Abstract
Background
Accumulating evidence has revealed the pivotal role of epigenetic regulation in the pathogenesis of liver disease. However, the epigenetic mechanism that accounts for hepatic stellate cells (HSCs) activation in liver fibrosis remains largely unknown.
Methods
Primary HSCs were used to screen the differentially expressed histone H3 lysine methyltransferases and demethylases during HSC activation. Loss-of-function experiments were applied to determine the cellular functions of KDM4D in HSCs. Transcriptome analysis was applied to explore the downstream targets of KDM4D. Real-time qPCR, western blotting, immunohistochemical staining, and chromatin immunoprecipitation were performed to uncover the underlying mechanism concerning KDM4D during liver fibrogenesis.
Findings
KDM4D was identified as a remarkable up-regulated histone H3 demethylase during HSC activation. The overexpression profile of KDM4D was confirmed in three fibrosis animal models and human fibrotic liver tissues. In vitro Kdm4d knockdown impaired the collagen gel contraction and migration capacity of primary HSCs. In established CCl4-induced mice model, Kdm4d knockdown inhibited fibrosis progression, and promoted fibrosis reversal, with enhanced thinning and splitting of fibrotic septa, as well as a dramatic decrease in collagen area. Whole gene transcriptome analysis showed the regulatory role of KDM4D in Toll-Like Receptor (TLR) signaling pathway. Mechanistically, KDM4D catalyzed histone 3 on lysine 9 (H3K9) di-, and tri-demethylation, which promoted TLR4 expression, and subsequently prompted liver fibrogenesis by activating NF-κB signaling pathways.
Interpretation
KDM4D facilitates TLR4 transcription through demethylation of H3K9, thus activating TLR4/NF-κB signaling pathways in HSCs, contributing to HSC activation and collagen crosslinking, further, hepatic fibrosis progression.
Fund
Shanghai New Hundred Talents Program, Shanghai Municipal Commission of Health and Family Planning, Key Developing Disciplines Program, Shanghai Key disciplines program of Health and Family Planning and Shanghai Sailing Program.
Keywords: JMJD2D, JHDM3D, Lysine demethylase 4D, KMT, KDM
Research in context.
Evidence before this study
Hepatic stellate cell (HSC) activation highlights the central theme of hepatic fibrosis, a common pathological process resulting from chronic liver injury of various etiologies that, without therapeutic intervention, can result in, irreversible cirrhosis and even deadly hepatocellular carcinoma.
Added value of this study
This study demonstrated that KDM4D is exclusively expressed in activated HSCs and fibrotic liver tissues in mice. The upregulation of KDM4D is required for liver fibrogenesis. KDM4D deficiency dramatically ameliorates experimental liver fibrosis in mice. Mechanistically, we uncovered that KDM4D promotes liver fibrosis by modulating TLR4/NF-κB signaling pathways.
Implications of all the available evidence
This finding expands our knowledge regarding the epigenetic mechanism that accounts for hepatic stellate cells activation and indicates that KDM4D is a potential target for anti-fibrotic therapy.
Alt-text: Unlabelled Box
1. Introduction
Hepatic fibrosis is caused by various harmful stimuli, characterized by the imbalance of extracellular matrix synthesis, degradation and deposition as well as intrahepatic connective tissue hyperplasia [[1], [2], [3], [4]]. Advanced liver fibrosis eventually leads to irreversible cirrhosis and even hepatocellular carcinoma (HCC), with liver transplantation remaining the only effective treatment for decompensated cirrhosis or advanced HCC [5,6]. Therefore, a better understanding of the molecular mechanisms underlying hepatic fibrogenesis would facilitate the development of preventive and therapeutic approaches for liver fibrosis and possibly for lethal HCC.
The central event during liver fibrogenesis is the activation of hepatic stellate cells (HSCs) [7]. Quiescent HSCs are similar to adipocytes, which can store lipid and retinoid A. Once stimulated, HSCs undergo notable phenotypic transitions, becoming more proliferative and contractile, while de novo expression of α-smooth muscle actin (α-SMA) as well as secretion of copious amounts of collagens, which disrupt liver anatomy, herald loss of liver function. This process proceeds to irreversible liver pathology and correlates with augmented mortality of patients with end-stage liver diseases [8,9]. Collagen crosslinking is an essential process for fibrotic matrix stability, which contributes to fibrosis progression and limits reversibility of liver fibrosis. Hence, inhibition of HSC activation and its collagen crosslinking ability are considered to be a promising candidate for halting or even reversing advanced fibrosis.
Epigenetic regulators such as DNA methyltransferases, methyl-DNA binding proteins, histone modifying enzymes and deregulated non-coding RNA have been identified as potential points of therapeutic intervention, which has garnered wide attention [[10], [11], [12]]. Specifically, histone methylation, a reversible process, is one of the most prominent histone posttranslational modifications in response to environmental cues. Accumulating evidence suggests that the methylation of histone lysine residues is a highly dynamic modification owing to the interplay between the epigenetic “writer” lysine methyltransferases (KMTs) and “eraser” lysine demethylases (KDMs) [[13], [14], [15]]. Several KMTs have been reported to be involved in the fibrogenic phenotype of HSC-derived myofibroblasts. For instance, ASH1 orchestrates the coordinated activation of pro-fibrogenic genes including Acta2, Col1A1, and Timp-1 [16]. EZH2 contributes to the transcriptional inhibition of the nuclear receptor PPARγ, which further reprograms the adipogenic HSC towards the myofibroblast phenotype [17,18]. However, the expression patterns and potential regulatory roles of KMTs and KDMs in liver fibrosis require further exploration.
The lysine (K)-specific demethylase 4 (KDM4) family is comprised of 4 isoforms, KDM4A to -D, also known as JMJD2A to -D. KDM4A, B, and C encode proteins consisting of a JmjC, a JmjN, two PHD, and two Tudor domains. KDM4D is unique within the KDM4 family in that it has neither PHD nor Tudor domains, therefore it is only half the size of KDM4A-C [19]. Previous studies showed that KDM4D can be swiftly recruited to DNA damage sites in a PARP1-dependent manner and facilitate double-strand break repair in human cells, which ensure efficient repair of DNA lesions to maintain genome stability [20,21]. KDM4D is also a novel cofactor of androgen receptor since it interacts with androgen receptor and stimulates its ability to up-regulate transcription, which plays an indispensable role in prostate cancer [22].
In this study, a large-scale screen was performed to identify the dysregulated H3 KMTs and KDMs involved during liver fibrosis pathophysiological process. As a result, the most differentially expressed KDM4D, which has never been evaluated in HSC, was selected for detailed investigation. Here we defined the function of KDM4D in HSC activation and liver fibrogenesis.
2. Materials and methods
2.1. Quantitative real-time PCR (qRT-PCR)
RNA extracted from liver tissues and cells was subjected to reverse transcription and subsequently underwent quantitative real-time PCR utilizing 7500 Real-time PCR system (Applied Biosystems, USA). Genes were normalized to β-actin. The relative expression level of each gene was calculated using the formula 2 (−ΔΔCt). Primer sequences used in qRT-PCR are listed in Supplementary Tables 1–3.
2.2. RNA interference and gene overexpression
Small interfering RNA (siRNA) oligonucleotides against KDM4D/Kdm4d, TLR4/Tlr4, and the scrambled sequences (si-NC) were synthesized by Tuoran Co., LTD (Shanghai, China). The expression vectors pcDNA4-FLAG-KDM4D and its inactive mutant pcDNA4-FLAG-KDM4D-H1122A, which contains histidine-to-alanine point mutation in the JmjC domain and does not possess histone-demethylase function, were synthesized by Sangon Co., LTD (Shanghai, China). For overexpressing TLR4, the pcDNA3.1-TLR4 vector was constructed by GenePharma Co., LTD (Shanghai, China). Constitutively active IKKβ cDNA was synthesized by Sangon Co., LTD (Shanghai, China) and subsequently cloned into pCDNA3.1-Hygro (+) for the experiments. For transfection, LX2 and T6 cells or HSCs were plated at 1 × 105 cells/well in 6-well plates and cultured overnight. Cells were transfected with siRNAs or overexpression vector using RNAimax or Lipofectamine 2000 following the manufacturer's protocol (Life Technologies, USA). At 72 h after transfection, cells were harvested for qRT-PCR and western blotting. Primer sequences used for transient interference are listed in Supplementary Tables 4–7.
2.3. Animal study
Six-week-old male C57BL/6J mice and Sprague Dawley rats were purchased from Shanghai Laboratory Animal Center, Chinese Academy of Sciences (SLAC, CAS). Mice and rats were housed and reared in specific pathogen-free and barrier conditions according to protocols approved by the East China Normal University Care Commission. All animals were housed in a controlled environment under a 12 h dark/light cycle with free access to food and water. All animals received humane care according to the criteria outlined in the “Guide for the Care and Use of Laboratory Animals” prepared by the National Academy of Sciences and published by the National Institutes of Health (NIH publication 86-23 revised 1985). All interventions were done during the light cycle.
2.4. CCl4, TAA-induced models of non-biliary fibrosis and BDL-induced model of biliary fibrosis
To generate animal models of hepatic fibrosis, 6-week-old male C57BL/6J mice were intraperitoneally (i.p.) injected with carbon tetrachloride (CCl4) (dissolved at 1:3 vol/vol in corn oil) (Sigma-Aldrich, St.Louis, MO, USA) or corn oil alone (3.0 ml/kg body weight) twice a week from 6 to 14 weeks of age. Alternatively, 6-week-old male Sprague Dawley rats were i.p. injected with thioacetamide (TAA) (dissolved at 1:3 vol/vol in corn oil) or corn oil alone (0.2 ml/kg body weight) twice a week from 6 to 14 weeks of age. Animals were then divided into test or control groups randomly. Another batch of C57BL/6J mice were subjected to bile duct ligation (BDL) or sham opening operation. They were sacrificed 2 weeks later after operation. All animals were fasted overnight before being sacrificed. Blood samples were harvested for biochemistry analysis. Part of liver tissues was fixed in 10% formalin for paraffin blocks, and the remaining fresh liver tissues were frozen at −80 °C for western blotting or qRT-PCR analysis.
2.5. Adeno-associated virus infection
To knockdown Kdm4d in vivo, we transduced mice with adeno-associated virus (AAV) serotype 9 that encoded a green fluorescent protein (GFP) reporter together with either short hairpin RNAs targeting Kdm4d in liver (shKdm4d) or empty vector (shNC). The short hairpin RNA (shRNA) sequences targeting the mouse Kdm4d gene was cloned into AAV by Genechem Co. LTD (Shanghai, China). Thirty- two C57BL/6J (male, 6-week-old) mice were obtained for this in vivo study. After 1 week of acclimatization, they were randomly divided into 4 groups (n = 8 per group) as follows: corn oil + sh-NC, corn oil+sh-Kdm4d, CCl4 + sh-NC, and CCl4 + sh-Kdm4d. The mice were given two i.p. administrations per week of CCl4 dissolved in corn oil (1:3 vol/vol) or corn oil alone (3.0 ml/kg) from 6 to 14 weeks of age. At week 12 of age, mice were injected AAV via tail vein (1 × 1011 viral particles/mouse) carrying shRNA targeting Kdm4d or vehicle alone at one time, followed by additional 2 weeks corn oil or CCl4 injection. Mice were sacrificed after overnight fasting, and blood and liver tissues were collected and stored for further analysis.
2.6. Gene expression array
Briefly, LX2 cells transfected with siRNA targeting KDM4D or scrambled siRNA were used. Total RNA (15 μg) was isolated from 3 biological replicates and reverse transcribed for hybridization to the whole human Genome Microarray (4 × 44 K, Agilent) using an Agilent Gene Expression Hybridization Kit as recommended by the manufacturer. The raw gene expression data were extracted from Agilent Feature Extraction Software (version 10.7) and imported into Agilent GeneSpring GX software (version 11.0) for further analysis. Background subtraction and normalization of probe set intensities were performed using Robust Multiarray Analysis (RMA). Genechip data used for analysis were shown in Supplementary Tables 8 and 9.
2.7. Protein extraction and western blotting
Western blotting analysis was performed as reported previously [23]. Whole cellular lysates and nuclear fractions were extracted from primary HSCs, activated HSC-T6 and LX2 cells. Nuclear proteins were prepared with the CelLytic NuCLEAR extraction kit (Sigma, NXTRACT, USA) following manufacturer's introductions. The protein content was determined using a BCA Protein Assay Kit (Beyotime Biotechnology, Shanghai, China). Proteins were subjected to SDS-PAGE and then transferred to a nitrocellulose membrane. After blocking with 5% skim milk for 1 h at room temperature, the membranes were respectively incubated overnight at 4 °C with indicated antibodies shown in Supplementary Table 10. After incubation with horseradish peroxidase-conjugated secondary antibodies for 2 h at room temperature, protein expression was detected by the enhanced chemiluminescent method and imaged with a Bio-Spectrum Gel Imaging System (UVP, USA).
2.8. Chromatin immunoprecipitation (ChIP)
ChIP assay was carried out by using antibodies listed in Supplementary Table 11 as well as 100 μg cross-linked native chromatin prepared from primary HSCs and LX2 cells (5 × 107) with or without KDM4D knockdown. Immunoprecipitated DNA was used as the template for quantitative PCR using primers specific for human TLR4 promoter. DNA enrichment was evaluated by average values of the eluate with immunoprecipitated DNA normalized to average values of input.
2.9. Collagen gel contraction assay
Cells at a density of 5 × 104/ml were seeded into 32 mm bacteriological plates (2 ml per dish) in DMEM supplemented with 10% FBS, 1% penicillin and streptomycin and 0.3 mg/ml of acid-extracted collagen I derived from Sprague Dawley rat tail. The cells were cultured at 37 °C for 60 min to allow collagen contraction. Then the gels were released from inner edges of plates by tilting plates slightly, and the gel contraction ability was monitored at time points up to 6 h. All assays were performed in triplicate.
2.10. Cell migration assay
Cell migration assays were performed using transwell chambers (Millipore, PIEP12R48, USA). A total of 5 × 104 indicated cells in 200 μl serum-free DMEM were seeded in the upper chamber and 700 μl medium with 10% FBS was added to the lower chamber. After incubation for 24 h, migrated cells were fixed and stained with 0.1% (w/v) crystal violet. Cells were photographed and counted in 5 independent random view fields.
2.11. Statistical analysis
Data are presented as the mean ± standard deviation. Comparisons between two groups were determined by two-sided, unpaired Student's t-test. Comparisons among multiple groups were analyzed by one-way ANOVA test. P values < .05 are considered statistically significant.
3. Results
3.1. KDM4D expression is up-regulated during HSC activation
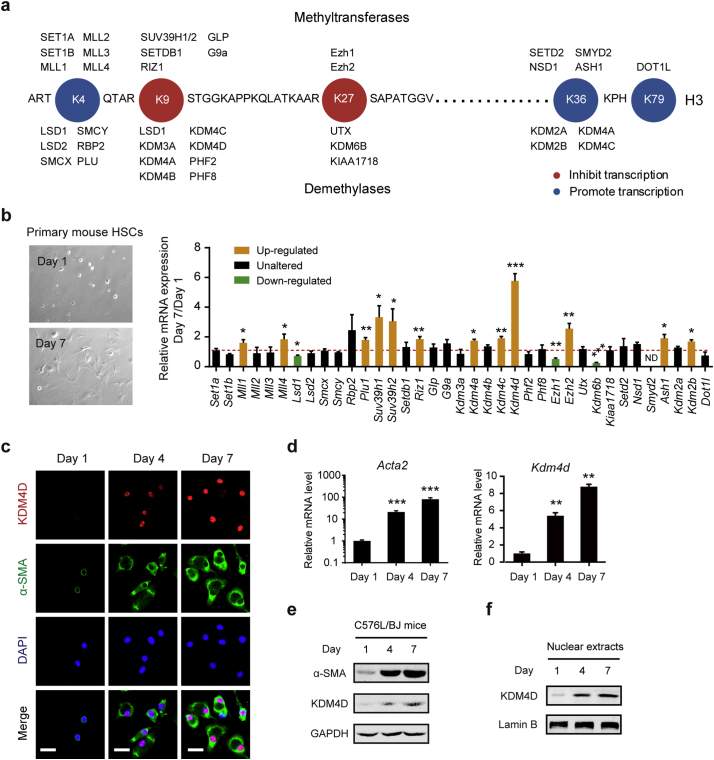
Primary HSCs isolated from normal C57BL/6J mice were used to determine the differentially expressed genes of histone H3 KMTs and KDMs during HSC activation. After resting for 24 h, quiescent (Day 1) HSCs were collected. After culturing for 7 consecutive days, activated (Day 7) HSCs were harvested and analyzed together with Day 1 cells by qRT-PCR. In this study, a total of 19 KMTs and 18 KDMs were tested (Fig. 1a). As a result, the mRNA level of Lsd1, Ezh1, and Kdm6b were significantly reduced, while the expression level of Mll1, Mll4, Plu1, Suv39h1, Suv39h2, Riz1, Kdm4a, Kdm4c, Kdm4d, Ezh2, Ash1, and Kdm2b were remarkably increased during HSC activation. Overview of the result, we found that H3K9 KMTs and KDMs were commonly altered. Notably, the mRNA expression level of Kdm4d was increased by more than five-fold in activated HSCs when compared with that in the quiescent ones (Fig. 1b). In activated HSCs, Kdm4d expression level was much higher than other H3K9 KMTs, including Suv9h1, Suv39H2 and Riz1. From the therapeutic point of view, Kdm4d was selected as a candidate gene for further investigation. By co-staining of KDM4D and α-SMA in primary HSCs derived from C57BL/6J mice, we further confirmed that activated HSCs gave rise to KDM4D nuclear expression (Fig. 1c). The qRT-PCR and western blotting also illustrated that KDM4D expression level augmented with HSC activation in a step-wise manner (Fig. 1d, e). Together, these data strongly suggested that KDM4D overexpression occurs gradually during HSC activation.
Fig. 1.
KDM4D expression is up-regulated during HSC activation. (a) Schematic of human histone H3 methyltransferases and demethylases. (b) Representative bright field images of C57BL/6J mouse primary HSCs on D1 and D7 (left panel). Relative mRNA expression levels of H3 methyltransferases and demethylases in primary HSCs (D7/D1) of C57BL/6J mouse (right panel). ND, not determined. (c) Cellular immunofluorescence staining of KDM4D and α-SMA in primary HSCs of C57BL/6J mice on day 1, 4, and 7. Scale bar, 25 μm. (d, e) The mRNA and protein expression level of Kdm4d and Acta2 in primary HSCs of the C57BL/6J mice on day 1, 4, and 7. (f) The nuclear KDM4D protein level in primary HSCs of the C57BL/6J mice on day 1, 4, and 7. Data were presented as mean ± SD, *p < .05, **p < .01, ***p < .001.
3.2. KDM4D expression is up-regulated during biliary and non-biliary fibrogenesis
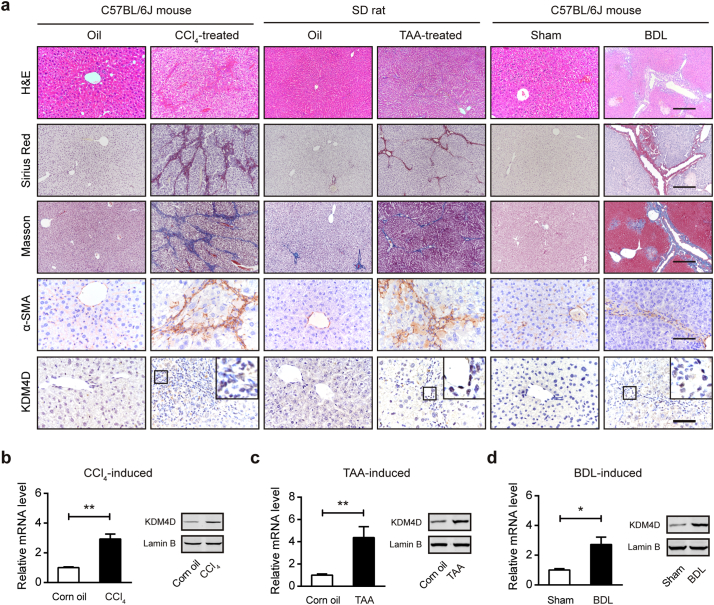
To further validate the expression pattern of KDM4D in liver fibrogenesis, biliary and non-biliary murine models induced by CCl4 (left), TAA (middle), and BDL (right) were generated (Fig. 2a). Repeated CCl4 and TAA administration led to robust and progressive scarring, characterized histologically as significantly pericentral fibrosis, while BDL contributed to biliary fibrosis. As shown in Fig. 2a, compared with their control liver sections, the fibrotic liver tissues had larger fibrotic areas (H&E and α-SMA staining), and severer collagen deposition (Sirius Red and Masson staining), respectively. As expected, IHC revealed that KDM4D, which was hardly detectable in non-fibrotic healthy livers, was strongly expressed after BDL operation or consecutive CCl4/TAA injection (Fig. 2a). The intense immunoreactivity of KDM4D in the fibrotic areas was widely observed in all the three animal models indicate of a common expression profile of KDM4D in both parenchymal and biliary fibrosis. Furthermore, RNA was isolated from primary HSCs from CCl4/TAA/oil or BDL/sham animal livers and we found that Kdm4d was commonly and highly expressed during liver fibrogenesis (Fig. 2b-d), implying the restricted expression of KDM4D in myofibroblast-like cells in liver fibrosis.
Fig. 2.
KDM4D is up-regulated during biliary and non-biliary fibrogenesis. (a) Representative images of H&E, Sirius red, Masson's trichrome staining, α-SMA, and KDM4D staining in liver tissues from CCl4-induced C57BL/6J mice, TAA-induced SD rat, and BDL-induced C57BL/6J mice as well as their corresponding counterparts. Scale bar, 200 μm for H&E, Sirius red, and Masson's trichrome staining; 100 μm for α-SMA and KDM4D staining. (b–d) Real-time qPCR and western blotting analysis of Kdm4d mRNA and protein level in quiescent or activated HSCs isolated from CCl4/TAA/BDL/Control animal livers. Data were presented as mean ± SD, *p < .05, and **p < .01.
3.3. Kdm4d deficiency ameliorates liver fibrosis in mice
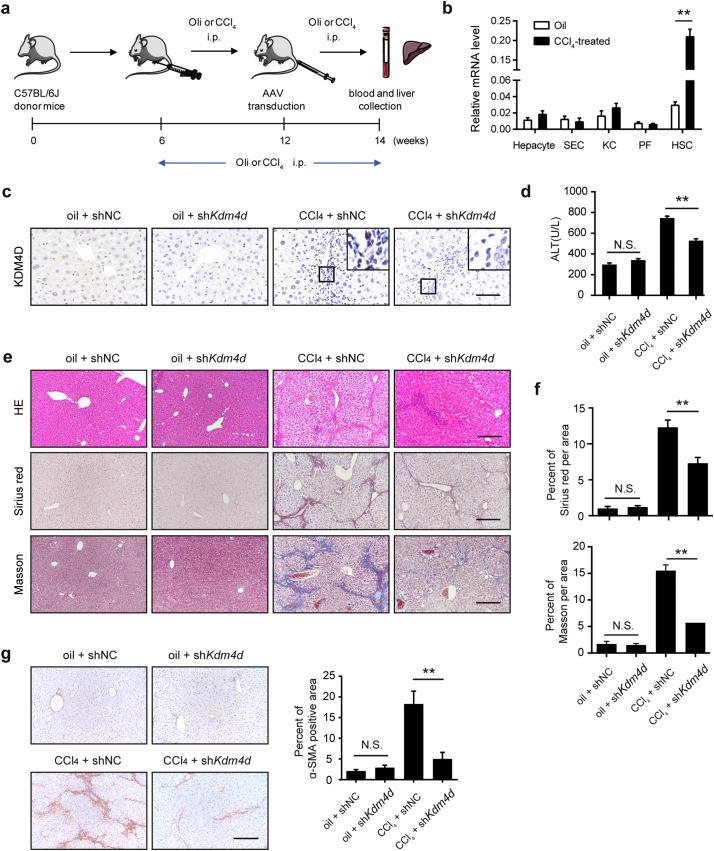
Next, we were prompted to investigate the in vivo role of Kdm4d knockdown in a CCl4-induced liver fibrosis mouse model (Fig. 3a). Kdm4d knockdown in liver tissue was then carried out using tail vein injection with a Kdm4d shRNA-expressing AAV9. AAV9 targets many tissues including central nervous system, heart, liver, and lung. We therefore tested the specificity of KDM4D knockdown in liver tissues. To address this, resident liver cells, including hepatocytes, sinusoidal endothelial cells (SEC), Kupffer cells (KC), portal myofibroblasts (PF), and HSCs from CCl4/oil mouse livers were isolated. RT-qPCR result showed that Kdm4d expression was mainly derived from activated HSCs, indicating the specificity of AAV9-mediated Kdm4d knockdown in HSCs (Fig. 3b). Indeed, AAV9-mediated Kdm4d deficiency led to significant reduction of KDM4D protein in fibrotic liver tissues (Fig. 3c). Kdm4d deficiency had no obvious deleterious effect on normal liver function, but significantly alleviated CCl4-induced liver injury (Fig. 3d) and release of pro-inflammatory cytokines (Supplementary Fig. 1). Specifically, Kdm4d knockdown exhibited a reduction in collagen accumulation, compared with their counterparts in the control group (Fig. 3e, f). Consistently, lower expression of α-SMA was found in Kdm4d-deficient mice than their control littermates as revealed by IHC analysis (Fig. 3g). Collectively, these in vivo data reinforced the notion that KDM4D served as an important driving force underlying the pathogenesis of liver fibrosis.
Fig. 3.
Kdm4d deficiency ameliorates liver fibrosis in mice. (a) The experiment scheme for Kdm4d knockdown in vivo. Liver fibrogenesis was induced by CCl4 injury using intraperitoneal injection twice a week from 6 to 14 weeks of age. Their counterparts were injected with oil. At week 12, shRNA expression vectors GFP carrying shRNA targeting Kdm4d or negative control were transfected into mouse liver through tail vein injection. Animals were humanely sacrificed, and their blood and liver samples were collected at week 14. (b) RT-qPCR analysis of Kdm4d mRNA level in HSCs, hepatocytes, sinusoidal endothelial cells (SEC), Kupffer cells (KC), and portal myofibroblasts (PF) isolated from the normal or CCl4-indced mouse liver tissues. (c) Representative IHC images of KDM4D in indicated groups. Scale bar, 200 μm. (d) The liver injury in each group was measured by ALT levels. (e) Representative histology images of H&E, Sirius red and Masson's trichrome staining in indicated groups. Scale bar, 200 μm. (f) Sirius red and Masson's trichrome positive areas were measured by Image J software. (g) Representative IHC images of α-SMA in indicated groups. Positive α-SMA staining area was measured by Image J software. Scale bar, 200 μm. Abbreviation: NS: not significant, shNC, empty vehicle alone. Data were presented as mean ± SD, **p < .01.
3.4. Kdm4d knockdown effectively suppresses HSC activation
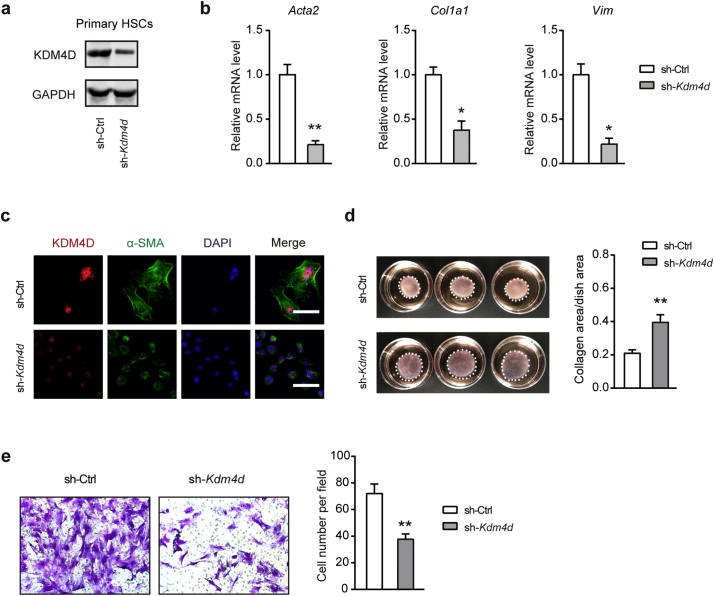
As HSCs are of vital importance in liver fibrogenesis, we further probed the impact of Kdm4d deficiency on HSC activation in intro. HSCs from shKdm4d mice had a pronounced depletion of KDM4D compared with that in the control mice as detected by western blotting (Fig. 4a). Likewise, transcriptional silencing of Kdm4d also dramatically diminished the expression of the pro-fibrogenic genes in primary HSCs including Acta2, Col1a1, and Vim as monitored by qRT-PCR (Fig. 4b). Moreover, immunofluorescence staining displayed that KDM4D knockdown attenuated the fibrogenic phenotype of activated HSCs (Fig. 4c). Given that activated HSCs are characterized by enhanced migratory and contractile capacities, we further investigated the impact of Kdm4d knockdown on these properties. Indeed, Kdm4d knockdown significantly impaired the collagen gel contraction and migratory capacities of primary HSCs as demonstrated by collagen gel contraction assay (Fig. 4d) and transwell cell migration assay (Fig. 4e), respectively. Meanwhile, loss-of-function studies in LX2 and T6 cells confirmed similar observations (Supplementary Fig. 2). These observations clearly demonstrate that KDM4D might be essential for HSC activation and Kdm4d deficiency could attenuate the crosslinking function and migration capacity of HSCs in mice.
Fig. 4.
Kdm4d knockdown effectively suppresses HSC activation in vitro. (a) Western blotting analysis of Kdm4d knockdown efficiency in activated C57BL/6J mice primary HSCs. (b) Effects of Kdm4d knockdown on the expression of pro-fibrogenic genes (Acta2, Col1a1, and Vim) were examined by qRT-PCR. (c) Representative immunofluorescence images of primary HSCs from sh-Ctrl and sh-Kdm4d mice. (d) Collagen gel contraction assay of primary HSCs from sh-Ctrl and sh-Kdm4d mice (n = 3 for each group). Statistical analysis of collagen area/dish area was shown in the right panel. (e) Representative migration images of primary HSCs from sh-Ctrl and sh-Kdm4d mice (n = 5 for each group). Statistical analysis of cell number per field was shown in the right panel. Data were presented as mean ± SD, *p < .05, and **p < .01.
3.5. KDM4D modulates TLR4 expression in HSCs
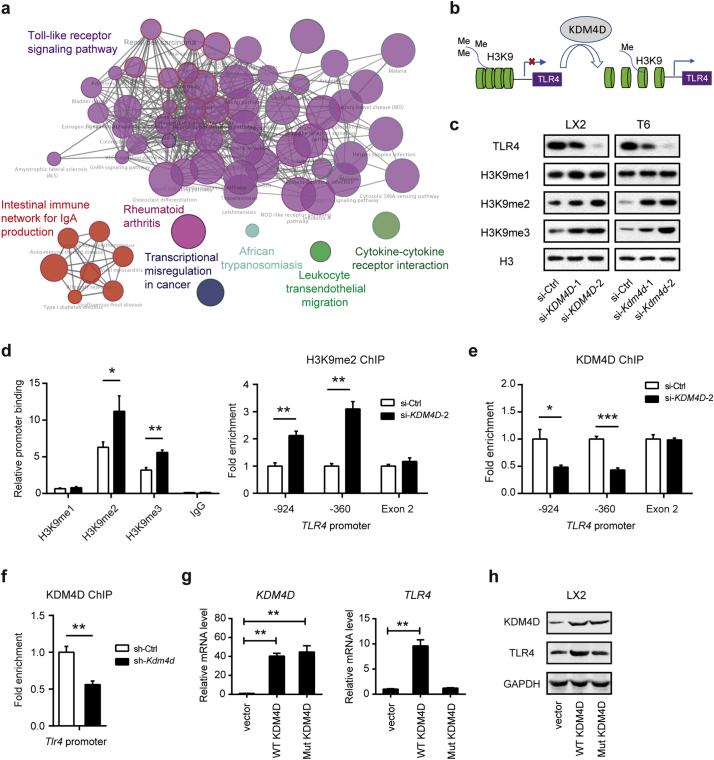
To uncover the mechanism by which KDM4D promotes liver fibrosis, a whole genome transcriptomic analysis was performed in si-NC or si-KDM4D treated LX2 cells. ClueGO analysis of the differentially downregulated genes (si-KDM4D vs. si-NC) revealed the predicted biological associations of these altered genes including the Toll-like receptor (TLR) signaling pathway, intestinal immune network for IgA production, rheumatoid arthritis, transcriptional deregulation in cancer, African trypanosomiasis, leukocyte transendothelial migration, and cytokine-cytokine interaction (Fig. 5a and Supplementary Fig. 3a). It is well known that TLR4 signaling is critically involved in the activation of HSCs during liver injury. Indeed, knockdown of TLR4 in LX2 cells dramatically inhibited the mRNA expression of pro-fibrogenic genes (Supplementary Fig. 3b, 3c). We therefore hypothesized that KDM4D might activate TLR4 expression by epigenetic modifications (Fig. 5b). Indeed, Kdm4d knockdown mainly led to dramatic reduction of TLR4 protein level and enhanced remarks of H3K9me2 and H3K9me3, but not H3K9me1 (Fig. 5c). The ChIP assay validated that compared with the negative control (mouse IgG), marked enrichment of the TLR4 promoter was found in the cistrome of H3K9me2 and H3K9me3 in KDM4D knockdown LX2 cells (Fig. 5d). Consistently, H3K9me2 ChIP analysis confirmed much higher H3K9me2 markers on the TLR4 promoter in KDM4D knockdown cells (Fig. 5d). Moreover, KDM4D ChIP analysis showed that KDM4D/Kdm4d knockdown markedly attenuated the occupancy of KDM4D on the TLR4/Tlr4 promoter in the LX2 cells (Fig. 5e) and primary HSCs (Fig. 5f), respectively. We also analyzed the implication of KDM4D overexpression on TLR4 expression by transient transfection of LX2 cells with extraneous expression vectors for KDM4D and its demethylation-defective mutant. As a result, overexpression of KDM4D but not the demethylation-defective mutant form significantly up-regulated the mRNA and protein expression of TLR4 (Fig. 5g, h). These observations clearly demonstrate the epigenetic regulatory role of KDM4D on the expression of TLR4 through modulating H3K9me2 and H3K9me3 status on its promoter in HSCs.
Fig. 5.
KDM4D modulates TLR4 expression in HSCs. (a) Expression profile chip analysis with scramble siRNA (NC) or si-KDM4D; ClueGO analysis of the differentially downregulated genes by CytoScape software. Enriched pathways are shown as nodes interconnected based on the κ score. (b) Model depicting the role of KDM4D in transcriptional regulation of TLR4 expression in liver fibrogenesis. (c) Immunoblotting of TLR4, H3K9me1, H3K9me2, H3K9me3 expression in LX2 and T6 cell lines transfected with specific siRNAs against KDM4D/Kdm4d. Histone H3 serves as the loading control. (d) ChIP assays were performed with indicated antibodies in LX2 cells with or without the KDM4D knockdown, and enrichment of target DNA was analyzed with qPCR using primers specific for the TLR4 promoter. Fold enrichments were calculated by DNA enrichment bound with H3K9me2 antibody compared with those bound with IgG. (e) KDM4D ChIP assays were performed in LX2 cells with or without KDM4D knockdown, and enrichment of target DNA was analyzed with qPCR using primers specific for the TLR4 promoter. (f) KDM4D ChIP assays were performed in primary HSCs isolated from mice with or without AAV-shKdm4d treatment, and enrichment of target DNA was analyzed with qPCR using primers specific for the Tlr4 promoter. (g, h) Effects of overexpressing of KDM4D and its demethylation-defective mutant vector on the mRNA (g) and protein expression (h) of TLR4 in LX2 cells. Data are presented as mean ± S.D. *p < .05, **p < .01, and ***p < .001.
3.6. KDM4D promotes liver fibrosis by activating TLR4/NF-κB signaling pathways
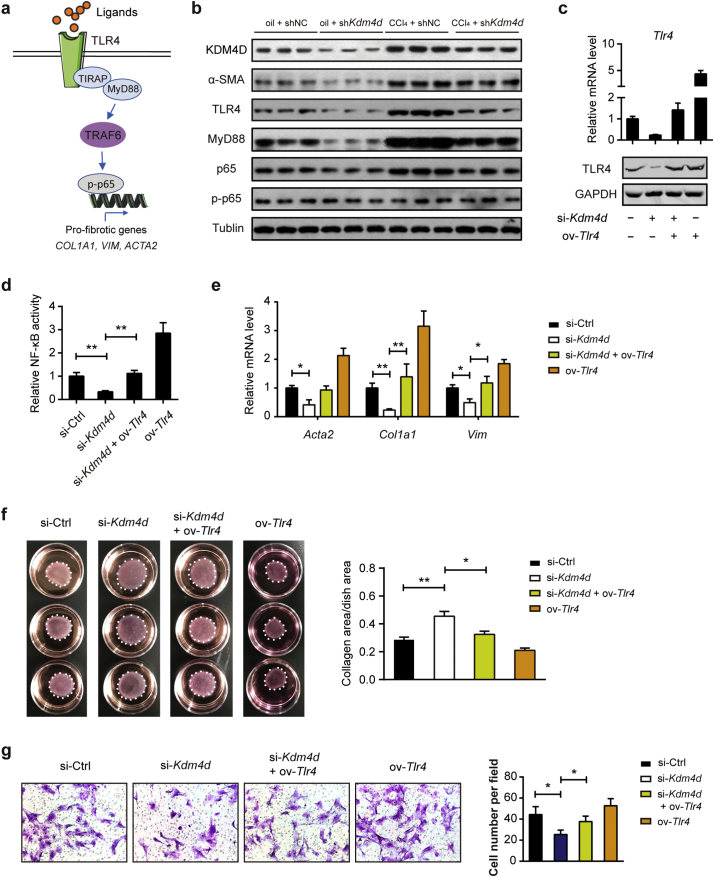
Upon stimulation with ligands, such as Lipopolysaccharides (LPS), TLR4 receptor triggers the myeloid differentiation factor 88 (MyD88)-dependent pathway and activates downstream nuclear factor κB (NF-κB) pathways (Fig. 6a) [24]. NF-κB is the master regulator for the transcription of pro-inflammatory mediators in the liver [25]. Consistently, the protein expression of MyD88 and phosphorylated p65 were significantly increased in isolated HSCs of CCl4-treated group, but notably declined when Kdm4d was knocked down in vivo (Fig. 6b). Additionally, KDM4D deficiency also significantly ameliorated the protein expression of α-SMA and TLR4 in isolated HSCs upon LPS stimulation as revealed by western blotting (Fig. 6b), suggesting the regulatory role of KDM4D in liver fibrosis might be mediated by TLR4-NF-κB signaling pathway. To confirm this hypothesis, we silenced Kdm4d and overexpressed Tlr4 in the primary isolated HSCs (Fig. 6c). Excitingly, we noticed that KDM4D deficiency in activated HSCs significantly decreased NF-κB transcriptional activity, which can be elevated by overexpression of Tlr4 (Fig. 6d). Rescue experiments also showed that decreased expression of pro-fibrotic genes (Fig. 6e), impaired contractile capacity (Fig. 6f), and migratory ability (Fig. 6g) of primary HSCs induced by Kdm4d knockdown can be largely restored by introduction of Tlr4. Likewise, similar observations were also found in LX2 cells (Supplementary Fig. 4). Together, these observations demonstrate that that KDM4D contributes to liver fibrosis possibly through modulating TLR4/NF-κB signaling pathways.
Fig. 6.
KDM4D promotes liver fibrosis by activating TLR4/NF-κB signaling pathways. (a) Schematic depicting TLR4/MyD88/NF-κB signaling pathway. (b) Proteins from primary HSCs in oil + shNC, oil + shKdm4d, CCl4 + shNC, and CCl4 + shKdm4d groups were harvested and assayed by western blotting (n = 3 representative mice for each group) with indicated antibodies. Tubulin was used as the loading control. (c) Certification of Kdm4d and Tlr4 expression at both mRNA and protein level in the presence of Kdm4d knockdown and/or Tlr4 overexpression in primary isolated HSCs. (d–g) The effects of Tlr4 overexpression on the NF-κB transcriptional activity, expression of pro-fibrotic genes, and contractile and migratory capacities in Kdm4d knockdown primary HSCs. Data are presented as mean ± S.D. *p < .05, **p < .01.
3.7. Expression pattern of KDM4D in human hepatic cirrhosis
To estimate the clinical relevance, we finally performed dual immunofluorescence staining of KDM4D expression in human fibrotic liver tissue samples. A commercial tissue microarray containing 30 normal liver and 40 hepatic cirrhosis tissues was used. As a result, KDM4D was specifically expressed in the fibrous septa and mainly co-localized with α-SMA in fibrotic liver tissues (Fig. 7a). Moreover, the immunoreactivity of KDM4D was more frequently observed in hepatic cirrhosis than normal liver tissues indicative of the potential clinical translational value of KDM4D in liver fibrosis (Fig. 7b).
Fig. 7.
Expression pattern of KDM4D in human hepatic cirrhosis. (a) Representative immunofluorescence con-focal images of α-SMA (green), KDM4D (red), and DAPI (blue) in tissue sections from human fibrotic liver. Scale bar, 50 μm. (b) Statistical analysis of the expression score of KDM4D in normal liver and hepatic cirrhosis tissues; Fisher's exact test, *p < .05.
4. Discussion
Hepatic fibrosis, characterized by HSC activation and excessive production and deposition of extracellular matrix in the liver, serves to disrupt normal liver structure and heralds more severe, irreparable liver pathologies such as hepatic failure and hepatocellular carcinoma. HSCs, the major source of fibrogenesis in the liver, locate in the Disse pace of the normal liver, and are activated into myofibroblasts by chronic stimulation. Once activated, HSCs show enhanced cell proliferation, express excessive α-SMA, and overproduce extracellular matrix. Post-translational regulations in mammals are dictated by the epigenetic mechanism, of which histone modifications constitute a key branch. They can influence chromatin structure, ultimately leading to gene expression alterations [26]. A series of epigenetic enzymes are actively involved in the addition or removal of covalent modifications, which include acetylation/deacetylation, methylation/demethylation, phosphorylation, ubiquitination, and sumoylation [27,28]. Deregulation of these processes is a hallmark of pathogenesis. The mechanism of epigenetic regulation in hepatic fibrogenesis has not been well elucidated, therefore, studies on the relationship between epigenetic modifications and liver fibrosis will refine our understanding of the fibrotic pathogenesis. The complexity of dynamic regulatory networks in the pathogenesis of liver fibrosis raises the importance of further exploration for novel factors and signaling pathways. Here, we uncovered that KDM4D, a histone demethylase, as a novel epigenetic regulator of HSC activation and thereby modulates hepatic fibrogenesis by altering methylation status of H3K9.
KDMs are versatile proteins that modulate multiple cellular processes, such as gene expression regulation, cell differentiation, embryonic stem cell renewal, and tumor development [29]. Our data portray KDM4D as a protein bridging H3K9 demethylation with HSC activation. Using H3K9me2 as a proxy for KDM4D activity is reasonable, as its level is coregulated by both histone methyltransferases and demethylases. Coincidently, the level of both histone H3 methyltransferases and demethylases are upregulated during HSC activation. Given KDM4D is the most abundant histone demethylase whose expression level is much higher than any of the other histone methylase or demethylase, we can say that it is histone demethylases rather than methyltransferases who predominantly contribute to the decline of H3K9me2 and H3K9me3 level during HSC activation. KDM4D consists of evolutionarily conserved JmjN and JmjC domains at its N terminus whereas the overall sequence of its C-terminal region contains no obvious characterized domain [10]. Apart from its well-known regulatory roles in the DNA damage response, KDM4D also stimulates p53-dependent transcriptional activity, which points to a pro-oncogenic implication [30]. In this study, a novel function of KDM4D in liver fibrogenesis was described. We uncovered that the expression of KDM4D greatly increased during trans-differentiation of quiescent HSCs to activated ones, also it was required for collagen contraction and migration capacity of HSCs. The specific change of KDM4D in HSC may represent an epigenetic alteration during fibrogenesis. Further, the severity of established liver fibrosis was alleviated as a result of Kdm4d knockdown in vivo. Kdm4d-deficient mice showed a notable reduction in not only hepatic injury severity. At the same time, Kdm4d depletion caused a significant suppression of crosslinked collagen deposition, which derived from HSC activation.
TLRs were originally identified as pathogen-associated molecular pattern recognition receptors that recognized exogenous ligands in response to infection [31]. In cirrhotic mice or patients, the gastrointestinal tract produces and absorbs considerable bacterial LPS with increased permeability of the intestinal mucosal barrier. It is well known that TLR4 signaling plays a pivotal role in liver inflammation and fibrosis. Activation of TLR4 signaling can down-regulate the bone morphogenic protein (BMP) and activin membrane-bound inhibitor (BAMBI), which enhances TGF-β1 signaling by capturing its activated ligands and subsequent amplifies fibrogenic signaling [32,33]. Therefore, targeting TLR4 signaling pathways represents an attractive strategy for liver fibrosis treatment [34]. In this study, our data portrayed that differentially expressed KDM4D modulated TLR4 level in HSCs by modulating chromatin structure. We found that genetic silencing of Kdm4d in HSCs resulted in a significant inhibition of TLR4 signaling pathway and KDM4D promoted TLR4 transcription through its demethylation activity. Engagement of ligands with the TLR4 receptor induces activation of intracellular signaling pathways through recruitment of the receptor adaptors MyD88, resulting in activation of the IκBα kinase complex and subsequent translocation of NF-κB (p65) [24]. Consistently, the expression level of TLR4 and the phosphorylated p65 as well as the fibrotic marker α-SMA were markedly decreased in Kdm4d-deficient HSC, indicating that KDM4D is indeed indispensable for HSC activation and liver fibrogenesis in a TLR4/MyD88/NF-κB-dependent manner. Apart from NF-κB, TLR4 signal transduction cascade can stimulate c-Jun N-terminal kinase (JNK), which is also essential for HSC activation [35]. Thus, targeting KDM4D provides an alternative approach against HSC activation, further, hepatic fibrogenesis.
We report that KDM4D might exert a pro-fibrotic role, which opens new horizons for hepatic fibrosis interference. It is conceivable that the presence of KDM4D drives fibrotic signaling through induction of TLR4. Certainly, the underlying mechanisms by which KDM4D facilitates liver fibrogenesis are much more complicated than we studied here. Therefore, we cannot fully exclude other targets or signaling pathways modulated by KDM4D in liver fibrosis and a ChIP-sequencing study is warranted to uncover more potential targets of KDM4D.
In conclusion, our current findings provide critical insights into the molecular mechanisms for the regulation of liver fibrosis in an epigenetic fashion. Our data highlight a previously unknown role for KDM4D, which shed light on how histone modification enzymes coordinate with transcription factors to regulate TLR4 expression and liver fibrosis. Thus, blockade of KDM4D-TLR4 signaling in HSCs may serve as a novel approach for the epigenetic interruption of liver fibrosis. Currently, there is no specific inhibitor for KDM4D, hampering the direct blockade of KDM4D-TLR4 signaling from the very beginning. However, our findings provide rationale for the screening of small-molecule compounds that target KDM4D as an interventional strategy to reverse liver fibrosis. Therefore, roles of KDM4D in the context of liver fibrosis may merit thorough investigation in follow-up studies.
Acknowledgments
Acknowledgements
We thank Asia-Vector Biotechnology Co. Ltd. (Shanghai, China), Shanghai Tuoran Co. Ltd. (Shanghai, China) and Shanghai Genechem Co. Ltd. (Shanghai, China) for their technical support. We thank Dr. Michael Patrick for manuscript polishing and Dr. Linli Yang for technical assistance with HSC isolation.
Funding sources
The research was supported by grants from Shanghai New Hundred Talents Program (No: XBR2013091), Shanghai Municipal Commission of Health and Family Planning, Key Developing Disciplines Program (No: 2015ZB0501), Shanghai Key disciplines program of Health and Family Planning (No: 2017ZZ02010) and Shanghai Sailing Program (No: 17YF1405200).
Conflicts of interest statement
The authors declare no conflicts of interest.
Authors' contributions
Zhijun Bao, Zhigang Zhang, and Shuheng Jiang conceived the study plan and contributed to the revision of the final manuscript. Fangyuan Dong, Xiaona Hu, Yiqin Huang, Xin Jiang performed the experiments, analyzed the data and finished the manuscript writing. Jun Li, Yahui Wang, and Lili Zhu contributed to the in vitro experiments. Fangyuan Dong and Shuheng Jiang performed analysis and interpretation of data.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ebiom.2018.11.055.
Contributor Information
Xiaona Hu, Email: huxnfd@hotmail.com.
Qi Zhou, Email: zhouqi@tjh.tjmu.edu.cn.
Zhigang Zhang, Email: zzhang@shsci.org.
Zhijun Bao, Email: xinyi8681@sina.com.
Appendix A. Supplementary data
Supplementary material
References
- 1.Bataller R., Brenner D.A. Liver fibrosis. J Clin Invest. 2005;115(2):209–218. doi: 10.1172/JCI24282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ellis E.L., Mann D.A. Clinical evidence for the regression of liver fibrosis. J Hepatol. 2012;56(5):1171–1180. doi: 10.1016/j.jhep.2011.09.024. [DOI] [PubMed] [Google Scholar]
- 3.Friedman S.L. Mechanisms of hepatic fibrogenesis. Gastroenterology. 2008;134(6):1655–1669. doi: 10.1053/j.gastro.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Koyama Y., Brenner D.A. Liver inflammation and fibrosis. J Clin Invest. 2017;127(1):55–64. doi: 10.1172/JCI88881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bosch F.X., Ribes J., Borras J. Epidemiology of primary liver cancer. Semin Liver Dis. 1999;19(3):271–285. doi: 10.1055/s-2007-1007117. [DOI] [PubMed] [Google Scholar]
- 6.Thiele M., Madsen B.S., Krag A. Is liver stiffness equal to liver fibrosis? Hepatology. 2017;65(2):749. doi: 10.1002/hep.28791. [DOI] [PubMed] [Google Scholar]
- 7.Chen L., Li J., Zhang J., Dai C., Liu X., Wang J. S100A4 promotes liver fibrosis via activation of hepatic stellate cells. J Hepatol. 2015;62(1):156–164. doi: 10.1016/j.jhep.2014.07.035. [DOI] [PubMed] [Google Scholar]
- 8.Senoo H., Yoshikawa K., Morii M., Miura M., Imai K., Mezaki Y. Hepatic stellate cell (vitamin A-storing cell) and its relative--past, present and future. Cell Biol Int. 2010;34(12):1247–1272. doi: 10.1042/CBI20100321. [DOI] [PubMed] [Google Scholar]
- 9.Mederacke I., Hsu C.C., Troeger J.S., Huebener P., Mu X., Dapito D.H. Fate tracing reveals hepatic stellate cells as dominant contributors to liver fibrosis independent of its aetiology. Nat Commun. 2013;4:2823. doi: 10.1038/ncomms3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Black J.C., Van Rechem C., Whetstine J.R. Histone lysine methylation dynamics: establishment, regulation, and biological impact. Mol Cell. 2012;48(4):491–507. doi: 10.1016/j.molcel.2012.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shi Y., Lan F., Matson C., Mulligan P., Whetstine J.R., Cole P.A. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119(7):941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 12.Tsukada Y., Fang J., Erdjument-Bromage H., Warren M.E., Borchers C.H., Tempst P. Histone demethylation by a family of JmjC domain-containing proteins. Nature. 2006;439(7078):811–816. doi: 10.1038/nature04433. [DOI] [PubMed] [Google Scholar]
- 13.Klose R.J., Zhang Y. Regulation of histone methylation by demethylimination and demethylation. Nat Rev Mol Cell Biol. 2007;8(4):307–318. doi: 10.1038/nrm2143. [DOI] [PubMed] [Google Scholar]
- 14.Teperino R., Schoonjans K., Auwerx J. Histone methyl transferases and demethylases; can they link metabolism and transcription? Cell Metab. 2010;12(4):321–327. doi: 10.1016/j.cmet.2010.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ortega-Molina A., Boss I.W., Canela A., Pan H., Jiang Y., Zhao C. The histone lysine methyltransferase KMT2D sustains a gene expression program that represses B cell lymphoma development. Nat Med. 2015;21(10):1199–1208. doi: 10.1038/nm.3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Atta H., El-Rehany M., Hammam O., Abdel-Ghany H., Ramzy M., Roderfeld M. Mutant MMP-9 and HGF gene transfer enhance resolution of CCl4-induced liver fibrosis in rats: role of ASH1 and EZH2 methyltransferases repression. PLoS One. 2014;9(11) doi: 10.1371/journal.pone.0112384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Beguelin W., Popovic R., Teater M., Jiang Y., Bunting K.L., Rosen M. EZH2 is required for germinal center formation and somatic EZH2 mutations promote lymphoid transformation. Cancer Cell. 2013;23(5):677–692. doi: 10.1016/j.ccr.2013.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Coward W.R., Feghali-Bostwick C.A., Jenkins G., Knox A.J., Pang L. A central role for G9a and EZH2 in the epigenetic silencing of cyclooxygenase-2 in idiopathic pulmonary fibrosis. FASEB J. 2014;28(7):3183–3196. doi: 10.1096/fj.13-241760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen Z., Zang J., Whetstine J., Hong X., Davrazou F., Kutateladze T.G. Structural insights into histone demethylation by JMJD2 family members. Cell. 2006;125(4):691–702. doi: 10.1016/j.cell.2006.04.024. [DOI] [PubMed] [Google Scholar]
- 20.Khoury-Haddad H., Guttmann-Raviv N., Ipenberg I., Huggins D., Jeyasekharan A.D., Ayoub N. PARP1-dependent recruitment of KDM4D histone demethylase to DNA damage sites promotes double-strand break repair. Proc Natl Acad Sci U S A. 2014;111(7):E728–E737. doi: 10.1073/pnas.1317585111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Khoury-Haddad H., Nadar-Ponniah P.T., Awwad S., Ayoub N. The emerging role of lysine demethylases in DNA damage response: dissecting the recruitment mode of KDM4D/JMJD2D to DNA damage sites. Cell Cycle. 2015;14(7):950–958. doi: 10.1080/15384101.2015.1014147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shin S., Janknecht R. Activation of androgen receptor by histone demethylases JMJD2A and JMJD2D. Biochem Biophys Res Commun. 2007;359(3):742–746. doi: 10.1016/j.bbrc.2007.05.179. [DOI] [PubMed] [Google Scholar]
- 23.Jiang S.H., Li J., Dong F.Y., Yang J.Y., Liu D.J., Yang X.M. Increased serotonin signaling contributes to the Warburg effect in pancreatic tumor cells under metabolic stress and promotes growth of pancreatic tumors in mice. Gastroenterology. 2017;153(1):277–291. doi: 10.1053/j.gastro.2017.03.008. [e19] [DOI] [PubMed] [Google Scholar]
- 24.Seki E., De Minicis S., Osterreicher C.H., Kluwe J., Osawa Y., Brenner D.A. TLR4 enhances TGF-beta signaling and hepatic fibrosis. Nat Med. 2007;13(11):1324–1332. doi: 10.1038/nm1663. [DOI] [PubMed] [Google Scholar]
- 25.Karin M., Lin A. NF-kappaB at the crossroads of life and death. Nat Immunol. 2002;3(3):221–227. doi: 10.1038/ni0302-221. [DOI] [PubMed] [Google Scholar]
- 26.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128(4):693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 27.Yamane K., Toumazou C., Tsukada Y., Erdjument-Bromage H., Tempst P., Wong J. JHDM2A, a JmjC-containing H3K9 demethylase, facilitates transcription activation by androgen receptor. Cell. 2006;125(3):483–495. doi: 10.1016/j.cell.2006.03.027. [DOI] [PubMed] [Google Scholar]
- 28.Whetstine J.R., Nottke A., Lan F., Huarte M., Smolikov S., Chen Z. Reversal of histone lysine trimethylation by the JMJD2 family of histone demethylases. Cell. 2006;125(3):467–481. doi: 10.1016/j.cell.2006.03.028. [DOI] [PubMed] [Google Scholar]
- 29.Shin S., Janknecht R. Diversity within the JMJD2 histone demethylase family. Biochem Biophys Res Commun. 2007;353(4):973–977. doi: 10.1016/j.bbrc.2006.12.147. [DOI] [PubMed] [Google Scholar]
- 30.Kim T.D., Oh S., Shin S., Janknecht R. Regulation of tumor suppressor p53 and HCT116 cell physiology by histone demethylase JMJD2D/KDM4D. PLoS One. 2012;7(4) doi: 10.1371/journal.pone.0034618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schwabe R.F., Seki E., Brenner D.A. Toll-like receptor signaling in the liver. Gastroenterology. 2006;130(6):1886–1900. doi: 10.1053/j.gastro.2006.01.038. [DOI] [PubMed] [Google Scholar]
- 32.Liu C., Chen X., Yang L., Kisseleva T., Brenner D.A., Seki E. Transcriptional repression of the transforming growth factor beta (TGF-beta) Pseudoreceptor BMP and activin membrane-bound inhibitor (BAMBI) by nuclear factor kappaB (NF-kappaB) p50 enhances TGF-beta signaling in hepatic stellate cells. J Biol Chem. 2014;289(10):7082–7091. doi: 10.1074/jbc.M113.543769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tao L., Xue D., Shen D., Ma W., Zhang J., Wang X. MicroRNA-942 mediates hepatic stellate cell activation by regulating BAMBI expression in human liver fibrosis. Arch Toxicol. 2018;92(9):2935–2946. doi: 10.1007/s00204-018-2278-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sun L., Dai J.J., Hu W.F., Wang J. Expression of toll-like receptors in hepatic cirrhosis and hepatocellular carcinoma. Genet Mol Res. 2016;15(2) doi: 10.4238/gmr.15027419. [DOI] [PubMed] [Google Scholar]
- 35.Paik Y.H., Schwabe R.F., Bataller R., Russo M.P., Jobin C., Brenner D.A. Toll-like receptor 4 mediates inflammatory signaling by bacterial lipopolysaccharide in human hepatic stellate cells. Hepatology. 2003;37(5):1043–1055. doi: 10.1053/jhep.2003.50182. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material