Abstract
Alterations in gut microbiota are known to affect intestinal inflammation and obesity. Antibiotic treatment can affect weight gain by elimination of histone deacetylase (HDAC) inhibitor–producing microbes, which are anti-inflammatory by augmenting regulatory T (Treg) cells. We asked whether mice that lack HDAC6 and have potent suppressive Treg cells are protected from microbiota-induced accelerated weight gain. We crossed wild-type and HDAC6-deficient mice and subjected the offspring to perinatal penicillin, inducing weight gain via microbiota disturbance. We observed that male HDAC6-deficient mice were not protected and developed profoundly accelerated weight gain. The antibiotic-exposed HDAC6-deficient mice showed a mixed immune phenotype with increased CD4+ and CD8+ T-cell activation yet maintained the enhanced Treg cell–suppressive function phenotype characteristic of HDAC6-deficient mice. 16S rRNA sequencing of mouse fecal samples reveals that their microbiota diverged with time, with HDAC6 deletion altering microbiome composition. On a high-fat diet, HDAC6-deficient mice were depleted in representatives of the S24-7 family and Lactobacillus but enriched with Bacteroides and Parabacteroides; these changes are associated with obesity. Our findings further our understanding of the influence of HDACs on microbiome composition and are important for the development of HDAC6 inhibitors in the treatment of human diseases.—Lieber, A. D., Beier, U. H., Xiao, H., Wilkins, B. J., Jiao, J., Li, X. S., Schugar, R. C., Strauch, C. M., Wang, Z., Brown, J. M., Hazen, S. L., Bokulich, N. A., Ruggles, K. V., Akimova, T., Hancock, W. W., Blaser, M. J. Loss of HDAC6 alters gut microbiota and worsens obesity.
Keywords: microbiome, HDAC, T-regulatory cells, inflammation
Obesity is a worldwide epidemic (1) and is associated with health disorders that include cardiovascular disease, type II diabetes, and metabolic syndrome (2). In addition to the effects of excessive food intake and altered genetic background, obesity is believed to be a composite phenotype affected by diet and certain intestinal microbiota compositions (3–5).
Germ-free mice cannot completely assimilate food nutrients, indicating the role of the microbiota in nutrition (6). Supplementing the feed of farm animals with low-dose antibiotics enhances growth rate and feeding efficiency (7, 8). Such effects are not observed in germ-free animals (9, 10) and thus are not side-effects of the antibiotics but are due to their effects on the microbiota (11). Our prior studies found similar growth promotion in mice by subtherapeutic dosing of several antibiotics, including penicillin (12), a phenotype that was enhanced by high-fat diet (HFD) (13, 14). Inoculation of germ-free mice with antibiotic-perturbed gut microbiota showed that the accelerated growth rate and adiposity phenotypes were transferable by the microbiota alone (13). This nondiet-based obesity was termed microbe-induced obesity (MIO) (3). More recent studies in mouse models given short antibiotic pulses at therapeutic levels showed profound effects on both the microbiota and the host (15, 16). As such, early-life antibiotic treatment has been postulated to cause shifts in the bacterial composition that in turn affect host development (17), and an inherited antibiotic-perturbed microbiota also might enhance inflammatory phenotypes and disease (18).
Ongoing low-grade inflammation may play a role in the pathogenesis of obesity (19, 20). Regulatory T (Treg) cells, a subset of T-helper lymphocytes, are critical to balancing pro- and anti-inflammatory immune responses (21). Treg cells express the transcription factor Forkhead box P3 (Foxp3), which has a key role in their development and suppressive functions (22, 23). Feeding an HFD to mice activates CD8+ T cells, promoting macrophage accumulation and activation and triggering an inflammatory cascade within visceral adipose tissues (24). By contrast, Treg cells and other suppressive immune cells can ameliorate inflammation within adipose tissue (25, 26). Female mice are better protected against insulin resistance in HFD-fed mice, possibly due to expanded Treg cell populations (27).
The Foxp3 protein is post-translationally regulated by lysine acetylation through histone/protein deacetylases (HDACs), and global HDAC inhibition (28–30) or deletion of HDAC6, -9, or -11 and Sirtuin-1 (31–37) augments Treg cell function, whereas deleting HDAC3 or -5 or Sirtuin-3 has the opposite effect (38–40). The gut microbiota, through production of short-chain fatty acids (SCFAs) that inhibit HDACs (41), thus may be involved in the activation of induced Treg cells in the intestine (42–45). HDAC6 is a class IIb HDAC of therapeutic interest for immunosuppression due to the availability of specific isoform-selective inhibitors (46). We asked whether mice lacking HDAC6 are protected from MIO in the setting of antibiotic perturbation of the microbiota, given the potential of HDAC6 to modulate inflammation due to enhanced Treg-cell suppressive function. Surprisingly, we found a strong paradoxical effect.
MATERIALS AND METHODS
Animal model
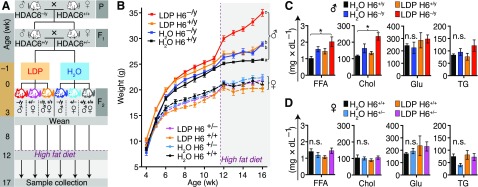
We purchased C57BL/6 mice from The Jackson Laboratory (Farmington, CT, USA) and housed HDAC6 knockout mice in our facility (47). Mice housed under specific pathogen–free conditions were studied using protocols approved by the Institutional Animal Care and Use Committees of the Children’s Hospital of Philadelphia (Protocol 14-001149). We mated female C57BL/6 with male HDAC6−/y mice (parental generation P, Fig. 1A) to generate female heterozygote HDAC6+/− and male HDAC6+/y offspring. The offspring were mated by pairing 20 female HDAC6+/− with 10 HDAC6+/y male mice (F1 in Fig. 1A). A week prior to the expected delivery, the pregnant HDAC6+/− mice were divided randomly to receive either low-dose penicillin (LDP) G potassium (21615, 113-98-4; Cayman Chemicals, Ann Arbor, MI, USA) added to the drinking water as described (12, 13) or standard water (pH 6.8). Based upon the observation that mice consume ∼15 ml of water daily per 100 g body weight (48), we added LDP G at 6.6 mg/L drinking water to achieve 1 μg/g/d/mouse. After the pregnant HDAC6+/− mice delivered, treatment was continued until weaning of the pups, at which time the offspring were genotyped (F2 in Fig. 1A), and the experiment was continued with 8 mice per group. All F2 mice were born within 3 d of each other. Just prior to 12 wk of life, normal chow (25% kcal from fat, 5015; Animal Specialties and Provisions, Quakertown, PA, USA) was changed to an HFD (45% kcal from fat, D12451; Research Diets, New Brunswick, NJ, USA). At 17 wk of life, the mice were euthanized (CO2, cervical dislocation), and samples were harvested. Throughout the experiment, mice had unrestricted access to food and water. Mice were weighed, and fecal samples were collected from weaning until the end of the experiment.
Figure 1.
Male mice lacking HDAC6 gain weight rapidly and develop hyperlipidemia. A) Experimental design and breeding strategy. Study groups were achieved by breeding WT female mice with HDAC6 (H6)-deficient hemizygous male mice (HDAC6−/y) in the parental generation (P). After delivery of the first filial generation (F1), the resulting heterozygous female mice (HDAC6+/−) were inbred with male littermate (HDAC6+/y) siblings. Six randomly chosen F1 dams received LDP (1 μg/g/d) through their drinking water 5 d before delivery and for 3 wk after delivery, and 6 others received water alone. The F2 progeny of each genotype received LDP or control water through their mother’s milk until weaning. After weaning, mice received normal chow for 8 wk and then were switched to an HFD until death at wk 17 of life. B) Mouse weight from weaning to death. The purple dashed line indicates the beginning of HFD. Letters (a–c) indicate subgroups with significantly different weight gain rates phenotype (refer to Supplemental Table 2–5 for details of the statistical analysis). C, D) Serum metabolites. Data from male (C) and female (D) mice. Chol, cholesterol; Glu, glucose; TG, triglycerides. Data shown as means ± sem. All groups were compared with each other. *P < 0.05 (Tukey's post hoc test).
T-cell function and flow cytometry
Leukocytes from spleen and mesenteric lymph nodes were isolated to single cell suspension, and T cells were isolated and cryopreserved as described (34, 49) or stained with flow cytometry antibodies (Supplemental Table 1). For Treg cell–suppression assays, purified CD4+CD25− conventional T cells were labeled with carboxyfluorescein succinimidyl ester (CFSE) (C34554; Thermo Fisher Scientific, Waltham, MA, USA) and stimulated with irradiated antigen-presenting cells plus CD3ε mAb (1 mg/3 ml, 553057, RRID:AB 394590; BD Biosciences, Franklin Lakes, NJ, USA). After 72 h, effector T-cell proliferation was determined by flow cytometric analysis of CFSE dilution. For intranuclear staining, we used a fixation/permeabilization buffer from Thermo Fisher Scientific (88-8823-88). All flow cytometry data were captured using Cyan (Dako, Carpinteria, CA, USA) and Cytoflex (Beckman Coulter, Brea, CA, USA) and analyzed using FlowJo 10.2 software (FlowJo, Ashland, OR, USA).
Serum analysis and metabolomics
Serum glucose, cholesterol, and triglycerides were measured using Infinity Glucose Hexokinase Reagent, Cholesterol Liquid Stable Reagent, and triglycerides reagent (Thermo Fisher Scientific), respectively. Free fatty acids were measured using the Wako nonesterified fatty acids, HR series kit (Wako Life Sciences, Richmond, VA, USA). In short, 2-μl serum samples and standards were placed in duplicate in 96-well microplates and incubated in the dark with the relevant reagent according to the manufacturers’ instructions. Concentrations were calculated based on standard curves. Insulin concentrations were determined by ELISA (EZRMI-13K; MilliporeSigma, Burlington, MA, USA). Stable isotope dilution liquid chromatography with tandem mass spectrometry (LC/MS-MS) was used to quantify circulating trimethylamine N-oxide, choline, carnitine, creatinine, and the branched-chain amino acids valine, leucine, and isoleucine in positive ion multiple reaction monitoring (MRM) mode using characteristic parent to daughter ion transitions: m/z 76.00 → 59.10, m/z 104.00 → 60.15, m/z 161.70 → 103.05, m/z 113.90 → 44.25, m/z 118.00 → 72.20, m/z 132.10 → 30.10, and m/z 132.10 → 69.10, respectively. Stable isotope–labeled internal standards, d9-trimethyl–trimethylamine N-oxide, d9-choline, d3-carnitine, d3-creatinine, u-13C5,15N-valine and u-13C6,15N-leucine (all from Cambridge Isotope Laboratories, Tewksbury, MA, USA) dissolved in methanol were spiked to serum samples to precipitate protein and were similarly monitored in MRM mode at m/z 85.00 → 66.25, m/z 113.10 → 69.20, m/z 165.00 → 103.05, m/z 117.00 → 47.20, m/z 124.00 → 77.15, and m/z 139.30 → 92.35, respectively, as described previously (50, 51). Varying concentrations of standards and a fixed amount of internal standards were spiked into control serum to prepare calibration curves for quantification of serum analytes. Supernatants were analyzed after injection onto a Luna silica column (2.0 × 150 mm, 5 μm; Phenomenex, Torrance, CA, USA) at a flow rate of 0.35 ml/min composed of solvent A, 0.1% propionic acid in water, and solvent B, 0.1% acetic acid in methanol, using a Shimadzu Nexera Ultra High Performance Liquid Chromatograph system interfaced with Shimadzu 8050 Triple Quadrupole Mass Spectrometer (Shimadzu, Kyoto, Japan). Liquid chromatography gradient starting from 100% A over 1 min, then to 10% B over 4 min, then to 50% B over 4 min, and then to 100% B over 0.5 min, followed by 100% B and A washing for 3 min was used to resolve analytes. Spectra were continuously acquired after the initial 2 min.
Histology
Liver specimens were fixed in 10% neutral buffered formalin, processed using standard histologic procedures, and embedded in paraffin. Histologic sections were embedded in paraffin, cut to a thickness of 4 µm, and stained with hematoxylin and eosin. All specimens were reviewed by a single pathologist (B.J.W.) blinded to the experimental conditions using a previously published semiquantitative (0–3) scoring system for mouse models of nonalcoholic fatty liver disease (52). Additional liver samples were processed to undergo staining as a single group with Oil Red O assessment of fat content.
Sample preparation for measuring SCFAs in mouse feces
Water solution of NaOH (1.0 ml, 5 mM) was added to weighed-out mouse fecal pellet in a screw-top tube with an O-ring cap (2 ml). Tubes were placed in a shaker and strongly agitated for 40 min at 4°C to extract SCFAs and centrifuged (17,000 g at 4°C for 20 min). An aliquot (50 µl) of sample supernatant was transferred to a GC vial with insert already containing water solution of NaOH (50 µl, 5 mM) and isotopically labeled internal standards (5 µl). Solution [a mixture of n-butanol/pyridine (3:2, v/v, 50 µl) and isobutyl chloroformate (10 µl)] was added to the samples, and the vials were capped, vortexed (1 min), and sonicated (2 min). Hexane (50 µl) was added, and liquid/liquid extraction was performed by vortexing the samples for 20 min. The samples were centrifuged (20 min, 5000 g), and the organic layer was transferred to another GC vial with insert.
GC/MS analysis
The quantitation of acetic acid, butyric acid, isovaleric acid, lactic acid, propionic acid, and succinic acid was performed using isotope dilution GC-MS/MS by using MRM mode. The absolute quantity of each SCFA was determined using calibration curves measured for each analyte. Measurements were normalized to sample dry weight. This was achieved by separating the stool samples into 2 pieces of equal weight (wet weight): 1 piece was processed for derivatization, and the other piece was dried overnight and then reweighed (dry weight). Dry weight normalization was calculated as (wet weight) × (dry weight/wet weight). Samples were analyzed by using the Thermo TSQ-Evo triple quadrupole in tandem with the Trace 1310 gas chromatograph (Thermo Fisher Scientific). Chromatographic separation was achieved by using an HP-5MS fused-silica capillary column (30 m × 0.250 mm × 0.25 µm; Agilent Technologies, Santa Clara, CA, USA) coated with 5% phenymethyl siloxane. Each extract (1 µl) was injected in split mode (10:1). Helium as carrier gas flow was 1 ml/min. The GC oven temperature program was as follows. The initial temperature of 40°C was held for 2 min after injection before it was increased up to 50°C at 3°C/min1, followed by increase to 110°C at 5°C/min, then 250°C at 30°C/min and 310°C at 70°C/min, and then held at 310°C for 3 min. Argon was used as collision gas. The injector, transfer line, and ion source temperature were set at 260, 290, and 230°C, respectively. The mass spectrometer was tuned to an electron impact ionization energy of 70 eV in the MRM mode with the following parent to daughter ion transitions: m/z 61.0 → 43.0 for acetic acid, m/z 63.0 → 45.0 for [13C2]-acetic acid, m/z 61.0 → 43.0 m/z 71.0 → 41.0 for butyric acid, m/z 78.1 → 46.1 for D7-butyric acid, m/z 85.1 → 57.1 for isovaleric acid, m/z 87.1 → 59.1 for D2-isovaleric acid, m/z 135.1 → 45.1 for lactic acid, m/z 138.1 → 48.0 for D3-lactic acid, m/z 75.1 → 57.0 for propionic acid, m/z 77.1 → 59.0 for D2-propionic acid, m/z 101.1 → 55.0 for succinic acid, and m/z 105.1 → 57.0 for D6-succinic acid.
Microbiome assessment
Most of the mouse fecal samples were collected between 9 am and 12 pm. Genomic DNA was extracted from frozen mouse fecal pellets using the Power-Soil-htp 96 Well Soil DNA Isolation Kit (MoBio, Carlsbad, CA, USA). The V4 region of the bacterial 16S rRNA gene was amplified by triplicate PCR using barcoded fusion primers (53). Amplified samples were quantified using the Quant-iT PicoGreen dsDNA Assay Kit (Thermo Fisher Scientific), pooled at equimolar ratios and purified using the Qiaquick PCR Purification Kit (Qiagen, Hilden, Germany). Libraries were submitted to the NYU Langone Medical Center Genome Technology Center for 151-bp paired-end sequencing on the MiSeq platform (Illumina, San Diego, CA, USA) in a single sequencing run.
Bioinformatic analysis of microbiome sequences
The forward and reverse paired-end reads were joined. Reads with a minimum overlap of 40 nt and with perfect matching of bases between reads were retained, demultiplexed, filtered, and analyzed using Quantitative Insights Into Microbial Ecology (QIIME) (54). Operational taxonomic units (OTUs) were assigned using QIIME’s uclust-based open-reference OTU-picking workflow (55, 56), with a threshold of 97% pairwise identity. To maximize species classification, reference-based OTU picking was performed using a representative subset of the Greengenes bacterial 16S rRNA database (13_8 release) (57, 58). Reads that did not identify to any reference were clustered de novo and aligned once more. Sequences failing alignment or identified as chimeric were removed prior to downstream analysis using ChimeraSlayer (59). Any OTU with <2 occurrences across samples was discarded. Differential taxonomic features were analyzed using the parametric Wald test in the DEseq2 package in R (60). For this analysis, only OTUs that appear at least among 25% of the samples were retained, as per the developer’s recommendations. The significance level cutoff used was 0.01. A principal coordinates analysis plot was generated using the vegan R package (61).
Data analysis
Data were analyzed using Prism 7 software (GraphPad Software, La Jolla, CA, USA). Data were displayed as mean ± sem. Measurements between 2 groups were done with a Student’s t test if normally distributed or otherwise with Mann-Whitney U test. Groups of 3 or more were analyzed by ANOVA if normally distributed or otherwise with the Kruskal-Wallis test. Multiple comparisons were corrected using the Holm approach. Weight gain longitudinal data were analyzed by linear mixed models with likelihood test using the lme4 package (v.1.1.13) (62) in R (v.3.3.2).
RESULTS
Accelerated weight gain in male HDAC6-deficient mice
We hypothesized that the enhanced Treg cell function of HDAC6-deficient mice (31, 33) might provide protection against the inflammatory effects of MIO. To test this hypothesis, we bred littermate HDAC6+/− heterozygote female mice with HDAC6+/y wild-type (WT) male mice (Fig. 1A). The pregnant HDAC6+/− mice, and subsequently their offspring, were subjected to either LDP or H2O control until weaning, initially through their mother’s milk and subsequently in their drinking water (13). The breeding strategy resulted in 3 genotypes: WT (female HDAC6+/+, male HDAC6+/y), female heterozygotes (HDAC6+/−), and male hemizygote (HDAC6−/y), originating from the same HDAC6+/− × HDAC6+/y crosses. Female HDAC6+/− are not heterozygote in the common sense but can develop cells with HDAC6null or WT due to random X-chromosomal inactivation. HDAC6 has been reported to escape X-chromosomal inactivation in some tissues (63). After weaning, mice received normal chow for 8 wk and then were switched to HFD (Fig. 1A).
Using linear mixed models, the first stage of mouse growth on a normal chow diet was fitted as a polynomial and the second stage on HFD as a linear model (Supplemental Tables 2–5). Male mice receiving normal chow showed a significantly higher rate of weight gain compared with female mice (P < 0.0001), with an additional fixed effect of 3.54 ± 0.31 g/wk (Fig. 1B), consistent with prior studies (13). The combined condition of LDP treatment with male sex was significantly enhanced for rate of weight gain in mice receiving normal chow (P < 0.0001), adding 1.8 ± 0.3 g/wk compared with the growth rate of the control group.
Deletion of HDAC6 did not provide protection from MIO; rather, HDAC6−/y mice showed accelerated weight gain independent of LDP treatment (P < 0.0001; Fig. 1B). In male mice receiving HFD, there was a significant interaction between time, treatment, and genotype (P < 0.0001 and Supplemental Table 3). HDAC6 deficiency or LDP treatment alone showed a significantly different weight gain rate over the control HDAC6+/y; LDP-treated HDAC6−/y showed the highest weight gain rate. In female mice, no interactions between time, treatment, and genotype were significant (Supplemental Table 5). Thus, as in prior studies (13, 14), LDP treatment combined with HFD was sufficient to induce accelerated weight gain. In summary, HDAC6−/y mice showed accelerated growth, which was augmented by LDP treatment and HFD.
Male HDAC6-deficient mice develop hyperlipidemia and hepatic steatosis
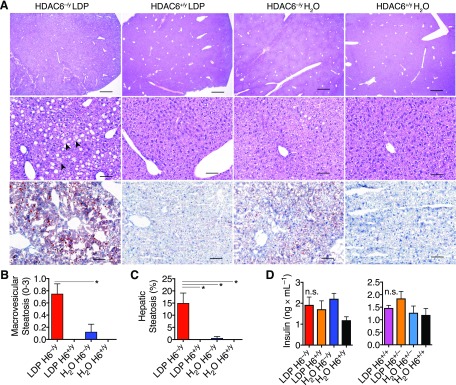
Because hepatic gene expression and metabolism can be altered with early-life antibiotic exposure and HFD (13) and because free fatty acids (FFAs) can regulate HDAC activities (41), we examined how combining exposure to antibiotics and to HFD affected serum metabolic markers. Male LDP-exposed HDAC6−/y mice had significantly higher cholesterol and FFA levels compared with the untreated WT mice 45 d after the switch to HFD (P = 0.032 and 0.019, respectively; Fig. 1C). HDAC6−/y mice not receiving LDP treatment showed a trend toward elevated FFAs and cholesterol. Among female mice, no significant differences were found across all measured metabolites (Fig. 1D). Histologic evaluation of liver samples obtained at the end of the experiment revealed increased macrovesicular hepatic steatosis in the LDP-exposed HDAC6−/y mice (Fig. 2A–C). Serum insulin (Fig. 2D), branched chain amino acids, and additional cardiovascular disease markers were not significantly different (Supplemental Fig. 1). In summary, LDP exposure enhanced the metabolic effects in male HDAC6-deficient mice, consistent with the accelerated growth phenotype.
Figure 2.
Male mice lacking HDAC6 develop hepatic steatosis after exposure to LDP. A–C) Liver histology samples obtained at the end of the experiment reported in Fig. 1A. Top and middle row: hematoxylin and eosin staining. Lower row: Oil Red O lipid staining. LDP-treated HDAC6−/y mice developed mild to moderate hepatic steatosis. At low-power magnification (top row), macrovesicular steatosis is notable. At high-power magnification, microvesicular steatosis is observed within hepatocytes (arrowheads, middle row). Scale bars: 100 µm (upper panels), 1000 µm (middle and lower panels). Representative (A) and quantitative (B, C) data are shown. D) Serum insulin measurements collected at the end of the experiment. Data shown as means ± sem. *P < 0.05 (Kruskal-Wallis test with Dunn’s multiple comparisons test).
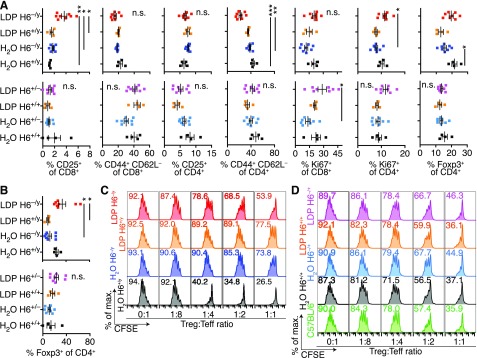
HDAC6−/y Treg cells are up-regulated in spleen and mesenteric lymphocyte populations
The finding that HDAC6−/y mice, which are known to exhibit an immunosuppressive phenotype with strong Treg-cell suppressive capacity (31, 33), developed an enhanced phenotype in an inflammation-related disease model compared with the WT control conditions was initially surprising to us. However, we observed an increase in IL-2 receptor (CD25) expression in CD8+ T cells as well as higher CD4+ T-cell proliferation (Ki67+) in the spleens of LDP-treated HDAC6−/y mice (Fig. 3A, upper panel). This finding is consistent with prior reports that associated accelerated weight gain with CD8+ T-cell tissue invasion and inflammation (64). However, in contrast to the higher T-cell proliferation and CD25 expression, activated (CD44+CD62L−) CD4+ T cells were lowest, and Foxp3+ Treg cells trended higher in the LDP-treated HDAC6−/y mice, despite LDP treatment in HDAC6+/y mice reducing the fraction of Foxp3+ Treg cells. These findings point to a mixed picture of T-cell activation and proliferation (CD25+, Ki67+) but also indicate some counter-regulatory suppression (high Foxp3+ Treg cells, low activated CD44+CD62L− T cells) in the HDAC6−/y mice. In the LDP-treated HDAC6−/y mice, CD8+ T-cell activation was correlated with weight gain (Supplemental Fig. 2). Among the female mice, LDP-treated HDAC6+/− mice showed a slight increase in activated and proliferating CD8+ T cells, but otherwise no differences were observed (Fig. 3A; lower panel). The increase in Foxp3+ Treg cells from LDP-treated HDAC6−/y mice also was noticeable in the mesenteric lymph node populations of HDAC6−/y mice (Fig. 3B). Next, we questioned whether the elevated Foxp3+ Treg cell populations observed in the LDP-treated HDAC6−/y mice had suppressive function. Treg cells were isolated from cryopreserved splenocytes and tested for their ability to suppress C57BL/6 effector T-cell proliferation. To reach sufficient Treg cell numbers for a suppressive function assay, we had to pool our samples into only 1 replicate per condition. This limitation precluded a quantitative comparison between the different groups. However, within the available data, Treg cell function appeared to decline with the increased weight gain phenotypes (Fig. 3C, D). All sampled Treg cells and even the LDP-treated HDAC6−/y mice with the most prominent accelerated weight gain phenotype retained at least some degree of suppressive function (Fig. 3C). In summary, we observed mixed signs of increased effector and cytotoxic T-cell proliferation but also evidence of T-cell suppression and increased Foxp3+ Treg cells in the HDAC6−/y mice, the latter consistent with the previously observed immunosuppressive phenotype in HDAC6-deficient mice.
Figure 3.
Foxp3+ Treg cells were undiminished in LDP-treated HDAC6−/y mice and retain their function. A) Percentage of CD25+, Ki67+, Foxp3+, and CD44+CD62L− cells in splenic CD4+ and CD8+ T cells is shown for male (upper panels) and female mice (lower panels) (see Fig. 1). LDP-treated HDAC6−/y mice exhibited a mixed phenotype of T-cell activation and proliferation (CD25+, Ki67+) but also counter-regulatory suppression (high Foxp3+ Treg cells, low activated CD44+CD62L− T cells). B) Percentage of Foxp3+ among the CD4 population within mesenteric lymph nodes is shown in male (upper panel) and female (lower panel) mice. C, D) CD4+CD25+ Treg cells isolated from the spleens of male (C) and female (D) mice were tested for their ability to suppress the proliferation of CFSE-labeled effector T cells (Teff) isolated from C57BL/6 mice; proliferation was induced using CD3ε mAb and antigen-presenting cells (APC). Flow plots show CD4+ Teff cells, and the number indicates the percentage of proliferating Teff (CFSE-dilution). Teff proliferation declined with increasing Treg/Teff ratios. Percentage of maximum (% of max.) shows normalization of overlaid data and represents the total number of cells in each bin divided by the number of cells in the bin with the most cells. Data shown as means ± sem. *P < 0.05, **P < 0.001, ***P < 0.001 (Kruskal Wallis test followed by Dunn’s multiple comparisons test).
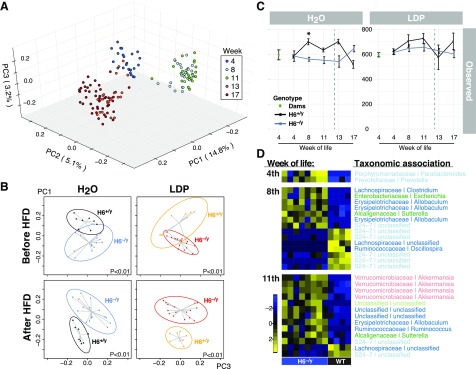
Deletion of HDAC6 is sufficient to alter gut microbiota composition
Initiating an energy-rich diet induces microbiota changes (65, 66). To assess whether LDP treatment or HFD resulted in different microbiota populations in WT or HDAC6-deficient mice, we characterized gut microbiota composition using 16S rRNA gene sequencing of fecal samples. Male mice of all subgroups showed major changes in gut microbial structure from weaning to 8 wk of age and the introduction of HFD (Fig. 4A). In addition, the HDAC6−/y and HDAC6+/y genotypes show distinct bacterial community compositions, preceding the onset of HFD (Fig. 4B). This finding indicates that the deletion of HDAC6 alters bacterial microbiome composition independent of LDP treatment and HFD. Bacterial richness of the 4-wk-old mice is similar to the richness of dams and did not change dramatically throughout the experiment (Fig. 4C). Tracking the appearance of differentiating taxa with time reveals that this phenotype developed with time and did not transfer from the dams in the F1 generation (Fig. 4D) but was acquired de novo in the F2 generation, with the defined genotypes. In female mice, the effects of maturation and diet change were similar to those observed in the male mice (Supplemental Fig. 3A). Untreated female mice receiving HFD did not substantially differ in bacterial community structure between the HDAC6+/+ and HDAC6+/− genotypes (Supplemental Fig. 3B). In summary, we conclude that HDAC6-deficiency was sufficient to induce gut microbiota changes, regardless of diet or antibiotic exposure.
Figure 4.
Bacterial community structure matures and is shaped by HFD. A) Male mice. Principal coordinates analysis (PCoA) representation of the unweighted UniFrac distances of fecal samples. Colors indicate different time points, which represent age of the progeny in week of life. Samples shown for time points 4, 8 and 11 wk of life reflect normal chow, and wk 13 and 17 are for samples collected after switch to HFD. B) The effect of genotype on the microbiota structure. Plots showing PCoA of unweighted UniFrac for male mice by treatment (control vs. LDP) and diet (NC vs. HFD). Ellipsoids represent 90% confidnce intervals. The P values are based on Adonis test. C) Bacterial population richness as “Observed OTUs.” Shown are F1 dams after the end of antibiotic exposure followed by their progeny at different time points. Left panel shows control mice; right panle shows LDP-treated mice. D) Differential abundance OTUs. Heat maps showing comparison between untreated H6+/y and H6−/y from weaning through the 8th and the 11th week of life. Labels show taxonomic association in the format: family genus and color coded by phylum; blue: Bacteroidetes, green: Firmicutes, light blue: Actinobacteria, reed: Tenericutes, pink: Proteobacteria, yellow: Verrucomicrobia. H, HDAC; NC, normal chow.
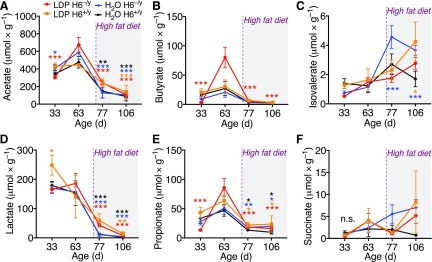
HFD, but not HDAC6 deletion, affects stool metabolite composition
To determine if the differences in weight phenotype and gut microbiota composition were also reflected in key stool metabolites with known immunomodulatory effects, we analyzed serially collected stool samples of male mice via GC/MS-MS. We included samples from male mice collected on d 30, 63, 77, and 106, with d 77 being 1 d after the switch to HFD (Fig. 1A). We observed that the introduction of HFD appeared to have a strong influence on the SCFA acetate, butyrate, isovalerate, propionate, as well as on lactate and succinate (Fig. 5). Acetate, butyrate, and lactate were much reduced after HFD. The alterations in stool metabolites match changes in stool microbial composition (Fig. 4). The loss of lactate matches with our previous reports linking lactate producing bacteria with lean body mass (14) and with the immunosuppressive properties of l- and d-lactate (67). Taken together, the stool metabolites suggest a proinflammatory change in bacterial composition after the introduction of HFD.
Figure 5.
Stool metabolites. GC-MS identifying key metabolites in stool samples collected from male mice: acetate (A), butyrate (B), isovalerate (C), lactate (D), propionate (E), and succinate (F). Introduction of HFD (purple dashed line) strongly affected stool metabolites, consistent with changes in bacterial composition. Asterisks indicate that the mean of this time point is significantly different from the mean of day 63 of the same group (color). Data shown as means ± sem. *P < 0.05, **P < 0.01, and ***P < 0.001 (2-way ANOVA).
Microbiota of HDAC6-deficient mice show a greater level of susceptibility to HFD shift
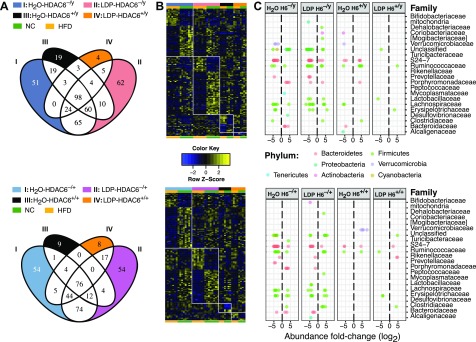
The switch from normal chow to HFD did not show a significant difference in bacterial richness (Fig. 4C); however, changes in stool SCFA, lactate, and succinate were apparent (Fig. 5). We observed major microbiota shifts after HFD with differential OTUs across both sexes and genotypes, involving 427 OTUs in total (Fig. 6 and Supplemental Table 6). One week after starting HFD, HDAC6+/y control mice showed differential abundance of 19 unique OTUs, with enrichment of several Oscillospira-associated OTUs, and relative depletion of Akkermansia, Lachnospiraceae, and Coriobacteriaceae. Bacteroidales family S24-7 OTUs show mixed patterns. The HFD-related decrease in Akkermansia we observed agrees with some (13, 68) but not other (65) prior studies. LDP-treated HDAC6−/y mice show 62 unique differentially abundant OTUs compared with all other studied groups (Fig. 6A, B; top). Among the over-represented OTUs were 3 Lachnospiraceae-associated and 2 Ruminococcaceae-associated OTUs. Under-represented OTUs consist of a wide consortium of bacteria, including many Oscillospira-associated OTUs (Fig. 6C, top). One week after starting HFD, HDAC6+/+ female mice show enrichment of several OTUs, among them Akkermansia and OTUs from the Ruminococcaceae and S24-7 (Fig. 6C; bottom). Immediately after weaning, the gut microbiota of male mice does not show differential OTUs compared with female mice; however, by 11 wk, control male and female mice have several differentiating OTUs (Fig. 7) and keep differentiating while receiving HFD. Exposure to LDP appears to blunt the extent of the differentiation. In summary, the differences in the microbiota due to the dietary shift were more prominent in the genetically modified mice, regardless of whether heterozygote or hemizygote HDAC6 deletion developed with time. Our data indicate that the modulation of HDAC6 dominates over the LDP effect of direct microbiota perturbation in governing the microbiota response to HFD.
Figure 6.
Differentially represented taxa in response to diet switch for all study conditions. A) Venn diagram highlighting the number of unique OTUs found for each study condition with respect to their response to diet switch. B) Heatmap showing comparison across all study conditions of z score normalized abundance in response to diet switch, corresponding to the Venn diagram as seen in A. Color scale emphasizes z score. C) Dot-plot showing the log2 fold-change of condition-specific OTUs for each family-level taxonomy association. OTU taxonomic classification is color coded by phylum. Upper and lower panels show data from male and female mice, respectively.
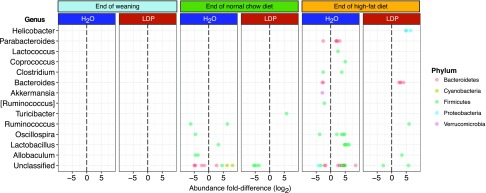
Figure 7.
Treatment-specific gender-differentiating taxa in WT mice receiving HFD. Dot-plot showing the log2 fold-differences in OTUs (within the indicated genera) that are significantly gender specific in their representation. The taxonomic classification of the OTUs/genera is color coded by phylum. In these comparisons, the reference sex is male.
DISCUSSION
We tested the hypothesis that MIO can be modulated through inhibition of HDAC6, expecting that HDAC6 targeting would attenuate inflammation due to augmented Foxp3+ Treg cell numbers and suppressive functions. We reasoned that, because Treg cell activation can ameliorate obesity (27, 69, 70), a strong Treg cell phenotype would protect from MIO. However, we found the opposite, with male mice lacking HDAC6 showing accelerated weight gain. Next, we studied the changes in the gut microbiota composition and gained new insights into the role of HDAC6 in shaping the gut microbiota and in obesity.
Considering the prominence of Foxp3+ Treg cells in HDAC6-deficient or HDAC6 inhibitor–treated mice (31, 33), we closely investigated immune phenotypes in our model. The HDAC6-deficient mice receiving LDP treatment showed numerous Foxp3+ Treg cells, but a high number does not necessarily imply functional suppression. Elimination of commensals such as Lactobacillus or Bifidobacter species promotes inflammation (71) because SCFA and lactate favor Foxp3 expression and acetylation as well as Treg cell development and function (44, 45, 67, 72). Assessing the function of Treg cells isolated from these mice showed that Treg cells isolated from LDP-treated HDAC6−/y mice retained at least some Treg cell–suppressive function. As such, the accelerated weight gain phenotype of LDP-treated HDAC6-deficient mice may not be explained by impaired Treg cell–suppressive function. Loss of HDAC6 and improved Foxp3 acetylation may have compensated for the secondary HDAC effects due to the gut microbiota alterations from LDP or HFD exposure. Therefore, we searched for additional mechanisms to explain the accelerated weight gain in the HDAC6-deficient male mice.
Recently, the role of HDAC6 in lipid metabolism was examined. Loss of HDAC6 in white adipose tissue leads to increased acetylation of the cell death–inducing DNA fragmentation factor subunit α-like effector C (CIDEC) protein, leading to increased lipid droplet storage (73), a change in lipid metabolism that could help explain the accelerated weight gain we saw. The combination of higher free fatty acids with overexpressed CIDEC in an earlier LDP model (13) plus its stabilization by HDAC6 absence provides a mechanism for enhanced lipid droplet accumulation (73), further supported by the hepatic steatosis observed in the HDAC6 deficient mice in this work. However, we also observed that HDAC6 loss alone was sufficient to change gut microbiota composition, independent of other factors. We have carefully controlled the microbiota in our experimental design, using 3 generations of inbreeding and having WT and HDAC6-deficient mice originate from the same mothers to reduce cage effects, which supports the finding that the alterations in the microbiome are HDAC6 dependent. Whether this is due to immunologic or metabolic shifts, such as changes in bile acid biosynthesis, must be considered in future studies. That HDAC6 affects regulation of antimicrobial peptides (13, 74) implies that innate immunity also may be involved. The commonality between early-life male and female pups showed that the later differences did not originate from inheritance of different bacterial communities.
Change to HFD had a further major effect on the microbiota composition, mainly in HDAC6-deficient mice, regardless antibiotic exposure. In prior studies (12, 13), LDP exposure raised intestinal Firmicutes levels during early life, which we confirm. Firmicute-rich microbial populations have been linked to obesity and to increased energy extraction from the diet (75). Erysipelotrichaceae, a family linked to protection against obesity (13), were underrepresented in HDAC6−/y regardless of treatment. After the switch to HFD, we observed several over-represented Oscillospira-associated OTUs, consistent with prior observations (66). Marked differences between WT male and female mice in Oscillospira are intriguing considering their proposed associations with human leanness (76).
This study has several limitations, especially the lack of HDAC6-deficient female mice. Although the 3-generation breeding strategy allowed us to carefully control for maternal microbiota sources, mating female HDAC6+/− with male HDAC6+/y mice only permitted obtaining HDAC6−/y and HDAC6−/+ pups. Alternative breeding in the future can overcome this limitation. Examining intestinal rather than splenic Treg cells may be a better future strategy to assess their function. Pooling spleens to generate sufficient Treg cell numbers for a functional Treg cell–suppression assay precluded any quantitative comparisons between the different experimental groups.
Despite these limitations, this study provides several important insights. We show that HDAC6 deficiency per se is sufficient to alter the gut microbiota and that, despite the enhanced Treg cell–suppression activity, HDAC6-deficient mice are not protected from MIO. Our results also indicate a strong interaction of perturbed obesogenic microbes with HDAC6 deficiency and are thus relevant for the evaluation of HDAC6 inhibitors in the treatment of human diseases.
Supplementary Material
This article includes supplemental data. Please visit http://www.fasebj.org to obtain this information.
ACKNOWLEDGMENTS
This work was supported by U.S. National Institutes of Health (NIH) National Institute of Allergy and Infectious Diseases (NIAID) Grants AI095353 (to U.H.B.), AI073489 and AI095276 (to W.W.H.), NIH National Institute of Diabetes and Digestive and Kidney Diseases Grant DK090989 and NIH NIAID Grant AI122285 (to M.J.B.); by funds from the Laffey McHugh Foundation and American Society of Nephrology (to U.H.B.); and by the C & D Fund and the Transatlantic Network of the Leducq Foundation (to M.J.B. and S.L.H.). The NYU Langone Medical Center Genome Technology Center, supported by the NIH National Cancer Institute (P30CA016087), provided expert DNA sequencing. The authors declare no conflicts of interest.
Glossary
- CFSE
carboxyfluorescein succinimidyl ester
- FFA
free fatty acid
- Foxp3
Forkhead box P3
- HDAC
histone/protein deacetylase
- HFD
high-fat diet
- LDP
low-dose penicillin
- MIO
microbe-induced obesity
- MRM
multiple reaction monitoring
- OTU
operational taxonomic unit
- SCFA
short-chain fatty acids
- Treg
regulatory T
- WT
wild type
Footnotes
This article includes supplemental data. Please visit http://www.fasebj.org to obtain this information.
AUTHOR CONTRIBUTIONS
U. H. Beier, N. A. Bokulich, W. W. Hancock, and M. J. Blaser designed the research; A. D. Lieber, U. H. Beier, H. Xiao, B. J. Wilkins, J. Jiao, X. S. Li, R. C. Schugar, C. M. Strauch, Z. Wang, and T. Akimova performed the research; A. D. Lieber, U. H. Beier, B. J. Wilkins, J. M. Brown, S. L. Hazen, K. V. Ruggles, W. W. Hancock, and M. J. Blaser analyzed the data; U. H. Beier and N. A. Bokulich wrote the study protocol; A. D. Lieber and U. H. Beier wrote the manuscript; C. M. Strauch and Z. Wang contributed text for methods; and W. W. Hancock and M. J. Blaser edited the manuscript.
REFERENCES
- 1.Popkin B. M., Doak C. M. (1998) The obesity epidemic is a worldwide phenomenon. Nutr. Rev. 56, 106–114 10.1111/j.1753-4887.1998.tb01722.x [DOI] [PubMed] [Google Scholar]
- 2.Kopelman P. G. (2000) Obesity as a medical problem. Nature 404, 635–643 10.1038/35007508 [DOI] [PubMed] [Google Scholar]
- 3.Cox L. M., Blaser M. J. (2013) Pathways in microbe-induced obesity. Cell Metab. 17, 883–894 10.1016/j.cmet.2013.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ley R. E., Bäckhed F., Turnbaugh P., Lozupone C. A., Knight R. D., Gordon J. I. (2005) Obesity alters gut microbial ecology. Proc. Natl. Acad. Sci. USA 102, 11070–11075 10.1073/pnas.0504978102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Turnbaugh P. J., Bäckhed F., Fulton L., Gordon J. I. (2008) Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host Microbe 3, 213–223 10.1016/j.chom.2008.02.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bäckhed F., Ding H., Wang T., Hooper L. V., Koh G. Y., Nagy A., Semenkovich C. F., Gordon J. I. (2004) The gut microbiota as an environmental factor that regulates fat storage. Proc. Natl. Acad. Sci. USA 101, 15718–15723 10.1073/pnas.0407076101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jukes T. H., Williams W. L. (1953) Nutritional effects of antibiotics. Pharmacol. Rev. 5, 381–420 [PubMed] [Google Scholar]
- 8.Taylor J. H., Gordon W. S. (1955) Growth-promoting activity for pigs of inactivated penicillin. Nature 176, 312–313 10.1038/176312a0 [DOI] [PubMed] [Google Scholar]
- 9.Coates M. E., Fuller R., Harrison G. F., Lev M., Suffolk S. F. (1963) A comparison of the growth of chicks in the Gustafsson germ-free apparatus and in a conventional environment, with and without dietary supplements of penicillin. Br. J. Nutr. 17, 141–150 10.1079/BJN19630015 [DOI] [PubMed] [Google Scholar]
- 10.Forbes M., Park J. T. (1959) Growth of germ-free and conventional chicks: effect of diet, dietary penicillin and bacterial environment. J. Nutr. 67, 69–84 10.1093/jn/67.1.69 [DOI] [PubMed] [Google Scholar]
- 11.Gaskins H. R., Collier C. T., Anderson D. B. (2002) Antibiotics as growth promotants: mode of action. Anim. Biotechnol. 13, 29–42 10.1081/ABIO-120005768 [DOI] [PubMed] [Google Scholar]
- 12.Cho I., Yamanishi S., Cox L., Methé B. A., Zavadil J., Li K., Gao Z., Mahana D., Raju K., Teitler I., Li H., Alekseyenko A. V., Blaser M. J. (2012) Antibiotics in early life alter the murine colonic microbiome and adiposity. Nature 488, 621–626 10.1038/nature11400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cox L. M., Yamanishi S., Sohn J., Alekseyenko A. V., Leung J. M., Cho I., Kim S. G., Li H., Gao Z., Mahana D., Zárate Rodriguez J. G., Rogers A. B., Robine N., Loke P., Blaser M. J. (2014) Altering the intestinal microbiota during a critical developmental window has lasting metabolic consequences. Cell 158, 705–721 10.1016/j.cell.2014.05.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mahana D., Trent C. M., Kurtz Z. D., Bokulich N. A., Battaglia T., Chung J., Müller C. L., Li H., Bonneau R. A., Blaser M. J. (2016) Antibiotic perturbation of the murine gut microbiome enhances the adiposity, insulin resistance, and liver disease associated with high-fat diet. Genome Med. 8, 48 10.1186/s13073-016-0297-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ruiz V. E., Battaglia T., Kurtz Z. D., Bijnens L., Ou A., Engstrand I., Zheng X., Iizumi T., Mullins B. J., Müller C. L., Cadwell K., Bonneau R., Perez-Perez G. I., Blaser M. J. (2017) A single early-in-life macrolide course has lasting effects on murine microbial network topology and immunity. Nat. Commun. 8, 518 10.1038/s41467-017-00531-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nobel Y. R., Cox L. M., Kirigin F. F., Bokulich N. A., Yamanishi S., Teitler I., Chung J., Sohn J., Barber C. M., Goldfarb D. S., Raju K., Abubucker S., Zhou Y., Ruiz V. E., Li H., Mitreva M., Alekseyenko A. V., Weinstock G. M., Sodergren E., Blaser M. J. (2015) Metabolic and metagenomic outcomes from early-life pulsed antibiotic treatment. Nat. Commun. 6, 7486 10.1038/ncomms8486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Blaser M. J. (2017) The theory of disappearing microbiota and the epidemics of chronic diseases. Nat. Rev. Immunol. 17, 461–463 10.1038/nri.2017.77 [DOI] [PubMed] [Google Scholar]
- 18.Schulfer A. F., Battaglia T., Alvarez Y., Bijnens L., Ruiz V. E., Ho M., Robinson S., Ward T., Cox L. M., Rogers A. B., Knights D., Sartor R. B., Blaser M. J. (2018) Intergenerational transfer of antibiotic-perturbed microbiota enhances colitis in susceptible mice. Nat. Microbiol. 3, 234–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cani P. D., Bibiloni R., Knauf C., Waget A., Neyrinck A. M., Delzenne N. M., Burcelin R. (2008) Changes in gut microbiota control metabolic endotoxemia-induced inflammation in high-fat diet-induced obesity and diabetes in mice. Diabetes 57, 1470–1481 10.2337/db07-1403 [DOI] [PubMed] [Google Scholar]
- 20.Henao-Mejia J., Elinav E., Jin C., Hao L., Mehal W. Z., Strowig T., Thaiss C. A., Kau A. L., Eisenbarth S. C., Jurczak M. J., Camporez J. P., Shulman G. I., Gordon J. I., Hoffman H. M., Flavell R. A. (2012) Inflammasome-mediated dysbiosis regulates progression of NAFLD and obesity. Nature 482, 179–185 10.1038/nature10809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Curotto de Lafaille M. A., Lafaille J. J. (2009) Natural and adaptive foxp3+ regulatory T cells: more of the same or a division of labor? Immunity 30, 626–635 10.1016/j.immuni.2009.05.002 [DOI] [PubMed] [Google Scholar]
- 22.Fontenot J. D., Gavin M. A., Rudensky A. Y. (2003) Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. Nat. Immunol. 4, 330–336 10.1038/ni904 [DOI] [PubMed] [Google Scholar]
- 23.Hori S., Nomura T., Sakaguchi S. (2003) Control of regulatory T cell development by the transcription factor Foxp3. Science 299, 1057–1061 10.1126/science.1079490 [DOI] [PubMed] [Google Scholar]
- 24.Nishimura S., Manabe I., Nagasaki M., Eto K., Yamashita H., Ohsugi M., Otsu M., Hara K., Ueki K., Sugiura S., Yoshimura K., Kadowaki T., Nagai R. (2009) CD8+ effector T cells contribute to macrophage recruitment and adipose tissue inflammation in obesity. Nat. Med. 15, 914–920 10.1038/nm.1964 [DOI] [PubMed] [Google Scholar]
- 25.Feuerer M., Herrero L., Cipolletta D., Naaz A., Wong J., Nayer A., Lee J., Goldfine A. B., Benoist C., Shoelson S., Mathis D. (2009) Lean, but not obese, fat is enriched for a unique population of regulatory T cells that affect metabolic parameters. Nat. Med. 15, 930–939 10.1038/nm.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Winer S., Chan Y., Paltser G., Truong D., Tsui H., Bahrami J., Dorfman R., Wang Y., Zielenski J., Mastronardi F., Maezawa Y., Drucker D. J., Engleman E., Winer D., Dosch H. M. (2009) Normalization of obesity-associated insulin resistance through immunotherapy. Nat. Med. 15, 921–929 10.1038/nm.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pettersson U. S., Waldén T. B., Carlsson P. O., Jansson L., Phillipson M. (2012) Female mice are protected against high-fat diet induced metabolic syndrome and increase the regulatory T cell population in adipose tissue. PLoS One 7, e46057 10.1371/journal.pone.0046057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tao R., de Zoeten E. F., Ozkaynak E., Chen C., Wang L., Porrett P. M., Li B., Turka L. A., Olson E. N., Greene M. I., Wells A. D., Hancock W. W. (2007) Deacetylase inhibition promotes the generation and function of regulatory T cells. Nat. Med. 13, 1299–1307 10.1038/nm1652 [DOI] [PubMed] [Google Scholar]
- 29.Akimova T., Ge G., Golovina T., Mikheeva T., Wang L., Riley J. L., Hancock W. W. (2010) Histone/protein deacetylase inhibitors increase suppressive functions of human FOXP3+ Tregs. Clin. Immunol. 136, 348–363 10.1016/j.clim.2010.04.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu Y., Wang L., Han R., Beier U. H., Hancock W. W. (2012) Two lysines in the forkhead domain of foxp3 are key to T regulatory cell function. PLoS One 7, e29035 10.1371/journal.pone.0029035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.De Zoeten E. F., Wang L., Butler K., Beier U. H., Akimova T., Sai H., Bradner J. E., Mazitschek R., Kozikowski A. P., Matthias P., Hancock W. W. (2011) Histone deacetylase 6 and heat shock protein 90 control the functions of Foxp3(+) T-regulatory cells. Mol. Cell. Biol. 31, 2066–2078 10.1128/MCB.05155-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.De Zoeten E. F., Wang L., Sai H., Dillmann W. H., Hancock W. W. (2010) Inhibition of HDAC9 increases T regulatory cell function and prevents colitis in mice. Gastroenterology 138, 583–594 10.1053/j.gastro.2009.10.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Beier U. H., Wang L., Han R., Akimova T., Liu Y., Hancock W. W. (2012) Histone deacetylases 6 and 9 and sirtuin-1 control Foxp3+ regulatory T cell function through shared and isoform-specific mechanisms. Sci. Signal. 5, ra45 10.1126/scisignal.2002873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Beier U. H., Wang L., Bhatti T. R., Liu Y., Han R., Ge G., Hancock W. W. (2011) Sirtuin-1 targeting promotes Foxp3+ T-regulatory cell function and prolongs allograft survival. Mol. Cell. Biol. 31, 1022–1029 10.1128/MCB.01206-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Levine M. H., Wang Z., Xiao H., Jiao J., Wang L., Bhatti T. R., Hancock W. W., Beier U. H. (2016) Targeting Sirtuin-1 prolongs murine renal allograft survival and function. Kidney Int. 89, 1016–1026 10.1016/j.kint.2015.12.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Akimova T., Xiao H., Liu Y., Bhatti T. R., Jiao J., Eruslanov E., Singhal S., Wang L., Han R., Zacharia K., Hancock W. W., Beier U. H. (2014) Targeting sirtuin-1 alleviates experimental autoimmune colitis by induction of Foxp3+ T-regulatory cells. Mucosal Immunol. 7, 1209–1220 10.1038/mi.2014.10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang J., Wang L., Dahiya S., Beier U. H., Han R., Samanta A., Bergman J., Sotomayor E. M., Seto E., Kozikowski A. P., Hancock W. W. (2017) Histone/protein deacetylase 11 targeting promotes Foxp3+ Treg function. Sci. Rep. 7, 8626 10.1038/s41598-017-09211-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xiao H., Jiao J., Wang L., O’Brien S., Newick K., Wang L. C., Falkensammer E., Liu Y., Han R., Kapoor V., Hansen F. K., Kurz T., Hancock W. W., Beier U. H. (2016) HDAC5 controls the functions of Foxp3(+) T-regulatory and CD8(+) T cells. Int. J. Cancer 138, 2477–2486 10.1002/ijc.29979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang L., Liu Y., Han R., Beier U. H., Bhatti T. R., Akimova T., Greene M. I., Hiebert S. W., Hancock W. W. (2015) FOXP3+ regulatory T cell development and function require histone/protein deacetylase 3. J. Clin. Invest. 125, 3304 10.1172/JCI83084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Beier U. H., Angelin A., Akimova T., Wang L., Liu Y., Xiao H., Koike M. A., Hancock S. A., Bhatti T. R., Han R., Jiao J., Veasey S. C., Sims C. A., Baur J. A., Wallace D. C., Hancock W. W. (2015) Essential role of mitochondrial energy metabolism in Foxp3+ T-regulatory cell function and allograft survival. FASEB J. 29, 2315–2326 10.1096/fj.14-268409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Davie J. R. (2003) Inhibition of histone deacetylase activity by butyrate. J. Nutr. 133 (7 Suppl), 2485S–2493S 10.1093/jn/133.7.2485S [DOI] [PubMed] [Google Scholar]
- 42.Arpaia N., Campbell C., Fan X., Dikiy S., van der Veeken J., deRoos P., Liu H., Cross J. R., Pfeffer K., Coffer P. J., Rudensky A. Y. (2013) Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature 504, 451–455 10.1038/nature12726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Furusawa Y., Obata Y., Fukuda S., Endo T. A., Nakato G., Takahashi D., Nakanishi Y., Uetake C., Kato K., Kato T., Takahashi M., Fukuda N. N., Murakami S., Miyauchi E., Hino S., Atarashi K., Onawa S., Fujimura Y., Lockett T., Clarke J. M., Topping D. L., Tomita M., Hori S., Ohara O., Morita T., Koseki H., Kikuchi J., Honda K., Hase K., Ohno H. (2013) Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells [Erratum]. Nature 504, 446–450 10.1038/nature12721 [DOI] [PubMed] [Google Scholar]
- 44.Smith P. M., Howitt M. R., Panikov N., Michaud M., Gallini C. A., Bohlooly-Y M., Glickman J. N., Garrett W. S. (2013) The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science 341, 569–573 10.1126/science.1241165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rooks M. G., Garrett W. S. (2016) Gut microbiota, metabolites and host immunity. Nat. Rev. Immunol. 16, 341–352 10.1038/nri.2016.42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Butler K. V., Kalin J., Brochier C., Vistoli G., Langley B., Kozikowski A. P. (2010) Rational design and simple chemistry yield a superior, neuroprotective HDAC6 inhibitor, tubastatin A. J. Am. Chem. Soc. 132, 10842–10846 10.1021/ja102758v [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang Y., Kwon S., Yamaguchi T., Cubizolles F., Rousseaux S., Kneissel M., Cao C., Li N., Cheng H. L., Chua K., Lombard D., Mizeracki A., Matthias G., Alt F. W., Khochbin S., Matthias P. (2008) Mice lacking histone deacetylase 6 have hyperacetylated tubulin but are viable and develop normally. Mol. Cell. Biol. 28, 1688–1701 10.1128/MCB.01154-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Harkness J. E., Wagner J. E. (1989) The Biology and Medicine of Rabbits and Rodents, Lea & Febiger, Philadelphia [Google Scholar]
- 49.Akimova T., Levine M. H., Beier U. H., Hancock W. W. (2016) Standardization, evaluation, and area-under-curve analysis of human and murine Treg suppressive function. Methods Mol. Biol. 1371, 43–78 10.1007/978-1-4939-3139-2_4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang Z., Klipfell E., Bennett B. J., Koeth R., Levison B. S., Dugar B., Feldstein A. E., Britt E. B., Fu X., Chung Y. M., Wu Y., Schauer P., Smith J. D., Allayee H., Tang W. H., DiDonato J. A., Lusis A. J., Hazen S. L. (2011) Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature 472, 57–63 10.1038/nature09922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li X. S., Wang Z., Cajka T., Buffa J. A., Nemet I., Hurd A. G., Gu X., Skye S. M., Roberts A. B., Wu Y., Li L., Shahen C. J., Wagner M. A., Hartiala J. A., Kerby R. L., Romano K. A., Han Y., Obeid S., Lüscher T. F., Allayee H., Rey F. E., DiDonato J. A., Fiehn O., Tang W. H. W., Hazen S. L. (2018) Untargeted metabolomics identifies trimethyllysine, a TMAO-producing nutrient precursor, as a predictor of incident cardiovascular disease risk. JCI Insight 3, pii: 99096 10.1172/jci.insight.99096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liang W., Menke A. L., Driessen A., Koek G. H., Lindeman J. H., Stoop R., Havekes L. M., Kleemann R., van den Hoek A. M. (2014) Establishment of a general NAFLD scoring system for rodent models and comparison to human liver pathology. PLoS One 9, e115922 10.1371/journal.pone.0115922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Caporaso J. G., Lauber C. L., Walters W. A., Berg-Lyons D., Huntley J., Fierer N., Owens S. M., Betley J., Fraser L., Bauer M., Gormley N., Gilbert J. A., Smith G., Knight R. (2012) Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 6, 1621–1624 10.1038/ismej.2012.8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Caporaso J. G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F. D., Costello E. K., Fierer N., Peña A. G., Goodrich J. K., Gordon J. I., Huttley G. A., Kelley S. T., Knights D., Koenig J. E., Ley R. E., Lozupone C. A., McDonald D., Muegge B. D., Pirrung M., Reeder J., Sevinsky J. R., Turnbaugh P. J., Walters W. A., Widmann J., Yatsunenko T., Zaneveld J., Knight R. (2010) QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336 10.1038/nmeth.f.303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Navas-Molina J. A., Peralta-Sánchez J. M., González A., McMurdie P. J., Vázquez-Baeza Y., Xu Z., Ursell L. K., Lauber C., Zhou H., Song S. J., Huntley J., Ackermann G. L., Berg-Lyons D., Holmes S., Caporaso J. G., Knight R. (2013) Advancing our understanding of the human microbiome using QIIME. Methods Enzymol. 531, 371–444 10.1016/B978-0-12-407863-5.00019-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Edgar R. C. (2010) Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461 10.1093/bioinformatics/btq461 [DOI] [PubMed] [Google Scholar]
- 57.Caporaso J. G., Bittinger K., Bushman F. D., DeSantis T. Z., Andersen G. L., Knight R. (2010) PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics 26, 266–267 10.1093/bioinformatics/btp636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.DeSantis T. Z., Hugenholtz P., Larsen N., Rojas M., Brodie E. L., Keller K., Huber T., Dalevi D., Hu P., Andersen G. L. (2006) Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 72, 5069–5072 10.1128/AEM.03006-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Haas B. J., Gevers D., Earl A. M., Feldgarden M., Ward D. V., Giannoukos G., Ciulla D., Tabbaa D., Highlander S. K., Sodergren E., Methé B., DeSantis T. Z., Petrosino J. F., Knight R., Birren B. W.; Human Microbiome Consortium (2011) Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res. 21, 494–504 10.1101/gr.112730.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Love M. I., Huber W., Anders S. (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jari Oksanen F. G. B., Friendly M., Kindt R., Legendre P., McGlinn D., Minchin P. R., O’Hara R. B., Simpson G. L., Solymos P., Stevens M. H. H., Szoecs E., Wagner H. (2017) Vegan: Community Ecology Package. Accessed December 31, 2017, at: https://CRAN.R-project.org/package=vegan
- 62.Bates D., Machler M., Bolker B. M., Walker S. C. (2015) Fitting linear mixed-effects models using lme4. J. Stat. Softw. 67, 1–48 10.18637/jss.v067.i01 [DOI] [Google Scholar]
- 63.Berletch J. B., Ma W., Yang F., Shendure J., Noble W. S., Disteche C. M., Deng X. (2015) Escape from X inactivation varies in mouse tissues. PLoS Genet. 11, e1005079 10.1371/journal.pgen.1005079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rausch M. E., Weisberg S., Vardhana P., Tortoriello D. V. (2008) Obesity in C57BL/6J mice is characterized by adipose tissue hypoxia and cytotoxic T-cell infiltration. Int. J. Obes. 32, 451–463 10.1038/sj.ijo.0803744 [DOI] [PubMed] [Google Scholar]
- 65.Carmody R. N., Gerber G. K., Luevano J. M., Jr., Gatti D. M., Somes L., Svenson K. L., Turnbaugh P. J. (2015) Diet dominates host genotype in shaping the murine gut microbiota. Cell Host Microbe 17, 72–84 10.1016/j.chom.2014.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Daniel H., Gholami A. M., Berry D., Desmarchelier C., Hahne H., Loh G., Mondot S., Lepage P., Rothballer M., Walker A., Böhm C., Wenning M., Wagner M., Blaut M., Schmitt-Kopplin P., Kuster B., Haller D., Clavel T. (2014) High-fat diet alters gut microbiota physiology in mice. ISME J. 8, 295–308 10.1038/ismej.2013.155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Angelin A., Gil-de-Gomez L., Dahiya S., Jiao J., Guo L., Levine M. H., Wang Z., Quinn W. J., 3rd, Kopinski P. K., Wang L., Akimova T., Liu Y., Bhatti T. R., Han R., Laskin B. L., Baur J. A., Blair I. A., Wallace D. C., Hancock W. W., Beier U. H. (2017) Foxp3 reprograms T cell metabolism to function in low-glucose, high-lactate environments. Cell Metab. 25, 1282–1293.e7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schneeberger M., Everard A., Gómez-Valadés A. G., Matamoros S., Ramírez S., Delzenne N. M., Gomis R., Claret M., Cani P. D. (2015) Akkermansia muciniphila inversely correlates with the onset of inflammation, altered adipose tissue metabolism and metabolic disorders during obesity in mice. Sci. Rep. 5, 16643 10.1038/srep16643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ilan Y., Maron R., Tukpah A. M., Maioli T. U., Murugaiyan G., Yang K., Wu H. Y., Weiner H. L. (2010) Induction of regulatory T cells decreases adipose inflammation and alleviates insulin resistance in ob/ob mice. Proc. Natl. Acad. Sci. USA 107, 9765–9770 10.1073/pnas.0908771107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Deiuliis J., Shah Z., Shah N., Needleman B., Mikami D., Narula V., Perry K., Hazey J., Kampfrath T., Kollengode M., Sun Q., Satoskar A. R., Lumeng C., Moffatt-Bruce S., Rajagopalan S. (2011) Visceral adipose inflammation in obesity is associated with critical alterations in tregulatory cell numbers. PLoS One 6, e16376 10.1371/journal.pone.0016376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Boulangé C. L., Neves A. L., Chilloux J., Nicholson J. K., Dumas M. E. (2016) Impact of the gut microbiota on inflammation, obesity, and metabolic disease. Genome Med. 8, 42 10.1186/s13073-016-0303-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Poutahidis T., Kleinewietfeld M., Smillie C., Levkovich T., Perrotta A., Bhela S., Varian B. J., Ibrahim Y. M., Lakritz J. R., Kearney S. M., Chatzigiagkos A., Hafler D. A., Alm E. J., Erdman S. E. (2013) Microbial reprogramming inhibits Western diet-associated obesity. PLoS One 8, e68596 10.1371/journal.pone.0068596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Qian H., Chen Y., Nian Z., Su L., Yu H., Chen F. J., Zhang X., Xu W., Zhou L., Liu J., Yu J., Yu L., Gao Y., Zhang H., Zhang H., Zhao S., Yu L., Xiao R. P., Bao Y., Hou S., Li P., Li J., Deng H., Jia W., Li P. (2017) HDAC6-mediated acetylation of lipid droplet-binding protein CIDEC regulates fat-induced lipid storage. J. Clin. Invest. 127, 1353–1369 10.1172/JCI85963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fischer N., Sechet E., Friedman R., Amiot A., Sobhani I., Nigro G., Sansonetti P. J., Sperandio B. (2016) Histone deacetylase inhibition enhances antimicrobial peptide but not inflammatory cytokine expression upon bacterial challenge. Proc. Natl. Acad. Sci. USA 113, E2993–E3001 10.1073/pnas.1605997113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Turnbaugh P. J., Ley R. E., Mahowald M. A., Magrini V., Mardis E. R., Gordon J. I. (2006) An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 444, 1027–1031 10.1038/nature05414 [DOI] [PubMed] [Google Scholar]
- 76.Konikoff T., Gophna U. (2016) Oscillospira: a central, enigmatic component of the human gut microbiota. Trends Microbiol. 24, 523–524 10.1016/j.tim.2016.02.015 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.