3p to activate the transcription of BMAL1 through the binding site of its 3′UTR, which was unexpected (Fig. 2d).
Fig. 2.
LIN28A directly regulated the transcription level of BMAL1, independent of let-7a. a, binding site of let-7a-3p on the 3′UTR region of BMAL1. b, qRT-PCR analysis of let-7a-3p and BMAL1 in AGS and BGC-823 cells transfected with mimics or inhibitor of let-7a-3p. c, Western blot analysis of BMAL1 and LIN28A in AGS and BGC-823 cells transfected with mimics or inhibitor of let-7a-3p. d, the dual luciferase assay detected the transcriptional regulation of let-7a-3p on the 3′UTR region of BMAL1. e and f, qRT-PCR and Western blot analysis of BMAL1 in AGS and BGC-823 cells transfected with LIN28A siRNA or overexpression plasmid. g, the binding sites of LIN28A in the BMAL1 promoter region, and the corresponding base mutation. h, ChIP was used to detect the binding between LIN28A and the two sites of the BMAL1 promoter region with and without LIN28A depletion. i, the transcriptional activation of the BMAL1 promoter region by LIN28A was detected by using a dual luciferase assay in case of binding sites mutation.
Based on the above results, we hypothesized the regulation of BMAL1 by let-7a-3p through LIN28A. After overexpression or depletion of LIN28A, BMAL1 showed corresponding up- or down-regulation at mRNA and protein levels (Fig. 2e and f). Although LIN28A is an RNA binding protein, it can directly bind to DNA, promoting transcription of the downstream target genes [40]. By analyzing the sequence of the BMAL1 promoter region, we found two potential LIN28A binding sites (Fig. 2g). Chromatin immunoprecipitation assay (ChIP) showed the direct binding of LIN28A to BMAL1's promoter, whereas after LIN28A suppression, the binding was accordingly weakened (Fig. 2h). To further confirm the transcriptional regulation of BMAL1 by LIN28A, we synthesized the wild type plasmid that contained the two binding sites, as well as the plasmids that the two sites were separately mutated. The subsequent dual luciferase assay was used to examine the effect of LIN28A on the transcriptional activation of BMAL1 (Fig. 2g). After overexpression or depletion of LIN28A, the transcriptional activity of the BMAL1 promoter regions was up- or down-regulated accordingly. However, these changes disappeared with the mutations of the binding sites (Fig. 2i). In summary, these results suggested the direct transcriptional regulation of BMAL1 by LIN28A by binding to the two DNA sites, independent of let-7a.
3.3. LIN28A enhanced H. pylori induced BMAL1 expression
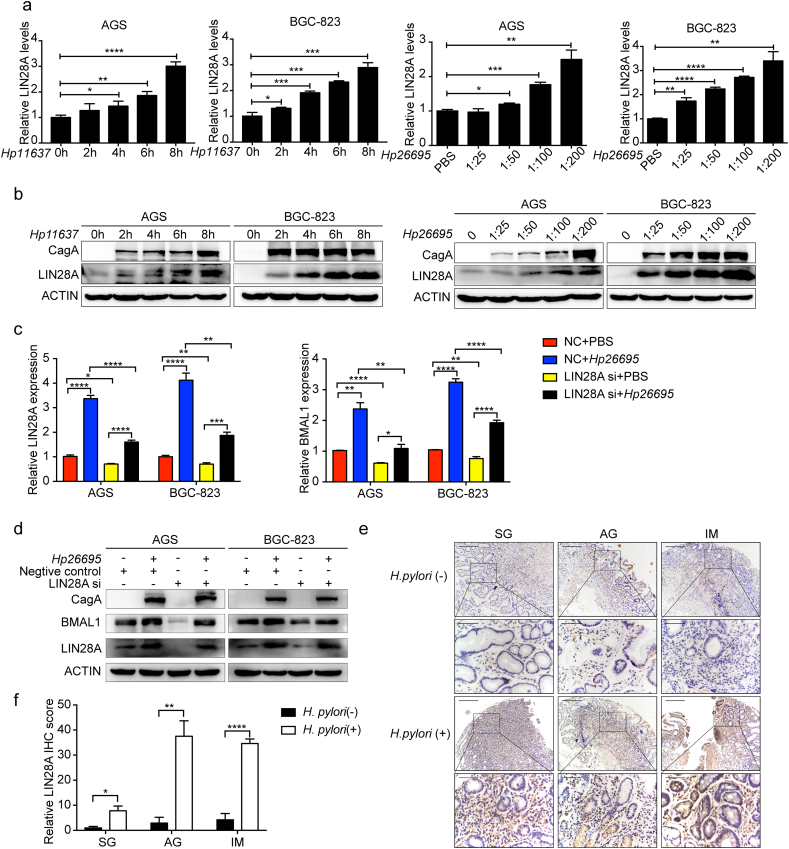
To explore whether LIN28A mediated the expression of BMAL1 caused by H. pylori, we detected the effect of H.pylori on LIN28A. Indeed H. pylori promoted LIN28A expression at both mRNA and protein levels in a time and concentration dependent way (Fig. 3a and b). Of note, comparing “LIN28A si + Hp26695” with “NC + Hp26695” group, the induction of BMAL1 by H. pylori was alleviated upon LIN28A suppression (Fig. 3c and d). In addition, the expression of LIN28A was higher in H. pylori-positive patients than that in H. pylori-negative patients, and the differences were reflected by the relative values of IHC optical density (Fig. 3e and f). The expression of LIN28A in specimen was very similar to the expression of BMAL1 in the same tissues. Therefore, LIN28A may enhance BMAL1 expression induced by H. pylori both in vitro and in vivo.
Fig. 3.
LIN28A enhanced BMAL1 expression induced by H. pylori. a and b, qRT-PCR and Western blot analysis of BMAL1 in AGS and BGC-823 cells infected with H. pylori. c and d, qRT-PCR and Western blot analysis of BMAL1 with LIN28A suppression on the basis of H. pylori infection. e and f, IHC staining for LIN28A in 37 cases of superficial gastritis(SG), atrophic gastritis(AG) and Intestinal metaplasia(IM) patients respectively with or without H. pylori infection, and the relative values of IHC optical density. Scale bars, 100 μm (insets 50 μm).
3.4. Effect of H. pylori-induced rhythm gene change on inflammatory factors
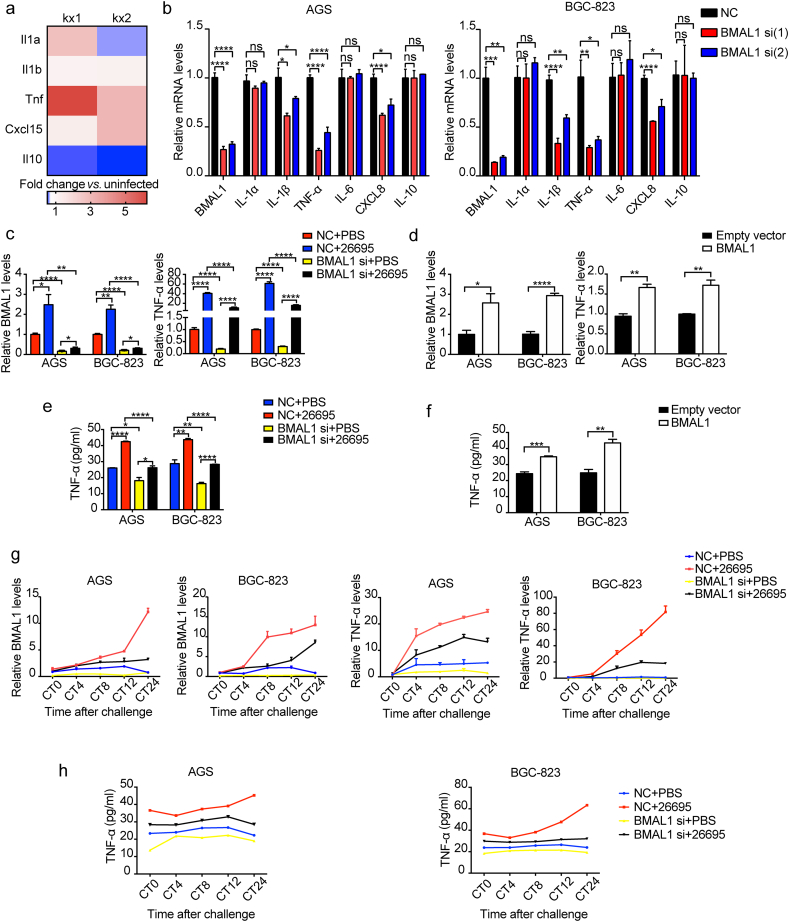
Circadian rhythm is involved in the development of inflammation, which prompted us to study the potential relationship between circadian rhythm and inflammatory factors upregulated by H. pylori. Based on previous results, we selected several inflammatory factors closely related to the pathogenesis of H. pylori. We found possible binding sites of BMAL1 (E-box region) in the promoter regions of all the selected molecules (Fig. S3a). H. pylori significantly activated Tnf expression upon analysis of the GEO database (GSE10262) (Fig. 4a). And decline of TNF-α was found to be the most pronounced among the selected factors by BMAL1 depletion (Fig. 4b). Therefore TNF-α may be a potential target of BMAL1. Comparing “BMAL1 si + Hp26695” with “NC + Hp26695” group, induction of TNF-α RNA expression by H. pylori was relived upon BMAL1 inhibition (Fig. 4c). Accordingly, overexpression of BMAL1 showed the opposite effect (Fig. 4d). The change of TNF-α secretion was align with that of its RNA (Fig. 4e and f). Here IL-6 and IL-8 were found to be up-regulated by H. pylori infection, however their expression was not mainly regulated by BMAL1 (Fig. S3b).
Fig. 4.
Effect of H. pylori-induced rhythm gene change on inflammatory factors. a, relative expression of inflammatory factors in mGEP. Three repeated experiments data were put in one box. The data were obtained from the GEO database(GSE10262). b, qRT-PCR analysis of inflammatory factors in AGS and BGC-823 cells transfected with BMAL1 siRNA. c, qRT-PCR analysis of BMAL1 and TNF-α with BMAL1 depletion on the basis of H. pylori infection. d, qRT-PCR analysis of BMAL1 and TNF-α in AGS and BGC-823 cells transfected with BMAL1 overexpression plasmid. e, ELISA analysis of TNF-α, after BMAL1 inhibition on the basis of H. pylori infection. f, ELISA analysis of TNF-α in AGS and BGC-823 cells transfected with BMAL1 overexpression plasmid. g, AGS and BGC-823 cells were reset circadian time, and BMAL1 was depleted on the basis of H. pylori infection. BMAL1 and TNF-α mRNA levels were detected at 0, 4, 8, 12, 24 h after the serum shock. h, AGS and BGC-823 cells were reset circadian time, and BMAL1 was inhibited on the basis of H. pylori infection. ELISA analysis of TNF-α was detected at 0, 4, 8, 12, 24 h after the serum shock.
Since BMAL1 functions as circadian rhythm gene, TNF-α is supposed to be rhythmic as its target. Similar to BMAL1, the rhythm of TNF-α in rhythm-synchronized cells was destroyed by infection of H. pylori. Specifically, its amplitude raised and the expression in most phases increased (Fig. S3c). Correspondingly, when BMAL1 was inhibited, the rhythm change of TNF-α caused by H. pylori was restored at RNA and protein levels (Fig. 4g and h). These results indicated that BMAL1 was involved in the regulation of inflammatory response through targeting TNF-α after H. pylori infection.
3.5. BMAL1 directly promoted TNF-α transcriptional expression
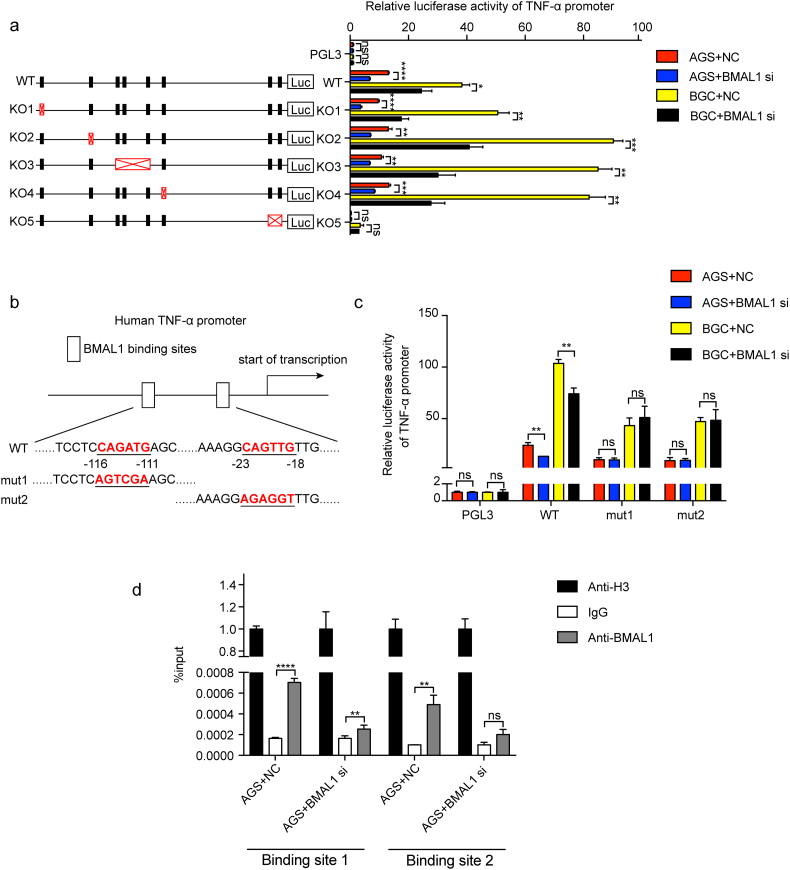
Based on the distribution of E-box elements in the TNF-α promoter region, we knocked them out separately and used double luciferase assay to detect the transcriptional activation of BMAL1 on wild-type plasmid and the five different knockout plasmids. It was found that when the E-box elements of the KO5 group were removed, the transcriptional activation of TNF-α by BMAL1 was not changed even though BMAL1 was inhibited. However, in the other four KO groups, transcriptional activation of TNF-α by BMAL1 was changed in line with the alterations of BMAL1, indicating the dispensableness of the these regions for TNF-α promoter activity regulated by BMAL1 (Fig. 5a). Since there are two E-box elements in the KO5 group, these two sites were mutated and tested separately in order to further determine the exact transcriptional regulation of TNF-α by BMAL1 (Fig. 5b). Both sites were found critical for BMAL1 activation since no alteration of promoter activity of TNF-α was observed upon BMAL1 suppression (Fig. 5c). Furthermore we confirmed the direct binding of BMAL1 to the above two sites in TNF-α promoter by ChIP with or without BMAL1 depletion (Fig. 5d). In summary, BMAL1 directly promoted TNF-α transcriptional expression.
Fig. 5.
BMAL1 promoted TNF-α expression induced by H. pylori. a, a map for knockout of the E-box elements in the promoter region of TNF-α. Correspondingly, the effect of BMAL1 on the transcriptional activity of TNF-α was examined using a dual luciferase assay. b, the binding sites of BMAL1 in the TNF-α promoter region, and the corresponding base mutation. c, the transcriptional activation of the TNF-α promoter region by BMAL1 was detected by using a dual luciferase assay in case of binding sites mutation. d, ChIP was used to detect the binding between BMAL1 and the sites of the TNF-α promoter region after BMAL1 suppression.
3.6. Rhythm disorder aggravated inflammatory response caused by H. pylori
We had confirmed the involvement of BMAL1 in the inflammatory response by modulating TNF-α under condition of H. pylori infection in vitro. However the relationship between H. pylori, gastric rhythm and inflammation in vivo was unknown. Since BMAL1 is centrally located in the regulatory loops of the rhythmic genes and its change was evident in in vitro studies, we still exploited the change of BMAL1 to represent the magnitude of the overall rhythm disorder of gastric epithelial cells in vivo.
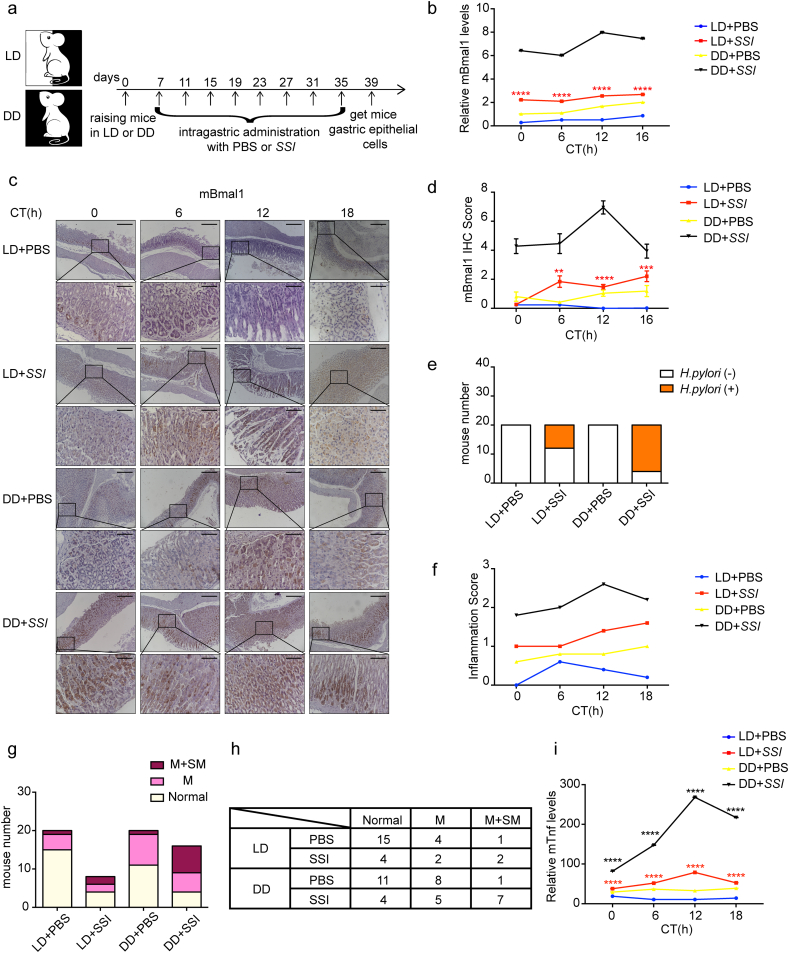
A mouse model was designed and generated as shown in Fig. 6a. Briefly there are 4 groups of mice, of which 2 groups were subjected to 12 h of light-12 h of dark (LD) cycle plus intragastric administration (IG) with PBS or H. pylori strain SSI at 12 pm respectively. The other 2 groups were raised under constant darkness (DD) plus intragastric administration (IG) with PBS or H. pylori strain SSI at 12 pm respectively. Uninfected mice were analyzed in “LD + PBS” and “DD + PBS” group, and only infected animals were analyzed in “LD + SSI” and “DD + SSI” group. Comparing “LD + SSI” with “LD + PBS” group, H. pylori infection up-regulated Bmal1 by two times at both mRNA and protein levels in vivo. Although this change was less than that in vitro, it still made sense (Fig. 6b-d). In order to prove the inflammation caused by H. pylori infection via rhythm change, we further aggravated the rhythm disorder by changing the light input. Comparing “DD + SSI” with “LD + SSI” group, in the case of the same conditions of H. pylori infection, the expression of Bmal1 in gastric epithelial cells increased more significantly when the external light input was artificially changed, indicating the aggravation of circadian rhythm disorder (Fig. 6b-d). Sydney system was used to define mild colonization which indicated less than one third of the sample surface was covered by H. pylori [41]. According to this standard, we found that although the degree of colonization of H. pylori in each mouse was basically the same, the number of mice colonized by H. pylori was significantly increased in the “DD + SSI” group with more severe rhythm disorder (Fig. S4a and Fig. 6e). This suggested the promoting role of circadian rhythm disorder on colonization of H. pylori. In addition, the detection and correlation analysis of the IHC scores of Bmal1 and Lin28a indicated the positive correlation between the two molecules in vivo, which was in line with our in vitro data (Fig. S4b-d).
Fig. 6.
Rhythm disorder aggravated inflammatory response caused by H. pylori. a, animal model were designed as described in the methods and materials b, qRT-PCT analysis of Bmal1 in gastric epithelial cells extracted from each mouse. c, IHC staining for Bmal1 in each group. Scale bars, 100 μm (insets 50 μm). d, IHC scores for Bmal1 in each group. e, quantification of mice with or without H. pylori infection in all the groups. Total number in each group was 20. H. pylori-positive mice number in “LD + SSI” group was 8, and in “DD + SSI” group was 16. f, inflammation scores of each group. g, histogram of each group with different depth of inflammatory cells infiltration. Normal: no inflammatory cells infiltration; M: inflammatory cells infiltrating only mucosal layer; M + SM: inflammatory cells infiltrating both mucosal layer and submucosa. h, specific data displayed in the G chart. i, qRT-PCT analysis of Tnf in gastric epithelial cells extracted from each mouse.
H. pylori infection caused infiltration of a variety of inflammatory cells in the stomachs of mice, and the stage of inflammation could be judged according to this (Fig. S4e). Inflammation indexes were scored according to H. pylori infection rate as well as the cell type, depth and location of inflammatory cell infiltration. Comparing with “LD + SSI” group, “DD + SSI” mice showed more severe inflammation, indicating the involvement of rhythm disorder in the process of inflammation induced by H. pylori (Fig. 6f). Our in vitro data demonstrated that the longer of the H. pylori infection, the higher of the expression of BMAL1 and the more severe of the rhythm disorder. This was further confirmed in vivo. Increasing severity of circadian rhythm disorder aggravated the inflammation caused by H. pylori. Besides, the “DD + SSI” group had the most cases and the highest proportion of mice with inflammatory cells infiltrating both the mucosal layer and the submucosa according to inflammatory response position (Fig. 6g and h). Similarly, comparing with “LD + SSI” group, Tnf expression from mouse epithelial cells stimulated by H. pylori was more significantly increased in the “DD + SSI” group (Fig. 6i). The above results strongly suggested the aggravation of H. pylori induced inflammatory response by circadian rhythm disorder.
4. Discussion
H. pylori infection was related to circadian gastric acidity, 24-h gastric pH, and methods of administration to treat acid-related diseases [14,42,43]. In addition, a variety of gastrointestinal diseases might result from unnatural circadian rhythms [44,45]. These studies highlighted the role of rhythm of the stomach in H. pylori induced diseases. We found that H. pylori promoted the expression of BMAL1 at the transcriptional level and disrupted its rhythm through LIN28A. Since BMAL1 is in a central regulatory position for the regulation of circadian rhythm genes, this result is instrumental for the understanding of effects of H. pylori on the overall rhythm.
Gastritis provides the microenvironment niche for malignant transformation. Most gastric adenocarcinomas start from chronic superficial gastritis, and then evolve to atrophic gastritis or intestinal metaplasia, and finally to dysplasia or adenocarcinoma. To provide insights into the interaction between circadian system and inflammation, we found that BMAL1 could directly bind to the E-box elements in the promoter of TNF-α, activating its transcription and increasing its synthesis and subsequent secretion. The elevated TNF-α level further promoted the development of inflammation. This finding is in line with the results of previous studies [46]. BMAL1 was found to regulate TNF-α expression in the pathological state of intestinal epithelial cells [46]. However the underlying mechanism remained elusive since then. In addition, TNF-α regulated the circadian clock in astrocytes and rheumatoid synovial cells, and it also enhanced the mRNA expression of BMAL1 and CRY1 [47,48]. And we found the transcriptional regulation of TNF-α by BMAL1. Combining these results, it may suggest a feedback loop between TNF-α and BMAL through their mutual regulation, There may be a cascade amplification of rhythm gene-mediated inflammatory response upon H. pylori infection, providing a new small field for future research. This also explains the fact that the expression of Lin28a is elevated correspondingly with the increase of rhythm disorder in animal experiments (Fig. S4b-c). Controversially, some studies showed a certain inhibitory effect of BMAL1 on inflammation in heart, lung, or macrophages and bone marrow cells [[49], [50], [51], [52], [53], [54], [55]]. This could be explained by different roles of circadian genes in different diseases. Or this may also be explained by differences in rhythm gene expression caused by different conditions. This is an open field and needs more work to illustrate in the future.
In the animal model of this study, we chose to administer the mice at 12 pm in order to investigate the effect of H. pylori infection on gastric rhythm in vivo. In future experiments, we will administer normal or related rhythm gene knockout mice with H. pylori at different time points. By observing the progress of gastric inflammation, immune cell aggregation and inflammatory factor release, we will confirm the regulation of epithelial circadian clock on inflammation induced by H. pylori. This will also help us find better treatment options and time points of administration in the future.
The in vitro data demonstrated that the longer of the H. pylori infection, the higher of the expression of BMAL1 and the more severe of the rhythm disorder. In order to prove that rhythm disorders aggravate inflammation induced by H. pylori in vivo, we can measure rhythm and inflammation changes at different infection times. We can also choose to change the external light input to aggravate the circadian rhythm disorder under the same infection conditions and measure inflammation scores. With the prolongation of H. pylori infection, many pathogenic factors of the bacteria may be aggravated. In order to exclude other pathogenic factors from judging the experimental results, we chose the latter experimental method. In Fig. 6, compared with “LD + PBS”, the increase in inflammation score was not obvious in “DD + PBS” group, indicating that only the change of external light had little effect on inflammation. In the case of the same light change, inflammation score of “DD + SSI” was significantly increased compared with “LD + SSI” group. It supports that the rhythm can aggravate inflammation under the condition of H. pylori infection. Since this experiment aims to find causes that aggravate the inflammatory response and gradually worsen the disease under the premise of H. pylori infection. Therefore, we did not explore in this experiment whether the circadian rhythm gene can cause the same degree of inflammation changes when the disorder is excessive under other conditions. We may explore the more complex effects of circadian rhythm disorders on inflammation in future studies.
Consistent with our results, studies have shown that inflammatory cytokines such as TNF-α in the gastric mucosa may be involved in the H. pylori induced gastric diseases [39,56]. In addition to BMAL1, H. pylori can also induce TNF-α through direct activation of the transcription factors NF-κB and AP-1 [57,58]. The genes encoding the NF-κB signaling pathway-related proteins belong to the clock-controlling gene family [59], and the activation of NF-κB is regulated by the rhythm loop represented by the clock genes BMAL1/CLOCK [60]. For example, in bone marrow cells, BMAL1 inhibits the activation of NF-κB [52]. CLOCK and NF-κB form a CLOCK/NF-κB complex that promotes NF-κB-mediated transcription and impedes the formation of the CLOCK/BMAL1 complex [61]. The above studies demonstrate the existence of mutual repulsion between NF-κB and BMAL1. Besides, in the presence of CLOCK, the RelB subunit of NF-κB binds to BMAL1, inhibiting or modulating the rhythmic target genes downstream of BMAL1/CLOCK in a more complex manner [62]. These results suggest that NF-κB and BMAL1 may have mutual regulation or binding and affect the expression of downstream genes such as TNF-α. However, the specific underlying mechanism requires a more in-depth study of various types of cells in different environments in vivo.
For the relationship between inflammatory response and circadian rhythm, only the unilateral regulation mechanism of inflammatory factors by rhythm genes was explored in our study. And H. pylori was found to stimulate epithelial cells or immune cells and promote the secretion of inflammatory factors through other mechanisms [22]. The inflammatory factors, such as TNF-α, may promote the progression of the disease by binding to receptors on the surface of epithelial cells and then activate the corresponding signaling pathways to influence the expression of circadian genes. In future, it is significant to explore the relationship between inflammation, circadian rhythm and disease progression in this direction, and further elucidate the underlying mechanisms.
There is an interaction between circadian rhythm and gut microbiota. Gut microbiota undergoes diurnal compositional and functional oscillations [63], while diurnal microbial behavior in turn drives host circadian transcriptional, epigenetic and metabolite oscillations [64]. The disruption of host circadian rhythm causes intestinal microbiota to lose the gut diurnally oscillates [65]. Furthermore, the rhythmic destruction of homeostatic microbiome not only eliminates normal chromatin and transcriptional oscillations, but also causes genome-wide de novo oscillations in both intestine and liver, thereby affecting the circadian fluctuations of physiology and disease susceptibility [64]. In addition, microbial metabolites directly affect circadian gene expression in the host [66]. Rhythmic genes, interfered by the disordered gut microbiota, disrupt body health by regulating downstream target genes [67]. The above results suggest the maintenance of homeostasis by interaction between rhythm genes and the gastrointestinal microbiota. In our study, we proved the aggravation of H. pylori-induced gastritis by rhythm disorders. It would be interesting to explore the specific components or metabolites of H. pylori to disrupt rhythm, the effects of rhythm disorder on H. pylori, and the influences of their interactions on the gastritis microenvironment. Since H. pylori is the most important and initial microbe to induce gastritis, the changes of other gut microbiota, and their relationship with rhythm disorder and progression of gastric diseases during the late stage of infection are worth to illustrating as well.
Circadian rhythm regulates physiological homeostasis of the gastrointestinal tract concerning gut motility, gastric acid secretion, maintenance and restoration of the protective mucosal barrier, production of digestive enzymes, and immunologic system of gastrointestinal tract. Disruption of circadian physiology, due to sleep disturbance or shift work, may result in various gastric diseases, such as gastroesophageal reflux disease (GERD), gastric dyspepsia, peptic ulcer disease, metabolic syndrome or cancer [68]. Circadian clock disruption results in similar deleterious effects as the chronic inflammation induced by H. pylori [69]. However, no relationship between GERD and H. pylori was found [70]. H. pylori may be one of the causes of functional dyspepsia. H. pylori infection significantly causes chronic mucosal inflammation in the stomach and duodenum, which in turn leads to abnormalities in gastroduodenal movement and sensitivity, affects various endocrine functions of the stomach, and aggravates functional dyspepsia symptom. This suggests the coordination of H. pylori infection and rhythm disorders to promote functional dyspepsia [71]. The main causes of peptic ulcer include H. pylori infection and long-term use of Nonsteroidal Anti-inflammatory Drugs (NSAIDs). The former is a major risk factor for gastric cancer, while the latter shows some preventing effects for gastric cancer. The presence of H. pylori infection and precancerous lesions affects the treatment and outcome of the ulcers associated with rhythm disorders [72]. Circadian rhythm disorder adversely affects food choices, hunger and appetite, and generates deleterious metabolic consequences that lead to obesity [73]. There are increased odds ratios or relative risks of several gastrointestinal complications of obesity: gastroesophageal reflux disease, erosive gastritis, and gastric cancer [74]. This suggests that rhythm disorder can promote the development of gastritis through adverse metabolic reactions, and the transformation of inflammation into cancer. Interestingly, a link between the molecular clock machinery and carcinogenesis has been revealed [75]. In the future, it would be a good point to study the relationship between H. pylori, rhythm disorder and the transformation from inflammation to cancer, and illustrate the underlying mechanisms through which they are connected.
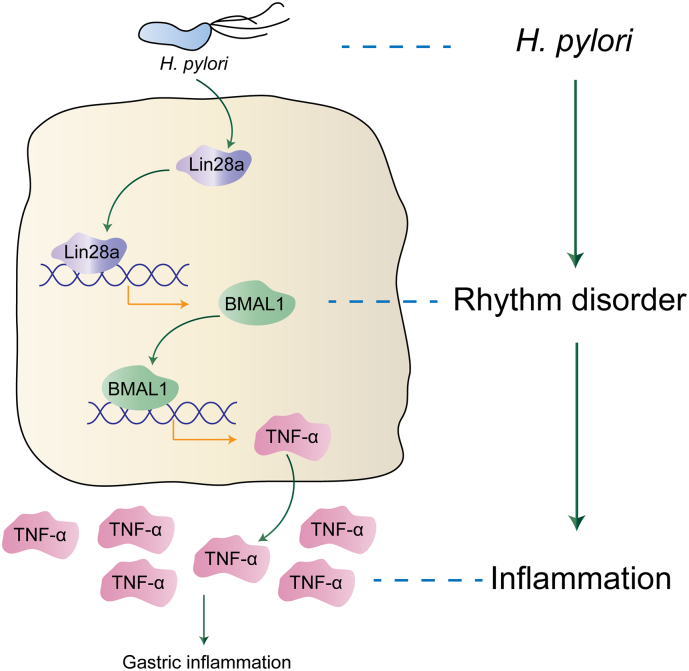
In summary, we found that H. pylori up-regulated the expression of LIN28A, and LIN28A directly bound to the BMAL1 promoter, activating the transcription of BMAL1 to increase its expression. BMAL1 in turn promoted transcription of TNF-α by directly binding to the E-box elements on its promoter to increase its secretion. Therefore, our study revealed the mechanism through which disorder of circadian rhythm aggravated the inflammatory response induced by H. pylori (Fig. 7). This research on circadian rhythm genes may provide new potential targets for the early diagnosis and treatment of diseases related to H. pylori infection.
Fig. 7.
Schematic model of the study. H. pylori up-regulated the expression of LIN28A, and LIN28A directly bound to the BMAL1 promoter, activating the transcription of BMAL1 to increase its expression. BMAL1 in turn promoted transcription of TNF-α by directly binding to the E-box elements on its promoter to increase its secretion. Therefore, our study revealed the mechanism through which disorder of circadian rhythm aggravated the inflammatory response induced by H. pylori.
Acknowledgments
Acknowledgements
Not applicable.
Declaration of interest
The authors indicate no potential conflicts of interest.
Author contributions
TL, XL and JJ contributed to the study conception and design. TL, WS, LZ and XL performed the experiment and contributed to data acquisition. TL, WS, LZ, XJ and PS contributed clinical specimens. TL, WS, SL, LM and LZ contributed to the analysis and interpretation of data. TL and XL wrote the manuscript.
Funding sources
This work was supported by the National Natural Science Foundation of China (Nos. 81571960, 81371781, 81372680, 81471991, 81501720, 81772151 and 81871620), and the Shandong Province Major Science and Technology Innovation Project (2018CXGC1208).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ebiom.2018.11.043.
Contributor Information
Xiuming Liang, Email: liangxm@sdu.edu.cn.
Jihui Jia, Email: jiajihui@sdu.edu.cn.
Appendix A. Supplementary data
Supplementary material
References
- 1.Storch K.F., Lipan O., Leykin I. Extensive and divergent circadian gene expression in liver and heart. Nature. 2002;417(6884):78–83. doi: 10.1038/nature744. [DOI] [PubMed] [Google Scholar]
- 2.Kelleher F.C., Rao A., Maguire A. Circadian molecular clocks and cancer. Cancer Lett. 2014;342(1):9–18. doi: 10.1016/j.canlet.2013.09.040. [DOI] [PubMed] [Google Scholar]
- 3.Bass J., Lazar M.A. Circadian time signatures of fitness and disease. Science. 2016;354(6315):994–999. doi: 10.1126/science.aah4965. [DOI] [PubMed] [Google Scholar]
- 4.Van Cutsem E., Sagaert X., Topal B., Haustermans K., Prenen H. Gastric cancer. Lancet. 2016;388(10060):2654–2664. doi: 10.1016/S0140-6736(16)30354-3. [DOI] [PubMed] [Google Scholar]
- 5.Correa P. Human gastric carcinogenesis: a multistep and multifactorial process--first American Cancer Society Award Lecture on Cancer Epidemiology and Prevention. Cancer Res. 1992;52(24):6735–6740. [PubMed] [Google Scholar]
- 6.Bik E.M., Eckburg P.B., Gill S.R. Molecular analysis of the bacterial microbiota in the human stomach. Proc Natl Acad Sci U S A. 2006;103(3):732–737. doi: 10.1073/pnas.0506655103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Andersson A.F., Lindberg M., Jakobsson H., Bäckhed F., Nyrén P., Engstrand L. Comparative analysis of human gut microbiota by barcoded pyrosequencing. PLoS One. 2008;3(7) doi: 10.1371/journal.pone.0002836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Theisen J., Nehra D., Citron D. Suppression of gastric acid secretion in patients with gastroesophageal reflux disease results in gastric bacterial overgrowth and deconjugation of bile acids. J Gastrointest Surg. 2000;4(1):50–54. doi: 10.1016/s1091-255x(00)80032-3. [DOI] [PubMed] [Google Scholar]
- 9.Ferreira R.M., Pereira-Marques J., Pinto-Ribeiro I. Gastric microbial community profiling reveals a dysbiotic cancer-associated microbiota. Gut. 2018;67(2):226–236. doi: 10.1136/gutjnl-2017-314205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rugge M., Pennelli G., Pilozzi E. Gastritis: the histology report. Dig Liver Dis. 2011;43(Suppl. 4):S373–S384. doi: 10.1016/S1590-8658(11)60593-8. [DOI] [PubMed] [Google Scholar]
- 11.Graham D.Y. History of Helicobacter pylori, duodenal ulcer, gastric ulcer and gastric cancer. World J Gastroenterol. 2014;20(18):5191–5204. doi: 10.3748/wjg.v20.i18.5191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rugge M., Capelle L.G., Cappellesso R., Nitti D., Kuipers E.J. Precancerous lesions in the stomach: from biology to clinical patient management. Best Pract Res Clin Gastroenterol. 2013;27(2):205–223. doi: 10.1016/j.bpg.2012.12.007. [DOI] [PubMed] [Google Scholar]
- 13.Scheving L.A., Russell W.E. It's about time: clock genes unveiled in the gut. Gastroenterology. 2007;133(4):1373–1376. doi: 10.1053/j.gastro.2007.08.068. [DOI] [PubMed] [Google Scholar]
- 14.Savarino V., Mela G.S., Zentilin P. Circadian gastric acidity in Helicobacter pylori positive ulcer patients with and without gastric metaplasia in the duodenum. Gut. 1996;39(4):508–512. doi: 10.1136/gut.39.4.508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hand L.E., Hopwood T.W., Dickson S.H. The circadian clock regulates inflammatory arthritis. FASEB J. 2016;30(11):3759–3770. doi: 10.1096/fj.201600353R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Man K., Loudon A., Chawla A. Immunity around the clock. Science. 2016;354(6315):999–1003. doi: 10.1126/science.aah4966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jahanban-Esfahlan R., Mehrzadi S., Reiter R.J. Melatonin in regulation of inflammatory pathways in rheumatoid arthritis and osteoarthritis: involvement of circadian clock genes. Br J Pharmacol. 2018;175(16):3230–3238. doi: 10.1111/bph.13898. 10.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu X., Yu R., Zhu L., Hou X., Zou K. Bidirectional Regulation of Circadian Disturbance and Inflammation in Inflammatory Bowel Disease. Inflamm Bowel Dis. 2017;23(10):1741–1751. doi: 10.1097/MIB.0000000000001265. [DOI] [PubMed] [Google Scholar]
- 19.Pagel R., Bär F., Schröder T. Circadian rhythm disruption impairs tissue homeostasis and exacerbates chronic inflammation in the intestine. FASEB J. 2017;31(11):4707–4719. doi: 10.1096/fj.201700141RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Steffens S., Winter C., Schloss M.J., Hidalgo A., Weber C., Soehnlein O. Circadian Control of Inflammatory Processes in Atherosclerosis and its Complications. Arterioscler Thromb Vasc Biol. 2017;37(6):1022–1028. doi: 10.1161/ATVBAHA.117.309374. [DOI] [PubMed] [Google Scholar]
- 21.Wang Z., Xiong F., Wang X. Nuclear receptor retinoid-related orphan receptor alpha promotes apoptosis but is reduced in human gastric cancer. Oncotarget. 2017;8(7):11105–11113. doi: 10.18632/oncotarget.14364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ricci V., Romano M., Boquet P. Molecular cross-talk between Helicobacter pylori and human gastric mucosa. World J Gastroenterol. 2011;17(11):1383–1399. doi: 10.3748/wjg.v17.i11.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Piao J.Y., Lee H.G., Kim S.J. Helicobacter pylori Activates IL-6-STAT3 Signaling in Human Gastric Cancer Cells: potential Roles for Reactive Oxygen Species. Helicobacter. 2016;21(5):405–416. doi: 10.1111/hel.12298. [DOI] [PubMed] [Google Scholar]
- 24.Beales I.L., Calam J. Stimulation of IL-8 production in human gastric epithelial cells by Helicobacter pylori, IL-1beta and TNF-alpha requires tyrosine kinase activity, but not protein kinase C. Cytokine. 1997;9(7):514–520. doi: 10.1006/cyto.1996.0195. [DOI] [PubMed] [Google Scholar]
- 25.Lin Q., Xu H., Chen X., Tang G., Gu L., Wang Y. Helicobacter pylori cytotoxin-associated gene a activates tumor necrosis factor-alpha and interleukin-6 in gastric epithelial cells through P300/CBP-associated factor-mediated nuclear factor-kappaB p65 acetylation. Mol Med Rep. 2015;12(4):6337–6345. doi: 10.3892/mmr.2015.4143. [DOI] [PubMed] [Google Scholar]
- 26.Hashiramoto A., Yamane T., Tsumiyama K. Mammalian clock gene Cryptochrome regulates arthritis via proinflammatory cytokine TNF-alpha. J Immunol. 2010;184(3):1560–1565. doi: 10.4049/jimmunol.0903284. [DOI] [PubMed] [Google Scholar]
- 27.Iliopoulos D., Hirsch H.A., Struhl K. An epigenetic switch involving NF-kappaB, Lin28, Let-7 MicroRNA, and IL6 links inflammation to cell transformation. Cell. 2009;139(4):693–706. doi: 10.1016/j.cell.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang T., Wang G., Hao D. Aberrant regulation of the LIN28A/LIN28B and let-7 loop in human malignant tumors and its effects on the hallmarks of cancer. Mol Cancer. 2015;14:125. doi: 10.1186/s12943-015-0402-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matsushima K., Isomoto H., Inoue N. MicroRNA signatures in Helicobacter pylori-infected gastric mucosa. Int J Cancer. 2011;128(2):361–370. doi: 10.1002/ijc.25348. [DOI] [PubMed] [Google Scholar]
- 30.Hayashi Y., Tsujii M., Wang J. CagA mediates epigenetic regulation to attenuate let-7 expression in Helicobacter pylori-related carcinogenesis. Gut. 2013;62(11):1536–1546. doi: 10.1136/gutjnl-2011-301625. [DOI] [PubMed] [Google Scholar]
- 31.Liu W.J., Xu Q., Sun L.P., Dong Q.G., He C.Y., Yuan Y. Expression of serum let-7c, let-7i, and let-7f microRNA with its target gene, pepsinogen C, in gastric cancer and precancerous disease. Tumour Biol. 2015;36(5):3337–3343. doi: 10.1007/s13277-014-2967-9. [DOI] [PubMed] [Google Scholar]
- 32.Chen W., Liu Z., Li T. Regulation of Drosophila circadian rhythms by miRNA let-7 is mediated by a regulatory cycle. Nat Commun. 2014;5:5549. doi: 10.1038/ncomms6549. [DOI] [PubMed] [Google Scholar]
- 33.Perales R., King D.M., Aguirre-Chen C., Hammell C.M. LIN-42, the Caenorhabditis elegans PERIOD homolog, negatively regulates microRNA transcription. PLoS Genet. 2014;10(7) doi: 10.1371/journal.pgen.1004486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nagoshi E., Saini C., Bauer C., Laroche T., Naef F., Schibler U. Circadian gene expression in individual fibroblasts: cell-autonomous and self-sustained oscillators pass time to daughter cells. Cell. 2004;119(5):693–705. doi: 10.1016/j.cell.2004.11.015. [DOI] [PubMed] [Google Scholar]
- 35.O'Brien D.P., Romero-Gallo J., Schneider B.G. Regulation of the Helicobacter pylori cellular receptor decay-accelerating factor. J Biol Chem. 2008;283(35):23922–23930. doi: 10.1074/jbc.M801144200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Franco A.T., Johnston E., Krishna U. Regulation of gastric carcinogenesis by Helicobacter pylori virulence factors. Cancer Res. 2008;68(2):379–387. doi: 10.1158/0008-5472.CAN-07-0824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Polk D.B., Peek R.M., Jr. Helicobacter pylori: gastric cancer and beyond. Nat Rev Cancer. 2010;10(6):403–414. doi: 10.1038/nrc2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Plummer M., van Doorn L.J., Franceschi S. Helicobacter pylori cytotoxin-associated genotype and gastric precancerous lesions. J Natl Cancer Inst. 2007;99(17):1328–1334. doi: 10.1093/jnci/djm120. [DOI] [PubMed] [Google Scholar]
- 39.Monstein H.J., Tiveljung A., Kraft C.H., Borch K., Jonasson J. Profiling of bacterial flora in gastric biopsies from patients with Helicobacter pylori-associated gastritis and histologically normal control individuals by temperature gradient gel electrophoresis and 16S rDNA sequence analysis. J Med Microbiol. 2000;49(9):817–822. doi: 10.1099/0022-1317-49-9-817. [DOI] [PubMed] [Google Scholar]
- 40.Zeng Y., Yao B., Shin J. Lin28A Binds active Promoters and Recruits Tet1 to Regulate Gene Expression. Mol Cell. 2016;61(1):153–160. doi: 10.1016/j.molcel.2015.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stolte M., Meining A. The updated Sydney system: classification and grading of gastritis as the basis of diagnosis and treatment. Can J Gastroenterol. 2001;15(9):591–598. doi: 10.1155/2001/367832. [DOI] [PubMed] [Google Scholar]
- 42.Savarino V., Mela G.S., Zentilin P. 24-hour gastric pH and extent of duodenal gastric metaplasia in Helicobacter pylori-positive patients. Gastroenterology. 1997;113(3):741–745. doi: 10.1016/s0016-5085(97)70166-5. [DOI] [PubMed] [Google Scholar]
- 43.Sugimoto M., Furuta T., Shirai N. Comparison of an increased dosage regimen of rabeprazole versus a concomitant dosage regimen of famotidine with rabeprazole for nocturnal gastric acid inhibition in relation to cytochrome P450 2C19 genotypes. Clin Pharmacol Ther. 2005;77(4):302–311. doi: 10.1016/j.clpt.2004.10.010. [DOI] [PubMed] [Google Scholar]
- 44.Erren T.C., Reiter R.J. Defining chronodisruption. J Pineal Res. 2009;46(3):245–247. doi: 10.1111/j.1600-079X.2009.00665.x. [DOI] [PubMed] [Google Scholar]
- 45.Hoogerwerf W.A. Role of biological rhythms in gastrointestinal health and disease. Rev Endocr Metab Disord. 2009;10(4):293–300. doi: 10.1007/s11154-009-9119-3. [DOI] [PubMed] [Google Scholar]
- 46.Stokes K., Cooke A., Chang H., Weaver D.R., Breault D.T., Karpowicz P. The Circadian Clock Gene BMAL1 Coordinates Intestinal Regeneration. Cell Mol Gastroenterol Hepatol. 2017;4(1):95–114. doi: 10.1016/j.jcmgh.2017.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Duhart J.M., Leone M.J., Paladino N. Suprachiasmatic astrocytes modulate the circadian clock in response to TNF-alpha. J Immunol. 2013;191(9):4656–4664. doi: 10.4049/jimmunol.1300450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yoshida K., Hashiramoto A., Okano T., Yamane T., Shibanuma N., Shiozawa S. TNF-alpha modulates expression of the circadian clock gene Per2 in rheumatoid synovial cells. Scand J Rheumatol. 2013;42(4):276–280. doi: 10.3109/03009742.2013.765031. [DOI] [PubMed] [Google Scholar]
- 49.Nguyen K.D., Fentress S.J., Qiu Y., Yun K., Cox J.S., Chawla A. Circadian gene Bmal1 regulates diurnal oscillations of Ly6C(hi) inflammatory monocytes. Science. 2013;341(6153):1483–1488. doi: 10.1126/science.1240636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gibbs J., Ince L., Matthews L. An epithelial circadian clock controls pulmonary inflammation and glucocorticoid action. Nat Med. 2014;20(8):919–926. doi: 10.1038/nm.3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hwang J.W., Sundar I.K., Yao H., Sellix M.T., Rahman I. Circadian clock function is disrupted by environmental tobacco/cigarette smoke, leading to lung inflammation and injury via a SIRT1-BMAL1 pathway. FASEB J. 2014;28(1):176–194. doi: 10.1096/fj.13-232629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Curtis A.M., Fagundes C.T., Yang G. Circadian control of innate immunity in macrophages by miR-155 targeting Bmal1. Proc Natl Acad Sci U S A. 2015;112(23):7231–7236. doi: 10.1073/pnas.1501327112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ingle K.A., Kain V., Goel M., Prabhu S.D., Young M.E., Halade G.V. Cardiomyocyte-specific Bmal1 deletion in mice triggers diastolic dysfunction, extracellular matrix response, and impaired resolution of inflammation. Am J Physiol Heart Circ Physiol. 2015;309(11):H1827–H1836. doi: 10.1152/ajpheart.00608.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yao H., Sundar I.K., Huang Y. Disruption of Sirtuin 1-Mediated Control of Circadian Molecular Clock and Inflammation in Chronic Obstructive Pulmonary Disease. Am J Respir Cell Mol Biol. 2015;53(6):782–792. doi: 10.1165/rcmb.2014-0474OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Early J.O., Menon D., Wyse C.A. Circadian clock protein BMAL1 regulates IL-1beta in macrophages via NRF2. Proc Natl Acad Sci U S A. 2018;115(36):E8460–E8468. doi: 10.1073/pnas.1800431115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yamaoka Y., Kita M., Kodama T., Sawai N., Kashima K., Imanishi J. Induction of various cytokines and development of severe mucosal inflammation by cagA gene positive Helicobacter pylori strains. Gut. 1997;41(4):442–451. doi: 10.1136/gut.41.4.442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Suganuma M., Watanabe T., Yamaguchi K., Takahashi A., Fujiki H. Human gastric cancer development with TNF-alpha-inducing protein secreted from Helicobacter pylori. Cancer Lett. 2012;322(2):133–138. doi: 10.1016/j.canlet.2012.03.027. [DOI] [PubMed] [Google Scholar]
- 58.Mitsuno Y., Yoshida H., Maeda S. Helicobacter pylori induced transactivation of SRE and AP-1 through the ERK signalling pathway in gastric cancer cells. Gut. 2001;49(1):18–22. doi: 10.1136/gut.49.1.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bozek K., Kiełbasa S.M., Kramer A., Herzel H. Promoter analysis of Mammalian clock controlled genes. Genome Inform. 2007;18:65–74. [PubMed] [Google Scholar]
- 60.Volt H., García J.A., Doerrier C. Same molecule but different expression: aging and sepsis trigger NLRP3 inflammasome activation, a target of melatonin. J Pineal Res. 2016;60(2):193–205. doi: 10.1111/jpi.12303. [DOI] [PubMed] [Google Scholar]
- 61.Spengler M.L., Kuropatwinski K.K., Comas M. Core circadian protein CLOCK is a positive regulator of NF-kappaB-mediated transcription. Proc Natl Acad Sci U S A. 2012;109(37):E2457–E2465. doi: 10.1073/pnas.1206274109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bellet M.M., Zocchi L., Sassone-Corsi P. The RelB subunit of NFkappaB acts as a negative regulator of circadian gene expression. Cell Cycle. 2012;11(17):3304–3311. doi: 10.4161/cc.21669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Farre N., Farre R., Gozal D. Sleep apnea morbidity: a consequence of microbial-immune cross-talk? Chest. 2018;154(4):754–759. doi: 10.1016/j.chest.2018.03.001. [DOI] [PubMed] [Google Scholar]
- 64.Thaiss C.A., Levy M., Korem T. Microbiota diurnal rhythmicity programs host transcriptome oscillations. Cell. 2016;167(6):1495–1510. doi: 10.1016/j.cell.2016.11.003. [e12] [DOI] [PubMed] [Google Scholar]
- 65.Thaiss C.A., Zeevi D., Levy M. Transkingdom control of microbiota diurnal oscillations promotes metabolic homeostasis. Cell. 2014;159(3):514–529. doi: 10.1016/j.cell.2014.09.048. [DOI] [PubMed] [Google Scholar]
- 66.Leone V., Gibbons S.M., Martinez K. Effects of diurnal variation of gut microbes and high-fat feeding on host circadian clock function and metabolism. Cell Host Microbe. 2015;17(5):681–689. doi: 10.1016/j.chom.2015.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Thaiss C.A., Nobs S.P., Elinav E. NFIL-trating the host circadian rhythm-microbes fine-tune the epithelial clock. Cell Metab. 2017;26(5):699–700. doi: 10.1016/j.cmet.2017.10.014. [DOI] [PubMed] [Google Scholar]
- 68.Konturek P.C., Brzozowski T., Konturek S.J. Gut clock: implication of circadian rhythms in the gastrointestinal tract. J Physiol Pharmacol. 2011;62(2):139–150. [PubMed] [Google Scholar]
- 69.Johns C.E., Newton J.L., Westley B.R., May F.E. The diurnal rhythm of the cytoprotective human trefoil protein TFF2 is reduced by factors associated with gastric mucosal damage: ageing, Helicobacter pylori infection, and sleep deprivation. Am J Gastroenterol. 2005;100(7):1491–1497. doi: 10.1111/j.1572-0241.2005.41859.x. [DOI] [PubMed] [Google Scholar]
- 70.Mungan Z., Pinarbasi Simsek B. Gastroesophageal reflux disease and the relationship with Helicobacter pylori. Turk J Gastroenterol. 2017;28(Suppl. 1):S61–S67. doi: 10.5152/tjg.2017.16. [DOI] [PubMed] [Google Scholar]
- 71.Suzuki H., Moayyedi P. Helicobacter pylori infection in functional dyspepsia. Nat Rev Gastroenterol Hepatol. 2013;10(3):168–174. doi: 10.1038/nrgastro.2013.9. [DOI] [PubMed] [Google Scholar]
- 72.O'Connor A., O'Morain C.A., Ford A.C. Population screening and treatment of Helicobacter pylori infection. Nat Rev Gastroenterol Hepatol. 2017;14(4):230–240. doi: 10.1038/nrgastro.2016.195. [DOI] [PubMed] [Google Scholar]
- 73.Broussard J.L., Van Cauter E. Disturbances of sleep and circadian rhythms: novel risk factors for obesity. Curr Opin Endocrinol Diabetes Obes. 2016;23(5):353–359. doi: 10.1097/MED.0000000000000276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Camilleri M., Malhi H., Acosta A. Gastrointestinal complications of obesity. Gastroenterology. 2017;152(7):1656–1670. doi: 10.1053/j.gastro.2016.12.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Qandeel H.G., Alonso F., Hernandez D.J. Role of vagal innervation in diurnal rhythm of intestinal peptide transporter 1 (PEPT1) J Gastrointest Surg. 2009;13(11):1976–1985. doi: 10.1007/s11605-009-0984-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material