Abstract
Background
Fungal infections represent a serious health problem especially in immunocompromised individuals. Candida albicans is the most common fungi that cause superficial and systemic infections with high mortality rates. Anti-fungal resistance of C. albicans may be attributed to its virulence. Biofilm formation and proteolytic activity are major virulence determents that may influence both pathogenicity and anti-fungal resistance of Candida albicans.
Objective
This work studied the relation between biofilm formation, proteolytic activity and prevalence of some Sap genes with reduced susceptibility of C. albicans to different anti-fungal agents.
Methods
Fifty three C. albicans strains isolated from patients with systemic infections, identified by germ tube, chromogenic agar and confirmed by PCR, were subjected to evaluate their proteolytic activity, the degree of biofilm production and the prevalence of Sap9 and Sap10 genes. The susceptibility of the isolates was determined by disk diffusion method against five antifungal drugs.
Results and conclusion
Four of the C. albicans isolates were resistant to 3 anti-fungal drugs, strong biofilm producer, have proteolytic activity and contain either Sap9 or Sap10 or both. Conclusively, although anti-fungal resistance among the isolates was rare, a relation between the anti-fungal resistance and some major virulence factors was evidently proved in this study.
Keywords: Candida albicans, resistance, biofilm, proteolytic, Sap
Introduction
Candida infections have a significant rise in morbidity and mortality, especially in recent years due to the continuous increase in the number of immunosuppressive patients1. Candida albicans is determined as the major human pathogen in the genus Candida2. C. albicans is classified as commensal fungi that present in many anatomical sites of the human body3. C. albicans can cause oral and vaginal infections as well as systemic diseases4. The rise in the incidence of Candida infections is complicated by the antimicrobial resistance and the limited number of available anti-fungal drugs5.
The ability of Candida to cause infection depends mainly on its intrinsic virulence attributes6. C. albicans cause critical problems because it has more virulence factors than non-Candida albicans isolates7. C. albicans has several virulence factors including phenotyping changes, biofilm formation, and production of harmful substances to cells, such as haemolysins, phospholipases and proteases as well as the ability to resist hydrogen peroxide8.
Among the hydrolytic enzymes that secreted by Candida spp. are the aspartic proteases which represent one of the major virulence determinants as they have a potential role in pathogenicity through facilitating the invasion and counteracting the host defense system9.
Secreted aspartic proteases (Saps), encoded by a family of 10 sap genes (sap1–sap10) which have a vital role in virulence of C. albicans by degrading host tissue proteins as well as adhere to epithelial host tissue10. Saps have a broad substrate specificity of human proteins such as albumin, haemoglobin, keratin, collagen, laminin, fibronectin, mucin and almost all immunoglobulins, including immunoglobulin A, which is resistant to the majority of bacterial proteases11. A correlation between the expansion of sap genes and the transition from commensal to pathogenic microorganisms has been reported12. Non-pathogenic Candida spp. usually has fewer genes encoding Sap than pathogenic species and this fact was confirmed by gene sequencing. However, this rule cannot be applied to species such as C. glabrata or C. krusei, which do not possess any sap genes12. On the other hand, there is a correlation reported between Sap proteinase production and biofilm formation13.
Biofilms are defined as self-derived extracellular matrix that produced by the microbial population attached to abiotic surface including biomedical devices or biotic surfaces including oral and vaginal mucosa14,15. C. albicans present in biofilm structure show a decrease in susceptibility to some anti-fungals as well as a reduction in killing by the host immune system16. Sap 9 and Sap10 enzymes maintain cell surface integrity of the Candida cell wall, and promote biofilm formation17.
This study aims to correlate between sap9 and sap10 prevalence and its role in biofilm formation and drug resistance among clinical isolates of C. albicans.
Materials and methods
Candida albicans isolates
All the clinical specimens were collected under ethical standards from different Departments at Mansoura University Hospitals, and identified according to Cheesbrough18. Briefly, the specimens were inoculated on Sabouraud dextrose agar (SDA) plates and incubated at 37 °C for 24–48 hr. The suspected colonies of C. albicans were examined for their colonial morphology, Gram staining, germ tube formation, culture characteristics on Candida chromogenic agar (Pronadisa Co., Madrid, Spain) and the identification was confirmed by PCR19, The standard strain of Candida albicans (ATCC 10231) was included in this study.
Determination of anti-fungal susceptibility by disk diffusion method
Candida albicans isolates were tested for their susceptibility to different anti-fungal agents by disk diffusion method according to CLSI guideline20. The tested disks that include; Amphotericin B (AMB, 10µg), Fluconazole (FLU, 25µg), Voriconazole (VOR, 1µg), Fluorocytosine (5FC, 1µg), Caspofungin (CASP, 5µg); were obtained from Bioanalyse, Turkey.
Phenotypic detection of aspartyl proteinase activity in C. albicans isolates
Briefly, Candida isolates were cultured in YEPD medium (2% glucose, 1% yeast extract, and 2% Bactopeptone) and then they induced to secrete proteinases onto the bovine serum albumin (BSA) agar. Filter paper disks, 6 mm diameter, were dipped into a suspension of Candida culture at a density of 107 cell mL−1 (0.5 McFarland) in YEPD medium and applied to BSA agar plate. A maximum of 4 disks was used for each 9 cm diameter plate. The plates were incubated at 28°C for 7 days. The plates were observed daily for opacity around the disks, the opacity caused by albumin precipitation was observed for subsequent clearing due to hydrolysis by the acid proteinases of the fungi. The millimetric zones were evaluated as negative (-) for no clearance, positive (+) for mild activity (lysis zone of 1–2 mm around the disk), and double-positive (++) for strong activity (lysis zone of 3–5 mm). The standard strain was used as positive controls, and the experiment was performed in triplicate21.
Genotypic detection of Sap9 and Sap10 in Candida albicans isolates using PCR
DNA of Candida albicans was extracted by colony PCR method22. The primers used for detection of sap9 and sap10 in C. albicans (SAP9F: 5′ ATTTACTCCACAGTTTATATCACTGAAGGT3′, SAP9R: 5′ CCACCAGAACCACCCTCAGTT 3′, SAP10F: 5′ CCCGGTATCCAATAGAATCGAA3′ and SAP10R: 5′ TCAGTGAATGTGACGAATTTGAAGA 3′) were purchased from Operon Biotechnologies GmbH Biocompus cologne, Germany23.
DNA samples were amplified in 25 reaction mixture containing 2.5 µL DNA, 12.5 µL my Taq red mix (Bioline Co., UK), 1 µL forward primer (10 µLM ), 1 µL reverse primer (10 µLM ) and nuclease free water to 25 µL. the cycling conditions include heating at 94°C for 5 min, then 35 cycles of 94°C for 10 sec, 59°C for 20 sec and 72°C for 30 sec and finally heating at 72°C for 3 min. The PCR products as well as GeneRuler 50 bp plus DNA ladder (Thermo scientific, USA) were separated on 1.5% agarose gel, stained with ethidium bromide, visualized by UV transilluminator and photographed.
Detection of biofilm formation in Candida albicans isolates
The biofilm formation was performed as described previously24. Briefly, a colony from each isolate was obtained from the overnight growth on SDA agar plate and inoculated into 5 mL of Sabouraud broth (SDB). Broths were incubated for 18–20 hr at 37°C. Yeast cells were twice centrifuged (5000 rpm for 5 min) and washed with 0.5 mL phosphate buffer saline (PBS). The cells were re-suspended in 1 mL SD broth and adjusted to concentration of 107cells/mL. Then, 200 µL of each isolate suspension was inoculated into individual well of polystyrene 96-well plates, planktonic cells were discarded through three rounds of washing with 200 µL sterile PBS buffer, and the plates dried at room temperature for 45 min. For staining, 150 µL of 0.4% Crystal Violet (CV), was added to each well, after 45 min, supernatant was discarded before adding 150 µL of 95% ethanol to dissolve and/or elute CV from the biofilm for 45 min. Finally, 100 µL of each well was transferred to a new 96-well microtiter plate and the absorbance at 540 nm was determined using a microtiter plate reader Synergy HT (BioTek Instruments, Winooski, VT, USA).
For each strain, the mean OD of four wells was calculated (ODt) and also cut-off OD (ODc) was defined as 3 standard deviations above the mean OD of the negative control. The level of biofilm production was determined as follows: non-biofilm producer (N) ODt ≤ ODc, weak biofilm producer (W) ODc < ODt ≤ 2×ODc, moderate biofilm producer (M) 2×ODc < ODt ≤ 4×ODc and strong biofilm producer (S) ODt > 4×ODc25.
Results
Identification of C. albicans isolates
Fifty three non-duplicate Candida albicans clinical isolates were identified in the present study. The isolates were identified as Candida albicans by PCR. Meanwhile, 51 isolates gave green colonies of Candida albicans on Chromogenic agar and 45 of Candida albicans isolates were germ tube positive. On the other hand, 8 isolates of C. albicans were germ tube negative (false negative). Among these isolates 30 were from the respiratory tract, 17 from urinary tract and 6 were from blood samples.
Susceptibility to anti-fungals
The data presented in table (1), showed the susceptibility of Candida albicans to the different anti-fungal disks. The data revealed that, out of 53 Candida albicans isolates five (9.4%) isolates were resistant to three anti-fungal drugs. Approximately, fluconazole and voriconazole show 9.4%, 11.3% resistance while 5 fluorocytosine show 100% resistance. On the other hand, both caspofungin and amphotericin B show no resistance.
Table 1.
| Antifungals | Number and percentage of C. albicans isolates (n=53) | |||||
| R | I | S | ||||
| N | % | N | % | N | % | |
| FLU | 5 | 9.4 | 2 | 3.8 | 46 | 86.8 |
| VOR | 6 | 11.3 | 3 | 5.7 | 44 | 83 |
| AMB | 0 | 0 | 2 | 3.8 | 51 | 96.2 |
| 5-FC | 53 | 100 | 0 | 0 | 0 | 0 |
| CASP | 0 | 0 | 1 | 1.9 | 52 | 98.1 |
Proteolytic and biofilm activity
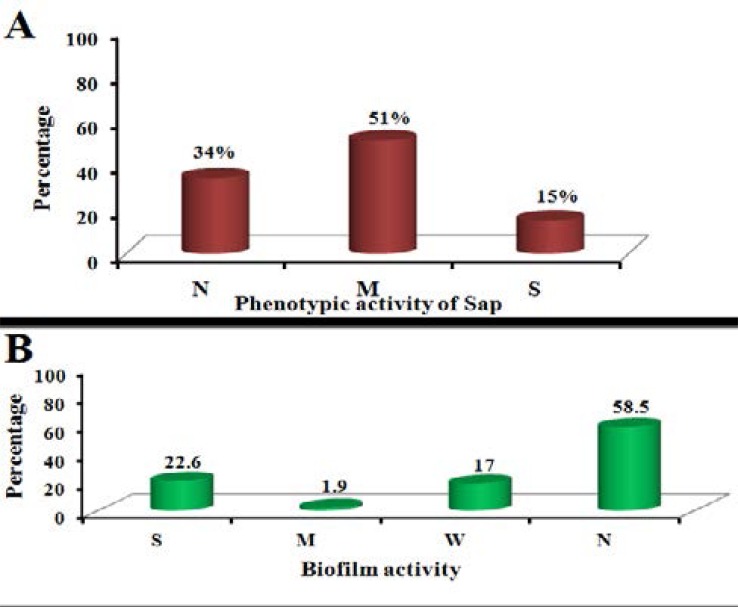
The data presented in Fig. (1A) showed that 8 out of 53 isolates of C. albicans (15%) have strong activity of aspartyl proteinase, 27 isolates have mild activity (51%), while 18 isolates (34%) revealed no aspartyl proteinase activity. The result of biofilm formation shown in Fig. (1B) revealed that 12 (22.6%) isolates out of 53 were strong biofilm forming. Meanwhile, 1 (1.9%) isolate was moderate and 9 (17%) were weak biofilm producers. In addition, 31 (58.5%) isolates were non-biofilm producers.
Fig. (1).
A: Phenotypic detection of secreted aspartyl protease activity in C. albicans isolates by culturing on bovine serum agar (BSA) agar and incubated at 28°C for 7 days and measuring the clearing zone around the growing colony, B: Detection of biofilm activity in C. albicans isolates after biofilm growth on microtiter plate containing Sabouraud broth (SDB) for 48 hours and measure the absorbance at 540 nm after staining with crystal violet.
Genotypic detection of Sap in C. albicans isolates using PCR
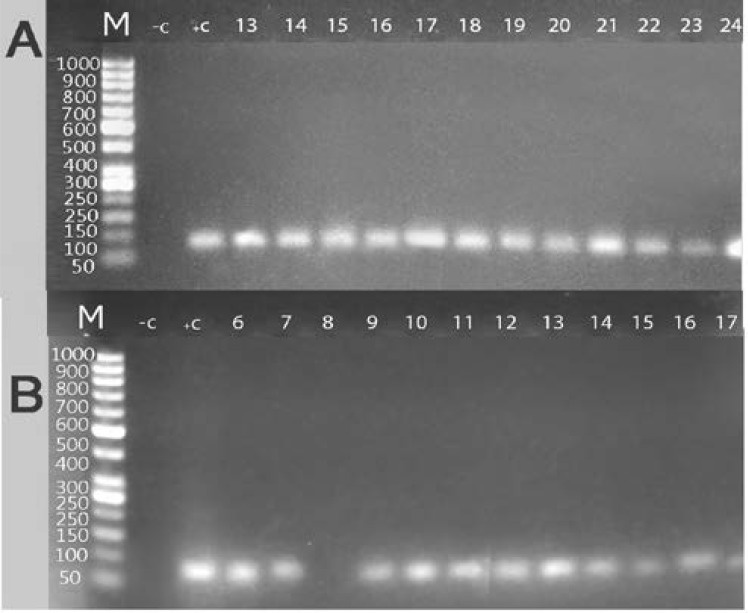
In the present study 53 isolates of C. albicans in addition to standard Candida albicans strain (ATCC10231) were tested for the presence of Sap9 and 10 genes. Sap9 gene was detected in 45 (84.9%) isolates, while Sap10 gene was detected in 42 (79.2%) isolates. Detection of Sap9 and Sap10 genes in the PCR products of Candida albicans isolates on agarose gel at approximately 80 bp, as shown in figure (2).
Fig. 2.
Electrophoretic graph of conventional PCR products on 1.5% agarose gel stained with ethidium bromide in some represented C. albicans isolates (A: detection of Sap9 gene (80 bp amplicon), B: detection of Sap10 gene (80 bp amplicon). Lane 1 (M): represent 50 bp DNA ladder, Lane 2 (-C): negative control (reagent control mixture without DNA), Lane 3 (+C): positive control (reagent control mixture with DNA of standard strain ATCC 10231), Lane 4 to lane 15: clinical C. albicans isolates. DNA was extracted from Clinical C. albicans isolated from patients with systemic Candida albicans infections (respiratory tract, urinary tract infections and candidemia).
The data presented in table (2) show that 5 isolates out of 53 isolates were resistant to 3 anti-fungal which have proteolytic activity with prevalence of either Sap9 or Sap10 genes or both; out of these isolates 4 isolates show strong biofilm formation. The rest of isolates (48 isolates) which were resistant to 2 or 1 anti-fungal drugs show strong biofilm formation in only 8 isolates (16%), while 30 isolates have proteolytic activity (65%).
Table 2.
| Isolate No. |
Source | Biofilm activity |
Sap activity |
Sap 9 gene | Sap 10 gene |
resistance profile |
| ST | ATCC 10231 |
W | + | + | + | 5-FC |
| 1 | R | N | + | + | + | 5-FC |
| 2 | R | N | − | + | + | 5-FC |
| 3 | R | N | ++ | + | + | 5-FC |
| 4 | R | S | ++ | + | + | 5-FC, FLU, VOR |
| 5 | R | N | + | − | + | 5-FC |
| 6 | R | N | − | − | + | 5-FC |
| 7 | R | N | ++ | + | + | 5-FC, FLU, VOR |
| 8 | R | N | + | + | − | 5-FC |
| 9 | R | N | − | + | + | 5-FC, VOR |
| 10 | R | N | + | + | + | 5-FC |
| 11 | B | N | ++ | + | + | 5-FC |
| 12 | R | N | − | + | + | 5-FC |
| 13 | R | N | − | + | + | 5-FC |
| 14 | R | N | − | + | + | 5-FC |
| 15 | R | N | + | + | + | 5-FC |
| 16 | R | W | − | + | + | 5-FC |
| 17 | R | N | − | + | + | 5-FC |
| 18 | R | W | + | + | + | 5-FC |
| 19 | R | N | + | + | + | 5-FC |
| 20 | R | W | − | + | + | 5-FC |
| 21 | B | N | + | + | + | 5-FC |
| 22 | R | N | − | + | + | 5-FC |
| 23 | R | N | − | + | + | 5-FC |
| 24 | B | N | + | + | + | 5-FC |
| 25 | R | N | − | + | + | 5-FC |
| 26 | R | W | + | + | + | 5-FC |
| 27 | R | N | + | + | + | 5-FC |
| 28 | R | S | ++ | + | − | 5-FC, FLU, VOR |
| 29 | R | W | − | + | − | 5-FC |
| 30 | B | N | + | + | − | 5-FC |
| 31 | R | N | + | + | − | 5-FC |
| 32 | R | N | − | + | − | 5-FC |
| 33 | R | N | + | + | + | 5-FC |
| 34 | R | N | − | − | + | 5-FC |
| 35 | B | N | − | + | + | 5-FC |
| 36 | B | S | + | + | + | 5-FC |
| 37 | U | W | + | + | + | 5-FC |
| 38 | U | S | + | + | − | 5-FC, FLU, VOR |
| 39 | U | S | + | + | + | 5-FC |
| 40 | U | M | + | + | + | 5-FC |
| 41 | U | S | + | + | + | 5-FC |
| 42 | U | S | + | + | + | 5-FC |
| 43 | U | W | − | − | − | 5-FC |
| 44 | U | S | + | − | − | 5-FC |
| 45 | U | S | + | − | + | 5-FC |
| 46 | U | S | ++ | + | + | 5-FC |
| 47 | U | N | ++ | + | + | 5-FC |
| 48 | U | N | + | + | + | 5-FC |
| 49 | U | W | + | − | + | 5-FC |
| 50 | U | N | − | + | − | 5-FC |
| 51 | U | W | + | − | − | 5-FC |
| 52 | U | S | ++ | + | + | 5-FC |
| 53 | U | S | + | + | + | 5-FC, FLU, VOR |
Discussion
The aim of this work was to determine the relationship between the anti-fungal susceptibility in one side, and the biofilm formation as well as the secretion of aspartic proteinases on the other side which represent major determinants associated with the pathogenicity of Candida species.
The available therapies against fungal diseases are limited and classified into five classes that have different targets in the fungal cells; polyenes, azoles and allylamines target cell membrane, while pyrimidine analogs target the fungal DNA and RNA. In contrast the new class echinocandins target the fungal cell wall26.
Prolonged usage of azole anti-fungals in treating infections caused by C. albicans has led to the emergence of resistance. The acquisition of azole resistance in clinical isolates of C. albicans generally results in Multi-Drug (MDR) Resistance27,28.
The presence of MDR in fungal pathogens is a serious complication during treatment of opportunistic fungal infections and poses a vital threat to the present therapeutic regimes by limiting the number of clinically useful anti-fungal drugs29.
Biofilm formation in C. albicans has a major rule in pathogenesis which allows Candida to adhere to mucosal cells and polymeric surfaces of medical devices leading to spread of nosocomial infections. Biofilm forming cells are characterized by a three-dimensional structure that can survive the immune system of the host and associated with increasing the resistance to anti-fungal drugs30. The mechanism of biofilm resistance to anti-microbial agents is not fully understood. One hypothesis supposed that the formation of polysaccharide matrix has negative effect on the drugs penetration to fungal cells by formation of strong barrier31, and only the outer layers are in contact with lethal doses of anti-fungals5.
Interestingly, a correlation between anti-fungal susceptibility and biofilm formation was observed in four out of the five resistant clinical isolates in this work. These isolates were strong biofilm producers and revealed resistant to 3 anti-fungal drugs which in agreement with that reported by Bitar et al.32, and Nobile and Mitchell33. They confirmed that the high resistance to fluconazole was usually associated with biofilm formation in C. albicans isolates. This correlation could be explained based on many mechanisms including active extrusion by efflux pumps34.
Severe candidiasis is correlated to the production of extracellular hydrolytic enzymes which have a vital role in the pathogenesis of the yeasts35,36.
Aspartyl proteases are one of the major virulence factors in C. albicans that reported to have a role in tissue invasion, hyphal formation, adherence, and phenotypic switching10. Many evidences prove the role of Sap enzymes in pathogenicity which include; the infected patients with C. albicans have higher proteolytic activity than asymptomatic carriers, in the other hand, the HIV infected patients with C. albicans have high proteolytic activity10. Severe candidiasis is correlated to the production of extracellular hydrolytic enzymes which have a vital role in the pathogenesis of the yeasts35,36.
Furthermore a correlation between reduced susceptibility to anti-fungal agents and aspartyl proteinase production was confirmed in this study where all resistant isolates to the anti-fungals under investigations were aspartyl proteinase producers. In addition 80% of these isolates were biofilm producers.
The current study revealed other correlation between prevalence of Sap9 and Sap10 genes and the strong biofilm producers by C. albicans isolates, as 66.7% of these isolates have both Sap9 and Sap10, while 25% have either Sap9 or Sap10. One isolate (8.3%) has neither Sap9 nor Sap10. Previous result was consistent with that reported by Chaffin37 and Schild et al.17, in which the Sap9 and Sap10 mediate biofilm formation. Silva et al.38, proved that pathogenicity is related to biofilm formation which is a likely indicator of growth, production of hydrolytic enzymes as well as resistance to anti-microbial activity. Furthermore, Rajendran et al.39 found a correlation between higher SAP productions and increase the adhesion to buccal cells as well as resistance to fluconazole of C. albicans isolates.
Another study made a correlation between the extracellular proteolytic activity in vitro and the virulence of Candida species and prove that only the most virulent species such as C. albicans, C. tropicalis and C. parapsilosis produce more proteinases in vitro than do less virulent species40.
Conclusion
Our study represents an advance in biomedical and health science by observing a link between the reduction in susceptibility to anti-fungal drugs and the virulence indicated by pathogenic C. albicans isolates mainly including the biofilm formation and Sap production.
Disclosure statement
No potential conflict of interest is reported by the authors.
References
- 1.Bassetti M, Righi E, Montravers P, Cornely O. What has changed in the treatment of invasive candidiasis? A look at the past 10 years and ahead. Journal of Antimicrobial Chemotherapy. 2018;73(1):i14–i25. doi: 10.1093/jac/dkx445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lai CC, Wang CY, Liu WL, Huang WL, Hsueh PR. Time to positivity of blood cultures of different Candida species causing fungaemia. Journal of medical microbiology. 2012;61:701–704. doi: 10.1099/jmm.0.038166-0. [DOI] [PubMed] [Google Scholar]
- 3.Pfaller MA, Diekema DJ. Epidemiology of invasive candidiasis: a persistent public health problem. Clinical Microbiology Reviews. 2007;20(1):133–163. doi: 10.1128/CMR.00029-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Odds FC, Gow NAR, Brown AJP. Toward a molecular understanding of Candida albicans virulence. In: Heitman J, Filler SG, Edwards JE Jr, Mitchell AP, editors. Molecular principles of fungal pathogenesis. Washington, DC: ASM Press; 2006. pp. 305–319. [Google Scholar]
- 5.Sardi JC, Scorzoni L, Bernardi T, Fusco-Almeida AM, Giannini MJS. Candida species: current epidemiology, pathogenicity, biofilm formation, natural antifungal products and new therapeutic options. Journal of medical Microbiology. 2013;62:10–24. doi: 10.1099/jmm.0.045054-0. [DOI] [PubMed] [Google Scholar]
- 6.White TC, Agabian N. Candida albicans secreted aspartyl proteinases: isoenzyme pattern is determined by cell type, and levels are determined by environmental factors. Journal of Bacteriology. 1995;177:5215–5221. doi: 10.1128/jb.177.18.5215-5221.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Calderone RA, Gow NAR. Host recognition by Candida species. In: Calderone RA, editor. Candida and candidiasis. Washington: ASM Press; 2002. pp. 67–86. PubMed. [Google Scholar]
- 8.Calderoni RA, Fonzi WA. Virulence factors of Candida albicans. Trends in Microbiology. 2001;9:327–335. doi: 10.1016/s0966-842x(01)02094-7. [DOI] [PubMed] [Google Scholar]
- 9.Schaller M, Schackert C, Korting HC, Januschke E, Hube B. Invasion of Candida albicans correlates with expression of secreted aspartic proteinases during experimental infection of human epidermis. Journal of Investigative Dermatology. 2000;114:712–717. doi: 10.1046/j.1523-1747.2000.00935.x. [DOI] [PubMed] [Google Scholar]
- 10.Naglik JR, Challacombe SJ, Hube B. Candida albicans secreted aspartyl proteinases in virulence and pathogenesis. Microbiology and Molecular Biology Reviews. 2003;67:400–428. doi: 10.1128/MMBR.67.3.400-428.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ray TL, Payne CD. Comparative production and rapid purification of Candida acid proteinase from protein-supplemented cultures. Infection and Immunity. 1990;58:508–514. doi: 10.1128/iai.58.2.508-514.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Parra-Ortega B, Cruz-Torres H, Villa-Tanaca L, Hernández-Rodríguez C. Phylogeny and evolution of the aspartyl protease family from clinically relevant Candida species. Memórias do Instituto Oswaldo Cruz. 2009;104:505–512. doi: 10.1590/s0074-02762009000300018. [DOI] [PubMed] [Google Scholar]
- 13.Sacristan B, Blanco MT, Galan-Ladero MA, Blanco J, Perez-Giraldo C, Gomez-Garcia AC. Aspartyl proteinase, phospholipase, hemolytic activities and biofilm production of Candida albicans isolated from bronchial aspirates of ICU patients. Medical Mycology. 2011;49(1):94–97. doi: 10.3109/13693786.2010.482947. [DOI] [PubMed] [Google Scholar]
- 14.Naglik JR, Moyes D, Markwana J, Kanzaria P, Tsichlaki E, Weindl G, et al. Quantitative expression of the Candida albicans secreted aspartyl proteinase gene family in human oral and vaginal candidiasis. Microbiology. 2008;154:3266–3280. doi: 10.1099/mic.0.2008/022293-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nailis H, Kucharikova S, Ricicova M, Van Dijck P, Deforce D, Nelis H, et al. Real-time PCR expression profiling of genes encoding potential virulence factors in Candida albicans biofilms: identification of model dependent and independent gene expression. BMC Microbiology. 2010;10:114. doi: 10.1186/1471-2180-10-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mathe L, Van Dijck P. Recent insights into Candida albicans biofilm resistance mechanisms. Current Genetics. 2013;59:251–264. doi: 10.1007/s00294-013-0400-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schild L, Heyken A, de Groot PW, Hiller E, Mock M, de Koster C, et al. Proteolytic cleavage of covalently linked cell wall proteins by Candida albicans Sap9 and Sap10. Eukaryotic Cell. 2011;10:98–109. doi: 10.1128/EC.00210-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cheesbrough M. Microbiological Tests. In: Cheesbrough M, editor. District Laboratory Practice in Tropical Countries, Part II. Low Priced Edition. Cambridge: Cambridge University Press; 2000. pp. 105–130. [Google Scholar]
- 19.Kadry A, El-Ganiny A, El-Baz A. Comparison of methods used in identification of Candida albicans. Research Journal of Pharmacy and Technology. (In press) [Google Scholar]
- 20.Clinical and Laboratory Standards Institute (CLSI), author Method for anti-fungal disk diffusion susceptibility testing of yeasts: approved guideline. 2nd edition. Wayne, Pa: Clinical and Laboratory Standards Instituite; 2009. M44-A2. [Google Scholar]
- 21.Al-Hedaithy SS. Spectrum and proteinase production of yeast causing vaginitis in Saudi Arabian woman. Medical Science Monitor. 2002;8:498–501. [PubMed] [Google Scholar]
- 22.Marinho SA, Teixeira AB, Santos OS, Cazanova RF, Ferreira CAS, Cherubini K, et al. Identification of candida spp. by phenotypic tests and PCR. Brazilian Journal of Microbiology. 2010;41:286–294. doi: 10.1590/S1517-83822010000200004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Monroy-Pérez E, Paniagua-Contreras G, Vaca-Paniagua F, Negrete-Abascal E, Vaca S. SAP Expression in Candida albicans Strains Isolated from Mexican Patients with Vaginal Candidosis. International Journal of Medical Medicine. 2013;4:25–31. [Google Scholar]
- 24.Younes S, Bahnan W, Dimassi HI, Khalaf RA. The Candida albicans Hwp2 is necessary for proper adhesion, biofilm formation and oxidative stress tolerance. Microbiological Research. 2011;166(5):430–436. doi: 10.1016/j.micres.2010.08.004. [DOI] [PubMed] [Google Scholar]
- 25.Stepanović S, Vuković D, Hola V, Di Bonaventura G, Djukic S, Cirkovic I, et al. Quantification of biofilm in microtiter plates: overview of testing conditions and practical recommendations for assessment of biofilm production by Staphylococci. Journal of Pathology, Microbiology and Immunology. 2007;115:891–899. doi: 10.1111/j.1600-0463.2007.apm_630.x. [DOI] [PubMed] [Google Scholar]
- 26.Denning DW, Hope WW. Therapy for fungal diseases: opportunities and priorities. Trends in Microbiology. 2010;18:195–204. doi: 10.1016/j.tim.2010.02.004. [DOI] [PubMed] [Google Scholar]
- 27.White TC, Marr KA, Bowden RA. Clinical, cellular, and molecular factors that contribute to antifungal drug resistance. Clinical Microbiology Reviews. 1998;11:382–402. doi: 10.1128/cmr.11.2.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Franz R, Ruhnke M, Morschhauser J. Molecular aspects of fluconazole resistance development in Candida albicans. Mycoses. 1999;42:453–458. doi: 10.1046/j.1439-0507.1999.00498.x. [DOI] [PubMed] [Google Scholar]
- 29.Cowen LE. The evolution of fungal drug resistance: modulating the trajectory from genotype to phenotype. Nature Reviews Microbiology. 2008;6:187–198. doi: 10.1038/nrmicro1835. [DOI] [PubMed] [Google Scholar]
- 30.Ingles DO, Skvzypek MS, Arnaudetal MB. Improved gene ontology annotation for biofilm form, filamentous growth, and phenotypic switching in Candida albicans. Eukaryotic Cell. 2013;12(1):101–108. doi: 10.1128/EC.00238-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kojic EM, Darouiche RO. Candida infections of medical devices. Clinical Microbiology Reviews. 2004;17:255–267. doi: 10.1128/CMR.17.2.255-267.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bitar I, Khalaf RA, Harastani H, Tokajian S. Identification, typing, antifungal resistance profile, and biofilm formation of Candida albicans isolates from Lebanese hospital patients. BioMed Research International. 2014;2014:931372. doi: 10.1155/2014/931372. 10 pages. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nobile CJ, Mitchell AP. Genetics and genomics of Candida albicans biofilm formation. Cellular Nicrobiology. 2006;8:1382–1391. doi: 10.1111/j.1462-5822.2006.00761.x. [DOI] [PubMed] [Google Scholar]
- 34.Ramage G, Rajendran R, Sherry L, Williams C. Fungal biofilm resistance. International Journal of Medical Microbiology. 2012;2012:528521. doi: 10.1155/2012/528521. 14 pages. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bramono K, Yamazaki M, Tsuboi R, Ogawa H. Comparison of proteinase, lipase and alpha-glucosidase activities from the clinical isolates of Candida species. Japanese Journal of Infectious Disease. 2006;59:73–76. [PubMed] [Google Scholar]
- 36.Ingham CJ, Boonstra S, Levels S, de Lange M, Meis JF, Schneeberger PM. Rapid susceptibility testing and microcolony analysis of Candida spp. cultured and imaged on porous aluminum oxide. PLoS ONE. 2012;7(3):e33818. doi: 10.1371/journal.pone.0033818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chaffin WL. Candida albicans cell wall proteins. Microbiology and Molecular Biology Reviews. 2008;72:495–544. doi: 10.1128/MMBR.00032-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Silva S, Henriques M, Oliveira R, Williams D, Azeredo J. In Vitro Biofilm Activity of Non-Candida albicans Candida Species. Current Microbiology. 2010;61:534–540. doi: 10.1007/s00284-010-9649-7. [DOI] [PubMed] [Google Scholar]
- 39.Rajendran R, Robertson DP, Hodge PJ, Lappin DF, Ramage G. Hydrolytic enzyme production is associated with Candida albicans biofilm formation from patients with type I diabetes. Mycopathologia. 2010;70:229–235. doi: 10.1007/s11046-010-9319-0. [DOI] [PubMed] [Google Scholar]
- 40.Ruchel R. Proteinase. In: Bennett J E, Hay R J, Peterson P K, editors. New strategies in fungal disease. Edinburgh, United Kingdom: Churchill Livingstone; 1992. pp. 17–31. [Google Scholar]