Abstract
Purpose
To investigate retinal structure in subjects with CNGA3-associated achromatopsia and evaluate disease symmetry and intrafamilial variability.
Methods
Thirty-eight molecularly confirmed subjects underwent ocular examination, optical coherence tomography (OCT), and nonconfocal split-detection adaptive optics scanning light ophthalmoscopy (AOSLO). OCT scans were used for evaluating foveal hypoplasia, grading foveal ellipsoid zone (EZ) disruption, and measuring outer nuclear layer (ONL) thickness. AOSLO images were used to quantify peak foveal cone density, intercell distance (ICD), and the coefficient of variation (CV) of ICD.
Results
Mean (±SD) age was 25.9 (±13.1) years. Mean (± SD) best corrected visual acuity (BCVA) was 0.87 (±0.14) logarithm of the minimum angle of resolution. Examination with OCT showed variable disruption or loss of the EZ. Seven subjects were evaluated for disease symmetry, with peak foveal cone density, ICD, CV, ONL thickness, and BCVA not differing significantly between eyes. A cross-sectional evaluation of AOSLO imaging showed a mean (±SD) peak foveal cone density of 19,844 (±13,046) cones/mm2. There was a weak negative association between age and peak foveal cone density (r = −0.397, P = 0.102), as well as between EZ grade and age (P = 0.086).
Conclusions
The remnant cone mosaics were irregular and variably disrupted, with significantly lower peak foveal cone density than unaffected individuals. Variability was also seen among subjects with identical mutations. Therefore, subjects should be considered on an individual basis for stratification in clinical trials. Interocular symmetry suggests that both eyes have comparable therapeutic potential and the fellow eye can serve as a valid control. Longitudinal studies are needed, to further examine the weak negative association between age and foveal cone structure observed here.
Keywords: adaptive optics, retinal phenotyping, retinal diseases, inherited retinal diseases, genetics, CNGA3, cone, cone dysfunction, achromatopsia
Achromatopsia (ACHM) presents from birth or early infancy, with poor visual acuity, pendular nystagmus, photophobia, and color vision loss in all three axes.1,2 Absent cone electroretinogram responses is the hallmark of disease, with normal rod function or uncommonly mildly reduced rod responses.3–5 ACHM is an autosomal recessive disorder with a heterogeneous genetic background. Disease-causing sequence variants have been reported in CNGA3 (ACHM2, Online Mendelian Inheritance in Man [OMIM] 600053),6,7 CNGB3 (ACHM3, OMIM 605080),8 GNAT2 (ACHM4, OMIM 139340),9,10 ATF6 (ACHM7, OMIM 616517),11 PDE6H (ACHM6, OMIM I610024),12 and PDE6C (ACHM5, OMIM 600827).1
CNGA3 encodes the α subunit of the cGMP-gated cation channel,13 located in cone outer segments, categorizing CNGA3-associated ACHM as a “channelopathy.”14 It has been shown in vitro that α subunits can create functional channels in the absence of β subunits, a property not shared by the β subunits.15 CNGA3 variants have a far greater allelic heterogeneity than CNGB3.16,17 In a large cross-sectional study of CNGA3-ACHM (n = 36), Zobor et al.18 reported a lack of robust association between age and morphologic (optical coherence tomography [OCT]) and/or functional (microperimetry) changes, in agreement with a previous cross-sectional study of ACHM by Sundaram et al.19 (n = 40, with 18 having CNGA3-ACHM). Aboshiha et al.,20 in a prospective longitudinal study (n = 40, with 18 having CNGA3-ACHM) using OCT and fundus autofluorescence imaging, identified a slow and subtle progression of disease in a minority of subjects. These studies were limited by the relatively low resolution of the imaging tools used.
Adaptive optics (AO) has been used for in-depth phenotyping in inherited retinal diseases for two decades,21 by allowing the noninvasive visualization of the photoreceptor mosaic22–24 through the correction of the eye's monochromatic aberrations.24,25 Confocal detection of reflected light has been shown to reflect the waveguiding ability of intact photoreceptors.23,26,27 Early investigations in ACHM with flood illumination-AO and confocal adaptive optics scanning light ophthalmoscopy (AOSLO) identified dark spaces in the cone mosaic, increased cone spacing, and/or decreased cone density (Fig. 1A).28–31 Marked variability in the cone mosaic has been observed across individuals; while no obvious difference has been reported between the two most common genotypes (CNGA3 and CNGB3), the rarer GNAT2 genotype is associated with a significantly less-disrupted photoreceptor mosaic.30,32 Reduced reflectivity (i.e., altered waveguiding) in the majority of remnant cones in CNGA3 and CNGB3 has also been noted, with relative preservation in GNAT2.30 Until the advent of nonconfocal split-detection AOSLO, it was unknown if these dark spaces harbored nonwaveguiding cones or indicated the loss of cones, an important question for defining the presence of a therapeutic target. Simultaneous confocal and split-detection AOSLO has allowed for the identification of cone inner segment structure in these spaces (Fig. 1B)33–36 and transformed our understanding of ACHM and participant selection for planned and ongoing CNGA3- and CNGB3-ACHM gene therapy trials. In confocal AOSLO imaging outside the foveal center, due to waveguiding rods surrounding the nonreflective dark cones, the cones can be identified (Fig. 1A). This is in contrast to the fovea, where the lack of surrounding rods makes the identification and quantification of cones impossible (Fig. 1C) and split-detection AOSLO is necessary (Fig. 1D). Given the potential disconnect between OCT and AOSLO measures of cone integrity37 and the ability of AOSLO to directly visualize the target cones for gene replacement, nonconfocal split-detection AOSLO imaging is the modality of choice to identify patients most likely to benefit from cone-directed rescue.
Figure 1.
Adaptive optics scanning light ophthalmoscopy imaging in CNGA3-associated ACHM. At 2° eccentricity: (A) confocal image, with red dots marking the dark cones. Cones are surrounded by waveguiding rods. (B) Split-detection image over the exact same region with overlying red dots showing the cones marked in panel A, which colocalize with cone inner segments, surrounded by rods. Foveal center: (C) confocal image at the foveal center, where the dark cones are not visible because no rods are present to delineate the dark spaces. (D) Split-detection image over the exact same region, with visible and quantifiable cone inner segments. All images are from the same retinal montage. Scale bar: 20 μm.
The current literature for CNGA3-ACHM and split-detection AOSLO imaging is limited, with most studies focusing on the more common CNGB3 phenotype and/or only using confocal AOSLO. The purpose of this study was to use OCT and split-detection AOSLO to explore the structural phenotype of patients with molecularly-confirmed CNGA3-ACHM, and lay the groundwork for a CNGA3 longitudinal AOSLO study to define the cellular natural history of this disorder, in light of the ongoing and upcoming gene therapy trials. In addition, we examined disease symmetry at the cellular level and investigated intrafamilial variability. We compared our observations with published CNGB3-ACHM data obtained using similar methodology.36
Materials and Methods
The study was approved by the Ethics Committees of Moorfields Eye Hospital and the Medical College of Wisconsin. Written informed consent was obtained from all subjects after explanation of the nature and possible consequences of the study. The research followed the tenets of the Declaration of Helsinki.
Participants
Thirty-eight ACHM subjects with likely disease-causing sequence variants in CNGA3 were recruited from Moorfields Eye Hospital, London, UK, and the Medical College of Wisconsin, Milwaukee, USA.
Ocular Examination
A full clinical history assessment and ocular examination was carried out in 32 of the subjects, including best corrected visual acuity (BCVA) using the Early Treatment Diabetic Retinopathy Study chart. Axial length was obtained with the Zeiss IOLMaster (Carl Zeiss Meditec, Jena, Germany) in order to scale the AOSLO retinal images and the OCT scans for all subjects.
Optical Coherence Tomography (OCT)
All subjects were imaged using the Bioptigen spectral domain OCT (Leica Microsystems, Research Triangle Park, NC, USA) bilaterally. Pupil dilation and cycloplegia were achieved by instilling one drop of phenylephrine hydrochloride (2.5%) and tropicamide (1%) in each eye prior to imaging (OCT/AOSLO). The scanning window covered a nominal 7 × 7-mm retinal area and vertical and horizontal scans centered at the fovea with a nominal 7-mm scan length (120 B scans) were recorded. In order to increase the signal-to-noise ratio, the maximum number (n > 5) of line scans including the fovea were aligned, registered, and averaged using ImageJ and the StackReg plugin (National Institutes of Health).38
The outer nuclear layer (ONL) thickness was calculated at the fovea for all subjects based on the longitudinal reflectivity profile (LRP), as described by Huang et al.39 The LRP was 5 pixels wide and positioned at the center of the foveal depression, and the foveal ONL thickness was calculated as the distance between the hyperreflective peaks of the posterior outer plexiform layer and external limiting membrane for subjects with foveal hypoplasia. In subjects with normal foveal excavation, the ONL thickness was measured as the distance between the hyperreflective peaks corresponding to the internal limiting membrane and the external limiting membrane. A similar approach has been used in several ACHM studies.19,36,40
The transfoveal line scans were then categorized into five grades according to the previously reported grading system by Sundaram et al.,19 based upon the integrity of the ellipsoid zone (EZ): (1) continuous EZ, (2) EZ disruption, (3) EZ absence, (4) presence of a hyporeflective zone (HRZ), and (5) outer retinal atrophy including retinal pigment epithelium (RPE) loss (Fig. 2). This grading allows meaningful comparisons with other recently published ACHM cohorts.18,36 The nomenclature used follows the International Nomenclature for Optical Coherence Tomography Consensus.41 Confirmation of the aforementioned grading was established by examining additional OCT scans acquired using the Spectralis spectral domain OCT system (Heidelberg Engineering Inc., Heidelberg, Germany). The grading was done by two observers independently (MG, CSL) and any discrepancies were resolved with consensus from a third grader (NH). The scans were also examined for the presence of foveal hypoplasia, defined as the presence of one or more inner retinal layers at the foveal center.19 Both eyes were imaged for each subject, though one eye was chosen randomly for quantitative analysis (except for EZ grade and hypoplasia, which were assessed bilaterally).
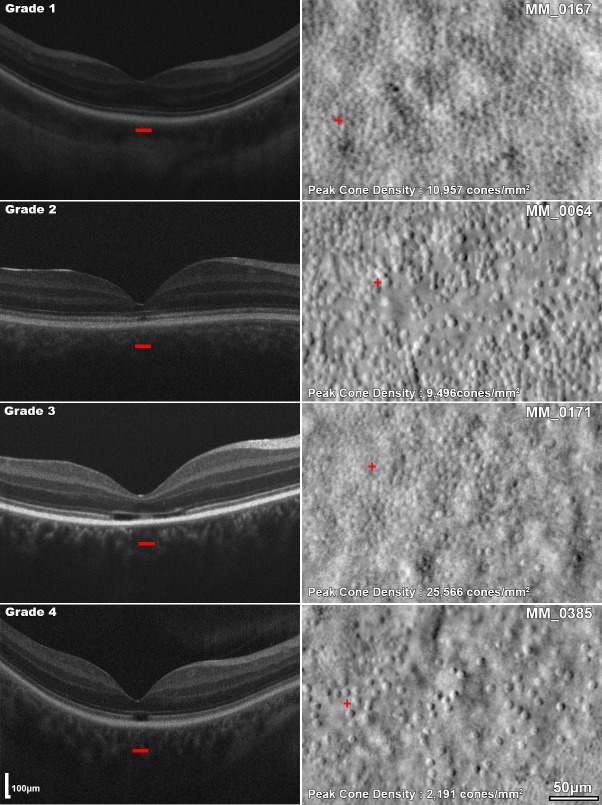
Figure 2.
Remnant cone structure in different OCT EZ grades of CNGA3-associated ACHM. Left column: horizontal transfoveal OCT scans of four subjects with different OCT EZ grades. Grade 1: Continuous EZ, grade 2: EZ disruption, grade 3: EZ absence, and grade 4: presence of a hyporeflective zone. The red horizontal bars mark the 300-μm area represented on the AOSLO images in the right column. Right column: split-detection AOSLO images for each subject. The red cross represents the location of peak foveal cone density.
AOSLO Imaging of the Photoreceptor Mosaic
All subjects were imaged using one of two identical AOSLO systems, as previously described,42 at Moorfields Eye Hospital, London, United Kingdom, and the Medical College of Wisconsin, Milwaukee, United States. Imaging with confocal and split-detector modes was performed, with simultaneous acquisition, in absolute spatial and temporal registration. Split-detection AOSLO allows for the visualization of cone inner segments in ACHM based on a differential phase technique.33
The imaging light source was a 790-nm super-luminescent diode (SLD; Superlum, Carrigtohill, Cork, Ireland). Image sequences were recorded as AVI files, of 150 frames, either at 1° and/or 1.5° square field of view (FOV). A desinusoiding algorithm was applied to each image sequence and individual frames were selected,43 registered,44 and averaged to increase the signal-to-noise ratio for subsequent analysis. Images were obtained over the fovea. The final images were combined into a single montage (Adobe Photoshop, Adobe Systems Inc., San Jose, CA, USA), with images from each modality overlaid on one another, manually and/or using a custom-built automated software.45 Images taken with a 1° FOV were analyzed preferentially, due to the higher resolution.
The scale in degrees per pixel of each FOV was determined using a Ronchi ruling of known spacing after each imaging session. Then, the value was linearly scaled using the subject's axial length and the final micron per pixel scale was obtained by multiplying by the retinal magnification factor of 291 μm/degree. The AOSLO montages were aligned to color fundus images and OCT images using common blood vessel patterns (Adobe Photoshop). The position of the anatomic foveal center was estimated on the AOSLO images using the aligned OCT images. The rod-free area surrounding the foveal center was used for further AOSLO analysis.
The eligibility criteria for AOSLO analysis were strict; low-quality images (due to involuntary eye motion/nystagmus or low signal-to-noise ratio), noncontiguous montages, and montages not covering the entire rod-free area were excluded from further analysis.
In some subjects (n = 7), bilateral AOSLO images were available for evaluating disease symmetry. The low number of subjects with adequate bilateral data was either due to time constraints or incomplete imaging of the fovea, in the second eye. For each subject, the eye with the better image quality (least eye motion, higher signal-to-noise ratio), was selected for cross-sectional evaluation of AOSLO data.
Cone coordinates from every given foveal image were extracted following manual annotation by a single grader (MG). The coordinate arrays were used to assess the peak foveal cone density (cones/mm2). A 55 × 55-μm sampling window was used to determine the density at each pixel of the coordinate array. The pixel location with the greatest value was the location of peak foveal cone density, by dividing the total number of cones by the area. In addition, the mean intercell distance (ICD, defined as the average distance of a cone from all its immediate neighbors) and its coefficient of variation (CV, defined as the standard deviation of ICD divided by the mean of ICD) were calculated for the rod-free area for all subjects with analyzable split-detection AOSLO, as metrics of remnant foveal cone packing.
Statistical Methods
Statistical analysis was performed with GraphPad Prism (version 8.0.1; GraphPad Software, San Diego, CA, USA). Parametric and nonparametric tests were used depending on normality tests (Shapiro-Wilk normality test), as well as correlation coefficients (either Pearson or Spearman, respectively).
Results
Demographics
Thirty-eight subjects from 30 families were recruited, including 21 males (55%) and 17 females (45%), with a mean (range, ±SD) age, in years, of 25.9 (10–64, ±13.1). Ten subjects were children (under 16 years old), with a mean (range, ±SD) age, in years, of 12.1 (10–14, ±1.7). Demographics are summarized in Table 1.
Table 1.
Demographics and Genotype Summary
|
Subject |
Sex |
Age, y |
Variant 1 |
Variant 2 |
||
|
cDNA Change |
AA Change |
cDNA Change |
AA Change |
|||
| MM_0002* | F | 41 | 1443-1444insC | Ile482His fs*6 | 1706G>A | Arg569His |
| MM_0008* | F | 11 | 536T>A | Val179Asp | ND† | ND† |
| MM_0009* | F | 14 | 536T>A | Val179Asp | ND† | ND† |
| MM_0014* | F | 35 | 848G>A | Arg283Gln | 667C>T | Arg223Trp |
| MM_0015* | F | 28 | 848G>A | Arg283Gln | 667C>T | Arg223Trp |
| MM_0016* | M | 30 | 848G>A | Arg283Gln | 667C>T | Arg223Trp |
| MM_0064* | F | 23 | 1694C>T | Thr565Met | 661C>T | Arg221Ter |
| MM_0164* | F | 35 | 1641C>A | Phe547Leu | 1641C>A | Phe547Leu |
| MM_0165* | M | 13 | 1641C>A | Phe547Leu | 1641C>A | Phe547Leu |
| MM_0167* | F | 28 | 847C>T | Arg283Trp | 1279C>T | Arg427Cys |
| MM_0168* | M | 39 | 661C>T | Arg221Ter | 848G>A | Arg283Gln |
| MM_0169* | M | 14 | 485A>T | Asp162Val | 485A>T | Asp162Val |
| MM_0170* | M | 16 | 1642G>A | Gly548Arg | 67C>T | Arg23Ter |
| MM_0171* | M | 22 | 1001C>T | Ser334Phe | 1360A>T | Lys454Ter |
| MM_0239* | M | 37 | 848G>A | Arg283Gln | 667C>T | Arg223Trp |
| MM_0251* | F | 33 | 661C>T | Arg221Ter | 661C>T | Arg221Ter |
| MM_0358 | M | 10 | 1641C>A | Phe547Leu | 1641C>A | Phe547Leu |
| MM_0385 | M | 37 | 1228C>G | Arg410Trp | 1228C>G | Arg410Trp |
| MM_0386 | F | 22 | 1580T>G | Leu527Arg | 1805G>A | Gly602Glu |
| MM_0387 | M | 18 | 1580T>G | Leu527Arg | 1805G>A | Gly602Glu |
| MM_0396 | M | 27 | 1306C>T | Arg436Trp | 1279C>T | Arg427Cys |
| MM_0397 | M | 18 | 1306C>T | Arg436Trp | 1279C>T | Arg427Cys |
| MM_0398 | F | 14 | 811C>T | Pro271Ser | 829C>T | Arg277Cys |
| MM_0412 | M | 13 | 1286T>C | Val429Ala | 608G>A | Trp203Cys fs*29 |
| MM_0418 | M | 18 | 661C>T | Arg221Ter | 661C>T | Arg221Ter |
| MM_0445 | M | 10 | 1641C>A | Phe547Leu | 1641C>A | Phe547Leu |
| MM_0446* | F | 50 | 67C>T | Arg23Ter | 67C>T | Arg23Ter |
| JC_0578 | M | 32 | 1687C>T | Arg563Cys | 778G>A | Asp260Asn |
| JC_0615 | M | 43 | 1306C>T | Arg436Trp | 1306C>T | Arg436Trp |
| JC_0626 | F | 20 | 1641C>A | Phe547Leu | 1641C>A | Phe547Leu |
| JC_0674 | M | 46 | 985G>T | Gly329Cys | 985G>T | Gly329Cys |
| JC_0698 | M | 43 | 848G>A | Arg283Gln | 1228C>T | Arg410Trp |
| JC_1240* | F | 24 | 1351dupG | Val451Gly fs*3 | 1279C>T | Arg427Cys |
| JC_10008 | F | 12 | 829C>T | Arg277Cys | 1641C>A | Phe547Leu |
| JC_10069* | M | 18 | 847C>T | Arg283Trp | 542A>G | Tyr181Cys |
| KS_10088* | F | 64 | 450-1G>A | splice defect | 1557G>A | Met519Ile |
| KS_10337‡ | F | 17 | 940_942delATC | Ile314del | 1114C>T | Pro372Ser |
| JC_10551 | M | 10 | 1228C>T | Arg410Trp | 1319G>C | Trp440Ser |
In bold are the previously unreported sequence variants. ND, no data; F, female; M, male; AA, amino acid.
Only one variant identified. Phenotype consistent with CNGA3-ACHM. No variants found in CNGB3.
Subject harbors an additional variant in CNGB3: c.473C>A: p.Pro158His.
Genetics
All subjects harbored likely disease-causing sequence variants in CNGA3, with 12 (32%) being homozygous and 24 (63%) compound heterozygous. The remaining two of the 38 subjects (MM_0008 and MM_0009, siblings) harbored a single heterozygous variant and were shown to have no variants in CNGB3, with a typical ACHM phenotype. In one subject (KS_10337), an additional CNGB3 sequence variant was identified. In total, 33 different variants were observed in our cohort, with 4 not previously reported to the best of our knowledge (Table 1, marked in bold).
The majority of the variants resulted in single amino acid substitutions (25/33, 76%). In addition, three different nucleotide changes introduced translation termination codons, three more introduced frame shift changes, one was an intronic change disrupting splicing, and one was a 3-base pair deletion. The most common was c.1641C>A (p.Phe547Leu; 11 alleles, three pedigrees), followed by c.848G>A (p.Arg283Gln; six alleles, three pedigrees) and c.661C>T (p.Arg221Ter; six alleles, four pedigrees). All sequence variants and protein alterations are reported in Table 1.
Ocular Examination
The mean (range, ±SD) axial length, in mm, for all 76 eyes was 24.21 (21.46–29.61, ±1.85); the mean (±SD) axial length, in mm, for the right and left eyes was 24.25 (±1.91) and 24.17 (±1.81), respectively. No significant difference was noted between eyes (P = 0.344, Wilcoxon matched-pairs test). The mean (±SD) axial length, in mm, for male and female subjects was 24.07 (±1.66) and 24.46 (±2.21), respectively. No significant difference in axial length was noted between sexes (P = 0.586, Mann-Whitney test). The age, in years, of male and female subcohorts also did not differ significantly (P = 0.508, Mann-Whitney test), with mean (±SD) 24.48 (±12.28) and 27.71 (±14.29), respectively.
BCVA was documented for 32 of 38 subjects. One subject (MM_0386) had congenital light perception in one eye, without the presence of other sequence variants or eye disease after thorough clinical investigation, had a BCVA of 0.98 logMAR in the other eye, and was excluded from further BCVA analysis. The mean (range, ±SD) BCVA was 0.87 logarithm of the minimum angle of resolution (LogMAR; 0.40–1.18, ±0.14). The mean (±SD) BCVA was 0.85 LogMAR (±0.14) and 0.88 LogMAR (±0.15) for right and left eyes, respectively; with no significant difference (P = 0.327, Wilcoxon matched-pairs test). BCVA and axial length for all subjects are shown in Table 2.
Table 2.
Retinal Phenotype–Optical Coherence Tomography and Adaptive Optics Imaging Summary
|
Subject |
Eye |
Axial Length, mm |
BCVA, LogMAR |
Foveal Hypoplasia |
EZ Grade |
ONL Thickness, μm |
PFD, Cones/mm2 |
ICD Mean, μm |
ICD SD (σ) |
ICD CV |
| MM_0002 | OD | 24.4 | 0.74 | Yes | 2 | 76.67 | 16,801 | 13.04 | 3.61 | 0.28 |
| OS | 24.38 | 0.82 | Yes | 2 | 82.56 | 13,148 | 12.85 | 3.46 | 0.27 | |
| MM_0008 | OD | 23.14 | 1.06 | No | 2 | 72.74 | ND | ND | ND | ND |
| OS | 23.08 | 1 | No | 2 | ND | ND | ND | ND | ND | |
| MM_0009 | OD | 21.53 | 0.98 | No | 2 | 72.74 | 22,644 | 9.31 | 2.63 | 0.28 |
| OS | 21.46 | 1 | No | 2 | ND | ND | ND | ND | ND | |
| MM_0014 | OD | 25.33 | 0.76 | Yes | 4 | 73.12 | 6,574 | 24.35 | 7.43 | 0.31 |
| OS | 25.54 | 0.72 | Yes | 4 | 75.48 | 7,305 | 25.24 | 8.06 | 0.32 | |
| MM_0015 | OD | 25.2 | 0.84 | Yes | 4 | 60.94 | 24,835 | 15.66 | 6.57 | 0.42 |
| OS | 24.75 | 0.78 | Yes | 4 | 49.15 | 32,140 | 10.19 | 4.32 | 0.42 | |
| MM_0016 | OD | 24.38 | 0.9 | Yes | 4 | ND | ND | ND | ND | ND |
| OS | 24.57 | 1.18 | Yes | 4 | 48.36 | 13,148 | 14.96 | 4.82 | 0.32 | |
| MM_0064 | OD | 22.46 | 0.78 | No | 2 | 27.52 | 9,496 | 11.41 | 2.88 | 0.25 |
| OS | 22.51 | 0.8 | No | 2 | 29.49 | 7,305 | 11.10 | 2.12 | 0.19 | |
| MM_0164 | OD | 25.56 | 0.92 | Yes | 2 | 123.85 | ND | ND | ND | ND |
| OS | 25.83 | 0.8 | Yes | 2 | ND | ND | ND | ND | ND | |
| MM_0165 | OD | 24.99 | 0.84 | Yes | 4 | 60.94 | 16,801 | 13.36 | 4.12 | 0.31 |
| OS | 24.88 | 0.7 | Yes | 4 | ND | ND | ND | ND | ND | |
| MM_0167 | OD | 24.96 | 1.06 | No | 1 | ND | ND | ND | ND | ND |
| OS | 25.04 | 1.16 | No | 1 | 110.09 | 10,957 | 12.89 | 6.62 | 0.51 | |
| MM_0168 | OD | 28.74 | 0.8 | No | 2 | 68.80 | ND | ND | ND | ND |
| OS | 28.24 | 0.92 | No | 2 | ND | ND | ND | ND | ND | |
| MM_0169 | OD | 24.77 | 0.74 | No | 1 | 92.39 | ND | ND | ND | ND |
| OS | 24.19 | 1.12 | No | 1 | ND | ND | ND | ND | ND | |
| MM_0170 | OD | 21.79 | 0.78 | Yes | 2 | 55.04 | 27,757 | 8.42 | 1.74 | 0.21 |
| OS | 21.71 | 0.76 | Yes | 2 | ND | ND | ND | ND | ND | |
| MM_0171 | OD | 22.87 | 0.7 | Yes | 3 | 47.18 | 25,566 | 12.18 | 6.01 | 0.49 |
| OS | 22.74 | 0.76 | Yes | 3 | ND | ND | ND | ND | ND | |
| MM_0239 | OD | 23.33 | 0.72 | Yes | 4 | 66.84 | 16,070 | 18.00 | 6.56 | 0.36 |
| OS | 23.23 | 0.9 | Yes | 4 | 58.40 | 13,148 | 17.09 | 6.44 | 0.38 | |
| MM_0251 | OD | 29.61 | 0.72 | Yes | 3 | 72.73 | ND | ND | ND | ND |
| OS | 29.51 | 0.66 | Yes | 3 | ND | ND | ND | ND | ND | |
| MM_0358 | OD | 23.62 | 0.82 | No | 2 | 94.36 | ND | ND | ND | ND |
| OS | 23.52 | 0.88 | No | 2 | ND | ND | ND | ND | ND | |
| MM_0385 | OD | 24.41 | 0.8 | No | 4 | ND | ND | ND | ND | ND |
| OS | 24.35 | 0.8 | No | 4 | 39.32 | 2,191 | 27.86 | 13.81 | 0.50 | |
| MM_0386 | OD | 24.1 | 0.98 | Yes | 2 | 94.36 | 13,148 | 12.93 | 3.85 | 0.30 |
| OS | 23.38 | LP | Yes | 2 | ND | ND | ND | ND | ND | |
| MM_0387 | OD | 22.89 | 0.84 | Yes | 4 | 74.7 | ND | ND | ND | ND |
| OS | 22.88 | 0.88 | Yes | 4 | ND | ND | ND | ND | ND | |
| MM_0396 | OD | 23.63 | 1 | Yes | 2 | ND | ND | ND | ND | ND |
| OS | 23.6 | 1.02 | Yes | 2 | 90.12 | ND | ND | ND | ND | |
| MM_0397 | OD | 22.15 | 0.96 | No | 2 | 96.33 | ND | ND | ND | ND |
| OS | 21.92 | 0.96 | No | 2 | ND | ND | ND | ND | ND | |
| MM_0398 | OD | 23.18 | 1 | Yes | 2 | 53.08 | 20,453 | 8.68 | 1.69 | 0.20 |
| OS | 23.49 | 1.12 | Yes | 2 | 49.15 | 19,722 | 8.44 | 1.42 | 0.17 | |
| MM_0412 | OD | 23.36 | 1 | Yes | 2 | 57.01 | ND | ND | ND | ND |
| OS | 23.37 | 1.02 | Yes | 2 | ND | ND | ND | ND | ND | |
| MM_0418 | OD | 24.2 | 0.9 | No | 2 | 64.04 | ND | ND | ND | ND |
| OS | 24.36 | 0.8 | No | 2 | ND | ND | ND | ND | ND | |
| MM_0445 | OD | 22.72 | 1 | Yes | 2 | 62.91 | ND | ND | ND | ND |
| OS | 22.87 | 1 | Yes | 2 | ND | ND | ND | ND | ND | |
| MM_0446 | OD | 24.81 | 0.76 | Yes | 4 | 70.76 | 6,574 | 18.90 | 6.45 | 0.34 |
| OS | 24.91 | 0.7 | Yes | 4 | 73.32 | 5,844 | 20.69 | 7.18 | 0.35 | |
| JC_0578 | OD | 27.21 | ND | Yes | 2 | 79.87 | ND | ND | ND | ND |
| OS | 26.47 | ND | Yes | 2 | ND | ND | ND | ND | ND | |
| JC_0615 | OD | 25.63 | ND | No | 3 | ND | ND | ND | ND | ND |
| OS | 25.65 | ND | No | 3 | 74.88 | ND | ND | ND | ND | |
| JC_0626 | OD | 28.85 | ND | Yes | 2 | 114.82 | ND | ND | ND | ND |
| OS | 28.35 | ND | Yes | 2 | ND | ND | ND | ND | ND | |
| JC_0674 | OD | 25.02 | ND | Yes | 4 | ND | ND | ND | ND | ND |
| OS | 24.36 | ND | Yes | 4 | 64.90 | ND | ND | ND | ND | |
| JC_0698 | OD | 24.27 | 0.9 | Yes | 4 | ND | ND | ND | ND | ND |
| OS | 23.79 | 0.9 | Yes | 4 | 73.63 | ND | ND | ND | ND | |
| JC_1240 | OD | 21.62 | 0.4 | No | 4 | 64.90 | ND | ND | ND | ND |
| OS | 21.72 | 0.54 | No | 4 | ND | ND | ND | ND | ND | |
| JC_10008 | OD | 22.62 | ND | Yes | 4 | 71.14 | ND | ND | ND | ND |
| OS | 22.76 | ND | Yes | 4 | ND | ND | ND | ND | ND | |
| JC_10069 | OD | 23.13 | 0.8 | Yes | 2 | 67.39 | 40,906 | 7.10 | 1.29 | 0.18 |
| OS | 23.59 | 0.8 | Yes | 2 | ND | ND | ND | ND | ND | |
| KS_10088 | OD | 25.19 | ND | Yes | 2 | 123.55 | 21,183 | 11.56 | 3.80 | 0.33 |
| OS | 25.34 | ND | Yes | 2 | ND | ND | ND | ND | ND | |
| KS_10337 | OD | 23.29 | 1.0 | No | 1 | 71.14 | 54,785 | 6.07 | 1.28 | 0.21 |
| OS | 23.78 | 0.88 | No | 1 | ND | ND | ND | ND | ND | |
| JC_10551 | OD | 22.46 | 0.9 | Yes | 1 | ND | ND | ND | ND | ND |
| OS | 22.39 | 0.9 | Yes | 1 | 97.34 | ND | ND | ND | ND |
ND, no data; BCVA, best corrected visual acuity; EZ, ellipsoid zone; PFD, peak foveal cone density; ICD, intercell distance; SD, standard deviation; CV, coefficient of variation; ONL, outer nuclear layer; LP, light perception.
OCT Imaging
Retinal Morphology
OCT revealed a variable degree of disruption or loss of the foveal EZ and was graded as previously published (Fig. 2).19 All subjects (n = 38) had the same EZ grade bilaterally. Four subjects (10.5%) had continuous EZ (grade 1), 19 subjects (50.0%) had EZ disruption (grade 2), 3 subjects (7.9%) had EZ absence (grade 3), and 12 subjects (31.6%) had presence of an HRZ (grade 4; Fig. 3A). None of the subjects included in the study had outer retinal atrophy including RPE loss (grade 5). In our cohort, the distribution of age was the same across the different OCT grades (P = 0.086, Kruskal-Wallis test).
Figure 3.
Results summary and graphical representation. (A) Distribution of our cohort according to OCT grading, with the vast majority having either grade 2 or 4. (B) Scatter plot of ONL thickness and age. No correlation was found. (C) Stacked scatterplot of ONL thickness and OCT EZ grade, showing the vast variability within and between grades, as well as a trend of decreasing ONL thickness with increasing grade. (D) Stacked scatterplot of peak cone density and OCT EZ showing the overlap between grades. In (C) and (D), individual data points within each EZ grade are displaced horizontally for better visibility. (E) Peak cone densities of all subjects with CNGA3-ACHM included in this study plotted against age. (F) Peak cone densities of all subjects with CNGB3-ACHM previously reported by Langlo et al.36 plotted against age on the same scale as (E). The CNGA3 cohort plotted on (E) shows a weak negative correlation between peak cone density and age (Pearson r = −0.397, P = 0.102).
Foveal ONL Thickness
The mean (±SD) ONL thickness, in μm, for grade 1 was 92.74 (±16.22), grade 2 was 78.69 (±25.12), grade 3 was 64.93 (±15.41), and grade 4 was 63.15 (±11.57). The mean (range, ±SD) ONL thickness, in μm, for all subjects was 74.18 (27.52–123.85, ± 21.80) (Table 2, Fig. 3B). Previously reported mean ± SD from unaffected controls for ONL thickness is 110.6 ± 15.7 μm, with an n = 38 (mean age ± SD; 28.8 ± 10.9 years);46 these values are significantly higher than our reported values for CNGA3-ACHM (P < 0.0001, t = 8.37, df = 74). ONL thickness at the fovea was measured with standard OCT. The residual Henle fiber layer (HFL) may have been included within the ONL measurement, but given the foveal location, only a small degree of error is expected. The HFL comprised only 17.5% of the measured HFL+ONL thickness (range, 11.4%–24.8%) at the fovea of healthy controls.47 Due to a small number of subjects with grade 1 and 3 EZ, the data were not further statistically analyzed. Grade 2 subjects had a significantly greater ONL thickness than grade 4 subjects (P = 0.054, t = 2.005, df = 29). Despite the trend toward decreasing ONL thickness with increasing EZ grade (i.e., increasing EZ disruption/loss), there is overlap between grades (Fig. 3C).
Foveal Hypoplasia
Hypoplasia was observed bilaterally in 25 subjects (65.8%) to variable degrees. There was no significant difference in ONL thickness between individuals with foveal hypoplasia and those with normal excavation of inner retinal layers (P = 0.861, Mann-Whitney test) (Table 2). No association was found between foveal hypoplasia and EZ grade (P = 0.199, chi-square test). Subjects with foveal hypoplasia (n = 19, BCVA = 0.86 LogMAR, SD = 0.13) had similar BCVA compared to subjects without foveal hypoplasia (n = 12, mean BCVA = 0.87 LogMAR, SD = 0.19; P = 0.924, t = 0.096, df = 29).
Photoreceptor Mosaic Assessed With AOSLO Imaging
AOSLO imaging was attempted in all recruited subjects (n = 38). Eighteen (47.4%) had good-quality images and met the aforementioned eligibility criteria. If bilateral data were available, the eye with the better image quality was included for further analysis in all subjects. Foveal hypoplasia did not correlate with successful AOSLO imaging (P = 0.506, Fisher's exact test), with the main limiting factor in AOSLO acquisition being the severity of nystagmus.
Peak Foveal Cone Density
The mean peak foveal cone density was 19,844 cones/mm2, ranging from 6,574 cones/mm2 to 54,785 cones/mm2 (Table 2). There was marked variability between subjects with a SD of 13,046 cones/mm2 (Table 2); however, as expected, these values do not overlap with previously reported normative values (range, 84,733–247,064 cones/mm2).48,49
There was no significant difference in peak foveal cone density between individuals with foveal hypoplasia and those with normal excavation of inner retinal layers (P = 0.974, t = 0.0334, df = 16), a finding that has also been previously reported for CNGB3-ACHM.36 Peak foveal cone density was available for two subjects with EZ grade 1 and only for one subject with EZ grade 3 and, therefore, were excluded from statistical evaluation. There was no significant difference in peak foveal cone density between individuals with a grade 2 EZ and those with a grade 4 EZ (P = 0.129, t = 1.619, df = 13). All peak cone density data are plotted in Figure 3D. In contrast, in the aforementioned CNGB3 study, peak foveal cone density, and EZ grade were significantly associated.36 Peak foveal cone density and ONL thickness were not correlated (P = 0.927, r = 0.023, Pearson correlation), in keeping with the previous CNGB3 study.36 There was a weak negative association between age and peak foveal cone density (P = 0.102, r = −0.397, Pearson correlation) (Figs. 3E).
Intercell Distance
The mean (μ) and SD (σ) of the ICD for each subject (n = 18) was evaluated, as well as the CV (CV = σ/μ), to measure the overall variability of ICD independent of the value, which, as expected, varied widely. The mean ICD (range, ±SD), in μm, was 13.40 (6.07–27.86, ±5.74). As expected, the mean ICD in CNGA3-ACHM was greater (P < 0.0001, Mann-Whitney test) than in unaffected controls (n = 6; ICD mean ± SD, 3.61 ± 0.22 μm).36
Our cohort had a CV (mean ± SD) of 0.32 ± 0.1, which was significantly higher (P = 0.0002, t = 4.537, df = 22) than healthy controls (n = 6; CV mean ± SD, 0.13 ± 0.014).36
Structural Disease Symmetry
In seven subjects, bilateral OCT and AOSLO imaging data were obtained (one male, six females; age (mean ± SD), 32.6 ± 11.9 years) (Table 2; Fig. 4). All subjects had the same bilateral EZ grade; three subjects had grade 2 and four had grade 4. Six (85.7%) subjects had bilateral foveal hypoplasia. The absolute difference in ONL thickness between right and left eyes (mean ± SD) was 4.16 ± 2.31 μm. ONL thickness did not differ significantly between eyes (P = 0.754, t = 0.329, df = 6), with mean (±SD) ONL thickness, in μm, of 59.03 (±17.92) and 59.65 (±18.68) for right and left eyes, respectively.
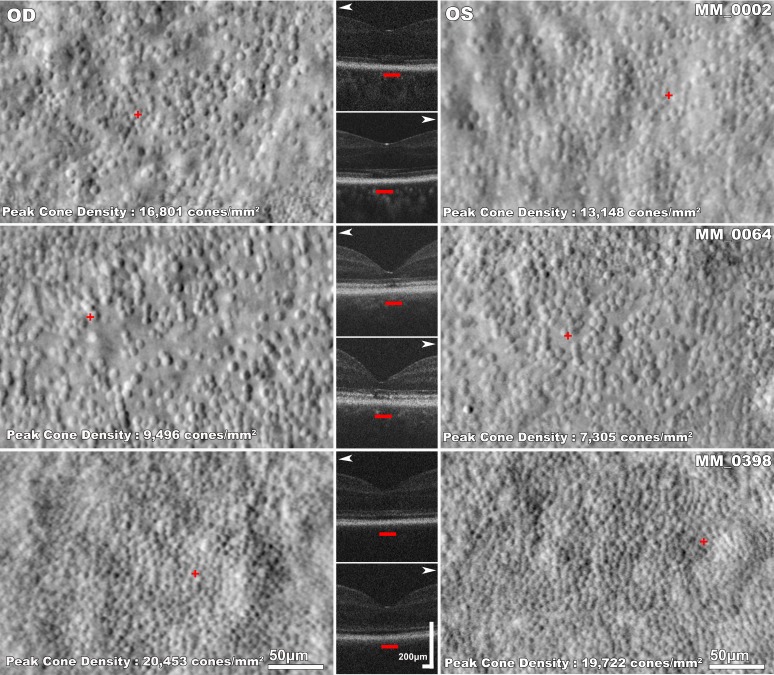
Figure 4.
Disease symmetry in CNGA3-associated ACHM. Left and right column: split-detection AOSLO images of the right and left eye respectively of three subjects with CNGA3-ACHM. Middle column: horizontal transfoveal OCT scans of the same subjects. White arrowheads point to the respective AOSLO image. The red horizontal bars mark the 300-μm area represented in the AOSLO images. There is substantial variability in foveal photoreceptor mosaic between the three subjects. There are structural similarities in the mosaic pattern of the right and left eyes of each subject; MM_0002 and MM_0064 have noncontiguous sparse foveal mosaics and MM_0398 has a relatively contiguous mosaic. MM_0002 has OCT grade 4 bilaterally with foveal hypoplasia. MM_0064 and MM_0398 have EZ grade 2 bilaterally.
Peak foveal cone density was also comparable bilaterally (P = 0.829, t = 0.225, df = 6), with mean ± SD of 14,400 ± 7,078 cones/mm2 and 14,087 ± 9,304 cones/mm2 for the right and left eyes, respectively. The difference in peak foveal cone density (mean ± SD) between eyes was 2,609 ± 2,380 cones/mm2. ICDs were also similar bilaterally (P = 0.496, t = 0.725, df = 6), with mean ± SD of 15.72 ± 5.25 μm and 15.09 ± 6.16 μm for right and left eyes, respectively. The mean ± SD of CV was 0.31 ± 0.07 and 0.30 ± 0.09 for right and left eyes, respectively; indicating that mosaic topography was similar between eyes (P = 0.448, t = 0.812, df = 6).
Intrafamilial Variability
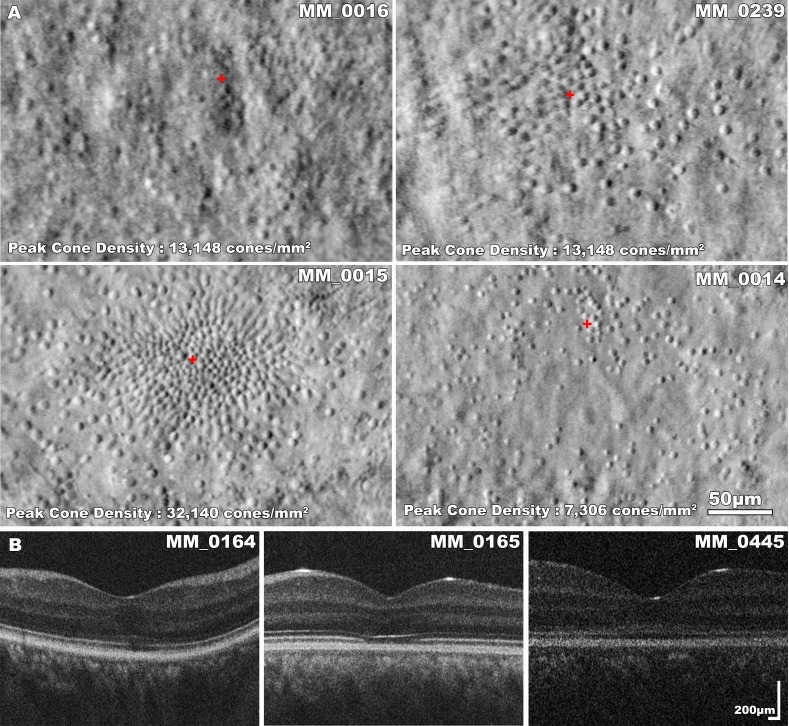
We recruited three pairs of siblings (Tables 1 and 2; MM_0008-MM_0009, MM_0386-MM_0387, and MM_0396-MM_0397), four siblings (MM_0014, MM_0015, MM_0016, and MM_0239), and a mother with two sons (MM_0164, MM_0165, and MM_0445, respectively) (Tables 1, 2). Two pairs of siblings (MM_0008-MM_0009 and MM_0396-MM_0397) had the same EZ grade and comparable ONL thickness (mean difference among siblings of 3 μm). AOSLO data were analyzable in only one of those four subjects. The four siblings (MM_0014, MM_0015, MM_0016, and MM_0239) had substantial variability of their cone mosaics even though their EZ grade was the same (grade 4). AOSLO imaging was available for all four siblings (three bilaterally) (Fig. 5A). The cone mosaic was variably disrupted, without any specific pattern among these subjects. Despite the differences in remnant cone structure, the subject with the lowest peak foveal cone density (right/left eye, 6,574 and 7,305 cones/mm2, respectively) had the best BCVA among his siblings. For the last pair of siblings and the family with the three affected members, only one cone mosaic per family could be analyzed and no conclusion about the intrafamilial variability of their mosaics could be drawn. However, in both families there was a notable difference in the disruption of the photoreceptor mosaic on OCT; the two siblings MM_0386 and MM_0387 had grade 2 and 4 EZs, respectively, and from the other pedigree, two subjects had grade 2 and one grade 4 EZs (Fig. 5B). BCVA was comparable among these subjects (Table 2).
Figure 5.
Intrafamilial variability in CNGA3-associated ACHM. (A) Split-detection AOSLO images of four siblings with CNGA3-ACHM. MM_0016 (VA, 0.86 LogMAR) and MM_0239 (VA, 0.90 LogMAR) are males, 30 and 37 years old, respectively. MM_0015 (VA, 0.84 LogMAR) and MM_0014 (VA, 0.74 LogMAR) are females, 28 and 35 years old, respectively. There is substantial variability in foveal photoreceptor mosaic across the four subjects, which is independent of sex or age. All four siblings have EZ grade 4 bilaterally with foveal hypoplasia. The identification of 40 cones within the 55 × 55-μm sampling window resulted in a density of 13,148 cones/mm2 for both siblings, MM_0016 and MM_0239. However, other parameters such as the ICD were different, reflecting the geometry of cones over the foveal center; MM_0239 had greater mean ICD due to a sparser mosaic in the inferior fovea. The red cross marks the location of peak foveal cone density. (B) Transfoveal OCT scans of subjects MM_0164, MM_0165, and MM_0445, from a single family. Female subject MM_0164 (VA, 0.92 LogMAR, mother of the male siblings MM_0165 and MM_0445) has a better preserved EZ than the 25 year younger subject MM_0165 (VA, 0.84 LogMAR); and subject MM_0445 (VA, 1.0 LogMAR) has a nearly continuous EZ.
Discussion
The foveal cone mosaic in CNGA3-ACHM was irregular, variably disrupted, and showed significantly lower peak foveal cone density than unaffected individuals. A striking structural feature that was clear in our cohort was the irregular size and shape of the remnant inner segments (Figs. 2, 4, 5). Irregularities in shape and size have been described in the limited number of histology reports in patients with ACHM.50–52 In all three reports, the patients were well phenotyped but not genotyped, because it was before the discovery of the ACHM genes. The unusual shape of the inner segments was attributed to the presence of ectopic nuclei.50–52 Using OCT imaging, Genead et al.29 previously reported a substantial variation in phenotype even in subjects with the same genotype. Our subjects with CNGA3-ACHM had a significantly thinner ONL than unaffected individuals, although there was significant variability between subjects. No correlation was found between peak foveal cone density and ONL thickness. Our findings are in keeping with previous reports for CNGB3-ACHM.36 There was a broad range of cone densities within each EZ grade. This dissociation between OCT and peak foveal cone density highlights the value of AOSLO for patient stratification and evaluation of retinal potential for therapy trials. Notable examples are subjects MM_0015 (Fig. 5) and MM_0171 (Fig. 2), who despite being graded as 4 and 3, respectively, have a peak foveal cone density higher than the average for the cohort and may thus be good candidates for an intervention. However, it is still unclear what should be considered as a threshold and which metric will best correlate with outcome. We will certainly be in a better position to address these questions when the results of the current gene therapy trials using AOSLO are released.
Despite the vast variability among subjects, interocular symmetry was observed in all aspects examined, including axial length, BCVA, EZ grade, presence of foveal hypoplasia, ONL thickness, peak foveal cone density, and mosaic geometry (ICD and CV). This information is of high value both for eye selection for an intervention, as well as for longitudinal evaluation of the outcome of ongoing and upcoming trials, in which the nontreated eye can serve as a control. Overall, CNGA3-ACHM appears to be a structurally symmetric disease in keeping with previous literature that did not use AOSLO.19 Many subjects had a variable degree of foveal hypoplasia, which is consistent with previous studies both for CNGA3 and CNGB3.19,29,36 No difference in BCVA was noted between eyes with and without foveal hypoplasia, in contrast to previous studies that suggested better19 or worse40 BCVA in the presence of foveal hypoplasia. As previously suggested by Thomas et al.,53 incomplete centrifugal displacement of the inner retinal layers is a common finding. No current literature has investigated the mechanism/molecular basis of foveal hypoplasia in ACHM.
The majority of subjects had missense sequence variants, suggesting little tolerance for substitutions, in agreement with the high degree of evolutionary conservation of the CNGA3 gene.17 There was marked genetic variability, although no genotype-phenotype correlation could be observed. There is a notable intrafamilial variability of the cone mosaic topography and of the peak foveal cone density (Fig. 5). EZ grade and ONL thickness were far less sensitive in detecting structural differences, which again illustrates a disconnect between OCT and AOSLO. In other cases, intrafamilial variability was also clear on OCT. The observed marked intrafamilial variability in CNGA3-ACHM shows a lack of genotype-phenotype correlation in ACHM caused by CNGA3 variants, with specific genetic variants not being predictive of remnant cone structure.
Thiadens et al.54 and Thomas et al.40 suggested an age-dependent ONL thinning.54 In contrast, Sundaram et al.19 and Aboshiha et al.20 did not identify such a relationship. Langlo et al.35 in a longitudinal CNGB3 assessment reported increasing ONL thickness over time (6–26 months). In our cohort, no correlation between ONL thickness and age was found. Previously Sundaram et al.19 suggested a wider window of opportunity for intervention in ACHM and no age-dependent association in retinal structure disruption, identifying the need for the specific measurement of remnant photoreceptor structure, with SD-OCT and AOSLO imaging for stratification of patients and evaluating intervention efficacy.
While there was no association between age and ONL thickness, we observed a weak negative association between age and peak foveal cone density (Fig. 3E) as well as age and EZ grade. While a similar cross-sectional study of CNGB3-ACHM showed an association between age and EZ grade,36 that study reported no relationship between age and peak foveal cone density (Fig. 3F). In the aforementioned CNGB3 study, the subcohort imaged successfully with AOSLO had a mean age (±SD), in years, of 23.4 (±10.3) and our cohort had a mean age (±SD) of 28.3 (±13.8), with the age distribution not differing significantly among the two studies (P = 0.186, t = 1.35, df = 42). The mean peak foveal cone density is also similar between the two studies (P = 0.71, Mann-Whitney test). No distinguishing phenotypic characteristics could be identified for our CNGA3 genotype compared to the current literature for CNGB3. Similarity in CNGA3- and CNGB3-associated ACHM was also observed in the data from microperimetry, fundus autofluorescence, and OCT by Aboshiha et al.20 and Sundaram et al.19
The proteins encoded by CNGA3 and CNGB3 have been well characterized, and differences in their function may relate to the age-dependent decline that we observed in our CNGA3 cohort, in contrast to the previously well characterized CNGB3 cohort using the same methodology.36 It has been shown than the α subunits expressed by CNGA3 are responsible for the ion-conducting activity of the cation channel and the β subunits modulate the activity.55,56 It has also been shown in CNGA3−/− mice that the loss of CNGA3 expression impairs the correct localization of cone opsins as well as the expression of other proteins of the visual cycle, with this inducing apoptotic cell death in cones.57 In the CNGB3−/− mice, a downregulation of CNGA3 gene expression of ∼90% has been observed.58 The residual CNGA3 expression may account for the preservation of remnant cones in CNGB3-ACHM. By extension, the complete absence/greater reduction of CNGA3 expression may lead to an age-dependent decline of remnant cones in CNGA3-ACHM (as we observe), also bearing in mind that α subunits in vitro can form channels in the absence of β subunits, a property not shared by β subunits.59
However, given the limitations of the cross-sectional nature of this study, a longitudinal assessment and further evaluation of the progressive or stationary nature of CNGA3-ACHM would be invaluable. Longitudinal studies are necessary to properly address the question of cone loss over time. The lack of studies using AOSLO in age-matched subjects in order to investigate possible age-related decline in healthy eyes makes it challenging to put our findings in context. The current literature in healthy eyes is inconclusive for possible age-dependent changes in peak foveal cone density, either using histology or AO retinal imaging.60–63 Nevertheless, in agreement with previous reports,29,36,59 all of the subjects in the present study had remnant cone structure (across the entire age range studied). However, no functional assessment of those cones was possible, which will be interesting both in terms of disease natural history and for assessing the outcomes of any intervention.64 Despite the observed decrease of peak foveal cone density with age, there were several notable exceptions, with high peak foveal cone density in older subjects and no clear age cutoff identified. This shows that despite the statistical relationship, there is still great variability in cone populations across all ages and, thus, the need for longitudinal studies.
Analyzable AOSLO was achieved in only 18/38 subjects and can be attributed to nystagmus in the vast majority of subjects, as well as to poor optical media and high myopia in some. Nystagmus (or absence thereof) has not been correlated with either lower or higher remnant photoreceptor structure in the current literature. Nystagmus in ACHM has previously been reported to improve over time,65,66 and given the stationary, or only slowly progressive, natural history of the condition, our reported peak foveal cone densities are likely representative of the CNGA3 genotype. In this study, subjects with strong nystagmus, in which AOSLO was not analyzable, had a range of EZ grades on OCT, further supporting that nystagmus is not representative of the structural severity of the disease or remnant cone architecture. We anticipate that the challenge nystagmus poses to imaging quality will be ameliorated with the use of eye tracking in the near future.67–69 In addition to the low successful acquisition rate, several other factors should be taken into account when considering using AOSLO in research studies or even clinical practice; acquisition time is considerable and requires the participation of two investigators for our custom-built system, with processing, montaging, and analysis being substantial bottlenecks. Newly developed software for simplifying the steps of the process are of high value for the field, such as montaging45,70 and cone counting.71–75
In conclusion, CNGA3-ACHM retinas can be successfully imaged with split-detection AOSLO, with most findings in keeping with CNGB3-ACHM. To the best of our knowledge, we report the largest CNGA3-ACHM study to include split-detection AOSLO imaging. There is a need for prospective longitudinal studies of large cohorts of molecularly confirmed CNGA3 subjects to improve our understanding of the cellular natural history.76 Given the large variability in cone mosaics, subjects with ACHM should be assessed on an individual basis for selection in future and ongoing clinical trials.
Acknowledgments
Supported by grants from the National Institute for Health Research, Biomedical Research Centre at Moorfields Eye Hospital National Health Service Foundation Trust, UCL Institute of Ophthalmology, MeiraGTx, Fight for Sight (UK), Onassis Foundation, Leventis Foundation, The Macular Society (UK), Moorfields Eye Hospital Special Trustees, Moorfields Eye Charity, Retinitis Pigmentosa Fighting Blindness, The Wellcome Trust (099173/Z/12/Z), Research to Prevent Blindness Departmental Challenge Award (Stanford), and the Foundation Fighting Blindness (USA). Research reported in this publication was supported in part by the National Eye Institute of the National Institutes of Health under award numbers R01EY017607, P30EY026877, F32EY029148, and U01EY025477. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Disclosure: M. Georgiou, None; K.M. Litts, None; A. Kalitzeos, None; C.S. Langlo, None; T. Kane, None; N. Singh, None; M. Kassilian, None; N. Hirji, None; N. Kumaran, None; A. Dubra, MeiraGTx (C), Boston Micromachines (C), P; J. Carroll, AGTC (F), OptoVue (F), MeiraGTx (C); M. Michaelides, MeiraGTx (C)
References
- 1.Aboshiha J, Dubis AM, Carroll J, Hardcastle AJ, Michaelides M. The cone dysfunction syndromes. Br J Ophthalmol. 2016;100:115–121. doi: 10.1136/bjophthalmol-2014-306505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hirji N, Aboshiha J, Georgiou M, Bainbridge J, Michaelides M. Achromatopsia: clinical features, molecular genetics, animal models and therapeutic options. Ophthalmic Genet. 2018;39:149–157. doi: 10.1080/13816810.2017.1418389. [DOI] [PubMed] [Google Scholar]
- 3.Maguire J, McKibbin M, Khan K, Kohl S, Ali M, McKeefry D. CNGB3 mutations cause severe rod dysfunction. Ophthalmic Genet. 2017;39:108–114. doi: 10.1080/13816810.2017.1368087. [DOI] [PubMed] [Google Scholar]
- 4.Khan NW, Wissinger B, Kohl S, Sieving PA. CNGB3 achromatopsia with progressive loss of residual cone function and impaired rod-mediated function. Invest Ophthalmol Vis Sci. 2007;48:3864–3871. doi: 10.1167/iovs.06-1521. [DOI] [PubMed] [Google Scholar]
- 5.Thiadens AA, Roosing S, Collin RW, et al. Comprehensive analysis of the achromatopsia genes CNGA3 and CNGB3 in progressive cone dystrophy. Ophthalmology. 2010;117:825–830.e1. doi: 10.1016/j.ophtha.2009.09.008. [DOI] [PubMed] [Google Scholar]
- 6.Wissinger B, Jagle H, Kohl S, et al. Human rod monochromacy: linkage analysis and mapping of a cone photoreceptor expressed candidate gene on chromosome 2q11. Genomics. 1998;51:325–331. doi: 10.1006/geno.1998.5390. [DOI] [PubMed] [Google Scholar]
- 7.Wissinger B, Gamer D, Jagle H, et al. CNGA3 mutations in hereditary cone photoreceptor disorders. Am J Hum Genet. 2001;69:722–737. doi: 10.1086/323613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mayer AK, Van Cauwenbergh C, Rother C, et al. CNGB3 mutation spectrum including copy number variations in 552 achromatopsia patients. Hum Mut. 2017;38:1579–1591. doi: 10.1002/humu.23311. [DOI] [PubMed] [Google Scholar]
- 9.Kohl S, Baumann B, Rosenberg T, et al. Mutations in the cone photoreceptor G-protein alpha-subunit gene GNAT2 in patients with achromatopsia. Am J Hum Genet. 2002;71:422–425. doi: 10.1086/341835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aligianis IA, Forshew T, Johnson S, et al. Mapping of a novel locus for achromatopsia (ACHM4) to 1p and identification of a germline mutation in the alpha subunit of cone transducin (GNAT2) J Med Genet. 2002;39:656–660. doi: 10.1136/jmg.39.9.656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kohl S, Zobor D, Chiang WC, et al. Mutations in the unfolded protein response regulator ATF6 cause the cone dysfunction disorder achromatopsia. Nat Genet. 2015;47:757–765. doi: 10.1038/ng.3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kohl S, Coppieters F, Meire F, et al. A nonsense mutation in PDE6H causes autosomal-recessive incomplete achromatopsia. Am J Hum Genet. 2012;91:527–532. doi: 10.1016/j.ajhg.2012.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fesenko EE, Kolesnikov SS, Lyubarsky AL. Induction by cyclic GMP of cationic conductance in plasma membrane of retinal rod outer segment. Nature. 1985;313:310–313. doi: 10.1038/313310a0. [DOI] [PubMed] [Google Scholar]
- 14.Yu WP, Grunwald ME, Yau KW. Molecular cloning, functional expression and chromosomal localization of a human homolog of the cyclic nucleotide-gated ion channel of retinal cone photoreceptors. FEBS Lett. 1996;393:211–215. doi: 10.1016/0014-5793(96)00889-7. [DOI] [PubMed] [Google Scholar]
- 15.Wissinger B, Muller F, Weyand I, et al. Cloning, chromosomal localization and functional expression of the gene encoding the alpha-subunit of the cGMP-gated channel in human cone photoreceptors. Eur J Neurosci. 1997;9:2512–2521. doi: 10.1111/j.1460-9568.1997.tb01680.x. [DOI] [PubMed] [Google Scholar]
- 16.Michaelides M, Hunt DM, Moore AT. The cone dysfunction syndromes. Br J Ophthalmol. 2004;88:291–297. doi: 10.1136/bjo.2003.027102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnson S, Michaelides M, Aligianis IA, et al. Achromatopsia caused by novel mutations in both CNGA3 and CNGB3. J Med Genet. 2004;41:e20. doi: 10.1136/jmg.2003.011437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zobor D, Werner A, Stanzial F, et al. The clinical phenotype of CNGA3related achromatopsia: pretreatment characterization in preparation of a gene replacement therapy trial. Invest Ophthalmol Vis Sci. 2017;58:821–832. doi: 10.1167/iovs.16-20427. [DOI] [PubMed] [Google Scholar]
- 19.Sundaram V, Wilde C, Aboshiha J, et al. Retinal structure and function in achromatopsia: implications for gene therapy. Ophthalmology. 2014;121:234–245. doi: 10.1016/j.ophtha.2013.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aboshiha J, Dubis AM, Cowing J, et al. A prospective longitudinal study of retinal structure and function in achromatopsia. Invest Ophthalmol Vis Sci. 2014;55:5733–5743. doi: 10.1167/iovs.14-14937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Georgiou M, Kalitzeos A, Patterson EJ, Dubra A, Carroll J, Michaelides M. Adaptive optics imaging of inherited retinal diseases. Br J Ophthalmol. 2018;102:1028–1035. doi: 10.1136/bjophthalmol-2017-311328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roorda A, Williams DR. The arrangement of the three cone classes in the living human eye. Nature. 1999;397:520–522. doi: 10.1038/17383. [DOI] [PubMed] [Google Scholar]
- 23.Dubra A, Sulai Y, Norris JL, et al. Noninvasive imaging of the human rod photoreceptor mosaic using a confocal adaptive optics scanning ophthalmoscope. Biomed Opt Express. 2011;2:1864–1876. doi: 10.1364/BOE.2.001864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rossi EA, Chung M, Dubra A, Hunter JJ, Merigan WH, Williams DR. Imaging retinal mosaics in the living eye. Eye (Lond) 2011;25:301–308. doi: 10.1038/eye.2010.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liang J, Williams DR, Miller DT. Supernormal vision and high-resolution retinal imaging through adaptive optics. J Opt Soc Am A Opt Image Sci Vis. 1997;14:2884–2892. doi: 10.1364/josaa.14.002884. [DOI] [PubMed] [Google Scholar]
- 26.Roorda A, Duncan JL. Adaptive optics ophthalmoscopy. Ann Rev Vis Sci. 2015;1:19–50. doi: 10.1146/annurev-vision-082114-035357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roorda A, Williams DR. Optical fiber properties of individual human cones. J Vis. 2002;2:404–412. doi: 10.1167/2.5.4. [DOI] [PubMed] [Google Scholar]
- 28.Carroll J, Choi SS, Williams DR. In vivo imaging of the photoreceptor mosaic of a rod monochromat. Vision Res. 2008;48:2564–2568. doi: 10.1016/j.visres.2008.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Genead MA, Fishman GA, Rha J, et al. Photoreceptor structure and function in patients with congenital achromatopsia. Invest Ophthalmol Vis Sci. 2011;52:7298–7308. doi: 10.1167/iovs.11-7762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dubis AM, Cooper RF, Aboshiha J, et al. Genotype-dependent variability in residual cone structure in achromatopsia: toward developing metrics for assessing cone health. Invest Ophthalmol Vis Sci. 2014;55:7303–7311. doi: 10.1167/iovs.14-14225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Merino D, Duncan JL, Tiruveedhula P, Roorda A. Observation of cone and rod photoreceptors in normal subjects and patients using a new generation adaptive optics scanning laser ophthalmoscope. Biomed Opt Express. 2011;2:2189–2201. doi: 10.1364/BOE.2.002189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ueno S, Nakanishi A, Kominami T, et al. In vivo imaging of a cone mosaic in a patient with achromatopsia associated with a GNAT2 variant. Jpn J Ophthalmol. 2017;61:92–98. doi: 10.1007/s10384-016-0484-7. [DOI] [PubMed] [Google Scholar]
- 33.Scoles D, Sulai YN, Langlo CS, et al. In vivo imaging of human cone photoreceptor inner segments. Invest Ophthalmol Vis Sci. 2014;55:4244–4251. doi: 10.1167/iovs.14-14542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Abozaid MA, Langlo CS, Dubis AM, Michaelides M, Tarima S, Carroll J. Reliability and repeatability of cone density measurements in patients with congenital achromatopsia. Adv Exp Med Biol. 2016;854:277–283. doi: 10.1007/978-3-319-17121-0_37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Langlo CS, Erker LR, Parker M, et al. Repeatability and longitudinal assessment of foveal cone structure in CNGB3associated achromatopsia. Retina. 2017;37:1956–1966. doi: 10.1097/IAE.0000000000001434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Langlo CS, Patterson EJ, Higgins BP, et al. Residual foveal cone structure in CNGB3associated achromatopsia. Invest Ophthalmol Vis Sci. 2016;57:3984–3995. doi: 10.1167/iovs.16-19313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Scoles D, Flatter JA, Cooper RF, et al. Assessing photoreceptor structure associated with ellipsoid zone disruptions visualized with optical coherence tomography. Retina. 2016;36:91–103. doi: 10.1097/IAE.0000000000000618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thevenaz P, Ruttimann UE, Unser M. A pyramid approach to subpixel registration based on intensity. IEEE Trans Image Process. 1998;7:27–41. doi: 10.1109/83.650848. [DOI] [PubMed] [Google Scholar]
- 39.Huang Y, Cideciyan AV, Papastergiou GI, et al. Relation of optical coherence tomography to microanatomy in normal and rd chickens. Invest Ophthalmol Vis Sci. 1998;39:2405–2416. [PubMed] [Google Scholar]
- 40.Thomas MG, Kumar A, Kohl S, Proudlock FA, Gottlob I. High-resolution in vivo imaging in achromatopsia. Ophthalmology. 2011;118:882–887. doi: 10.1016/j.ophtha.2010.08.053. [DOI] [PubMed] [Google Scholar]
- 41.Staurenghi G, Sadda S, Chakravarthy U, Spaide RF. Proposed lexicon for anatomic landmarks in normal posterior segment spectral-domain optical coherence tomography: the IN*OCT consensus. Ophthalmology. 2014;121:1572–1578. doi: 10.1016/j.ophtha.2014.02.023. [DOI] [PubMed] [Google Scholar]
- 42.Dubra A, Sulai Y. Reflective afocal broadband adaptive optics scanning ophthalmoscope. Biomed Opt Express. 2011;2:1757–1768. doi: 10.1364/BOE.2.001757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Salmon AE, Cooper RF, Langlo CS, Baghaie A, Dubra A, Carroll J. An automated reference frame selection (ARFS) algorithm for cone imaging with adaptive optics scanning light ophthalmoscopy. Trans Vis Sci Tech. 2017;6(2):9. doi: 10.1167/tvst.6.2.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dubra A, Harvey Z. Registration of 2D Images From Fast Scanning Ophthalmic Instruments. Berlin, Heidelberg: Springer Berlin Heidelberg;; 2010. pp. 60–71. [Google Scholar]
- 45.Davidson B, Kalitzeos A, Carroll J, et al. Fast adaptive optics scanning light ophthalmoscope retinal montaging. Biomed Opt Express. 2018;9:4317–4328. doi: 10.1364/BOE.9.004317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mastey R, Litts KM, Langlo CS, et al. Interocular symmetry and repeatability of foveal outer nuclear layer thickness in congenital achromatopsia. bioRxiV. 2018:367813. doi: 10.1101/367813. [DOI] [PMC free article] [PubMed]
- 47.Lee DJ, Woertz EN, Visotcky A, et al. The Henle fiber layer in albinism: comparison to normal and relationship to outer nuclear layer thickness and foveal cone density. Invest Ophthalmol Vis Sci. 2018;59:5336–5348. doi: 10.1167/iovs.18-24145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wilk MA, McAllister JT, Cooper RF, et al. Relationship between foveal cone specialization and pit morphology in albinism. Invest Ophthalmol Vis Sci. 2014;55:4186–4198. doi: 10.1167/iovs.13-13217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang T, Godara P, Blanco ER, et al. Variability in human cone topography assessed by adaptive optics scanning laser ophthalmoscopy. Am J Ophthalmol. 2015;160:290–300.e1. doi: 10.1016/j.ajo.2015.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Harrison R, Hoefnagel D, Hayward JN. Congenital total color blindness: a clincopathological report. Arch Ophthalmol. 1960;64:685–692. doi: 10.1001/archopht.1960.01840010687010. [DOI] [PubMed] [Google Scholar]
- 51.Glickstein M, Heath GG. Receptors in the monochromat eye. Vision Res. 1975;15:633–636. doi: 10.1016/0042-6989(75)90276-x. [DOI] [PubMed] [Google Scholar]
- 52.Falls HF, Wolter JR, Alpern M. Typical total monochromacy. A histological and psychophysical study. Arch Ophthalmol. 1965;74:610–616. doi: 10.1001/archopht.1965.00970040612005. [DOI] [PubMed] [Google Scholar]
- 53.Thomas MG, Kumar A, Mohammad S, et al. Structural grading of foveal hypoplasia using spectral-domain optical coherence tomography a predictor of visual acuity? Ophthalmology. 2011;118:1653–1660. doi: 10.1016/j.ophtha.2011.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Thiadens AA, Slingerland NW, Roosing S, et al. Genetic etiology and clinical consequences of complete and incomplete achromatopsia. Ophthalmology. 2009;116:1984–1989.e1. doi: 10.1016/j.ophtha.2009.03.053. [DOI] [PubMed] [Google Scholar]
- 55.Zagotta WN, Siegelbaum SA. Structure and function of cyclic nucleotide-gated channels. Ann Rev Neurosci. 1996;19:235–263. doi: 10.1146/annurev.ne.19.030196.001315. [DOI] [PubMed] [Google Scholar]
- 56.Gerstner A, Zong X, Hofmann F, Biel M. Molecular cloning and functional characterization of a new modulatory cyclic nucleotide-gated channel subunit from mouse retina. J Neurosci. 2000;20:1324–1332. doi: 10.1523/JNEUROSCI.20-04-01324.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Michalakis S, Geiger H, Haverkamp S, Hofmann F, Gerstner A, Biel M. Impaired opsin targeting and cone photoreceptor migration in the retina of mice lacking the cyclic nucleotide-gated channel CNGA3. Invest Ophthalmol Vis Sci. 2005;46:1516–1524. doi: 10.1167/iovs.04-1503. [DOI] [PubMed] [Google Scholar]
- 58.Ding XQ, Harry CS, Umino Y, Matveev AV, Fliesler SJ, Barlow RB. Impaired cone function and cone degeneration resulting from CNGB3 deficiency: down-regulation of CNGA3 biosynthesis as a potential mechanism. Hum Mol Genet. 2009;18:4770–4780. doi: 10.1093/hmg/ddp440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nishiguchi KM, Sandberg MA, Gorji N, Berson EL, Dryja TP. Cone cGMP-gated channel mutations and clinical findings in patients with achromatopsia, macular degeneration, and other hereditary cone diseases. Hum Mut. 2005;25:248–258. doi: 10.1002/humu.20142. [DOI] [PubMed] [Google Scholar]
- 60.Curcio CA, Millican CL, Allen KA, Kalina RE. Aging of the human photoreceptor mosaic: evidence for selective vulnerability of rods in central retina. Invest Ophthalmol Vis Sci. 1993;34:3278–3296. [PubMed] [Google Scholar]
- 61.Song H, Chui TY, Zhong Z, Elsner AE, Burns SA. Variation of cone photoreceptor packing density with retinal eccentricity and age. Invest Ophthalmol Vis Sci. 2011;52:7376–7384. doi: 10.1167/iovs.11-7199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chui TY, Song H, Clark CA, Papay JA, Burns SA, Elsner AE. Cone photoreceptor packing density and the outer nuclear layer thickness in healthy subjects. Invest Ophthalmol Vis Sci. 2012;53:3545–3553. doi: 10.1167/iovs.11-8694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Curcio CA. Photoreceptor topography in ageing and age-related maculopathy. Eye (Lond) 2001;15:376–383. doi: 10.1038/eye.2001.140. [DOI] [PubMed] [Google Scholar]
- 64.Domdei N, Domdei L, Reiniger JL, et al. Ultra-high contrast retinal display system for single photoreceptor psychophysics. Biomed Opt Express. 2018;9:157–172. doi: 10.1364/BOE.9.000157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kohl S, Jagle H, Wissinger B. Achromatopsia. In: Adam MP, Ardinger HH, Pagon RA, et al., editors. GeneReviews. Seattle, WA: University of Washington, Seattle;; 1993. [PubMed] [Google Scholar]
- 66.Zobor D, Zobor G, Kohl S. Achromatopsia: on the doorstep of a possible therapy. Ophthalmic Res. 2015;54:103–108. doi: 10.1159/000435957. [DOI] [PubMed] [Google Scholar]
- 67.Sheehy CK, Tiruveedhula P, Sabesan R, Roorda A. Active eye-tracking for an adaptive optics scanning laser ophthalmoscope. Biomed Opt Express. 2015;6:2412–2423. doi: 10.1364/BOE.6.002412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Privitera CM, Sabesan R, Winter S, Tiruveedhula P, Roorda A. Eye-tracking technology for real-time monitoring of transverse chromatic aberration. Optics Lett. 2016;41:1728–1731. doi: 10.1364/OL.41.001728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tam J, Roorda A. Speed quantification and tracking of moving objects in adaptive optics scanning laser ophthalmoscopy. J Biomed Opt. 2011;16:036002. doi: 10.1117/1.3548880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chen M, Cooper RF, Han GK, Gee J, Brainard DH, Morgan JI. Multi-modal automatic montaging of adaptive optics retinal images. Biomed Opt Express. 2016;7:4899–4918. doi: 10.1364/BOE.7.004899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Davidson B, Kalitzeos A, Carroll J, et al. Automatic cone photoreceptor localisation in healthy and stargardt afflicted retinas using deep learning. Sci Rep. 2018;8:7911. doi: 10.1038/s41598-018-26350-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bergeles C, Dubis AM, Davidson B, et al. Unsupervised identification of cone photoreceptors in non-confocal adaptive optics scanning light ophthalmoscope images. Biomed Opt Express. 2017;8:3081–3094. doi: 10.1364/BOE.8.003081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cunefare D, Langlo CS, Patterson EJ, et al. Deep learning based detection of cone photoreceptors with multimodal adaptive optics scanning light ophthalmoscope images of achromatopsia. Biomed Opt Express. 2018;9:3740–3756. doi: 10.1364/BOE.9.003740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cunefare D, Fang L, Cooper RF, Dubra A, Carroll J, Farsiu S. Open source software for automatic detection of cone photoreceptors in adaptive optics ophthalmoscopy using convolutional neural networks. Sci Rep. 2017;7:6620. doi: 10.1038/s41598-017-07103-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cunefare D, Cooper RF, Higgins B, et al. Automatic detection of cone photoreceptors in split detector adaptive optics scanning light ophthalmoscope images. Biomed Opt Express. 2016;7:2036–2050. doi: 10.1364/BOE.7.002036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hirji N, Georgiou M, Kalitzeos A, et al. Longitudinal assessment of retinal structure in achromatopsia patients with long-term follow-up. Invest Ophthalmol Vis Sci. 2018;59:5735–5744. doi: 10.1167/iovs.18-25452. [DOI] [PMC free article] [PubMed] [Google Scholar]