Abstract
Background
FOXP1, a transcriptional regulator of lymphocyte development, is abnormally expressed in some human tumors. This study investigated FOXP1-mediated regulation of tumor infiltrating lymphocytes (TIL) in untreated primary breast cancer (BC).
Methods
FOXP1 expression was analyzed in tissues from primary untreated breast tumors, BC cell lines and the METABRIC gene expression BC dataset. Cytokine and chemokine expression and lymphocyte migration in response to primary tumor supernatants (SN) was compared between FOXP1hi and FOXP1lo primary BC.
Finding
FOXP1 expression was higher in estrogen receptor positive compared to negative BC. FOXP1hi tumors were significantly associated with lower TIL and fewer tertiary lymphoid structures (TLS) compared to FOXP1lo BC. Silencing FOXP1 in BC cell lines positively impacted cytokine and chemokine expression with the inverse effect associated with overexpression. CXCL9, CXCL10, CXCL11, CXCL13, CX3CL, CCL20, IL2, IL21, GZMB and IFNG expression decreased while IL10 and TGFβ increased in FOXP1hi compared to FOXP1lo primary BC. Lymphocyte migration using primary BC supernatants detected decreased mobility toward FOXP1hi supernatants. FOXP1lo BC expresses higher levels of chemokines driving TIL migration. The METABRIC gene expression dataset analysis show FOXP1 expression is associated with unfavorable BC outcomes.
Interpretation
These data identify FOXP1 as an important negative regulator of immune responses in BC via its regulation of cytokine and chemokine expression.
Fund
Belgian Fund for Scientific Research (FNRS 3.4513.12F) and Opération Télévie (7.4636.13F and 7.4609.15F), Fonds J.C. Heuson and Fonds Lambeau-Marteaux.
Keywords: FOXP1, Breast cancer, Tumor infiltrating lymphocytes, Chemokines, Cytokines
Graphical abstract
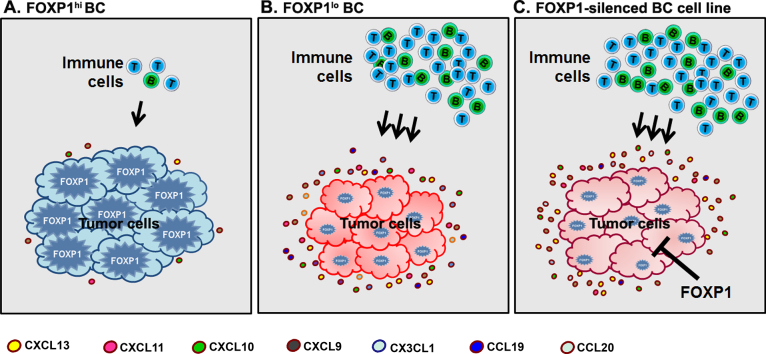
We identified (A) FOXP1 high (FOXP1hi) and (B) low (FOXP1lo) primary untreated breast tumors. FOXP1lo tumors express high levels of chemokine genes and have high TIL. Inversely, FOXP1hi tumors have repressed chemokine gene expression and low levels of TIL. (C) FOXP1 silencing induces chemokine gene expression in breast cancer cell lines which shows direct regulation by FOXP1. This study provides new insight by showing that FOXP1 repression can induce TIL recruitment to tumors, thereby arming the patient with competent immunological memory that can recognize residual tumor cells after treatment.
Research in context.
Evidence before this study
The published literature on FOXP1 has grown exponentially over the past few years. Initial studies of FOXP1 implicated this transcription factor in central nervous system disorders with recent work showing it can function as a key repressor of immune signaling and cytokine gene expression. Early on, FOXP1 was characterized as a tumor suppressor because its decreased expression was an unfavorable prognostic factor for several solid tumor types, including breast cancer. Alternatively, in B cell lymphomas it was perceived as an oncogene because FOXP1 overexpression was associated with poor patient prognosis. Recent studies have defined FOXP1 as a critical regulator or T cell and B cell differentiation revealing that it controls many steps in lymphocyte maturation. Currently, studies investigating the mechanisms whereby FOXP1 influences the interplay between malignant cells and immune cells in cancer are lacking.
Added value of the present study
The data presented in this study show that FOXP1 is an important negative regulator of immune infiltration in breast cancer via its control of chemokine expression. Specific chemokines, produced by normal and/or malignant cells in the tumor microenvironment, are needed to drive tumor infiltrating lymphocyte (TIL) migration and their organization in tertiary lymphoid structures (TLS). The anti-correlation we observed between FOXP1 expression and the extent of TIL is directly linked with the production of chemoattractants that recruit TIL. We show that breast tumors with high FOXP1 expression have low cytokine and chemokine production while the inverse was true for low FOXP1 tumors. Our previous work linked the B cell chemoattractant CXCL13 to extensive TIL infiltration and TLS organization in breast cancer. In this study, we show that CXCL13 is regulated by FOXP1, with incremental increases in FOXP1 gene expression associated with a decline in survival. These data suggest that FOXP1 expression creates and/or maintains an immunosuppressive tumor microenvironment by controlling critical immune gene expression. These data are, to our knowledge, the first to implicate FOXP1 in immune gene regulation and TIL migration in breast cancer.
Implications based on all available evidence
Published data show that FOXP1 is an important negative regulator of anti-tumor immune responses via its control of chemokine expression. The findings presented in this manuscript add novel insight into the regulation of immune migration and infiltration in breast cancer. A key regulator of immune activity in tumors associated with survival across multiple tumor types is the chemokine CXCL13. The present data extend our work on CXCL13 in breast cancer by demonstrating that CXCL13 expression is regulated by FOXP1.
Alt-text: Unlabelled Box
1. Introduction
Historically, breast cancer (BC) has not been viewed as an immunogenic tumor, primarily due to its intermediate mutational load [1] with limited data available on tumor-specific neoantigens in this malignancy. Recent clinical studies, however, reveal a strong link between patient prognosis, response to treatment and immune activities, including immune gene expression [2] and the extent of tumor infiltrating lymphocytes (TIL) at the tumor site [3]. Our recent work determined that TIL density in fresh BC tissues forms a continuum from TIL-negative (TILneg) to TIL-high (TILhi) [4]. Using thresholds defined by normal breast tissues, we identified 25% of tumors as TILneg while the remaining TIL-positive (75%; TILpos) tumors were equally divided into TIL-intermediate (TILint) and TILhi. We further identified a positive correlation between the extent of TIL and the level of immune organization in ectopic lymph node-like tertiary lymphoid structures (TLS). TLS, characterized by a B cell follicle surrounded by a T cell zone, function to generate humoral and cell-mediated immune responses at sites of chronic inflammation [5]; although, the sequence of events involved in their formation in tumors is currently unclear. A recent study demonstrated that induction of TLS formation in an experimental murine tumor model initiated an influx of T cells, which when combined with immunotherapy led to effector and memory cell generation [6].
Most cells, including epithelial cells, have the potential to modulate immune responses via their production and secretion of distinct immunomodulatory cytokines or chemokines. Cytokine/chemokine signaling can in turn affect downstream transcription factor (TF) activities. For example, interferon regulatory factors (IRF), NFκB and STAT, have all been shown to regulate TIL trafficking and TLS formation in BC [7]. Further, IRF5 was found to be a novel and direct regulator of CXCL13 expression in mammary epithelial tumor cells, a chemokine with important effects on T and B cell trafficking to the tumor [7,8] and TLS formation [9,10].
The TF forkhead box protein 1 (FOXP1) has been shown to regulate normal epithelial cell fate during lung development and regeneration [11]. Studies of FOXP1's role in the immune response have expanded exponentially over the past decade following an initial paper establishing its role as a critical regulator of early B cell development [12]. Subsequent work has shown that FOXP1 is also involved in T cell quiescence [13], monocyte differentiation and macrophage function [14].
Abnormal FOXP1 expression has been documented in various human cancers where it can either act as an oncoprotein or tumor suppressor depending on the tumor type. In hematological malignancies, FOXP1 was shown to function as an oncogene by suppressing immune gene expression and promoting tumor cell survival in B cell lymphoma [15] or a tumor suppressor due to the association of its overexpression with improved survival in T cell lymphoma [16]. Studies have identified oncogenic activities for FOXP1 in epithelial tumors, including ovarian cancer where it promotes stem cell features [17] and an association with reduced survival in oral squamous cell carcinoma [18], hepatocellular carcinoma [19] and glioblastoma [20]. FOXP1 has also been shown to act as a tumor suppressor in non-small cell lung, endometrial, colorectal and prostate cancer [reviewed in [21]].
In BC, FOXP1 has been detected in tumor cells where its increased expression was positively associated with the estrogen receptor (ER), including both ERα in a primary cohort [22] and ERβ in an invasive cohort [23]. Several studies analyzed global FOXP1 expression, independent from BC subtype, correlating its expression with an improved prognosis, again suggesting it functions as a tumor suppressor [[22], [23], [24], [25]]. Shigekawa et al. further showed that stimulating FOXP1 expression in the BC cell line MCF7, increases proliferation and elevates ER-mediated gene transcription [24]. They also correlated FOXP1 positivity with improved disease free survival in tamoxifen-treated BC patients. Recent experiments found FOXP1-depleted MCF7 cells had reduced growth while its overexpression enhanced proliferation, both supporting a pro-tumorigenic role in BC [26]. Finally, FOXP1 enhanced migration by repressing NFAT1 in the BC cell line MDA-MB-231 [27].
Overall, these findings indicate that further investigation of FOXP1's regulatory roles in BC may provide important insight on pro- and anti-tumor immune responses. The present study explores FOXP1 functionality in breast tumor cells and examines its effects on immune cell activity. We show that FOXP1 suppresses the expression of numerous cytokines/chemokines involved in T and B cell chemotaxis. We further demonstrate that FOXP1 has a direct negative impact on T and B cell migration and this anti-correlation parallels adverse clinical outcomes in early-stage BC.
2. Materials and methods
2.1. Human samples and cell lines
Formalin-fixed and paraffin-embedded (FFPE) tumor tissue from 104 untreated primary BC patients, diagnosed and treated in the early stage setting between 2012 and 2015 at the Institut Jules Bordet (part of the MIU prospective BC cohort [4]) were evaluated in this study with their clinicopathological parameters detailed in Tables S1 and S2. Peripheral blood mononuclear cells (PBMC) from healthy donors (HD) were also used. All specimens and clinical data were analyzed using procedures approved by the Institut Jules Bordet's Medical Ethics Committee (1981 CE) and the GZA Ziekenhuizen Medical Ethics Committee (CE 130909ACADEM). All patients and HD signed their informed consent.
MCF7 [Luminal or ER+/Progesterone Receptor (PR)+/ Human Epidermal Growth Factor Receptor 2 (HER2-)], BT474 (ER+/HER2+) and MDA-MB-231 [triple negative (TN; ER-/PR-/HER2-)] are human epithelial cell lines derived from breast tumors that were purchased from the American Type Culture Collection (ATCC; Manassas, VA, USA) and used at low passage numbers from the original vial. All cell lines routinely tested negative for Mycoplasma contamination.
2.2. Primary tumor supernatant and tumor cell line conditioned media
Primary tumor supernatants (SN) were prepared from fresh surgical tissues mechanically dissociated (2x30s; GentleMACS Dissociator) without enzymatic digestion in 3 ml of X-VIVO™ 20 (Lonza) as detailed in Garaud et al. [28]. Tumor cell line conditioned media (TCM) was collected from MCF7 cells (transfected or not) grown for 48 h before being refed with fresh medium containing 1% fetal bovine serum (FBS) to produce TCM collected after an additional 24 h. TCM was filtered through a 0.45 μm syringe filter (Millipore) and either used immediately or stored at −80 °C.
2.3. siRNA FOXP1 knock-down and FOXP1 overexpression
The FOXP1 gene was silenced in MCF7 cells using predesigned siRNA-targeted sequences directed against FOXP1 isoforms (Qiagen; SI00421120, SI03196655 and SI04954663). The FOXP1 gene was upregulated in MDA-MB-231 cells using a FOXP1 expression plasmid (pcDNA4-TO/FOXP1) for overexpression [control plasmid was the empty vector pcDNA4-TO; both kindly provided by Dr. M. Spaargaren (University of Amsterdam, Amsterdam, The Netherlands)]. This FOXP1 expression vector produces isoform 1, the full length 75 kDa FOXP1 protein, upon induction with tetracycline. Transfection of MCF7 and MDA-MB-231 was done using lipofectamine 3000 (Invitrogen) following the manufacturer's instructions with cells harvested after 48 h for total RNA extraction or 72 h for protein extraction. The empty vector and a universal scrambled sequence containing a FITC fluorescence marker (Santa Cruz Biotechnology) were used as transfection controls. Transfection efficiency was estimated by flow cytometry. RNA analysis was performed using RT-qPCR and protein analysis by immunoblotting to confirm FOXP1 repression or upregulation.
2.4. RNA extraction and analysis
Total RNA from cell lines was extracted using the TRIzol Reagent (Invitrogen) following standard protocols. Total RNA extracted from FFPE tissues (three 5 μm tissue sections per tumor) was isolated using RNeasy (Qiagen) following the manufacturers protocol. Adequate tissue for RNA extraction was available for 94/104 of the FFPE tissues. Isolated RNA was reverse transcribed (RT) using the High Capacity RNA-to-cDNA kit (Applied Biosystems) following standard procedures. cDNAs derived from FFPE tissues were subjected to 10 cycles of amplification in a cocktail of primers using a pre-amplification buffer (Life Technologies). Quantitative polymerase chain reactions (qPCR) were performed on an ABI 7900HT Prism sequence detector (Applied Biosystems).
FOXP1 primers (Invitrogen) were designed to target the different FOXP1 gene isoforms [13]. In this study, all analyses were performed for the FOXP1 isoform 1 with FOXP1 gene expression data shown in a log2 scale. The expression of cytokine and chemokine genes was performed using FFPE breast tumors (n = 50) from the MIU prospective BC cohort and sorted epithelial cells from fresh breast tissues (n = 8), with the primer sequences listed in Table S3. Relative mRNA expression levels were normalized using the mean expression for two reference genes, MLN51 and EF1α with fold changes calculated using the 2−ΔCt method. All RT-qPCR reactions were processed in duplicate. We used the median value of FOXP1 gene expression in the 94 FFPE BC tissues to divide our cohort into FOXP1 low (FOXP1lo) and FOXP1 high (FOXP1hi) tumors. FOXP1 expression in normal breast tissues (n = 10) was also analyzed.
2.5. Human cytokine/chemokine gene analysis in BC cell lines
Human cytokines and chemokines were analyzed using a human cytokine Taqman® array assay (Life Technologies; 4391139) with a 7900HT Prism sequence detector (Applied Biosystems). Data reflect the mean of three independent biological experiments and fold changes were calculated using the 2−ΔΔCt method.
2.6. METABRIC microarray and DNAseq data analysis
METABRIC dataset (available from the European Genome Archive; Accession number EGAS00000000083; discovery plus validation set, n = 1992) [2] were used to analyze global FOXP1 gene expression (log2 scale) and evaluate variation in mutations and copy number alterations among the BC molecular subtypes. The prognostic value [(disease specific survival (DSS) and overall survival (OS)] of global FOXP1 gene expression was evaluated using the same dataset (detailed in Supplementary Methods).
2.7. Flow cytometric analysis and epithelial cell sorting
Suspensions of PBMC or epithelial cells from fresh BC tissues or cell lines were incubated with the manufacturers' suggested dilution of fluorescently-labeled primary monoclonal antibodies (Table S4). Nuclear labeling of FOXP1 was accomplished using fixed and permeabilized cells (pre-labeled for membrane markers) with the BD Cytofix/Cytoperm™ Fixation/Permeabilization Solution Kit (BD 554714) following the manufacturer's protocol. Absolute cell count beads (123count eBeads; eBioscience) were used to quantify cellular subpopulations. Fluorescently-labeled cell acquired on a GALLIOS 10/3 cytometer and analyzed using Kaluza® 1.3 Flow Analysis Software (Beckman Coulter). Epithelial cell suspensions were prepared from fresh BC tissues by enzyme digestion using optimized conditions (detailed in Supplementary Methods) of Liberase DH enzyme (Roche) and sorted on a Moflo ASTRIOS EQ. 12/4 sorter with gating on EpCAM+ cells.
2.8. Immunohistochemical analysis and assessment
Consecutive FFPE tissue sections (n = 104) were immunohistochemically (IHC)-stained using a BenchMark XT IHC/ISH automated slide stainer (Ventana Mediated Systems using their reagents) for FOXP1 and dual IHC for CD3 (pan T cells; Dako) and CD20 (pan B cells; Dako) (Supplementary Table S4). Two experienced pathologists (RdW, GvE) scored the IHC-stained slides for: 1) the percentage of FOXP1 positive (FOXP1+) tumor cells; 2) TIL as a percentage in the tumor bed and/or peri-tumoral stroma areas; and 3) the number of TLS (dense aggregates of B cells with an adjacent T cell zone). The TLS score was normalized by calculating the number of TLS per tumor area (mm2). The threshold for FOXP1 nuclear positivity was set at ≥5% of positive cells in the tumor.
2.9. Immunoblot analysis
Protein extraction, immunoblotting and protein detection were done as previously described [13] using the antibodies detailed in Supplementary Table S4. Antibodies used are listed in Supplementary Table S4. The endogenous control used was mouse anti-beta actin (β-actin) antibody (Cell Signaling). Antibody labelling was identified using enhanced chemiluminescence reagents (Amersham) and quantified with a ChemiDoc™ XRS reader (Bio-Rad).
2.10. Confocal microscopy
Immunofluorescence (IF) staining was performed manually on FFPE tumor tissues sections (4 μm) [detailed in [13]]. Tonsils and normal breast tissues were used as controls. Antibodies used are listed in Table S4. After the final washing, slides were mounted with ProLong Gold antifade mounting medium with DAPI (Thermo Scientific) and visualized on a Zeiss LSM 710 confocal microscope equipped with an x20/0.8 and x63/0.8 Plan-Apochromat dry objective (Carl Zeiss, Oberkochen, Germany).
2.11. Migration assays
The migratory properties of fresh PBMC, isolated from HD blood using Lymphoprep™ density gradient centrifugation, were evaluated using a transwell system (24-well, 3.0 μm; Millipore) following the manufacturer's instructions. The bottom chambers contained either untreated or FOXP1-repressed MCF7 TCM or primary BC SN. 10% FBS was used as a positive control because it is a strong chemoattractant. Transwells were incubated at 37 °C in 5% CO2 for 24 h. The number of cells migrating into the bottom chamber was determined by flow cytometry.
2.12. Statistical analysis
The statistical significance for resulting data was calculated using an unpaired Student's t-test or a one-way ANOVA and the GraphPad Prism 6.0 software unless otherwise specified. A p value of <0.05 was considered statistically significant.
3. Results
3.1. FOXP1 gene expression in BC
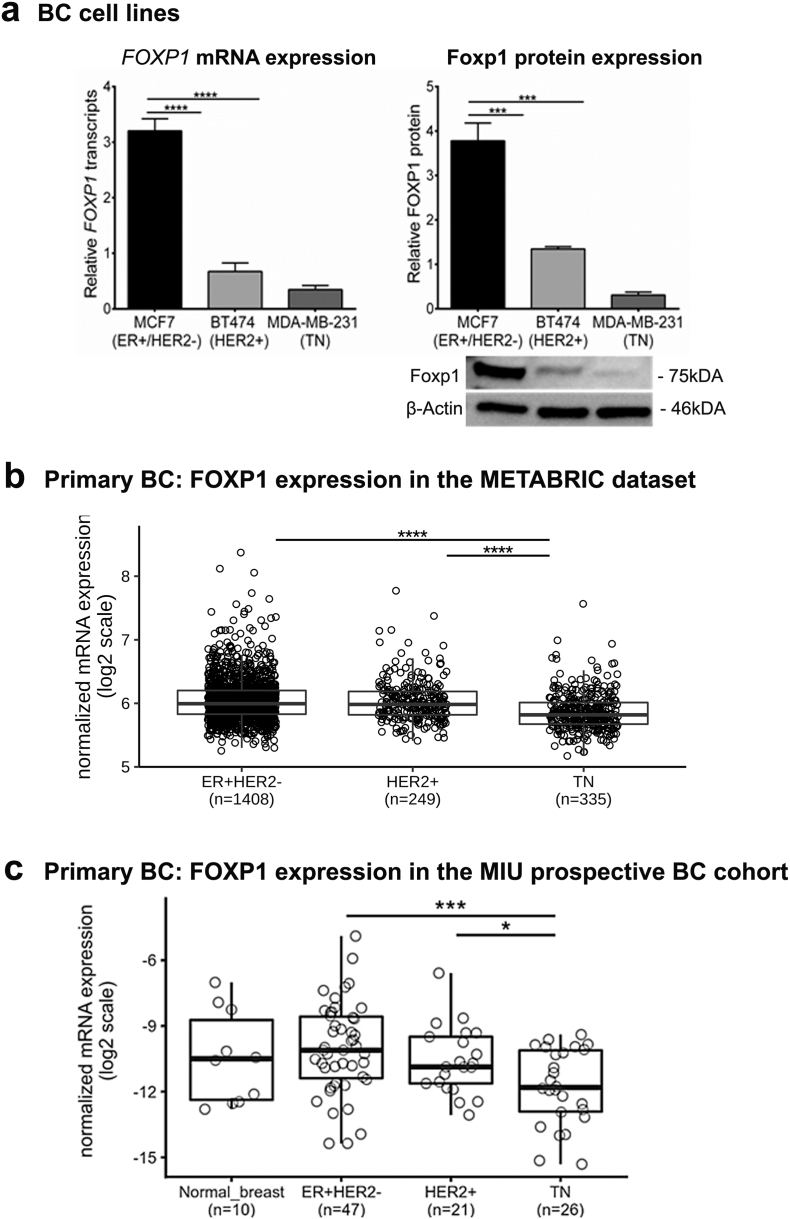
The full-length Foxp1 protein is encoded by isoform 1 (NCBI Gene ID: 27086), which is widely expressed in normal tissues but differentially expressed in epithelial cell malignancies including BC [29]. FOXP1 gene expression levels were determined for three BC cell lines: MCF7 (ER+/PR+/HER2-), BT474 (ER+/HER2+) and MDA-MB-231 (ER-/PR-/HER2-) with expression levels significantly lower in BT474 and MDA-MB-231 compared to MCF7 (Fig. 1A). Foxp1 protein analysis confirmed this substantial reduction in BT474 and MDA-MB-231 (Fig. 1A). Significant differences in the expression of smaller FOXP1 isoforms (transcripts or protein) were not detected between the three cell lines (Fig. S1A; some data not shown). All of the following functional experiments in this study therefore analyzed the FOXP1 isoform 1 and/or the full-length protein (~75 kDa; Fig. 1A).
Fig. 1.
FOXP1 expression in breast cancer cell lines and primary breast cancer.
(a) FOXP1 gene and protein expression in breast epithelial tumor cell lines [(MCF7 (ER+/HER2-), BT474 (HER2+) and MDA-MB-231 (triple negative: TN)] by RT-qPCR and immunoblotting. All data were obtained as the mean of three independent experiments. Blots were probed with anti-β actin as subcellular compartment control and isotype control for FOXP1 was also used.
(b) Microarray data from METABRIC dataset were analyzed for FOXP1 gene expression (log2) among the breast cancer molecular subtypes (n = 1992).
(c) Evaluation of FOXP1 gene expression using RT-qPCR was performed in a cohort of primary breast tumors (n = 94) from the tumor banks of Institut Jules Bordet. FOXP1 expression in normal breast also analyzed (n = 10). RT-qPCR data are relative to EIF1α and MLN51 expression (2-ΔCt) done in duplicates shown in log2 scale. Significant P values (<0.05) are marked by *. Degrees of significance: P < 0.05 (*), P < 0.001 (***) and P < 0.0001 (****), as assessed with ANOVA using Dunn's multiple comparisons test.
Data represent mean ± SEM.
Global FOXP1 mRNA expression in primary BC was analyzed using public microarray data from the METABRIC dataset (n = 1992; samples from untreated primary BC patients) and RNA extracted from tumors in the MIU prospective BC cohort (n = 94). Examination of the three BC molecular subtypes studied in METABRIC reveals that FOXP1 expression is significantly lower in HER2+ (ER- and ER+) and TN (ER-PR-HER2-) compared to Luminal A/B (ER + HER2-) (Fig. 1B and Fig. S1B). The METABRIC dataset was also used to determine if FOXP1 gene repression was associated with genetic alterations but no significant copy number alterations or mutations were detected in this gene (Fig. S2C and D). These data suggest that lower FOXP1 levels in HER2+ and TN are not due to genomic alterations in the FOXP1 gene. RT-qPCR quantification of FOXP1 mRNA in samples from the MIU prospective BC cohort reflect these data with significant reductions detected in TN and HER2+ BC (Fig. 1C). Some clinicopathological parameters from this cohort were significantly correlated with FOXP1 expression, including ER and PR positivity and low tumor grades (G1 & G2) (Table S2).
3.2. FOXP1 protein expression in BC
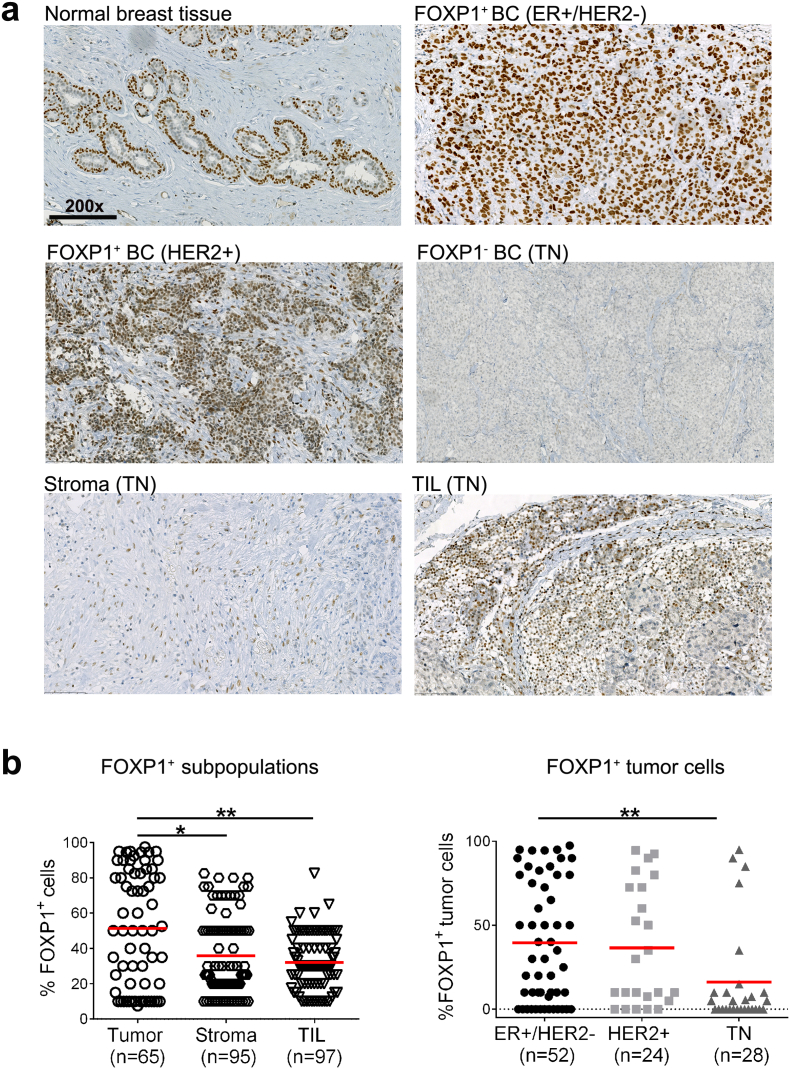
The localization of FOXP1 in the breast tumor microenvironment (TME) was determined using IHC-stained full-face tissue sections from patients in the MIU prospective BC cohort (n = 104) together with normal breast tissue controls. Prominent FOXP1 nuclear staining characterizes normal breast epithelial cells, tumor cells in some BC and TIL; while less intense staining typifies stromal cells and malignant cells in other tumors (images for different staining patterns are shown in Fig. 2A). Overall, fewer BC in our cohort contain FOXP1+ tumor cells (63%) than FOXP1+ TIL and/or stromal cells (>90%; Fig. S2A—C). However, within the FOXP1+ tumor cell group, the frequency of positive cells was significantly higher than stromal cells or TIL (i.e. the % of positive cells within a given subpopulation; Fig. 2B; left panel). Fewer FOXP1+ tumor cells were detected in HER2+ and TN compared with ER+/HER2- tumors (Fig. 2B; right panel). Overall, FOXP1+ tumors are more numerous in the ER+ BC subtype with the highest number of FOXP1+ cells found in the tumor cell subpopulation.
Fig. 2.
FOXP1 protein expression in primary breast cancer.
a) Representative FOXP1 staining in normal breast tissue, ER+/HER2-, HER2+ and triple negative breast cancer are shown. FOXP1 staining in stromal cells and TIL in a triple negative is shown in the bottom panel. All images are at magnifications of 200×.
(b) The frequency of FOXP1 protein expression in different cell subpopulations and frequency of FOXP1 tumor cells among breast cancer molecular subtypes using IHC. FOXP1 positivity in tumor cells, stromal and TIL have been considered while FOXP1 expression in tumor cells in ER+/HER2-, HER2+ and triple negative were analyzed. FOXP1 positivity was defined as ≥5% of any positive cell among all cells (n = 104). Significant P values (<0.05) are marked by *. Degrees of significance: P < 0.05 (*) and P < 0.01 (**), as assessed with ANOVA using Dunn's multiple comparisons test.
Data represent mean ± SD. IHC: Immunohistochemistry.
3.3. FOXP1 and TIL are anti-correlated in BC
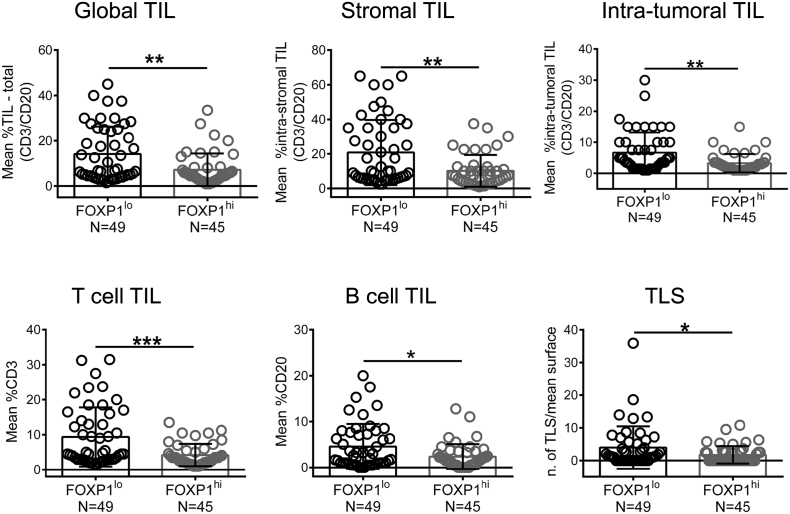
We next asked whether the lower FOXP1 expression observed in association with higher immune infiltration [the latter more frequent in HER2+ and TNBC, reviewed in [30]] was related to the number of TIL. Dual CD3/CD20 IHC-stained full-face tissue sections from the same 104 patients examined for FOXP1 expression (above) were scored for T and B cell TIL. Sequential tissue sections were cut for RNA extraction with FOXP1 mRNA expression used to divide the cohort in FOXP1lo and FOXP1hi groups based on the median. A comparison of CD3, CD20 or CD3 plus CD20 positivity for global, stromal and intra-tumoral TIL reveals significant decreases in FOXP1hi relative to FOXP1lo tumors for total lymphocytes as well as the individual T and B cell subpopulations (Fig. 3). Moreover, when we analyzed TIL and FOXP1 gene expression in all tumors, the data revealed significant anti-correlations with global TIL and TIL subpopulations (Fig. S3). Previous work in our laboratory identified TLS as a location and organizational feature of TIL in extensively-infiltrated BC [4,9,10,31]. The consistent anti-correlation detected between FOXP1 and TIL led us to compare TLS frequencies, and as anticipated, more TLS were detectable in TILhi FOXP1lo tumors (Fig. 3).
Fig. 3.
Relationship between FOXP1 and TIL infiltration.
Assessment of global TIL, stromal TIL, intra-tumoral TIL, T cell infiltration, B cell infiltration and TLS based on CD3/CD30 IHC in FOXP1lo (n = 49) and FOXP1hi (n = 45) breast cancer. Tumors (n = 94) were divided according to the median value of FOXP1 gene expression.
Significant P values (< 0.05) are marked by *. Degrees of significance: P < 0.05 (*), P < 0.01 (**), and P < 0.001 (***), as assessed with Mann Whitney test. Data represent mean ± SD.
Protein and transcript levels are frequently not well correlated, leading us to, evaluate FOXP1 transcripts relative to protein expression in the MIU cohort tumors. Interestingly, we found a significant correlation between the two (Fig. S2B) and therefore also divided our tumors (n = 94) into FOXP1lo and FOXP1hi groups based on the median value of Foxp1 protein expression. This analysis revealed a significant reduction in total TIL as well as individual T and B cell subpopulations in the FOXP1hi relative to FOXP1lo tumors, which was consistent with our transcript data (Fig. 3SC). Intra-tumoral TIL levels were unchanged in the two groups.
3.4. FOXP1 regulates cytokine and chemokine expression in BC cell lines
The observed inverse correlation between FOXP1 and TIL suggested cytokines and/or chemokines might be regulated by this TF in BC. Cytokine/chemokine gene expression was initially analyzed by using siRNA to silence its expression in FOXP1hi MCF7 cells and an expression vector for overexpression in FOXP1lo MDA-MB-231 BC cells. Validation of effective FOXP1 mRNA transcript and protein downregulation by siRNA as well as increased expression in transfected cells are shown in Fig. S4A and B.
The effect of FOXP1 repression on cytokine/chemokine expression was first analyzed using a 96-gene expression array. Twenty-eight cytokines/chemokines were reproducibly upregulated (>1.5-fold) in siRNA-silenced FOXP1hi MCF7 cells, including: the ligands CCL2, CCL3, CCL5, CCL17, CCL19, CCL20, CXCL9, CXCL10, CXCL11, CXCL13, IL6, IL8 and SLIT2; the adapter MYD88, the receptor TLR2 and the TF NFKB1 (p50 subunit) (Fig. S4C, D and Table S5; underlined genes were deregulated in both the repressed MCF7 and overexpressed MDA-MB-231). Four genes were downregulated (<0.5) in FOXP1-repressed MCF7 cells including the chemokine CXCL12 and its receptor CXCR4. Overexpressing FOXP1 protein in FOXP1lo MDA-MB-231 cells detected significant decreases in 34 genes (<0.5), including: the ligands CCL2, CCL3, CCL5, CCL17, CCL20, CCL25, CCL26, CXCL2, CXCL3, CXCL9, CXCL10, CXCL11, CXCL13, CXCL14, CX3CL1 and SLIT2; the adapter MYD88, the receptor TLR2 and the TF NFKB1. FOXP1 overexpression upregulated four genes (>1.5-fold): the negative regulator SOCS5, the cytokines IL10 and CKLF and the receptor CXCR4. The cytokine, chemokine, regulatory, receptor and transcription factor genes tested in this array all have the potential to direct a myriad of cellular activities in the TME. Stratification of this gene expression data on cell type-specific effects reveals a majority of the altered genes regulate immune cell functions and notably their migratory activities (Table S6).
3.5. FOXP1 and cytokine/chemokine gene expression in primary BC
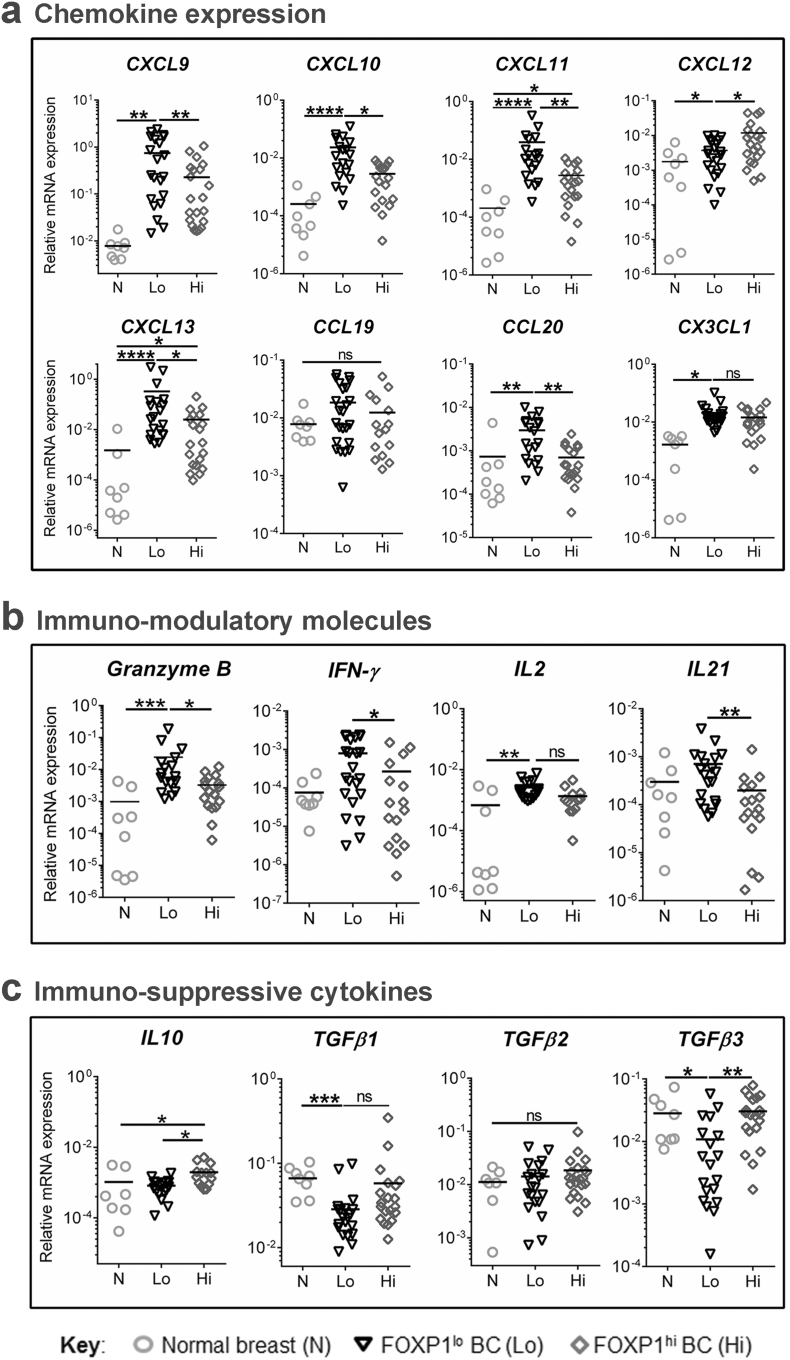
The modifications of cytokine and chemokine gene expression detected in the BC cell lines were explored in the MIU prospective BC cohort to determine how well these data reflect primary, untreated human tumors. FOXP1lo (n = 25) or FOXP1hi (n = 25) BC tissues together with normal breast tissues (control; n = 10) were analyzed by RT-qPCR to quantify selected cytokines and chemokines (known involvement in immune cell function or migration and TLS neogenesis). These data show that many chemokine (Fig. 4A) and cytokine (Fig. 4B and C) genes are differentially expressed in BC compared to normal breast. The expression of CXCL9, CXCL10,CXCL11, CXCL12, CXCL13, CX3CL1, IL2 and Granzyme B (GZMB) was significantly lower in normal breast compared to FOXP1lo or FOXP1hi BC (Fig. 4A and B; underlined genes were deregulated in in primary BC and one or both cell lines). A comparison between FOXP1lo and FOXP1hi BC reveals significant differences (decreases in the latter) in CXCL9, CXCL10, CXCL11, CXCL13, CXCL20, CXCL21, GZMB and IFNγ gene expression with CCL19, CX3CL1 and IL2 showing a trend for the same, although not statically significant. Paralleling the FOXP1-repressed MCF7 cell line, the chemokine CXCL12 was significantly upregulated in FOXP1hi compared to FOXP1lo tumors (Fig. 4A). The immune suppressive IL10 and TGFβ (particularly the TGFβ3 isoform) genes were expressed at significantly higher levels in FOXP1hi compared to FOXP1lo tumors (Fig. 4C). Interestingly, TGFβ1 and TGFβ3 were significantly lower in FOXP1lo BC relative to normal breast tissues.
Fig. 4.
FOXP1 is associated with chemokine and cytokine gene expression in breast cancer.
(a) Chemokines gene expression, (b) Immuno-modulatory molecules, and (c) immuno-suppressive cytokine gene expression in normal breast tissues (n = 10), FOXP1lo (n = 25) and FOXP1hi (n = 25) breast cancer. RT-qPCR data were normalized using EIF1α and MLN51 expression (2-ΔCt). Duplicate samples were analyzed. Significant P values (<0.05) are marked by * and ns: not significant. Degrees of significance: P < 0.05 (*), P < 0.01 (**) and P < 0.001 (***). Data represent mean ± SD.
The tumors in our cohort were screened for cellularity and only those containing >30% tumor cells were selected for analysis. Nevertheless, because the tumor microenvironment also contains stromal cells, endothelial cells and TIL, any or all of these subpopulations are potential producers of various cytokines and chemokines. EpCAM+ cells from a mixed group of BC were sorted (≥99% pure) and analyzed for FOXP1, CXCL9, CXCL10, CXCL11 and CXCL13 gene transcripts (Fig. S5). This analysis reveals that some tumors express high levels of the selected chemokines whereas in others they are undetectable or expressed only at low levels. The heterogeneity of chemokine expression is not significantly anti-correlated with FOXP1 expression for this small group of tumors; however, the highest FOXP1 tumors where those with undetectable chemokines suggesting a trend that needs further investigation.
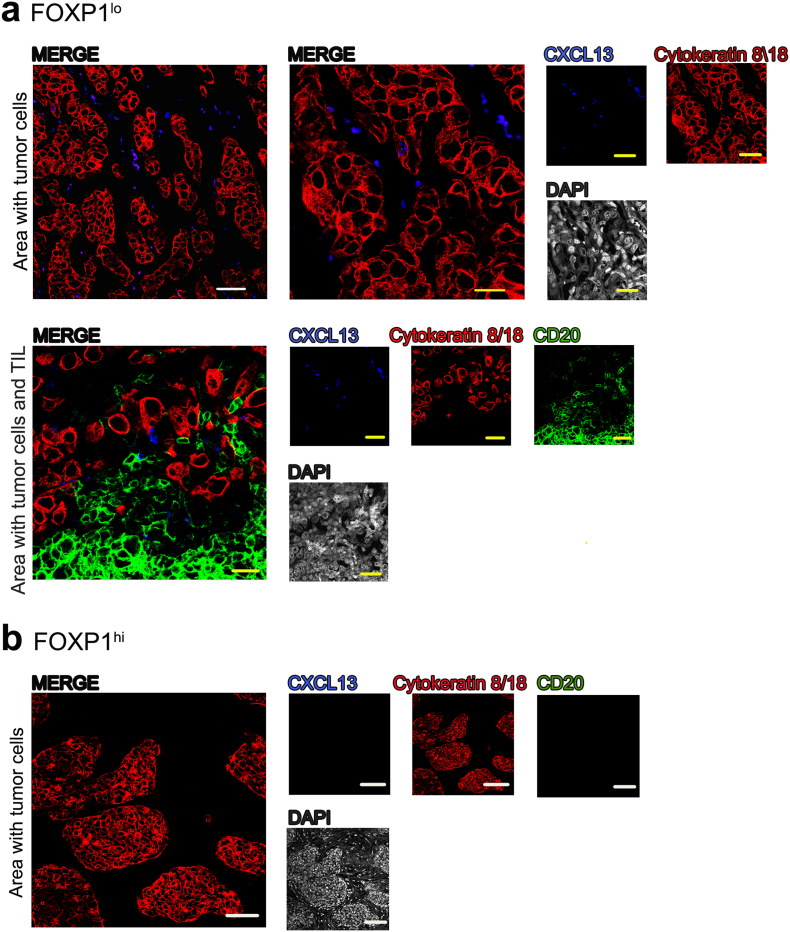
One of these chemokines analyzed, CXCL13, has been linked with higher TIL, TLS and better clinical outcomes in several tumor types including BC [9,10,32]. The sorted EpCAM+ cells showed significant heterogeneity in the expression of this important B cell chemoattractant. Immunofluorescence analysis of CXCL13 expression in tissue sections from FOXP1lo and FOXP1hi primary BC reveal that CXCL13 expression is associated with tumor cell areas where it is produced by malignant and non-malignant cells in FOXP1lo but not FOXP1hi BC, supporting the RT-qPCR data (Fig. 5). The FOXP1lo tumors also contain significant numbers of B cell TIL, again confirming this relationship. Overall, the cell line and primary BC data argue that FOXP1 can regulates a number of important cytokine and chemokine genes whose altered expression could reflect important events governing immune cell recruitment to the TME.
Fig. 5.
CXCL13 protein expression in breast cancer.
Representative images of CXCL13 protein expression in (a) FOXP1lo and (b) FOXP1hi primary breast cancer acquired by confocal microscopy.
Yellow scale bars: 50 μm and white scale bars: 100 μm. Contrast was enhanced by turning down DAPI (grey) intensity in merged images. CXCL13+ (blue), CD20+ (green) for B tumor infiltrating lymphocytes and cytokeratin 8/18 (red) for tumor epithelial cells were shown as single channel and merged images.
3.6. FOXP1 represses T and B cell migration
The cytokine/chemokine gene expression data suggest that FOXP1 has the ability to influence lymphocyte movement. We employed a transwell culture system to determine whether conditioned medium (TCM) from untreated or FOXP1-repressed MCF7 cells could affect T and B cell migration. These experiments found significant increases in PBMC migration (CD45+ cells from HD blood) in wells containing FOXP1-repressed TCM compared to control MCF7 cell TMC or medium alone (Fig. S6). Immune cell subpopulation analysis reveals this is characterized by a significant increase in CD4+ and CD8+ T cell and CD19+ B cell migration toward the FOXP1-repressed MCF7 cell TCM.
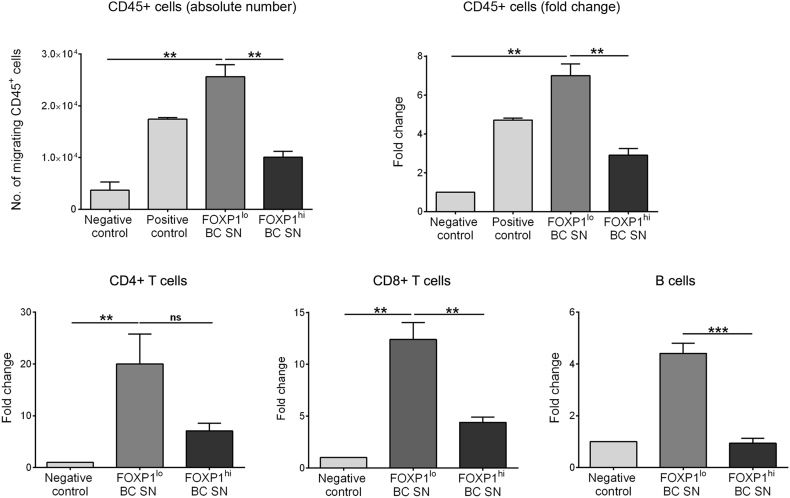
The MIU prospective BC cohort has the advantage that we routinely prepare and store primary tumor SN from the tissue homogenate on the day of surgery [28]. The validity of the TCM experiments was therefore tested in the migration assay using primary BC SN from five FOXP1lo and five FOXP1hi BC. Again, a significant decrease in total CD45+ lymphocyte migration was associated with FOXP1hi compared to FOXP1lo BC SN (Fig. 6). Similarly, separate analysis of migrating CD4+ and CD8+ T cells and CD19+ B cells detected significant decreases toward the FOXP1hi BC SN. These data support a regulatory role for FOXP1 via cytokines and/or chemokines produced in the breast tumor microenvironment.
Fig. 6.
FOXP1 regulates Lymphocyte migration in breast tumors.
Healthy donor PBMC migration in FOXP1lo and FOXP1hi tumor supernatants (SN) from primary breast cancer cohort. Top panel shows number of CD45+ cells migrated toward breast cancer SN and fold change. Bottom panel shows fold change of CD4+ T cells (CD3+CD4+); CD8+ T cells (CD3+CD8+) and B cells (CD19+) migrated toward BC SN after 24 h of incubation in the transwell assay. Data demonstrated as the mean of biological replicates done in duplicates of FOXP1lo (n = 5) and FOXP1hi (n = 5). Medium alone was used as a negative control and medium with 10% FBS was used a positive control. Significant P values (<0.05) are marked by * and ns: not significant. Degrees of significance: P < 0.05 (*), P < 0.01 (**) and P < 0.001 (***), as assessed with ANOVA using Dunn's multiple comparisons test.
Data represent mean ± SEM.
3.7. The clinical significance of FOXP1 expression in BC
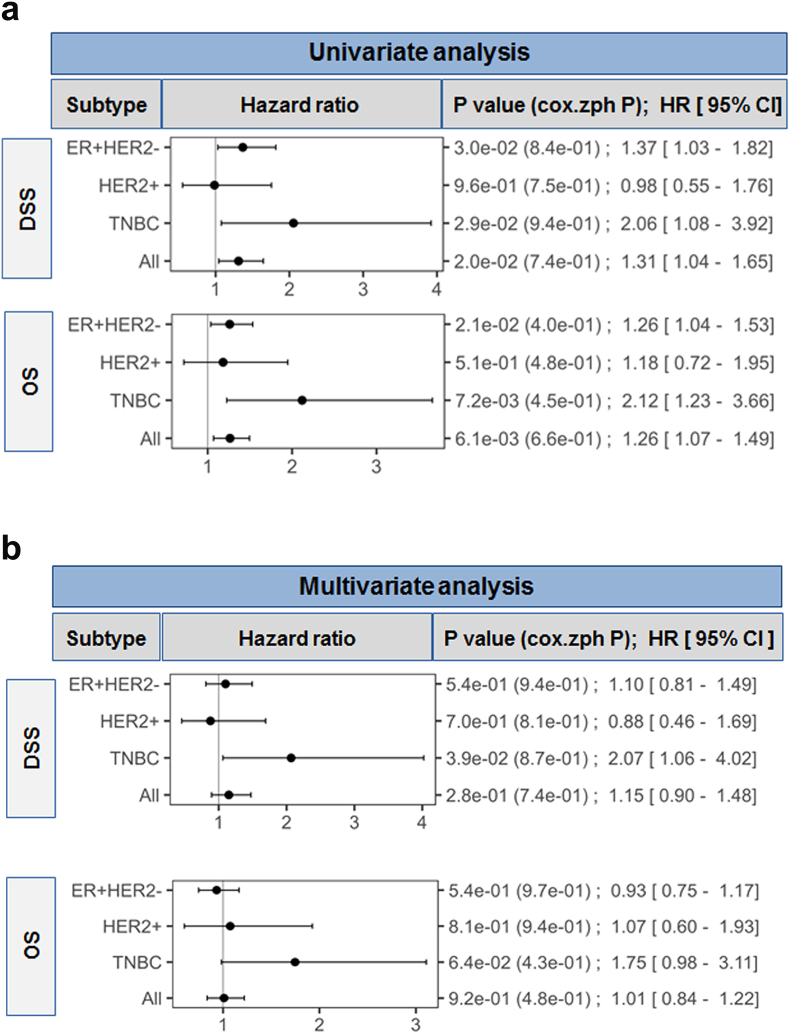
Recent studies of FOXP1 expression in BC suggest it can function as an oncogene [26,33]; however, its prognostic value among the BC subtypes is unclear with previous studies suggesting it may signal a better prognosis [[21], [22], [23], [24], [25]]. This study used the METABRIC dataset [2] to investigate associations between global FOXP1 expression and survival in ER+/HER-, HER2+ and TN. The METABRIC cohort includes ER+ patients that did not receive chemotherapy (nearly all) and ER- patients that were treated with chemotherapy, including HER2+ patients in the pre-trastuzumab era [2]. Univariate analysis found that a unit increase in FOXP1 expression (log2 scale) is significantly associated with: decreased DSS and OS in: 1) the ER+/HER2- subtype [(DSS: HR 1.37; 95% CI 1.03–1.82, P = .03) and (OS: HR 1.26; 95% CI 1.04–1.53, P = .021)]; 2) in the TN subtype [(DSS: HR 2.06; 95% CI 1.08–3.92, P = .029) and (OS: HR 2.12; 95% CI 1.23–3.66, P = .0072)]; and 3) in the pooled dataset [(DSS: HR 1.31; 95% CI 1.04–1.65, P = .02) and (OS: HR 1.26; 95% CI 1.07–1.49, P = .0061] (Fig. 7A and Fig. S7). Significant differences were not detected in the HER2+ subtype. After adjusting for clincopathological variables in the multivariate analysis only TNBC exhibited a significant association between high FOXP1 and decreased survival (DSS: HR 2.07; 95% CI 1.06–4.02, P = .039 and OS: HR 1.75; 95% CI 0.98–3.11, P = .064) (Fig. 7B). Overall, these data suggest that global FOXP1 expression is associated with worse BC prognosis, particularly in TN.
Fig. 7.
FOXP1 is associated with worst survival in breast cancer
(a) Univariate analysis for prognostic effect of FOXP1 gene expression in METABRIC dataset. Forest plots for disease specific survival (DSS) and overall survival (OS) based on FOXP1 gene expression in ER+/HER2- (n = 1394), HER2+ (n = 244), triple negative (TN) BC (n = 271) and all breast cancer (n = 1992).
(b) Multivariable model analyzed in the same dataset with adjustments for treatment (yes vs no), age (≤ 50 vs > 50), tumor size (T0, 1, 2 vs T3, 4), nodal status (negative vs positive), histologic grade (1, 2 vs 3), ER status (negative vs positive), and human epidermal growth factor receptor 2 (HER2) status (negative vs positive). P values <.05 are considered as significant. Proportional hazard assumptions were tested for P-values using cox.zph function.
4. Discussion
The data presented in this study show that FOXP1 plays an important role in the immune response to BC. We found that while FOXP1 is expressed in all molecular subtypes, it is enriched in ER+ compared to ER- BC. We further detected an anti-correlation between TIL and FOXP1 gene expression linked with tumor derived cytokine/chemokine expression. These data are, to our knowledge, the first to implicate FOXP1 in immune gene regulation and TIL migration in BC.
FOXP1 expression levels and subcellular localization vary greatly depending on the cancer tissue type and stage. Previous studies of epithelial cell malignancies detected opposing roles for FOXP1, acting either as a tumor suppressor or an oncogene depending upon the cell type. While FOXP1 functions are not well understood in BC, previous studies have promoted its role as a tumor suppressor based on the level and heterogeneity of its nuclear staining in various stages of tumor progression and its correlation with clinicopathological parameters [22,23,25]. In the present study, FOXP1 was shown to be significantly associated with ER/PR positivity and low grade tumors (grade 1 and 2 vs 3), which is consistent with previous findings [[22], [23], [24], [25]]. Analysis of gene or protein expression alone, however, is insufficient for understanding the functional role of FOXP1 in BC progression. Recent data in BC cell lines and mouse models suggest that FOXP1 can act as an oncogene [26,27]. Our data could also be interpreted as support for oncogenic function based on the significant negative association between FOXP1 gene expression and survival in the METABRIC dataset, but this remains to be validated.
FOXP1 was previously shown to have a significant positive association with ERα [22] and a positive correlation with nuclear ERβ in BC patients [23]. A possible explanation for these observations is that FOXP1 expression is regulated by estrogen. This idea is supported by upregulation of FOXP1 mRNA in MCF7 cells treated with estrogen together with the identification of ER binding sites in the FOXP1 gene using ChIP-on-chip analysis [24]. Our data link increased FOXP1 gene expression with ER+ BC and decreased with ER- BC; however, heterogeneity in the latter (METABRIC and our cohort) suggests that FOXP1 regulation can also be ER independent. FOXP1 expression in prostate cancer is positively correlated with the androgen receptor (AR), supporting the notion of ER independent mechanisms. FOXP1 has been shown to directly regulate AR-mediated transcription via its repressive effect on AR-induced transcriptional activity or histone modification in its enhancer regions [34]. AR is the most commonly expressed hormone receptor in BC, including a 25% to 75% prevalence in TN [35]. TN BC is increasingly recognized as a heterogeneous disease with a subset associated with AR signaling [35,36]. These data suggest that the level of FOXP1 expression could be another distinguishing parameter for TN BC subsets.
Our initial experiments using BC cell lines revealed that FOXP1 can directly modulate cytokines and chemokines, TF and receptors. FOXP1 is a repressor that functions by forming hetero- or homodimers with other molecules to suppress their activities [37]. Interestingly, a study where human FOXP1 was overexpressed in brain cells with a mutated Huntington protein detected robust downregulation of glial cell-associated immune genes that included a variety of cytokines and chemokines [38]. These authors suggested that FOXP1 functions as a transcriptional repressor of immune signaling in the central nervous system supporting the data we presented here for BC. FOXP1 has also been implicated in suppressing immune response signatures and MHC class II genes in B-cell lymphoma, highlighting the concept that FOXP1 repression could improve antigen presentation and immune surveillance [15].
Our data detected SOCS5 upregulation (suppressor of cytokine signaling-5) in MDA-MB-231 overexpressing FOXP1, similar to a previous study of human brain tissues [38]. SOCS5 is a known critical negative regulator of cytokine signaling under diverse conditions [39], suggesting that its increased expression in FOXP1hi cells may directly or indirectly modulate cytokine activities. FOXP1 upregulation also represses MYD88 (myeloid differentiation primary response gene 88) and SLIT2 (Slit homolog 2). MYD88 is a critical adapter protein in innate immune signal transduction through its involvement in Toll-like receptor and IL1 receptor signaling pathways, acting via IRAK1, IRAK2, IRF7 and TRAF6, which leads to NFƙB activation, cytokine secretion and inflammatory responses [40]. SLIT2 acts as a molecular guidance signal for cell migration and has been identified as a novel tumor suppressor gene via hyper-methylation of its promoter in tumor cells including BC [41]. The repression of these immune regulatory genes in FOXP1-overexpressing BC cell lines suggests that FOXP1hi BC is a non-permissive environment for immune cell recruitment.
The majority of chemokine and cytokine genes we found regulated by FOXP1 have documented activities in immune cell migration and immunity. They include CCL5, CXCL9, CXCL11 and CXCL13, all shown to impact anti-tumor immune responses and associated with good clinical outcomes in several tumor types [32,[42], [43], [44]]. A chemokine gene expression signature, including CCL17, CCL19, CCL21, CCL22 and CXCL13, has been associated with the presence of T cells in lung cancer TLS [45] suggesting that chemokine expression in the TME is a critical feature of TIL recruitment and TLS formation. These data suggest that chemokine expression in the TME is a critical feature of TIL recruitment and TLS formation. Our comparative analysis of cytokine and chemokine expression between FOXP1lo and FOXP1hi BC, both significantly reduced in the latter, indicate that FOXP1hi BC maintains an immunosuppressive TME. Patel et al. investigated gene expression related with resistance to T cell-based immunotherapy in human melanoma [46]. Among the genes identified, they found a reduction in apelin receptor (APLNR) expression associated with tumors from patient's refractory to immunotherapy. The present study found that APLNR is downregulated by FOXP1 overexpression in MDA-MB-231 cells, further supporting the notion that FOXP1 drives an immunosuppressive TME. We also found that FOXP1hi BC expresses high levels of TGFβ and IL10, both immunosuppressive cytokines known to favor tissue invasion and metastasis via their suppression of anti-tumor immune responses. Stephen et al. demonstrated that tumor-derived TGFβ can induce FOXP1 in CD8+ T cells leading to their unresponsiveness in mice [47]. We observed that human BC with low TIL had significantly higher levels of FOXP1 and TGFβ. We therefore reason that this tumor derived TGFβ, induced by FOXP1, could increase FOXP1 expression in T cell TIL and thereby direct their unresponsiveness. High FOXP1 was shown to reduce T cell proliferation and functionality [47] and could thus similarly affect BC TIL together impairment of their migration by tumor cells expressing high FOXP1.
The production and release of chemokines by normal and malignant cells in the TME can directly induce chemotaxis of specific TIL subpopulations to the tumor [48,49]. A study profiling cytokines in tumor interstitial fluid found that tumors containing high proportions of CD3+ TIL exhibited significantly higher levels of CXCL10 and CCL5 than samples from tumors with low CD3+ TIL [50]. We have shown here that FOXP1lo BC are more permissive to the expression of chemokines that drive TIL migration to the TME. The FOXP1-regulated chemokine, CXCL13, a B-cell chemoattractant, has been identified as one of the most robust predictors of improved survival in human cancer [51]. Our previous studies comparing extensive to minimally infiltrated BC found that CXCL13-producing CD4+ Tfh TIL, named TfhX13, distinguish TILhi tumors (a majority of HER2+ and TN), where they are principally located in TLS and linked with a good prognosis [9,10]. Based on the data presented here, we propose that TILhi tumors containing TfhX13 cells would primarily be FOXP1lo BC. This notion is supported by our data showing that CXCL13 expression is highest in FOXP1lo BC with tumor and non-tumor cells principally producing CXCL13 in the TME (Fig. 4, Fig. 5). Our previous work associated TfhX13 TIL with a role in guiding B cell TIL migration and promoting TLS formation suggesting that this could be a powerful factor in the TIL rich TME that characterizes FOXP1lo BC.
This study identifies FOXP1 as an important negative regulator of anti-tumor immune responses via its control of chemokine expression. The majority of genes we found regulated by FOXP1 play a role in T and B cell migratory activities with lower FOXP1 expression favoring TIL trafficking in BC. Thus, higher FOXP1 expression in ER+ compared to ER- BC offers a potential mechanism contributing to the lower immune infiltration levels that generally characterize ER+ tumors. Specific chemokines, produced by normal and/or malignant cells in the TME, are necessary to drive TIL migration in BC. The anti-correlation we demonstrated between FOXP1 expression and the extent of TIL in tumors is linked with production of the chemoattractants that guide TIL recruitment. Further, the association of incremental increases in FOXP1 gene expression with a decline in survival suggests this TF plays an important role in helping to create and/or maintain an immunosuppressive TME by controlling the expression of critical immune genes. The known association between higher TIL and survival in HER2+ and TN advocates for the importance of understanding the factors that regulate TIL recruitment in BC.
Financial support
This work was supported by grants from the Belgian Fund for Scientific Research (FNRS), FNRS-Opération Télévie, Fonds J.C. Heuson and Fonds Lambeau-Marteaux.
Declaration of interests
The authors declare no competing interests.
Author contributions
P.D.S., S.G., and K·W-G conceived the research and designed the experiments; P.D.S performed the majority of experiments with critical support from C.S., C.G-T., E.M., A.B., C.N., and H.D.; A.W. and G.V.E. performed the pathological evaluations; V.J. performed the public data analyses; L.C. and D.L. provided the patient samples; M.P-G. proposed important concepts; K·W-G. and S.G. supervised the research; P.D.S, S.G., and K·W-G. analyzed and interpreted the data and wrote the manuscript; All of the authors agree with the contents of this manuscript and consent to its publication.
Acknowledgements
We thank Dr. M. Spaargaren (University of Amsterdam, Amsterdam, The Netherlands) for providing the FOXP1 plasmid, Drs. Grégory Noel and Mireille Langouo-Fontsa for important practical support and critical advice, Jean-Nicolas Lodewyckx, Laurence Van Schoonwinkel, and Samira Majjaj for excellent technical assistance and Drs. David Brown and Sylvain Brohée for providing statistical insight for this study.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ebiom.2018.11.066.
Appendix A. Supplementary data
Supplementary material 1
Supplementary material 2
References
- 1.Schumacher T.N., Schreiber R.D. Neoantigens in cancer immunotherapy. Science. 2015;348(6230):69–74. doi: 10.1126/science.aaa4971. [DOI] [PubMed] [Google Scholar]
- 2.Curtis C., Shah S.P., Chin S.F., Turashvili G., Rueda O.M., Dunning M.J. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486(7403):346–352. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Salgado R., Denkert C., Demaria S., Sirtaine N., Klauschen F., Pruneri G. The evaluation of tumor-infiltrating lymphocytes (TILs) in breast cancer: recommendations by an International TILs Working Group 2014. Ann. Oncol. 2015;26(2):259–271. doi: 10.1093/annonc/mdu450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buisseret L., Garaud S., de Wind A., Van den Eynden G., Boisson A., Solinas C. Tumor-infiltrating lymphocyte composition, organization and PD-1/ PD-L1 expression are linked in breast cancer. Oncoimmunology. 2017;6(1) doi: 10.1080/2162402X.2016.1257452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sautes-Fridman C., Lawand M., Giraldo N.A., Kaplon H., Germain C., Fridman W.H. Tertiary lymphoid structures in cancers: prognostic value, regulation, and manipulation for therapeutic intervention. Front Immunol. 2016;7:407. doi: 10.3389/fimmu.2016.00407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johansson-Percival A., He B., Li Z.J., Kjellen A., Russell K., Li J. De novo induction of intratumoral lymphoid structures and vessel normalization enhances immunotherapy in resistant tumors. Nat Immunol. 2017;18(11):1207–1217. doi: 10.1038/ni.3836. [DOI] [PubMed] [Google Scholar]
- 7.Pimenta E.M., De S., Weiss R., Feng D., Hall K., Kilic S. IRF5 is a novel regulator of CXCL13 expression in breast cancer that regulates CXCR5(+) B- and T-cell trafficking to tumor-conditioned media. Immunol Cell Biol. 2015;93(5):486–499. doi: 10.1038/icb.2014.110. [DOI] [PubMed] [Google Scholar]
- 8.Garaud S., Willard-Gallo K. IRF5: a rheostat for tumor-infiltrating lymphocyte trafficking in breast cancer? Immunol Cell Biol. 2015;93(5):425–426. doi: 10.1038/icb.2015.39. [DOI] [PubMed] [Google Scholar]
- 9.Gu-Trantien C., Loi S., Garaud S., Equeter C., Libin M. Wind Ad, et al. CD4+ follicular helper T cell infiltration predicts breast cancer survival. J Clin Invest. 2013;123(7):2873–2892. doi: 10.1172/JCI67428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gu-Trantien C., Migliori E., Buisseret L., de Wind A., Brohee S., Garaud S. CXCL13-producing TFH cells link immune suppression and adaptive memory in human breast cancer. JCI Insight. 2017;2:11. doi: 10.1172/jci.insight.91487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fujino N., Ota C., Suzuki T., Suzuki S., Hegab A.E., Yamada M. Analysis of gene expression profiles in alveolar epithelial type II-like cells differentiated from human alveolar epithelial progenitor cells. Respir Investig. 2012;50(3):110–116. doi: 10.1016/j.resinv.2012.07.002. [DOI] [PubMed] [Google Scholar]
- 12.Hu H., Wang B., Borde M., Nardone J., Maika S., Allred L. Foxp1 is an essential transcriptional regulator of B cell development. Nat Immunol. 2006;7:819–826. doi: 10.1038/ni1358. [DOI] [PubMed] [Google Scholar]
- 13.Garaud S, Roufosse F, Silva PD, Gu-Trantien C, Lodewyckx J-N, Duvillier H, et al. Foxp1 is a key regulator of quiescence in healthy human CD3+CD4+ T-cells and constituatively repressed in CD3-CD4+ T-cells from patients with lymphoproliferative disorders. Manuscript in revision. [DOI] [PubMed]
- 14.Shi C., Sakuma M., Mooroka T., Liscoe A., Gao H., Croce K.J. Down-regulation of the forkhead transcription factor Foxp1 is required for monocyte differentiation and macrophage function. Blood. 2008;112(12):4699–4711. doi: 10.1182/blood-2008-01-137018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brown P.J., Wong K.K., Felce S.L., Lyne L., Spearman H., Soilleux E.J. FOXP1 suppresses immune response signatures and MHC class II expression in activated B-cell-like diffuse large B-cell lymphomas. Leukemia. 2016;30(3):605–616. doi: 10.1038/leu.2015.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yamada S., Sato F., Xia H., Takino H., Kominato S., Ri M. Forkhead box P1 overexpression and its clinicopathologic significance in peripheral T-cell lymphoma, not otherwise specified. Hum Pathol. 2012;43:1322–1327. doi: 10.1016/j.humpath.2011.10.013. [DOI] [PubMed] [Google Scholar]
- 17.Choi E.J., Seo E.J., Kim D.K., Lee S.I., Kwon Y.W., Jang I.H. FOXP1 functions as an oncogene in promoting cancer stem cell-like characteristics in ovarian cancer cells. Oncotarget. 2016;7(3):3506–3519. doi: 10.18632/oncotarget.6510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang M.H., Lin B.R., Chang C.H., Chen S.T., Lin S.K., Kuo M.Y. Connective tissue growth factor modulates oral squamous cell carcinoma invasion by activating a miR-504/FOXP1 signalling. Oncogene. 2012;31(19):2401–2411. doi: 10.1038/onc.2011.423. [DOI] [PubMed] [Google Scholar]
- 19.Wang X., Sun J., Cui M., Zhao F., Ge C., Chen T. Downregulation of FOXP1 inhibits cell proliferation in hepatocellular carcinoma by inducing G1/S phase cell cycle arrest. Int. J. Mol. Sci. 2016;17:9. doi: 10.3390/ijms17091501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gomez G.G., Volinia S., Croce C.M., Zanca C., Li M., Emnett R. Suppression of microRNA-9 by mutant EGFR signaling upregulates FOXP1 to enhance glioblastoma tumorigenicity. Cancer Res. 2014;74(5):1429–1439. doi: 10.1158/0008-5472.CAN-13-2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xiao J., He B., Zou Y., Chen X., Lu X., Xie M. Prognostic value of decreased FOXP1 protein expression in various tumors: a systematic review and meta-analysis. Sci Rep. 2016;6:30437. doi: 10.1038/srep30437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fox S.B., Brown P., Han C., Ashe S., Leek R.D., Harris A.L. Expression of the forkhead transcription factor FOXP1 is associated with estrogen receptor alpha and improved survival in primary human breast carcinomas. Clin Cancer Res. 2004;10(10):3521–3527. doi: 10.1158/1078-0432.CCR-03-0461. [DOI] [PubMed] [Google Scholar]
- 23.Bates G.J., Fox S.B., Han C., Launchbury R., Leek R.D., Harris A.L. Expression of the forkhead transcription factor FOXP1 is associated with that of estrogen receptor-beta in primary invasive breast carcinomas. Breast Cancer Res Treat. 2008;111(3):453–459. doi: 10.1007/s10549-007-9812-4. [DOI] [PubMed] [Google Scholar]
- 24.Shigekawa T., Ijichi N., Ikeda K., Horie-Inoue K., Shimizu C., Saji S. FOXP1, an estrogen-inducible transcription factor, modulates cell proliferation in breast cancer cells and 5-year recurrence-free survival of patients with tamoxifen-treated breast cancer. Hormones Cancer. 2011;2(5):286–297. doi: 10.1007/s12672-011-0082-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rayoo M., Yan M., Takano E.A., Bates G.J., Brown P.J., Banham A.H. Expression of the forkhead box transcription factor FOXP1 is associated with oestrogen receptor alpha, oestrogen receptor beta and improved survival in familial breast cancers. J Clin Pathol. 2009;62(10):896–902. doi: 10.1136/jcp.2009.065169. [DOI] [PubMed] [Google Scholar]
- 26.Chiang K., Zielinska A.E., Shaaban A.M., Sanchez-Bailon M.P., Jarrold J., Clarke T.L. PRMT5 is a critical regulator of breast cancer stem cell function via histone methylation and FOXP1 expression. Cell Rep. 2017;21(12):3498–3513. doi: 10.1016/j.celrep.2017.11.096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oskay Halacli S. FOXP1 enhances tumor cell migration by repression of NFAT1 transcriptional activity in MDA-MB-231 cells. Cell Biol Int. 2017;41(1):102–110. doi: 10.1002/cbin.10702. [DOI] [PubMed] [Google Scholar]
- 28.Garaud S., Gu-Trantien C., Lodewyckx J.-N., Boisson A., Silva P.D., Buisseret L. A simple and rapid protocol to non-enzymatically dissociate fresh human tissues for the analysis of infiltrating lymphocytes. J Vis Exp. 2014;94(e52392):1–9. doi: 10.3791/52392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Banham A.H., Beasley N., Campo E., Fernandez P.L., Fidler C., Gatter K. The FOXP1 winged helix transcription factor is a novel candidate tumor suppressor gene on chromosome 3p1. Cancer Res. 2001;61:8820–8829. [PubMed] [Google Scholar]
- 30.Solinas C., Carbognin L., De Silva P., Criscitiello C., Lambertini M. Tumor-infiltrating lymphocytes in breast cancer according to tumor subtype: current state of the art. Breast. 2017;35:142–150. doi: 10.1016/j.breast.2017.07.005. [DOI] [PubMed] [Google Scholar]
- 31.Solinas C., Garaud S., Silva P.D., Boisson A., GVD Eynden, Wind A.D. Immune checkpoint molecules on tumor-infiltrating lymphocytes and their association with tertiary lymphoid structures in human breast cancer. Front Immunol. 2017;8:1412. doi: 10.3389/fimmu.2017.01412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schmidt M., Weyer-Elberich V., Hengstler J.G., Heimes A.S., Almstedt K., Gerhold-Ay A. Prognostic impact of CD4-positive T cell subsets in early breast cancer: a study based on the FinHer trial patient population. Breast Cancer Res. 2018;20(1):15. doi: 10.1186/s13058-018-0942-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu B.H., Li B.Z., Zhou X.Y., Shi D.R., Yang W.T. Cytoplasmic FOXP1 expression is correlated with ER and calpain II expression and predicts a poor outcome in breast cancer. Diagn Pathol. 2018;13(1):36. doi: 10.1186/s13000-018-0715-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Takayama K., Suzuki T., Tsutsumi S., Fujimura T., Takahashi S., Homma Y. Integrative analysis of FOXP1 function reveals a tumor-suppressive effect in prostate cancer. Mol Endocrinol. 2014;28(12):2012–2024. doi: 10.1210/me.2014-1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Safarpour D., Tavassoli F.A. A targetable androgen receptor-positive breast cancer subtype hidden among the triple-negative cancers. Arch Pathol Lab Med. 2015;139(5):612–617. doi: 10.5858/arpa.2014-0122-RA. [DOI] [PubMed] [Google Scholar]
- 36.Lehmann B.D., Jovanovic B., Chen X., Estrada M.V., Johnson K.N., Shyr Y. Refinement of triple-negative breast cancer molecular subtypes: implications for neoadjuvant chemotherapy selection. PloS one. 2016;11(6) doi: 10.1371/journal.pone.0157368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Golson M.L., Kaestner K.H. Fox transcription factors: from development to disease. Development. 2016;143(24):4558–4570. doi: 10.1242/dev.112672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tang B., Becanovic K., Desplats P.A., Spencer B., Hill A.M., Connolly C. Forkhead box protein p1 is a transcriptional repressor of immune signaling in the CNS: implications for transcriptional dysregulation in Huntington disease. Hum Mol Genet. 2012;21(14):3097–3111. doi: 10.1093/hmg/dds132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Larsen L., Ropke C. Suppressors of cytokine signalling: SOCS. APMIS. 2002;110(12):833–844. doi: 10.1034/j.1600-0463.2002.1101201.x. [DOI] [PubMed] [Google Scholar]
- 40.Warner N., Nunez G. MyD88: a critical adaptor protein in innate immunity signal transduction. J Immunol. 2013;190(1):3–4. doi: 10.4049/jimmunol.1203103. [DOI] [PubMed] [Google Scholar]
- 41.Kim G.E., Lee K.H., Choi Y.D., Lee J.S., Lee J.H., Nam J.H. Detection of Slit2 promoter hypermethylation in tissue and serum samples from breast cancer patients. Virchows Archiv. 2011;459(4):383–390. doi: 10.1007/s00428-011-1143-5. [DOI] [PubMed] [Google Scholar]
- 42.Denkert Carsten, Loibl Sibylle, Noske Aurelia, Roller Marc, Berit Maria M., Komor Martina. Tumor-associated lymphocytes as an independent predictor of response to neoadjuvant chemotherapy in breast cancer. J Clin Oncol. 2010;28:1. doi: 10.1200/JCO.2009.23.7370. [DOI] [PubMed] [Google Scholar]
- 43.Fridman W.H., Pagès F., Sautès-Fridman C., Galon J. The immune contexture in human tumours: impact on clinical outcome. Nat Rev. 2012;12:298–306. doi: 10.1038/nrc3245. [DOI] [PubMed] [Google Scholar]
- 44.Araujo J.M., Gomez A.C., Aguilar A., Salgado R., Balko J.M., Bravo L. Effect of CCL5 expression in the recruitment of immune cells in triple negative breast cancer. Sci Rep. 2018;8(1):4899. doi: 10.1038/s41598-018-23099-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.de Chaisemartin L., Goc J., Damotte D., Validire P., Magdeleinat P., Alifano M. Characterization of chemokines and adhesion molecules associated with T cell presence in tertiary lymphoid structures in human lung cancer. Cancer Res. 2011;71(20):6391–6399. doi: 10.1158/0008-5472.CAN-11-0952. [DOI] [PubMed] [Google Scholar]
- 46.Patel S.J., Sanjana N.E., Kishton R.J., Eidizadeh A., Vodnala S.K., Cam M. Identification of essential genes for cancer immunotherapy. Nature. 2017;548(7669):537–542. doi: 10.1038/nature23477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stephen T.L., Rutkowski M.R., Allegrezza M.J., Perales-Puchalt A., Tesone A.J., Svoronos N. Transforming growth factor b-mediated suppression of antitumor T cells requires FoxP1 transcription factor expression. Immunity. 2014;41:427–439. doi: 10.1016/j.immuni.2014.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hojo S., Koizumi K., Tsuneyama K., Arita Y., Cui Z., Shinohara K. High-level expression of chemokine CXCL16 by tumor cells correlates with a good prognosis and increased tumor-infiltrating lymphocytes in colorectal cancer. Cancer Res. 2007;67(10):4725–4731. doi: 10.1158/0008-5472.CAN-06-3424. [DOI] [PubMed] [Google Scholar]
- 49.Muraki J., Tokue A., Nakazono M. Production of a lymphocyte chemotactic factor by human renal cancer cells. Jpn J Urol. 1994;85(11):1649–1655. doi: 10.5980/jpnjurol1989.85.1649. [DOI] [PubMed] [Google Scholar]
- 50.Espinoza J.A., Jabeen S., Batra R., Papaleo E., Haakensen V., Timmermans Wielenga V. Cytokine profiling of tumor interstitial fluid of the breast and its relationship with lymphocyte infiltration and clinicopathological characteristics. Oncoimmunology. 2016;5(12) doi: 10.1080/2162402X.2016.1248015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rachidi S.M., Qin T., Sun S., Zheng W.J., Li Z. Molecular profiling of multiple human cancers defines an inflammatory cancer-associated molecular pattern and uncovers KPNA2 as a uniform poor prognostic cancer marker. PloS One. 2013;8(3) doi: 10.1371/journal.pone.0057911. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material 1
Supplementary material 2