Abstract
Background:
In juvenile mammals, the epiphyses of long bones grow by chondrogenesis within the articular cartilage. A better understanding of the molecular mechanisms that regulate the growth of articular cartilage may give insight into the antecedents of joint disease, such as osteoarthritis.
Methods:
We used laser-capture microdissection to isolate chondrocytes from the superficial, middle, and deep zones of growing tibial articular cartilage in the 1-wk old mouse and then investigated expression patterns by microarray. To identify molecular markers for each zone of the growing articular cartilage, we found genes showing zone-specific expression and confirmed by real-time PCR and in situ hybridization.
Results:
Bioinformatic analyses implicated ephrin receptor signaling, Wnt signaling, and BMP signaling in the spatial regulation of chondrocyte differentiation during growth. Molecular markers were identified for superficial (e.g. Cilp, Prg4), middle (Cxcl14, Tnn), and deep zone (Sfrp5, Frzb). Comparison between juvenile articular and growth plate cartilage revealed that the superficial-to-deep zone transition showed similarity with the hypertrophic-to-resting zone transition.
Conclusion:
Laser capture microdissection combined with microarray analysis identified novel signaling pathways that are spatially regulated in growing mouse articular cartilage and revealed similarities between the molecular architecture of the growing articular cartilage and that of growth plate cartilage.
Introduction
Skeletal growth involves endochondral bone formation at the growth plate, which causes elongation of the metaphyses, intramembranous bone formation at the periosteal surface, which causes widening of the diaphysis, and chondrogenesis at the articular surface, which causes expansion of the epiphyses. This last process has received relatively little research attention but has important clinical implications because the articular cartilage is susceptible to degenerative diseases such as osteoarthritis due to its limited capacity for regeneration and repair (1). A better understanding of the molecular mechanisms that regulate the growth and maintenance of the articular cartilage may therefore facilitate the development of new therapeutic measures to prevent destruction and/or to achieve regeneration of articular cartilage.
Articular cartilage, like growth plate cartilage, is composed of chondrocytes surrounded by extracellular matrix and is spatially organized into three histologically distinct zones. The superficial zone (SZ) lies closest to the joint surface and consists of densely packed chondrocytes that are elongated parallel to the articular surface. In the middle zone (MZ), chondrocytes are scattered and more rounded in shape (MZ). In the deep zone (DZ), the chondrocytes are larger in size and arranged in columns perpendicular to the articular surface. These three zones differ not only in cellular morphology but also in their cellular metabolism (2) and extracellular matrix composition (3).
While cell proliferation occurs sparingly in adult articular cartilage, it is much more common in juvenile articular cartilage. Growth of the articular cartilage reportedly occurs appositionally from the articular surface (4), suggesting that the SZ may contain actively dividing chondrocytes, or a progenitor cell population that is capable of generating new chondrocytes (5). The mechanisms responsible for the distinctive properties of SZ are not well understood, but chondrocytes isolated from different zones of the articular cartilage, when cultured in monolayer, show substantially different response to growth factors, including insulin-like growth factor I, fibroblast growth factors (FGF), and transforming growth factor-β (6,7).
Thus, much remains unknown about the fundamental biological differences among the zones of articular cartilage and the mechanisms that initiate and maintain these differences. In the current study, we sought to elucidate the regulation of gene expression across different zones of the articular cartilage. We used laser capture microdissection (LCM) to isolate SZ, MZ, and DZ chondrocytes from juvenile mouse articular cartilage and compared gene expression across the three zones using microarray analysis. We then used bioinformatic approaches to identify spatially regulated signaling pathways that are likely to direct the zonal differences in the articular cartilage. Finally, we compared spatial regulation in articular cartilage and growth plate cartilage, which also contains three zones: resting zone (RZ), proliferative zone (PZ), and hypertrophic zone (HZ).
Results
Spatial Analysis of Articular Cartilage
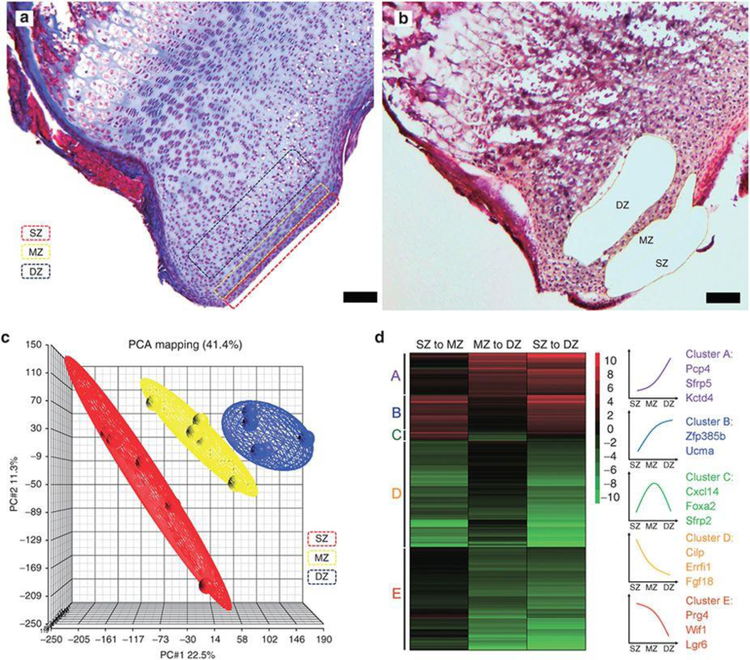
Articular cartilage from the proximal tibias of 1-wk-old mice was microdissected into the SZ, MZ, and DZ using LCM (Figure 1a,b). Microarray analysis was then used to identify spatial differences in gene expression. From SZ to MZ, expression of 47 genes was upregulated and expression of 142 genes was downregulated (≥twofold; false discovery rate (FDR) < 0.05; Supplementary Table S1 online). Ingenuity pathway analysis identified multiple signaling pathways that were overrepresented in these gene sets, including ephrin receptor signaling in the upregulated gene set and basal cell carcinoma signaling (which predominantly involves hedgehog signaling) in the downregulated gene set (Supplementary Table S1 online). This latter finding is consistent with our previous work showing that hedgehog signaling is active primarily on the articular surface (8). From MZ to DZ, 48 genes were upregulated and 161 genes were downregulated (≥twofold; FDR < 0.05; Supplementary Table S1 online). Pathway analysis showed that insulin-like growth factor I signaling was strongly overrepresented among downregulated genes (Supplementary Table S1 online). Comparison of SZ and DZ identified substantially more differentially expressed genes: 543 genes upregulated and 1,417 downregulated (Supplementary Table S1 online). Pathway analysis implicated protein kinase A signaling and Wnt signaling among upregulated genes and endocytosis signaling among downregulated genes (Supplementary Table S1 online).
Figure 1. Microarray analysis of laser capture microdissected articular cartilage.
(A) Mason Trichrome stained section of formalin fixed, decalcified 1-wk old mouse proximal tibial cartilage showing the location of zones in the articular cartilage that were targeted by laser capture microdissection (LCM). (B) Haemotoxylin & eosin stained frozen sections of 1-wk old mouse proximal tibial cartilage after LCM. (C) Principle component analysis (PCA) of microarray data from superficial, middle, and deep zone articular cartilage. (D) Heat map showing hierarchical clustering of spatially regulated genes (defined by false discovery rate < 0.05 SZ vs DZ). Red color represents upregulation, green color represents downregulation. Schematic line graphs on the right represent the typical pattern of changes in gene expression across the three zones and examples of genes in each cluster are listed. SZ, superficial zone; MZ, middle zone; DZ, deep zone. Scale bar, 100μm.
Principal component analysis suggested that MZ has a transitional expression profile between those of the SZ and DZ (Figure 1c). Similarly, hierarchical clustering of genes that showed a significant change across zones (FDR < 0.05) demonstrated that the majority of genes exhibited a progressive upregulation or downregulation across the three zones (Figure 1d), suggesting that the MZ primarily represents a transition between the SZ and DZ, rather than a functionally distinct region.
Wnt and Bmp signaling in Articular Cartilage
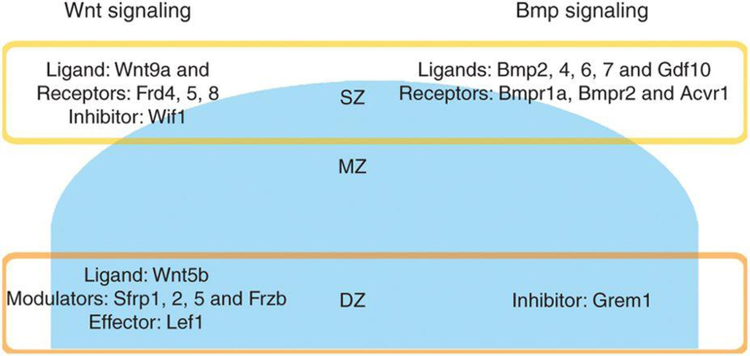
The bioinformatic analysis implicated Wnt signaling in the spatial regulation of articular cartilage (Supplementary Table S1 online). Interestingly, the expression data did not suggest a simple pattern of either increasing or decreasing Wnt signaling across the zones but a more complex pattern (Figure 2 and Supplementary Table S2 online). For example, Wnt5b was upregulated from SZ to DZ, while Wnt9a showed the reverse trend. Wif1, which encodes an inhibitor of Wnt signaling, was downregulated >80-fold from SZ to DZ, while four other soluble modulators/inhibitors of Wnt signaling, Sfrp1, 2, 5, and Frzb, were upregulated in the DZ. Receptors of Wnt signaling appeared to be more abundant on the articular surface, with downregulation of Fzd4, 5, and 8 from the SZ to DZ. Expression of Lef1, a transcriptional activator involved in Wnt signaling (9), was upregulated in the DZ. Consistently, β-catenin, which interacts with Lef1 and is critical for canonical Wnt signaling, appeared to be increased by immunohistochemistry at the peripheral part of the DZ (Supplementary Figure S1 online), suggesting cytoplasmic accumulation of β-catenin. However, there was not clear evidence of β-catenin nuclear translocation, which is needed for activation of the canonical Wnt signaling pathway.
Figure 2. Wnt/beta-catenin and Tgf-beta/Bmp signaling in articular cartilage.
Schematic diagram summarizing genes involved in Wnt signaling and BMP signaling that were significantly regulated across different zones of articular cartilage in 1-wk old mouse. Genes listed in each zone show substantially greater expression in that zone than in other zones. Fold changes and p-values are listed in Table S3. SZ, superficial zone; MZ, middle zone; DZ, deep zone.
In addition, multiple genes involved in bone morphogenetic protein (Bmp) signaling were differentially expressed in different zones of the articular cartilage. For example, several Bmps (Bmp2, 4, 5, 6, 7, and Gdf10) and their receptors (Bmpr1a, Bmpr2, and Acvr1) were significantly downregulated from SZ to DZ. Consistently, Grem1, which encodes a Bmp inhibitor, was upregulated in DZ (Figure 2 and Supplementary Table S3 online). Thus, the findings suggest that there exists a BMP signaling gradient across the articular cartilage, with greater signaling in SZ than that in DZ.
Identification of Spatial Markers for Articular Cartilage
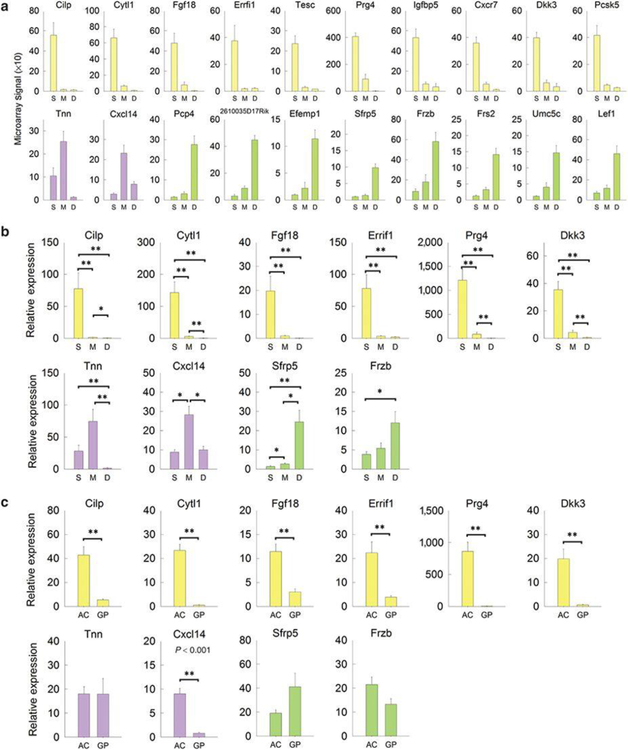
Next, we sought to identify useful molecular markers for each zone of the articular cartilage. We used an empirical formula (10) that gave high rank to genes that were expressed at high levels in a particular zone and at low levels in the other two zones. For SZ, the highest ranking marker was cartilage intermediate layer protein (Cilp, also known as Cilp-1) (expression in SZ 36-fold > MZ 42-fold > vs. DZ; Figure 3a and Supplementary Table S4 online). Other SZ markers included Fgf18, Prg4, Dkk3, Bmp2, and Bmp6. Only two genes, Tnn and Cxcl14, met the criterion for MZ markers, and these showed only moderate zonal specificity (Figure 3a and Supplementary Table S4 online) In DZ, three of the highest ranking markers were previously reported as RZ markers in the growth plate (10), including Pcp4, Efemp1, and Sfrp5 (Figure 3a and Supplementary Table S4 online), suggesting that DZ shares molecular similarities with RZ. Sfrp5 and another DZ marker, frizzled-related protein (Frzb), are both secreted frizzled-related proteins that antagonize Wnt signaling (11,12). These findings are in agreement with previous work showing that active Wnt signaling is important for SZ chondrocyte proliferation and Prg4 expression (13).
Figure 3. Differential gene expression (mean±SEM) of potential spatial markers across the tibial articular cartilage of 1-week old mice.
Different zones were isolated by laser capture microdissection and expression of potential spatial markers (yellow bars, superficial zone markers; purple bars, middle zone markers; green bars, deep zone markers) were analyzed by microarray (A) and real-time PCR (B). P-values in (B) represent pairwise comparison with Holm-Sidak correction following one-way ANOVA. To compare gene expression between overall articular cartilage and growth plate cartilage, manual microdissection of both regions from proximal tibias of 1-wk old mice was performed followed by real-time PCR (C). P-values in (C) represent t-test, articular cartilage versus growth plate cartilage. S, superficial zone; M, middle zone; D, deep zone; AC, articular cartilage; GP, growth plate cartilage.
The expression of specific markers in SZ, MZ, and DZ were evaluated using real-time PCR (Figure 3b), and their expression in whole articular cartilage and whole growth plate cartilage was also compared (Figure 3c). All six SZ markers showed zone-specific expression not just within the articular cartilage but more generally, in that expression levels were lower in growth plate than that in articular cartilage. In contrast, Tnn, Sfrp5, and Frzb showed similar expression levels in growth plate and articular cartilage (Figure 3c).
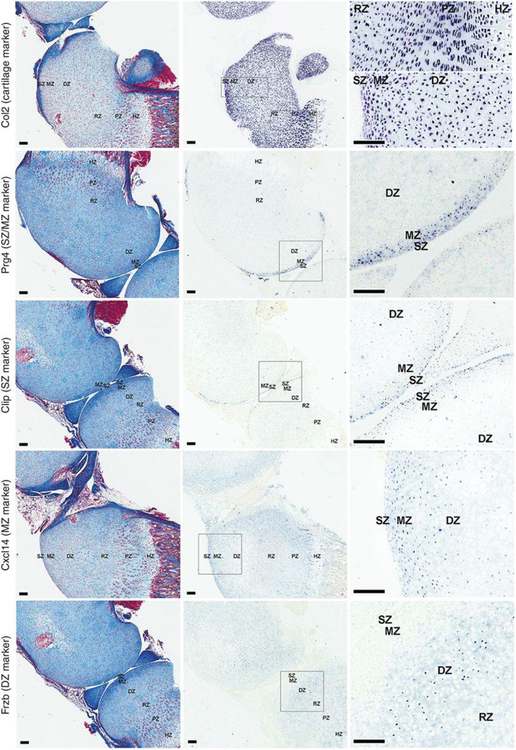
To further confirm our findings, the localization of several spatial markers in 1-wk-old mouse proximal tibia and distal femur was studied by in situ hybridization (Figure 4). Cilp expression was present primarily in the one- or two-cell layer closest to the articular surface, whereas signals for Prg4 were present more broadly in the SZ and part of the immediately adjacent MZ. Cxcl14 was preferentially detected in the MZ of the articular cartilage and also in HZ of the growth plate. Frzb was confirmed to be present specifically in DZ.
Figure 4. In situ hybridization of zonal markers of articular cartilage.
Formalin fixed, decalcified sections of 1-wk old mouse tibial cartilage were hybridized to DIG-labeled riboprobes, producing a purple color and were visualized by scanning the slides with a ScanScope CS digital scanner under bright field microscopy. Left panel, Mason-trichrome stained sections; middle panel, in situ hybridization without counterstaining; right panel, high magnification views taken from within the rectangular area indicated in the corresponding middle panel. SZ, superficial zone; MZ, middle zone; DZ, deep zone; RZ, resting zone; PZ, proliferative zone; HZ, hypertrophic zone.
Relationship between Spatial Regulation of Articular Cartilage and Growth Plate Cartilage
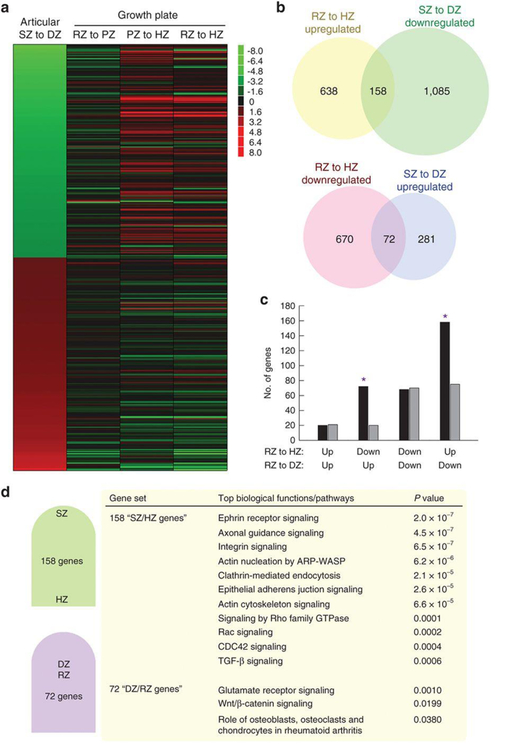
We hypothesized that the spatial regulation of gene expression in articular cartilage might be similar to growth plate cartilage, with the SZ corresponding to the RZ and the DZ corresponding to the HZ. This hypothesis was based on two prior observations. First, SZ and RZ reportedly contain progenitor cell populations (5,14). Second, chondrocytes gradually hypertrophy (enlarge) with the spatial transition from SZ to DZ and from PZ to HZ. To test this hypothesis, we combined the current articular cartilage analysis with our previous spatial analysis in 1-wk-old rat growth plate (10). Gene-set enrichment analysis (Supplementary Figure S2 online) showed that the genes most downregulated from SZ to DZ (greatest fold change; FDR < 0.05) are enriched with genes upregulated from RZ to HZ (P = 0.031), and conversely, genes upregulated from SZ to DZ are enriched with genes downregulated from RZ to HZ (P < 0.001). Similarly, among the 200 most strongly upregulated and 200 most strongly downregulated genes from SZ to DZ (greatest fold change; FDR < 0.05), there was a strong negative correlation between changes from SZ to DZ and changes from RZ to HZ (r = −0.4031; P < 0.0001, Pearson’s correlation; Table 1 and Figure 5a). This correlation was in large part attributable to the changes from PZ to HZ (r = −0.4595; P < 0.0001). Similarly, Changes from PZ to HZ showed a strong negative correlation with changes from MZ to DZ (r = −0.4567; P < 0.0001; Table 1), and, more weakly, with changes from SZ to MZ (r = −0.2489; P = 0.0004; Table 1). Additionally, we found a significant overlap between genes that were downregulated from SZ to DZ (≥twofold; FDR < 0.05) and genes upregulated from RZ to HZ (≥twofold; FDR < 0.05), and similarly, a significant overlap between genes that were upregulated from SZ to DZ and genes downregulated from RZ to HZ (P < 0.001, Pearson’s χ2; Figure 5b,c). Thus, multiple analyses suggest that SZ to DZ expression pattern is not, as we had hypothesized, similar to the RZ to HZ pattern. Instead, the correspondence is inverted so that the DZ to SZ expression pattern is similar to that of RZ to HZ.
Table 1.
Correlation Between Spatial Differences in Gene Expression of Articular and Growth Plate Cartilage
| Comparison: | Pearson’s r value | P value |
|---|---|---|
| 400 genes that differ most from SZ to DZ | ||
| SZ to DZ vs RZ to PZ | 0.0409 | 0.4149 |
| SZ to DZ vs PZ to HZ | −0.4595 | <0.0001 |
| SZ to DZ vs RZ to HZ | −0.4031 | <0.0001 |
| 200 genes that differ most from SZ to MZ | ||
| SZ to MZ vs RZ to PZ | 0.0251 | 0.7242 |
| SZ to MZ vs PZ to HZ | −0.2489 | 0.0004 |
| SZ to MZ vs RZ to HZ | −0.2121 | 0.0026 |
| 200 genes that differ most from MZ to DZ | ||
| MZ to DZ vs RZ to PZ | −0.0552 | 0.4376 |
| MZ to DZ vs PZ to HZ | −0.4567 | <0.0001 |
| MZ to DZ vs RZ to HZ | −0.4399 | <0.0001 |
Figure 5. Comparison of spatial regulation of gene expression in articular and growth plate cartilage.
Our current zonal analysis of 1-wk old tibial mouse articular cartilage was combined with our previous zonal analysis of 1-wk old tibial rat growth plate cartilage (26). (A) Heat maps were constructed for the 200 genes that were most strongly upregulated and 200 genes that were most strongly downregulated from SZ to DZ of articular cartilage. Red color represents upregulation, green color represents downregulation. Genes downregulated from SZ to DZ (green) tended to be upregulated from RZ to HZ (red), and vice versa. (B) Venn diagrams showing the overlap between RZ-to-HZ upregulation and SZ-to-DZ downregulation, and also the overlap between RZ-to-HZ downregulation and SZ-to-DZ upregulation. Numbers on Venn diagram indicate number of genes in each category. Significant regulation was defined as ≥ 2-fold and FDR < 0.05. (C) Observed (black bars) and expected (grey bars) overlap of RZ-to-HZ regulation in growth plate and SZ-to-DZ regulation in articular cartilage. Asterisks indicate observed overlap greater than expected by chance (P<0.001, Pearson’s chi-square test). (D) Ingenuity Pathway Analysis of the 158 that were both significantly upregulated from RZ to HZ, and also downregulated from SZ to DZ genes (“SZ/HZ genes”) and the 72 genes that were both significantly downregulated from RZ to HZ, and also upregulated from SZ to DZ (“DZ/RZ genes”). For SZ/HZ genes, the top 10 biological functions with the lowest p-value were listed. For DZ/RZ genes, all functions with P<0.05 were listed. SZ, superficial zone; MZ, middle zone; DZ, deep zone; RZ, resting zone; PZ, proliferative zone; HZ, hypertrophic zone.
Ingenuity pathway analysis identified multiple functional pathways that were overrepresented both among genes upregulated in SZ (vs. DZ) and also in HZ (vs. RZ), including ephrin receptor signaling, integrin signaling, and transforming growth factor-β signaling (Figure 5d). Ingenuity pathway analysis also identified functional pathways that were overrepresented both among genes commonly upregulated in the DZ and the RZ, including glutamate receptor signaling and Wnt signaling (Figure 5d).
Discussion
Using LCM and microarray analysis, we analyzed the spatial expression patterns of the articular cartilage in 1-wk-old mice. Bioinformatic analysis of these expression patterns implicated several signaling pathways that were spatially regulated across different zones of the articular cartilage, including Wnt/β-catenin signaling, which has been previously shown to play an important role in cartilage development (15). Our pathway analysis implicated Wnt/β-catenin signaling in the spatial regulation of articular cartilage (Supplementary Table S1 online) with a complex pattern. For example, DZ expressed a high level of Wnt5b, which promotes chondrogenesis and inhibits hypertrophy (16). However, the level of several soluble modulators/inhibitors of Wnt signaling, including Sfrp1, 2, 5, and Frzb, was also higher in DZ. Translocation of β-catenin to the nucleus, which is indicative of canonical Wnt signaling pathway activation, was not definitively demonstrated in the DZ by immunohistochemistry. Therefore, it remains unclear whether Wnt signaling is more active in the deeper regions of articular cartilage.
Another signaling pathway that is critical for cartilage development and function is transforming growth factor-β/BMPs signaling. Postnatal deletion of Bmpr1a (a BMPs receptor) in mice leads to accelerated degeneration of the articular cartilage (17), while a single nucleotide polymorphism (rs12593365) of GREM1 (a BMPs inhibitor) in humans is associated with 20% reduced risk of radiographic hip osteoarthritis (18), suggesting that BMPs contribute to articular cartilage maintenance. Our microarray analysis showed that Bmp2, 4, 6, 7, and Gdf10 (Bmp3b) were expressed at higher levels in the SZ, while the inhibitor Grem1 was expressed in the DZ, suggesting greater BMPs signaling at the articular surface, where it may serve to promote expression of Prg4 (19,20).
Our bioinformatic analysis also identified several other pathways that show differential expression patterns in the zones of the articular cartilage including integrin and ephrin receptor signaling. Integrins appear to play an important role in the interaction between cartilage extracellular matrix and the growth and differentiation of articular chondrocytes (21). Ephrin receptor signaling has been reported to be involved in cartilage morphogenesis of the limb bud in the chicken (22), but little is known about its role in articular cartilage.
We also identified mRNA markers for each zone of articular cartilage using microarray, real-time PCR, and in situ hybridization. We identified several novel markers for SZ, including Cytl1, Errfi1, and Fgf18, and confirmed previously reported markers Prg4 (ref. 23) and Dkk3 (ref. 24). We also found very specific expression of Cilp in the outermost one- to two-cell layers of the SZ. Although Cilp is known as cartilage intermediate layer protein and was initially thought to be specifically expressed in MZ (25,26,27), others have demonstrated Cilp primarily in SZ (28). Only two markers were identified for the MZ, both of which showed only moderate specificity, consistent with a transitional nature of the MZ. For the DZ, we identified multiple genetic markers, such as Sfrp5 and Efemp1, which we previously identified as RZ markers in the growth plate (10).
We also compared the spatial regulation between the articular cartilage and growth plate cartilage (10). Contrary to our hypothesis, the analysis suggested that the SZ shows an expression pattern similar to the HZ, and the DZ shows an expression pattern similar to the RZ. Part of this relationship could be attributable to the similarities between the DZ and RZ, which both arise from a common pool of round epiphyseal chondrocytes in the embryo which then become separated by the secondary ossification center. However, there were even more genes commonly expressed in the SZ and HZ (158 genes) than that in RZ and DZ (72 genes). Furthermore, bioinformatic analysis suggested that various signaling pathways, such as ephrin signaling, integrin signaling, and transforming growth factor-β signaling, were implicated by genes commonly upregulated in SZ and HZ (compared with DZ and RZ, respectively). For example, we found evidence for a BMP signaling gradient across the articular cartilage, with greater signaling in the SZ than in DZ, which corresponds to a BMP signaling gradient previously suggested across growth plate cartilage, with greater BMP action in HZ than in RZ. The results of the comparison are consistent with our previous study, which involved a less precise manual dissection of articular cartilage (29).
There have been other studies that used LCM and microarray analysis to investigate the zonal gene expression of the mammalian articular cartilage. For example, Fukui et al. (2,30) compared gene expression in the three zones of normal and osteoarthritic human articular cartilage. However, their study involved elderly subjects, whereas our study focused on spatial regulation in growing, early postnatal articular cartilage. Amanatullah et al. (31) studied gene expression in 1–3-mo-old bovine articular cartilage and obtained results somewhat similar to ours, including high Prg4 and Cilp expression in SZ. In contrast, the genes that we found to be specifically expressed in the DZ, such as Frzb, Sfrp5, and Efemp1, were reported by Amanatullah et al. to be expressed in the MZ, perhaps because of differences in defining the articular zones.
In summary, we used LCM to isolate different zones of the articular cartilage and used microarray to analyze global gene expression. Bioinformatic analysis corroborated previously reported signaling pathways, such as Wnt and Bmp signaling, and implicated novel pathways, such as ephrin and integrin signaling, for spatially associated articular chondrocyte differentiation. In addition, comparison of the spatial regulation of articular and growth plate cartilage revealed interesting similarities between the SZ of the articular cartilage and the HZ of the growth plate.
Materials and Methods
Animal procedures
All animal procedures were approved by the National Institute of Child Health and Human Development Animal Care and Use Committee. Although our previous spatial analysis of the growth plate cartilage was done in the rat, we chose to use the mouse in our current study of the articular cartilage based on two reasons. First, the mouse array is better annotated (39,000 transcripts) as compared with the rat array (30,000 transcripts). Second, many of the functional studies that give important insight into physiology and pathophysiology of the articular cartilage were done in the mouse using knockout models, and therefore, studying the spatial regulation of gene expression in the mouse articular cartilage will be more relevant to previous studies in the field. One-wk-old C57BL/6 male mice (Charles River Laboratory, Wilmington, MA) were killed by decapitation. Distal femur and proximal tibial epiphyses were excised. For laser capture and manual microdissection, cartilage was embedded in optimum cutting temperature compound (Electron Microscopy Sciences, Hatfield, PA), frozen on dry ice, and stored at −80 °C. For in situ hybridization and histology, cartilage was fixed overnight in 2% (v/v) paraformaldehyde (Electron Microscopy Sciences) and 0.2% (v/v) glutaraldehyde (Electron Microscopy Sciences) at 4 °C and decalcified in 10% (v/v) EDTA and 0.5% (v/v) paraformaldehyde, pH 7.4. Samples were then embedded in paraffin for sectioning.
Laser capture microdissection (LCM) of articular cartilage
Frozen longitudinal cartilage sections (10 μm) were placed on PEN membrane glass slides (MDS Analytical Technologies, Sunnyvale, CA) precoated with 0.01% (w/v) poly-D-lysine (Sigma-Aldrich, St Louis, MO). Slides were thawed and stained as previously described (32) with modifications (70% ethanol, 30 s; deionized water, 15 s; Mayer’s hematoxylin (Sigma-Aldrich), 30 s; deionized water, 15 s; Scott’s bluing (Sigma-Aldrich), 15 s; 70% ethanol, 15 s; eosin Y (Sigma-Aldrich), 1 s; 95% ethanol, 30 s; 100% ethanol, 30 s; and xylene, 30 s) to shorten staining time and minimize tissue detachment. SZ, MZ, and DZ were microdissected using the cut and capture method in the Arcturus LCM System (Thermo Scientific, Waltham, MA) using CapSure HS Caps (MDS Analytical Technologies) and dissolved in tissue extraction buffer (PicoPure RNA Isolation Kit; Thermo Scientific) on ice. For each zone, tissue dissected from 35 to 40 sections from a single animal was pooled, and RNA isolation was performed using the PicoPure RNA Isolation Kit (Thermo Scientific). RNA integrity and concentration were assessed using the Bioanalyzer 2100 (Agilent Biotechnologies, Palo Alto, CA). Five to 20 ng of total RNA was obtained from each zone with an RNA integrity number of 5–7.
Manual microdissection of cartilage
To compare whole articular cartilage and whole growth plate cartilage expression, we performed manual microdissection of 1-wk-old mouse proximal tibial sections (50 μm thickness) as previously described (33). To minimize cross-contamination, the lowermost articular cartilage DZ and the uppermost growth plate RZ were discarded. Tissue from 20 sections of a single animal was pooled prior to RNA isolation using the PicoPure RNA Isolation Kit. Approximately 1 μg of total RNA was obtained from the articular or growth plate cartilage of one animal, with a RNA integrity number of 7–8.
Microarray Analysis
For each animal, total RNA from SZ, MZ, or DZ (n = 5 each) was amplified using WT-Ovation Pico RNA Amplification System (NuGEN, San Carlos, CA) following the manufacturer’s protocol and hybridized to Mouse Genome 430 2.0 GeneChip Arrays (Affymetrix, Santa Clara, CA). Each array represents a single zone from a single animal, and a total of five animals were used. Affymetrix CEL files were imported into Partek Genomics Suite 6.6 (Partek, St Louis, MO) using the default normalization parameters. Probe-level data were preprocessed, including background correction, normalization, and summarization, using robust multiarray average analysis (34), and log2 transformed. Bioinformatic analysis was performed using Ingenuity Pathways Analysis Software (Ingenuity Systems, Redwood City, CA). Heat maps were constructed using JMP 9 software (SAS Institute, Cary, NC).
Spatial Markers Identification
To identify useful molecular markers for each zone of the articular cartilage, we employed an empirical formula that was previously used to identify growth plate zonal marker (10). This formula gives a low score to genes that are expressed at high levels in one zone and at low levels in both of the other two zones. For example, the formula for SZ markers was: score = (M + D + SA)/S, where S, M, and D represents expression level of a gene in the SZ, MZ, and DZ, respectively, and SA represents the geometric mean of expression of all probes on the microarray in the SZ. Genes were then ranked from the lowest to highest score, and an arbitrary cutoff score of 0.5 was used for genes to qualify as good zonal markers.
Quantitative real-time PCR
Total RNA, without amplification, from individual animals was reverse transcribed into complementary DNA, and expression was quantified by real-time PCR using an ABI Prism 7900 Fast Sequence Detector (Thermo Scientific), according to the manufacturer’s instructions. Prg4, Cilp, Fgf18, Cytl1, Errfi1, Dkk3, Tnn, Cxcl14, Sfrp5, and Frzb mRNA levels were measured by SYBR green–based assays using custom-designed primers (Supplementary Table S5 online), and the data were normalized to 18S ribosomal RNA measured with a predesigned Taqman assay (Thermo Scientific). Relative expression values were calculated as previously described (35).
In situ hybridization
cDNA from 1-wk-old mouse growth plate was amplified using Phusion DNA polymerase (Thermo Scientific) with primers that contained either a T7 promoter (for sense probes) or an SP6 promoter (for antisense probes) (Supplementary Table S6 online). Single stranded digoxigenin-labeled riboprobes for in situ hybridization were transcribed using DIG RNA Labeling Kit (Roche Diagnostics, Indianapolis, IN) following the manufacturer’s protocol. Riboprobes were purified by Micro Bio-Spin Columns P-30 Tris RNase free (Bio-Rad, Hercules, CA), followed by alkaline hydrolysis for 30 min as previously described (36). Paraffin-embedded sections of epiphyseal cartilage were hybridized to digoxigenin-labeled riboprobes as described by Bandyopadhyay et al. (37) and Murtaugh et al. (38). with some modifications. Notably, tissues were permeabilized with 300 mg/ml of proteinase K (Thermo Scientific) at 37 °C for 30 min prior to acetylation and prehybridization. For detection, tissue sections were incubated with anti-digoxigenin alkaline phosphatase Fab fragments (Roche Diagnostics) for 2 h at room temperature and then treated with nitro-blue tetrazolium chloride/5-bromo-4-chloro-3′-indolyphosphate p-toluidine salt (NBT/BCIP) (Sigma-Aldrich) in the dark until a colorimetric change was detected. The sections were visualized using a ScanScope CS digital scanner (Aperio Technologies, Vista, CA) under bright field microscopy.
Statistical analysis
Data are presented as mean ± SEM. Comparison of expression levels was performed by one-way ANOVA (SigmaPlot 10) using log-transformed relative expression data, followed by pairwise comparisons corrected for multiple comparisons using the Holm–Sidak method. All P values were two tailed, and significance was recognized at P < 0.05. For comparison between the spatial regulation of articular cartilage and growth plate cartilage, gene-set enrichment analysis (Broad Institute, Cambridge, MA) was used according to user’s manual (39), the probability of overlapping gene sets occurring by chance was determined using Pearson’s χ2 test (SigmaPlot 10).
Supplementary Material
Acknowledgments
Financial Support:
This work was supported by the Intramural Research Program of the Eunice Kennedy Shriver National Institute of Child Health and Human Development, The National Institute of Diabetes and Digestive and Kidney Diseases, and The National Cancer Institute, NIH.
References
- 1.Roelofs AJ, Rocke JP, De Bari C. Cell-based approaches to joint surface repair: a research perspective. Osteoarthritis Cartilage 2013;21:892–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fukui N, Ikeda Y, Ohnuki T, et al. Regional differences in chondrocyte metabolism in osteoarthritis: a detailed analysis by laser capture microdissection. Arthritis Rheum 2008;58:154–63. [DOI] [PubMed] [Google Scholar]
- 3.Pearle AD, Warren RF, Rodeo SA. Basic science of articular cartilage and osteoarthritis. Clin Sports Med 2005;24:1–12. [DOI] [PubMed] [Google Scholar]
- 4.Hayes AJ, MacPherson S, Morrison H, Dowthwaite G, Archer CW. The development of articular cartilage: evidence for an appositional growth mechanism. Anat Embryol (Berl) 2001;203:469–79. [DOI] [PubMed] [Google Scholar]
- 5.Hattori S, Oxford C, Reddi AH. Identification of superficial zone articular chondrocyte stem/progenitor cells. Biochem Biophys Res Commun 2007;358:99–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leipzig ND, Eleswarapu SV, Athanasiou KA. The effects of TGF-beta1 and IGF-I on the biomechanics and cytoskeleton of single chondrocytes. Osteoarthritis Cartilage 2006;14:1227–36. [DOI] [PubMed] [Google Scholar]
- 7.Darling EM, Athanasiou KA. Growth factor impact on articular cartilage subpopulations. Cell Tissue Res 2005;322:463–73. [DOI] [PubMed] [Google Scholar]
- 8.Chau M, Forcinito P, Andrade AC, et al. Organization of the Indian hedgehog–parathyroid hormone-related protein system in the postnatal growth plate. J Mol Endocrinol 2011;47:99–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Behrens J, von Kries JP, Kühl M, et al. Functional interaction of betacatenin with the transcription factor LEF-1. Nature 1996;382:638–42. [DOI] [PubMed] [Google Scholar]
- 10.Lui JC, Andrade AC, Forcinito P, et al. Spatial and temporal regulation of gene expression in the mammalian growth plate. Bone 2010;46: 1380–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang S, Krinks M, Lin K, Luyten FP, Moos M Jr. Frzb, a secreted protein expressed in the Spemann organizer, binds and inhibits Wnt-8. Cell 1997;88:757–66. [DOI] [PubMed] [Google Scholar]
- 12.Leyns L, Bouwmeester T, Kim SH, Piccolo S, De Robertis EM. Frzb-1 is a secreted antagonist of Wnt signaling expressed in the Spemann organizer. Cell 1997;88:747–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yasuhara R, Ohta Y, Yuasa T, et al. Roles of β-catenin signaling in phenotypic expression and proliferation of articular cartilage superficial zone cells. Lab Invest 2011;91:1739–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abad V, Meyers JL, Weise M, et al. The role of the resting zone in growth plate chondrogenesis. Endocrinology 2002;143:1851–7. [DOI] [PubMed] [Google Scholar]
- 15.Staines KA, Macrae VE, Farquharson C. Cartilage development and degeneration: a Wnt Wnt situation. Cell Biochem Funct 2012;30:633–42. [DOI] [PubMed] [Google Scholar]
- 16.Bradley EW, Drissi MH. Wnt5b regulates mesenchymal cell aggregation and chondrocyte differentiation through the planar cell polarity pathway. J Cell Physiol 2011;226:1683–93. [DOI] [PubMed] [Google Scholar]
- 17.Rountree RB, Schoor M, Chen H, et al. BMP receptor signaling is required for postnatal maintenance of articular cartilage. PLoS Biol 2004;2:e355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leijten JC, Emons J, Sticht C, et al. Gremlin 1, frizzled-related protein, and Dkk-1 are key regulators of human articular cartilage homeostasis. Arthritis Rheum 2012;64:3302–12. [DOI] [PubMed] [Google Scholar]
- 19.Yamane S, Reddi AH. Induction of chondrogenesis and superficial zone protein accumulation in synovial side population cells by BMP-7 and TGFbeta1. J Orthop Res 2008;26:485–92. [DOI] [PubMed] [Google Scholar]
- 20.Lee SY, Nakagawa T, Reddi AH. Induction of chondrogenesis and expression of superficial zone protein (SZP)/lubricin by mesenchymal progenitors in the infrapatellar fat pad of the knee joint treated with TGF-beta1 and BMP-7. Biochem Biophys Res Commun 2008;376:148–53. [DOI] [PubMed] [Google Scholar]
- 21.Loeser RF. Integrins and chondrocyte-matrix interactions in articular cartilage. Matrix Biol 2014;39:11–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wada N, Tanaka H, Ide H, Nohno T. Ephrin-A2 regulates position-specific cell affinity and is involved in cartilage morphogenesis in the chick limb bud. Dev Biol 2003;264:550–63. [DOI] [PubMed] [Google Scholar]
- 23.Schumacher BL, Block JA, Schmid TM, Aydelotte MB, Kuettner KE. A novel proteoglycan synthesized and secreted by chondrocytes of the superficial zone of articular cartilage. Arch Biochem Biophys 1994;311:144–52. [DOI] [PubMed] [Google Scholar]
- 24.Yamane S, Cheng E, You Z, Reddi AH. Gene expression profiling of mouse articular and growth plate cartilage. Tissue Eng 2007;13:2163–73. [DOI] [PubMed] [Google Scholar]
- 25.Karlsson C, Lindahl A. Articular cartilage stem cell signalling. Arthritis Res Ther 2009;11:121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lorenzo P, Bayliss MT, Heinegård D. A novel cartilage protein (CILP) present in the mid-zone of human articular cartilage increases with age. J Biol Chem 1998;273:23463–8. [DOI] [PubMed] [Google Scholar]
- 27.Yao Z, Nakamura H, Masuko-Hongo K, Suzuki-Kurokawa M, Nishioka K, Kato T. Characterisation of cartilage intermediate layer protein (CILP)- induced arthropathy in mice. Ann Rheum Dis 2004;63:252–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bernardo BC, Belluoccio D, Rowley L, Little CB, Hansen U, Bateman JF. Cartilage intermediate layer protein 2 (CILP-2) is expressed in articular and meniscal cartilage and down-regulated in experimental osteoarthritis. J Biol Chem 2011;286:37758–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chau M, Lui JC, Landman EB, et al. Gene expression profiling reveals similarities between the spatial architectures of postnatal articular and growth plate cartilage. PLoS One 2014;9:e103061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fukui N, Miyamoto Y, Nakajima M, et al. Zonal gene expression of chondrocytes in osteoarthritic cartilage. Arthritis Rheum 2008;58:3843–53. [DOI] [PubMed] [Google Scholar]
- 31.Amanatullah DF, Yamane S, Reddi AH. Distinct patterns of gene expression in the superficial, middle and deep zones of bovine articular cartilage. J Tissue Eng Regen Med 2014;8:505–14. [DOI] [PubMed] [Google Scholar]
- 32.Erickson HS, Albert PS, Gillespie JW, et al. Quantitative RT-PCR gene expression analysis of laser microdissected tissue samples. Nat Protoc 2009;4:902–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nilsson O, Parker EA, Hegde A, Chau M, Barnes KM, Baron J. Gradients in bone morphogenetic protein-related gene expression across the growth plate. J Endocrinol 2007;193:75–84. [DOI] [PubMed] [Google Scholar]
- 34.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 2003;19:185–93. [DOI] [PubMed] [Google Scholar]
- 35.Lui JC, Forcinito P, Chang M, Chen W, Barnes KM, Baron J. Coordinated postnatal down-regulation of multiple growth-promoting genes: evidence for a genetic program limiting organ growth. FASEB J 2010;24:3083–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cox KH, DeLeon DV, Angerer LM, Angerer RC. Detection of mrnas in sea urchin embryos by in situ hybridization using asymmetric RNA probes. Dev Biol 1984;101:485–502. [DOI] [PubMed] [Google Scholar]
- 37.Bandyopadhyay A, Kubilus JK, Crochiere ML, Linsenmayer TF, Tabin CJ. Identification of unique molecular subdomains in the perichondrium and periosteum and their role in regulating gene expression in the underlying chondrocytes. Dev Biol 2008;321:162–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Murtaugh LC, Chyung JH, Lassar AB. Sonic hedgehog promotes somitic chondrogenesis by altering the cellular response to BMP signaling. Genes Dev 1999;13:225–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 2005;102:15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.