Abstract
Background
Tetralogy of Fallot (TOF) is the most common cyanotic congenital heart defect (CHD) with an incidence of 1/3600 live births. This disorder was associated with mutations in the transcription factors involved in cardiogenesis, like Nk2 homeobox5 (NKX2-5), GATA binding protein4 (GATA4) and T-BOX1 (TBX1). GATA4 contributes particularly to heart looping and differentiation of the second heart field.
Objectives
The aim of this study was to screen a Moroccan cohort with tetralogy of Fallot for GATA4 mutations, and to assess environmental risk factors that could be involved in the occurrence of this disorder.
Methods
Thirty-one non-syndromic TOF patients, enrolled between 5th April 2014 and 18th June 2015, were screened for GATA4 mutations using direct sequencing of GATA4 coding exons. Statistical assessment of different risk factors, which is a retrospective study, was carried out using Chi-square and Fisher's exact tests.
Results
We identified seven exonic variants in nine patients (two missense and five synonymous variants); in addition of eight intronic variants. Assessment of environmental risk factors shows significant association of maternal passive smoking with TOF in the Moroccan population.
Conclusion
The present study allowed, for the first time, the molecular and environmental characterisation of Moroccan TOF population. Our findings emphasise particularly the strong association of passive smoking with the emergence of tetralogy of Fallot.
Keywords: Tetralogy of Fallot, GATA4, molecular screening, risk factors
Introduction
Tetralogy of Fallot (TOF) is the most common type of cyanotic congenital heart defect, affecting approximately 1/3,600 live births1. This cyanotic defect is characterised by the combination of four cardiac anomalies: ventricular septal defect; over-riding aorta; right ventricular outflow obstruction and right ventricular hypertrophy. Approximately 20–25% of TOF occurs within the context of syndromes, particularly with 22q11 microdeletion syndrome2.
TOF is one of the multifactorial disorders that results from interaction of environmental and genetic factors. Environmental factors include maternal toxic exposure (smoking, alcohol, organic solvents), maternal illness with use of medication, and consanguinity among others1. Whereas, genetic factors consists mainly of molecular alteration of genes involved in cardiogenesis1.
Several genes were proved to be involved in non-syndromic Tetralogy of Fallot. The most evident are those encoding the cardiac transcription factors, such as GATA binding protein 4 (GATA4), which is involved in the second heart field differentiation and heart looping during the cardiogenesis process3,4.
GATA4 (GATA-binding protein 4), member of GATA transcription factor family, is a zinc-finger protein that specifically bind to (T/A)GATA(A/G) motif in target genes. GATA4 interact with Nk2 homeobox 5 (NKX2-5) and T-BOX 5 (TBX5) within a complex regulatory network to regulate the down-stream genes involved in heart development process3.
In Lebanese and Chinese populations, GATA4 mutations are estimated to be present in 3 – 8% of patients with Tetralogy of Fallot5,6. Whereas, in other TOF populations, such as the American and Japanese, the GATA4 mutation rate seems to be very low, about 0.5 – 0.8%4,7.
Therefore, the aim of this study was to identify, for the first time, the spectrum of GATA4 variants and to assess the environmental risk factors that may contribute potentially in the emergence of tetralogy of Fallot in a non-syndromic Moroccan population.
Methods
Study subjects
In this retrospective study, thirty-six patients, with ages ranging from 2 months to 15 year-old, were enrolled between 5th April 2014 and 18th June 2015 from the cardiopediatics department. Each patient was evaluated via electrocardiography (ECG) and colour Doppler echocardiography, and confirmed to have tetralogy of Fallot. All patients were clinically evaluated for putative syndromic dysmorphology, and tested for chromosomal rearrangements (using karyotype) and 22q11 microdeletion by the Medical Genetics Department. Five subjects with confirmed or suggestive syndromic traits were excluded from the study. The included patients were interviewed to assess the personal and familial disease history.
Molecular screening
After obtaining informed consent as well as the Approval of the local ethic committee, the peripheral blood was collected from the 31 non-syndromic tetralogy of Fallot patients, then the DNA specimens were extracted from blood leukocytes. For each sample, we performed Polymerase chain reaction (PCR) to amplify GATA4 coding exons and their intron boundaries. Primers used were derived from previous study8.
PCR mix was prepared in a 25 µL volume comprising 10× PCR buffer (Invitrogen, California, CA, USA), 25 mM MgCl2, 10 mM dNTP, 1 U of Taq (Invitrogen, California, CA, USA), 10 pmol of each primer and 40 ng of genomic DNA. The PCR cycling conditions performed in the Veriti 96-well Thermal Cycler 9902 (Applied Biosystems, Massachusetts, MA, USA) were 94 °C for 5 min; 35 cycles of: 94 °C for 45 s, 59 – 62 °C (according to the specific annealing temperature relative to each pair of primers) for 40 s, and 72 °C for 45 s; and 72°C for 7 min. The purified PCR products underwent direct sequencing using the BigDye Terminator V1.1 Cycle Sequencing Kit (ABI Prism, Applied Biosystems, Massachusetts, MA, USA) and the Applied Biosystems 3500Dx Genetic Analyser, then chromatograms were analysed by Sequencing Analysis SeqA v.5.4 (Applied Biosystems, Massachusetts, MA, USA). The sequences thus obtained were analysed with the Bioinformatics analysis tool “Nucleotide Blast alignment program”, (http://blast.ncbi.nlm.nih.gov) (NCBI, Maryland, MD, USA).
Statistical analysis
To assess the significance of differences between the various rates through the study, we used chi-square tests for large cohort (>5) and Fisher's exact test for smaller cohort sizes. The statistical analysis was performed using the R software package version 3.2.
Results
Environmental features
A total of 31 patients with non-syndromic tetralogy of Fallot were recruited for GATA4 mutation screening. The cohort included 14 females and 17 males. The average age was 1 year-old at diagnosis, and 6 year-old at enrolment. All enrolled subjects were Moroccan with 80% of them from the Central North region of Morocco.
Patients born to consanguineous marriages represented 20.8% of the studied population. Averages of maternal and paternal ages were 28 and 38 years respectively. More than a third of patients (38%) had at least one incident of neonatal sibling death or spontaneous abortion in maternal obstetric history. Interestingly, we noticed a high frequency (47.6%) of second-hand smoking during the concerned pregnancies.
Molecular screening
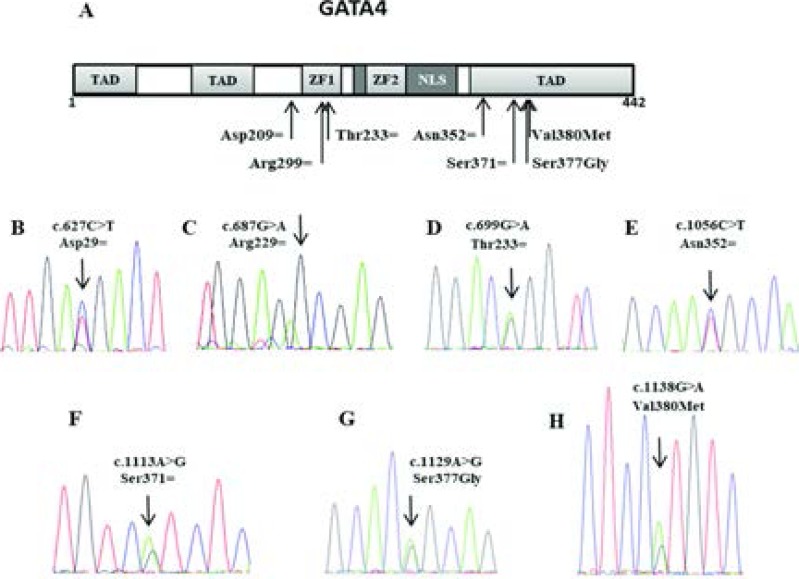
In this study, we investigated DNA samples of 31 non-syndromic TOF patients for GATA4 variants. Mutation screening revealed seven variants: two missense variants, rs114868912 and rs3729856; four synonymous variants that were previously reported, rs192122549, rs55788387, rs3729855 and rs112435835; in addition to a novel synonymous variant. We also identified eight polymorphisms spread over the intronic regions. Figure 1 and tables 1 and 2 show further details.
Figure 1.
(a) Organisation of GATA4 domains with position of the identified exonic variants. (b-h) Sequencing chromatograms showing the exonic variants detected in GATA4. NLS: nuclear localization signal, TAD: Transcriptional activation domain, ZF: zinc finger.
Table 1.
GATA4 exonic variants identified among 31 patients with non-syndromic TOF
| Variation ID | Chromosome localisation |
Nucleotide substitution |
Amino acid substitution |
Exon | Mutation type | Patients |
| rs192122549 | 8:11606438 | c.627C>T | Asp209Asp | 2 | Synonymous | P2 |
| Novel | 8:11606498 | c.687G>A | Arg229Arg | 2 | Synonymous | P7 |
| rs55788387 | 8:11606510 | c.699G>A | Thr233Thr | 2 | Synonymous | P21 |
| rs3729855 | 8:11614502 | c.1056C>T | Asn352Asn | 5 | Synonymous | P6 |
| rs112435835 | 8:11614559 | c.1113A>G | Ser371Ser | 5 | Synonymous | P6 |
| rs3729856 | 8:11614575 | c.1129A>G | Ser377Gly | 5 | Missense | P2, P4, P8, P10, P11 |
| rs114868912 | 8:11614584 | c.1138G>A | Val380Met | 5 | Missense | P4 |
Table 2.
GATA4 intronic variants identified among 31 patients with non-syndromic TOF
| Variation ID | Chromosome localisation |
Nucleotide substitution |
Exon/ intron | variant type | Patients | Frequency (%) |
| Novel | 8:11606365 | c.617-63C>A | Intron 1 | Non-coding | P7 | 3.2 |
| rs10503425 | 8:11606364 | c.617-64G>C | Intron 1 | Non-coding | P2, P4, P8, P10, P11 |
16 |
| rs3735819 | 8:11606312 | c.617-116T>C | Intron 1 | Non-coding | P5, P7, | 6.5 |
| rs200555437 | 8:11606610 | c.783+16G>A | Intron 2 | Non-coding | P31 | |
| rs804280 | 8:11612698 | c.997+56C>A | Intron 4 | Non-coding | P2, P4, P8, P9, P14, P15, P18, P20, P24, 26, P27, P29, P30 |
42 |
| rs76808439 | 8:11612665 | c.997+23A>T | Intron 4 | Non-coding | P13, P14, P15, P20 P29, |
16 |
| rs3729854 | 8:11614329 | c.998-115C>T | Intron 4 | Non-coding | P1, P3, | 6.5 |
| rs114345849 | 8:11615782 | c.1147-20G>A | Intron 4 | Non-coding | P2 | 3.2 |
Discussion
GATA4 is a zinc-finger transcription factor that plays a major role during different stages of embryonic heart development. Association of GATA4 with congenital heart disease was emphasised, for the first time, in 1999 by Pehlivan et al. who were studying a cohort with 8p23 microdeletion, a region that carries GATA49. Then, the subsequent studies confirmed the involvement of GATA4 mutations in different CHD types, with mutation rates varying across ethnic populations. Thus, the aim of the present study was to identify the spectrum and the rate of GATA4 mutations in a Moroccan population with non-syndromic TOF, and also to discuss the factors that may increase the risk of having tetralogy of Fallot.
Assessment of environmental risk factors
Sex-ratio in the studied TOF cohort was 1.2, reflecting a slightly higher proportion of males. An identical ratio has been reported by Sarikouch et al., 201110, Rauch et al., 201011 and Lindinger et al., 201012. In our cohort, consanguinity rate accounts for 20.8%. This rate does not seem to be significantly different from the general consanguinity rate in Moroccan population, that is reported to be around 15.25% (p = 0.4)13. Indeed, the association between consanguinity and CHDs differs from a subset to another, and also across populations. Thus, our data is consistent with a previous study conducted in Saudi population14, in which there was no significant relationship between consanguinity and tetralogy of Fallot sub-group, but different from Lebanese15,16 and Indian17 studies that reported a significant association. The non-association observed in our cohort suggests probably a different mode of inheritance or sporadic events.
According to previous studies, advanced parental age is said to have no particular association with tetralogy of Fallot alongside several CHD subsets18,19. We noticed in our cohort high rates of previous miscarriage in maternal obstetric histories and sibling's neonatal sudden death, respectively 28.5% and 9.5%. According to the National Survey on Population and Family Health, 2011 (ENPSF – 2011) conducted by Moroccan ministry of health in the general population, the rate of perinatal death is limited to 2.8%20, which does not constitute a significant difference to our data (p = 0.12). Our conclusion confirms the studies conducted by Hassan et al.21 and Roodpeyma et al.22 respectively in a Pakistani and Iranian populations that attempted to assess the relationship between previous maternal abortion or perinatal sudden death among sibling and CHDs, but did not find a significant association. In contrast, other studies prove that previous abortions may be associated with the likelihood of having a child with CHD23,24.
Considering the sociocultural and religious characteristics of the Moroccans, maternal smoking or alcohol exposure is very unusual, unlike the paternal's. Thus, to assess the relationship between this well-known cardiovascular risk factor and TOF, we rather checked the maternal passive smoking during the concerned pregnancy. Interestingly, we noticed in our cohort that 47.6% of mothers were exposed to passive smoking during the concerned pregnancy. This rate is significantly higher compared to that described in a Moroccan ministry of health report based on the Global Youth Tobacco Survey (GITS), in which the passive smoking rate in families of Moroccan general population was about 19.7% (p = 0.003)25. These findings lead us to conclude that exposure to passive smoking during pregnancy could be a potential risk factor for tetralogy of Fallot. This conclusion is consistent with the Heart Disease and Stroke Statistics -2016 Update26.
GATA4 variants
GATA4 mutation screening of 31 non-syndromic TOF patients allowed us to identify two heterozygous missense variants. The first variant c.1138G>A (rs114868912), found in P4, leads to the Valine-to-Methionine amino acid change at the position 380. While the second missense variant c.1129A>G (rs3729856) found in five patients (16%) results in Serine-to-Glycine amino acid substitution at the position 377 (Table 1, Figure 1).
These two variants are located at the carboxy-terminal segment that plays an essential role in regulating the GATA4 transcription activity8. Indeed, Gallagher et al. proved that CDK4, a crucial protein required to activate theCyclinD2-GATA4 transcriptional synergy in cardiogenesis interacts directly with GATA4 via its C-terminal region (362-440 aa) , which shows the potential role of GATA4 C-terminal through the heart development process27.
The non-synonymous variant S377G is located near to the nuclear localization signal (NLS), a domain responsible of transporting the GATA4 protein to the nucleus. In order to determine whether this mutation affects the NLS function, Wang et al. carried out immunofluorescence staining for GFP-tagged GATA4 in human cardiomyocytes cells. The result shows that a small amount of S377G GATA4 mutants, among other NLS adjacent mutants, was detected in the cytoplasmic region. Such localisation pattern was considered a partially abnormal, compared to the wildtype pEGFP-GATA4 that was exclusively localised in the nucleus. This finding suggests that S377G may partially impair the GATA4 protein trafficking28.
However, since several studies reported that the variant Ser377Gly was frequently observed in both affected and healthy groups, often with no significant difference6,29,30, it could be considered as possibly benign.
On the other hand, the non-synonymous variant Val-380Met was proven to have no impact in the protein secondary structure and was detected among healthy groups in several studies. Moreover, the codon Val380 seems to be weakly conserved through species4,31–33. These data lead us to conclude that this variant is likely benign.
Mutation screening allowed us to identify five additional variants (c.627C>T, c.687G>A, c.699G>A, c.1056C>T, c.1113A>G), one of them constitutes a non-previously-reported variant, c.687G>A. These variants were identified in one patient each, and found to be synonymous mutations (Asp209Asp, Arg229Arg, Thr233Thr, Asn352Asn, Ser371Ser) (table 1, Figure 1).
As these silent mutations have obviously no damaging effect on protein conformation, we checked their possible earlier effect on splicing function during the post-transcriptional process34–36. We tested hence whether or not those variants are located in a critical exonic splicing enhancer (ESE) or silencer (ESS) sites using Human Splicing Finder tool (HSF 3.0)37.
Interestingly we found that except the variant c.1113A>G (Ser371Ser) that has no particular impact, the remaining four variants have an interesting effect on splicing function. Indeed, the novel synonymous variant c.687G>A (Arg229Arg) leads to a potential alteration of splicing function by altering an exonic splicing enhancer (ESE) site. The synonymous variant c.1056C>T (Asn352Asn) results in a creation of anexonic splicing silencer (ESS) site, while the synonymous variants c.627C>T (Asp209Asp) and c.699G>A (Thr233Thr) lead to a creation of an ESS site, simultaneously with an alteration of an ESE site, which constitutes a potential source of splicing impairment. Disruption of splicing function with such variants can result in skipping the involved exons and thus forming alternative isoforms that may be non-functional, or at least contribute in destabilising the GATA4 isoforms balance7.
Given that GATA4 is a highly dosage-sensitive protein, a slight change in the level of protein expression by such isoform balance disruption, may lead to potential cardiac morphogenesis impairment, involving life-threatening embryonic complications38.
This significant impact is related to the pivotal role that GATA4 plays in proliferation of cardiomyocytes, development of right ventricle and endocardial cushion, and in septation of the outflow tract39,40.
Tomita et al. noticed in TOF subgroup a significantly higher rate of silent variants compared to the non-synonymous variants (p = 0.03), and thus suggested that silent variants may be a potential genetic cause involved specifically in TOF subset7. In our cohort, we noticed a high number of synonymous mutations, but the difference was not significant.
In addition to these exonic variants, we identified several non-coding polymorphisms that were clustered mainly in introns 1 and 4 (Table 2). We used the Human Splicing Finder (HSF 3.0)37 to check the impact of these non-coding variants on splicing machinery, but we did not found a particular altering effect.
It is worth to mention that 71% of the studied patients carry at least one variant that is, in 90% of cases, non-coding polymorphism. This data matches the conclusion made by Tomita and al. who noticed that amongst all CHD patients, those with conotruncal carry a substantial number of non-coding variants, unlike control groups that did not harbour such amounts. This may suggest a possible CHD-predisposing effect, though the molecular mechanism has yet to be elucidated7.
Taken together, this data shows the absence of pathogenic or likely pathogenic mutations of GATA4 gene in the studied Moroccan population. This prompts further molecular investigations in the other genetic factors of cardiogenesis pathways, particularly, those involved in the second heart field differentiation and the heart looping process (e.g. NKX2.5, TBX1 …).
Conclusion
The present study emphasises particularly the association of GATA4 silent mutations with conotruncal defects, and supports the critical role of GATA4 with its highly sensitive dosage in the developmental process of the outflow-tract. On the other hand, this study confirms the strong involvement of maternal passive smoking in increasing the risk of TOF, and underlines notably the heterogeneity of the cardiac defects aetiology. Finally, to have a full overview about the aetiology of TOF in Morocco, it would be pertinent to conduct a case-control study in a larger cohort; to screen GATA4 gene in a control group as well, in order to compare variant frequency between healthy and affected cohorts; and also to consider more additional genetic and environmental factors.
Acknowledgments
We thank the TOF patients and their families who agreed to participate in this study. This work was supported by the Hassan II University Hospital and the Faculty of Medicine and Pharmacy of the University of Sidi Mohamed Ben Abdellah, Fez, Morocco.
Conflict of interest
The authors declare no conflict of interest.
References
- 1.Anderson RH, Baker EJ, Macartney FJ, Rigby ML, Shinebourne EA, Tynan M, editors. Paediatric cardiology. London, England: Churchill Livingstone; 2002. pp. 1213–1502. [Google Scholar]
- 2.Davis S. Tetralogy of Fallot with and without pulmonary atresia. In: Nichols DG, Ungerleider RM, Spevall PJ, editors. Critical Heart Disease in Infants and Children. Philadelphia, PA, USA: Mosby; 2006. pp. 755–766. [Google Scholar]
- 3.Tomita-Mitchell A, Maslen CL, Morris CD, Garg V, Goldmuntz E. GATA4 sequence variants in patients with congenital heart disease. J Med Genet. 2007;44:779–783. doi: 10.1136/jmg.2007.052183. PubMed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Garg V, Kathiriya IS, Barnes R, Schluterman MK, King IN, Butler CA, Rothrock CR, Eapen RS, Hirayama-Yamada K, Joo K, et al. GATA4 mutations cause human congenital heart defects and reveal an interaction with TBX5. Nature. 2003;424:443–447. doi: 10.1038/nature01827. PubMed. [DOI] [PubMed] [Google Scholar]
- 5.Nemer G, Fadlalah F, Usta J, Nemer M, Dbaibo G, Obeid M, Bitar F. A novel mutation in the GATA4 gene in patients with Tetralogy of Fallot. Hum Mutat. 2006;27:293–294. doi: 10.1002/humu.9410. [DOI] [PubMed] [Google Scholar]
- 6.Zhang W, Li X, Shen A, Jiao W, Guan X, Li Z. GATA4 mutations in 486 Chinese patients with congenital heart disease. Eur J Med Genet. 2008;51:527–535. doi: 10.1016/j.ejmg.2008.06.005. PubMed. [DOI] [PubMed] [Google Scholar]
- 7.Kodo K, Nishizawa T, Furutani M, Arai S, Ishihara K, Oda M, Makino S, Fukuda K, Takahashi T, Matsuoka R. Genetic analysis of essential cardiac transcription factors in 256 patients with non-syndromic congenital heart defects. Circ J. 2012;76(7):1703–1711. doi: 10.1253/circj.cj-11-1389. PubMed. [DOI] [PubMed] [Google Scholar]
- 8.Yang YQ, Wang J, Liu XY, Chen XZ, Zhang W, Wang XZ, Liu X, Fang WY. Novel GATA4 mutations in patients with congenital ventricular septal defects. Med Sci Monit. 2012;18(6):CR344–CR350. doi: 10.12659/MSM.882877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pehlivan T, Pober BR, Brueckner M, Garrett S, Slaugh R, Van Rheeden R, Wilson DB, Watson MS, Hing AV. GATA4 haploinsufficiency in patients with interstitial deletion of chromosome region 8p23.1 and congenital heart disease. Am J Med Genet. 1999;83:201–206. [PubMed] [Google Scholar]
- 10.Sarikouch S, Koerperich H, Dubowy KO, Boethig D, Boettler P, Mir TS, Peters B, Kuehne T, Beerbaum P. Impact of Gender and Age on Cardiovascular Function Late After Repair of Tetralogy of Fallot Percentiles Based on Cardiac Magnetic Resonance. Circ Cardiovasc Imaging. 2011;4(6):703–711. doi: 10.1161/CIRCIMAGING.111.963637. PubMed. [DOI] [PubMed] [Google Scholar]
- 11.Rauch R, Hofbeck M, Zweier C, Koch A, Zink S, Trautmann U, Hoyer J, Kaulitz R, Singer H, Rauch A. Comprehensive genotype-phenotype analysis in 230 patients with tetralogy of Fallot. J Med Genet. 2010;47(5):321–331. doi: 10.1136/jmg.2009.070391. [DOI] [PubMed] [Google Scholar]
- 12.Lindinger A, Schwedler G, Hense HW. Prevalence of congenital heart defects in newborns in Germany: results of the first registration year of the PAN Study (July 2006 to June 2007) Klin Padiatr. 2010;222:321–326. doi: 10.1055/s-0030-1254155. [DOI] [PubMed] [Google Scholar]
- 13.Jaouad IC, Elalaoui SC, Sbiti A, Elkerh F, Belmahi L, Sefiani A. Consanguineous marriages in Morocco and the consequence for the incidence of autosomal recessive disorders. J Biosoc sci. 2009;41(05):575–581. doi: 10.1017/S0021932009003393. [DOI] [PubMed] [Google Scholar]
- 14.Becker SM, Al Halees Z, Molina C, Paterson RM. Consanguinity and congenital heart disease in Saudi Arabia. Am J Med Genet. 2001;99:8–13. doi: 10.1002/1096-8628(20010215)99:1<8::aid-ajmg1116>3.0.co;2-u. PubMed. [DOI] [PubMed] [Google Scholar]
- 15.Chehab G, Chedid P, Saliba Z, Bouvagnet P. Congenital cardiac disease and inbreeding: specific defects escape higher risk due to parental consanguinity. Cardiol Young. 2007;17:414–422. doi: 10.1017/S1047951107000704. PubMed. [DOI] [PubMed] [Google Scholar]
- 16.Nabulsi MM, Tamim H, Sabbagh M, Obeid MY, Yunis KA, Bitar FF. Parental consanguinity and congenital heart malformations in a developing country. Am J Med Genet. 2003;116A:342–347. doi: 10.1002/ajmg.a.10020. [DOI] [PubMed] [Google Scholar]
- 17.Gnanalingham MG, Gnanalingham KK, Singh A. Congenital heart disease and parental consanguinity in South India. Acta Paediatr. 1999;88:473–474. doi: 10.1080/08035259950169936. [DOI] [PubMed] [Google Scholar]
- 18.Su XJ, Yuan W, Huang GY, Olsen J, Li J. Paternal Age and Offspring Congenital Heart Defects: A National Cohort Study. PLoS ONE. 2015;10(3):e0121030. doi: 10.1371/journal.pone.0121030. PubMed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Olshan AF, Schnitzer PG, Baird PA. Paternal age and the risk of congenital heart defects. Teratology. 1994;50:80–84. doi: 10.1002/tera.1420500111. PubMed. [DOI] [PubMed] [Google Scholar]
- 20.Ministère de la Santé, author. Rapport de l'Enquête Nationale sur la Population et la Santé Familiale. 2011. p. 189. [Google Scholar]
- 21.Hassan I, Haleem AA, Bhutta ZA. Profile and risk factors for congenital heart disease. J Pak Med Assoc. 1997;47:78–81. [PubMed] [Google Scholar]
- 22.Roodpeyma S, Kamali Z, Afshar F, Naraghi S. Risk factors in congenital heart disease. Clin pediatr. 2002;41(9):653–658. doi: 10.1177/000992280204100903. [DOI] [PubMed] [Google Scholar]
- 23.Clark EB. Etiology of congenital cardiovascular malformations: Epidemiology and genetics. In: Allen HD, Gutgesell HP, Clark EB, Driscoll DJ, editors. MOS and Adams' Heart Disease in Infants, Children, and Adolescents Including the Fetus and Young Adult. Philadelphia, PA, USA: Williams & Wilkins; 2001. pp. 73–74. [Google Scholar]
- 24.Correa-Villasenor A, Ferencz C, Neill CA, Wilson PD, Boughman JA. Ebsteins' malformation of the tricuspid valve: genetic and environmental factors. The Baltimore Washington infants study group. Teratology. 1994;50:137–147. doi: 10.1002/tera.1420500208. PubMed. [DOI] [PubMed] [Google Scholar]
- 25.Sabiri Ahmed. Rapport de l'enquête épidémiologique sur le tabagisme chez les jeunes scolarises de 13 à 15 ans au Maroc. Direction de l'Epidémiologie et de Lutte contre les Maladies, Ministère de la Santé, Royaume du Maroc. 2012. Nov, p. 16. [Google Scholar]
- 26.Mozaffarian D, Benjamin EJ, Go AS, Arnett DK, Blaha MJ, Cushman M, Das SR, de Ferranti S, Després J-P, Fullerton HJ, et al. on behalf of the American Heart Association Statistics Committee and Stroke Statistics Subcommittee Heart disease and stroke statistics-2016 update: a report from the American Heart Association. Circulation. 2016;133:000–000. [Google Scholar]
- 27.Gallagher JM, Yamak A, Kirilenko P, Black S, Bochtler M, Lefebvre C, Nemer M, Latinkić BV. Carboxy terminus of GATA4 transcription factor is required for its cardiogenic activity and interaction with CDK4. Mech Dev. 2014;134:31–41. doi: 10.1016/j.mod.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang E, Sun S, Qiao B, Duan W, Huang G, An Y, Xu S, Zheng Y, Su Z, Gu X, et al. Identification of functional mutations in GATA4 in patients with congenital heart disease. PloS One. 2013;8(4):e62138. doi: 10.1371/journal.pone.0062138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Butler TL, Esposito G, Blue GM, Cole AD, Costa MW, Waddell LB, Walizada G, Sholler GF, Kirk EP, Feneley M. GATA4 mutations in 357 unrelated patients with congenital heart malformation. Genet Test Mol Biomarkers. 2010;14(6):797–802. doi: 10.1089/gtmb.2010.0028. PubMed. [DOI] [PubMed] [Google Scholar]
- 30.Posch MG, Perrot A, Schmitt K, Mittelhaus S, Esenwein E-M, Stiller B, Geier C, Dietz R, Geßner R, Özcelik C, et al. Mutations in GATA4, NKX2.5, CRELD1, andBMP4 are infrequently found in patients with congenital cardiac septal defects. Am J Med Genet Part A. 2008;146A:251–253. doi: 10.1002/ajmg.a.32042. [DOI] [PubMed] [Google Scholar]
- 31.Manjegowda DS, Karunakar P, Ramachandra NB. Effect of structural changes in proteins derived from GATA4 nonsynonymous single nucleotide polymorphisms in congenital heart disease. Indian J Pharm Sci. 2015;77:735–741. doi: 10.4103/0250-474x.174988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dinesh SM, Lingaiah K, Savitha MR, Krishnamurthy B, Narayanappa D, Ramachandra NB. GATA4 specific nonsynonymous single-nucleotide polymorphisms in congenital heart disease patients of Mysore, India. Genet Test Mol Biomarkers. 2011;15(10):715–720. doi: 10.1089/gtmb.2010.0278. PubMed. [DOI] [PubMed] [Google Scholar]
- 33.Schluterman MK, Krysiak AE, Kathiriya IS, Abate N, Chandalia M, Srivastava D, Garg V. Screening and biochemical analysis of GATA4 sequence variations identified in patients with congenital heart disease. Am J Med Genet A. 2007;43:817–823. doi: 10.1002/ajmg.a.31652. PubMed. [DOI] [PubMed] [Google Scholar]
- 34.Modrek B, Lee C. A genomic view of alternative splicing. Nat Genet. 2002;30:13–19. doi: 10.1038/ng0102-13. PubMed. [DOI] [PubMed] [Google Scholar]
- 35.Modrek B, Lee CJ. Alternative splicing in the human, mouse and rat genomes is associated with an increased frequency of exon creation and/or loss. Nat Genet. 2003;34:177–180. doi: 10.1038/ng1159. PubMed. [DOI] [PubMed] [Google Scholar]
- 36.Cartegni L, Chew SL, Krainer AR. Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nat Rev Genet. 2002;3:285–298. doi: 10.1038/nrg775. PubMed. [DOI] [PubMed] [Google Scholar]
- 37.Desmet FO, Hamroun D, Lalande M, Collod-Beroud G, Claustres M, Beroud C. Human Splicing Finder: an online bioinformatics tool to predict splicing signals. Nucleic Acids Res. 2009;37(9):e67. doi: 10.1093/nar/gkp215. PubMed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pu WT, Ishiwata T, Juraszek AL, Ma Q, Izumo S. GATA4 is a dosage-sensitive regulator of cardiac morphogenesis. Dev Biol. 2004;275:235–244. doi: 10.1016/j.ydbio.2004.08.008. [DOI] [PubMed] [Google Scholar]
- 39.Rojas A, Kong SW, Agarwal P, Gilliss B, Pu WT, Black BL. GATA4 is a direct transcriptional activator of cyclin D2 and Cdk4 and is required for cardiomyocyte proliferation in anterior heart field-derived myocardium. Mol Cell Biol. 2008;28:5420–5431. doi: 10.1128/MCB.00717-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zeisberg EM, Ma Q, Juraszek AL, Moses K, Schwartz RJ, Izumo S, Pu WT. Morphogenesis of the right ventricle requires myocardial expression of Gata4. J Clin Invest. 2005;115:1522–1531. doi: 10.1172/JCI23769. PubMed. [DOI] [PMC free article] [PubMed] [Google Scholar]