Abstract
Aims: Ferroportin (FPN) is an iron exporter that plays an important role in cellular and systemic iron metabolism. Our previous work has demonstrated that FPN is decreased in prostate tumors. We sought to identify the molecular pathways regulated by FPN in prostate cancer cells.
Results: We show that overexpression of FPN induces profound effects in cells representative of multiple histological subtypes of prostate cancer by activating different but converging pathways. Induction of FPN induces autophagy and activates the transcription factors tumor protein 53 (p53) and Kruppel-like factor 6 (KLF6) and their common downstream target, cyclin-dependent kinase inhibitor 1A (p21). FPN also induces cell cycle arrest and stress-induced DNA-damage genes. Effects of FPN are attributable to its effects on intracellular iron and can be reproduced with iron chelators. Importantly, expression of FPN not only inhibits proliferation of all prostate cancer cells studied but also reduces growth of tumors derived from castrate-resistant adenocarcinoma C4-2 cells in vivo.
Innovation: We use a novel model of FPN expression to interrogate molecular pathways triggered by iron depletion in prostate cancer cells. Since prostate cancer encompasses different subtypes with a highly variable clinical course, we further explore how histopathological subtype influences the response to iron depletion. We demonstrate that prostate cancer cells that derive from different histopathological subtypes activate converging pathways in response to FPN-mediated iron depletion. Activation of these pathways is sufficient to significantly reduce the growth of treatment-refractory C4-2 prostate tumors in vivo.
Conclusions: Our results may explain why FPN is dramatically suppressed in cancer cells, and they suggest that FPN agonists may be beneficial in the treatment of prostate cancer.
Keywords: prostate cancer, iron, ferroportin, cell cycle, autophagy, chelator
Introduction
Ferroportin (FPN) is a cellular iron efflux pump that is critical to maintenance of systemic iron homeostasis (23, 75). When expressed on the apical surface of enterocytes, FPN mediates efflux of dietary iron into the circulation. FPN is also expressed on the surface of macrophages and hepatocytes, where it participates in recycling of iron from cellular stores and senescent red blood cells. Since FPN is the only known protein with an iron export function in vertebrates, loss of its activity is not compensated for by functionally redundant proteins. Germline FPN mutations result in the iron overload disease hemochromatosis (52, 66).
Apart from its expression in tissues that regulate systemic iron balance, FPN is also expressed in peripheral tissues and is markedly reduced in a number of tumors (70). Our laboratory and others have reported that FPN is decreased in prostate cancer transcriptionally, post-transcriptionally by hepcidin, a negative regulator of FPN (69), and epigenetically through methylation of CpG islands in the prostate promoter (69, 77). Induction of hepcidin, a negative regulator of FPN, also contributes to the decrease in FPN in prostate cancer. The consequence of decreased FPN is increased retention of labile iron in tumors, which facilitates tumor growth (70).
Innovation.
Pathways triggered by iron depletion have been traditionally explored by using iron chelators. Here, we use expression of ferroportin (FPN), an endogenous iron efflux pump markedly downregulated in prostate cancer, as a novel approach to interrogate molecular responses to iron depletion. Focusing on cells representative of three different, clinically important, histolopathological subtypes of prostate cancer, we observe that iron depletion induces DNA damage, autophagy, cell cycle arrest, and a stress response in all subtypes of prostate cancer by activating different but converging molecular pathways. Using microarray analysis, we further show that FPN-mediated iron depletion induces additional pathways that are both overlapping and cell-type specific. We demonstrate that FPN-mediated iron depletion is sufficient to significantly reduce the growth of treatment-refractory C4-2 prostate tumors in vivo. These results may provide a molecular explanation for why FPN is suppressed in prostate cancer tissue.
Prostate cancer is the most common malignancy and the second most frequent cause of cancer death in men (64). The vast majority of prostate cancers are adenocarcinomas, with varying degrees of luminal organization, glandular formation, and expression of the androgen receptor (AR) (11, 13). Targeting of the AR is a successful therapy for prostatic adenocarcinoma; however, most patients become resistant to such therapies (castrate-resistant disease), which leads to disease progression (35). A smaller fraction of prostatic epithelial malignancies are of other histological types, including small cell (neuroendocrine) carcinoma (SCNC), which occurs in approximately 0.5–2% of men with prostate cancer (28). SCNC do not express AR, are clinically more aggressive than adenocarcinoma, and are treated with different therapeutic regimens (28).
Cell culture models of these various types of prostate cancer have been developed. LNCaP cells, which express AR and prostate-specific antigen, are among the most widely used cell line to model androgen-responsive prostate adenocarcinoma (65). Derivatives of LNCaP cells that are androgen independent, such as C4-2 cells, have been created to model castrate-resistant adenocarcinoma (65). The PC3 cell line, historically used to model aggressive castrate-resistant adenocarcinoma, is now believed to derive from a neuroendocrine subtype of prostate cancer (67).
Here, we use LNCaP, C4-2, and PC3 cells to probe the mechanisms by which modulation of FPN expression affects prostate cancer. We show that enhanced iron efflux mediated by overexpression of FPN is sufficient to trigger substantial iron depletion and induce profound effects in prostate cancer cells by activating different but convergent pathways.
Results
FPN is a key regulator of iron metabolism in prostate cancer
FPN is a cell surface iron efflux pump that is expressed in a variety of tissues (23, 75). It is downregulated in several tumor types, including prostate cancer (69, 77). To determine whether FPN affects iron homeostasis and whether its effects are similar in prostate adeonocarcinoma and SCNC prostate cancer, we constructed a lentiviral vector carrying a CMV promoter-driven FPN expression cassette. This virus was used to infect LNCaP prostate adenocarcinoma and PC3 SCNC prostate cancer cells to generate cells with overexpression of FPN (referred to as FPN OE cells) (Fig. 1A and Supplementary Fig. S1).
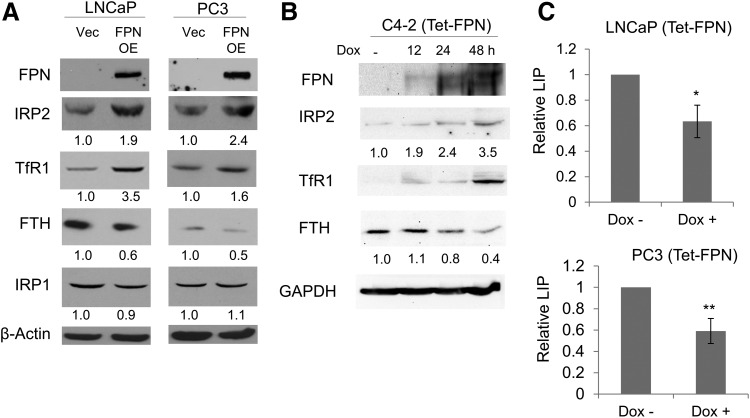
FIG. 1.
FPN affects iron homeostasis in prostate cancer cells. (A) Western blot of FPN, IRP2, TFRC1, IRP1, FTH, and β-actin (loading control) in LNCaP and PC3 cells expressing a control vector (Vec) or FPN OE. (B) Western blot of proteins in C4-2 cells expressing doxycycline-inducible FPN (Tet-FPN). Cells were untreated or treated with 1 μg/mL doxycycline for 12, 24, or 48 h. GAPDH was used as a loading control. (C) LIP in LNCaP (Tet-FPN) and PC-3 (Tet-FPN) cells ±1 μg/mL doxycycline for 48 h. Experiments were repeated at least three times. Uncropped blots are shown in Supplementary Figure S1. *p < 0.05; **p < 0.01. FPN, ferroportin; FPN OE, ferroportin overexpression vector; FTH, ferritin H; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; IRP1, iron regulatory protein 1; IRP2, iron regulatory protein 2; LIP, labile iron pool; (Tet-FPN), cells infected with a tetracycline-inducible FPN; TFRC1, transferrin receptor 1.
To determine the consequences of FPN expression on cellular iron metabolism, we measured levels of iron regulatory protein 2 (IRP2) and its downstream targets transferrin receptor 1 (TfR1) and ferritin (45). IRP2 is a master regulator of cellular iron that post-transcriptionally controls levels of ferritin, an intracellular iron storage protein, and transferrin receptor, the primary protein responsible for iron uptake (56). When levels of intracellular iron are low, IRP activity increases, which represses ferritin and stabilizes transferrin receptor, resulting in the accumulation of labile iron in cells and the restoration of levels of intracellular iron.
Since FPN drives iron efflux, FPN overexpression should lower intracellular iron, resulting in increased IRP2, increased TfR1, and decreased ferritin. Consistent with these predictions, overexpression of FPN increased IRP2 (but not iron regulatory protein 1 [IRP1]) in both LNCaP and PC3 cells (Fig. 1A and Supplementary Fig. S1). As anticipated, TfR1 and ferritin H (FTH), downstream targets of IRP2, were increased and reduced in the FPN-overexpressing cells, respectively.
To substantiate these findings, we generated prostate cancer cells that conditionally expressed FPN driven by a tetracycline-inducible promoter [hereafter referred to as (Tet-FPN)]. Consistent with data from FPN OE cells (Fig. 1A and Supplementary Fig. S1), LNCaP (Tet-FPN) cells exhibited increased IRP2 and TfR1 after Tet-mediated induction of FPN (Supplementary Fig. S2). To determine whether C4-2 cells, the androgen-insensitive derivative of LNCaP cells, had a similar response to FPN, we generated stable C4-2 (Tet-FPN) cells and examined IRP2, TfR1, and FTH. Similarly, FPN induction increased IRP2 and TfR1 and reduced FTH in C4-2 (Tet-FPN) cells (Fig. 1B and Supplementary Fig. S1).
Next, we directly measured the intracellular labile iron pool (LIP) in control and FPN-expressing cells. Induction of FPN significantly reduced LIP levels in both LNCaP (Tet-FPN) and PC3 (Tet-FPN) cells (Fig. 1C). Collectively, these results indicate that overexpression of FPN reduces intracellular iron in both adenocarcinoma and SCNC prostate cancer cells.
FPN induces autophagy in prostate cancer cells
Having determined that levels of FPN, an iron-specific efflux pump, could markedly alter iron homeostasis in prostate cancer cells, we next sought to understand the downstream consequences of iron depletion.
Autophagy is a physiological process that is upregulated by numerous stimuli, including nutrient deprivation, and can function to preserve cell viability under unfavorable environmental conditions (46). Increasing evidence indicates that autophagy contributes to maintenance of iron balance by promoting the turnover of iron storage proteins (19, 43). Supporting this concept, iron chelators have been shown to induce autophagy in malignant plasma cells and breast cancer cells (25, 54). Whether upregulation of iron efflux is sufficient to trigger autophagy, and whether histopathological subtypes of prostate cancer cells differ in their response to enhanced iron efflux, is not known.
To test whether iron depletion induced by FPN triggered autophagy, we first examined the effect of FPN on conversion of the microtubule-associated protein light chain 3 beta (LC3B-I), a marker of autophagosomes, to its phosphatidylethanolamine-conjugated form (LC3B-II), a widely used indicator of autophagy (34, 54). We found a substantive accumulation of LC3B-II and a slight increase of LC3B-I in FPN-overexpressing LNCaP cells (Fig. 2A and Supplementary Fig. S3), consistent with an induction of autophagosome synthesis. LC3B-I and LC3B-II were likewise upregulated in cells with tet-inducible FPN in the presence of doxycycline (Fig. 2B and Supplementary Fig. S3). As a positive control, we treated cells with the iron chelator desferoxamine (DFO), and observed a similar effect on LC3B-I and LC3B-II in both LNCaP and PC3 cells (Fig. 2C and Supplementary Fig. S3).
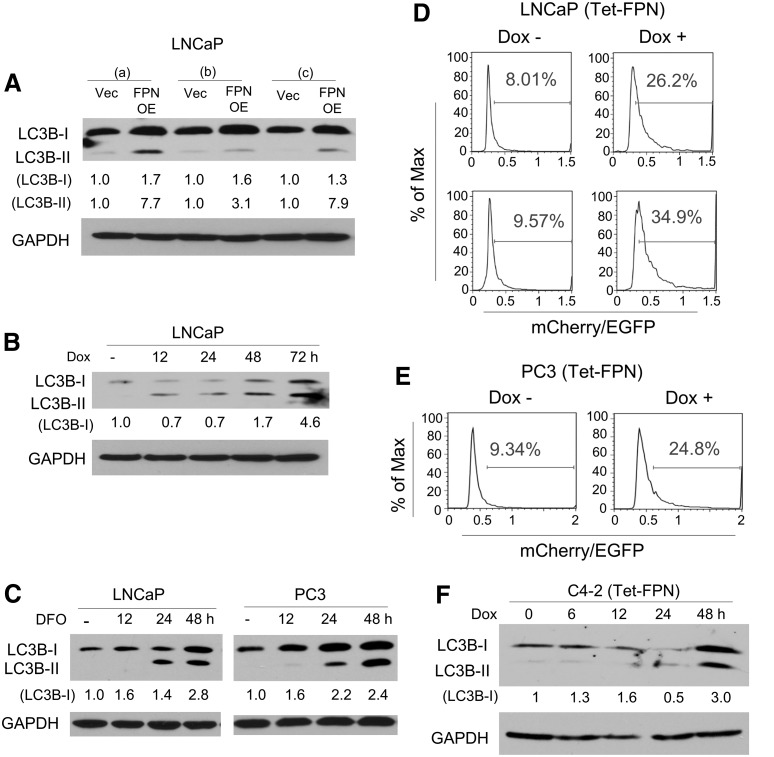
FIG. 2.
FPN overexpression induces autophagy. (A) Western blot of LC3B-I and LC3B-II in three independent infections (a–c) of LNCaP cells containing a control vector (Vec) or FPN OE. (B) Western blot of LC3B-I and LC3B-II in LNCaP cells expressing doxycycline-inducible FPN (Tet-FPN). Cells were untreated or treated with 1 μg/mL doxycycline for 12, 24, 48, or 72 h. (C) Western blot of LC3B-I and LC3B-II in LNCaP and PC3 cells untreated or treated with 100 μM DFO for 12, 24, or 48 h. (D, E) Ratio of mCherry/EGFP fluorescence intensity in cells expressing an mCherry-EGFP-LC3B reporter as determined by flow cytometry in (D) LNCaP (Tet-FPN) and (E) PC3 (Tet-FPN) cells treated ± 1 μg/mL doxycycline for 3 days (D top and E) or 4 days (Panel D bottom). Data were analyzed with the FlowJo software (TreeStar, Inc.). (F) Western blot of LC3B-I and LC3B-II in C4-2 (Tet-FPN) cells untreated or treated with 1 μg/mL doxycycline for 6, 12, 24, or 48 h. (A–C, F) GAPDH was used as a loading control. Experiments were repeated at least three times. Uncropped blots are shown in Supplementary Figure S3. DFO, desferoxamine, an iron chelator; EGFP, enhanced green fluorescent protein; LC3B-I, microtubule-associated protein light chain 3 beta; LC3B-II, phosphatidylethanolamine-conjugated microtubule-associated protein light chain 3 beta.
To confirm that the increase in LC3B-I and LC3B-II reflected an increase in autophagy (rather than a blockade in autophagosome degradation), we measured autophagic flux, the fusion of autophagosomes with lysosomes, and the degradation of autophagic substrates, using the mCherry-enhanced green fluorescent protein (EGFP)-LC3B reporter (33), which measures autophagic flux as the ratio between mCherry and EGFP fluorescence (24, 46). The reporter was introduced into tet-inducible LNCaP FPN cells, and fluorescence was monitored before and after induction of FPN expression with doxycycline. FPN induction with doxycycline addition notably increased the ratio of mCherry/EGFP in LNCaP cells (Fig. 2D). FPN induction similarly induced autophagy in C4-2 (Tet-FPN) cells as well as in PC3 (Tet-FPN) cells (Fig. 2E, F and Supplementary Fig. S3). Collectively, these data indicate that FPN-mediated iron depletion induces autophagy in multiple prostate cancer cell types.
FPN inhibits prostate cancer cell proliferation through its effect on iron efflux
Another survival strategy that cells utilize at times of nutrient deprivation is to limit cellular proliferation (9, 26). We tested whether iron depletion affected cell proliferation in LNCaP, PC3, and C4-2 cells. Vector and FPN-OE cells were seeded at the same density, and cell proliferation was evaluated by counting cells after 6–7 days. As illustrated in Figure 3A and B, FPN overexpression reduced cell number compared with vector control cells in both LNCaP and PC3 cells. Similar results were observed by using a tet-inducible system (Fig. 3C). Doxycycline treatment did not affect control tet-inducible luciferase (Luc) cells (Fig. 3D).
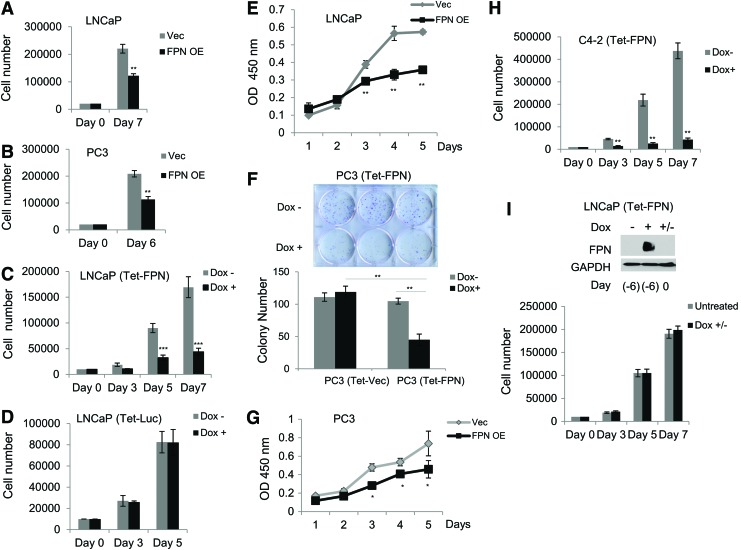
FIG. 3.
FPN overexpression inhibits prostate cancer cell proliferation and colony formation. (A, B) Cell count by hemocytometer of (A) LNCaP and (B) PC3 cells expressing a control vector (Vec) or FPN OE. Cells were plated in six well-plates at a density of 20,000 cells/well and counted 6 or 7 days after seeding. (C, D) Cell count by hemocytometer of LNCaP cells expressing (C) doxycycline-inducible FPN (Tet-FPN) or (D) doxycycline-inducible luciferase (Tet-Luc). Cells were plated in six well-plates at a density of 10,000 cells/well and treated with ±1 μg/mL doxycycline for 3, 5, or 7 [LNCaP (Tet-FPN) only] days. (E) WST-1 assay of cell proliferation of LNCaP cells expressing a control vector (Vec) or FPN OE. (F) Representative plate and quantification of clonogenic assays for PC3 (Tet-FPN) and vector control (Tet-Vec) cells. Three hundred cells were plated in six-well plates for 12 days with ±1 μg/mL doxycycline. (G) WST-1 assay for cell proliferation of PC3 cells expressing a control vector (Vec) or FPN OE. (H) Cell count by hemocytometer of C4-2 (Tet-FPN) cells plated in six-well plates at a density of 10,000 cells/well and treated with ±1 μg/mL doxycycline for 3, 5, or 7 days. (I) Cell count by hemocytometer of LNCaP (Tet-FPN) cells untreated or pre-treated with 1 μg/mL doxycycline for 2 days before doxycycline withdrawal for 6 days. Cells were counted 0, 3, 5, and 7 days post-doxycycline withdrawal. Means and standard deviations are shown. Western blot demonstrates FPN protein levels relative to GAPDH at indicated times. Uncropped blot is shown in Supplementary Figure S4. Experiments were repeated at least three times with the exception of (I) (n = 2). *p ≤ 0.05; **p ≤ 0.01; ***p ≤ 0.001. Luc, luciferase; (Tet-Vec), cells infected with a tetracycline-inducible vector control; WST-1, water-soluble tetrazolium salt-1. Color images are available online.
We confirmed these results by using an independent (water-soluble tetrazolium salt-1 [WST-1]) assay, which also demonstrated an inhibition of proliferation by FPN in LNCaP cells (Fig. 3E). Similarly, FPN overexpression significantly reduced the proliferation of PC3 cells as measured by both a clonogenic and metabolic (WST-1) assay (Fig. 3F, G), as well as C4-2 cells (Fig. 3H). We tested whether FPN-mediated growth arrest was permanent or transient by measuring the rate of cell proliferation after doxycycline withdrawal in LNCaP (Tet-FPN) versus control cells. As shown in Figure 3I, doxycycline-treated LNCaP (Tet-FPN) cells proliferated at a rate indistinguishable from control cells after doxycycline removal, indicating that FPN-mediated proliferative arrest is transient.
We asked whether the iron efflux function of FPN was directly related to its observed ability to inhibit proliferation. To accomplish this, we used FPN-A77D (cells constitutively overexpressing a functionally impaired mutant FPN driven by the cytomegalovirus promoter), a well-characterized FPN mutant with impaired iron efflux function (40, 62). The A77D mutation, present in the predicted transmembrane domain 2 of FPN, results in mislocalization of FPN and reduced display on the cell surface, and thus attenuates FPN's iron transport function (40, 62). To confirm the preservation of this phenotype in LNCaP cells, we created the FPN A77D mutant by using site-directed mutagenesis, and we generated stable LNCaP cell lines expressing either constitutive or tet-inducible FPN (A77D) (Fig. 4A, B and Supplementary Fig. S4).
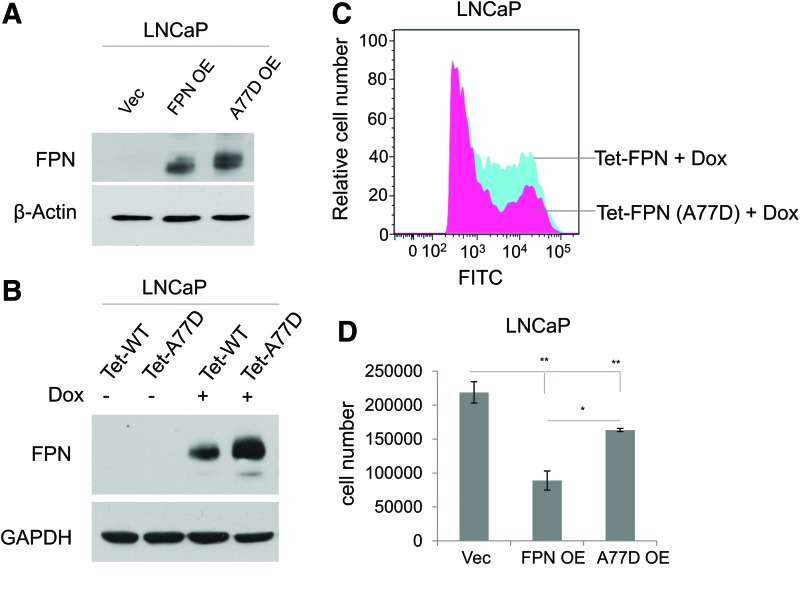
FIG. 4.
A77D mutation compromises the inhibitory function of FPN in cell proliferation. (A) Western blot of FPN in LNCaP cells expressing a control vector (Vec), FPN OE, or a vector overexpressing a functionally impaired FPN mutant (A77D). β-actin was used as a loading control. (B) Western blot of FPN in LNCaP cells expressing doxycycline-inducible FPN (Tet-FPN) or doxycycline-inducible A77D mutant FPN (Tet-A77D). Cells were treated with ±1 μg/mL doxycycline for 24 h. GAPDH was used as a loading control. (C) Detection of cell surface expression of FPN, or mutated FPN, in LNCaP (Tet-FPN) and LNCaP (Tet-A77D) cells induced with 1 μg/mL doxycycline. (D) Cell count by hemocytometer of LNCaP (Vec), LNCaP (FPN OE), and LNCaP (A77D OE) cells at 7 days. Experiments were repeated at least three times. Uncropped blots are shown in Supplementary Figure S4. *p ≤ 0.05; **p ≤ 0.01. (Tet-A77D), cells infected with a tetracycline inducible, functionally impaired mutant FPN. Color images are available online.
We then used flow cytometry to compare the level of cell surface FPN in cells expressing wildtype or mutant FPN. Consistent with previous results (40), display of FPN (A77D) on the cell surface was substantially reduced when compared with wildtype FPN (Fig. 4C), despite higher overall expression of the FPN mutant (Fig. 4A, B and Supplementary Fig. S4). We then tested the effect of the FPN A77D mutant on proliferation by expressing both mutant and wildtype in LNCaP cells and assessing effects on cell number. As shown in Figure 4D, FPN A77D was less effective in reducing proliferation than wildtype. Thus, the ability of FPN to inhibit proliferation is directly related to its iron efflux capacity, and does not represent off-target effects of this protein.
FPN decreases prostate cancer cell proliferation by affecting cell cycle regulators
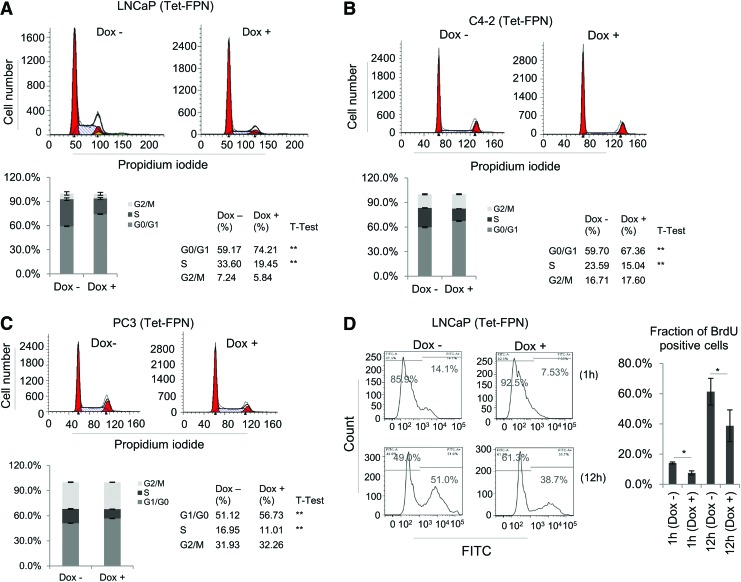
To determine the mechanism by which FPN affects prostate cancer cell proliferation, we first used flow cytometry to ask whether expression of FPN affects a particular phase of the cell cycle. As shown in Figure 5A and B, FPN expression increased the number of LNCaP and C4-2 cells in G0/G1 but reduced the number in S phase. A similar trend of decrease in S phase and increase in G1 phase was observed in PC3 cells, although the magnitude of the effect was modest (Fig. 5C). Consistent with the ability of FPN to attenuate DNA replication, a pronounced decrease in the fraction of 5-bromo-2′-deoxy (BrdU) positive cells was observed in a BrdU incorporation assay (Fig. 5D). Collectively, these findings demonstrate that FPN is a potent suppressor of proliferation that acts, at least in part, by inhibiting cell cycle progression, most dramatically in LNCaP and C4-2 prostate adenocarcinoma cells.
FIG. 5.
FPN overexpression inhibits cell cycle progression. (A) LNCaP, (B) C4-2, and (C) PC3 cells expressing doxycycline-inducible FPN (Tet-FPN) were treated ±1 μg/mL doxycycline for 48 h and stained with propidium iodide. DNA content was assessed by flow cytometry, and cell cycle distribution was analyzed by ModFit LT software. (D) BrdU incorporation assay for LNCaP (Tet-FPN) cells treated ±1 μg/mL doxycycline. Experiments were repeated at least three times. *p < 0.05; **p ≤ 0.01. BrdU, 5-bromo-2′-deoxy. Color images are available online.
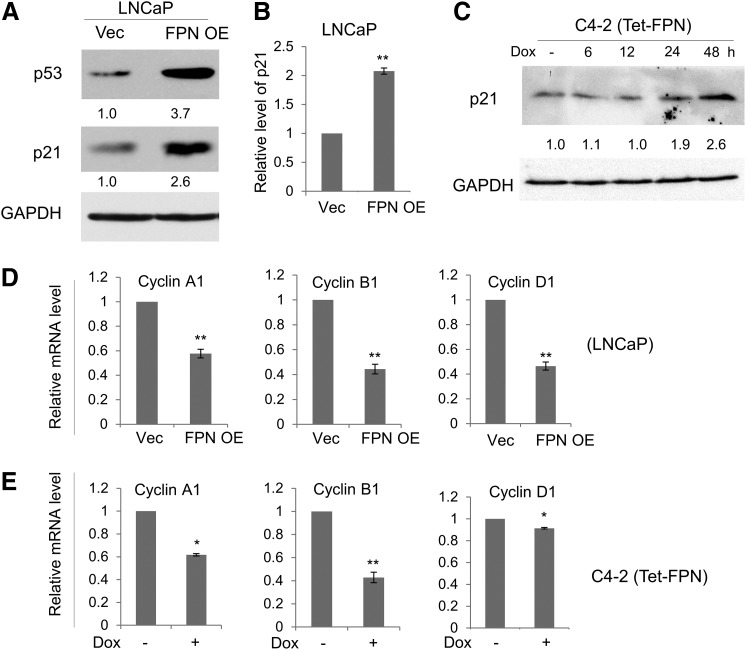
We next evaluated the mechanisms responsible for FPN-mediated inhibition of proliferation. A major G1/S checkpoint pathway involves tumor protein 53 (p53)-mediated activation of cyclin-dependent kinase inhibitor 1A (p21) (29). Since LNCaP cells express functional p53, we hypothesized that p21 might be involved in FPN-mediated inhibition of proliferation. As shown in Figure 6A (Supplementary Fig. S5), FPN overexpression did, indeed, activate p53 and its downstream target p21 (Fig. 6A and Supplementary Figs. S5 and S6B and S6C) in LNCaP cells. Elevated levels of p21 protein were associated with increased levels of p21 messenger RNA (mRNA) (Fig. 6B).
FIG. 6.
FPN induces p21 and inhibits expression of cyclin genes. (A) Western blot of p53 and p21 in LNCaP cells expressing a control vector (Vec) or FPN OE. GAPDH was used as a loading control. (B) RT-qPCR of p21 mRNA transcripts in LNCaP (Vec and FPN OE) cells. (C) Western blot of p21 in C4-2 cells expressing doxycycline-inducible FPN (Tet-FPN) cells untreated or treated with 1 μg/mL doxycycline for 6, 12, 24, or 48 h. (D, E) RT-qPCR of cyclin genes in (D) LNCaP (Vec and FPN OE) cells 3 days after infection and (E) C4-2 (Tet-FPN) cells treated with ±1 μg/mL doxycycline for 48 h. Experiments were repeated at least three times. Uncropped blots are shown in Supplementary Figure S5. *p < 0.05; **p ≤ 0.01. mRNA, messenger RNA; p21, cyclin-dependent kinase inhibitor 1A; p53, tumor protein 53; RT-qPCR, quantitative reverse transcription polymerase chain reaction.
Analysis of LNCaP and C4-2 (Tet-FPN) cells similarly showed p21 induction after doxycycline-mediated induction of FPN (Fig. 6C and Supplementary Figs. S5 and S7). We also assessed the effects of FPN on levels of cyclins. FPN overexpression decreased mRNA levels of Cyclin A1, Cyclin B1, and Cyclin D1 in LNCaP and C4-2 (Tet-FPN) cells, demonstrating a profound impact of FPN on drivers of the cell cycle in these cells (Fig. 6D, E).
We tested whether the more modest G0/G1 cell cycle arrest seen in PC3 cells could be traced to a lack of induction of p21, since PC3 cells lack functional p53, a major upstream inducer of p21 (Supplementary Fig. S6A) (12). Surprisingly, similar to LNCaP cells, FPN overexpression in PC3 cells induced p21 and downregulated expression of cyclin A1 and B1 (Fig. 7A).
FIG. 7.
KLF6-dependent induction of p21 and downregulation of cyclin genes by FPN in PC3 cells. (A) RT-qPCR of p21, cyclin A1, and cyclin B1 in PC3 cells expressing a control vector (Vec) or FPN OE. (B) RT-qPCR and Western blots of KLF6 in PC3 (Vec and FPN OE) cells as well as PC3 cells expressing doxycycline-inducible FPN (Tet-FPN). (C) RT-qPCR of p21 in PC3 (Vec and FPN OE) cells in the presence of either KLF6 knockdown or scrambled shRNA control. (A–C) Cells were analyzed 3 days after infection. Experiments were repeated at least three times. Uncropped blots are shown in Supplementary Figure S8. **p ≤ 0.01; ***p ≤ 0.001. KLF6, Kruppel-like factor 6; shRNA, short hairpin RNA.
To explore mechanisms of FPN-mediated induction of p21 in PC3 p53 null cells, we analyzed known transcriptional regulators of p21 (1). We observed that Kruppel-like factor 6 (KLF6), a zinc finger transcription factor, tumor suppressor, and regulator of p21 in prostate cancer cells (49), was induced in PC3 cells after overexpression of FPN (Fig. 7B and Supplementary Fig. S8). Transcript levels of other known transcriptional regulators of p21, including cAMP response element-binding protein binding protein, E2F transcription factor 1, AP4, SP1, SP3, cMyc, and Kruppel-like factor 4, were unaffected by FPN overexpression (not shown). Knockdown of KLF6 attenuated FPN-mediated induction of p21, confirming the involvement of KLF6 in p21 induction in PC3 cells (Fig. 7C and Supplementary S9).
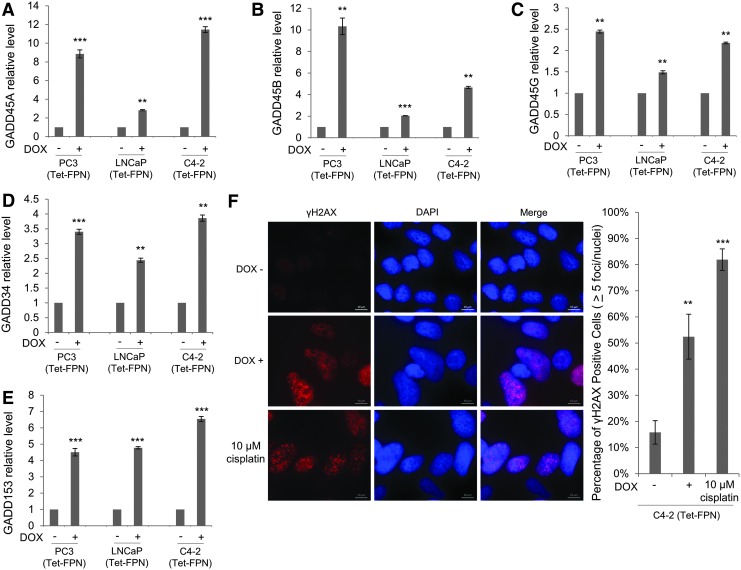
In addition to p21, p53 induces other proteins that inhibit the cell cycle. In particular, members of the DNA damage-induced (GADD) gene family are upregulated by p53 (74) and are induced by DNA damage (22), nutrition deprivation (21), or iron chelation (14, 61). Further, members of the GADD gene family, GADD34 and GADD153, promote autophagy (71). As seen in Figure 8, FPN overexpression induced GADD genes GADD45A, GADD45B, GADD45G, GADD34, and GADD153 in all three prostate cancer cell lines, including p53 null PC3 cells (Fig. 8A–E).
FIG. 8.
FPN induces GADD genes and DNA damage. (A–E) RT-qPCR of (A) GADD45A, (B) GADD45B, (C) GADD45G, (D) GADD34, and (E) GADD153 in LNCaP, C4-2, and PC3 cells expressing doxycycline-inducible FPN (Tet-FPN) treated with ±1 μg/mL doxycycline for 72 h. (F) C4-2 (Tet-FPN) cells were treated with ±1 μg/mL doxycycline for 72 h or treated with 10 μM cisplatin for 24 h as a positive control. Cells were stained with antibody to γH2AX and DAPI. Left: representative images are shown. Scale bar = 10 μm. Right: Nuclei with greater than five γH2AX foci were counted. Experiments were repeated at least three times. **p ≤ 0.01; ***p ≤ 0.001.
These data suggest that induction of GADD genes is a widespread response to FPN-mediated iron depletion, independent of p53 status. Considering that both p53 (31) and GADD genes (22) are induced by DNA damage, we asked whether FPN overexpression led to the accumulation of DNA damage. Indeed, FPN overexpression significantly increased γH2AX staining (36), a marker of DNA double-stranded breaks, in C4-2 (Tet-FPN) prostate cancer cells (Fig. 8F).
FPN overexpression induces cell-specific gene changes as well as changes that converge on similar pathways
Our observations to this point suggested that the three prostate cancer cell lines we tested activated converging pathways in response to FPN-mediated iron depletion, despite their derivation from different histopathological subtypes of prostate cancer. To more fully explore the similarities and differences in the response of these cells to FPN-mediated iron depletion, we performed a microarray analysis of LNCaP (Tet-FPN), C4-2 (Tet-FPN), and PC3 (Tet-FPN) cells as well as a vector control [LNCaP (Tet-VEC)] in the presence and absence of doxycycline, and we identified genes that were significantly differentially expressed after overexpression of FPN (false discovery rate [FDR] <0.05). The number of FPN-regulated genes varied among these cell lines from 113 in LNCaP cells, 486 in PC-3 cells, to 2916 in C4-2 Tet-FPN cells (Supplementary Table S1).
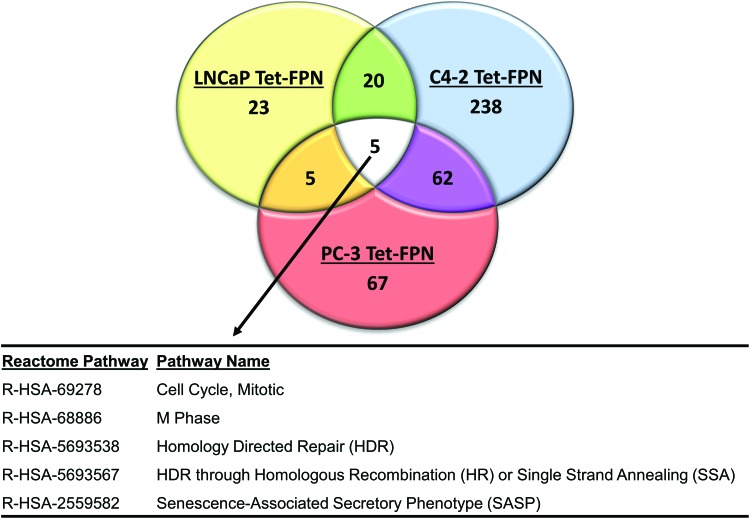
To map these genes to pathways, we first removed genes that were differentially expressed in the doxycycline control from the analysis (n = 30), and then mapped the remaining differentially expressed genes to Reactome pathways using ReactomePA (79). As expected, pathways perturbed in all three prostate cell lines converged on the cell cycle and DNA repair (Fig. 9). FPN-mediated changes were also observed in unanticipated pathways, notably the senescence-associated secretory phenotype (SASP). In addition to alterations in these shared pathways, numerous pathways were perturbed in only two of the three cell lines (e.g., chromatin modification, regulation of ribosomal RNA (rRNA) expression, HOX genes, sumolyation) or were cell-type specific, such as glycolysis, Wnt signaling, and mitochondrial translation (Supplementary Table S2). Thus, FPN-mediated iron depletion induces divergent as well as convergent changes in gene expression in prostate cancer cells.
FIG. 9.
FPN overexpression regulates both diverse and converging pathways. Venn diagram depicting the number of differentially expressed Reactome pathways in LNCaP (Tet-FPN), C4-2 (Tet-FPN), and PC3 (Tet-FPN) cells after 48 h of FPN overexpression (1 μg/mL doxycycline). Common pathways differentially expressed in all three cells are shown. See Supplementary Table S2 for details of other differentially expressed pathways. Color images are available online.
FPN inhibits the growth of prostate tumor xenografts in vivo
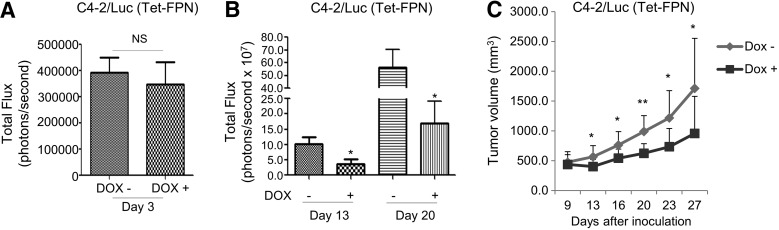
Given the ability of FPN overexpression to induce cell cycle arrest and inhibit proliferation of prostate cancer cells in vitro, we used mouse xenografts to assess the effect of FPN on growth of prostate tumors in vivo. For these experiments we utilized C4-2 adenocarcinoma cells, since they represent a clinically relevant model of hormone refractory disease. To facilitate tracking of tumor cells in vivo, we created a stable C4-2/Luc (Tet-FPN) cell line containing a luciferase reporter. Two days after implantation of C4-2/Luc (Tet-FPN) cells in mice, we measured luciferase activity and divided mice into experimental and control groups with comparable average luciferase activity (Fig. 10A). Doxycycline was added to the drinking water to induce FPN in the experimental group, whereas the control group was provided with drinking water without doxycycline. Tumor growth was monitored over time.
FIG. 10.
FPN inhibits growth of C4-2 tumor xenografts in vivo. (A) Bioluminescence of tumor xenografts formed from C4-2 cells expressing luciferase and doxycycline-inducible FPN (C4-2/luc Tet-FPN) before treatment (measured 2 days after cell inoculation). (B) Bioluminescence of C4-2/luc (Tet-FPN) xenografts in mice treated with and without doxycycline for 13 and 20 days. (C) Tumor volume calculated from caliper measurements in mice treated with and without doxycycline for up to 27 days. Data are presented as mean ± standard error of mean (n = 12), and significant differences are indicated. The experiment was conducted once. NS, p > 0.05; *p < 0.05; **p ≤ 0.01.
In vivo measures of luciferase demonstrated significantly lower activity in the doxycycline-treated mice (Fig. 10B). Consistent with these results, direct measures of tumor volume revealed a substantial inhibition of tumor growth after doxycycline treatment at a series of time points (Fig. 10C). Importantly, FPN remained overexpressed and FTH was decreased in these tumors (Supplementary Fig. S10), consistent with a reduced iron phenotype in tumors harvested from doxycycline-treated mice. As anticipated, doxycycline had no effect on tumor growth in mice implanted with control C4-2/Luc cells (Supplementary Fig. S11). These data indicate that FPN suppresses growth of prostate adenocarcinoma cells in vivo.
Discussion
A large body of evidence suggests that iron is critical to malignant growth, and that cancer cells acquire and retain more iron than their non-malignant counterparts (70). Repression of FPN, an iron efflux pump, is a mechanism of enhanced iron retention that has been observed in prostate cancer cells and tumors (69, 77). The use of iron chelators to deprive cancer cells of this essential nutrient has, therefore, become the subject of clinical interest and has engendered a number of ongoing clinical trials (30). However, fundamental questions remain regarding the pathways triggered by iron depletion, whether similar pathways are activated in different types of cancer cells, and the mechanisms that trigger these pathways.
In this study, we focused on mechanisms triggered by iron depletion in prostate cancer, a major cause of suffering and death in older men. We used cell lines to compare three important histological and biological subtypes of this disease: LNCaP cells, representative of AR positive adenocarcinoma; C4-2 cells, representative of castrate-resistant adenocarcinoma; and PC3 cells, representative of a neuroendocrine carcinoma of the prostate, a rare but highly aggressive malignancy (67). We used overexpression of FPN, an endogenous iron efflux pump, to identify and compare pathways triggered by iron depletion in these cell types.
Manipulation of FPN exerted a substantial effect on iron metabolism in all three prostate cancer cell types. Iron is an essential element required for cellular viability, and iron metabolism is characterized by a delicate feedback system that preserves iron homeostasis. When cells are depleted of iron, IRP activity increases, which, in turn, simultaneously increases levels of the iron import protein TFR1 and decreases levels of the iron storage protein ferritin to restore iron homeostasis (20).
We observed this physiologic feedback response in prostate cancer cells with overexpression of FPN: As seen in Figure 1, expression of FPN activated IRP2, induced TFR1, repressed ferritin, and decreased the LIP. However, the response was insufficient to prevent the antiproliferative effects of FPN overexpression (Fig. 3). Thus, sustained expression of the iron exporter FPN is able to override the controls put in place by the iron regulatory system, and therefore results in cellular iron depletion and growth inhibition. These findings establish FPN as a key lever in regulating iron balance and proliferation in multiple prostate cancer cell types.
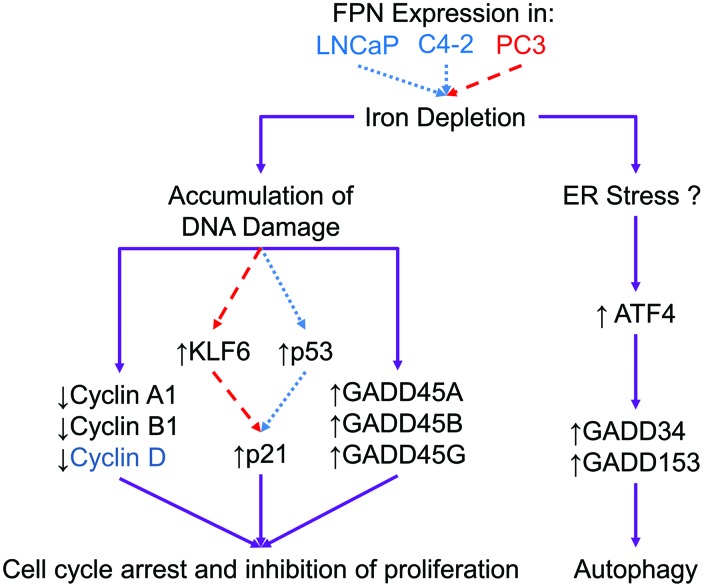
We studied downstream pathways affected by FPN-mediated iron depletion. We observed a profound effect on three stress-response pathways: autophagy, cell cycle arrest, and environmental stress (Fig. 11).
FIG. 11.
Mechanistic model of FPN-mediated induction of cell cycle arrest and autophagy. FPN was overexpressed in biologically diverse prostate cancer cell lines. Cells with functional p53 activate different mechanisms (shown in dotted lines) than cells lacking functional p53 (shown in dashed lines). Despite differing mechanisms, the effects of FPN overexpression in these cells converge (shown in solid lines), resulting in autophagy and cell cycle arrest. FPN overexpression in prostate cancer cells causes intracellular iron depletion. Subsequently, iron depletion results in DNA damage and cell cycle arrest, in part, by repression of cyclin genes (Cyclin A1, Cyclin B1; Cyclin D1 is also repressed in C4-2 and LNCaP cells) and activation of GADD genes (GADD45A, GADD45B, and GADD45G). FPN overexpression also causes cell cycle arrest through induction of p21 by p53, or KLF6 in cells lacking functional p53. FPN-mediated iron depletion induces autophagy and upregulation of ATF4, GADD34, and GADD153, possibly by causing ER stress. ATF4, activating transcription factor 4; ER, endoplasmic reticulum. Color images are available online.
We first tested the effect of FPN overexpression on autophagy. Overexpression of FPN induced autophagy in LNCaP, C4-2, and PC3 cells, eliciting a response similar to that induced by DFO, a potent iron chelator (Fig. 2). Supporting earlier studies (4, 32), recent work has identified a specific cargo receptor for degradation of the iron storage protein ferritin in the autophagolysosome, and suggested that autophagy is important for recycling of intracellular iron (19, 43). Our observation that FPN induces autophagy in all prostate cancer cells tested is consistent with this model, and it further suggests that degradation of subcellular components is a general survival mechanism by which prostate cancer cells attempt to re-allocate iron in response to iron deprivation.
We next investigated the effect of FPN-mediated iron depletion on proliferation and cell cycle progression. In all three prostate cancer cell types, FPN significantly decreased proliferation (Fig. 3). Cell cycle analyses revealed that FPN expression caused an accumulation of cells in G0/G1 (Fig. 5), particularly in the prostate adenocarcinoma cells LNCaP and C4-2. This was associated with an induction of the tumor suppressor p53 and its downstream target p21CIP/WAF1, a key cyclin-dependent kinase inhibitor (1) (Fig. 6 and Supplementary Fig. S6).
Intriguingly, PC3 cells, which do not express functional p53 [(12); Supplementary Fig. S6A], also exhibited an increase in p21 mRNA (Fig. 7), suggesting the existence of an alternative mechanism through which iron depletion induces p21. We observed that in PC3 cells, FPN induced KLF6 (Fig. 7), a tumor suppressor that has been specifically associated with prostate cancer (49). Knockdown of KLF6 prevented FPN-mediated p21 induction, directly implicating KLF6 in p21 induction in PC3 cells (Fig. 7). KLF6 has not previously been linked to iron depletion. Other targets of KLF6 include genes involved in motility, invasion, growth factor signaling, and angiogenesis (17), and it will be of interest to determine to what extent these pathways are activated in iron-depleted cells.
We also examined the effect of FPN overexpression on expression of GADD genes, since these are induced by, and play a role in the response to, nutrient and environmental stress (39). GADD34, GADD45A, GADD45B, GADD45G, and GADD153 were induced in all prostate cancer cells that overexpressed FPN (Fig. 8). Although the GADD genes were named based on their similar routes of discovery (21), subsequent work has revealed that they play very different roles (39).
GADD45 genes are induced by p53, trigger cell cycle arrest, and have a tumor suppressive function, in part through interaction with p21 (8). Induction of GADD45A and GADD45B may, therefore, functionally augment the anti-proliferative activity of p21 in cells expressing FPN. Interestingly, induction of GADD45A was also observed in PC3 cells that do not express functional p53, indicating that iron depletion can induce GADD45A through a mechanism that is p53 independent (Fig. 11). We speculate that an increase in DNA damage (Fig. 8F) may underpin the upregulation of GADD45A and GADD45B gene expression in these cells. Further, since DNA damage induces p53 (44, 63) as well as KLF6 (5), DNA damage may also underlie the induction of p53 (Fig. 6A) and KLF6 (Supplementary Fig. S9) after induction of FPN.
In contrast to the GADD45 genes, GADD34 and GADD153 are upregulated by agents that trigger endoplasmic reticulum (ER) stress, such as nutrient deprivation and protein misfolding, and play a role in rectifying ER stress (24). ER stress can be induced by iron chelation (37) and can trigger autophagy (24, 55), which we observed in all prostate cancer cells we studied (Fig. 2). We, therefore, speculate that FPN-mediated iron depletion may induce an ER stress response, and that this may contribute to the induction of autophagy in prostate cancer cells. Indeed, two markers of ER stress (50), spliced X-box binding protein 1, sXBP1, and activating transcription factor 4 (ATF4), were transcriptionally upregulated after FPN overexpression (Supplementary Fig. S12). ATF4 is a known activator of GADD34 and GADD153 (CHOP) genes (42) and is likely involved in upregulation of these genes after FPN overexpression. This model is presented in Figure 11.
Pathways induced by FPN overexpression are similar to those activated by iron chelators. In addition to their use as medicinal agents, iron chelators have been used to elucidate mechanisms of iron trafficking and regulation. However, like any drug, iron chelators have the potential to exhibit off-target effects, and their ability to abstract intracellular iron in a manner that truly mimics physiological iron depletion has occasionally been questioned (51, 56).
Although we did not directly address this issue, we observed that pathways activated by overexpression of FPN, an endogenous iron efflux pump, show remarkable similarity to pathways triggered in cells treated with iron chelators (41). This includes induction of p53 (41), induction of p21 (41), induction of GADD genes (61), induction of autophagy (54), and inhibition of proliferation (38), all of which have been observed in cells treated with iron chelators. Since FPN is presumably unable to liberate iron from non-physiological targets, our findings not only confirm that upregulation of this efflux pump has far-reaching physiological consequences but also reinforce the notion that iron chelators can be used as effective probes of intracellular iron pathways.
FPN-mediated iron depletion activated different molecular pathways in prostate cancer cells representative of different histopathological subtypes of prostate cancer. For example, in LNCaP cells, FPN overexpression induced p21 via a p53-mediated pathway; whereas in p53 null PC3 cells, KLF6 subsumed this role (Figs. 6 and 7). KLF6 is a tumor suppressor that exhibits both p53-dependent and p53-independent activities. KLF6 suppresses proliferation through p53-independent p21 transactivation (49) and cyclin D1 sequestration (7), but it also stabilizes p53 through repression of mdm2 (68). p53 and KLF6 can also interact directly to control transcription of genes that are important in prostate cancer such as IGFI (59). Thus, the role(s) played by each of these multifaceted tumor suppressors in the response to iron depletion is likely to be multifactorial, and modulated by the extent to which each is expressed in a given tumor.
Although the details of the molecular interaction between KLF6 and p53 in response to iron depletion remain to be elucidated, in our experiments we observed that pathways modulated by these proteins converged on a similar phenotype characterized by G1 arrest, inhibition of DNA synthesis, induction of autophagy, and induction of GADD genes involved in cell cycle arrest (GADD45A, GADD45B, and GADD45G) and ER stress (GADD34 and GADD153).
We performed a microarray analysis to gain additional insight into pathways perturbed by FPN in prostate cancer cells (Fig. 9 and Supplementary Tables S1 and S2). This analysis revealed five pathways that were significantly perturbed by FPN overexpression in all three prostate cancer cell lines tested (Fig. 9). Of these five pathways, four mapped to alterations in the cell cycle or DNA damage (Fig. 9), consistent with other data presented here. However, our microarray analysis also uncovered a new pathway perturbed by FPN overexpression: the SASP, a pathway that has not been previously linked to iron depletion (Fig. 9).
SASP refers to the altered secretome exhibited by senescent cells. It includes survival factors, cytokines, and growth factors (3, 72, 76, 78). SASP is induced by several senescent triggers, including DNA damage (57), consistent with our finding of DNA damage after FPN overexpression. Further, p53 and p21, which are increased after FPN overexpression (Figs. 6 and 7 and Supplementary Fig. S5), play a role in coordinating DNA damage-mediated SASP (2). Interestingly, SASP is known to induce autophagy, which we observed after FPN overexpression (Fig. 2), as well as ER stress (18, 60). Thus, SASP may represent a bridge between induction of DNA damage, autophagy, and ER stress in cells that overexpress FPN. Further work will be required to test this hypothesis.
In addition to the induction of common pathways by FPN overexpression, microarray analysis also revealed cell-type specific responses to FPN, perhaps reflecting the different histopathological origin of the cell types studied. These pathways included glycolysis in LNCaP cells, mitochondrial translation in C4-2 cells, Wnt signaling in PC3 cells, and several others (Supplementary Table S2). The extent to which these reinforce or amplify FPN-mediated effects on cell proliferation will require further investigation. Nevertheless, the profound effects of FPN-mediated iron depletion on a variety of key intracellular targets suggests that iron may be a broadly useful target in the treatment of prostate cancer, despite the wide variability in pathology and clinical course exhibited by prostate cancer subtypes.
FPN-mediated effects on iron metabolism were sufficient to inhibit growth of castrate-resistant prostate tumors in vivo (Fig. 10). This finding underscores the critical contribution of iron metabolism to tumor growth, and in particular demonstrates that manipulation of a single nutrient—iron—is sufficient to dramatically affect tumor growth. It is also notable that therapeutic options for castrate-resistant prostate cancer are limited (28). The ability of iron depletion to effectively retard growth of this prevalent and treatment-refractory tumor type suggests that deeper exploration of anti-tumor strategies targeting iron may prove clinically useful in the treatment of prostate cancer.
Our results also support the further study of iron depletion as an anti-tumor strategy. Synthetic iron chelators represent one strategy for inducing iron depletion that has shown considerable promise (27), and remains an area of active investigation (6). Our work suggests that other approaches to targeting tumor iron dependence, such as FPN agonists, may also merit exploration. For example, pharmacological agents that inhibit hepcidin, a negative regulator of FPN, might be modified and repurposed for use in cancer therapy. The recent interest in the development of multiple hepcidin inhibitors may accelerate this process (10).
Materials and Methods
Cells and cell culture
The prostate cancer cell lines LNCaP and PC3 were obtained from American Type Culture Collection (ATCC). Cells were grown in RPMI medium (GIBCO Life Technologies, Gaithersburg, MD) supplemented with 10% fetal bovine serum (FBS; Gemini) and incubated at 37°C in a humidified atmosphere with 5% CO2. The C4-2 cell line was a gift of Dr. Robert Sikes at the University of Delaware. The cell lines were authenticated by ATCC.
DNA vectors, viral package, and infection
A human FPN complementary DNA clone was obtained from Open Biosystems (Biosystems/GE Dharmacon, Lafayette, CO), and it was used as a polymerase chain reaction (PCR) template to introduce FPN into pSL2 lentiviral overexpression vector (16) and pLVX-TetOne-puro, a lentiviral tetracycline (tet) inducible vector (Clontech Laboratories, Mountain View, CA). The FPN A77D mutant was created by using site-directed mutagenesis and cloned into pSL2 and pLVX-TetOne-puro vectors.
The retroviral construct pBABE-puro-mCherry-EGFP-LC3B, an autophagic reporter (48), was obtained from Addgene (Catalog No. 22418). The LUC2 gene (from the pGL4 vector [Promega Co., Madison, WI]) and zeocin resistance gene (from the pcDNA4/His vector [Invitrogen, Carlsbad, CA]) were subcloned into the pLKO.1-shCtr vector (Addgene Catalog No. 10879) (47) to create a luciferase lentiviral expression vector. To do this, the short hairpin RNA (shRNA) expression cassette was first deleted. Next, the puromycin resistance gene was replaced with the LUC2 coding region, an internal ribosome entry site, and the zeocin resistance gene. These modifications resulted in expression of luciferase and zeocin driven by the original phosphoglycerate kinase promoter.
Lentiviral shRNAs against KLF6 gene were obtained from Sigma-Aldrich (St. Louis, MO). To prepare lentiviral particles, the lentiviral expression vector was cotransfected with third-generation packaging plasmids (pVSV-G, pRSV-REV, and pMDLg/pRRE) into 293FT cells as previously described (15). Lentiviral particles were concentrated by ultracentrifugation and incubated with cells in the presence of polybrene (8 μg/mL). Retrovirus was prepared by transfecting GP2-293 cells (Catalog No. 631458; Clontech Laboratories, Inc., Palo Alto, CA) with pBABE-mCherry-EGFP-LC3B vector and the packaging plasmid PCL10A-1. Three days after transfection, virus-containing medium was harvested and filtered with 0.45 μM filter. Retroviral infection was performed by adding virus-containing medium to cells grown in six-well plates. A stably infected cell population was acquired by fluorescence-activated cell sorting, FACS, on BD FacsAria II.
Cell cycle analysis
Cells were harvested by trypsinization and washed twice with pre-chilled wash buffer (1 × phosphate-buffered saline [PBS] containing 0.1% FBS). After centrifugation, cells were resuspended at about 1 × 106 cells/mL, fixed by dropwise addition of three volumes of cold (−20°C) absolute ethanol while vortexing, and stored at −20°C for at least 1 h. After washing twice with PBS, fixed cells were stained with FxCycle™ PI/RNase Staining Solution (Catalog No. F10797; Life Technologies, Carlsbad, CA) according to the manufacturer's instruction. Cell cycle analysis was performed by using MACSQuant flow cytometer (Miltenyi Biotec, Bergisch Gladbach, Germany). The data were analyzed with ModFit LT 3.0 software (Verity Software House, Topsham, ME).
Cell growth assay
Cell proliferation assays were conducted as previously described (73). To determine whether FPN-mediated inhibition of cell proliferation was permanent or transient, cells were first treated with 1 μg/mL doxycycline for 2 days. The media were then replaced with doxycycline-free media for 6 days. Cells were then replated at 10,000 cells per well, and cell proliferation was measured by counting cells with a hemocytometer after 3, 5, and 7 days. As a control, cell proliferation was measured at the same time in cells that had not been pre-treated with doxycycline.
Cell surface localization of FPN and A77D
Cell surface proteins were determined by flow cytometry. Antibodies used were Ab-FPN 38C8 (Amgen) (58) and Alexa Fluor® 488 AffiniPure Goat Anti-Human IgG (Catalog No. 109-545-098; Jackson ImmunoResearch Laboratories, West Grove, PA).
BrdU incorporation assay
BrdU incorporation was measured by using an FITC BrdU flow kit (Catalog No. 559619; BD Biosciences, San Diego, CA) by essentially following the manufacturer's protocol, except that labeling was performed only with BrdU and nuclear DNA was not further stained. The labeled cell populations were analyzed by flow cytometry.
LIP assay
The labile iron level in cells was determined by calcein-acetoxymethyl ester (CA-AM)-based measurement as previously described (53).
Immunofluorescence imaging
Cells were plated on an 8-chamber slide (BD Falcon) and treated with ±1 μg/mL doxycycline (Sigma-Aldrich) for 72 h or with 10 μM cisplatin (cis-Diamineplatinum(II) dichloride, 479306; Sigma-Aldrich). Cells were fixed with 4% paraformaldehyde for 15 min and blocked with 5% bovine serum albumin at 4°C for 2 h. Anti-human phospho-histone H2A.X (9718S; Cell Signaling) was applied overnight followed by Alexa Fluor 488-conjugated secondary antibody (ab150077; Abcam) incubation for 1 h. Coverslips were mounted to chamber slides with ProLong™ Gold Antifade reagent with DAPI (P36935; Life Technologies). Fluorescent images were acquired by using an Axio Vert.A1 microscope (Zeiss) and ZEN software (Zeiss). Nuclear γH2AX foci were detected by using Fuji.
Real-time PCR and Western blotting
Total RNA was extracted by using the High Pure RNA Isolation Kit (Roche), and the concentration of RNA was quantified with a Nanodrop ND-2000 spectrophotometer (Thermo Scientific, Rockford, IL). Taqman reverse transcription kit (Thermofisher Scientific) was used for reverse transcription. Amplification was performed on an Applied Biosystems ViiA™ 7 Real-Time PCR system. Primers used in the real-time PCR are detailed in Supplementary Table S3.
To detect FPN by Western blotting, protein samples were prepared in sodium dodecyl sulfate (SDS) reducing buffer without boiling and run on 10% SDS polyacrylamide gel. Antibodies used in Western blotting were glyceraldehyde-3-phosphate dehydrogenase (GAPDH; Fitzgerald), FPN (Amgen), TfR1 (Invitrogen-Life Technologies), FTH (73), IRP1 (Medimabs, Montreal, Canada), IRP2 (Santa Cruz Biotechnology, Inc., Santa Cruz, CA), KLF6 (Thermofisher Scientific), and LC3B (Novus Biological, Littleton, CO). Blots shown in the figures were cropped for clarity. Uncropped blots are shown in Supplementary Figures S1, S3–S5, S8, and S13.
Xenograft experiments
All animal procedures were approved by the UConn Health Institutional Animal Care and Use Committee (IACUC). C4-2/Luc cells were generated by infection with a LUC2 lentiviral expression vector. C4-2/Luc (Tet-FPN) cells were generated by infection with a LUC2 lentiviral expression vector in which a tet-on FPN expression cassette had been introduced through infection with pLVX-TetOne-puro-FPN lentivirus. Cells were subcutaneously injected into the lower flank of 8-week-old male immunodeficient nude mice [Crl:NU(NCr)-Foxn1nu; Charles River; 2 × 106 cells/mouse]. The mice were divided into mock and doxycycline-treated groups (8–12 mice per group). They were provided with water containing 4% sucrose with or without 2 mg/ml doxycycline 3 days after cell implantation until the termination of the experiment at 6 weeks. Tumor size was measured twice a week by using digital calipers. In vivo bioluminescent imaging was performed on an IVIS imaging system (PerkinElmer) and analyzed by using Living Image software (version 4.4).
Microarray analysis
LNCaP (Tet-Vec), LNCaP (Tet-FPN), C4-2 (Tet-FPN), and PC3 (Tet-FPN) cells were treated with ±1 μg/mL doxycycline for 48 h. RNA was isolated from harvested cells by using the High Pure RNA Isolation Kit (11828665001; Roche). Three biological replicates were sent to the Yale Center for Genome Analysis (Yale University, West Haven, CT) for analysis on human Clariom D microarray chips (902923; Thermofisher Scientific). Data were normalized by using Expression Console software (version 1.4.1.46; Affymetrix). Differential expression analysis was performed in R with the Bioconductor package limma. A gene was considered differentially expressed if the FDR <0.05. Pathway analysis was performed in R with the Bioconductor package ReactomePA (79). Differentially expressed genes in LNCaP (Tet-Vec) cells were removed from pathway analysis of Tet-FPN cells if the direction of differential expression was consistent between Tet-Vec and all Tet-FPN cell lines. Data analysis was limited to fully annotated genes.
Statistics
All in vitro experiments were performed independently at least three times. Data used for quantitative analyses were collected from representative triplicate experiments, which are presented as mean ± standard deviation, unless indicated otherwise. Student's t-test was used to test for a significant difference between the means in two independent groups by using GraphPad Prism software (Graphpad Software, Inc., San Diego, CA). A p-value <0.05 was considered statistically significant.
Supplementary Material
Acknowledgments
The authors are grateful to laboratory members Dr. David Lemler, PhD and Erica Lemler for their generous assistance in measurements of the LIP and tumor growth. This work was supported in part by grants F30 DE026380 and T90 DE021989 from the NIDCR (D.H.M.), and NCI grants R01 CA188025 (S.V.T), and R01 CA171101 (F.M.T).
Abbreviations Used
- AR
androgen receptor
- ATCC
American Type Culture Collection
- ATF4
activating transcription factor 4
- BrdU
5-bromo-2′-deoxy
- DFO
desferoxamine, an iron chelator
- EGFP
enhanced green fluorescent protein
- ER
endoplasmic reticulum
- FBS
fetal bovine serum
- FDR
false discovery rate
- FPN
ferroportin
- FPN A77D
cells constitutively overexpressing a functionally impaired mutant FPN driven by the cytomegalovirus promoter
- FPN OE cells
cells constitutively overexpressing FPN driven by the cytomegalovirus promoter
- FTH
ferritin H
- GADD
growth arrest DNA damage proteins
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- IRP1
iron regulatory protein 1
- IRP2
iron regulatory protein 2
- KLF6
Kruppel-like factor 6
- LC3B-I
microtubule-associated protein light chain 3 beta
- LC3B-II
phosphatidylethanolamine-conjugated microtubule-associated protein light chain 3 beta
- LIP
labile iron pool
- Luc
luciferase
- mRNA
messenger RNA
- p21
cyclin-dependent kinase inhibitor 1A
- p53
tumor protein p53
- PBS
phosphate-buffered saline
- PCR
polymerase chain reaction
- RT-qPCR
quantitative reverse transcription polymerase chain reaction
- SASP
senescence-associated secretory phenotype
- SCNC
small cell (neuroendocrine) carcinoma
- SDS
sodium dodecyl sulfate
- shRNA
short hairpin RNA
- (Tet-FPN)
cells infected with a tetracycline-inducible FPN
- (Tet-Vec)
cells infected with a tetracycline-inducible vector control
- TfR1
transferrin receptor 1
- WST-1
water-soluble tetrazolium salt-1
Author Disclosure Statement
No competing financial interests exist.
References
- 1. Abbas T. and Dutta A. p21 in cancer: intricate networks and multiple activities. Nat Rev Cancer 9: 400–414, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Acosta JC, Banito A, Wuestefeld T, Georgilis A, Janich P, Morton JP, Athineos D, Kang TW, Lasitschka F, Andrulis M, Pascual G, Morris KJ, Khan S, Jin H, Dharmalingam G, Snijders AP, Carroll T, Capper D, Pritchard C, Inman GJ, Longerich T, Sansom OJ, Benitah SA, Zender L, and Gil J. A complex secretory program orchestrated by the inflammasome controls paracrine senescence. Nat Cell Biol 15: 978–990, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Acosta JC, O'Loghlen A, Banito A, Guijarro MV, Augert A, Raguz S, Fumagalli M, Da Costa M, Brown C, Popov N, Takatsu Y, Melamed J, d'Adda di Fagagna F, Bernard D, Hernando E, and Gil J. Chemokine signaling via the CXCR2 receptor reinforces senescence. Cell 133: 1006–1018, 2008 [DOI] [PubMed] [Google Scholar]
- 4. Asano T, Komatsu M, Yamaguchi-Iwai Y, Ishikawa F, Mizushima N, and Iwai K. Distinct mechanisms of ferritin delivery to lysosomes in iron-depleted and iron-replete cells. Mol Cell Biol 31: 2040–2052, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Banck MS, Beaven SW, Narla G, Walsh MJ, Friedman SL, and Beutler AS. KLF6 degradation after apoptotic DNA damage. FEBS Lett 580: 6981–6986, 2006 [DOI] [PubMed] [Google Scholar]
- 6. Basu A, Sohn YS, Alyan M, Nechushtai R, Domb AJ, and Goldblum A. Discovering novel and diverse iron-chelators in silico. J Chem Inf Model 56: 2476–2485, 2016 [DOI] [PubMed] [Google Scholar]
- 7. Benzeno S, Narla G, Allina J, Cheng GZ, Reeves HL, Banck MS, Odin JA, Diehl JA, Germain D, and Friedman SL. Cyclin-dependent kinase inhibition by the KLF6 tumor suppressor protein through interaction with cyclin D1. Cancer Res 64: 3885–3891, 2004 [DOI] [PubMed] [Google Scholar]
- 8. Berkers CR, Maddocks OD, Cheung EC, Mor I, and Vousden KH. Metabolic regulation by p53 family members. Cell Metab 18: 617–633, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bieging KT, Mello SS, and Attardi LD. Unravelling mechanisms of p53-mediated tumour suppression. Nat Rev Cancer 14: 359–370, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Blanchette NL, Manz DH, Torti FM, and Torti SV. Modulation of hepcidin to treat iron deregulation: potential clinical applications. Expert Rev Hematol 9: 169–186, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bostwick DG. The pathology of early prostate cancer. CA Cancer J Clin 39: 376–393, 1989 [DOI] [PubMed] [Google Scholar]
- 12. Carroll AG, Voeller HJ, Sugars L, and Gelmann EP. p53 Oncogene mutations in three human prostate cancer cell lines. Prostate 23: 123–134, 1993 [DOI] [PubMed] [Google Scholar]
- 13. Culig Z, Hobisch A, Bartsch G, and Klocker H. Androgen receptor—an update of mechanisms of action in prostate cancer. Urol Res 28: 211–219, 2000 [DOI] [PubMed] [Google Scholar]
- 14. Darnell G, and Richardson DR. The potential of iron chelators of the pyridoxal isonicotinoyl hydrazone class as effective antiproliferative agents III: the effect of the ligands on molecular targets involved in proliferation. Blood 94: 781–792, 1999 [PubMed] [Google Scholar]
- 15. Deng Z, Sui G, Rosa PM, and Zhao W. Radiation-induced c-Jun activation depends on MEK1-ERK1/2 signaling pathway in microglial cells. PLoS One 7: e36739, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Deng Z, Wan M, Cao P, Rao A, Cramer SD, and Sui G. Yin Yang 1 regulates the transcriptional activity of androgen receptor. Oncogene 28: 3746–3757, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. DiFeo A, Martignetti JA, and Narla G. The role of KLF6 and its splice variants in cancer therapy. Drug Resist Updat 12: 1–7, 2009 [DOI] [PubMed] [Google Scholar]
- 18. Dorr JR, Yu Y, Milanovic M, Beuster G, Zasada C, Dabritz JH, Lisec J, Lenze D, Gerhardt A, Schleicher K, Kratzat S, Purfurst B, Walenta S, Mueller-Klieser W, Graler M, Hummel M, Keller U, Buck AK, Dorken B, Willmitzer L, Reimann M, Kempa S, Lee S, and Schmitt CA. Synthetic lethal metabolic targeting of cellular senescence in cancer therapy. Nature 501: 421–425, 2013 [DOI] [PubMed] [Google Scholar]
- 19. Dowdle WE, Nyfeler B, Nagel J, Elling RA, Liu S, Triantafellow E, Menon S, Wang Z, Honda A, Pardee G, Cantwell J, Luu C, Cornella-Taracido I, Harrington E, Fekkes P, Lei H, Fang Q, Digan ME, Burdick D, Powers AF, Helliwell SB, D'Aquin S, Bastien J, Wang H, Wiederschain D, Kuerth J, Bergman P, Schwalb D, Thomas J, Ugwonali S, Harbinski F, Tallarico J, Wilson CJ, Myer VE, Porter JA, Bussiere DE, Finan PM, Labow MA, Mao X, Hamann LG, Manning BD, Valdez RA, Nicholson T, Schirle M, Knapp MS, Keaney EP, and Murphy LO. Selective VPS34 inhibitor blocks autophagy and uncovers a role for NCOA4 in ferritin degradation and iron homeostasis in vivo. Nat Cell Biol 16: 1069–1079, 2014 [DOI] [PubMed] [Google Scholar]
- 20. Eisenstein RS. Iron regulatory proteins and the molecular control of mammalian iron metabolism. Annu Rev Nutr 20: 627–662, 2000 [DOI] [PubMed] [Google Scholar]
- 21. Fornace AJ, Jr., Jackman J, Hollander MC, Hoffman-Liebermann B, and Liebermann DA. Genotoxic-stress-response genes and growth-arrest genes. gadd, MyD, and other genes induced by treatments eliciting growth arrest. Ann N Y Acad Sci 663: 139–153, 1992 [DOI] [PubMed] [Google Scholar]
- 22. Fornace AJ, Jr., Nebert DW, Hollander MC, Luethy JD, Papathanasiou M, Fargnoli J, and Holbrook NJ. Mammalian genes coordinately regulated by growth arrest signals and DNA-damaging agents. Mol Cell Biol 9: 4196–4203, 1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ganz T. Cellular iron: ferroportin is the only way out. Cell Metab 1: 155–157, 2005 [DOI] [PubMed] [Google Scholar]
- 24. Gump JM. and Thorburn A. Sorting cells for basal and induced autophagic flux by quantitative ratiometric flow cytometry. Autophagy 10: 1327–1334, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gutierrez E, Richardson DR, and Jansson PJ. The anticancer agent di-2-pyridylketone 4,4-dimethyl-3-thiosemicarbazone (Dp44mT) overcomes prosurvival autophagy by two mechanisms: persistent induction of autophagosome synthesis and impairment of lysosomal integrity. J Biol Chem 289: 33568–33589, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gwinn DM, Shackelford DB, Egan DF, Mihaylova MM, Mery A, Vasquez DS, Turk BE, and Shaw RJ. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol Cell 30: 214–226, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hatcher HC, Singh RN, Torti FM, and Torti SV. Synthetic and natural iron chelators: therapeutic potential and clinical use. Future Med Chem 1: 1643–1670, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hotte SJ. and Saad F. Current management of castrate-resistant prostate cancer. Curr Oncol 17 Suppl 2: S72–S79, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Johnson DG. and Walker CL. Cyclins and cell cycle checkpoints. Annu Rev Pharmacol Toxicol 39: 295–312, 1999 [DOI] [PubMed] [Google Scholar]
- 30. Kalinowski DS, Stefani C, Toyokuni S, Ganz T, Anderson GJ, Subramaniam NV, Trinder D, Olynyk JK, Chua A, Jansson PJ, Sahni S, Lane DJ, Merlot AM, Kovacevic Z, Huang ML, Lee CS, and Richardson DR. Redox cycling metals: pedaling their roles in metabolism and their use in the development of novel therapeutics. Biochim Biophys Acta 1863: 727–748, 2016 [DOI] [PubMed] [Google Scholar]
- 31. Kastan MB, Onyekwere O, Sidransky D, Vogelstein B, and Craig RW. Participation of p53 protein in the cellular response to DNA damage. Cancer Res 51: 6304–6311, 1991 [PubMed] [Google Scholar]
- 32. Kidane TZ, Sauble E, and Linder MC. Release of iron from ferritin requires lysosomal activity. Am J Physiol Cell Physiol 291: C445–C455, 2006 [DOI] [PubMed] [Google Scholar]
- 33. Kimura S, Noda T, and Yoshimori T. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy 3: 452–460, 2007 [DOI] [PubMed] [Google Scholar]
- 34. Klionsky DJ, Abdalla FC, Abeliovich H, Abraham RT, Acevedo-Arozena A, Adeli K, Agholme L, Agnello M, Agostinis P, Aguirre-Ghiso JA, Ahn HJ, Ait-Mohamed O, Ait-Si-Ali S, Akematsu T, Akira S, Al-Younes HM, Al-Zeer MA, Albert ML, Albin RL, Alegre-Abarrategui J, Aleo MF, Alirezaei M, Almasan A, Almonte-Becerril M, Amano A, Amaravadi R, Amarnath S, Amer AO, Andrieu-Abadie N, Anantharam V, Ann DK, Anoopkumar-Dukie S, Aoki H, Apostolova N, Arancia G, Aris JP, Asanuma K, Asare NY, Ashida H, Askanas V, Askew DS, Auberger P, Baba M, Backues SK, Baehrecke EH, Bahr BA, Bai XY, Bailly Y, Baiocchi R, Baldini G, Balduini W, Ballabio A, Bamber BA, Bampton ET, Banhegyi G, Bartholomew CR, Bassham DC, Bast RC, Jr., Batoko H, Bay BH, Beau I, Bechet DM, Begley TJ, Behl C, Behrends C, Bekri S, Bellaire B, Bendall LJ, Benetti L, Berliocchi L, Bernardi H, Bernassola F, Besteiro S, Bhatia-Kissova I, Bi X, Biard-Piechaczyk M, Blum JS, Boise LH, Bonaldo P, Boone DL, Bornhauser BC, Bortoluci KR, Bossis I, Bost F, Bourquin JP, Boya P, Boyer-Guittaut M, Bozhkov PV, Brady NR, Brancolini C, Brech A, Brenman JE, Brennand A, Bresnick EH, Brest P, Bridges D, Bristol ML, Brookes PS, Brown EJ, Brumell JH, Brunetti-Pierri N, Brunk UT, Bulman DE, Bultman SJ, Bultynck G, Burbulla LF, Bursch W, Butchar JP, Buzgariu W, Bydlowski SP, Cadwell K, Cahova M, Cai D, Cai J, Cai Q, Calabretta B, Calvo-Garrido J, Camougrand N, Campanella M, Campos-Salinas J, Candi E, Cao L, Caplan AB, Carding SR, Cardoso SM, Carew JS, Carlin CR, Carmignac V, Carneiro LA, Carra S, Caruso RA, Casari G, Casas C, Castino R, Cebollero E, Cecconi F, Celli J, Chaachouay H, Chae HJ, Chai CY, Chan DC, Chan EY, Chang RC, Che CM, Chen CC, Chen GC, Chen GQ, Chen M, Chen Q, Chen SS, Chen W, Chen X, Chen X, Chen X, Chen YG, Chen Y, Chen Y, Chen YJ, Chen Z, Cheng A, Cheng CH, Cheng Y, Cheong H, Cheong JH, Cherry S, Chess-Williams R, Cheung ZH, Chevet E, Chiang HL, Chiarelli R, Chiba T, Chin LS, Chiou SH, Chisari FV, Cho CH, Cho DH, Choi AM, Choi D, Choi KS, Choi ME, Chouaib S, Choubey D, Choubey V, Chu CT, Chuang TH, Chueh SH, Chun T, Chwae YJ, Chye ML, Ciarcia R, Ciriolo MR, Clague MJ, Clark RS, Clarke PG, Clarke R, Codogno P, Coller HA, Colombo MI, Comincini S, Condello M, Condorelli F, Cookson MR, Coombs GH, Coppens I, Corbalan R, Cossart P, Costelli P, Costes S, Coto-Montes A, Couve E, Coxon FP, Cregg JM, Crespo JL, Cronje MJ, Cuervo AM, Cullen JJ, Czaja MJ, D'Amelio M, Darfeuille-Michaud A, Davids LM, Davies FE, De Felici M, de Groot JF, de Haan CA, De Martino L, De Milito A, De Tata V, Debnath J, Degterev A, Dehay B, Delbridge LM, Demarchi F, Deng YZ, Dengjel J, Dent P, Denton D, Deretic V, Desai SD, Devenish RJ, Di Gioacchino M, Di Paolo G, Di Pietro C, Diaz-Araya G, Diaz-Laviada I, Diaz-Meco MT, Diaz-Nido J, Dikic I, Dinesh-Kumar SP, Ding WX, Distelhorst CW, Diwan A, Djavaheri-Mergny M, Dokudovskaya S, Dong Z, Dorsey FC, Dosenko V, Dowling JJ, Doxsey S, Dreux M, Drew ME, Duan Q, Duchosal MA, Duff K, Dugail I, Durbeej M, Duszenko M, Edelstein CL, Edinger AL, Egea G, Eichinger L, Eissa NT, Ekmekcioglu S, El-Deiry WS, Elazar Z, Elgendy M, Ellerby LM, Eng KE, Engelbrecht AM, Engelender S, Erenpreisa J, Escalante R, Esclatine A, Eskelinen EL, Espert L, Espina V, Fan H, Fan J, Fan QW, Fan Z, Fang S, Fang Y, Fanto M, Fanzani A, Farkas T, Farre JC, Faure M, Fechheimer M, Feng CG, Feng J, Feng Q, Feng Y, Fesus L, Feuer R, Figueiredo-Pereira ME, Fimia GM, Fingar DC, Finkbeiner S, Finkel T, Finley KD, Fiorito F, Fisher EA, Fisher PB, Flajolet M, Florez-McClure ML, Florio S, Fon EA, Fornai F, Fortunato F, Fotedar R, Fowler DH, Fox HS, Franco R, Frankel LB, Fransen M, Fuentes JM, Fueyo J, Fujii J, Fujisaki K, Fujita E, Fukuda M, Furukawa RH, Gaestel M, Gailly P, Gajewska M, Galliot B, Galy V, Ganesh S, Ganetzky B, Ganley IG, Gao FB, Gao GF, Gao J, Garcia L, Garcia-Manero G, Garcia-Marcos M, Garmyn M, Gartel AL, Gatti E, Gautel M, Gawriluk TR, Gegg ME, Geng J, Germain M, Gestwicki JE, Gewirtz DA, Ghavami S, Ghosh P, Giammarioli AM, Giatromanolaki AN, Gibson SB, Gilkerson RW, Ginger ML, Ginsberg HN, Golab J, Goligorsky MS, Golstein P, Gomez-Manzano C, Goncu E, Gongora C, Gonzalez CD, Gonzalez R, Gonzalez-Estevez C, Gonzalez-Polo RA, Gonzalez-Rey E, Gorbunov NV, Gorski S, Goruppi S, Gottlieb RA, Gozuacik D, Granato GE, Grant GD, Green KN, Gregorc A, Gros F, Grose C, Grunt TW, Gual P, Guan JL, Guan KL, Guichard SM, Gukovskaya AS, Gukovsky I, Gunst J, Gustafsson AB, Halayko AJ, Hale AN, Halonen SK, Hamasaki M, Han F, Han T, Hancock MK, Hansen M, Harada H, Harada M, Hardt SE, Harper JW, Harris AL, Harris J, Harris SD, Hashimoto M, Haspel JA, Hayashi S, Hazelhurst LA, He C, He YW, Hebert MJ, Heidenreich KA, Helfrich MH, Helgason GV, Henske EP, Herman B, Herman PK, Hetz C, Hilfiker S, Hill JA, Hocking LJ, Hofman P, Hofmann TG, Hohfeld J, Holyoake TL, Hong MH, Hood DA, Hotamisligil GS, Houwerzijl EJ, Hoyer-Hansen M, Hu B, Hu CA, Hu HM, Hua Y, Huang C, Huang J, Huang S, Huang WP, Huber TB, Huh WK, Hung TH, Hupp TR, Hur GM, Hurley JB, Hussain SN, Hussey PJ, Hwang JJ, Hwang S, Ichihara A, Ilkhanizadeh S, Inoki K, Into T, Iovane V, Iovanna JL, Ip NY, Isaka Y, Ishida H, Isidoro C, Isobe K, Iwasaki A, Izquierdo M, Izumi Y, Jaakkola PM, Jaattela M, Jackson GR, Jackson WT, Janji B, Jendrach M, Jeon JH, Jeung EB, Jiang H, Jiang H, Jiang JX, Jiang M, Jiang Q, Jiang X, Jiang X, Jimenez A, Jin M, Jin S, Joe CO, Johansen T, Johnson DE, Johnson GV, Jones NL, Joseph B, Joseph SK, Joubert AM, Juhasz G, Juillerat-Jeanneret L, Jung CH, Jung YK, Kaarniranta K, Kaasik A, Kabuta T, Kadowaki M, Kagedal K, Kamada Y, Kaminskyy VO, Kampinga HH, Kanamori H, Kang C, Kang KB, Kang KI, Kang R, Kang YA, Kanki T, Kanneganti TD, Kanno H, Kanthasamy AG, Kanthasamy A, Karantza V, Kaushal GP, Kaushik S, Kawazoe Y, Ke PY, Kehrl JH, Kelekar A, Kerkhoff C, Kessel DH, Khalil H, Kiel JA, Kiger AA, Kihara A, Kim DR, Kim DH, Kim DH, Kim EK, Kim HR, Kim JS, Kim JH, Kim JC, Kim JK, Kim PK, Kim SW, Kim YS, Kim Y, Kimchi A, Kimmelman AC, King JS, Kinsella TJ, Kirkin V, Kirshenbaum LA, Kitamoto K, Kitazato K, Klein L, Klimecki WT, Klucken J, Knecht E, Ko BC, Koch JC, Koga H, Koh JY, Koh YH, Koike M, Komatsu M, Kominami E, Kong HJ, Kong WJ, Korolchuk VI, Kotake Y, Koukourakis MI, Kouri Flores JB, Kovacs AL, Kraft C, Krainc D, Kramer H, Kretz-Remy C, Krichevsky AM, Kroemer G, Kruger R, Krut O, Ktistakis NT, Kuan CY, Kucharczyk R, Kumar A, Kumar R, Kumar S, Kundu M, Kung HJ, Kurz T, Kwon HJ, La Spada AR, Lafont F, Lamark T, Landry J, Lane JD, Lapaquette P, Laporte JF, Laszlo L, Lavandero S, Lavoie JN, Layfield R, Lazo PA, Le W, Le Cam L, Ledbetter DJ, Lee AJ, Lee BW, Lee GM, Lee J, Lee JH, Lee M, Lee MS, Lee SH, Leeuwenburgh C, Legembre P, Legouis R, Lehmann M, Lei HY, Lei QY, Leib DA, Leiro J, Lemasters JJ, Lemoine A, Lesniak MS, Lev D, Levenson VV, Levine B, Levy E, Li F, Li JL, Li L, Li S, Li W, Li XJ, Li YB, Li YP, Liang C, Liang Q, Liao YF, Liberski PP, Lieberman A, Lim HJ, Lim KL, Lim K, Lin CF, Lin FC, Lin J, Lin JD, Lin K, Lin WW, Lin WC, Lin YL, Linden R, Lingor P, Lippincott-Schwartz J, Lisanti MP, Liton PB, Liu B, Liu CF, Liu K, Liu L, Liu QA, Liu W, Liu YC, Liu Y, Lockshin RA, Lok CN, Lonial S, Loos B, Lopez-Berestein G, Lopez-Otin C, Lossi L, Lotze MT, Low P, Lu B, Lu B, Lu B, Lu Z, Luciano F, Lukacs NW, Lund AH, Lynch-Day MA, Ma Y, Macian F, MacKeigan JP, Macleod KF, Madeo F, Maiuri L, Maiuri MC, Malagoli D, Malicdan MC, Malorni W, Man N, Mandelkow EM, Manon S, Manov I, Mao K, Mao X, Mao Z, Marambaud P, Marazziti D, Marcel YL, Marchbank K, Marchetti P, Marciniak SJ, Marcondes M, Mardi M, Marfe G, Marino G, Markaki M, Marten MR, Martin SJ, Martinand-Mari C, Martinet W, Martinez-Vicente M, Masini M, Matarrese P, Matsuo S, Matteoni R, Mayer A, Mazure NM, McConkey DJ, McConnell MJ, McDermott C, McDonald C, McInerney GM, McKenna SL, McLaughlin B, McLean PJ, McMaster CR, McQuibban GA, Meijer AJ, Meisler MH, Melendez A, Melia TJ, Melino G, Mena MA, Menendez JA, Menna-Barreto RF, Menon MB, Menzies FM, Mercer CA, Merighi A, Merry DE, Meschini S, Meyer CG, Meyer TF, Miao CY, Miao JY, Michels PA, Michiels C, Mijaljica D, Milojkovic A, Minucci S, Miracco C, Miranti CK, Mitroulis I, Miyazawa K, Mizushima N, Mograbi B, Mohseni S, Molero X, Mollereau B, Mollinedo F, Momoi T, Monastyrska I, Monick MM, Monteiro MJ, Moore MN, Mora R, Moreau K, Moreira PI, Moriyasu Y, Moscat J, Mostowy S, Mottram JC, Motyl T, Moussa CE, Muller S, Muller S, Munger K, Munz C, Murphy LO, Murphy ME, Musaro A, Mysorekar I, Nagata E, Nagata K, Nahimana A, Nair U, Nakagawa T, Nakahira K, Nakano H, Nakatogawa H, Nanjundan M, Naqvi NI, Narendra DP, Narita M, Navarro M, Nawrocki ST, Nazarko TY, Nemchenko A, Netea MG, Neufeld TP, Ney PA, Nezis IP, Nguyen HP, Nie D, Nishino I, Nislow C, Nixon RA, Noda T, Noegel AA, Nogalska A, Noguchi S, Notterpek L, Novak I, Nozaki T, Nukina N, Nurnberger T, Nyfeler B, Obara K, Oberley TD, Oddo S, Ogawa M, Ohashi T, Okamoto K, Oleinick NL, Oliver FJ, Olsen LJ, Olsson S, Opota O, Osborne TF, Ostrander GK, Otsu K, Ou JH, Ouimet M, Overholtzer M, Ozpolat B, Paganetti P, Pagnini U, Pallet N, Palmer GE, Palumbo C, Pan T, Panaretakis T, Pandey UB, Papackova Z, Papassideri I, Paris I, Park J, Park OK, Parys JB, Parzych KR, Patschan S, Patterson C, Pattingre S, Pawelek JM, Peng J, Perlmutter DH, Perrotta I, Perry G, Pervaiz S, Peter M, Peters GJ, Petersen M, Petrovski G, Phang JM, Piacentini M, Pierre P, Pierrefite-Carle V, Pierron G, Pinkas-Kramarski R, Piras A, Piri N, Platanias LC, Poggeler S, Poirot M, Poletti A, Pous C, Pozuelo-Rubio M, Praetorius-Ibba M, Prasad A, Prescott M, Priault M, Produit-Zengaffinen N, Progulske-Fox A, Proikas-Cezanne T, Przedborski S, Przyklenk K, Puertollano R, Puyal J, Qian SB, Qin L, Qin ZH, Quaggin SE, Raben N, Rabinowich H, Rabkin SW, Rahman I, Rami A, Ramm G, Randall G, Randow F, Rao VA, Rathmell JC, Ravikumar B, Ray SK, Reed BH, Reed JC, Reggiori F, Regnier-Vigouroux A, Reichert AS, Reiners JJ, Jr., Reiter RJ, Ren J, Revuelta JL, Rhodes CJ, Ritis K, Rizzo E, Robbins J, Roberge M, Roca H, Roccheri MC, Rocchi S, Rodemann HP, Rodriguez de Cordoba S, Rohrer B, Roninson IB, Rosen K, Rost-Roszkowska MM, Rouis M, Rouschop KM, Rovetta F, Rubin BP, Rubinsztein DC, Ruckdeschel K, Rucker EB, 3rd, Rudich A, Rudolf E, Ruiz-Opazo N, Russo R, Rusten TE, Ryan KM, Ryter SW, Sabatini DM, Sadoshima J, Saha T, Saitoh T, Sakagami H, Sakai Y, Salekdeh GH, Salomoni P, Salvaterra PM, Salvesen G, Salvioli R, Sanchez AM, Sanchez-Alcazar JA, Sanchez-Prieto R, Sandri M, Sankar U, Sansanwal P, Santambrogio L, Saran S, Sarkar S, Sarwal M, Sasakawa C, Sasnauskiene A, Sass M, Sato K, Sato M, Schapira AH, Scharl M, Schatzl HM, Scheper W, Schiaffino S, Schneider C, Schneider ME, Schneider-Stock R, Schoenlein PV, Schorderet DF, Schuller C, Schwartz GK, Scorrano L, Sealy L, Seglen PO, Segura-Aguilar J, Seiliez I, Seleverstov O, Sell C, Seo JB, Separovic D, Setaluri V, Setoguchi T, Settembre C, Shacka JJ, Shanmugam M, Shapiro IM, Shaulian E, Shaw RJ, Shelhamer JH, Shen HM, Shen WC, Sheng ZH, Shi Y, Shibuya K, Shidoji Y, Shieh JJ, Shih CM, Shimada Y, Shimizu S, Shintani T, Shirihai OS, Shore GC, Sibirny AA, Sidhu SB, Sikorska B, Silva-Zacarin EC, Simmons A, Simon AK, Simon HU, Simone C, Simonsen A, Sinclair DA, Singh R, Sinha D, Sinicrope FA, Sirko A, Siu PM, Sivridis E, Skop V, Skulachev VP, Slack RS, Smaili SS, Smith DR, Soengas MS, Soldati T, Song X, Sood AK, Soong TW, Sotgia F, Spector SA, Spies CD, Springer W, Srinivasula SM, Stefanis L, Steffan JS, Stendel R, Stenmark H, Stephanou A, Stern ST, Sternberg C, Stork B, Stralfors P, Subauste CS, Sui X, Sulzer D, Sun J, Sun SY, Sun ZJ, Sung JJ, Suzuki K, Suzuki T, Swanson MS, Swanton C, Sweeney ST, Sy LK, Szabadkai G, Tabas I, Taegtmeyer H, Tafani M, Takacs-Vellai K, Takano Y, Takegawa K, Takemura G, Takeshita F, Talbot NJ, Tan KS, Tanaka K, Tanaka K, Tang D, Tang D, Tanida I, Tannous BA, Tavernarakis N, Taylor GS, Taylor GA, Taylor JP, Terada LS, Terman A, Tettamanti G, Thevissen K, Thompson CB, Thorburn A, Thumm M, Tian F, Tian Y, Tocchini-Valentini G, Tolkovsky AM, Tomino Y, Tonges L, Tooze SA, Tournier C, Tower J, Towns R, Trajkovic V, Travassos LH, Tsai TF, Tschan MP, Tsubata T, Tsung A, Turk B, Turner LS, Tyagi SC, Uchiyama Y, Ueno T, Umekawa M, Umemiya-Shirafuji R, Unni VK, Vaccaro MI, Valente EM, Van den Berghe G, van der Klei IJ, van Doorn W, van Dyk LF, van Egmond M, van Grunsven LA, Vandenabeele P, Vandenberghe WP, Vanhorebeek I, Vaquero EC, Velasco G, Vellai T, Vicencio JM, Vierstra RD, Vila M, Vindis C, Viola G, Viscomi MT, Voitsekhovskaja OV, von Haefen C, Votruba M, Wada K, Wade-Martins R, Walker CL, Walsh CM, Walter J, Wan XB, Wang A, Wang C, Wang D, Wang F, Wang F, Wang G, Wang H, Wang HG, Wang HD, Wang J, Wang K, Wang M, Wang RC, Wang X, Wang X, Wang YJ, Wang Y, Wang Z, Wang ZC, Wang Z, Wansink DG, Ward DM, Watada H, Waters SL, Webster P, Wei L, Weihl CC, Weiss WA, Welford SM, Wen LP, Whitehouse CA, Whitton JL, Whitworth AJ, Wileman T, Wiley JW, Wilkinson S, Willbold D, Williams RL, Williamson PR, Wouters BG, Wu C, Wu DC, Wu WK, Wyttenbach A, Xavier RJ, Xi Z, Xia P, Xiao G, Xie Z, Xie Z, Xu DZ, Xu J, Xu L, Xu X, Yamamoto A, Yamamoto A, Yamashina S, Yamashita M, Yan X, Yanagida M, Yang DS, Yang E, Yang JM, Yang SY, Yang W, Yang WY, Yang Z, Yao MC, Yao TP, Yeganeh B, Yen WL, Yin JJ, Yin XM, Yoo OJ, Yoon G, Yoon SY, Yorimitsu T, Yoshikawa Y, Yoshimori T, Yoshimoto K, You HJ, Youle RJ, Younes A, Yu L, Yu L, Yu SW, Yu WH, Yuan ZM, Yue Z, Yun CH, Yuzaki M, Zabirnyk O, Silva-Zacarin E, Zacks D, Zacksenhaus E, Zaffaroni N, Zakeri Z, Zeh HJ, 3rd, Zeitlin SO, Zhang H, Zhang HL, Zhang J, Zhang JP, Zhang L, Zhang L, Zhang MY, Zhang XD, Zhao M, Zhao YF, Zhao Y, Zhao ZJ, Zheng X, Zhivotovsky B, Zhong Q, Zhou CZ, Zhu C, Zhu WG, Zhu XF, Zhu X, Zhu Y, Zoladek T, Zong WX, Zorzano A, Zschocke J, and Zuckerbraun B. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy 8: 445–544, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Koivisto P, Kolmer M, Visakorpi T, and Kallioniemi OP. Androgen receptor gene and hormonal therapy failure of prostate cancer. Am J Pathol 152: 1–9, 1998 [PMC free article] [PubMed] [Google Scholar]
- 36. Kuo LJ. and Yang LX. Gamma-H2AX—a novel biomarker for DNA double-strand breaks. In Vivo 22: 305–309, 2008 [PubMed] [Google Scholar]
- 37. Lane DJ, Mills TM, Shafie NH, Merlot AM, Saleh Moussa R, Kalinowski DS, Kovacevic Z, and Richardson DR. Expanding horizons in iron chelation and the treatment of cancer: role of iron in the regulation of ER stress and the epithelial-mesenchymal transition. Biochim Biophys Acta 1845: 166–181, 2014 [DOI] [PubMed] [Google Scholar]
- 38. Le NT. and Richardson DR. The role of iron in cell cycle progression and the proliferation of neoplastic cells. Biochim Biophys Acta 1603: 31–46, 2002 [DOI] [PubMed] [Google Scholar]
- 39. Liebermann DA. and Hoffman B. Myeloid differentiation (MyD)/growth arrest DNA damage (GADD) genes in tumor suppression, immunity and inflammation. Leukemia 16: 527–541, 2002 [DOI] [PubMed] [Google Scholar]
- 40. Liu XB, Yang F, and Haile DJ. Functional consequences of ferroportin 1 mutations. Blood Cells Mol Dis 35: 33–46, 2005 [DOI] [PubMed] [Google Scholar]
- 41. Lui GY, Kovacevic Z, Richardson V, Merlot AM, Kalinowski DS, and Richardson DR. Targeting cancer by binding iron: dissecting cellular signaling pathways. Oncotarget 6: 18748–18779, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ma Y. and Hendershot LM. Delineation of a negative feedback regulatory loop that controls protein translation during endoplasmic reticulum stress. J Biol Chem 278: 34864–34873, 2003 [DOI] [PubMed] [Google Scholar]
- 43. Mancias JD, Wang X, Gygi SP, Harper JW, and Kimmelman AC. Quantitative proteomics identifies NCOA4 as the cargo receptor mediating ferritinophagy. Nature 509: 105–109, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Meek DW. The p53 response to DNA damage. DNA Repair (Amst) 3: 1049–1056, 2004 [DOI] [PubMed] [Google Scholar]
- 45. Meyron-Holtz EG, Ghosh MC, Iwai K, LaVaute T, Brazzolotto X, Berger UV, Land W, Ollivierre-Wilson H, Grinberg A, Love P, and Rouault TA. Genetic ablations of iron regulatory proteins 1 and 2 reveal why iron regulatory protein 2 dominates iron homeostasis. EMBO J 23: 386–395, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Mizushima N, Yoshimori T, and Levine B. Methods in mammalian autophagy research. Cell 140: 313–326, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Moffat J, Grueneberg DA, Yang X, Kim SY, Kloepfer AM, Hinkle G, Piqani B, Eisenhaure TM, Luo B, Grenier JK, Carpenter AE, Foo SY, Stewart SA, Stockwell BR, Hacohen N, Hahn WC, Lander ES, Sabatini DM, and Root DE. A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell 124: 1283–1298, 2006 [DOI] [PubMed] [Google Scholar]
- 48. N'Diaye EN, Kajihara KK, Hsieh I, Morisaki H, Debnath J, and Brown EJ. PLIC proteins or ubiquilins regulate autophagy-dependent cell survival during nutrient starvation. EMBO Rep 10: 173–179, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Narla G, Heath KE, Reeves HL, Li D, Giono LE, Kimmelman AC, Glucksman MJ, Narla J, Eng FJ, Chan AM, Ferrari AC, Martignetti JA, and Friedman SL. KLF6, a candidate tumor suppressor gene mutated in prostate cancer. Science 294: 2563–2566, 2001 [DOI] [PubMed] [Google Scholar]
- 50. Oslowski CM. and Urano F. Measuring ER stress and the unfolded protein response using mammalian tissue culture system. Methods Enzymol 490: 71–92, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Petrat F, de Groot H, Sustmann R, and Rauen U. The chelatable iron pool in living cells: a methodically defined quantity. Biol Chem 383: 489–502, 2002 [DOI] [PubMed] [Google Scholar]
- 52. Pietrangelo A. The ferroportin disease. Blood Cells Mol Dis 32: 131–138, 2004 [DOI] [PubMed] [Google Scholar]
- 53. Pinnix ZK, Miller LD, Wang W, D'Agostino R, Jr., Kute T, Willingham MC, Hatcher H, Tesfay L, Sui G, Di X, Torti SV, and Torti FM. Ferroportin and iron regulation in breast cancer progression and prognosis. Sci Transl Med 2: 43ra56, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Pullarkat V, Meng Z, Donohue C, Yamamoto VN, Tomassetti S, Bhatia R, Krishnan A, Forman SJ, and Synold TW. Iron chelators induce autophagic cell death in multiple myeloma cells. Leuk Res 38: 988–996, 2014 [DOI] [PubMed] [Google Scholar]
- 55. Rashid HO, Yadav RK, Kim HR, and Chae HJ. ER stress: autophagy induction, inhibition and selection. Autophagy 11: 1956–1977, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Richardson DR. and Ponka P. Development of iron chelators to treat iron overload disease and their use as experimental tools to probe intracellular iron metabolism. Am J Hematol 58: 299–305, 1998 [DOI] [PubMed] [Google Scholar]
- 57. Rodier F, Coppe JP, Patil CK, Hoeijmakers WA, Munoz DP, Raza SR, Freund A, Campeau E, Davalos AR, and Campisi J. Persistent DNA damage signalling triggers senescence-associated inflammatory cytokine secretion. Nat Cell Biol 11: 973–979, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Ross SL, Tran L, Winters A, Lee KJ, Plewa C, Foltz I, King C, Miranda LP, Allen J, Beckman H, Cooke KS, Moody G, Sasu BJ, Nemeth E, Ganz T, Molineux G, and Arvedson TL. Molecular mechanism of hepcidin-mediated ferroportin internalization requires ferroportin lysines, not tyrosines or JAK-STAT. Cell Metab 15: 905–917, 2012 [DOI] [PubMed] [Google Scholar]
- 59. Rubinstein M, Idelman G, Plymate SR, Narla G, Friedman SL, and Werner H. Transcriptional activation of the insulin-like growth factor I receptor gene by the Kruppel-like factor 6 (KLF6) tumor suppressor protein: potential interactions between KLF6 and p53. Endocrinology 145: 3769–3777, 2004 [DOI] [PubMed] [Google Scholar]
- 60. Salama R, Sadaie M, Hoare M, and Narita M. Cellular senescence and its effector programs. Genes Dev 28: 99–114, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Saletta F, Suryo Rahmanto Y, Siafakas AR, and Richardson DR. Cellular iron depletion and the mechanisms involved in the iron-dependent regulation of the growth arrest and DNA damage family of genes. J Biol Chem 286: 35396–35406, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Schimanski LM, Drakesmith H, Merryweather-Clarke AT, Viprakasit V, Edwards JP, Sweetland E, Bastin JM, Cowley D, Chinthammitr Y, Robson KJ, and Townsend AR. In vitro functional analysis of human ferroportin (FPN) and hemochromatosis-associated FPN mutations. Blood 105: 4096–4102, 2005 [DOI] [PubMed] [Google Scholar]
- 63. Shieh SY, Ikeda M, Taya Y, and Prives C. DNA damage-induced phosphorylation of p53 alleviates inhibition by MDM2. Cell 91: 325–334, 1997 [DOI] [PubMed] [Google Scholar]
- 64. Siegel RL, Miller KD, and Jemal A. Cancer statistics, 2016. CA Cancer J Clin 66: 7–30, 2016 [DOI] [PubMed] [Google Scholar]
- 65. Sobel RE. and Sadar MD. Cell lines used in prostate cancer research: a compendium of old and new lines—part 1. J Urol 173: 342–359, 2005 [DOI] [PubMed] [Google Scholar]
- 66. Subramaniam VN, Wallace DF, Dixon JL, Fletcher LM, and Crawford DH. Ferroportin disease due to the A77D mutation in Australia. Gut 54: 1048–1049, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Tai S, Sun Y, Squires JM, Zhang H, Oh WK, Liang CZ, and Huang J. PC3 is a cell line characteristic of prostatic small cell carcinoma. Prostate 71: 1668–1679, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Tarocchi M, Hannivoort R, Hoshida Y, Lee UE, Vetter D, Narla G, Villanueva A, Oren M, Llovet JM, and Friedman SL. Carcinogen-induced hepatic tumors in KLF6+/− mice recapitulate aggressive human hepatocellular carcinoma associated with p53 pathway deregulation. Hepatology 54: 522–531, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Tesfay L, Clausen KA, Kim JW, Hegde P, Wang X, Miller LD, Deng Z, Blanchette N, Arvedson T, Miranti CK, Babitt JL, Lin HY, Peehl DM, Torti FM, and Torti SV. Hepcidin regulation in prostate and its disruption in prostate cancer. Cancer Res 75: 2254–2263, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Torti SV. and Torti FM. Iron and cancer: more ore to be mined. Nat Rev Cancer 13: 342–355, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Uddin MN, Ito S, Nishio N, Suganya T, and Isobe K. Gadd34 induces autophagy through the suppression of the mTOR pathway during starvation. Biochem Biophys Res Commun 407: 692–698, 2011 [DOI] [PubMed] [Google Scholar]
- 72. Wajapeyee N, Serra RW, Zhu X, Mahalingam M, and Green MR. Role for IGFBP7 in senescence induction by BRAF. Cell 141: 746–747, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Wang W, Deng Z, Hatcher H, Miller LD, Di X, Tesfay L, Sui G, D'Agostino RB, Jr., Torti FM, and Torti SV. IRP2 regulates breast tumor growth. Cancer Res 74: 497–507, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Wang X, Simpson ER, and Brown KA. p53: Protection against tumor growth beyond effects on cell cycle and apoptosis. Cancer Res 75: 5001–5007, 2015 [DOI] [PubMed] [Google Scholar]
- 75. Ward DM. and Kaplan J. Ferroportin-mediated iron transport: expression and regulation. Biochim Biophys Acta 1823: 1426–1433, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Welford SM. and Giaccia AJ. Hypoxia and senescence: the impact of oxygenation on tumor suppression. Mol Cancer Res 9: 538–544, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Xue D, Zhou CX, Shi YB, Lu H, and He XZ. Decreased expression of ferroportin in prostate cancer. Oncol Lett 10: 913–916, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Young AR. and Narita M. SASP reflects senescence. EMBO Rep 10: 228–230, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Yu G. and He QY. ReactomePA: an R/Bioconductor package for reactome pathway analysis and visualization. Mol Biosyst 12: 477–479, 2016 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.