Abstract
Background
Gene-microbiome interactions are important in aetiology and pathogenesis of inflammatory bowel disease, a chronic inflammatory disorder of the gastrointestinal tract consisting of Crohn’s disease and ulcerative colitis. Scarce studies on gene-microbiome interactions show very little overlap in their results. Therefore, it is of utmost importance that gene-microbiome studies are repeated. We aimed to replicate the association between the SLC39A8 [Thr]391 risk allele and gut microbiome composition in patients with inflammatory bowel disease and healthy controls.
Methods
We collected faecal samples, peripheral blood and extensive phenotype data from 291 patients with inflammatory bowel disease and 476 healthy controls. Carrier status information was obtained from whole exome sequencing data, generated using the Illumina HiSeq. The gut microbiome composition was determined by tag-sequencing the 16S rRNA gene. Associations between carrier status and disease were tested using the Wilcoxon-Mann-Whitney test. Associations between carriers and gut microbiome composition were determined using principal coordinate analyses, variance explained, alpha diversity and additive general linear models in inflammatory bowel disease, healthy controls and all groups combined.
Results
Crohn’s disease patients were more often carriers of the missense variant (21/171, 12.3%) than controls (30/476, 6.3%) (OR = 2.1, P = 0.01). We could not identify associations between carrier status and overall gut microbiome composition and microbial richness in all tested groups after correcting for potential confounding factors. We did identify 37 different operational taxonomical units to be associated with carrier status among the tested groups. Two of these 37 were identified before in the discovery study.
Conclusions
We could confirm the genetic association of the SLC39A8 [Thr]391 risk allele with Crohn’s disease but we could only limited replicate the association in gut microbiome composition. Independent replication of gene-microbiome studies is warranted to identify true biological mechanisms.
Introduction
Inflammatory bowel disease (IBD) is a common, chronic disorder of the gastrointestinal tract. Patients with this disease experience periods of inflammation alternated by periods of remission. The most common subgroups of IBD are Crohn’s disease (CD), ulcerative colitis (UC) and inflammatory bowel disease undetermined (IBDU)[1].
In the last ten years, large efforts have been made to identify the genomic landscape of patients with IBD by conducting genome-wide association studies (GWAS). These studies have resulted in the discovery of over 200 genomic regions associated with IBD [2–4]. The genes in these genomic regions indicate that gene-microbe interactions underlie key parts of the pathogenesis of IBD. As a result, studies have now begun to unravel the gut microbiome composition in IBD patients [5,6], and IBD research is moving towards integration of the associated genomic regions with the associated changes in gut microbiome composition.
Recently, three independent studies have reported on the integration of the host genome and gut microbiome composition in the general population in Nature Genetics [7–9]. The results of these studies showed very little overlap, probably due to the complexity and variability of the gut microbiome composition. This already starts at the beginning of fecal sampling collection. Differences in collecting methods of these samples or differences in DNA extraction techniques could already lead to differences in results [10]. On top of that, different sequencing techniques were used in these studies; two of them used tag sequencing of the 16S rRNA gene to determine the gut microbiota composition and one study used metagenomics sequencing. Furthermore, over a 100 factors have been identified to be of influence of the gut microbiome composition [11]. In order to make different studies comparable to one another, the same factors should be taken into account. Thus, genome-microbiome associations should be assessed with caution and replication of gene-microbiome associations is certainly warranted [12].
In 2016, Li et al reported the identification of a novel exonic missense variant in the SLC39A8 gene (alanine 391 threonine, rs13107325) that was associated with Crohn’s disease (CD) [13]. Importantly, they also reported that the SLC39A8 [Thr]391 risk variant for CD was associated with the gut microbiome composition in patients with CD and in healthy controls (HC), using microbiome data of 338 mucosal lavage samples from 171 individuals (including patients with CD and HC) [13].
Given the importance of verifying gene-microbiome associations, we aimed to replicate Li et al’s association between the SLC39A8 [Thr]391 risk allele and gut microbiome composition in faecal samples in our Dutch cohort of patients with IBD and HC which is 4.5 times larger than the original cohort [5,13]. Despite the differences in sample collection methods (lavage vs stool samples) we hypothesize that the influence of the SLC39A8 missense variant is also present in our faecal samples, because of the large effect of this missense variant on the gut microbiome composition identified earlier by Li et al [13].
Materials and methods
Cohorts
We included 767 individuals, comprising 171 patients with CD, 104 patients with UC and 16 patients with IBDU from the University Medical Center Groningen (UMCG) IBD cohort and 476 HC from the LifeLines DEEP general population cohort in the Netherlands [5,14]. Faecal samples, peripheral blood, and extensive phenotype data, including age, sex, BMI, and current medication use, were available for all participants. The Institutional Review Board of the University Medical Center Groningen approved both the IBD cohort and the LifelinesDEEP cohort (Institutional Review Board number 2008.338 for the IBD cohort and document number M12.113965 for the LifeLinesDEEP cohort) [5,14].
Genotyping
The determination of the presence of the SLC39A8 missense variant was obtained by using whole exome sequencing (WES). For each participant, peripheral blood samples stored in EDTA tubes (BD Vacutainer) were available. DNA extraction was performed using the Qjagon Autopure LS with Puregene chemistry (Qiagen NV, Venlo, Netherlands). Sample preparation was done by using Illumina Nextera prep kit and enrichment of the exonic sequences was performed by hybrid capture using Illumina rapid Capture Enrichment (37 mb target). The illumina hiSeq platform with 150 bp paired reads was used for sequencing. The mapping of reads to the human genome reference sequence (GRCh37) was performed by using BWA-MEM. An average sequencing depth of 20x covering 80 percent was used in the sequencing of all samples. The average of our obtained reads is 90,655,419 (range 51,590,508–201,639,082) for all the samples. Around 94 percent of the targeted regions were covered by 10 times or more and in the case of 20 times or more this percentage is 87 percent. An extensive quality control and variant calling process has been performed on all samples and is described in a previously published study [15].
Gut microbiome composition
The gut microbiome composition of stool samples was determined using tag-sequencing of the 16S rRNA gene as described previously [5,14]. In short: participants were asked to produce and freeze a stool sample within 15 minutes after production. The samples were collected from the patients’ homes on dry ice and stored in our –80°C freezer. Faecal DNA was isolated by making aliquots and for the isolation of microbial DNA the Qiagen AllPrep DNA/RNA Mini Kit cat #80204 was used. Illumina MiSeq paired-end sequencing of the V4 region of the 16S rRNA gene was performed. The forward primer 515F [GTGCCAGCMGCCGCGGTAA] and the reverse primer 806R [GGACTACHVGGGTWTCTAAT] was used for this step. The microbiome taxonomy was determined by operational taxonomic unit (OTU) picking using QIIME and Usearch (V.7.0.1090) based on similarity of 97%, and Greengenes (V.13.8) was used as a reference database [5]. This led to the identification of 12,556 OTUs. Samples with less than 10,000 counts were removed. In comparison with the discovery paper, the OTUs were filtered based on availability in at least 10 percent of the samples. Lastly, these OTUs were classified into 250 different taxa by summing the read counts and transform them into relative abundances. The codes used for these analyses are publicly available at:
https://github.com/WeersmaLabIBD/Microbiome/blob/master/16s_qiime2_pipeline.md
Overall gut microbiome composition and microbial richness
All statistical analyses were performed in each phenotypic cohort (CD, UC and HC) and all groups combined (all phenotypic cohorts including patients with IBDU). The Wilcoxon-Mann-Whitney test was used to identify the possible association between the presence of the SLC39A8 [Thr]391 risk allele and disease status and the odds ratio was used to determine the quantity of this effect. Overall microbiome composition (beta diversity) was assessed by Bray-Curtis, Jensen-Shannon, Jaccard unweighted Unifrac and weighted Unifrac measurements. Associations between the SLC39A8 [Thr]391 risk allele and the overall composition of the gut microbiome were estimated by calculating the proportion of variance explained by the mutation on the different beta diversity distance matrix, using a PERMANOVA test as implemented in the adonis function of the vegan package in R [16]. Additionally, we analysed the alpha diversity by calculating the Shannon, Chao1 and Simpson index as well as the number of observed species per sample using vegan and phyloseq R packages. Differences in alpha diversity between carriers and non-carriers were analysed through the non-parametric Wilcoxon-Mann-Whitney test. All associations were evaluated with and without taking into account the microbiome confounding factors: age, sex, sequencing depth and for the disease cohort also the disease duration (measured as: years having IBD = fecal sample date—first diagnosis date). For all these tests, p-values of <0.05 were considered statistically significant. The codes are available at:
Association to taxa and OTUs
Possible associations between the individual operational taxonomical units (OTUs) and the SLC39A8 [Thr]391 risk allele status were determined in the described groups by using univariate and multivariate additive general linear models in the software tool MaAsLin as described previously [5]. Covariates in the multivariate models comprised 11 confounding factors known to influence the gut microbiome composition: age, sex, body mass index, proton-pump inhibitor use, as well as antibiotic use and IBD medication (mesalazines, steroids, thiopurines, methotrexate and TNF-α inhibitors) [5]. For patients with CD and UC, the amount of years having the diagnosis IBD was also added as a covariate in the linear models. All analyses were corrected for multiple testing by using the false discovery rate (FDR, Benjamini Hochberg method) incorporated in the Q-package in R. An FDR of <0.05 was considered statistically significant. In the discovery papers, only analyses in individual OTUs correcting for less confounding factors was performed [13]. In this study, we corrected for more factors, to prevent identifying false-positives by the influence of confounding factors.
Results
Clinical characteristics and genetic association
We identified 21 carriers of the SLC39A8 missense variant in patients with CD (12.3%), 7 carriers in patients with UC (6.7%), 1 carrier in patients with IBDU (6.3%) and 30 carriers in HC (6.3%). Because only 1 patient with IBDU was carrier of the missense variant, the differences in the clinical characteristics between carriers and non-carriers of the SLC39A8 [Thr]391 risk allele was not assessed in this group as depicted in Table 1. Smoking is defined as current smoker at time of faecal sampling. Active disease is defined as a score of higher than 4 of the Harvey-Bradshaw index (disease severity measure for CD) or a score higher than 2.5 of the Simple Clinical Colitis Activity Index (disease severity measure for UC). There were no statistically significant differences between carriers and non-carriers of the SLC39A8 missense variant and the clinical characteristics in the described groups (Table 1). We did identify a trend in which carriers of the missense variant have had longer the diagnosis IBD at time of faecal sampling than non-carriers in patients with CD (means of 15 vs 12 years, P = 0.066, as depicted in Table 1). Therefore, we took this factor into account in the analyses of the gene-microbiome interaction. Patients with CD were more often carriers of the SLC39A8 risk allele than HC (OR = 2.1, P = 0.01). This was not the case for patients with UC, nor for patients with IBDU compared to HC (P = 0.8271 and P = 0.9949 respectively).
Table 1. Clinical characteristics of patients with CD, UC and healthy controls.
| Crohn’s disease (n = 171) | Ulcerative colitis (n = 104) | Healthy controls (n = 476) | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Factors | SLC39A8 risk allele carrier | No SLC39A8 risk allele carrier | P-Value | SLC39A8 risk allele carrier | No SLC39A8 risk allele carrier | P-Value | SLC39A8 risk allele carrier | No SLC39A8 risk allele carrier | P-Value |
| Number | 21 | 150 | NA | 7 | 97 | NA | 30 | 446 | NA |
| Age in years (SD) | 42.1 (15.3) | 41.0 (14.1) | 0.7047 | 55 (19.0) | 46.7 (14.3) | 0.159 | 44.4 (13.0) | 45.8 (13.5) | 0.6157 |
| Males (%) | 6 (28.6) | 51 (34.0) | 0.8048 | 3 (42.9) | 47 (48.5) | 1 | 12 (40) | 211 (47.3) | 0.5568 |
| BMI (SD) | 25.7 (5.1) | 24.7 (4.7) | 0.5267 | 26.5 (5.2) | 26.5 (4.4) | 0.9534 | 24.4 (4.0) | 24.9 (3.8) | 0.4268 |
| PPI (%) | 8 (38.1) | 31 (20.1) | 0.09497 | 2 (28.6) | 11 (11.3) | 0.2111 | 1 (3.3) | 19 (4.3) | 1 |
| Antibiotics (%) | 6 (28.6) | 33 (22.0) | 0.5787 | 2 (28.6) | 13 (13.4) | 0.2653 | 0 (0) | 0 (0) | 1 |
| Mesalazines (%) | 0 (0) | 12 (8.0) | 0.3651 | 6 (85.7) | 76 (78.4) | 1 | 0 (0) | 0 (0) | 1 |
| Steroids (%) | 6 (28.6) | 29 (19.3) | 0.3853 | 1 (14.3) | 23 (23.7) | 1 | 0 (0) | 0 (0) | 1 |
| Thiopurines (%) | 7 (33.3) | 50 (33.3) | 1 | 2 (28.6) | 29 (29.9) | 1 | 0 (0) | 0 (0) | 1 |
| Methotrexate (%) | 3 (14.3) | 19 (12.7) | 0.7369 | 0 (0) | 1 (1.0) | 1 | 0 (0) | 0 (0) | 1 |
| Anti-TNFα (%) | 9 (42.9) | 64 (42.7) | 1 | 0 (0) | 10 (10.3) | 1 | 0 (0) | 0 (0) | 1 |
| CRP mean (range) | 2.2 (1–14) | 1.8 (1–13) | 0.3223 | 1.4 (1–3) | 1.4 (1–8) | 0.7179 | NA | NA | NA |
| Fcal mean (range) | 12 (1–67) | 9 (1–89) | 0.665 | 14 (1–62) | 15 (1–390) | 0.7141 | NA | NA | NA |
| Active disease (%) | 8 (38) | 31 (22) | 0.1875 | 2 (29) | 25 (26) | 1 | NA | NA | NA |
| Disease location | |||||||||
| Ileum | 9 (43) | 51 (38) | 0.7163 | 0 (0) | 0 (0) | 1 | NA | NA | NA |
| Colon | 5 (24) | 25 (19) | 0.6782 | 7 (100) | 97 (100) | 1 | NA | NA | NA |
| Both | 7 (33) | 59 (44) | 0.6859 | 0 (0) | 0 (0) | 1 | NA | NA | NA |
| Smoking (%) | 8 (38) | 43 (30) | 0.5916 | 2 (29) | 13 (13.4) | 0.5849 | 14 (24) | 140 (20) | 0.618 |
| Disease duration mean (range) | 15 (1–29) | 12 (1–48) | 0.066 | 10 (2–17) | 11 (1–37) | 0.8301 | NA | NA | NA |
SD standard deviation; BMI body mass index; PPI proton pump inhibitors; Anti-TNFα tumour-necrosis-factor-α inhibitors; CRP C-reactive protein; Fcal Fecal calprotectin; NA not applicable.
Overall gut microbiome composition and microbial richness
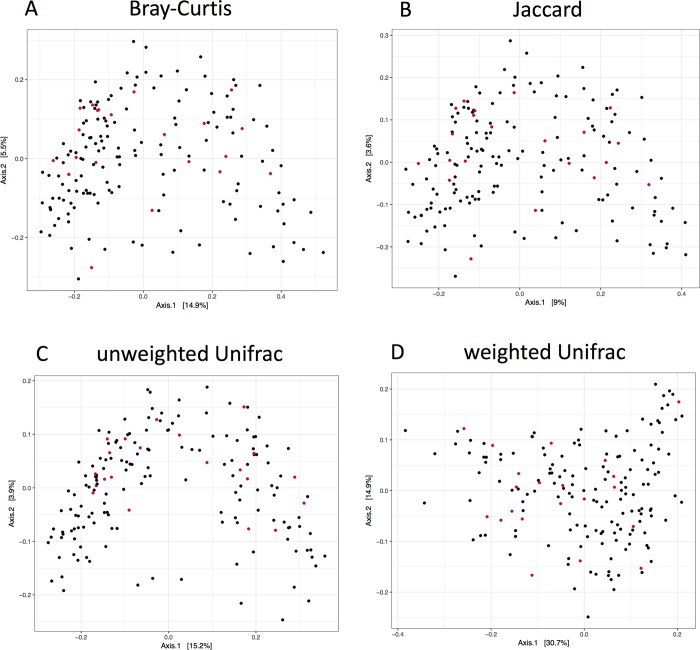
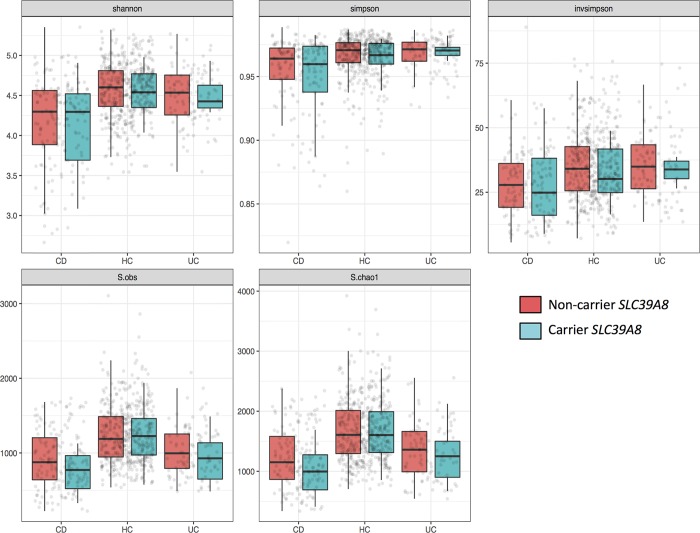
In overall gut microbiome composition, none of the tested groups showed any differential clustering or distribution between mutation and non-mutation carriers in the principal coordinate analyses of beta diversity (Fig 1 for within CD, S1–S3 Figs for within UC, HC and all groups combined). We did identify a statistically significant difference between carrier status and beta diversity in all groups combined in the unweighted Unifrac calculations of beta diversity, without correcting for any potentially confounding factors (P = 0.024, S1 Table). Since the missense variant is enriched in CD, we also added diagnosis in the corrected analysis of beta diversity and carrier status. After correction, carrier status was not statistically significant anymore on beta-diversity in all groups combined using unweighted Unifrac (P = 0.464, S2 Table). Furthermore, we could not identify any statistically significant changes between carrier status of the missense variant and alpha diversity in all tested groups, both in corrected and uncorrected analyses (Fig 2 for within CD, UC and HC, S4 Fig for all groups combined and S3 Table).
Fig 1. Beta diversity within Crohn’s disease by using four methods.
Principal coordinate analysis of gut microbiome composition generated using 16S rRNA sequencing of stool samples of 171 patients with CD. Depicted are four different methods to identify the beta diversity of these samples: A) Bray-Curtis distances, B) Jaccard, C) unweighted Unifrac and D) weighted Unifrac. The 21 SLC39A8 [Thr]391 risk carriers are shown by red dots and 150 non-carriers by black dots. There was no statistically significant association between the SLC39A8 [Thr]391 risk allele and beta diversity identified in CD, nor in the different methods used.
Fig 2. Alpha diversity within CD, UC and HC by using five methods.
Alpha diversity calculated by five different methods, from left to right: Shannon Index, Simpson, inversed Simpson, observed species and Chao1. Carrier status does not show statistically significant differences in non-carriers and carriers of the SLC39A8 missense variants in Crohn’s disease, healthy controls and ulcerative colitis.
Associations individual OTUs and taxa
We were able to identify associations between carrier status of the SLC39A8 missense variant and individual OTUs; 2 OTUs in the univariate analyses and 37 in the multivariate analyses (S4 Table). In the univariate analyses we identified in both CD and UC 1 associated OTU (OTU-IDs = 2210025 and 4374663, FDR = 0.002 and 0.01 respectively) and none in HC or all groups combined. In the multivariate analyses we identified 5 OTUs in CD, 5 in UC, 14 in HC and 14 in all groups combined (S4 Table). For example, the family Lachnospiraceae was statistically significantly increased in all tested groups in SLC39A8 missense variant carriers compared to non-carriers, represented by 10 different OTUs divided over the tested groups. Only 2 of these 10 OTUs (OTU-ID 329703 and 4343184, FDR = 0.02 and 0.03, respectively), were also statistically significantly associated with carrier status in non-overweight healthy controls in the discovery paper [13]. Another example is in patients with UC, HC and all groups combined, in which the family Ruminococcaceae was increased in carriers compared to non-carriers. Ruminococcaceae was represented in 12 different OTUs divided over the mentioned phenotypic cohorts, in which 1 OTU (OTU-ID 195950, FDR = 0.048) showed overlap with the findings of the discovery paper13. From the 877 associated OTUs from the discovery paper [13], we were only able to identify 5 of them to be statistically significantly associated with carrier status of the SLC39A8 missense variant in our cohort (S4 Table). However, we could not identify any statistically significant associations between the SLC39A8 [Thr]391 risk allele and the individual microbial taxa in the tested groups, in either the univariate- or multivariate analysis in which we corrected for covariates.
Discussion
The aim of this study was to replicate the finding of Li et al’s association between the SLC39A8 [Thr]391 risk allele and gut microbiome composition in our independent cohort of patients with CD, UC, IBDU and HC, which was 4.5 times larger than the original cohort. This provides an increased power in order to detect true associations between carriers and microbial changes. The SLC39A8 gene is known as a transporter of Zinc [17]. Zinc deficiency has been associated before with a boost of inflammatory responses and with the increase of oxidative stress, indicating the role of Zinc in immune functions [18,19]. The role of Zinc has also been studied in the context of IBD, in which an in vitro study has shown that Zinc affects the integrity of the intestinal mucosa [20]. In our previous study we have shown multiple factors to be associated with the gut microbiome composition in the context of IBD [5]. Given the large effects of the missense variant on the gut microbiome composition observed by the discovery paper, the association of the missense variant to Crohn’s disease and the role of Zinc on the immune system, we hypothesized that this missense variant could also be of influence in the altered gut microbiome composition in IBD we observed earlier [5,13,17–20].
In this study we could identify the genetic variant to be associated with CD. In addition, the impact on the microbiome composition was limited to a few OTUs, which due differences between boths studies could not be directly compared. However, in OTU identification, we could only replicate 5 OTUs to be associated with carrier status from the discovery paper [13]. When we restrict our analysis to taxonomical level, we could not identify any associations.
Microbial richness and overall gut microbiome composition changes previously reported, could not be replicated. Although the mutation carrier status was statistically significantly associated with beta diversity in the uncorrected analysis of the group all combined by using unweighted Unifrac, after correction for IBD diagnosis, this was not statistically significant anymore. Since the missense variant is enriched in CD, the difference was most likely to be explained by the diagnosis IBD, instead of carrier status of the missense variant. This highlights the importance of considering other factors when performing association studies in IBD context.
We have observed in our results that all identified OTUs were characterized by low mean read counts, ranging from 0–125 (S4 Table). This is also observed in the discovery paper, in which 85% of their identified OTUs also had mean read counts ranging between these values [13]. Therefore, it is very hard to determine if the reported results are indeed positive findings or false positive results. It seems that on top of filtering the minimum prevalence of each taxon (done at 10% in discovery and replication cohort), filtering for minimum abundance of each OTU could reduce the amount of variation between cohorts but also reduce the amount of potentially false positive associations. However, the observed differences between our study and the discovery paper could also be due to the different experimental design [13]. The discovery paper used mucosal lavage samples while the presented study uses faecal content. Although both methods target the same ecosystem, the gut microbiota, different collection methods as well as the use of intestinal preparation for the mucosal lavage can introduce significant changes in the proportion of microbes characterized [21]. In addition, we should also consider the possibility that the findings are cohort specific due to geographical and cultural differences. Furthermore, some extra variation could be introduced by discrepancies in the computational processing and analyses of the samples. Finally, the effect of individual OTUs could be biologically that small, that these effects cannot be detected in alpha and beta diversity.
In the discovery paper of Li et al, large effects of the SLC39A8 missense variant on gut microbiome composition has been described, indicated by the large numbers of associations and effect sizes identified (more than 800 individual OTUs) [13]. In the past, strong signals on the gut microbiota has been replicated, despite different methodologies used. One example of these associations are the one between CD and the increased abundance of the family Enterobacteriaceae. This association has been identified in both adults and paediatric patients with CD, in different sample types (faecal samples and gut mucosal biopsies) and in multiple disease locations [22–28]. On top of that, our sample size is 4.5 times as large as the discovery study. Taken these considerations into account, we hypothesized that despite different sampling collection methods used, we would expect that the positive gene-microbiome signal could also be reproduced in our faecal samples.
Since IBD research is still in the early phase of discovering genome-microbiome associations, the lack of replication is not uncommon in IBD cohorts or in general population studies. Previous studies on IBD patients, in which functional variants in the mucus layer gene FUT2, the bacterial antigen receptor gene NOD2, and the autophagy gene ATG16L1 were associated with the gut microbiome [25,29,30], could also not be replicated [5]. Nor could the interactions between the variant in NOD2 or the variant in the vitamin D receptor (VDR) and the gut microbiome in the general population be replicated [7–9].
Genome-microbiome associations are hard to discover, since the presumed effect of individual genomic variants on the gut microbiome is small, whereas the effect of environmental factors on the gut microbiome can be much larger. Recently, we reported that the variance of the gut microbiome is partly explained by over 100 phenotypes and environmental factors, including medication use and diet [11]. In addition, correcting for multiple testing in genome-wide, microbiome-wide association studies is complex, since the number of tests is hundreds of times larger than in GWAS because of the addition of hundreds more microbial features.
Conclusions
Therefore, we argue that in future gene-microbiome studies much larger sample sizes, more stringent statistical analyses (especially with regard to mean counts of OTUs and correcting for confounding factors), replication in independent cohorts and elaborate descriptions of the methods used are needed to pinpoint genome-microbiome associations in both IBD and HC.
Supporting information
Principal coordinate analysis of gut microbiome composition generated using 16S rRNA sequencing of stool samples of 104 patients with UC. Depicted are four different methods to identify the beta diversity of these samples: A) Bray-Curtis distances, B) Jaccard, C) unweighted Unifrac and D) weighted Unifrac. The 7 SLC39A8 [Thr]391 risk carriers are shown by red dots and 97 non-carriers by black dots. There was no statistically significant association between the SLC39A8 [Thr]391 risk allele and beta diversity identified in UC, nor in the different methods used.
(TIFF)
Principal coordinate analysis of gut microbiome composition generated using 16S rRNA sequencing of stool samples of 476 healthy controls. Depicted are four different methods to identify the beta diversity of these samples: A) Bray-Curtis distances, B) Jaccard, C) unweighted Unifrac and D) weighted Unifrac. The 30 SLC39A8 [Thr]391 risk carriers are shown by red dots and 446 non-carriers by black dots. There was no statistically significant association between the SLC39A8 [Thr]391 risk allele and beta diversity identified in HC, nor in the different methods used.
(TIFF)
Principal coordinate analysis of gut microbiome composition generated using 16S rRNA sequencing of stool samples of all 767 participants. Depicted are four different methods to identify the beta diversity of these samples: A) Bray-Curtis distances, B) Jaccard, C) unweighted Unifrac and D) weighted Unifrac. The 59 SLC39A8 [Thr]391 risk carriers are shown by red dots and 708 non-carriers by black dots. After correction, there was no statistically significant association between the SLC39A8 [Thr]391 risk allele and beta diversity identified in all groups combined, nor in the different methods used.
(TIFF)
Alpha diversity calculated by five different methods, from left to right: Shannon Index, Simpson, inversed Simpson, observed species and Chao1. Carrier status does not show statistically significant differences in non-carriers and carriers of the SLC39A8 missense variants in all groups combined.
(TIFF)
These analyses were performed in all tested groups: Patients with CD, patients with UC, HC and all combined. A statistically significant difference was only identified between carrier status and beta diversity in all groups combined by using the method unweighted Unifrac without correction of disease status. In patients with CD and UC these analyses were also performed in patients with a BMI < 25.
(XLSX)
These analyses were performed in all tested groups: Patients with CD, patients with UC, healthy controls and all combined. In patients with CD and UC, also disease duration was added in the analyses. By correcting for different factors, carrier status was not associated to changes in beta diversity in all tested groups. This was also the case for all different methods used.
(XLSX)
Carrier status was not associated to changes in alpha diversity. This was the case for all tested groups: Patients with CD, patients with UC, HC and all combined. Also the different methods used for calculating alpha diversity, and correcting or not correcting for potential confounding factors led to the same result.
(XLSX)
A total of 2 OTUs in the univariate and 37 OTUs in the multivariate analyses were identified to be associated to the SLC39A8 missense variant and individual OTUs. The asterisk indicates OTUs which have also been identified in the discovery paper.
(XLSX)
Acknowledgments
We thank Jackie Senior, science editor at the Department of Genetics, UMCG, Groningen, for editing the final manuscript.
Data Availability
All data used for this study is publicly available for the IBD UMCG cohort. This data can be obtained at the European Genome-Phenome Archive, available at: https://ega-archive.org, study number: EGAS00001002702. Data transfer requests could be send to the contact person: Ruggero Barbieri (r.barbieri@umcg.nl). For the LifeLinesDeep cohort, raw sequencing reads are publicly available upon request for all 16S rRNA sequenced stool samples used in this study. This data can also be obtained at the European Genome-Phenome Archive, available at: https://ega-archive.org, study number: EGAD0001003453. Request for data transfer could be send to the contact person: Jackie Dekens (j.a.m.dekens@umcg.nl). Other data from the LifeLinesDEEP cohort used in this study is not publicly available since this data belongs to LifeLines (third party data). This can only be retrieved from the LifeLines Research Office after writing a proposal and sending it to LLscience@umcg.nl. The website http://www.lifelines.net contains more information about this application process. We hereby confirm that those interested would be able to access these datasets via the directions we have provided and we confirm that others would be able to access these data in the same manner as we did. Lastly we confirm that we did not have any special access privileges that others would not have.
Funding Statement
This work was supported by VIDI grants (grant numbers 016.136.308 and 864.13.013 to RKW and JF) from the Netherlands Organization for Scientific Research (NWO). Sequencing of the control cohort was funded by the Dutch Top Institute Food and Nutrition (grant number GH001 to CW); CW is further supported by an ERC advanced grant (grant number ERC-671274) and a Spinoza award (grant number NWO SPI 92-266). AZ holds a Rosalind Franklin fellowship (University of Groningen) and an ERC Starting Grant (grant number ERC-715772). JF and AZ are also supported by a CardioVasculair Onderzoek Nederland (grant number CVON 2012-03).
References
- 1.Abraham C, Cho JH. Inflammatory bowel disease. N Engl J Med 2009;361:2066–2078 10.1056/NEJMra0804647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jostins L, Ripke S, Weersma RK, Duerr RH, McGovern DP, Hui KY, et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491:119–124 10.1038/nature11582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu J, van Sommeren S, Huang H, Ng SC, Alberts R, Takahashi A, et al. Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat Genet. 2015;47:979–986 10.1038/ng.3359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.De Lange KM, Moutsianas L, Lee JC, Lamb CA, Luo Y, Kennedy NA, et al. Genome-wide association study implicates immune activitation integrin genes in inflammatory bowel disease. Nat Genet. 2017;49:256–261 10.1038/ng.3760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Imhann F, Vich Vila A, Bonder MJ, Fu J, Gevers D, Visschedijk MC, et al. Interplay of host genetics and gut microbiota underlying the onset and clinical presentation of inflammatory bowel disease. Gut. 2018;67:108–119 10.1136/gutjnl-2016-312135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Knights D, Silverberg MS, Weersma RK, Gevers D, Dijkstra G, Huang H, et al. Complex host genetics influence the microbiome in inflammatory bowel disease. Genome Med. 2014;6:107 10.1186/s13073-014-0107-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bonder MJ, Kurilshikov A, Tigchelaar EF, Mujagic Z, Imhann F, Vich Vila A, et al. The effect of host genetics on the gut microbiome. Nat Genet. 2016;48:1407–1412 10.1038/ng.3663 [DOI] [PubMed] [Google Scholar]
- 8.Turpin W, Espin-Garcia O, Xu W, Silverberg MS, Kevans D, Smith MI, et al. Association of host genome with intestinal microbial composition in a large healthy cohort. Nat Genet. 2016;48:1413–1417 10.1038/ng.3693 [DOI] [PubMed] [Google Scholar]
- 9.Wang J, Thingholm LB, Skiecevičienė, Rausch P, Kummen M, Hov JR, et al. Genome-wide association analysis identifies variation in vitamin D receptor and other host factors influencing the gut microbiota. Nat Genet. 2016;48:1396–1406 10.1038/ng.3695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sinha R, Abnet CC, White O, Knight R, Huttenhower C. The microbiome quality control project: baseline study design and future directions. Genome Biol. 2015;16:276 10.1186/s13059-015-0841-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhernakova A, Kurilshikov A, Bonder MJ, Tigchelaar EF, Schirmer M, Vatanen T, et al. Population-based metagenomics analysis reveals markers for gut microbiome composition and diversity. Science. 2016;352:565–569 10.1126/science.aad3369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Benson AL. The gut microbiome–an emerging complex trait. Nat Genet. 2016;48:1301–1302 10.1038/ng.3707 [DOI] [PubMed] [Google Scholar]
- 13.Li D, Achkar JP, Haritunians T, Jacobs JP, Hui KY, D’Amato M, et al. A pleiotropic missense variant in SLC39A8 is associated with Crohn’s disease and human gut microbiome composition. Gastroenterology. 2016;151:724–732 10.1053/j.gastro.2016.06.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tigchelaar EF, Zhernakova A, Dekens JA, Hermes G, Baranska A, Mujagic Z, et al. Cohort profile: LifeLines DEEP, a prospective, general population cohort study in the northern Netherlands: study design and baseline characteristics. BMJ Open. 2015;5:e006772 10.1136/bmjopen-2014-006772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rivas MA, Avila BE, Koskela J, Huang H, Stevens C, Pirinen M, et al. Insights into the genetic epidemiology of Crohn's and rare diseases in the Ashkenazi Jewish population. 2018;14:e1007329 10.1371/journal.pgen.1007329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oksanen J, Blanchet FG, Friendly M, Kindt R, Legendre P, McGlinn D, et al. Vegan: Community Ecology Package (2016)
- 17.Zang ZS, Xu YM, Lau ATY. Molecular and pathophysiological aspects of metal ion uptake by the zinc transporter ZIP8 (SLC39A8). Toxicol Res (Camb). 2016;5:987–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wessels I, Maywald M, Rink L. Zinc as a gatekeeper of immune function. Nutrients. 2017;9:E1286 10.3390/nu9121286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bonaventura P, Benedetti G, Albarède F, Miossec P. Zinc and its role in immunity and inflammation. Autoimmun Rev. 2015;14:277–285 10.1016/j.autrev.2014.11.008 [DOI] [PubMed] [Google Scholar]
- 20.Finamore A, Massimi M, Conti Devirgiliis L, Mengheri E. Zinc deficiency induces membrane barrier damage and increases neutrophil transmigration in Caco-2 cells. J Nutr 2008;138:1664–1670 10.1093/jn/138.9.1664 [DOI] [PubMed] [Google Scholar]
- 21.Tropini C, Moss EL, Merrill BD, Ng KM, Higginbottom SK, Casavant EP, et al. Transient Osmotic Perturbation Causes Long-Term Alteration to the Gut Microbiota. Cell. 2018;173:1742–1754 10.1016/j.cell.2018.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eun CS, Kwak M, Han DS, Lee AR, Park DI, Yang SK, et al. Does the intestinal microbial community of Korean Crohn’s disease patients differ from that of western patients? BMC Gastroen terol. 2016;16:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Quince C, Ijaz UZ, Loman N, Eren AM, Saulnier D, Russell J, et al. Extensive modulation of the fecal metagenome in children with Crohn’s disease during exclusive enteral nutrition. Am J Gastroenterol. 2015;110:1718–1729 10.1038/ajg.2015.357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haberman Y, Tickle TL, Dexheimer PJ, Kim MO, Tang D, Karns R, et al. Pediatric Crohn disease patients exhibit specific ileal transcriptome and microbiome signature. J Clin Invest. 2014;124:3617–3633 10.1172/JCI75436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gevers D, Kugathasan S, Denson LA, Vázquez-Baeza Y, Van Treuren W, Ren B, et al. The treatment-naive microbiome in new-onset Crohn’s disease. Cell Host Microbe. 2014;15:382–392 10.1016/j.chom.2014.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaakoush NO, Day AS, Huinao KD, Leach ST, Lemberg DA, Dowd SE, et al. Microbial dysbiosis in pediatric patients with Crohn’s disease. J Clin Microbiol. 2012;50:3258–3266 10.1128/JCM.01396-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morgan XC, Tickle TL, Sokol H, Gevers D, Devaney KL, Ward DV, et al. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol. 2012;13:R79 10.1186/gb-2012-13-9-r79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Willing BP, Dicksved J, Halfvarson J, Andersson AF, Lucio M, Zheng Z, et al. A pyrosequencing study in twins shows that gastrointestinal microbial profiles vary with inflammatory bowel disease phenotypes. Gastroenterology. 2010;139:1844–1854 10.1053/j.gastro.2010.08.049 [DOI] [PubMed] [Google Scholar]
- 29.Rausch P, Rehman A, Künzel, Häsler R, Ott SJ, Schreiber S, et al. Colonic mucosa-associated microbiota is influenced by an interaction of Crohn disease and FUT2 (Secretor) genotype. Proc Natl Acad Sci U S A. 2011;108:19030–19035 10.1073/pnas.1106408108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sadaghian Sadabad M, Regeling A, de Goffau MC, Blokzijl T, Weersma RK, Penders J, et al. The ATG16L1-T300A allele impairs clearance of pathosymbionts in the inflamed ileal mucosa of Crohn’s disease patients. Gut. 2015;64:1546–1552 10.1136/gutjnl-2014-307289 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Principal coordinate analysis of gut microbiome composition generated using 16S rRNA sequencing of stool samples of 104 patients with UC. Depicted are four different methods to identify the beta diversity of these samples: A) Bray-Curtis distances, B) Jaccard, C) unweighted Unifrac and D) weighted Unifrac. The 7 SLC39A8 [Thr]391 risk carriers are shown by red dots and 97 non-carriers by black dots. There was no statistically significant association between the SLC39A8 [Thr]391 risk allele and beta diversity identified in UC, nor in the different methods used.
(TIFF)
Principal coordinate analysis of gut microbiome composition generated using 16S rRNA sequencing of stool samples of 476 healthy controls. Depicted are four different methods to identify the beta diversity of these samples: A) Bray-Curtis distances, B) Jaccard, C) unweighted Unifrac and D) weighted Unifrac. The 30 SLC39A8 [Thr]391 risk carriers are shown by red dots and 446 non-carriers by black dots. There was no statistically significant association between the SLC39A8 [Thr]391 risk allele and beta diversity identified in HC, nor in the different methods used.
(TIFF)
Principal coordinate analysis of gut microbiome composition generated using 16S rRNA sequencing of stool samples of all 767 participants. Depicted are four different methods to identify the beta diversity of these samples: A) Bray-Curtis distances, B) Jaccard, C) unweighted Unifrac and D) weighted Unifrac. The 59 SLC39A8 [Thr]391 risk carriers are shown by red dots and 708 non-carriers by black dots. After correction, there was no statistically significant association between the SLC39A8 [Thr]391 risk allele and beta diversity identified in all groups combined, nor in the different methods used.
(TIFF)
Alpha diversity calculated by five different methods, from left to right: Shannon Index, Simpson, inversed Simpson, observed species and Chao1. Carrier status does not show statistically significant differences in non-carriers and carriers of the SLC39A8 missense variants in all groups combined.
(TIFF)
These analyses were performed in all tested groups: Patients with CD, patients with UC, HC and all combined. A statistically significant difference was only identified between carrier status and beta diversity in all groups combined by using the method unweighted Unifrac without correction of disease status. In patients with CD and UC these analyses were also performed in patients with a BMI < 25.
(XLSX)
These analyses were performed in all tested groups: Patients with CD, patients with UC, healthy controls and all combined. In patients with CD and UC, also disease duration was added in the analyses. By correcting for different factors, carrier status was not associated to changes in beta diversity in all tested groups. This was also the case for all different methods used.
(XLSX)
Carrier status was not associated to changes in alpha diversity. This was the case for all tested groups: Patients with CD, patients with UC, HC and all combined. Also the different methods used for calculating alpha diversity, and correcting or not correcting for potential confounding factors led to the same result.
(XLSX)
A total of 2 OTUs in the univariate and 37 OTUs in the multivariate analyses were identified to be associated to the SLC39A8 missense variant and individual OTUs. The asterisk indicates OTUs which have also been identified in the discovery paper.
(XLSX)
Data Availability Statement
All data used for this study is publicly available for the IBD UMCG cohort. This data can be obtained at the European Genome-Phenome Archive, available at: https://ega-archive.org, study number: EGAS00001002702. Data transfer requests could be send to the contact person: Ruggero Barbieri (r.barbieri@umcg.nl). For the LifeLinesDeep cohort, raw sequencing reads are publicly available upon request for all 16S rRNA sequenced stool samples used in this study. This data can also be obtained at the European Genome-Phenome Archive, available at: https://ega-archive.org, study number: EGAD0001003453. Request for data transfer could be send to the contact person: Jackie Dekens (j.a.m.dekens@umcg.nl). Other data from the LifeLinesDEEP cohort used in this study is not publicly available since this data belongs to LifeLines (third party data). This can only be retrieved from the LifeLines Research Office after writing a proposal and sending it to LLscience@umcg.nl. The website http://www.lifelines.net contains more information about this application process. We hereby confirm that those interested would be able to access these datasets via the directions we have provided and we confirm that others would be able to access these data in the same manner as we did. Lastly we confirm that we did not have any special access privileges that others would not have.