Abstract
Background
Sickle cell anaemia (SCA) is a hereditary blood disorder caused by a single mutation in the haemoglobin gene. The disease burden of SCA is highest in Nigeria. The allele specific polymerase chain reaction (ASPCR) method is applicable for the direct detection of known single nucleotide polymorphisms (SNPs).
Objective
To investigate the use of the single tube ASPCR as an accurate and affordable method for SCA screening in Nigeria.
Methods
DNA was extracted from study subjects with normal haemoglobin, HbAA (20), sickle cell anaemia, HbSS (20) and carriers, HbAS (1). Haemoglobin was genotyped by ASPCR using two primer sets that amplifies the wildtype and mutant haemoglobins in each sample. Amplicon sizes were analyzed by gel electrophoresis.
Results
Amplicons were visible after electrophoresis at regions 517 base pair (bp) for HbA and 267 bp for HbS. ASPCR correctly and unambiguously detected the presence or absence of haemoglobins A and S from all samples collected, demonstrating its accuracy and precision for the screening of SCA.
Conclusion
This study validates ASPCR as an effective, low cost approach for the clinical screening of SCA in Nigeria. ASPCR is also applicable for other genetic diseases, paternity testing, and forensics where more expensive fluorescence-based approaches are not obtainable.
Keywords: Sickle cell anaemia, polymerase chain reaction, molecular diagnostic techniques
Introduction
Sickle cell anaemia (SCA) is a chronic blood disorder initiated by inheriting a faulty (sickle) haemoglobin with chronic haemolytic anaemia and recurrent vaso-occlusion as its key pathological features1,2. The molecular explanation of the sickle hemoglobin is a single nucleotide polymorphism (SNP) or point mutation (GAG → GTG) in the β-haemoglobin gene (HBB). The homozygosity for the mutated S haemoglobin (HbS) leads to a substitution of a hydrophobic valine amino acid for the normal hydrophilic glutamic acid residue at the sixth amino acid of the β-globin chain (glu6val) and the resulting HbS β-globin chains are substituted for normal HbA β-globin chains3,4. This single-point mutation is associated with complex genetic interactions evident by the multiple phenotypic expressions and disease modifiers that are not well understood5,6. The clinical heterogeneity of SCA varies widely from a mild clinical course to very severe complications with significant organ damage and early mortality7,8. Haemoglobin A also termed the wild type haemoglobin, is the normal adult form of haemoglobin and its homozygosity (HbAA) indicates the normal genotype while heterozygosity (HbAS) denotes a carrier status but individuals do not have symptoms.
Sickle cell anaemia is predominant in Africa, the Middle East and Mediterranean axis, the Indian sub-continent, the Caribbean and South America. Sub-Saharan Africa records the highest burden and incidence of SCA. Annually over 250,000 affected babies are born in this region, representing 1–2% of new-borns in some countries9,10. In Nigeria alone, about 25–30% of the population are trait carriers and SCA occurs in approximately 2% of all births representing over 150,000 babies born yearly with the disease11,12.
In Nigeria, current diagnosis and management of SCA is still sub-optimal. Screening is limited to the sickling solubility test and haemoglobin electrophoresis. Sickling solubility test can detect the presence of haemoglobin S, but these tests cannot distinguish SCA from sickle cell trait (SCT)13. Haemoglobin electrophoresis is a useful screening method that can determine the phenotype of SCA but is not a reliable method for the determination of genotype in infants less than 6 months since high levels of fetal haemoglobin (haemoglobin F) that persists from birth is the predominant haemoglobin at this age. Thus haemoglobin electrophoresis is not suitable for prenatal and neonatal screening14.
More accurate methods like thin-layer isoelectric focusing (IEF) or high performance liquid chromatography (HPLC) are rare in Nigeria. Molecular methods for the SCA diagnosis are not common due to the paucity of skilled personnel. In addition molecular laboratories are few and mostly sequestered in federal institutions. In this study, the single tube ASPCR genotyping of SCA as a low cost method of molecular diagnosis of SCA in developing countries was investigated.
Materials and methods
Study sites and patient recruitment: Study subjects were recruited from Lagos State in Nigeria. A total of 41 participants were pooled from two clinics. Twenty (20) sickle cell anaemia (HbSS) subjects were recruited from the Haematology Unit, Lagos State University Teaching Hospital, while twenty (20) non-carriers (HbAA) and one (1) trait carrier (HbAS) were recruited from the Health Centre, University of Lagos Akoka Yaba, Lagos State. A questionnaire was administered and participants were enrolled after signing the consent forms approved by the Ethics Committees.
Sample collection: 5ml of whole blood from each patient was collected into EDTA tubes. Blood samples were stored in ice coolers and transported to the postgraduate laboratory at the Department of Cell Biology and Genetics, Faculty of Science, University of Lagos for genetic analysis.
DNA extraction: The DNA extraction was carried out using E.Z.N.A. Blood DNA Mini Kit Omega Biotek USA following the outlined protocol.
DNA quality and quantity: DNA quality and quantity was measured using an ultraviolet (UV) spectrophotometer at 260nm and 280nm wavelength.
Bi-directional Allele Specific Amplification: The genotype results obtained from patient files of all volunteers enrolled for this study was confirmed by abi-directional allele specific amplification ASPCR method described by Waterfall and Cobb (2001). Primer sequences to detect the wild-type and mutant alleles are as follows:
WT-AS (5′-ATG GTG CAC CTG ACT CCT GA-3′) and WT-CP517 (5′-CCC CTT CCT ATG ACA TGA ACT-3′) for amplification of a 517 bp fragment from the normal β-globin gene (wild-type primer set).
MUT-AS (5′-CAG TAA CGG CAG ACT TCT CCA-3′) and MUT-CP267 (5′-GGG TTT GAA GTC CAA CTC CTA-3′) for amplification of a 267 bp fragment from homozygous mutant DNA confiming sickle cell disease (mutant primer set).
Amplification was performed using a Thermos Scientific Arktik Thermal Cycler (Thermo Fisher Scientific Oy, Finland) through an initial denaturation at 95°C for 2 min, followed by 30 cycles of denaturation at 95°C for 30s, primer annealing at 60°C for 30s, extension at 72°C for 35s and a final extension at 72°C for 10 mins.
Analysis of pcr products
An aliquot (10µl) of each PCR product was loaded with 6× loading dye buffer (Promega) onto a 2% agarose gel stained with Red Safe (Molecular Probes, UK) and run in 0.5X tris borate EDTA (TBE) buffer at 75volts for 1 hr. Amplification products were visualised by UV transillumination (305 nM). The sizes of expected amplicons, 517 bp indicating the presence of the normal β-globin gene (HBA/A) and 267 bp indicating presence of the mutant gene (HbS/S) were read against the 100 bp DNA molecular weight ladder and genotype results were obtained.
Ethical approval
Ethical approvals obtained for this study are from University of Lagos Teaching Hospital (LUTH) HREC with approval number (LREC/10/86/479) and Lagos State University Teaching Hospital (LASUTH) Research Ehtics Committee with approval number (ADM/DCST/HREC/1510).
Results
Experimental design
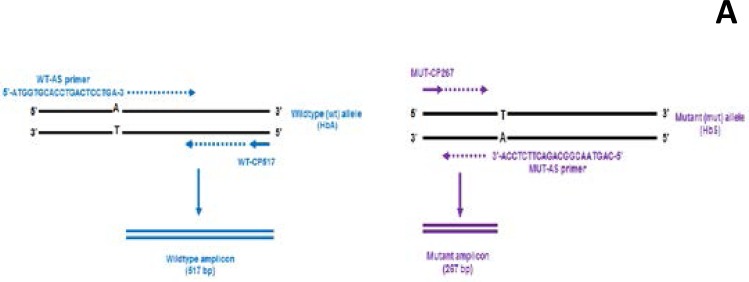
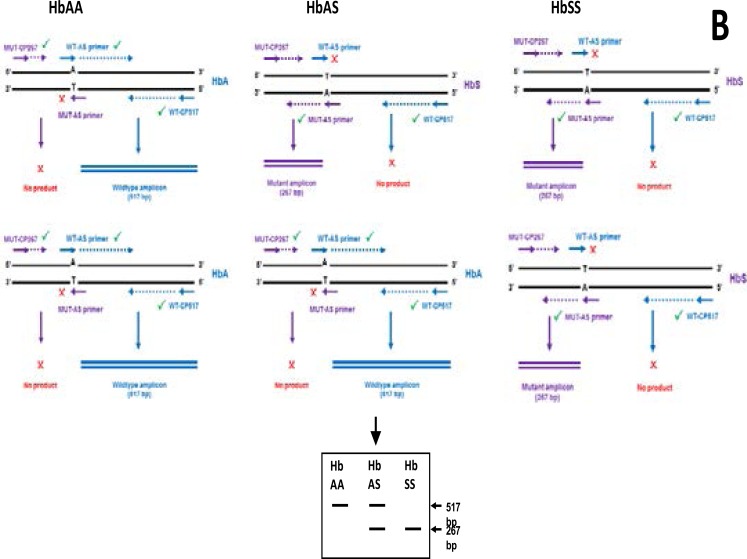
The primers used in this study are designed to amplify both mutant and wild-type alleles present in a single reaction tube. To achieve this, a wild-type primer (WT-AS) which is complimentary to the non-coding strand of the wild-type allele, is designed to terminate at the polymorphic site where the mutation occurs. In the presence of this primer, only wild-type DNA will extend. The mutant primer (MUT-AS) on the other hand is complimentary to the coding strand of a mutant allele, it terminates at the SNP site where the mutation (A T) has occurred and will extend only in the presence of a mutant allele. In order to achieve molecular conformation, two outer primers WTCP517and MUT-CP267 are placed at a predefined number of bases on the opposite strands of the allele specific pair. Figure 1 shows an illustration of the bi-directional ASPCR as described by Waterfall and Cobb 2001.
Figure 1.
An illustration of the bi-directional ASPCR as described by Waterfall and Cobb 2001. The wild-type primer set (WTAS/WT-CP517) is shown in blue and mutant primer set (MUT-AS/ MUT-CP267) is in purple.
A -Illustrates that purple amplicons are formed only when the mutant primers are extended and
B- Illustrates that blue amplicons are formed only when the wild-type primers are extended.
The presence of both mutant and wildtype amplicons identifies the heterozygous individual.
Optimization of the sickle cell anaemia ASPCR conditions
Reaction conditions were optimised by altering the magnesium chloride (MgCl2) concentration in the PCR master- mix and the input DNA template on samples that were positive for HbAA, HbSS and HbAS. Table 1. Shows optimized conditions arrived at in this study for the ASPCR.
Table 1.
Optimized concentration of reactants in PCR master-mix
| Reactants | WT-AS/ WT-CP517 |
MUT-AS/ MUT-CP267 |
MgCl2 | dNTPs | DNA template |
| Concentration | 0.8µM | 0.8µM | 1.5mM | 80 µM | 4ng |
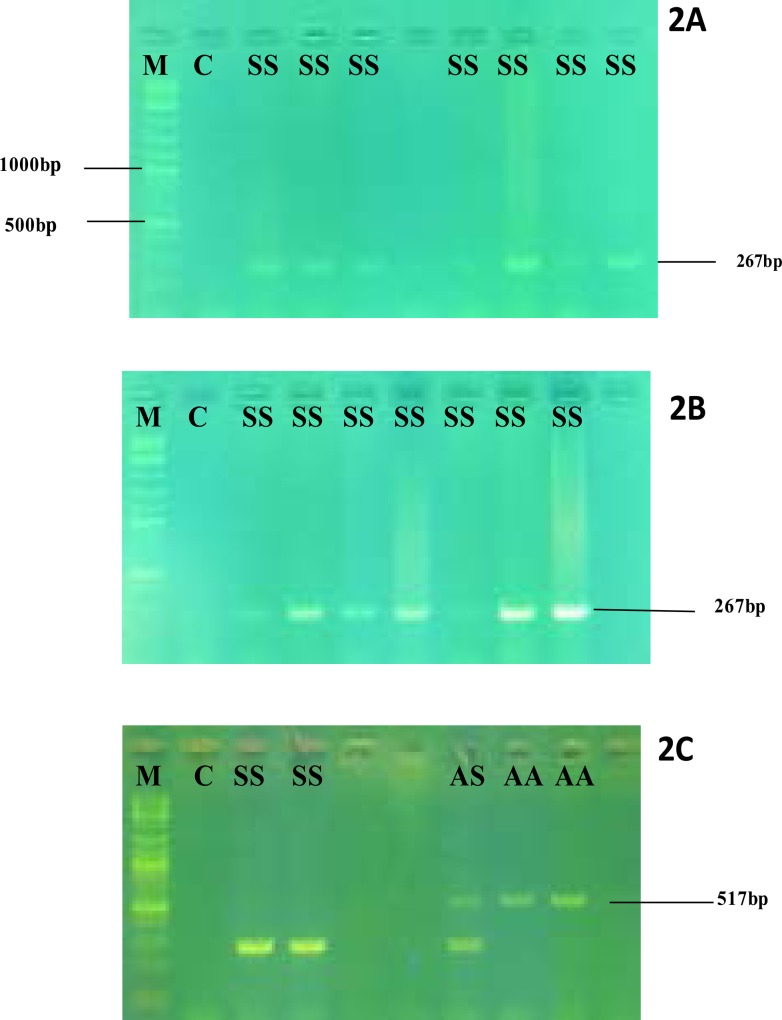
Optimization results, indicate that up to 4ng of DNA template and 1.5mM MgCl2 concentrations increased amplicon yield especially of the wild-type amplicon. Higher MgCl2 concentrations did not have much effect on product yield. Figure 2.
Figure 2.
Gel electrophoresis after optimization of reactants
2A-Gel electrophoresis results with 2ng DNAand 1mM MgCl2
2B-Gel electrophoresis results with 4ng DNAtemplate and 1.5mM MgCl2
2C-Gel electrophoresis results with 4ng DNAtemplate and 2.5mM MgCl2
M- Molecular ladder, C-negative control, SS- reactions with HbSS DNA template, AS- reactions with HbAS DNA template, AA- reactions with HbAA DNA template
Genotyping results
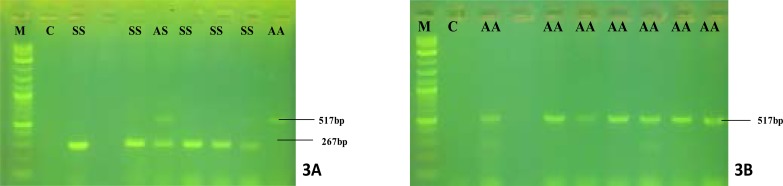
A total of 41 subjects participated in this study; 20 HbSS (13 female and 7 male) with; 20 HbAA (5 females and 15 males) and 1 trait carrier HbAS (female). Amplification results of all homozygous (HbSS) patients revealed amplicon sizes at 267 bp only and heterozygous (HbAS) revealed amplicons at both 267 and 517 bp (Fig.3a), while healthy individuals' (HbAA) revealed amplicon sizes at 517 bp only (Fig. 3b).
Fig 3.
Gel electrophoresis of PCR amplifications.
M-100bp Molecular marker and C- negative control.
Set 3A- lanes 3, 5, 6, 7 and 8 are reactions with mutant DNA template (HbSS), lane 6 represents a reaction with both alleles (HbAS) and lane 10 is reaction with wild-type (HbAA).
Set 3B- lanes 3, 5, 6, 7, 8, 9 and 10 are reactions with wild-type (HbAA).
According to participants' clinical records, all patients were genotyped using the alkaline haemoglobin electrophoresis method. The results of Hb electrophoresis and ASPCR based single tube genotyping were in full agreement for the sickle cell trait, sickle cell individuals and the healthy controls (Table 2).
Table 2.
Results of haemoglobin electrophoresis compared with ASPCR
| Nos of samples |
Haemoglobin Electrophoresis results from patients records |
Alleles detected by ASPCR |
Percentage match |
| 20 | Hb SS | Hb SS | 100% |
| 20 | Hb AA | Hb AA | 100% |
| 1 | Hb AS | Hb AS | 100% |
Discussion
This study investigated the use of ASPCR in screening/diagnosing SCA in a Nigerian setting. This method is adapted from the allele specific amplification (ASA) methods. With ASPCR, the bi-directional nature of primers enable the use of a single tube for the diagnosis of a two allele system15. The results indicate that, ASPCR can be used for screening both for healthy, SCT and SCA. Furthermore, the use of a single tube to detect variant haemoglobin alleles present in each patient sample projects this technique as potentially fast and affordable by reducing the use of scarce PCR master-mixes, consumables and other reagents. Akanni et al.14 reportedly employed the ASPCR method in their study however their approach differed by the use of conventional separate tubes and separate PCR master-mixes each requiring different primers to amplify the different haemoglobin types present per patient sample.
Thin-layer isoelectric focusing (IEF) and high performance liquid chromatography (HPLC) are routine techniques employed in the accurate diagnosis of SCA in more developed countries. Challenges that have limited the use of these up-to-date methods in Nigeria include the huge cost of equipment acquisition, additional costs of consumables and regular equipment maintenance. A recent survey carried out by the Nigerian Sickle Cell Disease Network (NSCDN) on the available facilities and the prevalent management practices in SCD clinics in Nigeria revealed that the haemoglobin electrophoresis method is commonly used for screening and diagnosis of SCA; In addition, only three teaching Hospitals in Nigeria had HPLC machines installed but were not routinely used, and none had IEF facilities 2 (Galadanci et al2. The quest for more affordable approach to accurately screen or diagnose SCA in Nigeria is of serious concern.
Several molecular methods for diagnosis of haemoglobin disorders have been described. The availability of additional equipment such as sequencers and fluorescence detectable devices will affect the method of choice.16 Methods such as ASPCR and PCR- restriction fragment length polymorphism (RFLP) may offer an affordable alternative to HPLC and IEF in Nigeria as it provides the ability to detect alleles of the different globin genes directly, and if necessary results can be confirmed via other methodology like DNA sequencing. These molecular methods are also suitable for new-born sickle cell screening programs as they offer several potential advantages, such as greater reliability, specimen stability, and amenability to automation and large-scale screening.
While the cost of haemoglobin electrophoresis ($3–$4) remains the most affordable method of screening for SCA, it is saddled with limitations and still requires additional tests for accurate diagnosis. The cost of SCA screening using HPLC in a government laboratory in Lagos Nigeria is estimated at $50 to $60 per test. In such laboratories where HPLC test is done, the charge for haemoglobin electrophoresis is absorbed in the cost for HPLC test. Besides the initial set up costs for molecular laboratory, screening using molecular methods will vary with the approach and can range from $25 to $70 per test or more if sequencing of fragment DNA is desired. The single tube ASPCR method is estimated at $20 to $25 per test based on our study. The cost of IEF techniques in Nigeria is not known at the time of this study probably due to its non-availability.
Improving SCA diagnosis in Nigeria and sub-Saharan Africa is very vital in reducing the mortality levels attributed to SCA. It is well reported that most deaths from SCA in Africa occur before the diagnosis is ever made. Many children and sometimes adults die of pneumonia, bacterial sepsis, and acute anaemia without identifying SCA as the critical underlying cause of death. Lack of early and accurate diagnosis also makes it difficult to proffer adequate interventions and treatment for children whose true underlying disease remains unknown13. Similarly, many adults are not aware of their accurate sickle cell status. In a study by Burnham-Marusich et al17 in South-Eastern Nigeria, about half of parental self-reports on sickle cell status or genotypes status were wrongly reported while enrolling for the study. This calls for an urgent need and approach to accurately diagnose the sickle cell status of adults or entire families in the Nigerian population. Introducing the ASPCR method in the Nigerian will significantly increase the awareness of adult accurate sickle cell status and be of great benefit to couples before getting married or starting families. This will further reduce the population of new-borns affected with sickle cell anaemia in Nigeria.
Conclusion
As many research and clinical laboratories in Nigeria embrace the use of molecular biology techniques, it is hoped that early and accurate diagnosis of sickle cell anaemia becomes easily accessible. The ASPCR method is also applicable for other genetic diseases, paternity testing, forensic and pharmacogenomic research where more expensive fluorescence-based approaches are not obtainable.
Acknowledgement
The molecular biology assays were all carried out in the Post-Graduate Molecular Laboratory, Department of Cell Biology and Genetics, University of Lagos, Akoka, Yaba, Lagos. The authors are grateful to the University of Calgary's Global Health Program for the donation of primers and technical assistance.
Individual Author Contributions
Conception: ETT, WH
Design: ETT, WH, G vanMarle
Execution: ETT, OA
Interpretation: ETT, G vanMarle, WH
Writing the paper: ETT, G vanMarle, JO.
Conflict of interest
The authors declared that there is no conflict of interest in this research study.
References
- 1.Kato GJ, Gladwin MT, Steinberg MH. Deconstructing sickle cell disease: reappraisal of the role of haemolysis in the development of clinical sub-phenotypes. Blood Reviews. 2007;21(1):37–47. doi: 10.1016/j.blre.2006.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Galadanci N, Wudil BJ, Balogun TM, Ogunrinde GO, Akinsulie A, Kehinde MO, et al. Current sickle cell disease management practices in Nigeria. International Health. 2014;6(1):23–28. doi: 10.1093/inthealth/iht022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Steinberg MH. Sickle cell anaemia, the first molecular disease: overview of molecular etiology, pathophysiology, and therapeutic approaches. The Scientific World Journal. 2008;(8):1295–1324. doi: 10.1100/tsw.2008.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Driss A, Kwaku OA, Hibbert JM, Gee BE, Adamkiewicz TV, Stiles JK. Sickle Cell Disease in the Post Genomic Era: A Monogenic Disease with a Polygenic Phenotype. Genomics Insights. 2009;(2):23–48. 2009. [PMC free article] [PubMed] [Google Scholar]
- 5.Sebastiani P, Solovieff N, Hartley SW, Milton JN, Riva A, Dworkis DA, et al. Genetic modifiers of the severity of sickle cell anaemia identified through a genome-wide association study. American Journal of Haematology. 2010;85(1):29–35. doi: 10.1002/ajh.21572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Steinberg MH. Genetic etiologies for phenotypic diversity in sickle cell anaemia. The Scientific World Journal. 2009;(9):46–67. doi: 10.1100/tsw.2009.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kutlar A. Sickle cell disease: a multigenic perspective of a single gene disorder. Haemoglobin. 2007;31(2):209–224. doi: 10.1080/03630260701290233. [DOI] [PubMed] [Google Scholar]
- 8.Ballas SK, Kesen MR, Goldberg MF, Lutty GA, Dampier C, Osunkwo I, et al. Beyond the Definitions of the Phenotypic Complications of Sickle Cell Disease: An Update on Management. Scientific World Journal. 2012:1–55. doi: 10.1100/2012/949535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Segal JB, Strouse JJ, Beach MC, Haywood C, Witkop C, Park H, et al. Hydroxyurea for the treatment of sickle cell disease. Evidence Report Technology Assessment. 2008;(165):1–95. [PMC free article] [PubMed] [Google Scholar]
- 10.Anie KA, Egunjobi FE, Akinyanju OO. Psychosocial impact of sickle cell disorder: perspectives from a Nigerian setting. Globalization and Health. 2010;6(1):2. doi: 10.1186/1744-8603-6-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sickle cell anaemia: Report by the Secretariat. Provisional agenda item 11.4. 59th World Health Assembly WHA 59.20 World Health Organization. 2006. http://apps.who.int/gb/archive/pdf_files/WHA59/A59_9-en.pdf. [Google Scholar]
- 12.McGann PT, Tshilolo L, Santos B, Tomlinson GA, Stuber S, Latham T, et al. Hydroxyurea Therapy for Children with Sickle Cell Anaemia in Sub-Saharan Africa: Rationale and Design of the REACH Trial. Pediatric Blood and Cancer. 2016;63(1):98–104. doi: 10.1002/pbc.25705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McGann PT, Hernandez AG, Ware RE. Sickle cell anemia in sub-Saharan Africa: advancing the clinical paradigm through partnerships and research. Blood. 2007;129(2):155–161. doi: 10.1182/blood-2016-09-702324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Akanni EO, Alli OAT, Mabayoje VO. Molecular diagnosis of hemoglobinopathies using allele-specific polymerase chain reaction in Nigeria. Am J Biotechnol Mol Sci. 2013;3(1):24–28. [Google Scholar]
- 15.Waterfall CM, Cobb BD. Single tube genotyping of sickle cell anaemia using PCR-based SNP analysis. Nucleic Acids Research. 2001;29(23):e119. doi: 10.1093/nar/29.23.e119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clark BE, Thein SL. Molecular diagnosis of haemoglobin disorders. Clin Lab Haematol. 2004;26:159–176. doi: 10.1111/j.1365-2257.2004.00607.x. [DOI] [PubMed] [Google Scholar]
- 17.Burnham-Marusich AR, Ezeanolue CO, Obiefune M C, Yang W, Osuji A, Ogidi AG, et al. Prevalence of Sickle Cell Trait and Reliability of Self-Reported Status among Expectant Parents in Nigeria: Implications for Targeted Newborn Screening. Public Health Genomics. 2016;19(5):298–306. doi: 10.1159/000448914. [DOI] [PMC free article] [PubMed] [Google Scholar]