Introductory paragraph
Cell motility is essential for viral dissemination1. Vaccinia virus (VACV), a close relative of smallpox virus, is thought to exploit cell motility as a means to enhance the spread of infection1. A single viral protein, F11L, contributes to this by blocking RhoA signalling to facilitate cell retraction2. However, F11L alone is not sufficient for VACV induced cell motility, indicating that additional viral factors must be involved. Here we show that the VACV epidermal growth factor homolog, VGF, promotes infected cell motility and the spread of viral infection. We found that VGF secreted from early infected cells is cleaved by ADAM10 whereupon it acts largely in a paracrine fashion to direct cell motility at the leading edge of infection. Real-time tracking of cells infected in the presence of EGFR/MAPK/FAK/ADAM10 inhibitors, or with VGF and F11 deleted viruses, revealed defects in radial velocity and directional migration efficiency leading to impaired cell-to-cell spread of infection. Furthermore, intravital imaging showed that virus spread and lesion formation are attenuated in the absence of VGF. Our results demonstrate how poxviruses hijack epidermal growth factor receptor induced cell motility to promote rapid and efficient spread of infection in vitro and in vivo.
Cell motility is fundamental for the development, maintenance and immune integrity of multicellular organisms. Its dysregulation is linked to defects in embryonic development, immune cell trafficking and wound healing as well as the induction of metastasis and tumour cell invasion3, 4. Cell motility is a repetitive multistep process under the control of G-protein coupled- or growth factor- receptors5. Receptor activation initiates leading edge extension, followed by adhesion to the cell-substrate, cell body translocation and finally rear edge retraction. Underlying this complex set of movements are cell adhesion molecules and the Rho GTPases, Rac1/Cdc42 and RhoA, which regulate actin dynamics for leading edge extension and rear edge retraction, respectively3, 5. Viruses, including Kaposi sarcoma herpesvirus, herpes simplex virus, Eppstein-Barr virus, human papillomavirus and the poxvirus VACV are known to activate this essential cell process,1, 2, 6–9. For poxviruses, a single viral protein, F11, is known to be needed for virus-induced cell motility2, 10, 11. F11 has been shown to inhibit RhoA signalling by bringing together RhoA and the Rho GTPase-activating protein, Myosin-9a, using its functional PDZ-like domain12. The subsequent loss of RhoA signalling to ROCK and mDia facilitates infected cell detachment and tail retraction2. Viruses lacking F11 display reduced infected cell motility, a reduction in virus spread, and are attenuated in murine infection models2, 10, 11. Surprisingly, given the critical importance of VACV-induced cell motility for efficient virus spread, to date, the viral factor(s), cell receptor(s) and downstream effectors required for activating virus-mediated cell motility have not been identified. In addition, the dynamics of infected cell migration, how this is influenced by the loss of viral and cellular factors that control motility, and how this impacts virus spread has not been determined.
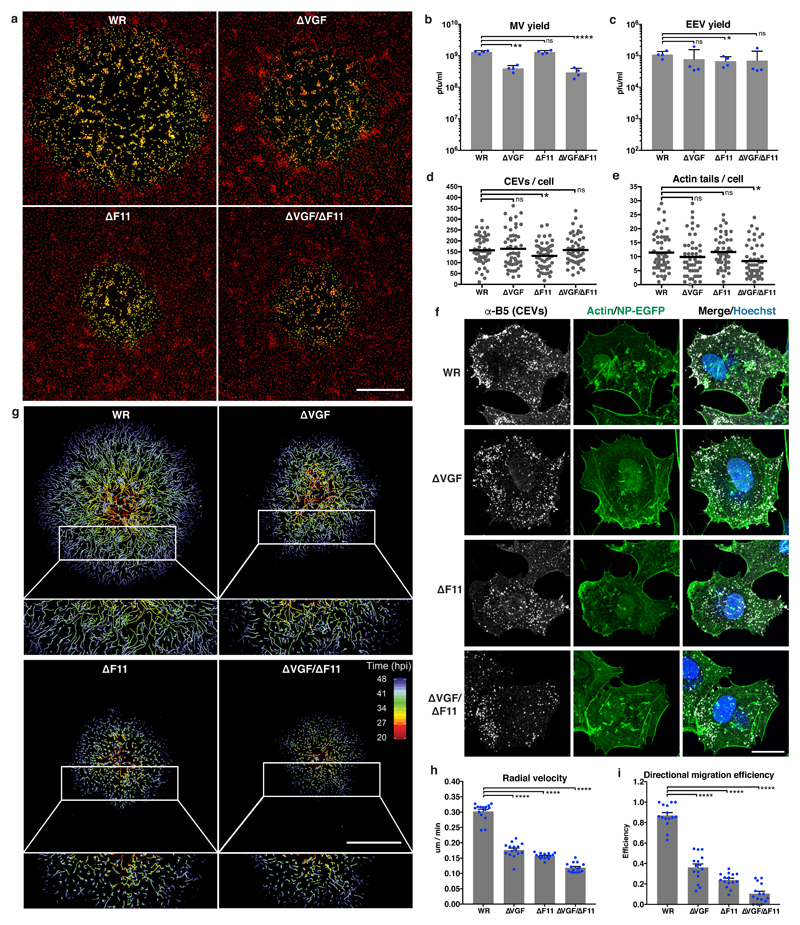
Many poxviruses encode homologues of epidermal growth factor (EGF)13 (Supplementary Fig. 1a). VACV encodes two copies of vaccinia growth factor (VGF) which, when cleaved from infected cells by an unknown protease, can activate EGFR14–17. VACV deleted for both copies of VGF can no longer induce cell proliferative responses in chicken eggs and is attenuated in mice18–20. To determine if VGF is needed for VACV-induced cell motility and virus spread, we generated viruses deleted for VGF (ΔVGF), F11 (ΔF11), or VGF and F11 (ΔVGF/ΔF11) in a parental western reserve (WR) strain expressing a nuclear targeting NP-EGFP21. With its role in virus-induced cell motility well established2, 10, 11, ΔF11 virus was used as the control. Plaque assays, that measure the efficiency of cell-to-cell spread, indicated that ΔVGF, ΔF11 and ΔVGF/ΔF11 viruses were attenuated compared to WR (Fig. 1a). Measurements of plaque diameters indicated that ΔVGF, ΔF11 and ΔVGF/ΔF11 plaques are respectively 38.6%, 53.1% and 62.6% smaller than those of WR (Supplementary Fig. 1b, c). VACV produces two types of infectious particles: single membrane mature virions (MVs) and double membrane enveloped virions (EVs). MVs are released from host cells by lysis and EVs by membrane wrapping and exocytosis of MVs. EVs are released from the cell as extracellular enveloped virions (EEVs), or remain associated with the plasma membrane as cell-associated enveloped virions (CEVs), a subset of which can direct the formation of actin tails22. MVs are thought to mediate host-to-host transmission, EEVs to facilitate long-range spread in an organism, while CEVs and actin tails contribute to cell-to-cell spread. Analysis of virus production over a 24 h period showed a 3.3- and 4.4-fold defect in MV yield with ΔVGF and ΔVGF/ΔF11 viruses, respectively (Fig.1b). More relevant to virus spread, no defect in the formation of EEVs, CEVs or actin tails was seen with ΔVGF (Fig. 1c-f). Minor reductions in EEV (1.6-fold) and CEV (1.2-fold) formation with ΔF11, and actin tails (1.4-fold) with ΔVGF/ΔF11 were observed (Fig. 1c-f). These results indicate that the dramatic attenuation of virus plaque formation and spread of infection could not be attributed to a major defect in virus production. To assess virus-induced cell motility, live-cell imaging and single cell tracking of WR, ΔVGF, ΔF11 and ΔVGF/ΔF11 plaque formation was performed (Supplementary Video 1). Tracks of WR plaques showed kinetically synchronized waves of cell migration out from the plaque centre, while ΔVGF, ΔF11 and ΔVGF/ΔF11 cells appeared to cover less distance and to move more sporadically (Fig. 1g). To quantify this, we measured the distance cells moved from the plaque origin over time (radial velocity), and the degree to which cells migrate in the straightest possible fashion between their origin and endpoint (directional migration efficiency). When assessed relative to WR, both the radial velocity and directional migration efficiency of cells infected with mutant viruses was impaired, with ΔVGF/ΔF11 being the most severe (Fig. 1h,i). These results identify VGF as a new viral factor that facilitates VACV-induced cell motility to enhance cell-to-cell virus spread.
Figure 1. VGF is required for VACV-induced cell motility and virus spread.
a, Plaque formation by VACV WR and mutants (ΔVGF, ΔF11 and ΔVGF/ΔF11). Nuclei (red) and infected cells (green). b, c, 24 h MV and EEV yields from WR and mutant VACVs. d, e, CEVs and actin tails per cell during WR and mutant VACV infections. f, Representative images of cells infected with WR or VACV mutants for 10 h. g, Single cell tracking of WR and mutant VACV plaque formation (20 - 48 hpi). Tracks are colour-coded by time (hpi). h,i, The radial velocity and directional migration efficiency of cells migrating from the centre of plaques in g. Data represents 3 or more biological replicates (a-i). Images are representative of 3 biological replicates (a, f, g). Scale bars are 500 μm (a, g) or 20 μm (f). Bars represent means + SD (b, c), or means + SEM of n=5 plaques per condition/replicate (h, i). Lines represent means of 15-20 cells per condition/replicate (d, e). Paired (b, c) or unpaired (d, e, h, i) t-test was applied (**** P< 0.0001, ** P< 0.01, * P< 0.05, ns = not significant). See Supplementary Table 1 for exact statistics.
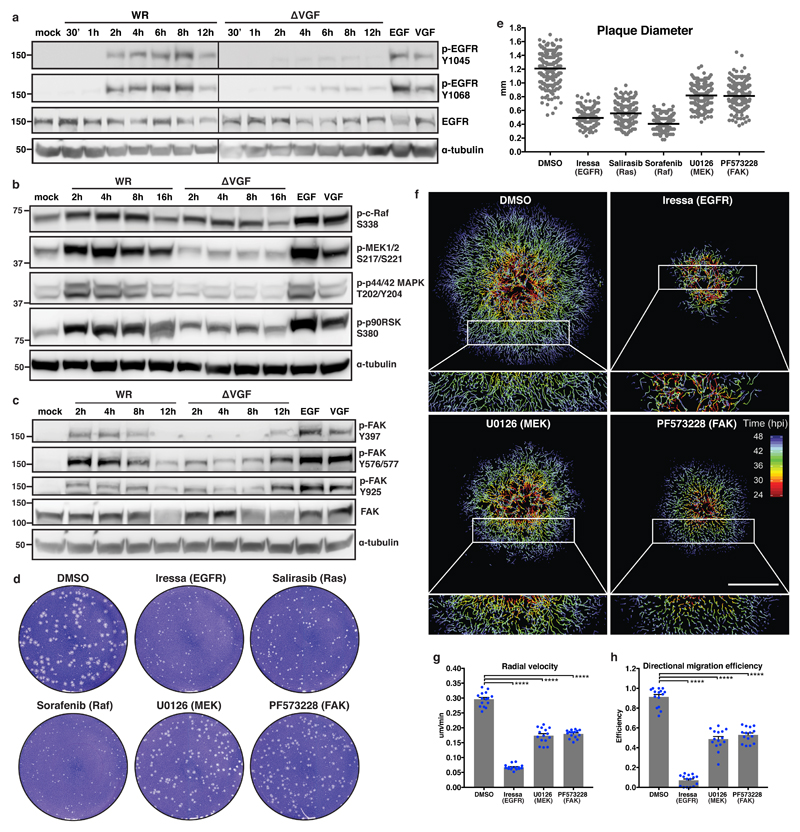
To simultaneously stimulate cell growth and prevent cell death, VGF activates EGFR, and the down-stream signalling molecules PLC-γ1 and Erk1/223–27. Given the diversity of EGFR signalling, activation arrays were employed to analyse the full compendium of EGFR signalling in WR or ΔVGF infected cells, as well as cells treated with purified VGF or EGF (Supplementary Fig. 2a). Significant increase in VGF-dependent steady-state phosphorylation of EGFR, MEK1/2, Akt, and Erk1/2 was seen (Supplementary Fig. 2b-d). This suggested that VGF-mediated activation of EGFR may be triggering Erk/MAPK-mediated cell migration28. To extend these findings, the phosphorylation of EGFR, c-Raf, MEK1/2, p44/42 MAPK (Erk1/2), p90RSK and FAK were compared during WR and ΔVGF infections. As opposed to WR, which showed robust activation of this pathway between 2 and 8 hours post infection (hpi), phosphorylation of each of these proteins was markedly lower in ΔVGF infected cells (Fig. 2a-c). Addition of exogenous VGF confirmed that it could effectively mimic EGF by activating EGFR/MEK/FAK signalling (Fig. 2a-c).
Figure 2. VGF activates cell motility through EGFR/MAPK/FAK signalling.
a-c, Immunoblot analysis of EGFR, MAPK and FAK phosphorylation during WR and ΔVGF infections. d, Plaque formation in the presence of VGF signalling inhibitors. e, Diameter of plaques from d. f, Single cell tracking of VACV plaque formation in the presence of EGFR, MEK or FAK inhibitors (24 - 48 hpi). Tracks are colour-coded by hpi. g, h, The radial velocity and directional migration efficiency of cells migrating from the centre of plaques in f. Data represent 3 or more biological replicates (a-h). Images are representative of 3 biological replicates (a-d, f). Lines represent means of 100 plaques per condition/replicate (e). Scale bar=500 μm (f). Bars represent means + SEM of n=5 plaques per condition/replicate (g, h). Unpaired t-test was applied (**** P< 0.0001). See Supplementary Table 1 for exact statistics.
To determine if activation of this pathway is critical for cell-to-cell virus spread and cell motility, inhibitors of EGFR (Iressa), Ras (Salirasib), Raf (Sorafenib), MEK1/2 (U0126) and FAK (PF573228) were assessed for their ability to retard plaque formation (Fig. 2d). Inhibitors of EGFR, Ras and Raf reduced plaque size by ≥ 50.0% and inhibitors of MEK and FAK by 33.3% (Fig. 2e). Reduced plaque size with EGFR and MEK1/2 inhibitors is in accordance with previous findings29, 30. As these inhibitors may disrupt various stages of the virus life cycle resulting in attenuated virus spread, their impact on WR MV, EEV, CEV and actin tail formation was assessed. No major defect in MV production was seen in the presence of any inhibitor tested, while minor defects in EEV formation was seen upon Raf (1.6-fold) and MEK1/2 (1.4-fold) inhibition (Fig. S2e,f). Most pronounced, inhibition of EGFR resulted in a 1.6-fold decrease in CEVs and 1.6-fold decrease in actin tail formation, and inhibition of FAK reduced actin tail formation by 2.8-fold (Fig. S2g,h). Due to their impact on virus production and/or actin tail formation, inhibitors of EGFR, MEK and FAK were directly assessed for their impact on VACV-induced cell motility by live cell imaging and single cell tracking (Fig. 2f, Supplementary Video 2). Despite their respective defects in virus formation, both radial velocity and directional migration efficiency of infected cells were impaired by more than 4-fold upon inhibition of EGFR and nearly 2-fold upon inhibition of MEK and FAK (Fig. 2g,h). Collectively, these results indicate that activation of the EGFR/Ras/Raf/MEK/FAK signalling pathway by VGF is critical for VACV-induced cell motility and virus spread.
VACV-mediated cell motility was first described using a classical scratch assay, whereby infection of an entire monolayer facilitated cell crawling into the wound1. To investigate this process during formation of a plaque, cell monolayers were scratched and single-cell tracking performed on plaques adjacent to wounds (Fig. 3a, Supplementary Video 3). Vector field analysis showed that motile cells within a plaque that approach a wound had a high propensity to halt or turn and move parallel to the scratch, while cells that did not encounter a wound tended to maintain their kinetically synchronized outward movement (Fig. 3a). This phenotype led us to hypothesise that VGF expressed in early infected cells at the leading edge of a plaque might establish a chemotactic gradient toward which virus producing cells crawl. If this were the case, with no cells in the wound, and thus a lack of VGF signal to follow, motile virus producing cells would either halt or follow their closest early infected neighbor. To test the chemoattractant activity of VGF, transwell migration assays were employed. Neither soluble VGF nor VGF-secreting infected cells were capable of directing chemotaxis of ΔVGF infected cells (Supplementary Fig. 3a, b).
Figure 3. ADAM10-mediated VGF release triggers cell motility in a paracrine fashion.
a, Analysis of directional motility by live cell imaging, single cell tracking and vector field analysis during plaque formation adjacent to wounds. b, Spatial analysis of VGF expression within a plaque using immunofluorescence. c, Supernatant and cell transfer assays to investigate paracrine and juxtacrine mediated activation of EGFR signalling by VGF. d, Metalloprotease inhibitors of ADAM10 (GI) and ADAM10/17 (GW), prevent VGF shedding and VGF paracrine signalling activity. e, RNAi-mediated silencing of ADAM10, but not ADAM17, prevents VGF shedding and VGF paracrine signalling activity. f, Single cell tracking of VACV plaque formation in the presence of ADAM10 inhibitor (GI) (24 - 48 hpi). Tracks are colour-coded by hpi. g, h, The radial velocity and directional migration efficiency of cells migrating from the centre of plaques in f. i, j, 24 h MV and EEV yields from DMSO- or GI-treated WR infected cells. k, l, CEVs and actin tails per cell during WR infections in the presence of GI for 10 h. Data represents 3 or more biological replicates (a-l). Images are representative of 3 biological replicates (a-f). Scale bars=500 μm (b, f). Bars represent means + SD (i, j), or means + SEM of n=5 plaques per condition/replicate (g, h). Lines represent means of 15-20 cells per condition/replicate (k, l). Paired (i, j) or unpaired (g, h, k, l) t-test was applied (**** P< 0.0001, ns = not significant). See Supplementary Table 1 for exact statistics.
As VGF appeared to harbour no chemoattractant activity we asked which cells, in the context of a VACV plaque, were expressing VGF. Immunofluorescence staining of WR or ΔVGF plaques showed that VGF was expressed only in infected cells at the leading edge where cell motility is on-going, and not in cells at the plaque centre where motility has ceased (Fig. 3b). We reasoned that the expression of VGF in early infected cells at the leading edge of plaques serves to spatially and temporally regulate infected cell motility. Consistent with this notion, the activation pattern of EGFR within VACV plaques, a hallmark of VGF-mediated cell motility, mirrors VGF expression29.
Cellular growth factors exert their effects by activating their cognate receptors on cells from which they are produced (autocrine), on direct neighbouring cells (juxtacrine), or on cells in the vicinity (paracrine)31. VGF is an early gene product of VACV infection produced between 2 and 8 hpi as a 25 kDa precursor, and released from cells as a 22 kDa soluble ligand 14 (Supplementary Fig. 3c-e). To test if VGF could induce cell motility in an autocrine fashion, individual uninfected, WR or ΔVGF infected cells were sorted into single wells, monitored by live cell imaging and subjected to single cell tracking (Supplementary Fig. 3f). Infection with WR significantly increased the radial velocity and directional migration efficiency of single cells, while cells infected with ΔVGF showed no radial velocity increase, and only a minor increase in directional migration efficiency, over uninfected cells (Supplementary Fig. 3g). This indicates that VGF secreted from an infected cell can trigger its motility. As both autocrine and paracrine signalling are directed by soluble ligands, this result is consistent with supernatant transfer experiments showing that secreted VGF can activate EGFR in a paracrine fashion15.
However, as juxtacrine signalling is mediated by membrane-bound ligand, and infected cells within a viral plaque are in close proximity, we sought to determine the relative contribution of juxtacrine and paracrine signalling in VGF-mediated EGFR activation. For this, WR or ΔVGF infected supernatants and cells were collected and separately transferred to naive cells which were then probed for activation of EGFR and MEK1/2. While control ΔVGF supernatants and cells showed neither paracrine nor juxtacrine activity, WR-infected supernatants showed potent paracrine activity and WR-infected cells no juxtacrine activity (Fig. 3c). These results indicate that VGF can only promote motility of infected cells in an autocrine and paracrine fashion once it is shed from infected cells.
With one exception, shedding of EGFR ligands from the cell surface is mediated by a disintegrin and metalloproteases 10 or 17 (ADAM10 / ADAM17)32. To assess if VGF is released from cells by one of these, cells were infected in the presence of ADAM inhibitors GW280264X (GW) or GI254023X (GI). GW is an inhibitor of ADAM10 and ADAM17, whereas GI inhibits ADAM10 with 100-fold more potency33. Both GI and GW effectively blocked the shedding of VGF into the supernatant resulting in its accumulation on infected cells (Fig. 3d, left). Transfer experiments showed that supernatants from GI- or GW-treated infected cells could not trigger EGFR and MEK1/2 signalling (Fig. 3d, right). RNAi-mediated silencing of ADAM10 and ADAM17 followed by supernatant transfer experiments, confirmed that ADAM10, and not ADAM17, was largely responsible for VGF shedding and subsequent activation of EGFR and MEK1/2 signalling (Fig. 3e). These results identify ADAM10 as the metalloprotease responsible for shedding of VGF. In line with this, the addition of GI resulted in reduced plaque size (Supplementary Fig. 4a,b), which correlated with shortened tracks, a 1.3-fold reduction in cell velocity and a 3.9-fold reduction in directional migration efficiency (Fig. 3f-h, Supplementary Video 4). No defects in WR MV, EEV, CEV or actin tail formation were seen in the presence of GI (Fig. 3i-l, Supplementary Fig. 4c). Thus, ADAM10-mediated shedding of VGF is a pre-requisite for its autocrine/paracrine activation of EGFR signalling, virus-induced cell motility and virus spread.
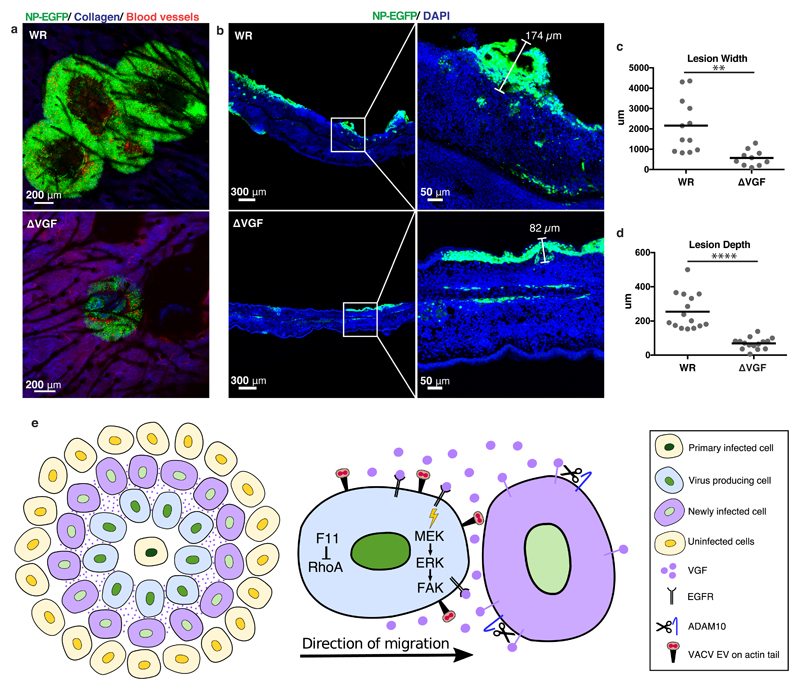
An in vivo hallmark of poxvirus infection is the formation of cutaneous lesions. As plaque formation may serve as a 2-D in vitro surrogate for this, the role of VGF in VACV lesion formation was addressed. Mice ear pinnae were epicutaneously infected with WR or ΔVGF viruses, and lesions visualised using multiphoton microscopy. By six days post infection WR had formed large multi-foci lesions, while ΔVGF lesions were less numerous and 3.8-fold smaller (Fig. 4a,c). Analysis of lesion cross-sections revealed that the depth of ΔVGF lesions was also reduced by 3.7-fold (Fig. 4b,d). That ΔVGF displays no major defects in virus production (Fig. 1b-d), strongly suggests that the reduction in lesion size is due to the observed attenuation of virus-induced cell motility.
Figure 4. VGF is required for lesion formation in vivo.
a, Multiphoton microscopy of WR and ΔVGF lesions in mice ear pinnae 6 days pi. Infected cells (green), collagen (blue), blood vessels (red). b, Confocal imaging of WR and ΔVGF lesions in cross section. Infected cells (green), nuclei (blue) c, d Quantification of lesion widths and depths from a and b, respectively. e, Model of VGF mediated VACV induced cell motility (refer to text for details). Representative data from 2 mice/virus in biological triplicates (a. b). Lines represent means of 10-15 lesions per condition from 5 mice/virus (3 cross-section, 2 frontal sections). Unpaired t-test was applied (**** P< 0.0001, ** P< 0.01) (c, d). See Supplementary Table 1 for exact statistics.
The cell-to-cell spread of poxvirus infection is a complex process relying on the production of extracellular virus, actin tail formation, cell motility and superinfection repulsion2, 34, 35. Blocking of any one of these reduces VACVs ability to spread cell-to-cell, results in a concomitant reduction in plaque size, and attenuation in mice. That loss of each of these processes results in a diminution, but not complete ablation, suggests that each provides a relative contribution to VACVs ability to spread. Future studies should be aimed at determining this relative contribution, whether these processes are additive or cooperative, and if their contribution differs depending on whether virus spread is occurring in a cell monolayer (2D) or in an infected host tissue (3D).
Here we show that the VACV epidermal growth factor homologue, VGF, contributes to this process through activation of EGFR-mediated cell motility. As illustrated in Figure 4e, our data suggests that within the context of a VACV plaque VGF expression is relegated to newly infected cells (purple) at the leading edge. The short window of VGF expression, from 2-8 hours post infection, in combination with ADAM10-mediated shedding appears to spatially and temporally restrict VGF paracrine activity. This invokes a model whereby autocrine/paracrine activation of EGFR/MEK/ERK/FAK signalling promotes rapid and efficient motility of infected cells in the vicinity, including those infected for longer than 6 hours and in the process of producing virus (blue cells). Thus, it reasons that VGF-mediated cell motility increases the efficiency of spread by assuring increased cell-to-cell contact between virus-producing cells and their more distant uninfected neighbours. This model is supported by the appearance of kinetically synchronised waves of infected cell motility away from the plaque origin. Our previous simulations of VACV plaque formation indicate that the average diameter of uninfected and infected cells is 43.5 +/-13.4μm and 34.7 +/-14.6 μm, respectively36. Based on radial velocity measurements in WR plaques (0.3μm/min), infected cells can traverse on average 10 uninfected cells within 24 h. By providing new molecular understanding of VACV-induced cell migration, using virus mutants and inhibiting EGFR signalling at multiple levels we show that infected cell motility is directly linked to the ability of VACV to spread. The induction of cell motility through hijacking of the EGFR/MAPK/FAK signalling axis by VGF is reminiscent of metastatic transformation37. Perhaps motile infected cells harbouring cell-associated virus rather than free virus particles38 act as mediators of tissue-to-tissue virus spread. This would explain the ability of EGFR-targeting chemotherapeutic compounds to prevent poxvirus spread in vitro29, and block VACV-mediated lethality in vivo39, thus encouraging the potential repurposing of other chemotherapeutics as antiviral agents.
Methods
Cells
HeLa (ATCC CCL-2) and BSC40 (from Paula Traktman, Medical University of South Carolina) cells were maintained in Dulbecco's Modified Eagle Medium (DMEM) supplemented with 10% FBS, 2 mM Glutamax, and Penicillin-Streptomycin at 37.0 °C and 5.0% CO2. BSC40 medium was supplemented with 100 μM non-essential amino acids and 1 mM sodium pyruvate (Thermo Fischer Scientific). HeLa (ATCC CCL-2) cells have been authenticated by ATCC and BSC40 cells have not been authenticated. Both cell lines were tested regularly, and remained mycoplasma free, throughout this study.
Inhibitors and antibodies
Salirasib (5 μM, #SML1166; Sigma), Sorafenib (5 μM, #8705) and U0126 (20 μM, #9903; Cell Signaling Technology), Gefitinib (10 μM, Iressa, #S1025; Selleck-Chem), GI254023X (20 μM, #3995) and PF573228 (10 μM, #3239; Tocris), and GW280264X (20 μM, AOB3632; Aobious) were all used at the indicated concentrations. Monoclonal anti-B5 (VMC-20) was a kind gift of Gary H. Cohen and Roselyn J Eisenberg (University of Pennsylvania)40. Phospho-EGF receptor antibody sampler kit (#9922, containing #4267, 3777, 2237), phospho-Erk1/2 pathway sampler kit (#9911, containing #9427, 9154, 4370, 11989), FAK antibody sampler kit (#9330, containing #3281, 3284, 8556, 13009), alpha-tubulin DM1A antibody (#3873), and anti-rabbit and anti-mouse IgG HRP-linked antibodies (#7074, 7076) were purchased from Cell Signaling Technology and used at 1:1000. Anti-ADAM10 (#ab124695) and anti-ADAM17 (#ab2051) were purchased from Abcam and used at 1:1000. IRDye-coupled secondary antibodies were purchased from Licor and used at 1:10'000. Alexa Fluor-conjugated secondary antibodies and phalloidin were purchased from Invitrogen/Thermo Fischer Scientific and used at 1:1000 or 1:100, respectively.
Construction of recombinant viruses
Recombinant vaccinia viruses were generated using homologous recombination as previously described41. Briefly, VACV infected BSC40 cells were transfected with linearised plasmid 4 hours post infection (hpi) and harvested 48 hpi. Plaques were selected by fluorescence through four rounds. Final plaques were sequenced to confirm the correct insertion of the construct. The VGF double knockout virus, vSC20, was a generous gift from Bernard Moss (NIAID; NIH Bethesda MD)18. WR lacZ NP-SIINFEKL-EGFP (WR NP-EGFP) and vSC20 lacZ NP-SIINFEKL-EGFP (ΔVGF NP-EGFP) virus were constructed by inserting pNP-SIINFEKL-EGFP21 into the TK locus of WR or vSC20. The WR ΔF11 NP-EGFP and ΔVGF/ΔF11 NP-EGFP viruses were generated by replacing amino acids 1-320 of the F11L locus with mCherry under the control of a late VACV promoter.
MV/EEV 24h yield
Confluent BSC40 cells in 6-well plates were infected with virus at MOI 10 and fed with 1 ml full medium. At 24 hpi the supernatant containing EEVs was collected and cleared of cells by 2x centrifugation at 400 g for 10 min. For MVs, cells were collected by scraping, centrifugation, and resuspension in 100 μl 1 mM Tris (pH 9.0), prior to 3x freeze-thaw. The plaque forming units/milliliter (pfu/ml) were determined by crystal violet staining of plaques, 48 hpi after serial dilution on confluent monolayers of BSC40 cells.
CEV/actin tail analysis
BSC40 cells were seeded on fibronectin-coated coverslips. Cells were infected with WR or mutant viruses at MOI 5. For inhibitor treatment, cells were infected with WR mCherry-A4 virus and inhibitors added at 1 hpi. At 10 hpi cells were fixed with methanol-free formaldehyde for 20 min before two PBS washes. Fixed cells were blocked with 2% BSA in PBS for 30 min before incubation with anti-B5 (VMC-20, 1:10'000) in PBS (1% BSA) for 1.5 h. After 3 PBS washes, cells were incubated with anti-mouse-Alexa647 secondary antibody (1:1,000) for 1 h. Cells were then permeabilised with 0.1% Triton in PBS for 20 min prior to staining with Phalloidin-Alexa488 (1:100) and Hoechst (1:10,000). Samples were mounted with Immu-Mount and stacks acquired using a Leica TCS SPE confocal microscope. CEVs were counted for each cell using the spot detection function of Imaris (version 7.6.5, Bitplane). Identical settings were used for all samples (0.6μm spot size, quality above 40). Actin tails were counted manually.
Plaque assays
Confluent BSC40 monolayers were infected with 100 plaque forming units (PFU). After 1 h, cells were fed with DMEM(-) without supplements. For inhibitor experiments, compounds were added at 22 hpi. For assays with GI254023X, the drug was added at 1 hpi and topped up every 12 h. Plaque diameters were measured manually using fiji. 100 plaques per condition/experiment were measured.
Live cell imaging of VACV plaques
BSC40 cells infected with VACV were imaged from 24-48 hpi in an environmental chamber maintained at 37°C and 5% CO2. Two different time-lapse microscope settings were used: A Zeiss Axiovert 200M with a Hamamatsu Orca AG camera, and a Nikon Ti inverted microscope with a Nikon DS-Qi2 high sensitivity scientific CMOS camera. A 4x objective was used for the Zeiss Axiovert, and a 10x objective was used for the Nikon Ti. For each condition, 10-15 plaques were imaged every 10 min. For plaques next to wounds, each well was scratched at 22 hpi with a 200 μl pipette tip, and the well was washed 2x with DMEM(-) before imaging as above.
Single cell tracking
To obtain the single infected cell trajectories, live cell time-lapse movies of plaque formation were analyzed using TrackMate version v3.5.1 or lower42. To detect cells the Laplacian of Gaussian (LoG) filter was applied, and estimated blob diameter and threshold were fitted for optimal detection of cells in each dataset. For tracking, the Linear Assignment Problem (LAP) Tracker was used with a maximum frame-to-frame linking of 30 μm, maximum gap-closing distance of 30 μm, and maximum gap-closing frame gap of 1.
Measurement of radial velocity and directional migration efficiency in plaques and single cell experiments
To determine the radial velocity of cells within plaques the coordinate system zero was defined at the plaque origin. Coordinates of the plaque origin were obtained using the algorithm described in43. The coordinates of each cell track were then converted from a Cartesian to a polar coordinates system. The radial component of each cell track was then used to compute an average radial velocity (RV) of the cells within a plaque using Equation 1.
| (1) |
where RV - is radial velocity, Δρ - is maximum radial component of trajectory, t – is time from experiment start.
Following the RV measurement, the directional migration efficiency (DME) of infected cells within plaques was determined using Equation 2.
| (2) |
where DME – is directional migration efficiency, ωΔρ - is the minmax normalized RV, and ωΔθ – is the maximum range of the normalized angular polar component of each track relative to the origin. Values were averaged to obtain a representative value for each plaque.
To measure radial velocity and directional migration efficiency in single cell experiments, live-cell, time-lapse phase contrast images were collected. Images were processed by pixel classification using a Random Forest44 machine learning algorithm in Weka software45 to ensure compatibility with TrackMate42. Similar to cell tracking in plaques, TrackMate with a spot size parameter of 80 pixels was used. The RV and DME of single cell tracks was computed using Equation 1 and 2. To overcome under-sampling bias in radial velocity and directional migration efficiency measurements associated with down-scaling from plaques to single cells we performed a Monte-Carlo based bootstrapping46 resampling of the experimental data with 100,000 permutations. Reciprocal hypothesis testing was performed using permutation tests.
Vector field analysis of directional cell motility
To determine the general directional tendency of motile infected cells, the spatio-temporal tensor of live-cell, time-lapse tracks of plaque formation were fitted to a vector field. For this, the Vector Field K-means clustering algorithm47 was applied to the trajectory data. To ensure background-to-signal separation, prior to application of the algorithm the cell tracking data was appended with synthetic background trajectories of constant radial velocity, distance and direction.
VGF antibody production
Anti-VGF was produced by GenScript USA Inc. The peptide DSGNAIETTSPEITC, previously used by Chang et al.14, corresponding to residues 1-14 of the cleaved VGF including an additional cysteine at the C-terminus was conjugated to KLH. The peptide-KLH conjugate was used to immunise one rabbit and anti-VGF antibody was affinity purified after three immunisations.
Expression and purification of recombinant VGF/EGF
The sequence of cleaved VGF was amplified from VACV genomic DNA and inserted into the pQE30 vector, resulting in 6xHis-VGF. The sequence of fully cleaved EGF was codon-optimised for expression in bacteria, ordered as gblock from IDT, and inserted into the pQE30 vector using Gibson cloning, resulting in 6xHis-EGF. Transformed XL1 Blue bacteria were inoculated and grown overnight with antibiotics. 500 ml of LB medium was inoculated with the cultures and grown at 30°C. At OD 0.4-0.6 gene expression was induced with 1 mM IPTG. After 4 hours cells were harvested by centrifugation at 4,000 rpm for 15 min at 4°C. Cell pellets were resuspended in 30 ml suspension buffer (500 mM NaCl, 50 mM Na2HPO4, 10 mM Imidazole, 0.1% Tween-20, pH 8.0), and sonicated on ice (15 pulses of 15 seconds). Crude extracts were filtered through a 0.22 μm filter. Protein was purified on Qiagen Ni-NTA agarose columns. Briefly, columns were washed with 5 column volumes of suspension buffer, followed by a 3ml elution with 125 mM imidazole and a 12 ml elution with 250 mM imidazole. Fractions of 1ml were collected and analysed by SDS-PAGE. The most concentrated fractions were pooled and dialysed overnight in suspension buffer without imidazole, using a membrane with MWCO 3.5 kDa. Samples were then adjusted to 25% glycerol, aliquoted, and stored at -80°C. A BCA assay was performed to determine protein concentration.
Western blot
For each sample confluent HeLa cells (10 cm dish) were maintained without serum overnight prior to infection or stimulation. To harvest, cells were transferred to ice, washed with ice cold PBS, and 150 ul lysis buffer (25 mM Tris-HCl pH 7.5, 100 mM NaCl, 1% Triton X-100, 0.5% NP40, 1 mM EDTA, 1 mM EGTA) with protease/phosphatase inhibitor (#5872, NEB) was added. Cells were harvested by scraping and lysates were incubated on ice for 20 minutes prior to centrifugation at 20'000 x g (4 °C). Samples were boiled in reducing sample buffer, run on 4-12% Bis-Tris gels and transferred to nitrocellulose. Membranes were incubated with primary antibodies (1:1000) overnight in either 5% BSA or 5% milk in TBS-T according to manufacturer’s recommendations. IRDye-coupled secondary antibodies were used for detection on a Licor Odyssey, or HRP-coupled secondary antibodies with HRP Substrate (Merck Millipore) were used for detection on a GE Healthcare ImageQuant.
EGFR PathScan activation array
Confluent HeLa cells were starved overnight prior to infection with WR or ΔVGF at MOI 5 for 6 h or treatment with EGF or VGF (2.5 μg/ml) for 1 h. Subsequently, cells were put on ice, washed with cold PBS and lysed with 200 μl lysis Buffer (#9803, Cell Signaling). Supernatants were collected by scraping, cleared by centrifugation, and the EGFR PathScan activation array (#12622, Cell Signaling) was coated with a 1:2 dilution of lysate in array buffer. The array was performed according to the manufactures protocol and a GE Healthcare ImageQuant was used for detection. For quantification, images were masked to retain only spot related intensities within the image. The total intensity per spot was quantified and compared to array controls.
Supernatant/cell transfer experiment
For supernatant/cell transfer experiments, HeLa cells were kept in serum-free media overnight, then infected in the presence or absence of indicated inhibitors. At 6 hpi supernatants were removed and cleared of cells by centrifugation (2x 300 x g). Cells were washed, gently scraped in 1 ml medium, and separated by gentle pipetting. Cells and supernatant were then diluted in fresh serum-free medium as indicated and added to starved HeLa cells for 1 h. For cell transfer, GI254023X was added during activation to prevent further cleavage of VGF. Cells were harvested for western blotting as described above. Input supernatant was concentrated using a 10 kDa cutoff Amicon centrifugal filter before analysis. Input supernatant and cell samples were adjusted to equal volumes and immunoblot analyses were performed.
siRNA silencing of ADAM10/ADAM17
HeLa cells were reversed transfected with scrambled, ADAM10 or ADAM17 siRNA at a final concentration of 20 nM. 72 h post transfection cells were infected with WR at MOI 2 for 4 h and supernatant transfer experiments performed as described above. ON-TARGETplus SMARTpool siRNA for human ADAM10 (L-004503-00-0005) and ADAM17 (L-003453-00-0005) were purchased from Dharmacon. The sequences for ADAM10 siRNA were CAUCUGACCCUAAACCAAA, CAAGGGAAGGAAUAUGUAA, GAACUAUGGGUCUCAUGUA, CGAGAGAGUUAUCAAAUGG. The sequences for ADAM17 siRNA were GAAGAACACGUGUAAAUUA, GCACAAAGAAUUAUGGUAA, UAUGGGAACUCUUGGAUUA, GGAAAUAUGUCAUGUAUCC. AllStars negative control siRNA was purchased from Qiagen.
VGF Immunofluorescence
WR and ΔVGF plaques were fixed at 36 h with 4% Formaldehyde (FA) and stained with anti-VGF (1:500), anti-rabbit-Alexa594 secondary antibody (1:1000) and Hoechst (1:10,000). Samples were imaged with a 10x objective on the Nikon TI microscope.
Transwell migration assay
BSC40 and HeLa cells were seeded in the top well of 6.5 mm transwells with 8 μm pore size polyester membrane inserts (Corning). The bottom well was filled with 650 μl serum-free DMEM and cells incubated overnight. Cells in the top chamber were infected with ΔVGF NP-EGFP (MOI 1) and inserts were transferred into wells containing 650 μl of serum-free DMEM, 5% FBS, 2 μg/ml VGF, uninfected cells or WR infected cells (MOI 1). After 20 h, the top side of transwell inserts were swabbed with a cotton bud to remove remaining cells. Wells were then fixed with 4% FA and stained with Hoechst to visualize cells which had migrated through the transwell membrane. The membrane was excised with a scalpel and mounted in Vectashield. Membranes were imaged using the 10x objective of the Nikon Ti inverted microscope. Cells were counted using the spot detection function of the fiji plugin TrackMate.
Single cell infection, sorting and imaging
HeLa cells were left uninfected or were infected with WR LacZ NP SIINFEKL (MOI=1). At 1 hpi cells were washed with PBS and detached from plates with 200 μl of 0.25% trypsin per well. For sorting, 400 μl of medium (5% FCS) was added and individual cells sorted into single wells of a collagen coated 384-well plate using the scatter of a 488 nm laser of a BD FACSAriaTM III (Becton Dickinson). Individual wells were imaged at 37°C and 5.0% CO2 on an inverted microscope (TI-E Eclipse, Nikon) using a 10x DIC, Plan Apo objective and a 0.7x TV Adapter in front of an Orca Flash 4.0 Camera (Hamamatsu). The field of view encompassed one well and images were acquired every 10 min for 27 h.
Inoculation of mice
Pathogen-free C57BL/6 and albino B6(Cg)-Tyrc-2J/J (used for MP imaging) were acquired from Taconic or from The Jackson Laboratory. 6–12 week old adult female mice were used in all experiments. Mice were housed in pathogen–free conditions (including MNV, MPV, and MHV) and maintained on standard rodent chow and water supplied ad libitum. All animal experiments were conducted in accordance with the Animal Welfare Act and the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. NIAID animal facilities have full accreditation from the Association for Assessment and Accreditation of Laboratory Animal Care and are PHS-assured (Assurance Number: # A4149-01). All animal procedures were approved by the NIAID Animal Care and Use Committee. For epicutaneous infection, approximately 20 μl of rVV stock solution (1 x 108 pfu/ml) was placed on the ear skin and gently poked 5 times per ear with a bifurcated needle (similar to the human vaccination protocol). Virus stocks were grown and titered in house.
Intravital multiphoton microscopy (MPM) imaging
MPM imaging was performed as described48. Briefly, images were acquired on an upright Leica SP5 MP microscope (Leica Microsystems) equipped with two Mai Tai Ti:Sapphire lasers (Spectra Physics) and 10-Watt pumps or on an inverted Leica TCS SP8 MP microscope (Leica Microsystems). Ears were immobilized on an imaging platform and bathed in warm saline. All images were acquired using a 20x objective (NA 1.00). Emitted fluorescence was collected with a four-channel non-descanned detector. For blue/green channels, wavelength separation was accomplished with a dichroic mirror at 495 nm followed by emission filters of 460/50 nm bandpass and 525/50 nm bandpass. Second harmonic generation of collagen in the dermis is shown in blue according to convention. Tile-scans of 8 fields of view were collected covering approximately 2.8 mm of the ear. Sequences of image stacks were transformed into maximum intensity projections using Imaris software (Bitplane).
Confocal imaging of frozen sections
Ears were fixed in periodate-lysine-paraformaldehyde overnight as reported 49, cryoprotected in 15% sucrose, embedded in OCT medium (Electron Microscopy Sciences) and frozen in dry-ice cooled isopentane. Eighteen-micron cross-sections were cut on a Leica cryostat (Leica Microsystems), blocked with 5% goat or donkey serum, then stained with a combination of DAPI (Perkin Elmer), CD11b-Alexa 647 (clone M1/70, eBioscience), and VGF peptide antibody followed by anti-rabbit Alexa 568 (Invitrogen). Images were acquired on an inverted SP5 confocal microscope (Leica Microsystems) using identical PMT (photomultiplier tube) and laser settings. Lesion depth was scored manually using Imaris software.
Supplementary Material
Supplementary Information is available in the online version of the paper.
Acknowledgements
We thank Krzysztof Gonciarz and Ivo F. Sbalzarini for helpful discussions. We thank Beat Siegenthaler and Joshua Crouse for initial work on the project. We thank all members of the Mercer lab for helpful discussions and comments on the manuscript. We thank A. Vaughan, V. Jäggin, T. Lopes, T. Lummen, T. Horn, and D. Loeffler for technical support. We thank Dr. Bernard Moss (NIAID) for vSC20 virus. This research was supported by core funding to the MRC Laboratory for Molecular Cell Biology, University College London (MC_UU12018/7) (J.M.), the European Research Council (649101—UbiProPox) (J.M.), ETH Zurich (G.F. and D.J.M), and the Division of Intramural Research of the National Institute of Allergy and Infectious Diseases (G.V.R. and H.D.H.). C.B. is funded by the Medical Research Council LMCB PhD programme.
Footnotes
Data availability
All data and reagents will be made available upon reasonable request.
Code availability
All custom code used in this study will be made available upon reasonable request.
Conflict of Interest Statement
All authors declare no conflict of interest.
Author Contributions C.B, S.K., and J.M. conceived the project. C.B., A.Y. S.K. and J.M. designed the experiments. G.V.R and H.D.H. designed, performed and analyzed the intra-vital imaging. G.F., and D.J.M. designed and performed the cell sorting and single cell imaging. C.B., A.Y., and J.M. analysed the data. C.B., A.Y., H.D.H, G.F., and J.M. wrote the manuscript. All authors discussed the experiments, read and approved the manuscript.
References
- 1.Sanderson CM, Way M, Smith GL. Virus-induced cell motility. Journal of virology. 1998;72:1235–1243. doi: 10.1128/jvi.72.2.1235-1243.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Valderrama F, Cordeiro JV, Schleich S, Frischknecht F, Way M. Vaccinia virus-induced cell motility requires F11L-mediated inhibition of RhoA signaling. Science. 2006;311:377–381. doi: 10.1126/science.1122411. [DOI] [PubMed] [Google Scholar]
- 3.Ridley AJ. Rho GTPase signalling in cell migration. Curr Opin Cell Biol. 2015;36:103–112. doi: 10.1016/j.ceb.2015.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Friedl P, Wolf K. Tumour-cell invasion and migration: diversity and escape mechanisms. Nat Rev Cancer. 2003;3:362–374. doi: 10.1038/nrc1075. [DOI] [PubMed] [Google Scholar]
- 5.Reig G, Pulgar E, Concha ML. Cell migration: from tissue culture to embryos. Development. 2014;141:1999–2013. doi: 10.1242/dev.101451. [DOI] [PubMed] [Google Scholar]
- 6.Cheng F, et al. KSHV-initiated notch activation leads to membrane-type-1 matrix metalloproteinase-dependent lymphatic endothelial-to-mesenchymal transition. Cell host & microbe. 2011;10:577–590. doi: 10.1016/j.chom.2011.10.011. [DOI] [PubMed] [Google Scholar]
- 7.Abaitua F, Zia FR, Hollinshead M, O'Hare P. Polarized cell migration during cell-to-cell transmission of herpes simplex virus in human skin keratinocytes. Journal of virology. 2013;87:7921–7932. doi: 10.1128/JVI.01172-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dawson CW, Laverick L, Morris MA, Tramoutanis G, Young LS. Epstein-Barr virus-encoded LMP1 regulates epithelial cell motility and invasion via the ERK-MAPK pathway. Journal of virology. 2008;82:3654–3664. doi: 10.1128/JVI.01888-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kivi N, Greco D, Auvinen P, Auvinen E. Genes involved in cell adhesion, cell motility and mitogenic signaling are altered due to HPV 16 E5 protein expression. Oncogene. 2008;27:2532–2541. doi: 10.1038/sj.onc.1210916. [DOI] [PubMed] [Google Scholar]
- 10.Irwin CR, Evans DH. Modulation of the myxoma virus plaque phenotype by vaccinia virus protein F11. Journal of virology. 2012;86:7167–7179. doi: 10.1128/JVI.06936-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cordeiro JV, et al. F11-mediated inhibition of RhoA signalling enhances the spread of vaccinia virus in vitro and in vivo in an intranasal mouse model of infection. PloS one. 2009;4:e8506. doi: 10.1371/journal.pone.0008506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Handa Y, Durkin CH, Dodding MP, Way M. Vaccinia virus F11 promotes viral spread by acting as a PDZ-containing scaffolding protein to bind myosin-9A and inhibit RhoA signaling. Cell host & microbe. 2013;14:51–62. doi: 10.1016/j.chom.2013.06.006. [DOI] [PubMed] [Google Scholar]
- 13.McFadden G, Moyer R. Poxvirus Growth Factors Related to Epidermal Growth Factor. Academic Press; 2000. [Google Scholar]
- 14.Chang W, Lim JG, Hellstrom I, Gentry LE. Characterization of vaccinia virus growth factor biosynthetic pathway with an antipeptide antiserum. Journal of virology. 1988;62:1080–1083. doi: 10.1128/jvi.62.3.1080-1083.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.King CS, Cooper JA, Moss B, Twardzik DR. Vaccinia virus growth factor stimulates tyrosine protein kinase activity of A431 cell epidermal growth factor receptors. Molecular and cellular biology. 1986;6:332–336. doi: 10.1128/mcb.6.1.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Twardzik DR, Brown JP, Ranchalis JE, Todaro GJ, Moss B. Vaccinia virus-infected cells release a novel polypeptide functionally related to transforming and epidermal growth factors. Proceedings of the National Academy of Sciences of the United States of America. 1985;82:5300–5304. doi: 10.1073/pnas.82.16.5300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stroobant P, et al. Purification and characterization of vaccinia virus growth factor. Cell. 1985;42:383–393. doi: 10.1016/s0092-8674(85)80133-1. [DOI] [PubMed] [Google Scholar]
- 18.Buller RM, Chakrabarti S, Cooper JA, Twardzik DR, Moss B. Deletion of the vaccinia virus growth factor gene reduces virus virulence. Journal of virology. 1988;62:866–874. doi: 10.1128/jvi.62.3.866-874.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lai AC, Pogo BG. Attenuated deletion mutants of vaccinia virus lacking the vaccinia growth factor are defective in replication in vivo. Microbial pathogenesis. 1989;6:219–226. doi: 10.1016/0882-4010(89)90071-5. [DOI] [PubMed] [Google Scholar]
- 20.Buller RM, Chakrabarti S, Moss B, Fredrickson T. Cell proliferative response to vaccinia virus is mediated by VGF. Virology. 1988;164:182–192. doi: 10.1016/0042-6822(88)90635-6. [DOI] [PubMed] [Google Scholar]
- 21.Hickman HD, et al. Direct priming of antiviral CD8+ T cells in the peripheral interfollicular region of lymph nodes. Nat Immunol. 2008;9:155–165. doi: 10.1038/ni1557. [DOI] [PubMed] [Google Scholar]
- 22.Bidgood SR, Mercer J. Cloak and Dagger: Alternative Immune Evasion and Modulation Strategies of Poxviruses. Viruses. 2015;7:4800–4825. doi: 10.3390/v7082844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Postigo A, Martin MC, Dodding MP, Way M. Vaccinia-induced epidermal growth factor receptor-MEK signalling and the anti-apoptotic protein F1L synergize to suppress cell death during infection. Cellular microbiology. 2009;11:1208–1218. doi: 10.1111/j.1462-5822.2009.01327.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Andrade AA, et al. The vaccinia virus-stimulated mitogen-activated protein kinase (MAPK) pathway is required for virus multiplication. The Biochemical journal. 2004;381:437–446. doi: 10.1042/BJ20031375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tzahar E, et al. Pathogenic poxviruses reveal viral strategies to exploit the ErbB signaling network. The EMBO journal. 1998;17:5948–5963. doi: 10.1093/emboj/17.20.5948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim HS, et al. Tyrosine phosphorylation of phospholipase C-gamma 1 by vaccinia virus growth factor. Virology. 1995;214:21–28. doi: 10.1006/viro.1995.9958. [DOI] [PubMed] [Google Scholar]
- 27.Bonjardim CA. Viral exploitation of the MEK/ERK pathway - A tale of vaccinia virus and other viruses. Virology. 2017;507:267–275. doi: 10.1016/j.virol.2016.12.011. [DOI] [PubMed] [Google Scholar]
- 28.Huang C, Jacobson K, Schaller MD. MAP kinases and cell migration. Journal of cell science. 2004;117:4619–4628. doi: 10.1242/jcs.01481. [DOI] [PubMed] [Google Scholar]
- 29.Langhammer S, Koban R, Yue C, Ellerbrok H. Inhibition of poxvirus spreading by the anti-tumor drug Gefitinib (Iressa) Antiviral research. 2011;89:64–70. doi: 10.1016/j.antiviral.2010.11.006. [DOI] [PubMed] [Google Scholar]
- 30.Schweneker M, et al. The vaccinia virus O1 protein is required for sustained activation of extracellular signal-regulated kinase 1/2 and promotes viral virulence. Journal of virology. 2012;86:2323–2336. doi: 10.1128/JVI.06166-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Singh AB, Harris RC. Autocrine, paracrine and juxtacrine signaling by EGFR ligands. Cellular signalling. 2005;17:1183–1193. doi: 10.1016/j.cellsig.2005.03.026. [DOI] [PubMed] [Google Scholar]
- 32.Blobel CP. ADAMs: key components in EGFR signalling and development. Nature reviews. Molecular cell biology. 2005;6:32–43. doi: 10.1038/nrm1548. [DOI] [PubMed] [Google Scholar]
- 33.Ludwig A, et al. Metalloproteinase inhibitors for the disintegrin-like metalloproteinases ADAM10 and ADAM17 that differentially block constitutive and phorbol ester-inducible shedding of cell surface molecules. Comb Chem High Throughput Screen. 2005;8:161–171. doi: 10.2174/1386207053258488. [DOI] [PubMed] [Google Scholar]
- 34.Cudmore S, Cossart P, Griffiths G, Way M. Actin-based motility of vaccinia virus. Nature. 1995;378:636–638. doi: 10.1038/378636a0. [DOI] [PubMed] [Google Scholar]
- 35.Doceul V, Hollinshead M, van der Linden L, Smith GL. Repulsion of superinfecting virions: a mechanism for rapid virus spread. Science. 2010;327:873–876. doi: 10.1126/science.1183173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yakimovich A, et al. Infectio: a Generic Framework for Computational Simulation of Virus Transmission between Cells. mSphere. 2016;1 doi: 10.1128/mSphere.00078-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang M, Huang CZ. Mitogen-activated protein kinase signaling pathway and invasion and metastasis of gastric cancer. World J Gastroenterol. 2015;21:11673–11679. doi: 10.3748/wjg.v21.i41.11673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Payne LG. Significance of extracellular enveloped virus in the in vitro and in vivo dissemination of vaccinia. The Journal of general virology. 1980;50:89–100. doi: 10.1099/0022-1317-50-1-89. [DOI] [PubMed] [Google Scholar]
- 39.Yang H, et al. Antiviral chemotherapy facilitates control of poxvirus infections through inhibition of cellular signal transduction. J Clin Invest. 2005;115:379–387. doi: 10.1172/JCI23220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Aldaz-Carroll L, et al. Epitope-mapping studies define two major neutralization sites on the vaccinia virus extracellular enveloped virus glycoprotein B5R. Journal of virology. 2005;79:6260–6271. doi: 10.1128/JVI.79.10.6260-6271.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mercer J, Helenius A. Vaccinia virus uses macropinocytosis and apoptotic mimicry to enter host cells. Science. 2008;320:531–535. doi: 10.1126/science.1155164. [DOI] [PubMed] [Google Scholar]
- 42.Tinevez JY, et al. TrackMate: An open and extensible platform for single-particle tracking. Methods. 2017;115:80–90. doi: 10.1016/j.ymeth.2016.09.016. [DOI] [PubMed] [Google Scholar]
- 43.Yakimovich A, et al. Plaque2.0-A High-Throughput Analysis Framework to Score Virus-Cell Transmission and Clonal Cell Expansion. PloS one. 2015;10:e0138760. doi: 10.1371/journal.pone.0138760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Breiman L. Random Forests. Machine Learning. 2001;45:5–32. [Google Scholar]
- 45.Arganda-Carreras I, et al. Trainable Weka Segmentation: a machine learning tool for microscopy pixel classification. Bioinformatics. 2017;33:2424–2426. doi: 10.1093/bioinformatics/btx180. [DOI] [PubMed] [Google Scholar]
- 46.Efron B. Bootstrap Methods: Another Look at the Jackknife. The Annals of Statistics. 1979;7:1–26. [Google Scholar]
- 47.Ferreira N, Klosowski JT, Scheidegger CE, Silva CT. Vector Field k-Means: Clustering Trajectories by Fitting Multiple Vector Fields. Computer Graphics Forum. 2013;32:201–210. [Google Scholar]
- 48.Cush SS, et al. Locally Produced IL-10 Limits Cutaneous Vaccinia Virus Spread. PLoS pathogens. 2016;12:e1005493. doi: 10.1371/journal.ppat.1005493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pieri L, Sassoli C, Romagnoli P, Domenici L. Use of periodate-lysine-paraformaldehyde for the fixation of multiple antigens in human skin biopsies. Eur J Histochem. 2002;46:365–375. doi: 10.4081/1749. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.