Abstract
Aberrant expression of epigenetic regulators of gene expression contributes to initiation and progression of cancer. During recent years, considerable research efforts have focused on the role of histone acetyltransferases (HATs) and histone deacetylases (HDACs) in cancer cells, and the identification of pharmacologic agents that modulate gene expression via inhibition of HDACs. The following review highlights recent studies pertaining to HDAC expression in cancer cells, the plieotropic mechanisms by which HDAC inhibitors (HDACi) mediate antitumor activity, and the potential clinical implications of HDAC inhibition as a strategy for cancer therapy.
Keywords: Epigenetics, cancer, HDAC inhibitor
Introduction
During recent years, considerable research efforts have focused on potentially reversible alterations in chromatin structure, which modulate gene expression during malignant transformation. The basic structure of chromatin is the nucleosome, which is composed of ~146 bp of DNA wrapped twice around an octamer of core histones (H3-H4 tetramer, and two H2A-H2B dimers). Core histone proteins contain a basic N-terminal tail region, a histone fold, and a carboxy-terminal region. All of these regions-particularly the positively charged N terminal tails protruding from the DNA helix, are sites for a variety of covalent modifications such as acetylation, methylation, phosphorylation, ubiquitination, biotinylation, ADP ribosylation, sumoylation, glycosylation, and carbonylation (1). These dynamic alterations modulate interactions between DNA, histones, multiprotein chromatin remodeling complexes and transcription factors, thereby enhancing or repressing gene expression (2;3).
The emerging delineation of histone alterations that coincide with aberrant gene expression and malignant transformation provides impetus for the development of agents that target histone modifiers for cancer therapy. The following discussion will focus on recent insights regarding the mechanisms by which histone deacetylase (HDAC) inhibitors mediate cytotoxicity in cancer cells.
Histone Acetyltransferases and Histone Deacetylases
Acetylation of core histones is governed by opposing actions of a variety of histone acetyl transferases (HAT) and histone deacetylases (HDACs). Histone acetylases mediate transfer of an acetyl group from acetyl-co-A to the ε-amino site of lysine, and are divided into two groups. Type A HATs are located in the nucleus, and acetylate nucleosomal histones as well as other chromatin-associated proteins; as such, these HATs directly modulate gene expression. In contrast, Type B HATs are localized in the cytoplasm, and acetylate newly synthesized histones, thus facilitating their transport into the nucleus and subsequent association with newly synthesized DNA (4;5). Type A HATs typically are components of high-molecular complexes and comprise five families; GNAT, P300/CBP, MYST, nuclear receptor coactivators, and general transcription factors (4). Some HATs, notably p300 and CBP, associate with a variety of transcriptional regulators including Rb and p53, and may function as tumor suppressors. In addition, HATs acetylate a variety of non-histone proteins including p53, E2F1, Rb, p73, HDACs, and heat shock protein (Hsp) 90(6;7) (Table 1).
Table 1.
Non-histone Cellular Proteins Targeted by HATS and HDACs
| p53, p73, Hsp 90, C-MYC, H2A-2, E2F1, RUNX 3, Amod-7, STAT-3, p50, p65, HMG-A1, PLAGL2, p300, ATM, MYO-D, Sp1, β-catenin, pRb, GATA-1, YY-1, HIF-1α, STAT-1, FOX01, FOX04 |
HDACs are currently divided into four classes based on phylogenetic and functional criteria (reviewed in ref (7)). Class I HDACs (1, 2, 3, and 8), which range in size from ~40–55 Kd, are structurally similar to yeast transcription factor, Rpd-3, and typically associate with multi-protein repressor complexes containing sin3, Co-REST, Mi2/NuRD, N-COR/SMRT and EST1B (8). HDACs 1, 2, and 3 are localized in the nucleus, and target multiple substrates including p53, myo-D, STAT-3, E2F1, Rel-A, and YY1 (9;10). HDAC 8 is localized in the nucleus as well as the cytoplasm; no substrates of this Class I HDAC have been defined to date.
Class II HDACs (4, 5, 6, 7, 9, 10), which range in size from ~70 – 130 Kd, are structurally similar to yeast HDA1 deacetylase and are subdivided into two classes. Class IIA HDACs (4, 5, 7, and 9) contain large N-terminal domains that regulate DNA binding, and interact in a phosphorylation-dependent manner with 14–3-3 proteins, which mediate movement of these HDACs between cytoplasm and nucleus in response to mitogenic signals (7). Class IIB HDACs (6 and 10) are localized in the cytoplasm. HDAC 6 is unique in that it contains two deacetylase domains and a zinc finger region in the c-terminus. HDAC 10 is similar to HDAC 6, but contains an additional inactive domain (7;10).
In contrast to Class I HDACs, Class II HDACs exhibit family-restricted interactions with a variety of proteins including ANKRA, RFXANK, estrogen receptor (ER), REA, HIF1α, Bcl-6, and Fox3P. These HDACs have a variety of non-histone target substrates including GATA-1, GCMa, HP-1, and SMAD-7, as well as FLAG-1 and FLAG-2 (9;10). Relatively little information is available regarding binding partners for HDAC 6 and HDAC 10 (11;12). Notably, HDAC 6 has emerged as a major deacetylase of α-tubulin as well as Hsp90 ; as such, HDAC 6 mediates cell motility, and stability of oncoproteins such as EGFR, RAF1, and ABL, that are client proteins of Hsp90 (13). Additionally, HDAC 6 can interact via its zinc finger with ubiquitin to modulate aggressesome function and autophagy (14). Recent studies suggest that HDAC 10 may also function to modulate acetylation of Hsp90 (15).
Class I and Class II HDACs are zinc-dependent enzymes containing catalytic pockets that can be inhibited by zinc chelating compounds such as hydroxamic acid. HDAC 11, the only known Class IV HDAC, exhibits conserved residues within the catalytic domain that are shared with Class I and Class II HDACs; binding partners and target substrates for this HDAC have not been defined to date (10;16). Class I, Class II and Class IV HDACs are referred to as “classical” HDACs, and are targeted by histone deacetylace inhibitors (HDACi) currently in clinical development. In contrast, Class III HDACs (sirtuins) are structurally similar to yeast SirT2, and require NAD+ as a co-factor for enzymatic activity (17). Recent studies suggest that sirtuins are critical regulators of energy-dependent transcription. These latter HDACs, which are rapidly emerging as potential novel targets for cancer therapy will not be further discussed here due to the limited clinical experience with sirtuins inhibitors. Several recent reviews have focused on the biochemistry and potential clinical significance of Class III HDACs (18;19).
HDAC Activity in Normal and Malignant Tissues
Despite structural similarities, HDACs exhibit non-redundant functions during embryogenesis, and aberrant HDAC expression in cancers may be a manifestation of tissue-specific epigenetic reprogramming events (9;10). Knock-out of HDACs 1, 2, 3, or 7 results in embryonic lethality in mice due to aberrant cell cycle regulation or abnormal blood vessel development; in contrast, mice lacking HDACs 4, 5, 6, or 9 are viable, yet exhibit markedly abnormal cardiovascular, bone and muscle development (reviewed in ref (10)). The unique roles of individual HDACs regarding embryonic development and maintenance of organ function may account, in part, for activities and potential systemic toxicities of HDACi currently evaluated in clinical settings.
In light of the complex roles of HDACs during embryogenesis and their expression profiles in normal tissues, it is not surprising that the effects of targeted inhibition of HDACs in cancer cells appear tissue-dependent. For example, knock-down of HDAC 1 inhibits proliferation of cultured colon cancer cells, and induces apoptosis in osteosarcoma and breast cancer cells (20;21). Knock-down of HDAC 2 induces growth arrest in colon cancer cells, but has no such effects in osteosarcoma or breast cancer cells (20;21). However, inhibition of HDAC 2 down-regulates ER/PR expression, and potentiates tamoxifen-induced apoptosis in ER/PR positive breast cancer cells (22). Knock-down of HDAC-2 enhances p53-dependent gene activation/repression, and inhibits proliferation of cultured breast cancer cells (23). Knock-down of HDAC 8, which modulates telomerase function by inhibiting ubiquitin-mediated degradation of hEST1B (24), inhibits proliferation of lung, colon, cervical carcinoma and neuroblastoma cells, and induces apoptosis in cultured lymphoma/leukemia cells (25–27).
To date, targeted inhibition of Class II HDACs has not been systematically examined. However, knock-down of HDAC 6 or HDAC 10 enhances acetylation of Hsp90 in various cancer cell lines, resulting in destabilization of client oncoproteins such as Bcr-Ab1, and VEGF-R (15;28).
Clinical Manifestation of HDAC Expression in Cancer
A number of studies have been performed recently to examine expression and potential relevance of HDAC expression in cancer tissues. The majority of reports have focused on Class I HDACs, and suggest the clinical manifestations of aberrant HDAC expression may be histology dependent (29). Nakagawa, et al (30) systematically examined expression levels of HDACs 1, 2, 3, and 8 in a variety of cultured cancer lines and a broad panel of primary human lung, esophageal, gastric, colon, pancreas, breast, ovary and thyroid cancers. Seventy-five per cent of esophageal, gastric, colon and prostate cancers, as well as corresponding adjacent “normal” tissues exhibited “high-level” Class I HDAC expression. Although HDAC expression in tumors often was not higher than corresponding normal tissues, 5 – 40% of these cancers exhibited HDAC over-expression; esophageal and prostate cancers tended to exhibit more consistent over-expression of Class I HDACs. Additional studies suggest that high level HDAC 1 expression correlates with advanced stage of disease in lung cancer patients(31), as well as aggressive tumor histology, advanced stage of disease, and poor prognosis in patients with pancreatic carcinoma(32). In contrast, HDAC 1 expression in breast cancer is associated with ER/PR expression, earlier stage of disease (T as well as N classifications), and improved patient survival(33;34).
In a large retrospective study, Weichert, et al (35) observed simultaneous over-expression of HDACs 1, 2, and 3 in approximately 30 % of 150 gastric cancers. An additional 30% of tumors exhibited very low or undetectable expression of these HDACs; global HDAC 1 over-expression in primary cancers correlated significantly with nodal metastases and diminished patient survival. In an additional study, high-level expression of HDACs 1, 2, and 3, was observed in 36%, 58% and 73% of 140 colon carcinomas. HDAC expression correlated significantly with proliferation index, poorly-differentiated histology, and diminished patient survival; HDAC 2 expression was an independent prognosticator of poor outcome. In a related study (36), high level expression of HDAC 1, 2, and 3 was observed in 70%, 74% and 95% of 192 prostate cancers. Over-expression of HDAC 1 and/or HDAC 2 correlated with poorly differentiated tumors, and diminished prostate specific antigen-associated disease free survival. Simultaneous over-expression of all three Class I HDACs coincided with increased proliferation index. HDAC 2 over-expression was an independent prognosticator of poor outcome in prostate cancer patients. Over-expression of HDAC 2 also correlates with advanced stage of disease and diminished survival of oropharyngeal carcinoma patients (37). HDAC 8 expression correlates with aggressive histology and advanced stage of pediatric neuroblastomas, as well as diminished survival of patients with these neoplasms; spontaneous regressions of neuroblastomas coincide with down-regulation of this HDAC (38).
Relatively limited information is available regarding the frequency and clinical relevance of Class II HDAC expression in human cancers. Over-expression of HDAC 4 has been observed in breast cancers, relative to renal, colorectal, or bladder cancers, whereas colon cancers appear to have relatively higher levels of HDACs 5 and 7 (29). Decreased expression of several Class II HDACs – particularly HDACs 5 and 10, appears to correlate with advanced stage of disease and diminished survival of lung cancer patients (39). HDAC 6 expression correlates with advanced stage of disease in oropharyngeal cancers (40). In contrast, HDAC 6 expression in breast cancers coincides with early stage tumors, ER/PR expression, response to tamoxifen, and in some cases, improved patient survival (41).
Cytotoxic Effects of HDAC Inhibitors
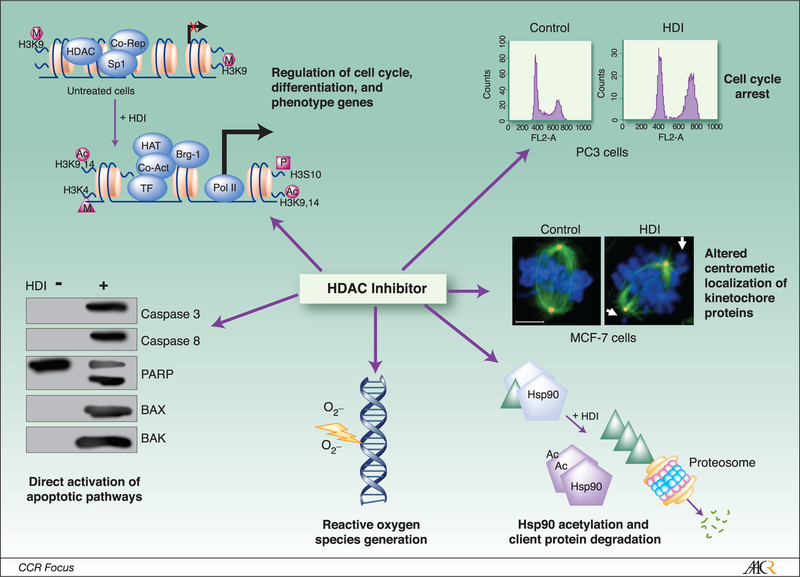
During recent years, intense efforts have focused on the development of HDACi for cancer therapy. These initiatives have been prompted by considerable pre-clinical evidence of plieotropic cytotoxic effects of HDACi of diverse structural classes in cultured cancer cells and various human tumor xenografts (Figure 1), as well as encouraging results of early phase trials in cancer patients.
Figure 1:
Plieotropic mechanisms of cytotoxicity mediated by HDAC inhibitors in cancer cells. The kinetochore panel is reproduced with the permission of Landes Bioscience, from Robbins et al., Cell Cycle, 2005 (69).
Effects on Gene Expression:
HDACi mediate complex effects on global gene expression by directly modulating chromatin structure via acetylation of core histones, as well as “marking” chromatin for subsequent recruitment of chromatin remodeling complexes (42). Equally and perhaps more importantly, these agents influence gene expression via acetylation of numerous non-histone proteins involved in signal transduction and transcription (6)(Table 1). In general, acetylation increases the negative charge of core histones, resulting in relaxation of chromatin structure; whereas chromatin de-condensation often enhances gene expression, the net effect of histone acetylation regarding transcriptional activity of different genes is influenced by concomitant alterations in chromatin structure mediated by DNA as well as histone methylation, and the summation of activators and repressors recruited to the respective promoters (43;44). These issues account for the fact that only ~10% of genes are modulated by HDACi, with approximately equal if not more numbers repressed as induced by these agents (reviewed in ref (9). Gene expression profiles in cancer cells mediated by HDACi of diverse structural classes including sodium butyrate, vorinostat, MS-275, TSA, and FK228 are time and dose dependent; while many similarities have been observed regarding effects of various HDACi on gene expression, some profiles appear agent specific (45–49).
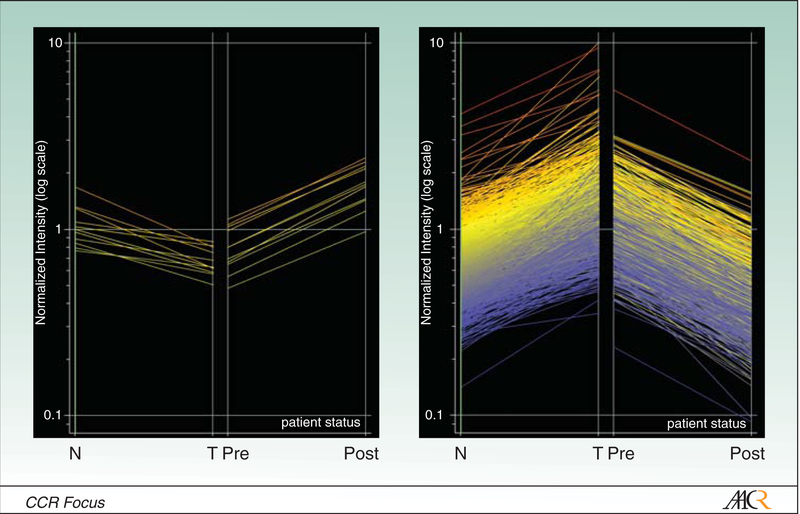
The majority of such micro-array studies pertain to analysis of gene expression in cultured cancer lines; limited information is available regarding gene expression alterations in primary tumors from patients receiving HDACi. In a recent clinical trial, long oligo array techniques were used to examine global gene expression profiles in laser-captured tumor cells from pre- and post-treatment biopsies from lung cancer patients receiving FK228 infusions. Pre-treatment RNA was used as the reference for each respective post-treatment array. Considerable heterogeneity was detected in baseline as well as post-treatment gene expression profiles. Only 16 genes were induced 2 fold or more in one or more patients following FK228 treatment. In contrast, more than 1000 genes were repressed 2 fold or more in one or more patients following FK228 infusion (50). Results of these arrays were compared to a large, robust data set pertaining to gene expression profiles in laser captured lung cancer cells and adjacent histologically normal bronchial epithelia from patients undergoing potentially curative resections. Those genes which were induced or repressed twofold or more by FK228 appeared to be down-regulated or over-expressed, respectively, in the resected primary lung cancers relative to adjacent, histologically normal bronchial epithelial cells (Figure 2).
Figure 2:
Comparison of gene expression in pre/post treatment biopsies relative to paired lung cancer/normal bronchial epithelia. FK228 appears to “normalize” gene expression in lung cancer cells.
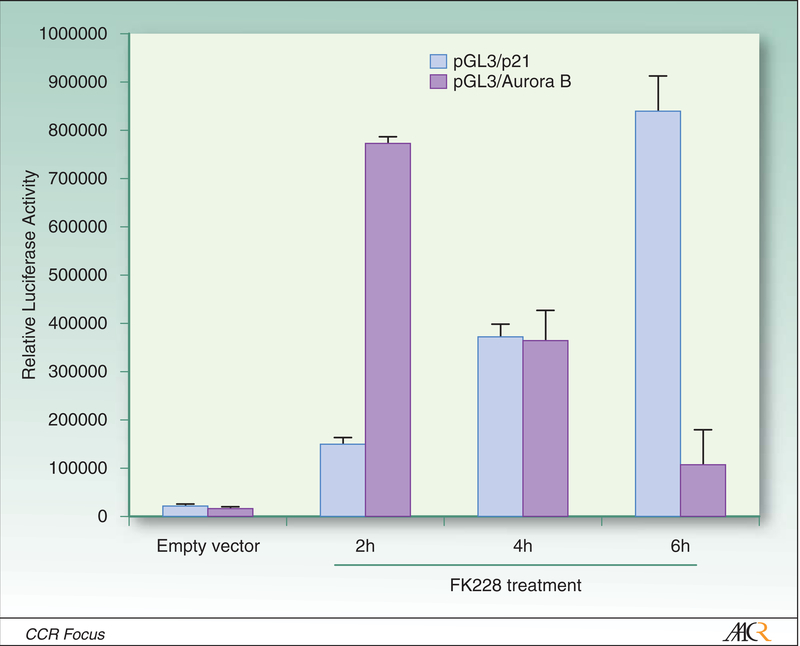
One of the genes consistently induced by HDACi in vitro and in-vivo is p21, which is up-regulated via p53 dependent as well as p53 independent mechanisms (51–53). Activation of p21 coincides with acetylation of histones H3 and H4, methylation of several histone sites within the p21 promoter, and alterations of multi-protein complexes that regulate p21 transcription. Vorinostat as well as TSA-mediated activation of p21 coincides with dissociation of HDAC 1 and c-myc, and recruitment of RNA polymerase II within the p21 promoter (54;55). Other genes, such as Aurora B, are directly down-regulated via HDACi mediated recruitment of repressor complexes (Figure 3) (44).
Figure 3:
Modulation of p21 and Aurora B expression in lung cancer cells following FK228 exposure. Calu-6 lung cancer cells were transiently transfected with p21 or Aurora B luciferase promoter-reporter constructs. Twenty-four hours later, cells were exposed to FK228 (25ng/ml). FK228 increased p21 reporter activity, while diminishing Aurora B promoter-reporter activity in a time-dependent manner. Full details and additional experiments contained in reference 44.
It is well established that HDACi enhance activation of aberrantly methylated tumor suppressor gene promoters in cancer cells by DNA demethylating agents such as 5-azacytidine (5-AC) and 5-aza-2’deoxycytidine (DAC) (56;57). In addition, HDACi potentiate de novo induction of germ-cell restricted genes such as NY-ESO-1 and MAGE family members in cancer cells by DNA demethylating agents (58;59). Although these phenomena have been attributed to acetylation of core histones, more recent studies suggest that potentiation of 5-AC or DAC-mediated gene induction by HDACi may be more complex. For example, Xiong et al (60), observed that TSA decreases stability of DNMT3b mRNA, resulting in diminished de novo methylation activity in human endometrial cancer cells. You et al (61), observed that apicidin down-regulates DNMT1 in Hela cells; repression of DNMT1 coincided with localized deacetylation of histones H3 and H4 at the E2F1 binding site with recruitment of Rb and HDAC1, dissociation of RNA pol II, and trimethylation of H3K9 and K3K27 (repressive histone marks) within the DNMT1 promoter. Additional studies have indicated that TSA destabilizes DNMT 1 mRNA in leukemia cells (62). Zhou, et al (63), observed that vorinostat and panobinostat mediate intranuclear acetylation of Hsp90, leading to destabilization of Hsp90-DNMT 1-HDAC 1 complex, with subsequent depletion of HDAC 1, proteosomal degradation of DNMT1, and up-regulation of ER gene expression in cultured breast cancer cells. Knock-down of HDAC 1 - but not HDAC 6, induced depletion of DNMT 1 in these cancer cells. Wu et al (64) observed that FK228 and the structurally related cyclic peptide apicidin mediate demethylation of a variety of tumor suppressor gene promoters including p16, SALL-3, and GATA-4 in lung, colon, and pancreatic cancer lines. FK228 and apicidin- but not TSA, inhibited expression of G9A and SUV39H1 histone methyltransferases, thereby decreasing di- and tri-methylation of H3K9, and diminishing binding of repressive heterochromatin protein (HP) 1α and 1β, as well as DNMT1 to these promoters. Collectively, these recent studies highlight the complexity of mechanisms by which HDACi mediate epigenetic regulation of gene expression in cancer cells.
Modulation of Cell Cycle Progression:
Depending on exposure conditions, HDACi of various structural classes induce G1/S and/or G2/M arrest, and disrupt mitotic progression in normal as well as malignant cells. Cell cycle arrest mediated by HDACi coincides with decreased expression of cyclins A, B, D, and E, as well as their respective cyclin-dependent kinases, hypophosphorylation of Rb, and induction of p21 and p27 (65).
Presently, the mechanisms contributing to aberrant mitotic progression in cancer cells following exposure to HDACi are less fully defined. HDAC3 is critical for maintaining deacetylated histone tails that become phosphorylated by Aurora B as cells enter mitosis, and inactivation of HDAC 3 induces mitotic delay and apoptosis in murine embryonic fibroblasts(66;67). Ma et al (68), observed that TSA induced prometaphase arrest in Hela cells, characterized by aberrant microtubule-kinetochore attachments, and HP1 localization at pericentromeric heterochromatin, as well as disruption of the chromosome passenger complex. TSA as well as FK228 deplete levels of several kinetochore proteins including HBUB1, CENP-E, and CENP-F, and decrease pre-mitotic phosphorylation of histone H3 in pericentromeric heterochromatin during G2, resulting in deficient assembly of kinetochores(68;69). In addition, TSA disrupts localization of the kinetochore protein BubR1, and decreases phoso-histone H3 after paclitaxel treatment(70). Park et al(71) observed that LAQ824 depletes Aurora-A in gastric cancer cells via inhibition of HDAC 6 mediated de-acetylation of Hsp90. Although Aurora B also associates with Hsp90, LAQ824 did not appear to destabilize this complex. Zhang et al (44) observed that FK228, TSA, and vorinostat inhibit transcription of Aurora A, Aurora B, and survivin in a panel of cultured lung cancer cells. Transcriptional repression mediated by these HDACi was more pronounced in cells expressing wild-type p53. Depletion of Aurora A and survivin protein levels preceded depletion of Aurora B, possibly due to combinatorial effects of these agents on transcription as well as post-translational stabilization of these proteins. Additional experiments revealed that down-regulation of Aurora B expression coincided with increased total acetylation of histone H3, decreased levels of acetylated H3K9, and dimethyl H3K4, and recruitment of MBD1, MBD2, and MBD3 to the Aurora B promoter. Diminished expression of Aurora A, Aurora B, and survivin in lung cancer cells exposed to FK228 or TSA resulted in apparent mitotic catastrophe. More recent studies indicate that panobinostat induces proteosomal degradation of Aurora A and Aurora B in renal cancer cells via inhibition of HDAC 3 and HDAC 6. Degradation of Aurora A and Aurora B coincided with G2/M arrest and apoptosis in these cancer cells (72).
Autophagic/Apoptic Effects of HDAC Inhibitors in Cancer Cells:
Tremendous research efforts have focused on molecular pathways regulating HDACi-mediated cytotoxicity in cancer cells (9;65). Depending on exposure conditions, these agents mediate caspase-independent autophagy as well as caspase-dependent apoptosis in cancer cells of diverse histologies.
Autophagy is complex process by which proteins and organelles are sequestered in autophagosomes, and subsequently degraded following fusion with lysosomes. Autophagy is induced by nuclear (but not cytoplasmic) p53 via upregulation of damage-regulated autophagy modulator (DRAM), as well as p73 in response to cellular stress(73;74). Recent studies indicate that mTOR regulates autophagy by inhibiting p73-mediated activation of a variety of genes including ATG5, ATG7, and UVRAG(75;76), and that p53 can inhibit mTOR via activation of AMPK(75).
Autophagy induced by HDACi appears related to inhibition of HDAC 1 as well as HDAC 6 (77;78). Shao et al(79) observed that sodium butyrate and vorinostat mediated autophagy as well as apoptosis in HeLa cells; constitutive over-expression of Bcl-Xl inhibited caspase activation, but did not appear to diminish cell death mediated by these HDACi. Hrzenjak et al(80) observed that vorinostat diminished mTOR expression, and mediated caspase-independent cytotoxicity in endometrial sarcoma cells via autophagic mechanisms. Furthermore, Watanabe et al(81) observed that FK228-mediated autophagy in rhabdomyosarcoma cells coincided with nuclear translocation of apoptosis-inducing factor (AIF); knock-down of AIF abrogated autophagy following FK228 exposure. Chloroquine, an inhibitor of autophagy, enhanced FK228-mediated apoptosis in these cells. Carew, et al (82) examined the relative contributions of autophagy and apoptosis regarding vorinostat-mediated cytotoxicity in cultured CML cells. Chloroquine exposure dramatically increased reactive oxygen species formation and enhanced vorinostat-mediated apoptosis in these cells. The relative contributions of autophagy and apoptosis with regard to tumor regressions in clinical settings have yet to be fully elucidated. Of particular concern are observations that depending on tissue histology/context, autophagy may be cytoprotective (83). For example, autophagy enhances anti-estrogen resistance in cultured breast cancer cells (84), and protects cancer cells from hypoxia (85).
Cancer cells exhibit a variety of defects in caspase-mediated apoptotic pathways due to upregulation of decoy receptors such as TRAIL-R3 and TRAIL-R4 that inhibit activation of death receptors by ligands such TNF or TRAIL, as well as aberrant expression of anti-apoptotic proteins including Bcl-2, Bcl-xl, and XIAP family members, which inhibit caspase activation (9;65). HDACi including TSA, FK228, vorinostat, and panobinostat, decrease expression of Bcl-2, Bcl-xl, and XIAP, and enhance expression of pro-apoptotic proteins such as BAX and BAK, thereby enhancing TRAIL-mediated cytotoxicity in a variety of cancer cells via amplification of intrinsic as well as extrinsic apoptotic pathways (86;87). Agents such as flavopiridol that potentiate mitochondrial injury enhance apoptosis mediated by HDACi in cancer cells(88). In addition, HDAC inhibitors augment apoptosis mediated by a variety of conventional chemotherapeutic agents by up-regulating death receptor 5 or other components of apoptotic pathways such as AIF, thereby enhancing caspase activation(89;90). Recently, Xu, et al (91) observed that MS275 as well as vorinostat induce TRAIL expression without altering DR4 or DR5 levels in breast cancer cells; HDACi induction of TRAIL by was mediated via SP1, and markedly enhanced adriamycin cytotoxicity in these cells.
Additional studies have examined the effects of HDACi on nuclear receptor signaling in cancer cells; results have varied depending on agents and exposure conditions, and hormone receptor status of cells used for these experiments(92). For example, TSA potentiates de-repression of ER- α mediated by DNA demethylating agents, and enhances response to tamoxifen in ER-negative breast cancer cells(93); TSA, as well as vorinostat, and valproic acid alone induce only modest up-regulation of ER-α in these breast cancer cells. Jang et al(94) observed that TSA markedly induced ER-β but not ER-α expression, and enhanced nuclear transport of ER-β, resulting in activation of ER target genes and increased tamoxifen sensitivity in ER negative cells. Fiskus et al (95) observed that LAQ824, vorinostat, and panobinostat depleted ER-α via acetylation of Hsp90, thereby diminishing response to E2, and enhancing tamoxifen sensitivity in ER-positive breast cancer cells. Bicaku, et al (22) observed that co-treatment of cultured breast cancer cells with vorinostat or valproic acid depleted ER as well as progesterone receptor (PR), and synergistically enhanced tamoxifen-mediated cytotoxicity in ER+ or ER+/PR+ breast cancer cells. Knock-down experiments revealed that depletion of ER and PR, and potentiation of tamoxifen-mediated cytotoxicity by these HDACi was attributable to inhibition of HDAC 2 (but not HDAC 1 or HDAC 6) activity in these cells.
HDACi of various classes modulate androgen receptor (AR) expression, and enhance activity of AR-mediated blockade in prostate cancer cells(96). TSA, vorinostat and MS-275 inhibit expression of TMPRSS2-ERG fusion transcripts, and enhance apoptosis mediated by flutamide in androgen responsive prostate cancer cells in part by inhibiting translocation of AR from cytoplasm to the nucleus(97); in contrast, HDACi do not potentiate androgen blockade in AR-negative prostate cancer cells(98). Welsbie, et al comprehensively examined the mechanisms underlying the synergistic cytotoxic activity of AR blockade and HDAC inhibition in prostate cancer cells. Vorinostat and panobinostat inhibited AR-mediated activation of downstream target genes including TMPRSS2. Knock-down of HDAC 1 or HDAC 3 suppressed expression of androgen regulated genes, and mimicked the effects of HDACi exposure in these cancer cells. Interestingly, inhibition of AR signaling by HDACi was independent of AR protein depletion. Additional experiments indicated that HDACi prevent assembly of co-activator/ RNA pol II complexes after AR binds to enhancer elements of target genes.
A variety of studies have been performed to examine the effects of HDACi on retinoic acid signaling in cancer cells(100). Elegant functional genetic screening experiments revealed that apoptosis of p53-deficient murine embryonic fibroblasts expressing oncogenic ras exposed to PXD101, vorinostat, MS-275, sodium butyrate and spiruchostatin A could be inhibited by constitutive expression of RARα or the preferentially expressed antigen of melanoma (PRAME), which represses retinoic acid (RA) signaling(101;102). The anti-apoptotic effects of RARα and PRAME did not appear attributable to modulation of histone acetylation. Additional studies have demonstrated that combined valproic acid/RA or MS-275/RA regimens enhance apoptosis and decrease in-vivo growth of neuroblastoma and renal carcinoma cells(103;104).
Effects on Anti-tumor Immunity:
HDACi can potentially modulate anti-tumor immunity via numerous mechanisms including up-regulation of tumor antigens, enhancement of cellular immune recognition and lysis of tumor targets by T cells and NK cells, and alteration of T cell subsets as well as inflammatory cytokine profiles. FK228 as well as TSA enhance deoxyazacytidine-mediated activation of genes encoding cancer testis antigens such as NY-ESO-1 and MAGE family members, and augment NY-ESO-1 and MAGE expression in cancer cells exhibiting de-repression of these germ-cell restricted genes; up-regulation of antigen expression facilitates recognition of cancer cells by cytolyic T cells in vitro(105;106). Whereas HDACi such as TSA and FK228 enhance HLA expression(107), exposure to these agents does not appear to restore antigen presentation in cancer cells deficient in antigen processing(105;106).
Several recent studies indicate that HDACi can modulate TH1/TH2 effector function(108), and enhance the activity of Foxp3-positive T regulatory cells, which contribute to immune tolerance in cancer patients(109). Furthermore, TSA abrogates interferon-gamma (IFN-γ)-mediated inhibition of TNF-α-induced activation of inflammatory cytokine genes such as IL-6 and IL-8, which enhance metastatic potential of cancer cells(110;111).On the other hand, FK228 enhances NK cell- mediated lysis of tumor cells of various histologies by up-regulating DR5 (TRAIL-R2) expression without altering expression of MHC-class I, DR-4 (TRAIL-R1), MIC A/B, or FAS (CD95) on tumor cells(112). Consistent with these findings, Deirmayr et al(113) observed that valproic acid enhances expression of NKG2D ligands on AML cells, thereby enhancing their susceptibility to NK cell mediated lysis.
Sensitivity and Mechanisms of Resistance:
Whereas HDACi induce cell cycle arrest in normal as well as non-transformed cells, the proapoptotic effects of these agents are observed primarily in cancer cells. Preferential tumoricidal activity mediated by HDACi appears related, at least in part, to differential responses of transformed and normal cells to oxidative stress. HDACi such as vorinostat decrease expression of thioreduxin (TRX) in transformed but not normal cells. TRX is a scavenger of reactive oxygen species (ROS), and a hydrogen donor for numerous proteins involved in DNA synthesis and transcription. In addition, TRX inhibits apoptosis signaling regulating kinase-1 (ASK-1). Vorinostat also increases expression of TRX binding protein, an inhibitor of TRX, thereby increasing expression ofASK-1. The net result is an accumulation of ROS, which triggers apoptosis/autophagy in cancer cells(114).
Generation of ROS cannot fully account for sensitivity of cancer cells to HDACi. Increasing evidence indicates that even within given histologies, cancer lines or xenografts exhibit differential sensitivities to these agents. Several studies have been performed recently to define gene expression profiles that correlate with response to HDACi in cancer cells. Susakawa et al(115) identified a 76 gene expression signature that coincided with sensitivity of human tumor xenografts to FK228. Miyanaga et al(116) identified a nine gene expression signature that correlated with response of cultured lung cancer cells to TSA and vorinostat; modulation of three of these genes (NOG1, SEC23A: upregulated; PSNE2: down-regulated) markedly correlated with sensitivity to these HDACi. More recently, Dejligbjerg, et al(117) observed that modulation four genes (ODC1, SKI, STAT1, TYMS) correlated with belinostat sensitivity in a broad panel of cultured cell lines. Dokamanovic et al(117) observed that depletion of HDAC 7 coincided with sensitivity of cultured cancer cells of various cytologies to vorinostat and FK228; interestingly, knock-down of HDAC 7 only modestly inhibited cancer cell proliferation.
Stapnes et al(118), examined responses of cultured AML cells from 59 patients to multiple HDACi, including valproic acid, TSA, PDX101, and sodium butyrate. At high concentrations, all of these HDACi mediated dose-dependent apoptosis. However, exposure to low or intermediate doses of these agents paradoxically increased proliferation in a subset of cell lines. Expression of 25 genes with fold change ≥3.0 discriminated between FLT3-ITD+ AML cells with and without growth enhancement mediated by intermediate doses of HDACi.
An important issue regarding these studies is that HDACi sensitivity often was determined by proliferation rather than cytoxicity assays. Diminished proliferation in response to HDACi appears related, at least in part, to induction of p21, which may be cytoprotective in cancer cells exposed to chemotherapeutic agents and HDACi. Indeed, abrogation of p21 expression by agents such as flavopiridol, markedly enhances apoptosis mediated by HDACi in cultured cancer cells(119). Furthermore, HDACi increase expression of NFκB, which mediates a variety of pro-survival pathways in cancer cells(120); the relevance of NFκB activation regarding sensitivity of cancer cells to these agents is highlighted by the fact that parthenolide which inhibits NFκB function, as well as proteosome inhibitors such as bortezamib, which prevent degradation of IκB markedly enhance cytotoxicity of TSA, vorinostat, valproic acid, and FK228 in cancer cells(121–123).
Recently, Fantin et al(124) observed that activation of signal transducer and activation of transcription (STAT)-1,−3, and −5 correlated with vorinostat resistance in cultured lymphoma cells. Janus-activated kinase inhibition enhanced vorinostat-mediated cytotoxicity in these cells. Subsequent studies revealed that nuclear accumulation of STAT1 and increased levels of nuclear phospho-STAT3 in skin biopsies correlated with lack of response to vorinostat in patients with CTCL.
Observations that HDACi modulate apoptosis thresholds in cancer cells have prompted considerable interest in utilizing these agents to potentiate the effects of standard chemotherapetic regimens or radiation therapy in clinical settings(125–127). However, HDACi may induce resistance that may be clinically relevant. For instance, FK228 is a substrate for P-glycoprotein (Pgp) and multi-drug resistance-associated protein-1 (MRP1); up-regulation of Pgp appears to be a major mechanism of resistance to FK228 as well as apicidin in cultured cancer cells(128;129). Robey et al(130) observed an eight-fold increase in expression of MDR-1, which encodes Pgp, in circulating tumor cells from patients with hematologic malignancies receiving FK228. Additional studies have demonstrated that FK228 induces expression of ABCG2; chromatin alterations within the ABCG2 promoter induced by FK228 are similar to those observed in drug-resistant cells(131).
To date, the mechanisms mediating resistance to other HDAC inhibitors in cancer cells have not been fully defined. Vorinostat and valproic acid-induced resistance appears irreversible and unrelated to MDR-1 expression, and does not appear to alter sensitivity of cultured colon cancer cells to standard chemotherapeutic agents(132). Fiskus et al(133) observed that HL-60 cells selected for resistance to vorinostat, sodium butyrate, LAQ824, and panobinosat exhibited increased expression of HDACs 1, 2, and 4, yet lacked expression of HDAC 6. HL-60 cells resistant to HDACi were also resistant to etoposide, cytarabine and TRAIL, and exhibited increased proliferation in vitro and in vivo, suggesting that HDACi exposure may select for outgrowth of cancer cells with a more aggressive phenotype.
Conclusions and Future Directions
HDACi have emerged as major pharmacologic agents for cancer therapy. In all likelihood, these agents will be used in combination with standard treatment regimens. Efforts to further develop these agents should be focused on thorough evaluation of HDAC expression in different human cancers, comprehensive analysis of the mechanisms of action of various classes of HDACi in vitro using array-based profiling techniques, and validation of recently identified prognosticators of response in clinical settings.
Synthesis of HDACi that selectively target HDACs relevant to cancer initiation/progression may enhance the anti-tumor effects while decreasing systemic toxicities of HDAC inhibition in cancer patients. For example, HDAC6 enhances oncogenic transformation(134), and modulates epithelial-mesenchymal transition in cancer cells(11); as such, selective inhibitors of HDAC6 may prove highly effective for cancer therapy. As HDAC inhibitors are further evaluated in cancer patients, it will be important for investigators remain cognizant of the potential immunosuppressive effects of these agents, given their ability to perturb T cell function and alter expression of inflammatory cytokines mediating innate antiviral and anti-tumor immunity(108;135–137).
Reference List
- (1).Munshi A, Shafi G, Aliya N, Jyothy A. Histone modifications dictate specific biological readouts. J Genet Genomics 2009;36:75–88. [DOI] [PubMed] [Google Scholar]
- (2).Kouzarides T Chromatin modifications and their function. Cell 2007;128:693–705. [DOI] [PubMed] [Google Scholar]
- (3).Bernstein BE, Meissner A, Lander ES. The mammalian epigenome. Cell 2007;128:669–81. [DOI] [PubMed] [Google Scholar]
- (4).Keppler BR, Archer TK. Chromatin-modifying enzymes as therapeutic targets--Part 1. Expert Opin Ther Targets 2008;12:1301–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Keppler BR, Archer TK. Chromatin-modifying enzymes as therapeutic targets--Part 2. Expert Opin Ther Targets 2008;12:1457–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Spange S, Wagner T, Heinzel T, Kramer OH. Acetylation of non-histone proteins modulates cellular signalling at multiple levels. Int J Biochem Cell Biol 2009;41:185–98. [DOI] [PubMed] [Google Scholar]
- (7).Brandl A, Heinzel T, Kramer OH. Histone deacetylases: salesmen and customers in the post-translational modification market. Biol Cell 2009;101:193–205. [DOI] [PubMed] [Google Scholar]
- (8).Yang XJ, Seto E. The Rpd3/Hda1 family of lysine deacetylases: from bacteria and yeast to mice and men. Nat Rev Mol Cell Biol 2008;9:206–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Xu WS, Parmigiani RB, Marks PA. Histone deacetylase inhibitors: molecular mechanisms of action. Oncogene 2007;26:5541–52. [DOI] [PubMed] [Google Scholar]
- (10).Witt O, Deubzer HE, Milde T, Oehme I. HDAC family: What are the cancer relevant targets? Cancer Lett 2009;277:8–21. [DOI] [PubMed] [Google Scholar]
- (11).Shan B, Yao TP, Nguyen HT, et al. Requirement of HDAC6 for transforming growth factor-beta1-induced epithelial-mesenchymal transition. J Biol Chem 2008;283:21065–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Bali P, Pranpat M, Bradner J, et al. Inhibition of histone deacetylase 6 acetylates and disrupts the chaperone function of heat shock protein 90: a novel basis for antileukemia activity of histone deacetylase inhibitors. J Biol Chem 2005;280:26729–34. [DOI] [PubMed] [Google Scholar]
- (13).Banerji U Heat shock protein 90 as a drug target: some like it hot. Clin Cancer Res 2009;15:9–14. [DOI] [PubMed] [Google Scholar]
- (14).Kawaguchi Y, Kovacs JJ, McLaurin A, Vance JM, Ito A, Yao TP. The deacetylase HDAC6 regulates aggresome formation and cell viability in response to misfolded protein stress. Cell 2003;115:727–38. [DOI] [PubMed] [Google Scholar]
- (15).Park JH, Kim SH, Choi MC, et al. Class II histone deacetylases play pivotal roles in heat shock protein 90-mediated proteasomal degradation of vascular endothelial growth factor receptors. Biochem Biophys Res Commun 2008. April 4;368:318–22. [DOI] [PubMed] [Google Scholar]
- (16).Glaser KB. HDAC inhibitors: clinical update and mechanism-based potential. Biochem Pharmacol 2007;74:659–71. [DOI] [PubMed] [Google Scholar]
- (17).Vaquero A, Sternglanz R, Reinberg D. NAD+-dependent deacetylation of H4 lysine 16 by class III HDACs. Oncogene 2007;26:5505–20. [DOI] [PubMed] [Google Scholar]
- (18).Zschoernig B, Mahlknecht U. SIRTUIN 1: regulating the regulator. Biochem Biophys Res Commun 2008;376:251–5. [DOI] [PubMed] [Google Scholar]
- (19).Liu T, Liu PY MG. The critical role of class III histone deacetylase SIRT-1 in cancer 2009. p. 1702–5. [DOI] [PubMed]
- (20).Weichert W, Roske A, Niesporek S, et al. Class I histone deacetylase expression has independent prognostic impact in human colorectal cancer: specific role of class I histone deacetylases in vitro and in vivo. Clin Cancer Res 2008;14:1669–77. [DOI] [PubMed] [Google Scholar]
- (21).Senese S, Zaragoza K, Minardi S, Muradore I, Ronzoni S, Passafaro A, et al. Role for histone deacetylase 1 in human tumor cell proliferation. Mol Cell Biol 2007;27:4784–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Bicaku E, Marchion DC, Schmitt ML, Munster PN. Selective inhibition of histone deacetylase 2 silences progesterone receptor-mediated signaling. Cancer Res 2008;68:1513–9. [DOI] [PubMed] [Google Scholar]
- (23).Harms KL, Chen X. Histone deacetylase 2 modulates p53 transcriptional activities through regulation of p53-DNA binding activity. Cancer Res 2007;67:3145–52. [DOI] [PubMed] [Google Scholar]
- (24).Lee H, Sengupta N, Villagra A, Rezai-Zadeh N, Seto E. Histone deacetylase 8 safeguards the human ever-shorter telomeres 1B (hEST1B) protein from ubiquitin-mediated degradation. Mol Cell Biol 2006;26:5259–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Vannini A, Volpari C, Filocamo G, et al. Crystal structure of a eukaryotic zinc-dependent histone deacetylase, human HDAC8, complexed with a hydroxamic acid inhibitor. Proc Natl Acad Sci U S A 2004;101:15064–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Oehme I, Deubzer HE, Wegener D, et al. Histone deacetylase 8 in neuroblastoma tumorigenesis. Clin Cancer Res 2009;15:91–9. [DOI] [PubMed] [Google Scholar]
- (27).Balasubramanian S, Ramos J, Luo W, Sirisawad M, Verner E, Buggy JJ. A novel histone deacetylase 8 (HDAC8)-specific inhibitor PCI-34051 induces apoptosis in T-cell lymphomas. Leukemia 2008;22:1026–34. [DOI] [PubMed] [Google Scholar]
- (28).Bali P, Pranpat M, Bradner J, et al. Inhibition of histone deacetylase 6 acetylates and disrupts the chaperone function of heat shock protein 90: a novel basis for antileukemia activity of histone deacetylase inhibitors. J Biol Chem 2005;280:26729–34. [DOI] [PubMed] [Google Scholar]
- (29).Ozdag H, Teschendorff AE, Ahmed AA, Hyland SJ, Blenkiron C, Bobrow L, et al. Differential expression of selected histone modifier genes in human solid cancers. BMC Genomics 2006;7:90:90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Nakagawa M, Oda Y, Eguchi T, et al. Expression profile of class I histone deacetylases in human cancer tissues. Oncol Rep 2007;18:769–74. [PubMed] [Google Scholar]
- (31).Sasaki H, Moriyama S, Nakashima Y, et al. Histone deacetylase 1 mRNA expression in lung cancer. Lung Cancer 2004;46:171–8. [DOI] [PubMed] [Google Scholar]
- (32).Miyake K, Yoshizumi T, Imura S, et al. Expression of hypoxia-inducible factor-1alpha, histone deacetylase 1, and metastasis-associated protein 1 in pancreatic carcinoma: correlation with poor prognosis with possible regulation. Pancreas 2008;36:e1–e9. [DOI] [PubMed] [Google Scholar]
- (33).Krusche CA, Wulfing P, Kersting C, et al. Histone deacetylase-1 and −3 protein expression in human breast cancer: a tissue microarray analysis. Breast Cancer Res Treat 2005;90:15–23. [DOI] [PubMed] [Google Scholar]
- (34).Zhang Z, Yamashita H, Toyama T, et al. Quantitation of HDAC1 mRNA expression in invasive carcinoma of the breast*. Breast Cancer Res Treat 2005. ;94:11–6. [DOI] [PubMed] [Google Scholar]
- (35).Weichert W, Roske A, Gekeler V, et al. Association of patterns of class I histone deacetylase expression with patient prognosis in gastric cancer: a retrospective analysis. Lancet Oncol 2008;9:139–48. [DOI] [PubMed] [Google Scholar]
- (36).Weichert W, Roske A, Gekeler V, et al. Histone deacetylases 1, 2 and 3 are highly expressed in prostate cancer and HDAC2 expression is associated with shorter PSA relapse time after radical prostatectomy. Br J Cancer 2008;98:604–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Chang HH, Chiang CP, Hung HC, Lin CY, Deng YT, Kuo MY. Histone deacetylase 2 expression predicts poorer prognosis in oral cancer patients. Oral Oncol 2008. October 23. [DOI] [PubMed]
- (38).Oehme I, Deubzer HE, Wegener D, et al. Histone deacetylase 8 in neuroblastoma tumorigenesis. Clin Cancer Res 2009;15:91–9. [DOI] [PubMed] [Google Scholar]
- (39).Osada H, Tatematsu Y, Saito H, Yatabe Y, Mitsudomi T, Takahashi T. Reduced expression of class II histone deacetylase genes is associated with poor prognosis in lung cancer patients. Int J Cancer 2004;112:26–32. [DOI] [PubMed] [Google Scholar]
- (40).Sakuma T, Uzawa K, Onda T, et al. Aberrant expression of histone deacetylase 6 in oral squamous cell carcinoma. Int J Oncol 2006;29:117–24. [PubMed] [Google Scholar]
- (41).Zhang Z, Yamashita H, Toyama T, et al. HDAC6 expression is correlated with better survival in breast cancer. Clin Cancer Res 2004;10:6962–8. [DOI] [PubMed] [Google Scholar]
- (42).Choi JK, Howe LJ. Histone acetylation: truth of consequences? Biochem Cell Biol 2009;87:139–50. [DOI] [PubMed] [Google Scholar]
- (43).Ellis DJ, Lawman ZK, Bonham K. Histone acetylation is not an accurate predictor of gene expression following treatment with histone deacetylase inhibitors. Biochem Biophys Res Commun 2008;367:656–62. [DOI] [PubMed] [Google Scholar]
- (44).Zhang XH, Rao M, Loprieato JA, et al. Aurora A, Aurora B and survivin are novel targets of transcriptional regulation by histone deacetylase inhibitors in non-small cell lung cancer. Cancer Biol Ther 2008;7:1388–97. [DOI] [PubMed] [Google Scholar]
- (45).Glaser KB, Staver MJ, Waring JF, Stender J, Ulrich RG, Davidsen SK. Gene expression profiling of multiple histone deacetylase (HDAC) inhibitors: defining a common gene set produced by HDAC inhibition in T24 and MDA carcinoma cell lines. Mol Cancer Ther 2003;2:151–63. [PubMed] [Google Scholar]
- (46).Gray SG, Qian CN, Furge K, Guo X, Teh BT. Microarray profiling of the effects of histone deacetylase inhibitors on gene expression in cancer cell lines. Int J Oncol 2004;24:773–95. [DOI] [PubMed] [Google Scholar]
- (47).Peart MJ, Smyth GK, van Laar RK, et al. Identification and functional significance of genes regulated by structurally different histone deacetylase inhibitors. Proc Natl Acad Sci U S A 2005;102:3697–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Mitsiades CS, Mitsiades NS, McMullan CJ, et al. Transcriptional signature of histone deacetylase inhibition in multiple myeloma: biological and clinical implications. Proc Natl Acad Sci U S A 2004;101:540–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Crabb SJ, Howell M, Rogers H, et al. Characterisation of the in vitro activity of the depsipeptide histone deacetylase inhibitor spiruchostatin A. Biochem Pharmacol 2008;76463–75. [DOI] [PubMed]
- (50).Schrump DS, Fischette MR, Nguyen DM, et al. Clinical and molecular responses in lung cancer patients receiving Romidepsin. Clin Cancer Res 2008;14:188–98. [DOI] [PubMed] [Google Scholar]
- (51).Zhao Y, Lu S, Wu L, et al. Acetylation of p53 at lysine 373/382 by the histone deacetylase inhibitor depsipeptide induces expression of p21(Waf1/Cip1). Mol Cell Biol 2006;26:2782–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Liu PY, Chan JY, Lin HC, et al. Modulation of the cyclin-dependent kinase inhibitor p21(WAF1/Cip1) gene by Zac1 through the antagonistic regulators p53 and histone deacetylase 1 in HeLa Cells. Mol Cancer Res 2008;6:1204–14. [DOI] [PubMed] [Google Scholar]
- (53).Wilson AJ, Byun DS, Nasser S, et al. HDAC4 promotes growth of colon cancer cells via repression of p21. Mol Biol Cell 2008;19:4062–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Gui CY, Ngo L, Xu WS, Richon VM, Marks PA. Histone deacetylase (HDAC) inhibitor activation of p21WAF1 involves changes in promoter-associated proteins, including HDAC1. Proc Natl Acad Sci U S A 2004;101:1241–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Li H, Wu X. Histone deacetylase inhibitor, Trichostatin A, activates p21WAF1/CIP1 expression through downregulation of c-myc and release of the repression of c-myc from the promoter in human cervical cancer cells. Biochem Biophys Res Commun 2004;324:860–7. [DOI] [PubMed] [Google Scholar]
- (56).Cameron EE, Bachman KE, Myohanen S, Herman JG, Baylin SB. Synergy of demethylation and histone deacetylase inhibition in the re-expression of genes silenced in cancer. Nat Genet 1999;21:103–7. [DOI] [PubMed] [Google Scholar]
- (57).Steiner F, Hong J, Fischette M, et al. Sequential 5-Aza 2′-deoxycytidine/depsipeptide FK228 treatment induces tissue factor pathway inhibitor 2 (TFPI-2) expression in cancer cells. Oncogene 2005;24:2386–97. [DOI] [PubMed] [Google Scholar]
- (58).Hong JA, Kang Y, Abdullaev Z, et al. Reciprocal binding of CTCF and BORIS to the NY-ESO-1 promoter coincides with derepression of this cancer-testis gene in lung cancer cells. Cancer Res 2005;65:7763–74. [DOI] [PubMed] [Google Scholar]
- (59).Schrump DS, Hong JA, Nguyen DM. Utilization of chromatin remodeling agents for lung cancer therapy. Cancer J 2007;13:56–64. [DOI] [PubMed] [Google Scholar]
- (60).Xiong Y, Dowdy SC, Podratz KC, et al. Histone deacetylase inhibitors decrease DNA methyltransferase-3B messenger RNA stability and down-regulate de novo DNA methyltransferase activity in human endometrial cells. Cancer Res 2005;65:2684–9. [DOI] [PubMed] [Google Scholar]
- (61).You JS, Kang JK, Lee EK, et al. Histone deacetylase inhibitor apicidin downregulates DNA methyltransferase 1 expression and induces repressive histone modifications via recruitment of corepressor complex to promoter region in human cervix cancer cells. Oncogene 2008;27:1376–86. [DOI] [PubMed] [Google Scholar]
- (62).Januchowski R, Dabrowski M, Ofori H, Jagodzinski PP. Trichostatin A down-regulate DNA methyltransferase 1 in Jurkat T cells. Cancer Lett 2007;246:313–7. [DOI] [PubMed] [Google Scholar]
- (63).Zhou Q, Agoston AT, Atadja P, Nelson WG, Davidson NE. Inhibition of histone deacetylases promotes ubiquitin-dependent proteasomal degradation of DNA methyltransferase 1 in human breast cancer cells. Mol Cancer Res 2008;6:873–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (64).Wu LP, Wang X, Li L, et al. Histone deacetylase inhibitor depsipeptide activates silenced genes through decreasing both CpG and H3K9 methylation on the promoter. Mol Cell Biol 2008;28:3219–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).Emanuele S, Lauricella M, Tesoriere G. Histone deacetylase inhibitors: apoptotic effects and clinical implications (Review). Int J Oncol 2008;33:637–46. [PubMed] [Google Scholar]
- (66).Li Y, Kao GD, Garcia BA, et al. A novel histone deacetylase pathway regulates mitosis by modulating Aurora B kinase activity. Genes Dev 2006;20:2566–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (67).Bhaskara S, Chyla BJ, Amann JM, et al. Deletion of histone deacetylase 3 reveals critical roles in S phase progression and DNA damage control. Mol Cell 2008;30:61–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (68).Ma Y, Cai S, Lu Q, et al. Inhibition of protein deacetylation by trichostatin A impairs microtubule-kinetochore attachment. Cell Mol Life Sci 2008;65:3100–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (69).Robbins AR, Jablonski SA, Yen TJ, et al. Inhibitors of histone deacetylases alter kinetochore assembly by disrupting pericentromeric heterochromatin. Cell Cycle 2005;4:717–26. [DOI] [PubMed] [Google Scholar]
- (70).Dowling M, Voong KR, Kim M, Keutmann MK, Harris E, Kao GD. Mitotic spindle checkpoint inactivation by trichostatin a defines a mechanism for increasing cancer cell killing by microtubule-disrupting agents. Cancer Biol Ther 2005;4:197–206. [PubMed] [Google Scholar]
- (71).Park JH, Jong HS, Kim SG, et al. Inhibitors of histone deacetylases induce tumor-selective cytotoxicity through modulating Aurora-A kinase. J Mol Med 2008;86:117–28. [DOI] [PubMed] [Google Scholar]
- (72).Cha TL, Chuang MJ, Wu ST, et al. Dual degradation of aurora A and B kinases by the histone deacetylase inhibitor LBH589 induces G2-M arrest and apoptosis of renal cancer cells. Clin Cancer Res 2009;15:840–50. [DOI] [PubMed] [Google Scholar]
- (73).Crighton D, Wilkinson S, O’Prey J, et al. DRAM, a p53-induced modulator of autophagy, is critical for apoptosis. Cell 2006;126:121–34. [DOI] [PubMed] [Google Scholar]
- (74).Crighton D, O’Prey J, Bell HS, Ryan KM. p73 regulates DRAM-independent autophagy that does not contribute to programmed cell death. Cell Death Differ 2007;14:1071–9. [DOI] [PubMed] [Google Scholar]
- (75).Rosenbluth JM, Mays DJ, Pino MF, Tang LJ, Pietenpol JA. A gene signature-based approach identifies mTOR as a regulator of p73. Mol Cell Biol 2008. October;28(19):5951–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (76).Rosenbluth JM, Pietenpol JA. mTOR regulates autophagy-associated genes downstream of p73. Autophagy 2009;5:114–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (77).Oh M, Choi IK, Kwon HJ. Inhibition of histone deacetylase1 induces autophagy. Biochem Biophys Res Commun 2008;369:1179–83. [DOI] [PubMed] [Google Scholar]
- (78).Rodriguez-Gonzalez A, Lin T, Ikeda AK, Simms-Waldrip T, Fu C, Sakamoto KM. Role of the aggresome pathway in cancer: targeting histone deacetylase 6-dependent protein degradation. Cancer Res 2008;68:2557–60. [DOI] [PubMed] [Google Scholar]
- (79).Shao Y, Gao Z, Marks PA, Jiang X. Apoptotic and autophagic cell death induced by histone deacetylase inhibitors. Proc Natl Acad Sci U S A 2004;101:18030–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (80).Hrzenjak A, Kremser ML, Strohmeier B, Moinfar F, Zatloukal K, Denk H. SAHA induces caspase-independent, autophagic cell death of endometrial stromal sarcoma cells by influencing the mTOR pathway. J Pathol 2008;216:495–504. [DOI] [PubMed] [Google Scholar]
- (81).Watanabe M, Adachi S, Matsubara H, et al. Induction of autophagy in malignant rhabdoid tumor cells by the histone deacetylase inhibitor FK228 through AIF translocation. Int J Cancer 2009;124:55–67. [DOI] [PubMed] [Google Scholar]
- (82).Carew JS, Nawrocki ST, Kahue CN, et al. Targeting autophagy augments the anticancer activity of the histone deacetylase inhibitor SAHA to overcome Bcr-Abl-mediated drug resistance. Blood 2007;110:313–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (83).Eisenberg-Lerner A, Kimchi A. The paradox of autophagy and its implication in cancer etiology and therapy. Apoptosis 2009. January 27. [DOI] [PubMed]
- (84).Schoenlein PV, Periyasamy-Thandavan S, Samaddar JS, Jackson WH, Barrett JT. Autophagy facilitates the progression of ERalpha-positive breast cancer cells to antiestrogen resistance. Autophagy 2009;5. [DOI] [PubMed]
- (85).Jaakkola PM, Pursiheimo JP. p62 degradation by autophagy: Another way for cancer cells to survive under hypoxia. Autophagy 2009;5(3). [DOI] [PubMed] [Google Scholar]
- (86).Reddy RM, Yeow WS, Chua A, et al. Rapid and profound potentiation of Apo2L/TRAIL-mediated cytotoxicity and apoptosis in thoracic cancer cells by the histone deacetylase inhibitor Trichostatin A: the essential role of the mitochondria-mediated caspase activation cascade. Apoptosis 2007;12:55–71. [DOI] [PubMed] [Google Scholar]
- (87).Fulda S Modulation of TRAIL-induced apoptosis by HDAC inhibitors. Curr Cancer Drug Targets 2008;8:132–40. [DOI] [PubMed] [Google Scholar]
- (88).Dai Y, Rahmani M, Dent P, Grant S. Blockade of histone deacetylase inhibitor-induced RelA/p65 acetylation and NF-kappaB activation potentiates apoptosis in leukemia cells through a process mediated by oxidative damage, XIAP downregulation, and c-Jun N-terminal kinase 1 activation. Mol Cell Biol 2005;25:5429–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (89).Elrod HA, Sun SY. Modulation of death receptors by cancer therapeutic agents. Cancer Biol Ther 2008;7:163–73. [DOI] [PubMed] [Google Scholar]
- (90).Hajji N, Wallenborg K, Vlachos P, Nyman U, Hermanson O, Joseph B. Combinatorial action of the HDAC inhibitor trichostatin A and etoposide induces caspase-mediated AIF-dependent apoptotic cell death in non-small cell lung carcinoma cells. Oncogene 2008;27:3134–44. [DOI] [PubMed] [Google Scholar]
- (91).Xu J, Zhou JY, Wei WZ, Philipsen S, Wu GS. Sp1-mediated TRAIL induction in chemosensitization. Cancer Res 2008;68:6718–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (92).Thomas S, Munster PN. Histone deacetylase inhibitor induced modulation of anti-estrogen therapy. Cancer Lett 2009. January 29. [DOI] [PubMed]
- (93).Fan J, Yin WJ, Lu JS, et al. ER alpha negative breast cancer cells restore response to endocrine therapy by combination treatment with both HDAC inhibitor and DNMT inhibitor. J Cancer Res Clin Oncol 2008;134:883–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (94).Jang ER, Lim SJ, Lee ES, et al. The histone deacetylase inhibitor trichostatin A sensitizes estrogen receptor alpha-negative breast cancer cells to tamoxifen. Oncogene 2004;23:1724–36. [DOI] [PubMed] [Google Scholar]
- (95).Fiskus W, Ren Y, Mohapatra A, et al. Hydroxamic acid analogue histone deacetylase inhibitors attenuate estrogen receptor-alpha levels and transcriptional activity: a result of hyperacetylation and inhibition of chaperone function of heat shock protein 90. Clin Cancer Res 2007;13:4882–90. [DOI] [PubMed] [Google Scholar]
- (96).Abbas A, Gupta S. The role of histone deacetylases in prostate cancer. Epigenetics 2008;3:300–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (97).Bjorkman M, Iljin K, Halonen P, et al. Defining the molecular action of HDAC inhibitors and synergism with androgen deprivation in ERG-positive prostate cancer. Int J Cancer 2008;123:2774–81. [DOI] [PubMed] [Google Scholar]
- (98).Marrocco DL, Tilley WD, Bianco-Miotto T, Evdokiou A, Scher HI, Rifkind RA, et al. Suberoylanilide hydroxamic acid (vorinostat) represses androgen receptor expression and acts synergistically with an androgen receptor antagonist to inhibit prostate cancer cell proliferation. Mol Cancer Ther 2007;6:51–60. [DOI] [PubMed] [Google Scholar]
- (99).Welsbie DS, Xu J, Chen Y, et al. Histone deacetylases are required for androgen receptor function in hormone-sensitive and castrate-resistant prostate cancer. Cancer Res 2009;69:958–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (100).Epping MT, Bernards R. Molecular basis of the anti-cancer effects of histone deacetylase inhibitors. Int J Biochem Cell Biol 2009;41:16–20. [DOI] [PubMed] [Google Scholar]
- (101).Epping MT, Wang L, Plumb JA, et al. A functional genetic screen identifies retinoic acid signaling as a target of histone deacetylase inhibitors. Proc Natl Acad Sci U S A 2007;104:17777–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (102).Epping MT, Wang L, Edel MJ, Carlee L, Hernandez M, Bernards R. The human tumor antigen PRAME is a dominant repressor of retinoic acid receptor signaling. Cell 200;122:835–47. [DOI] [PubMed] [Google Scholar]
- (103).Hahn CK, Ross KN, Warrington IM, et al. Expression-based screening identifies the combination of histone deacetylase inhibitors and retinoids for neuroblastoma differentiation. Proc Natl Acad Sci U S A 2008;105:9751–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (104).Wang XF, Qian DZ, Ren M, et al. Epigenetic modulation of retinoic acid receptor beta2 by the histone deacetylase inhibitor MS-275 in human renal cell carcinoma. Clin Cancer Res 2005;11:3535–42. [DOI] [PubMed] [Google Scholar]
- (105).Weiser TS, Guo ZS, Ohnmacht GA, et al. Sequential 5-Aza-2 deoxycytidine-depsipeptide FR901228 treatment induces apoptosis preferentially in cancer cells and facilitates their recognition by cytolytic T lymphocytes specific for NY-ESO-1. J Immunother 2001;24:151–61. [DOI] [PubMed] [Google Scholar]
- (106).Wargo JA, Robbins PF, Li Y, et al. Recognition of NY-ESO-1+ tumor cells by engineered lymphocytes is enhanced by improved vector design and epigenetic modulation of tumor antigen expression. Cancer Immunol Immunother 2009;58:383–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (107).Gialitakis M, Kretsovali A, Spilianakis C, et al. Coordinated changes of histone modifications and HDAC mobilization regulate the induction of MHC class II genes by Trichostatin A. Nucleic Acids Res 2006;34:765–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (108).Brogdon JL, Xu Y, Szabo SJ, et al. Histone deacetylase activities are required for innate immune cell control of Th1 but not Th2 effector cell function. Blood 2007;109:1123–30. [DOI] [PubMed] [Google Scholar]
- (109).Tao R, de Zoeten EF, Ozkaynak E, et al. Deacetylase inhibition promotes the generation and function of regulatory T cells. Nat Med 2007;13:1299–307. [DOI] [PubMed] [Google Scholar]
- (110).Ara T, Song L, Shimada H, et al. Interleukin-6 in the bone marrow microenvironment promotes the growth and survival of neuroblastoma cells. Cancer Res 2009;69:329–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (111).Charafe-Jauffret E, Ginestier C, Iovino F, et al. Breast cancer cell lines contain functional cancer stem cells with metastatic capacity and a distinct molecular signature. Cancer Res 2009;69:1302–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (112).Lundqvist A, Abrams SI, Schrump DS, et al. Bortezomib and depsipeptide sensitize tumors to tumor necrosis factor-related apoptosis-inducing ligand: a novel method to potentiate natural killer cell tumor cytotoxicity. Cancer Res 2006;66:7317–25. [DOI] [PubMed] [Google Scholar]
- (113).Diermayr S, Himmelreich H, Durovic B, et al. NKG2D ligand expression in AML increases in response to HDAC inhibitor valproic acid and contributes to allorecognition by NK-cell lines with single KIR-HLA class I specificities. Blood 2008;111:1428–36. [DOI] [PubMed] [Google Scholar]
- (114).Butler LM, Zhou X, Xu WS, et al. The histone deacetylase inhibitor SAHA arrests cancer cell growth, up-regulates thioredoxin-binding protein-2, and down-regulates thioredoxin. Proc Natl Acad Sci U S A 2002;99:11700–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (115).Sasakawa Y, Naoe Y, Sogo N, et al. Marker genes to predict sensitivity to FK228, a histone deacetylase inhibitor. Biochem Pharmacol 2005;69:603–16. [DOI] [PubMed] [Google Scholar]
- (116).Miyanaga A, Gemma A, Noro R, et al. Antitumor activity of histone deacetylase inhibitors in non-small cell lung cancer cells: development of a molecular predictive model. Mol Cancer Ther 2008;7:1923–30. [DOI] [PubMed] [Google Scholar]
- (117).Dejligbjerg M, Grauslund M, Christensen IJ, Tjornelund J, Buhl JP, Sehested M. Identification of predictive biomarkers for the histone deacetylase inhibitor belinostat in a panel of human cancer cell lines. Cancer Biomark 2008;4:101–9. [DOI] [PubMed] [Google Scholar]
- (118).Stapnes C, Ryningen A, Hatfield K, et al. Functional characteristics and gene expression profiles of primary acute myeloid leukaemia cells identify patient subgroups that differ in susceptibility to histone deacetylase inhibitors. Int J Oncol 2007;31:1529–38. [PubMed] [Google Scholar]
- (119).Nguyen DM, Schrump WD, Chen GA, et al. Abrogation of p21 expression by flavopiridol enhances depsipeptide-mediated apoptosis in malignant pleural mesothelioma cells. Clin Cancer Res 2004;10:1813–25. [DOI] [PubMed] [Google Scholar]
- (120).Graham B, Gibson SB. The two faces of NFkappaB in cell survival responses. Cell Cycle 2005;4:1342–5. [DOI] [PubMed] [Google Scholar]
- (121).Law AY, Lai KP, Lui WC, Wan HT, Wong CK. Histone deacetylase inhibitor-induced cellular apoptosis involves stanniocalcin-1 activation. Exp Cell Res 2008;314:2975–84. [DOI] [PubMed] [Google Scholar]
- (122).Miller CP, Ban K, Dujka ME, et al. NPI-0052, a novel proteasome inhibitor, induces caspase-8 and ROS-dependent apoptosis alone and in combination with HDAC inhibitors in leukemia cells. Blood 2007;110:267–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (123).Dai Y, Chen S, Kramer LB, Funk VL, Dent P, Grant S. Interactions between bortezomib and romidepsin and belinostat in chronic lymphocytic leukemia cells. Clin Cancer Res 2008;14:549–58. [DOI] [PubMed] [Google Scholar]
- (124).Fantin VR, Loboda A, Paweletz CP, et al. Constitutive activation of signal transducers and activators of transcription predicts vorinostat resistance in cutaneous T-cell lymphoma. Cancer Res 2008;68:3785–94. [DOI] [PubMed] [Google Scholar]
- (125).Lee MJ, Kim YS, Kummar S, Giaccone G, Trepel JB. Histone deacetylase inhibitors in cancer therapy. Curr Opin Oncol 2008;20:639–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (126).Camphausen K, Tofilon PJ. Inhibition of histone deacetylation: a strategy for tumor radiosensitization. J Clin Oncol 2007;25:4051–6. [DOI] [PubMed] [Google Scholar]
- (127).Chinnaiyan P, Cerna D, Burgan WE, et al. Postradiation sensitization of the histone deacetylase inhibitor valproic acid. Clin Cancer Res 2008;14:5410–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (128).Xiao JJ, Foraker AB, Swaan PW, et al. Efflux of depsipeptide FK228 (FR901228, NSC-630176) is mediated by P-glycoprotein and multidrug resistance-associated protein 1. J Pharmacol Exp Ther 2005;313:268–76. [DOI] [PubMed] [Google Scholar]
- (129).Kim YK, Kim NH, Hwang JW, et al. Histone deacetylase inhibitor apicidin-mediated drug resistance: involvement of P-glycoprotein. Biochem Biophys Res Commun 2008;368:959–64. [DOI] [PubMed] [Google Scholar]
- (130).Robey RW, Zhan Z, Piekarz RL, Kayastha GL, Fojo T, Bates SE. Increased MDR1 expression in normal and malignant peripheral blood mononuclear cells obtained from patients receiving depsipeptide (FR901228, FK228, NSC630176). Clin Cancer Res 2006;12:1547–55. [DOI] [PubMed] [Google Scholar]
- (131).To KK, Polgar O, Huff LM, Morisaki K, Bates SE. Histone modifications at the ABCG2 promoter following treatment with histone deacetylase inhibitor mirror those in multidrug-resistant cells. Mol Cancer Res 2008;6:151–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (132).Fedier A, Dedes KJ, Imesch P, Von Bueren AO, Fink D. The histone deacetylase inhibitors suberoylanilide hydroxamic (Vorinostat) and valproic acid induce irreversible and MDR1-independent resistance in human colon cancer cells. Int J Oncol 2007;31:633–41. [PubMed] [Google Scholar]
- (133).Fiskus W, Rao R, Fernandez P, et al. Molecular and biologic characterization and drug sensitivity of pan-histone deacetylase inhibitor-resistant acute myeloid leukemia cells. Blood 2008;112:2896–905. [DOI] [PubMed] [Google Scholar]
- (134).Lee YS, Lim KH, Guo X, et al. The cytoplasmic deacetylase HDAC6 is required for efficient oncogenic tumorigenesis. Cancer Res 2008;68:7561–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (135).Nusinzon I, Horvath CM. Positive and negative regulation of the innate antiviral response and beta interferon gene expression by deacetylation. Mol Cell Biol 2006;26:3106–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (136).Nusinzon I, Horvath CM. Unexpected roles for deacetylation in interferon- and cytokine-induced transcription. J Interferon Cytokine Res 2005;25:745–8. [DOI] [PubMed] [Google Scholar]
- (137).Khan AN, Tomasi TB. Histone deacetylase regulation of immune gene expression in tumor cells. Immunol Res 2008;40:164–78. [DOI] [PMC free article] [PubMed] [Google Scholar]