Abstract
Understanding the formation and properties of self-assembled peptide nanostructures is the basis for the design of new architectures for various applications. Here we show the potential of fluorescence and super resolution imaging to unveil the structural and dynamic features of peptide nanofibers with high spatiotemporal resolution.
Nanostructures obtained by self-assembly of short peptides play a pivotal role in nanotechnology and materials chemistry due to the interesting properties of these architectures and the ease of synthesis.1–3 Despite the minimalistic size of short peptides, a plethora of materials varying in size, shape and physicochemical properties can be obtained, including nanoparticles, nanofibers and nanotubes.4,5 The properties of the final assembly are encoded in the amino acid composition of the building block, allowing to explore a remarkably extended chemical diversity by simply changing the primary sequence of the peptides. Various materials with application-relevant properties have thus been obtained by simple synthesis and self-organization in water or in organic solvents. Di-peptide self-assembled materials have been reported to show good mechanical properties,6–8 high thermal and chemical stability,9,10 and optical and electrical properties.11–13 Despite the simple chemical nature of the building blocks, the rational design of such materials is still highly challenging due to the complexity of the self-assembly process. Moreover, the same properties that make self-assembled materials highly interesting, e.g. size, hierarchical structure and dynamic nature, pose a serious challenge for their characterization and understanding. Peptide assemblies have so far been studied by ensemble methods, such as circular dichroism and IR spectroscopy,14 and by imaging methods, such as atomic force microscopy (AFM)15 and electron microscopy (EM).16 Despite the many advantages of fluorescence microscopy, including multicolour imaging, minimal invasiveness and good spatiotemporal resolution, its use in the study of nanomaterials is still limited. Notably, the field of optical imaging has been recently revolutionized by the advent of novel techniques such as super resolution microscopy or nanoscopy.17 Super resolution techniques retain many of the advantages of classical fluorescence microscopy, while providing a resolution below the diffraction limit. A wide range of nanoscopy methods are currently available, including stimulated emission depletion (STED),18 structured illumination microscopy (SIM)19 and single molecule localization microscopy (SMLM).20–22 Initially developed for cell biology, super resolution microscopy is now emerging as a powerful tool to study (nano)materials such as supramolecular gels,23 block-copolymers assemblies24 and DNA origami.25 We recently show the potential of SMLM to image supramolecular materials in vitro and in the biological environment, paving the way towards the understanding of self-assembly in water and to rational design of novel nanostructures.26–30 Here, we report the use of fluorescence microscopy and super resolution SMLM imaging for the study of diphenylalanine-based nanostructures. Diphenylalanine (FF) is a widely-used self-assembling building block, allowing to obtain nanofibers with remarkable optical and electrical properties. FF nanofiber structures have so far been studied mostly by AFM and EM.5 In this framework, the optical microscopy methods presented here represent an array of complementary tools for the study of FF structural and dynamic properties.
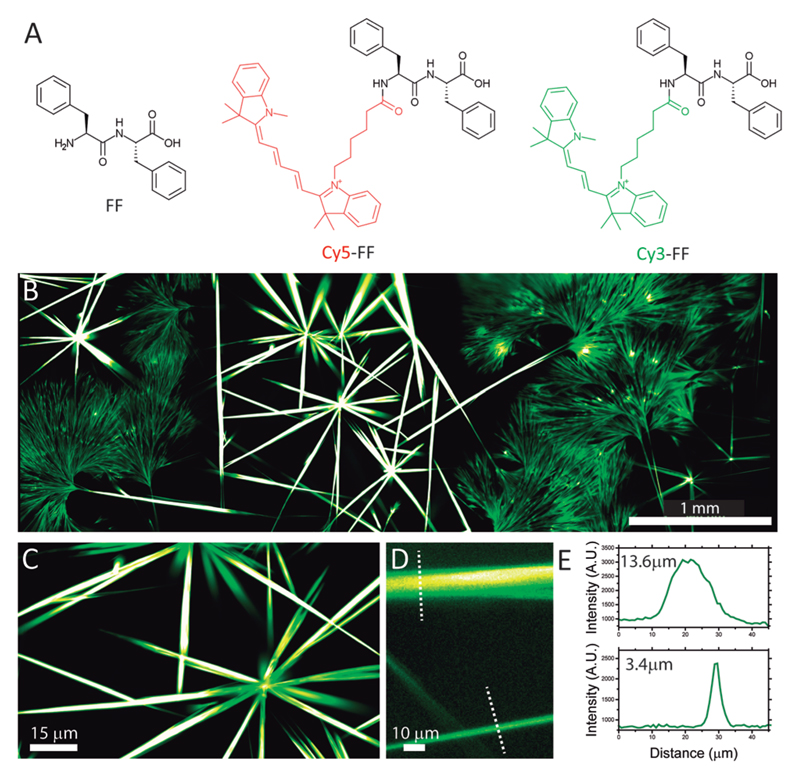
In order to perform fluorescent measurements, we designed two probes comprising a FF peptide N-terminally labelled with Cy3 and Cy5 (Fig. 1A), two spectrally separated fluorescent dyes; see ESI† for the synthesis procedure and materials characterization. Co-assembly of FF dipeptides with the Cy3-FF probe facilitated fluorescent imaging of the assembled nanofibers. The design of the probe is crucial given the very small size of FF as the dye can interfere with the self-assembly process. We designed our probes so that the dyes are exposed to the outside and do not interfere with the self-assembly process (Fig. S3, ESI†). The co-assembly resulted in the formation of 1-dimensional objects with different morphologies (Fig. 1B). Large tubular structures 10–15 μm in diameter were observed together with nanofibers of smaller diameter (<1–4 μm), both several hundred μm in length (Fig. 1D and E). This is in good agreement with previous reports describing the formation of both nanotubes and nanofibers upon FF self-assembly in water.31 Fig. 1B shows a large fluorescence image obtained by rapid acquisition and stitching of over 300 high magnification images. This allowed to image with a good spatial resolution and high signal-to-noise ratio over a very large field of view, which is crucial for imaging assemblies displaying a hierarchical structure and interesting features both at the nanometer and the μm-to-mm scale (Fig. 1C–E). Notably, these images were acquired in native conditions, i.e., in solution without the need for sample drying, freezing or metal coating, thus highlighting the potential of fluorescence microscopy to image peptide nanoassemblies in native conditions.
Fig. 1.
(A) Chemical structure of the self-assembly building block (FF) and the two fluorescent probes. (B) Epifluorescence image of Cy3-FF nanofibers obtained by stitching of 324 frames. Scale Bar = 1 mm. (C and D) Zoom-in images from panel B. Scale Bars = 15 μm and 10 μm. (E) Profile intensity for two fibers highlighted in panel D.
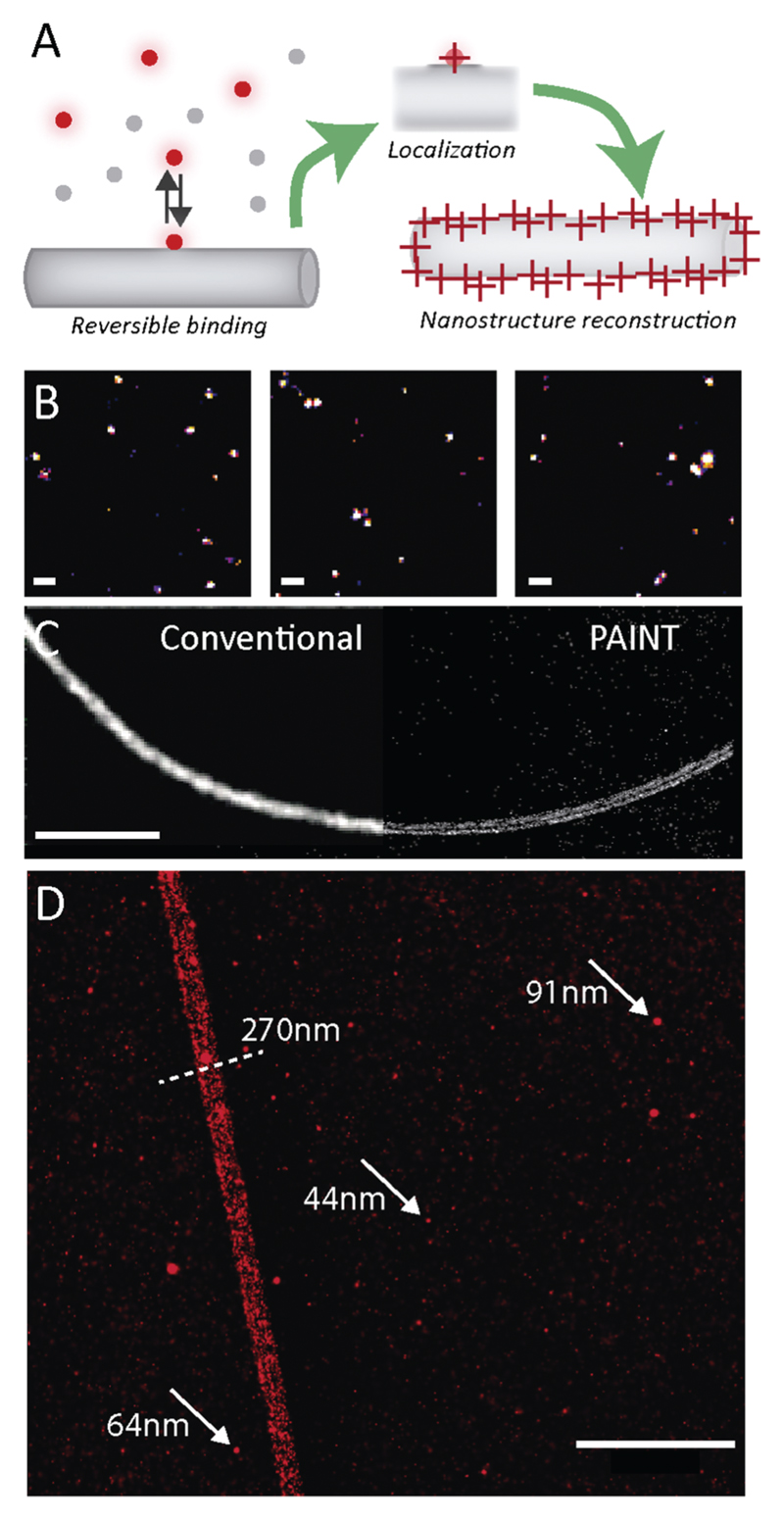
A main drawback of optical imaging is the limited resolution compared to AFM and EM. To overcome this obstacle, super resolution imaging approaches have been proposed.32 We recently reported the use of stochastic optical reconstruction microscopy (STORM) to image several self-assembled nanofibers.26,33,34 Here, we introduced the use of point accumulation for imaging in nanoscale topography (PAINT) to image peptide-based assemblies. While STORM relies on the photoswitching of fluorescent probes to achieve single molecule imaging and localization, PAINT is based on the reversible binding of fluorescent probes to the structure of interest.35 In every individual binding event, a molecule is localized with high accuracy and a super resolution map of the object of interest constructed based on the localizations. As shown in Fig. 2A, we used Cy5-FF as a PAINT probe for FF self-assembly, thus facilitating super resolution imaging of the FF assembly structure, as well as obtaining preliminary data on the surface dynamics of FF nanostructures. Several single imaging frames highlighting individual molecules appearing and disappearing with millisecond kinetics are shown in Fig. 2B. This indicates that the Cy5-FF probe is capable of binding and unbinding the structures, suggesting that the surface of FF assemblies is still exchanging monomers with the solution. Fig. 2C shows the super resolution PAINT image of FF nanofibers (right) compared to conventional diffraction-limited imaging (left), demonstrating the enhancement of resolution using PAINT, allowing to detect the fine details of the structure. As shown in Fig. 2D, features below the diffraction limits, such as the diameter of the nanofibers, can be identified and quantified. Notably, the Cy5-FF probes adsorbed only at the surface, as expected from the structure of FF-assemblies (Fig. 2C). Intriguingly, along with the nanofibers, PAINT revealed the presence of small spherical aggregates (see arrows in Fig. 2D) with a size smaller than 100 nm. Size quantification over several images revealed an average diameter of 64 nm (see Fig. S4, ESI†). It has been proposed that the process of FF self-assembly comprises several steps, starting with the formation of spherical peptide “dots” that hierarchically assemble into nanofibers36 (see also time-resolved AFM characterization in Fig. S5, ESI†). However, characterization of these dots has been elusive due to their small size and poor contrast for optical microscopy and EM. Thus, the coexistence of larger nanofibers and small dots has never been imaged before. Here we show that super resolution PAINT microscopy can visualize nanoscopic features of molecular assemblies and simultaneously resolve both dots and fibers, demonstrating the potential to image hidden features of peptide self-assembly.
Fig. 2.
(A) Schematic representation of PAINT super resolution imaging of FF nanofibers. (B) Representative frames from a PAINT acquisition highlighting single molecule localizations. Scale Bar = 2 μm. (C) Comparison of a conventional TIRF imaging (left) and PAINT imaging (right) of a FF nanofiber. Scale Bar = 5 μm. (D) PAINT imaging highlighting the co-existence of nanofibers and peptide dots. Scale Bar = 1 μm.
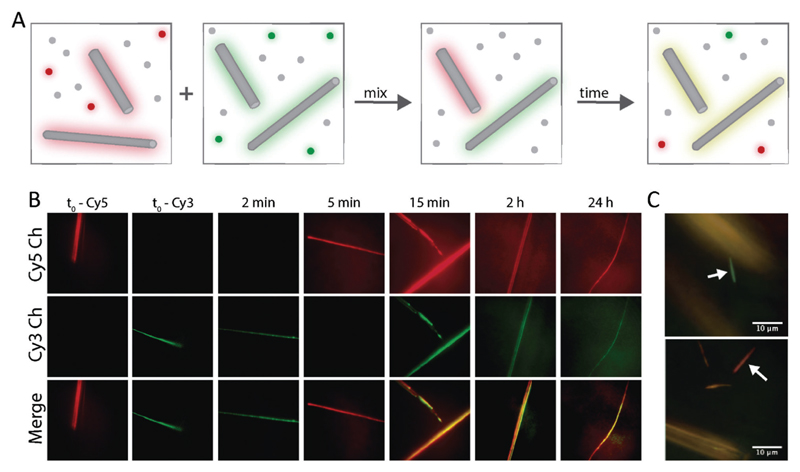
Finally, we used the multicolour ability of fluorescence microscopy to investigate the dynamic nature of FF nanofibers. The ability to continuously polymerize and depolymerize is a key feature of supramolecular structures and it is at the basis of several unique material properties, such as adaptivity, responsivity and self-healing.37 Peptide-based materials have been shown to be highly dynamic38 or completely kinetically trapped,27 depending on the monomer design. However, there are very few reports regarding minimalistic small peptides, and unveiling their dynamic properties is therefore an important step towards their application. To this aim we designed a 2-color kinetic experiment, as schematically represented in Fig. 3A. First, FF was separately co-assembled with either Cy3-FF or Cy5-FF, resulting in “green” and “red” assemblies, respectively. Subsequently, taking advantage of the sedimentation of the large assemblies, we removed the fluorescent monomers from the solution and exchanged them for non-fluorescent FF building blocks, in order to track only monomers that are already part of a nanofiber. Finally, the two samples were mixed and the signal from Cy3 and Cy5 was imaged over time (Fig. 3B). The fibers initially appeared to be either green or red, indicating that no exchange took place. However, in the time scale of minutes, more and more aggregates were visible in both channels, until after about 15 min, the fibers were indistinguishable as they showed strong fluorescence in both channels. These results indicate that the fibers are dynamic and exchange monomers over time, until reaching a thermodynamic equilibrium, where the two probes are equally incorporated.
Fig. 3.
(A) Schematic representation of the imaging experiment to probe assembly dynamics. (B) Fluorescent images of FF assemblies at different time points in the Cy3 (green) and Cy5 (red) channels. (C) Representative images showing co-existence of fully exchanged fibers (yellow) and kinetically trapped fibers (red or green, see arrows).
Surprisingly, minority small fraction of the fibers (<5%) remained single-coloured even after a long mixing time, indicating that no monomer exchange took place. This interesting heterogenous behaviour is very relevant for the technological application of FF materials and can only be identified using optical microscopy, by virtue of its multicolour and single fiber imaging abilities. Recent reports highlighted that depending on the self-assembly building block, it is possible to obtain fully dynamic objects (e.g. benzenetricarboxamide fibers26), fully kinetically trapped aggregates (e.g. polypeptides-based nanofibers27) and an intermediate behaviour. FF nanofibers resemble the heterogenous behaviour of peptide amphiphiles, where fully exchanged fibers are present together with non-exchanged static aggregates.33
In conclusion, we present methods for the fluorescent labelling and imaging of self-assembled nanofibers based on a minimalistic peptide. We show that by incorporation of cyanine-labelled probes, it is possible to image FF nanofibers using epifluorescence, TIRF imaging and PAINT super resolution imaging, thus allowing to visualize the nanostructures from mm-scale to the nanometric level. This allowed us to unveil interesting phenomena, such as the co-existence of different assembly topologies and the properties of monomer exchange, crucial information for clarifying the mechanism underlying bioinspired self-assembly and for the technological application of peptide-based self-assembled materials, thereby paving the way towards the use of fluorescence and super resolution microscopy for the study of a variety of self-assembled peptide structures.
Supplementary Material
† Electronic supplementary information (ESI) available. See DOI: 10.1039/c7cc02176c
Acknowledgments
This work was financially supported by the AXA Research Fund (L. A.) and by the Spanish Ministry of Economy, Industry and Competitiveness through the Centro de Excelencia Severo Ochoa Award (L. A., A. T. and S. P.), by the Generalitat de Catalunya through the CERCA program. Moreover, this work was supported by the Spanish Ministry of Economy and Competitiveness through the project SAF2016-75241-R (MINECO/FEDER).
Footnotes
Kai Tao: 0000-0003-3899-5181
Notes and references
- 1.Adler-Abramovich L, Gazit E. Chem Soc Rev. 2014;43:6881–6893. doi: 10.1039/c4cs00164h. [DOI] [PubMed] [Google Scholar]
- 2.Fleming S, Ulijn RV. Chem Soc Rev. 2014;43:8150–8177. doi: 10.1039/c4cs00247d. [DOI] [PubMed] [Google Scholar]
- 3.Zhang S. Nat Biotechnol. 2003;21:1171–1178. doi: 10.1038/nbt874. [DOI] [PubMed] [Google Scholar]
- 4.Kim S, Kim JH, Lee JS, Park CB. Small. 2015;11:3623–3640. doi: 10.1002/smll.201500169. [DOI] [PubMed] [Google Scholar]
- 5.Yan X, Zhu P, Li J. Chem Soc Rev. 2010;39:1877–1890. doi: 10.1039/b915765b. [DOI] [PubMed] [Google Scholar]
- 6.Adler-Abramovich L, Kol N, Yanai I, Barlam D, Shneck RZ, Gazit E, Rousso I. Angew Chem, Int Ed. 2010;49:9939–9942. doi: 10.1002/anie.201002037. [DOI] [PubMed] [Google Scholar]
- 7.Levin A, Michaels TCT, Adler-Abramovich L, Mason TO, Müller T, Zhang B, Mahadevan L, Gazit E, Knowles TPJ. Nat Phys. 2016;12:926–930. [Google Scholar]
- 8.Ikezoe Y, Washino G, Uemura T, Kitagawa S, Matsui H. Nat Mater. 2012;11:1081–1085. doi: 10.1038/nmat3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Adler-Abramovich L, Reches M, Sedman VL, Allen S, Tendler SJB, Gazit E. Langmuir. 2006;22:1313–1320. doi: 10.1021/la052409d. [DOI] [PubMed] [Google Scholar]
- 10.Ryu J, Park CB. Biotechnol Bioeng. 2010;105:221–230. doi: 10.1002/bit.22544. [DOI] [PubMed] [Google Scholar]
- 11.Fan Z, Sun L, Huang Y, Wang Y, Zhang M. Nat Nanotechnol. 2016;11:388–394. doi: 10.1038/nnano.2015.312. [DOI] [PubMed] [Google Scholar]
- 12.Amdursky N, Molotskii M, Aronov D, Adler-Abramovich L, Gazit E, Rosenman G. Nano Lett. 2009;9:3111–3115. doi: 10.1021/nl9008265. [DOI] [PubMed] [Google Scholar]
- 13.Kim JH, Lee M, Lee JS, Park CB. Angew Chem, Int Ed. 2012;51:517–520. doi: 10.1002/anie.201103244. [DOI] [PubMed] [Google Scholar]
- 14.Hamley IW. Angew Chem, Int Ed. 2007;46:8128–8147. doi: 10.1002/anie.200700861. [DOI] [PubMed] [Google Scholar]
- 15.Sedman V, Chen X, Allen S, Roberts C, Korolkov V, Tendler S. J Microsc. 2013;249:165–172. doi: 10.1111/jmi.12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tao K, Levin A, Jacoby G, Beck R, Gazit E. Chem – Eur J. 2016;22:15237–15241. doi: 10.1002/chem.201603882. [DOI] [PubMed] [Google Scholar]
- 17.Schermelleh L, Heintzmann R, Leonhardt H. J Cell Biol. 2010;190:165–175. doi: 10.1083/jcb.201002018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hell SW, Wichmann J. Opt Lett. 1994;19:780–782. doi: 10.1364/ol.19.000780. [DOI] [PubMed] [Google Scholar]
- 19.Gustafsson MG. J Microsc. 2000;198:82–87. doi: 10.1046/j.1365-2818.2000.00710.x. [DOI] [PubMed] [Google Scholar]
- 20.Betzig E, Patterson GH, Sougrat R, Lindwasser OW, Olenych S, Bonifacino JS, Davidson MW, Lippincott-Schwartz J, Hess HF. Science. 2006;313:1642–1645. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- 21.Bates M, Dempsey GT, Chen KH, Zhuang X. ChemPhysChem. 2012;13:99–107. doi: 10.1002/cphc.201100735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hess ST, Girirajan TPK, Mason MD. Biophys J. 2006;91:4258–4272. doi: 10.1529/biophysj.106.091116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Onogi S, Shigemitsu H, Yoshii T, Tanida T, Ikeda M, Kubota R, Hamachi I. Nat Chem. 2016;8:743–752. doi: 10.1038/nchem.2526. [DOI] [PubMed] [Google Scholar]
- 24.Qiu H, Gao Y, Boott CE, Gould OEC, Harniman RL, Miles MJ, Webb SED, Winnik MA, Manners I. Science. 2016;352:697–701. doi: 10.1126/science.aad9521. [DOI] [PubMed] [Google Scholar]
- 25.Iinuma R, Ke Y, Jungmann R, Schlichthaerle T, Woehrstein JB, Yin P. Science. 2014;344:65–69. doi: 10.1126/science.1250944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Albertazzi L, van der Zwaag D, Leenders CMA, Fitzner R, van der Hofstad RW, Meijer EW. Science. 2014;344:491–495. doi: 10.1126/science.1250945. [DOI] [PubMed] [Google Scholar]
- 27.Leun LH, Albertazzi L, van der Zwaag D, de Vries R, Cohen Stuart MA. ACS Nano. 2016;10:4973–4980. doi: 10.1021/acsnano.6b01017. [DOI] [PubMed] [Google Scholar]
- 28.van der Zwaag D, Vanparijs N, Wijnands S, De Rycke R, De Geest BG, Albertazzi L. ACS Appl Mater Interfaces. 2016;8:6391–6399. doi: 10.1021/acsami.6b00811. [DOI] [PubMed] [Google Scholar]
- 29.Jungmann R, Steinhauer C, Scheible M, Kuzyk A, Tinnefeld P, Simmel FC. Nano Lett. 2010;10:4756–4761. doi: 10.1021/nl103427w. [DOI] [PubMed] [Google Scholar]
- 30.Boott CE, Laine RF, Mahou P, Finnegan JR, Leitao EM, Webb SED, Kaminski CF, Manners I. Chem – Eur J. 2015;21:18539–18542. doi: 10.1002/chem.201504100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nguyen V, Zhu R, Jenkins K, Yang R. Nat Commun. 2016;7 doi: 10.1038/ncomms13566. 13566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schermelleh L, Heintzmann R, Leonhardt H. J Cell Biol. 2010;190:165–175. doi: 10.1083/jcb.201002018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.da Silva RMP, van der Zwaag D, Albertazzi L, Lee SS, Meijer EW, Stupp SI. Nat Commun. 2016;7 doi: 10.1038/ncomms11561. 11561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aloi A, Vargas Jentzsch A, Vilanova N, Albertazzi L, Meijer EW, Voets IK. J Am Chem Soc. 2016;138:2953–2956. doi: 10.1021/jacs.5b13585. [DOI] [PubMed] [Google Scholar]
- 35.Jungmann R, Avendaño MS, Woehrstein JB, Dai M, Shih WM, Yin P. Nat Methods. 2014;11:313–318. doi: 10.1038/nmeth.2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Amdursky N, Molotskii M, Gazit E, Rosenman G. J Am Chem Soc. 2010;132:15632–15636. doi: 10.1021/ja104373e. [DOI] [PubMed] [Google Scholar]
- 37.Webber MJ, Appel EA, Meijer EW, Langer R. Nat Mater. 2016;15:13–26. doi: 10.1038/nmat4474. [DOI] [PubMed] [Google Scholar]
- 38.Baker MB, Gosens RPJ, Albertazzi L, Matsumoto NM, Palmans ARA, Meijer EW. ChemBioChem. 2016;17:207–213. doi: 10.1002/cbic.201500606. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.