Abstract
Carboxypeptidase E (CPE), an exopeptidase involved in proneuropeptide processing, is also a neurotrophic factor, named neurotrophic factor-α1 (NF-α1) and has important roles in neuroprotection, stem cell differentiation, and neurite outgrowth, independent of enzymatic activity. Additionally, an N-terminal–truncated CPE/NF-α1 variant, (CPE/NF-α1)-ΔN, proposed from bioinformatic analysis of GenBank (National Center for Biotechnology Information, Bethesda, MD, USA) DNA sequences and encoding a 40-kDa protein, has been found to be exclusively expressed in embryonic neurons. To investigate the function of (CPE/NF-α1)-ΔN in neurodevelopment, we first cloned (CPE/NF-α1)-ΔN transcripts from an embryonic mouse brain. A rapid amplification of cDNA ends assay, DNA sequencing, and Northern blot revealed 1.9- and 1.73-kb transcripts, which encoded 47- and 40-kDa (CPE/NF-α1)-ΔN proteins, respectively. Those proteins were expressed in embryonic mouse brain. Expression of the 2 (CPE/NF-α1)-ΔN mRNAs surged at embryonic d 10.5, correlating with the time of neurogenesis in the developing brain and also at postnatal d 1. HT22 cells, a mouse hippocampal cell line, transduced with 40 kDa (CPE/NF-α1)-ΔN up-regulated expression of genes involved in embryonic neurodevelopment: insulin-like growth factor binding protein 2 (IGFBP2), death-associated protein 1, and ephrin A1, which regulate proliferation, programmed cell death, and neuronal migration, respectively. HT22 cells and embryonic cortical neurons overexpressing 40 kDa (CPE/NF-α1)-ΔN exhibited enhanced proliferation, which was inhibited by IGFBP2 short interfering RNA treatment. Thus, 40 kDa (CPE/NF-α1)-ΔN has an important, enzymatically independent role in the regulation of genes critical for neurodevelopment.—Xiao, L., Yang, X., Sharma, V. K., Loh, Y. P. Cloning, gene regulation, and neuronal proliferation functions of novel N-terminal–truncated carboxypeptidase E/neurotrophic factor-αl variants in embryonic mouse brain.
Keywords: neurodevelopment, insulin-like growth factor binding protein 2, death associated protein-1, ephrin A1
Neurodevelopment involves many steps including differentiation of progenitors to neurons and glia, axon outgrowth, synaptogenesis, and programmed neuronal cell death. Identifying players that mediate these processes is important for understanding the development of the embryonic nervous system. A molecule that has been shown to be involved in multiple ways in the development of the nervous system is carboxypeptidase E (CPE), also known as neurotrophic factor-α1 (NF-α1) (1). CPE was first found as an exopeptidase that is highly expressed in neuronal and endocrine systems (2, 3). It is a 476-aa protein and contains a signal peptide, a catalytic domain, and a highly acidic C-terminal domain. Intracellularly, CPE has a critical role in prohormone processing by cleaving C-terminal basic amino acids from peptide intermediates to produce bioactive neuropeptides. It also acts as a regulated secretory-pathway sorting receptor for prohormones, including proopiomelanocortin, proinsulin, proenkephalin, and pro–brain-derived neurotrophic factor (2, 4). Interestingly, emerging studies have indicated that CPE also exerts potent neurotrophic effects extracellularly, independent of its enzymatic activity. For example, CPE protected rat hippocampal primary neurons against hydrogen peroxide– and glutamate-induced cell death by activating ERK and AKT signaling cascades, which increased BCL2 and decreased caspase-3 activation (1, 5). In addition, CPE blocked the inhibitive effect of wingless-related integration site 3a (Wnt3a) on nerve growth factor–induced neurite outgrowth by down-regulating β-catenin and rho in both PC12 cells and primary mouse cortical neurons (6). CPE also promoted embryonic neural stem cell differentiation to astrocytes during neurodevelopment via up-regulation of the ERK1/2–Sox9 (SRY-box 9) signaling cascade (7). These effects were observed when a rat nonenzymatically active, mutant CPE form, E300Q, was applied exogenously to cells (5), suggesting that CPE exerts neurotrophic effects independent of its enzymatic activity and has a critical role in neurodevelopment.
Although the functions of CPE have been extensively studied in cell lines and animal models for more than a decade ago, the first clinical case with a CPE mutation was reported, to our knowledge, only in 2015. The 20-yr-old woman, who carried a homozygous mutation of CPE was diagnosed with several medical conditions, including obesity, type 2 diabetes, hypogonadotropic hypogonadism, and intellectual disability characterized by her inability to read or write despite adequate schooling (8). Furthermore, a mutation in the CPE gene was found in a patient with Alzheimer disease, and a mouse model revealed neurodegeneration and hyperphosphorylation of τ in the brain (9). Consistent with the clinical symptoms in humans with CPE mutation, absence of CPE in animal models also produced similar phenotypes. CPE-knockout (KO) mice exhibited obesity, hyperproinsulinemia, infertility, anxiety and depression, and memory deficits (2). The most striking finding in the CPE-KO mouse is the degeneration in the pyramidal neurons in the hippocampal CA3 region after stress (10). Behaviorally, CPE-KO mice displayed profound deficits in memory and learning as evidenced by poor performance in Morris water maze, object preference, and social transmission of food preference (10). In mutant CPEfat/fat mice, lacking CPE, anxiety and depression-like behaviors are very prominent compared with control mice, in addition to obesity and diabetes. Interestingly, studies revealed that rosiglitazone can enhance hippocampal CPE level and neurogenesis in mice, through increasing fibroblast growth factor 2 (FGF2) expression to alleviate the depression-like behavior during stress in mice (11). Furthermore, other neurologic symptoms, such as poor muscle tone was evident in CPE-KO mice (12), and abnormal glutamate-mediated b wave was observed with aging in electroretinography in both CPEfat/fat and CPE-KO mice (13). An increased level of CPE was found in the hippocampal CA1 and CA3 regions in the rat brain after global ischemia (14). Vice versa, mice lacking CPE had exacerbated brain injury induced by ischemia (15). These studies from both clinical patients and animal models suggest that CPE has a critical role in brain function, and mutation of the human CPE gene contributes to neurologic and psychiatric disorders, such as decreased cognitive function and depression.
In humans and other mammals, the CPE gene has 9 exons (16, 17). Alternatively, spliced variants of CPE mRNA have been found in humans (18). One of those variants, has 198 nt spliced out in the first exon and encodes a truncated protein lacking the N-terminal region named 40-kDa CPE-ΔN. The mRNA encoding that 40-kDa CPE-ΔN has been cloned in hepatocellular carcinoma (HCC) cells. Clinical studies have shown that the 40-kDa CPE-ΔN protein is found in cancer cells, such as HCC, but not in healthy tissues. Furthermore, its expression is correlated with high metastatic status (19–21). The 40-kDa CPE-ΔN has been shown to promote proliferation and invasion of HCC cells by translocating them into the nucleus and interacting with the histone deacetylase 1 and 2 to enhance the expression of the metastatic protein neural precursor cell expressed developmentally down-regulated gene 9 (NEDD9) (20).
In mice, a similar transcript, derived from bioinformatic analysis of expressed sequence tags in GenBank (National Center for Biotechnology Information, Bethesda, MD, USA), which encodes a 40-kDa CPE-ΔN protein, has been reported (22). That protein appears to be expressed only in embryonic, but not adult, brain. Transduction of that transcript into primary rat embryonic cortical neurons protected them from glutamate and H2O2-induced apoptosis by up-regulating FGF2 expression (22). That 40-kDa CPE-ΔN was localized to the nucleus of rat E18 cortical and hippocampal neurons transduced with that transcript, suggesting that it may function as a nuclear gene regulator during neurodevelopment. Despite previous studies indicating the existence of a 40-kDa N-terminal–truncated CPE variant that might have important roles in neuroprotection, full characterization of the CPE-ΔN mRNA variants by direct molecular cloning from embryonic mouse brain to define the function of the 40-kDa CPE-ΔN in proliferation during neurodevelopment has not yet, to our knowledge, been performed.
In the present study we cloned mouse wild-type (WT)-CPE, a 40-kDa CPE-ΔN, and a 47-kDa CPE-ΔN mRNA into E14.5 mouse embryonic brain. We detected the 2 CPE-ΔN mRNA variants (1.9 and 1.73 kb in size) by rapid amplification of cDNA ends (RACE) assay and DNA sequencing. Northern blot analysis indicated that the 1.73-kb mRNA encoding the 40-kDa CPE-ΔN protein was more abundant than the 1.9-kb mRNA was. Unlike its human counterpart, the mouse CPE-ΔN mRNAs appeared to be generated by alternative transcription start sites, which encode N-terminal truncated versions of CPE. The function of 40-kDa CPE-ΔN protein in gene regulation and neuronal proliferation was investigated in mouse hippocampal HT22 cells and mouse primary cortical neurons. Several genes involved in neurodevelopment were up-regulated, including insulin-like growth factor binding protein 2 (IGFBP2) in 40-kDa CPE-ΔN–transduced neurons. Moreover, those neurons exhibited enhanced proliferation in a IGFBP2-dependent manner, indicating an important nonenzymatic role for 40-kDa CPE-ΔN in neurodevelopment.
MATERIALS AND METHODS
Animals
All animals were given food and water ad libitum in a humidity- and temperature-controlled room under a 12/12-h light/dark cycle. All animal procedures were approved by the Animal Care and Use Committee, National Institute of Child Health and Human Development, National Institutes of Health (Bethesda, MD, USA). The timed pregnant mice were generated by mating C57BL6 mice in our animal facility.
Cell cultures
HT22 cells were cultured in DMEM (MilliporeSigma, Billerica, MA, USA), supplemented with 10% fetal bovine serum (FBS; Thermo Fisher Scientific, Waltham, MA, USA), 100 U/ml penicillin (Thermo Fisher Scientific), 100 μg/ml streptomycin (Thermo Fisher Scientific), 0.25 μg/ml fungizone (Thermo Fisher Scientific) and 25 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES; Thermo Fisher Scientific) at 37°C in a 5%-CO2 environment with constant humidity. Mouse primary embryonic d 13.5 (E13.5) cortical neurons were prepared, as previously described (22). Timed pregnant mice at gestational d 13.5 were sacrificed by cervical dislocation. Embryos were removed, and the brain cortices were dissected. The cortices were digested with 1 ml of 0.05% trypsin (Thermo Fisher Scientific) for 5 min at 37°C, followed by neutralizing with an equal volume of soybean trypsin inhibitor (MilliporeSigma). The digested cortices were strained with a 40-μm cell filter (Falcon; BD Biosciences, San Jose, CA, USA) and then centrifuged at 1800 rpm for 5 min. The cell pellets were resuspended and cultured in DMEM (MilliporeSigma) with 1× penicillin–streptomycin (Thermo Fisher Scientific) and 10% FBS (Thermo Fisher Scientific), and 24 h later, culture medium was replaced with neurobasal medium with 2% B27 (Thermo Fisher Scientific).
RNA preparation
Total RNAs of mouse tissues and heads of embryos were prepared with Trizol reagent (Thermo Fisher Scientific) and mRNA were isolated with a NucleoTrap mRNA kit (Takara Bio, Kusatsu, Japan), following the manufacturers’ protocol.
Northern blot analysis
A 236-bp mouse CPE probe, covering the middle region (694–929 nt) of mouse CPE mRNA (GenBank accession: NM_001873.2) was labeled with a DIG kit (11585550910; Roche, Mannheim, Germany) by PCR. PCR amplification was performed in a 50-μl volume consisting of 30 ng of WT-mCPE plasmid DNA, 2′-deoxynucleoside 5′-triphosphates (200 μM of dATP, dGTP, dCTP, 130 μM dTTP, and 70 μM of DIG: 11 deoxyuridine triphosphate), 1 U of Takara Hot Start DNA polymerase (R007A; Takara Bio) and 10 pmol of each primer (F694, 5′‑TGAGAAAGAAGGCGGTCCTA‑3′; R929: 5′‑TTTGGAAAATTGCGTCATCA‑3′). The amplification cycles involved an initial “hot start” at 95 °C for 5 min, followed by 31 cycles of amplification (94°C for 30 s; 56°C for 30 s; and 72°C for 40 s) with a final extension step at 72°C for 5 min. Labeled CPE probe was purified by using the Qiaex II Kit (20051; Qiagen, Hilden, Germany), according to manufacturer’s protocol. mRNA samples and DIG-labeled RNA markers (11526529910; F. Hoffmann-La Roche) were run on a denaturing formaldehyde gel and blotted to a nylon membrane with the Northern Max Kit (Ambion brand AM1940; Thermo Fisher Scientific. Membranes were probed overnight at 50°C with the DIG-labeled mCPE probe directed to the mCPE open reading frame (ORF). After 6 washes (described in the Ambion protocol), the membranes were processed for immunodetection by using an anti-DIG-AP antibody (1:10,000; F. Hoffmann-La Roche) and visualized with disodium 3-(4-methoxyspiro {1,2-dioxetane-3,2′-(5′-chloro)tricyclo [3.3.1.13,7]decan}-4-yl)phenyl phosphate (CSPD; 1:100, 11655884001; Roche) against X-film.
R5′/3′–RACE assay
The RACE assay was performed with 5 μg of mRNA isolated from E14.5 mouse brains with the Clontech Smarter RACE kit (Takara Bio), according to manufacturer’s protocol with minor modifications. Briefly, 5 μg of mRNA samples were heated at 70°C for 5 min and quickly cooled on ice before mixing with RT reaction mixture. RT was performed with SMARTScribe Reverse Transcriptase (Takara Bio), a genetically modified Moloney murine leukemia virus in a 20-μl volume containing 5 μg of mRNA, olio dT primer, and 15 U of Smarter II oligonucleotide A (5′-RACE only) at 42°C for 2 h. cDNA was diluted with 240 μl of Tricine-EDTA buffer, and touch-down PCR protocol was used to amplify the 5′ end of mCPE transcript(s) using a kit that provided universal long primer, paired with mCPE specific primer 5′-883 or 3′-634 (5′-883, 5′‑GATTACGCCAAGCTTACCGCTCCGTGTCTCATCATATGGGTA‑3′; 3′-634, 5′‑GATTACGCCAAGCTTCGCCCAGGGAATAGATCTGAACCGTAA‑3′). PCR amplification was performed in a 50-μl volume consisting of 5 μl of diluted cDNA, 1 U of SeqAmp DNA polymerase (Takara Bio), 1 μM of each primer. Touch-down PCR was used to amplify the 5′-end of the CPE gene; cycles involved an initial “hot start” at 95°C for 5 min, followed by 5 cycles of amplification 1 (94°C for 30 s, then 72°C for 3 min), 5 cycles of amplification 2 (94°C for 30 s, 70°C for 30 s, and 72°C for 3 min), and 30 cycles of amplification 3 (94°C for 30 s, 68°C for 30 s, and 72°C for 3 min), with a final extension step of 72°C for 10 min. PCR products were analyzed on 1.8% agarose gels. Bands from 5′/3′-RACE were excised, purified, inserted into the pRACE vector provided in the kit, and sequenced.
Transduction and treatment of HT22 cells and mouse primary cortical neurons
To study the effect of 40-kDa CPE-ΔN on gene regulation, HT22 cells or primary cortical neurons were transduced with adenovirus carrying the 40-kDa CPE-ΔN or the LacZ construct at 10 or 20 multiplicity of infection (MOI) for 24 h. On d 1, 2, 3, and 4 after transduction, RNA and protein were extracted from those cells and evaluated for death-associated protein 1 (DAP1), IGFBP2, and EFNA1 mRNA and protein expression by quantitative RT-PCR (qRT-PCR) and Western blot, respectively. In other experiments, the CPE-ΔN–transduced cells were also treated with IGFBP2 short interfering RNA (siRNA; GE Healthcare Life Sciences, Little Chalfont St Giles, United Kingdom) or scramble RNA using Lipofectamine RNAiMax transfection reagent (Thermo Fisher Scientific) according to manufacturers’ protocols for 24 h and then evaluated for changes in proliferation by 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide (MTT) and Ki-67 assays (see below).
qRT-PCR
Total RNA from HT22 was extracted and purified with the RNeasy Mini Kit (Qiagen), according to the manufacturer’s protocol and then quantified. First-strand cDNA was prepared from 500 ng RNA using the SensiFAST cDNA Synthesis Kit (Bioline, Eveleigh, NSW, Australia). The cDNA was then used for qRT-PCR. The qPCR cycling conditions were denaturation at 95°C for 10 min and 40 cycles of DNA synthesis at 95°C for 15 s and 60°C for 1 min. Primer sequences are as follows: DAP1 forward, 5′‑GGAGATGGGAAGGAAGAGAGA‑3′, reverse, 5′‑TTGTCACCCCGAGCAATAA‑3′; EFNA1 forward, 5′‑ACCAGGTCCGTTGGAATTG‑3′, reverse 5′‑CAAGCACTGGGATTCCTGAT‑3′; IGFBP2 forward, 5′‑CCTTGCCAGCAGGAGTTG‑3′, reverse, 5′‑TCCGTTCAGAGACATCTTGC‑3′; and 18S forward, 5′‑CTCTTAGCTGAGTGTCCCGC‑3′, reverse, 5′‑CTGATCGTCTTCGAACCTCC‑3′. The results were analyzed by SDS 1.9.1 software (Thermo Fisher Scientific). All qPCR experiments were performed in triplicates, and the relative amount of mRNA was normalized to 18S rRNA.
Western blot
For Western blots of embryonic brain, 4–8 brains were collected and pooled for each developmental stage, and postnatal d 1 (P1) brain was extracted in ice-cold lysis buffer (RIPA; Thermo Fisher Scientific) supplemented with protease inhibitor cocktail (MilliporeSigma). Mouse cortical neurons and HT22 cells were extracted in a similar manner. The lysates were collected, centrifuged at 15,000 rpm for 10 min at 4°C, and 20 μg of protein was analyzed by Western blot, according to a protocol previously described by Cheng et al. (5). The membrane was incubated with primary antibodies for CPE-WT and variants: rabbit CPE antibody 6135 (1:2000 dilution), rabbit C-terminal (last 11 aa of CPE) antibody CPH 6–8 (1:2000 dilution), and rabbit N-terminal (1:2000, first 13 aa of mature CPE: QEDGISFEYHRYP) antibody, all generated in our laboratory; and mouse CPE mAb (1:2000; BD, Franklin Lakes, NJ, USA), rabbit anti-IGFBP2, rabbit anti-DAP1 (1:2000; Abcam, , Cambridge, United Kingdom), rabbit anti-EFNA1 antibody (1:2000; St. John’s Laboratory, London, United Kingdom), or anti-tubulin (1:5000; Cell Signaling Technology, Danvers, MA, USA) overnight at 4°C. For absorption control for 6135 antibody, 20 μg of antigen peptide was incubated with 10 μl antibody at 4°C overnight. The membrane was then incubated with donkey anti-rabbit or anti-mouse antibodies conjugated with IRDye 700 or 800, respectively (1:5000; Rockland Immunochemicals, Limerick, PA, USA), and protein bands were visualized and quantified by the Odyssey Infrared Imaging System and software v.2.1 (Li-Cor Biosciences, Lincoln, NE, USA). The protein expression level for each sample was normalized to β-actin (1:10,000; MilliporeSigma).
CPE/CPE-ΔN protein expression, purification, and enzymatic assay
To check the enzymatic activity of 40-kDa CPE-ΔN, COS-7 cells were infected with His-tagged CPE-WT (20 MOI), His-tagged 40-kDa CPE-ΔN (40 MOI), or control adenovirus (20 MOI) for 48 h. These cells were lysed in 50 mM sodium acetate, pH 5.5, 0.01% Triton X-100, and 1 mM PMSF (MilliporeSigma). The samples were centrifuged at 100,000 rpm for 15 min, and the supernatants were saved in fresh tubes. Western blot was performed to confirm the expression of CPE-WT and CPE-ΔN protein in extracts of cells infected with those proteins, but not in cells infected with adenovirus alone (data not shown). The COS-7 lysates, containing ∼500 ng protein, were incubated with 2 μg ACTH (1–17) peptide (Bachem, Bubendorf, Switzerland) for 24 h at 37°C in 50 mM sodium acetate, pH 5.5, with, and without, cobalt chloride (2 mM) and/or the CPE inhibitor guanidinoethylmercaptosuccinic acid (GEMSA; 10 μM; MilliporeSigma). The elution of cleaved ACTH peptide products (ACTH 1–16, 1–15, 1–14 standard peptides; Bachem) were analyzed on a reverse-phase Jupiter C18 HPLC column (Phenomenex, Torrance, CA, USA) by monitoring the absorbance at 214 nm. Buffer A (0.1% TFA) and buffer B (80% acetonitrile/0.1% TFA) were used, and the gradient was run 30–34% B in 12 min (for details of the assay see Cong et al. (23). These experiments were also performed at pH 7.4 in Tris-HCL buffer.
For quantitative comparison of the enzymatic activity of CPE-WT and 40-kDa CPE-ΔN, these proteins were purified from COS-7 cells infected with His-tagged CPE-WT (20 MOI) or 40-kDa CPE-ΔN (40 MOI) adenovirus for 72 h. The infected cells were harvested and washed with PBS and lysed in binding buffer (20 mM sodium phosphate, 300 mM NaCl, 10 mM imidazole, 10% glycerol, pH 7.0) by sonication. Lysates were centrifuged at 17,000 g for 30 min, and the supernatants were incubated with Ni-NTA resin (Qiagen) for 1 h at 4°C and then put into a column. Columns were washed with 20 column volumes of wash buffer (20 mM sodium phosphate, 20 mM imidazole, pH 7.0) to remove any unbound proteins. His-tagged proteins were eluted with (20 mM sodium phosphate, 250 mM imidazole, pH 7.0) elution buffer. Imidazole buffer was removed from eluted proteins by using 3-kDa Centricon centrifugal filter units (Qiagen). SDS-PAGE and Western blotting confirmed expression and purification of CPE-WT and 40-kDa CPE-ΔN. Enzymatic activity of purified His-tagged CPE-WT and 40-kDa CPE-ΔN was tested using water-soluble (chloroform-insoluble) dansyl-Phe-Ala-Arg (custom synthesized from Biomatik, Cambridge, ON, Canada) as substrate, a method developed by Fricker (24). CPE converts the substrate into the chloroform-soluble dansyl-Phe-Ala product. The fluorescence generated by that chloroform phase represents the enzymatic activity as relative fluorescence units. In our experiments, 40 ng purified CPE-WT or 100 ng CPE-ΔN proteins were incubated with 100 µl of dansyl-Phe-Ala-Arg substrate (0.5 mM) in 100 mM sodium acetate buffer (pH 5.5) for 16 h at 37°C. Similar experiments in the presence of 1 mM CoCl2 or 1 µM GEMSA were also performed, and fluorescence was recorded at excitation (360/40 nm filter) and emission (528/20 nm filter).
MTT assay
Mouse primary cortical neurons or HT22 cells were plated on a 96-well poly-l-lysine–coated plate and transduced with adenovirus containing CPE-ΔN or LacZ constructs (10 and 20 MOI, respectively) for 24 h. That transduction was followed by IGFBP2 siRNA or scramble RNA treatment for 24 h. On d 1, 2, 3, 4, and 5 after siRNA or scramble RNA treatment, 10 µl of 5 mg/ml Thiazolyl Blue Tetrazolium Bromide (MTT; MilliporeSigma) solution was added in each well, followed by incubation at 37°C for 3 h. Then, the medium was removed, and 100 µl/well DMSO (MilliporeSigma) was added and incubated at room temperature for 10 min. Absorbance value was measured at 490 nm using a Synergy HT microplate reader (BioTek Instruments, Winooski, VT, USA).
Ki-67 assay
Because Ki-67 is exclusively expressed during active phases of the cell cycle, it is well known as a marker for cell proliferation (25). For the Ki-67 assay, mouse primary cortical neurons or HT22 cells were plated on poly-l-lysine–coated chamber slides, transduced with adenovirus containing CPE-ΔN or LacZ constructs (10/20 MOI), and treated with IGFBP2 siRNA or scramble RNA. Those cells were gently washed with 1× PBS, followed by fixation with 4% paraformaldehyde in PBS for 15 min. Cells were then washed with PBS and blocked with PBS containing 10% donkey serum and 0.3% Triton X-100. After removing fixation solution, the cells were incubated with rabbit anti-Ki67 antibody (1:2000; Abcam) diluted in 3% serum-PBS at 4°C overnight. The cells were then rinsed and incubated with secondary Alexa Fluor 594 antibody (1:2000; Thermo Fisher Scientific) for 1 h at room temperature. After washing with PBS, the cells were stained with DAPI and then mounted with mounting medium (Thermo Fisher Scientific). Images of primary cortical neurons or HT22 were visualized and acquired under a fluorescence microscope (×20; Eclipse 80i; Nikon, Tokyo, Japan). Nine fields for each experimental condition were imaged, and ∼12 cells in each field were counted, with total of ∼108 cells counted in each group.
Microarray analysis of HCCL cells transfected with mouse CPE-ΔN vs. HCCL control
Cell lines
The human HCC cell line, MHCC97L was obtained from the Liver Cancer Institute, Fudan University (Shanghai, China). HCC cells were plated at a density of 40–50% and allowed to grow to 75% confluence overnight in DMEM (ATCC, Manassas, VA, USA) supplemented with 10% FBS.
Transfection and RNA isolation
A plasmid construct containing CPE-ΔN ORF was transiently overexpressed in low-metastatic HCC cells (MHCC97L). Transfected and control MHCC97L were harvested, and total RNA was isolated using SV Total RNA Isolation kits (Promega, Madison, WI, USA), which included a DNase treatment. RNA was quantified by nanodrop spectrophotometer, and RNA integrity was analyzed with an 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). Samples having a registrant identification number >9.0 were used for genome-wide gene expression analysis performed by SABiosciences-Qiagen (Fredrick, MD, USA). All samples were in triplicates.
Microarray analysis
Genome-wide gene expression levels in transfected and untransfected HCC cells were quantified with the HumanHT-12 v4 Expression BeadChip (Illumina, San Diego, CA, USA). Probe intensity and gene expression data were analyzed with Illumina GenomeStudio software (version 2011.1; Gene Expression Module, v.1.9.0). In brief, 250 ng total RNA was used for cDNA synthesis, followed by an amplification/labeling step (in vitro transcription) to synthesize biotin-labeled cRNA according to the MessageAmp II aRNA Amplification kit (Thermo Fisher Scientific). An RNA Nano Chip Assay on an Agilent 2100 Bioanalyzer was used for quality control of cRNA. A Bead Station Array Scanner (Illumina) was used for microarray scanning at a setting adjusted to a scaling factor of 1 with photomultiplier tube (PMT) settings at 430. Data extraction was performed for all beads individually, and outliers are removed when >2.5 median absolute deviation. All remaining data points were used for the calculation of the mean average signal for a given probe, and the sd for each probe was calculated. All arrays were quantile-normalized without background subtraction with the Illumina BeadStudio software. The Illumina probes were annotated with the custom mappings from the Bioconductor (Seattle, WA, USA) R package (R Foundation for Statistical Computing, Vienna, Austria) Illumina HumanHT-12 (v.4) annotation data. Probes that were flagged as “bad,” that is, matching repeat sequences, intergenic, or intronic regions, and those having “No match,” that is, without match for any genomic region or transcript, according to the Illumina Human (v.4) Probe Quality mapping, were discarded. The remaining probes were annotated to Entrez identifications provided by the Illumina Human (v.4) Entrezre Annotated map. The “find largest” function from the R Bioconductor package genefilter resolved the problem if multiple probes matched the same Entrez gene.
Statistical analysis
Data are representative of 3 separate experiments with each experiment performed in triplicate to obtain the means ± sem. Data were analyzed by 2-tail Student’s t test and 1-way ANOVA, followed by Tukey’s post hoc multiple comparisons tests, where noted. Significance was set at P < 0.05.
RESULTS
Novel CPE transcripts revealed by Northern blotting analysis
To determine an overall expression profile of CPE mRNAs in the CNS of embryos during embryonic development and to search for mCPE transcript variant(s), Northern blotting analysis (Fig. 1A) was performed with RNAs from the head of embryos at various developmental stages (E8.5, E10.2, E12.5, E14.5, and E16.5) as well as postnatal d 1 (P1) cortex. For comparison, adult mouse hippocampus, heart, lung, and liver were also analyzed. We observed 3 transcripts with sizes ranging from ∼1.73 to 2.1 kb in heads of embryos and P1 cortex. Among the 3 transcripts, the major one appears to be ∼2.1 kb in size, which is approximately the same size as recorded in GenBank database (accession NM_013494.4), and we regarded this one as the CPE-WT band; the other 2 smaller bands, present in much lower amounts, however, were not, to our knowledge, described previously, and we hypothesized that they were novel CPE transcript variants. Consistent with previous studies, CPE-WT is expressed abundantly in embryonic and adult brain, whereas, in other organs, such as adult liver, lung, and heart, its expression is weak or undetectable. Furthermore, a negative control using P1 cortex from CPE-KO mice showed no CPE-WT or variant transcripts (Supplemental Fig. S1). We observed that levels of CPE transcripts varied among different embryonic stages, the first surge of CPE-WT expression appeared at E10.5, and it gradually decreased and resurged at P1; the 2 smaller transcripts displayed a very similar expression pattern as CPE-WT (as shown in Fig. 1A).
Figure 1.
Identification of CPE transcript variants in embryonic mouse brain. A) Northern blot analysis of RNA isolated from the head of embryos at various developmental stages (E8.5, E10.5, E12.5, E14.5, and E16.5) and from P1 cortex, P1 hippocampus, and adult mouse hippocampus, heart, lung, and liver. The membrane was hybridized to a digoxigenin (DIG)-labeled CPE probe. Lanes 1 and 13, DIG-labeled RNA marker; lane 2, E8.5 hippocampus; lane 3, E10.5; lane 4, E12.5; lane 5, E14.5; lane 6, E16.5; lane 7, P1 cortex; lane 8, P1 hippocampus; lane 9, adult liver; lane 10, adult lung; lane 11, adult heart; and lane 12, adult hippocampus. Three CPE transcripts (2.1, 1.9, and 1.73 kb) are detected as indicated by the arrows. B, Upper) Schematic illustration of the relationships of primers used in the RACE PCRs. Numbers refer to the position of the primers. B, Lower) 5′/3′-RACE PCRs were performed with mRNA isolated from E14.5 mouse brain. Lane 1, 100-bp DNA marker; lane 2, 5′-883 RACE PCR; lane 3, 1-kb DNA marker; and lane 4, 3′-634 RACE PCR. C) The nucleotide sequences of the full-length mRNAs and the deduced proteins of mouse CPE-WT and the 2 variants. Left) Nucleotide sequences of the full-length mRNAs, boldface italic, shaded letters indicated with arrowheads are transcription starting sites of CPE-WT, 1.9 and 1.73 kb CPE transcripts. Arrows above boxed letters indicate potential translation start sites at 110, 296, and 446 nt used by CPE-WT, 1.9 and 1.73 kb CPE transcript variant, respectively. Right) Deduced amino acid sequences of the 3 CPE transcripts.
RACE assay reveals WT-CPE and 2 CPE-ΔN transcripts in embryonic mouse brain
To obtain full length cDNA of the mCPE gene, RACE PCR was performed with mouse E14.5 brain cDNA as a template. The strategy employed for RACE-PCR cloning is shown in Fig. 1B; there was a 250-bp overlap between the PCR products. Specific 3′-RACE-PCR product was amplified with primer 3′-634, which showed a single band on an agarose gel. This result indicates that the 3 CPE transcripts share the same C terminus. However, when the 5′-RACE-PCR products were amplified with primer 5′-883, 3 distinct bands, ranging from 600–1000 bp, were detected. Sequencing results from 5′/3′-RACe products indicate that the 3′-end of mCPE is intact, whereas the 5′-RACE-PCR products differ only in size (as shown in Fig. 1C). Sizes of these combined 5′/3′-RACE fragments (2.1, 1.9, and 1.73 kb) approximately match the 3 CPE transcripts observed in the Northern analysis. This suggests that different 5′ transcription-initiation sites might be used to generate the 3 CPE transcripts.
To understand the coding potential of these transcripts identified for the mouse CPE gene, we analyzed the potential ORFs using the online tool, ORF finder (https://www.ncbi.nlm.nih.gov/orffinder/). The consensus sequence of the 3 transcripts contains 3 potential ORFs that overlap with each other. The 2.1-kb transcript encodes a 476-aa polypeptide which is identical to the WT mCPE. The 1.73-kb transcript encodes 364-aa polypeptide, which is a truncated form of CPE lacking the N-terminal signal peptide with a theoretical MW of 40 kDa. The 1.9-kb CPE transcript, however, might use an alternative CTG starting codon (26) to encode a 418-aa polypeptide with a theoretical MW of 47 kDa; we, therefore, designated the 2 CPE variants as 47-kDa CPE-ΔN and 40-kDa CPE-ΔN, respectively (Fig. 1A).
CPE-ΔN variants expressed in mouse embryonic brain and P1 cortex
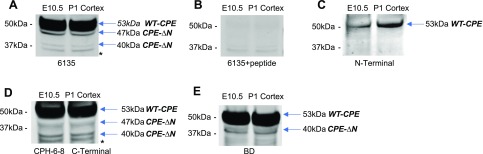
To determine whether the CPE mRNA variants, are translated into proteins, Western blots were performed. Because Northern blot studies showed expression of the highest levels of CPE mRNA variants at E10.5 and P1 cortex, CPE proteins at those 2 time points were evaluated. The Western blots (Fig. 2A) showed a 53-, a 47-, and a 40-kDa CPE immunoreactive band using the 6135 rabbit antibody, corresponding to the size of CPE-WT and the 2 CPE-ΔN variants expected from the estimation of MW from the RACE and Northern blot data (Fig. 1). Those bands disappeared when the 6135 antiserum that was absorbed by the antigen peptide was used, verifying the specificity of the immunoreactive CPE bands (Fig. 2B). The 53-, 47-, and 40-kDa immunoreactive CPE bands were also observed with a C-terminal–specific antibody (CPH6–8) in E10.5 brain and P1 cortex (Fig. 2D), indicating they have the same C-terminal sequence. In addition, a N-terminal antibody detected only in the 53-kDa CPE-WT (Fig. 2C), consistent with the identification of the 47- and 40-kDa protein as CPE-ΔN variants. The 53- and 40-kDa bands were detected by a third antibody (BD) (Fig. 2E). A negative control using P1 cortex from CPE-KO mice showed no expression of CPE proteins (Supplemental Fig. S2). These results suggest that CPE mRNAs detected in the Northern blot and RACE assays are translated into proteins consistent with the estimated sizes of 53, 47, and 40 kDa, representing WT-CPE and 2 CPE-ΔN forms, respectively.
Figure 2.
Expression of CPE and CPE-∆N protein in mouse E10.5 brain and P1 cortex. Western blots were performed with several different antibodies. A) The 6135 rabbit antibody showed CPE-WT at 53 kDa and CPE-∆N variants at 47 and 40 kDa. B) All these bands were blocked in the 6135 peptide absorption control. C) The N-terminal antibody revealed only 53-kDa CPE-WT. D) The CPH6–8 rabbit C-terminal antibody also revealed the 53-kDa CPE-WT and the 2 CPE-∆N variants at 47 and 40 kDa. E) The BD mouse mAb showed 53-kDa CPE-WT and the 40-kDa CPE-∆N variant.
40-kDa CPE-ΔN has weak enzymatic activity at pH 5.5
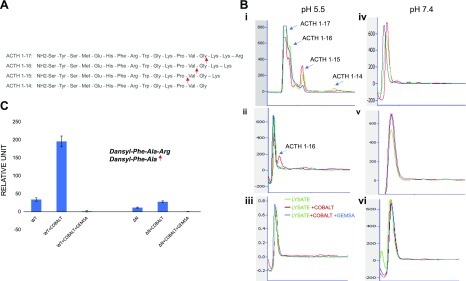
Because 40-kDa CPE-ΔN contains the enzymatic pocket, we determined whether it has enzymatic activity. Cell lysates containing CPE-WT, CPE-ΔN, and control cell lysates were incubated with ACTH (1–17), with and without cobalt or GEMSA (Fig. 3). In sodium acetate buffer at pH 5.5, lysates containing CPE-WT generated ACTH peptides (1–16), (1–15), and (1–14) representing sequential C-terminal cleavages of the basic residues of the substrate [ACTH (1–17), (Fig. 3A)], which was stimulated by cobalt and inhibited by GEMSA, a known inhibitor for CPE (Fig. 3Bi). In contrast, lysates containing 40-kDa CPE-ΔN generated only 1 cleavage product with cobalt stimulation, showing that CPE-ΔN has some enzymatic activity (Fig. 3Bii). No enzymatic activity was observed in the lysate from control adenovirus-infected cells (Fig. 3Biii). At pH 7.4 (Tris-HCL buffer), no activity was observed with CPE-WT or 40-kDa CPE-ΔN, as shown in Fig. 3Biv–vi. To compare the enzymatic activity between CPE-WT and 40-kDa CPE-ΔN quantitatively, we assayed the activity of the purified enzymes using dansyl-Phe-Ala-Arg as substrate. The enzymatic activity of CPE-ΔN (11.66 fluorescence units/100 ng protein) was found to be 34.01% of CPE-WT (34.16 fluorescence units/100 ng protein), and it was stimulated by cobalt and inhibited by GEMSA (Fig. 3C). Assay of buffer alone as a blank showed <2 fluorescence units. These results indicate that 40-kDa CPE-ΔN is unlikely to function as an enzyme because it has low enzymatic activity and only at pH 5.5, and it is localized in the cytoplasm and nucleus, where the pH is neutral. Instead, CPE-WT is localized in secretory vesicles that have an acidic internal pH to carry out neuropeptide processing.
Figure 3.
The enzymatic activity of CPE-ΔN was analyzed by HPLC. A) Sequence of ACTH (1–17)–(1–14) showing cleavage sites for CPE at C-terminal basic amino acid residues. B) Lysates from CPE-WT, CPE-ΔN, or control adenovirus-infected COS-7 cells were incubated for 24 h with ACTH (1–17), with and without cobalt and/or GEMSA, and the enzymatically processed products were analyzed by HPLC. At pH 5.5, CPE-WT generated different peptide products: ACTH (1–16), ACTH (1–15), and ACTH (1–14), which were induced by cobalt and inhibited by GEMSA (i). CPE-ΔN generated only 1 peptide product, ACTH (1–16) (ii); whereas control lysate did not show any peptide product (iii). At pH 7.4, no product peaks were observed for any lysate from CPE-WT (iv), ΔN–CPE (v), or control adenovirus-infected COS-7 cells (vi). C) Enzymatic activity of purified CPE-WT or CPE-ΔN was measured with dansyl-Phe-Ala-Arg as the substrate. CPE-WT (40 ng) and CPE-ΔN (100 ng) were incubated with dansyl-Phe-Ala-Arg (0.5 mM) for 16 h at 37°C in the presence or absence of CoCl2 and GEMSA, and fluorescence was recorded as excitation (360/40 nm filter) and emission (528/20 nm filter). Values shown in bar graph are corrected for 100-ng protein (CPE-WT). means ± sd, n = 2.
40-kDa CPE-ΔN up-regulates expression of IGFBP2, DAP1, and EFNA1, genes in HT22 neurons
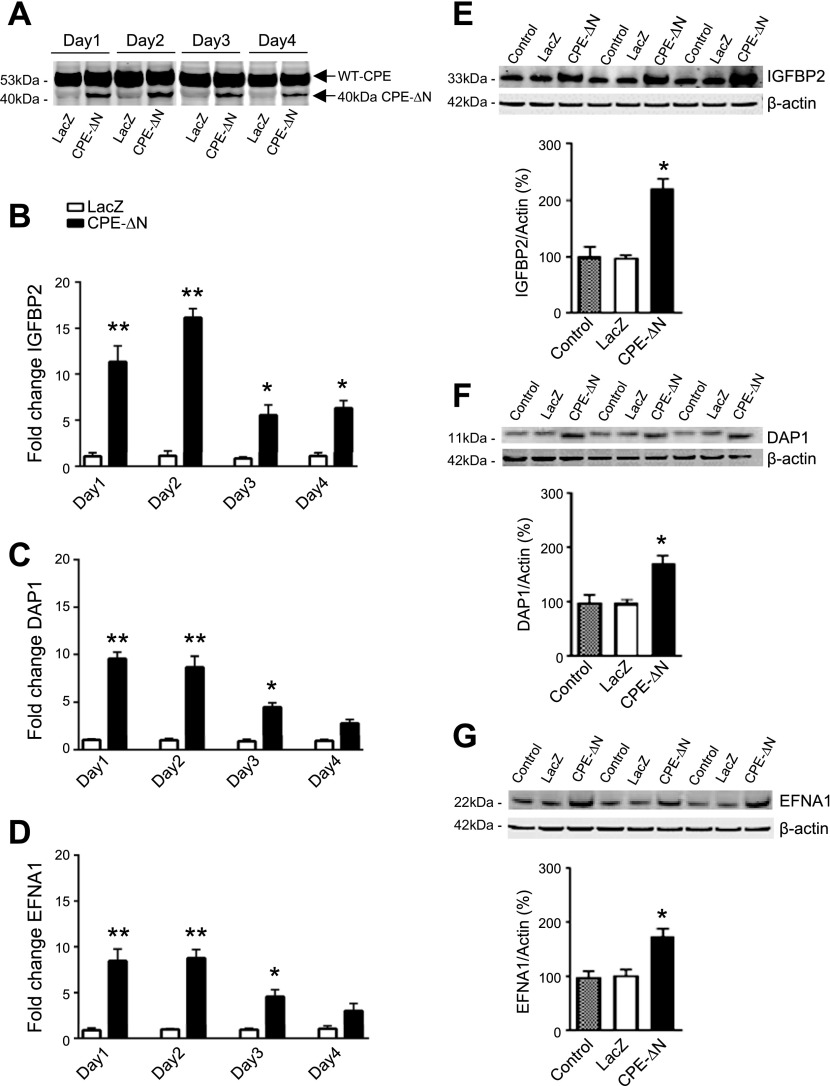
Previous studies have indicated that CPE-ΔN functions by binding to a nuclear complex to regulate gene expression in cancer cells (20). Because it is well known that oncology recapitulates ontology, we performed a microarray study comparing HCC cells transfected with mouse 40-kDa CPE-ΔN vs. control cells, to screen for neurodevelopmentally important genes that might be regulated by 40-kDa CPE-ΔN. The genes that were up-regulated or down-regulated by at least 2-fold in the microarray analysis are shown by the bar graphs in Supplemental Figs. S3 and S4. The 2 most highly up-regulated genes were IGFBP2 and DAP1, both known to have a role in neurodevelopment. Of the genes that were down-regulated (Supplemental Fig. S4), the EFNA1 or Ephrin 1 is known to bind to the EphA2 receptor to mediate neuronal migration during neurodevelopment. Interestingly, the EphA2 mRNA was up-regulated in HCC cells (Supplemental Fig. S3). We, therefore, chose to determine whether expression of IGFBP2, DAP1, and EFNA1 genes was regulated by 40-kDa CPE-ΔN in the hippocampal neuronal cell line HT22. Western blot in Fig. 4A shows that HT22 cells transduced with 40-kDa CPE-ΔN adenovirus at 10 MOI exhibited an increase in the level of 40-kDa CPE-ΔN protein from d 1 to 4 compared with cells transduced with LacZ adenovirus. The qRT-PCR assay showed that the expression of IGFBP2, DAP1, and EFNA1 mRNAs was up-regulated on d 1 and 2, and then gradually returned to normal levels in the 40-kDa CPE-ΔN-transduced HT22 cells (Fig. 4B–D). The protein levels of IGFBP2 (Fig. 4E), DAP1 (Fig. 4F), and EFNA1 (Fig. 4G) were also significantly increased by 123.71, 72.95, and 68.53% on d 1 in those 40-kDa CPE-ΔN-transduced HT22 cells, in parallel with the changes in mRNA levels of those molecules.
Figure 4.
The 40-kDa CPE-∆N up-regulates expression of IGFBP2, DAP1, and EFNA1 genes in HT22 neurons. A) Western blot showing 40-kDa CPE-∆N expression and LacZ control in HT22 neurons transduced with 10 MOI virus. B–D) Bar graphs showing fold-change of mRNA determined by qRT-PCR for IGFBP2 (B), DAP1 (C), and EFNA1 (D), on d 1–4 in HT22 cells after CPE-∆N transduction with 10 MOI virus. E–G) Western blots of protein expression and bar graphs showing quantification of IGFBP2 (E), DAP1 (F), and EFNA1 (G) protein expression, on d 1 after transduction of 10 MOI 40-kDa CPE-∆N virus. Transduction of 40-kDa CPE-∆N significantly enhanced protein expression of IGFBP2, DAP1, and EFNA1 on d 1. Quantification of protein expression was normalized against β-actin. Values are means ± sem. Student’s t test, n = 3. *P < 0.05, **P < 0.01 compared with control.
40-kDa CPE-ΔN promotes HT22 cell proliferation via up-regulation of IGFBP2 expression
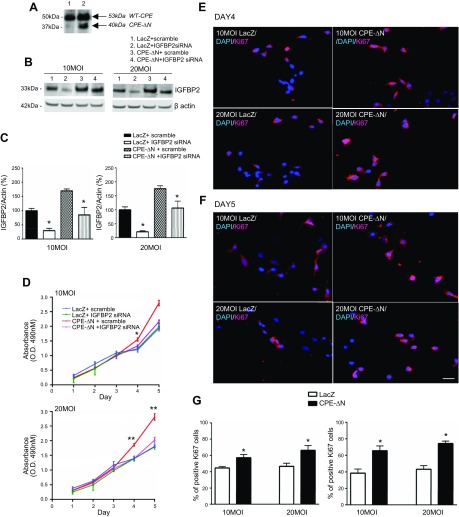
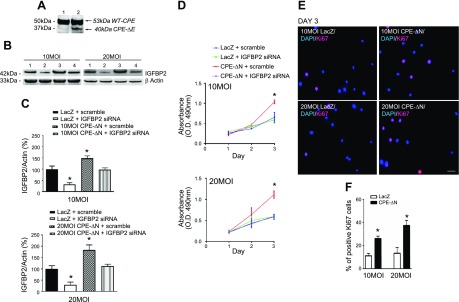
To investigate whether 40-kDa CPE-ΔN has any effect on cell proliferation and whether IGFBP2 is involved, HT22 cells were transduced with 40-kDa CPE-ΔN (Fig. 5A, lane 2) or LacZ adenovirus (Fig. 5A, lane 1) for 24 h. The cells were then treated with IGFBP2 siRNA or scramble RNA. Proliferation was evaluated by MTT and Ki-67 assays. Western blot analysis showed an increase in expression of 40-kDa CPE-ΔN in HT22 cells that were transduced with 10 MOI of virus (Fig. 5A). The MTT assay showed that transduction of 40-kDa CPE-ΔN at both 10 and 20 MOI concentrations of virus significantly enhanced proliferation of HT22 cells on d 4 (F3,8 = 12.31, P < 0.05 for 10 MOI; F3,8 = 31.10, P < 0.01 for 20 MOI) and d 5 (F3,8 = 35.69, P < 0.01 for 10 MOI; F3,8 = 47.27, P < 0.01 for 20 MOI) (in Fig. 5D, compare red- and blue-line graphs for both 10 and 20 MOI). This enhanced proliferation of HT22 cells by 40-kDa CPE-ΔN was further confirmed by immunocytochemistry of Ki-67, a marker for proliferation (Fig. 5E–G). To determine whether the promotion of proliferation by 40-kDa CPE-ΔN was mediated by IGFBP2, HT22 cells were treated with IGFBP2 siRNA, which effectively suppressed IGFBP2 expression in those cells (Fig. 5B, C). Concomitantly, the enhanced proliferation by 40-kDa CPE-ΔN transduction was inhibited by IGFBP2 siRNA treatment in HT22 cells (Fig. 5D, compare red- and pink-line graphs). This result indicates that the increased proliferation of HT22 cells induced by 40-kDa CPE-ΔN is mediated through up-regulation of IGFBP2 expression.
Figure 5.
CPE-∆N increased proliferation in HT22 cells via up-regulation of IGFBP2. A) Western blot showing enhanced expression of CPE-∆N in HT22 cells transduced with 10 MOI of virus (lane 2) vs. control cells (lane 1). B) Western blot showing expression of IGFBP2 in HT22 cells transduced with 10 and 20 MOI CPE-∆N virus. CPE-∆N transduced at both 10 and 20 MOI enhanced IGFBP2 protein levels, whereas IGFBP2 siRNA significantly reduced the protein expression of IGFBP2. C) Bar graphs showing the quantification of the Western blots in B. D) MTT assay for proliferation. CPE-∆N transduced at 10 and 20 MOI increased proliferation on d 4 and 5; that effect was blocked by IGFBP2 siRNA. One-way ANOVA with Tukey’s post hoc test. *P < 0.05, **P < 0.01, compared with controls. E, F) Immunocytochemistry of Ki-67 (the nuclear marker for cell proliferation)-positive cells in CPE-∆N transduced mouse primary cortical neurons. Scale bar, 100 μm. G) Bar graphs showing that CPE-∆N transduced at 10 and 20 MOI increased Ki-67+ cells on d 4 and 5 compared with control. Values are means ± sem. Student’s t test, n = 3. *P < 0.05, compared with control.
40 kDa CPE-ΔN promotes embryonic cortical neuron proliferation via up-regulation of IGFBP2 expression
To determine whether CPE-ΔN has effects on proliferation of mouse primary cortical neurons, experiments similar to those carried out in HT22 cells were performed. Embryonic cortical neurons when transduced with 40-kDa CPE-ΔN (Fig. 6A, lane 2) or LacZ adenovirus (Fig. 6A, lane 1) at 10 MOI for 24 h. The neurons showed an increase in expression of the 40-kDa CPE-ΔN compared with the LacZ control. When CPE-ΔN adenovirus was transduced into those cortical neurons at 10 or 20 MOI, there was an concomitant increase in IGFBP2 expression, whereas IGFBP2 siRNA significantly reduced it (Fig. 6B, C). In addition, CPE-ΔN enhanced proliferation on d 3 in primary cortical neurons, as revealed by MTT assay (Fig. 6D compare red- and blue-line graphs). IGFBP2 siRNA treatment reversed that proliferation effect (Fig. 6D, compare red- and pink-line graphs; F3,8 = 6.453, P < 0.05 for 10 MOI; F3,8 = 16.51, P < 0.01 for 20 MOI). Ki-67 immunocytochemistry also showed an increase in Ki-67–expressing cells at 10 and 20 MOI, respectively, on d 3 (Fig. 6E, F). This result indicates that CPE-ΔN enhances proliferation of primary cortical neurons by up-regulating IGFBP2 expression.
Figure 6.
CPE-∆N increased proliferation via IGFBP2 in mouse primary cortical neurons. A) Western blot showing enhanced expression of CPE-∆N in cortical neurons transduced with 10 MOI of virus (lane 2) vs. control cells (lane 1) for 24 h. B) Western blot showing expression of IGFBP2 protein in mouse primary cortical neurons transduced with 10 and 20 MOI CPE-∆N adenovirus. Both 10 and 20 MOI CPE-∆N enhanced IGFBP2 protein levels, whereas IGFBP2 siRNA significantly reduced the protein expression of IGFBP2. C) Bar graphs showing the quantification of the Western blots in A. *P < 0.05. D) MTT assay for proliferation. CPE-∆N transduced at 10 and 20 MOI increased proliferation on d 3; that effect was blocked by IGFBP2 siRNA. One-way ANOVA with Tukey’s post hoc test. *P < 0.05, compared with controls. E) Immunocytochemistry of Ki-67+ cells in CPE-∆N–transduced mouse primary cortical neurons. Scale bar, 100 μm. *P < 0.05. F) Bar graph showing that CPE-∆N transduced at 10 and 20 MOI increased Ki-67+ cells on d 3 compared with control. Values are means ± sem.Student’s t test, n = 3. *P < 0.05, compared with control.
DISCUSSION
Development of the embryonic nervous system is a complex process and requires carefully orchestrated temporal–spatial expression of many genes involved in cellular differentiation, proliferation, migration, and apoptosis. Identifying “master” genes that can activate the expression of multiple genes to mediate those processes is critical to understanding the mechanisms underlying the development of the nervous system. Our previous studies indicate that the CPE/NFα-1 gene might be an important regulatory gene in neurodevelopment. WT CPE/NFα-1 acts as a trophic factor in differentiation of embryonic neuronal stem cells to astrocytes through up-regulating glial fibrillary acidic protein expression, whereas an N-terminal–truncated 40-kDa (CPE/NFα-1)-ΔN variant acts in the nucleus to promote mouse embryonic cortical neuronal survival through up-regulation of FGF2 gene expression (22). Hence, in the present study, we have investigated whether the 40-kDa (CPE/NFα-1)-ΔN protein regulates expression of multiple genes involved in neurodevelopment. To that end, we first cloned the 40-kDa (CPE/NFα-1)-ΔN mRNA from embryonic mouse brain to obtain the complete mRNA sequence because the transcript studied previously was derived from bioinformatic analysis of expressed sequence tagged DNA from GenBank. In addition, we cloned the mouse CPE-WT mRNA as well because it has not, to our knowledge, been previously cloned.
The RACE assay, followed by DNA sequencing, revealed the expression of 2 different (CPE/NFα-1)-ΔN transcripts in embryonic mouse brain. Unlike the proposed human CPE-ΔN transcript, in which splicing takes place within exon 1 (18, 20), the use of alternative promoters appears to be the mechanism to generate transcripts with different 5′-ends for the mouse CPE variants, which yields 3 products of differing lengths. Indeed, our bioinformatic analysis suggests that there are several potential promoters upstream of the mouse CPE gene locus (data not shown). A previous work by Thouennon et al. (27) demonstrated that rosiglitazone, an antidiabetic drug in the thiazolidinedione class, activated PPARγ (peroxisome proliferation-activated receptor-γ) directly to regulate CPE expression in neurons through binding to the CPE promoter. It remains to be elucidated how expression of these CPE transcripts are regulated. Given the key roles epigenetic mechanisms, such as methylation and acetylation, have in transcriptional regulation, it is of great importance to determine whether CPE promoter methylation or acetylation status at different embryonic stages could regulate expression of CPE transcripts.
Northern blot analysis of mRNA from E8.5 to P1 mouse brains indicate that besides WT CPE/NFα-1, there were expression of 2 (CPE/NFα-1)-ΔN mRNAs, 1.9 and 1.73 kb in size, encoding 47- and 40-kDa proteins, respectively. These 2 mRNAs were expressed in E8.5 embryonic brain and peaked at E10.5, with the 1.73-kb mRNA being more abundant than the 1.9-kb transcript. Expression of these transcripts decreased at E12.5, E14.5, and E16.5 and increased again at P1, but none was found in adult hippocampus or other organs. Western blot analysis confirmed the expression of the 47- and 40-kDa (CPE/NFα-1)-ΔN protein in E10.5 and P1 cortex. The peak of expression of these 2 mRNAs at E10.5 coincides with the period of massive neurogenesis (28). Hence, we focused on the function of the 40-kDa (CPE/NFα-1)-ΔN protein in regulating expression of genes that might be important in neurodevelopment. We used cancer cells as a model to screen for such genes because cancer represents development gone awry. Genes regulated by (CPE/NFα-1)-ΔN in cancer cells to enhance proliferation and migration are likely to be regulated in embryonic neurons as well because studies suggest that the molecular mechanisms underlying those events in both those cell types are similar.
From the microarray analysis of genes that were up-regulated in HCC cells transfected with mouse 40-kDa (CPE/NFα-1)-ΔN, we identified 2 genes that were the most highly up-regulated, IGFBP2 and DAP1 (see Supplemental Figs. S3 and S4), and investigated their expression in HT22 cells, a hippocampal neuronal cell line. Our data showed that IGFBP2 gene was significantly up-regulated in expression in HT22 cells and embryonic cortical neurons transduced with 40 kDa (CPE/NFα-1)-ΔN. Moreover, these neurons showed enhanced proliferation that was inhibited by IGFBP2 siRNA. Studies have shown that IGFBP2, a member of a highly conserved family of 6 IGFBPs, is the predominant IGFBP found in neonatal rat serum and rat amniotic fluid (29). It is also highly expressed in the CNS (29, 30). The known functions of IGFBP2 is to keep IGF-1 from binding to its receptor, but it also modulates cellular processes, independent of IGF-1 binding, in several tissues and cancer cells (31–33). Recently, IGFBP2 has been shown to stimulate proliferation and differentiation of neural stem cells to astrocytes through activation of the ERK signaling (34). Thus 40-kDa (CPE/NFα-1)-ΔN may promote proliferation and differentiation of embryonic neuronal progenitors and stem cells through up-regulation of IGFBP2 expression during development.
Another gene that was significantly up-regulated by 40-kDa (CPE/NFα-1)-ΔN in HT22 hippocampal neurons was DAP1, a member of a family of 5 DAP proteins (35). DAP1, a 15-kDa protein, is a novel substrate of mechanistic target of rapamycin that negatively regulates autophagy and serves as an antioncogenic molecule in some cancers (35, 36). During embryonic development and remodeling of the nervous system, there is programmed cell death and removal of axonal and synaptic debris (37, 38). DAP1 is a mediator of programmed cell death, although not much is known about its role in the nervous system, and it warrants further study (39, 40).
Ephrins and EPH receptors have important roles in neurodevelopment. They have been implicated in the guidance of cell migration and axon movements during neural development. The EFNA1 gene, also known as EphrinA1, was down-regulated in HCC cells transduced with 40-kDa (CPE/NFα-1)-ΔN in the microarray analysis. However, EPHA2, the receptor for Ephrin A1, was up-regulated in those cells. In HT22 cells, 40-kDa (CPE/NFα-1)-ΔN increased the expression of EFNA1 mRNA and protein levels. That finding suggests that 40-kDa (CPE/NFα-1)-ΔN may function to regulate the spatial organization of neurons during development through up-regulation of ephrinA1 and EPHA2 receptors.
Interestingly, 40-kDa (CPE/NFα-1)-ΔN has been shown to enter the nucleus and interact with histone decarboxylase 1/2 to up-regulate the expression of Nedd9, a metastatic gene, to induce invasion in HCC cells (20). Nedd9 was first identified as a neural precursor cell expressed developmentally down-regulated gene from embryonic mouse brain and implicated in mediating cell migration in neurodevelopment (41). Whether 40-kDa (CPE/NFα-1)-ΔN up-regulates Nedd9 in primary neurons remains to be studied.
In conclusion, the 40-kDa (CPE/NFα-1)-ΔN variant, which is expressed only in embryonic mouse brain to postnatal d 1 may have a critical role in neurodevelopment through up-regulating the expression of many genes that promote proliferation, differentiation, axonal guidance, and cell migration, as well as programmed cell death, all events that take place during the formation of the nervous system. Previous studies have shown that 40-kDa (CPE/NFα-1)-ΔN is translocated into the nucleus and binds to a nuclear complex to activate gene transcription. Future work to study the regulation of expression of 40-kDa (CPE/NFα-1)-ΔN will provide a deeper understanding of the hierarchy this molecule has in building the embryonic nervous system.
Supplementary Material
This article includes supplemental data. Please visit http://www.fasebj.org to obtain this information.
ACKNOWLEDGMENTS
The authors thank Dr. Constantine Stratakis [National Institute of Child Health and Human Development (NICHD)/U.S. National Institiutes of Health (NIH), Bethesda, MD, USA] for critical reading of the manuscript and Dr. Vincent Schram (NICHD Microscopy Core Facility) for his assistance in the confocal microscopy. This work was supported by the Intramural Research Program of the Eunice Kennedy Shriver NICHD, NIH. The authors declare no conflicts of interest.
Glossary
- BCL2
B cell lymphoma
- CPE/NF-α1
carboxypeptidase E
- CSPD
disodium 3-(4-methoxyspiro {1,2-dioxetane-3,2′-(5′-chloro)tricyclo [3.3.1.13,7]decan}-4-yl)phenyl phosphate
- DAP1
death-associated protein 1
- EFNA1
ephrin A1
- FBS
fetal bovine serum
- FGF2
fibroblast growth factor 2
- GEMSA
guanidinoethylmercaptosuccinic acid
- HCC
hepatocellular carcinoma
- IGFBP2
insulin-like growth factor binding protein 2
- KO
knockout
- MOI
multiplicity of infection
- MTT
3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide
- NEDD9
neural precursor cell expressed developmentally down-regulated gene 9
- NF-α1
neurotrophic factor-α1
- ORF
open reading frame
- P1
postnatal d 1
- qRT-PCR
quantitative RT-PCR
- RACE
rapid amplification of cDNA ends
- siRNA
short interfering RNA
- WNT3a
wingless-related integration site 3a
- WT
wild type
Footnotes
This article includes supplemental data. Please visit http://www.fasebj.org to obtain this information.
AUTHOR CONTRIBUTIONS
L. Xiao, X. Yang, and Y. P. Loh designed the research; L. Xiao, X. Yang, and V. K. Sharma performed the research; and L. Xiao, X. Yang, V. K. Sharma, and Y. P. Loh analyzed the data and wrote the paper.
REFERENCES
- 1.Cheng Y., Cawley N. X., Loh Y. P. (2014) Carboxypeptidase E (NF-α1): a new trophic factor in neuroprotection. Neurosci. Bull. 30, 692–696 10.1007/s12264-013-1430-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cawley N. X., Wetsel W. C., Murthy S. R., Park J. J., Pacak K., Loh Y. P. (2012) New roles of carboxypeptidase E in endocrine and neural function and cancer. Endocr. Rev. 33, 216–253 10.1210/er.2011-1039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fricker L. D. (1988) Carboxypeptidase E. Annu. Rev. Physiol. 50, 309–321 10.1146/annurev.ph.50.030188.001521 [DOI] [PubMed] [Google Scholar]
- 4.Cool D. R., Normant E., Shen F., Chen H. C., Pannell L., Zhang Y., Loh Y. P. (1997) Carboxypeptidase E is a regulated secretory pathway sorting receptor: genetic obliteration leads to endocrine disorders in Cpe(fat) mice. Cell 88, 73–83 10.1016/S0092-8674(00)81860-7 [DOI] [PubMed] [Google Scholar]
- 5.Cheng Y., Cawley N. X., Loh Y. P. (2013) Carboxypeptidase E/NFα1: a new neurotrophic factor against oxidative stress-induced apoptotic cell death mediated by ERK and PI3-K/AKT pathways. PLoS One 8, e71578 10.1371/journal.pone.0071578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Selvaraj P., Huang J. S., Chen A., Skalka N., Rosin-Arbesfeld R., Loh Y. P. (2015) Neurotrophic factor-α1 modulates NGF-induced neurite outgrowth through interaction with Wnt-3a and Wnt-5a in PC12 cells and cortical neurons. Mol. Cell. Neurosci. 68, 222–233 10.1016/j.mcn.2015.08.005 [DOI] [PubMed] [Google Scholar]
- 7.Selvaraj P., Xiao L., Lee C., Murthy S. R., Cawley N. X., Lane M., Merchenthaler I., Ahn S., Loh Y. P. (2017) Neurotrophic factor-α1: a key Wnt-β-catenin dependent anti-proliferation factor and ERK-Sox9 activated inducer of embryonic neural stem cell differentiation to astrocytes in neurodevelopment. Stem Cells 35, 557–571 10.1002/stem.2511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alsters S. I., Goldstone A. P., Buxton J. L., Zekavati A., Sosinsky A., Yiorkas A. M., Holder S., Klaber R. E., Bridges N., van Haelst M. M., le Roux C. W., Walley A. J., Walters R. G., Mueller M., Blakemore A. I. (2015) Truncating homozygous mutation of Carboxypeptidase E (CPE) in a morbidly obese female with type 2 diabetes mellitus, intellectual disability and hypogonadotrophic hypogonadism. PLoS One 10, e0131417 10.1371/journal.pone.0131417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheng Y., Cawley N. X., Yanik T., Murthy S. R., Liu C., Kasikci F., Abebe D., Loh Y. P. (2016) A human carboxypeptidase E/NF-α1 gene mutation in an Alzheimer’s disease patient leads to dementia and depression in mice. Transl. Psychiatry 6, e973 10.1038/tp.2016.237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Woronowicz A., Koshimizu H., Chang S. Y., Cawley N. X., Hill J. M., Rodriguiz R. M., Abebe D., Dorfman C., Senatorov V., Zhou A., Xiong Z. G., Wetsel W. C., Loh Y. P. (2008) Absence of carboxypeptidase E leads to adult hippocampal neuronal degeneration and memory deficits. Hippocampus 18, 1051–1063 10.1002/hipo.20462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheng Y., Rodriguiz R. M., Murthy S. R., Senatorov V., Thouennon E., Cawley N. X., Aryal D. K., Ahn S., Lecka-Czernik B., Wetsel W. C., Loh Y. P. (2015) Neurotrophic factor-α1 prevents stress-induced depression through enhancement of neurogenesis and is activated by rosiglitazone. Mol. Psychiatry 20, 744–754 10.1038/mp.2014.136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cawley N. X., Zhou J., Hill J. M., Abebe D., Romboz S., Yanik T., Rodriguiz R. M., Wetsel W. C., Loh Y. P. (2004) The carboxypeptidase E knockout mouse exhibits endocrinological and behavioral deficits. Endocrinology 145, 5807–5819 10.1210/en.2004-0847 [DOI] [PubMed] [Google Scholar]
- 13.Zhu X., Wu K., Rife L., Cawley N. X., Brown B., Adams T., Teofilo K., Lillo C., Williams D. S., Loh Y. P., Craft C. M. (2005) Carboxypeptidase E is required for normal synaptic transmission from photoreceptors to the inner retina. J. Neurochem. 95, 1351–1362 10.1111/j.1471-4159.2005.03460.x [DOI] [PubMed] [Google Scholar]
- 14.Jin K., Graham S. H., Nagayama T., Goldsmith P. C., Greenberg D. A., Zhou A., Simon R. P. (2001) Altered expression of the neuropeptide-processing enzyme carboxypeptidase E in the rat brain after global ischemia. J. Cereb. Blood Flow Metab. 21, 1422–1429 10.1097/00004647-200112000-00006 [DOI] [PubMed] [Google Scholar]
- 15.Zhou A., Minami M., Zhu X., Bae S., Minthorne J., Lan J., Xiong Z. G., Simon R. P. (2004) Altered biosynthesis of neuropeptide processing enzyme carboxypeptidase E after brain ischemia: molecular mechanism and implication. J. Cereb. Blood Flow Metab. 24, 612–622 10.1097/01.WCB.0000118959.03453.17 [DOI] [PubMed] [Google Scholar]
- 16.Manser E., Fernandez D., Loo L., Goh P. Y., Monfries C., Hall C., Lim L. (1990) Human carboxypeptidase E. Isolation and characterization of the cDNA, sequence conservation, expression and processing in vitro. Biochem. J. 267, 517–525 10.1042/bj2670517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rodríguez C., Brayton K. A., Brownstein M., Dixon J. E. (1989) Rat preprocarboxypeptidase H. Cloning, characterization, and sequence of the cDNA and regulation of the mRNA by corticotropin-releasing factor. J. Biol. Chem. 264, 5988–5995 [PubMed] [Google Scholar]
- 18.Yang X., Cong L., Lou H., Loh Y. P. (2017) Abstract 1967: A novel 40kDa CPE-ΔN isoform promotes proliferation and invasion in pancreatic cancer cells. AACR Annual Meeting 2017, Washington, DC [Google Scholar]
- 19.Fan S., Li X., Li L., Wang L., Du Z., Yang Y., Zhao J., Li Y. (2016) Silencing of carboxypeptidase E inhibits cell proliferation, tumorigenicity, and metastasis of osteosarcoma cells. OncoTargets Ther. 9, 2795–2803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee T. K., Murthy S. R., Cawley N. X., Dhanvantari S., Hewitt S. M., Lou H., Lau T., Ma S., Huynh T., Wesley R. A., Ng I. O., Pacak K., Poon R. T., Loh Y. P. (2011) An N-terminal truncated carboxypeptidase E splice isoform induces tumor growth and is a biomarker for predicting future metastasis in human cancers. J. Clin. Invest. 121, 880–892 10.1172/JCI40433 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 21.Sun J., Meng D., Li L., Tian X., Jia Y., Wang H., Yu H., Sun T., Qu A., Shen H., Bao J., Zhang G. (2016) N-terminal truncated carboxypeptidase E expression is associated with poor prognosis of lung adenocarcinoma. Oncol. Lett. 12, 4659–4664 10.3892/ol.2016.5283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qin X. Y., Cheng Y., Murthy S. R., Selvaraj P., Loh Y. P. (2014) carboxypeptidase E-ΔN, a neuroprotein transiently expressed during development protects embryonic neurons against glutamate neurotoxicity. PLoS One 9, e112996 10.1371/journal.pone.0112996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cong L., Cheng Y., Cawley N. X., Murthy S. R., Loh Y. P. (2017) A novel single nucleotide T980C polymorphism in the human carboxypeptidase E gene results in loss of neuroprotective function. PLoS One 12, e0170169 10.1371/journal.pone.0170169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fricker L. D. (1995) Methods for studying carboxypeptidase E. Methods in Neurosciences 23, 237–250 10.1016/S1043-9471(06)80124-2 [DOI] [Google Scholar]
- 25.Scholzen T., Gerdes J. (2000) The Ki-67 protein: from the known and the unknown. J. Cell. Physiol. 182, 311–322 [DOI] [PubMed] [Google Scholar]
- 26.Touriol C., Bornes S., Bonnal S., Audigier S., Prats H., Prats A. C., Vagner S. (2003) Generation of protein isoform diversity by alternative initiation of translation at non-AUG codons. Biol. Cell 95, 169–178 10.1016/S0248-4900(03)00033-9 [DOI] [PubMed] [Google Scholar]
- 27.Thouennon E., Cheng Y., Falahatian V., Cawley N. X., Loh Y. P. (2015) Rosiglitazone-activated PPARγ induces neurotrophic factor-α1 transcription contributing to neuroprotection. J. Neurochem. 134, 463–470 10.1111/jnc.13152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Van den Ameele J., Tiberi L., Vanderhaeghen P., Espuny-Camacho I. (2014) Thinking out of the dish: what to learn about cortical development using pluripotent stem cells. Trends Neurosci. 37, 334–342 10.1016/j.tins.2014.03.005 [DOI] [PubMed] [Google Scholar]
- 29.Ocrant I. (1991) Insulin-like growth factor binding proteins in the nervous system. Adv. Exp. Med. Biol. 293, 471–482 10.1007/978-1-4684-5949-4_42 [DOI] [PubMed] [Google Scholar]
- 30.Ocrant I., Fay C. T., Parmelee J. T. (1990) Characterization of insulin-like growth factor binding proteins produced in the rat central nervous system. Endocrinology 127, 1260–1267 10.1210/endo-127-3-1260 [DOI] [PubMed] [Google Scholar]
- 31.Aizenman Y., de Vellis J. (1987) Brain neurons develop in a serum and glial free environment: effects of transferrin, insulin, insulin-like growth factor-I and thyroid hormone on neuronal survival, growth and differentiation. Brain Res. 406, 32–42 10.1016/0006-8993(87)90766-9 [DOI] [PubMed] [Google Scholar]
- 32.Milanesi E., Zanardini R., Rosso G., Maina G., Barbon A., Mora C., Minelli A., Gennarelli M., Bocchio-Chiavetto L. (2017) Insulin-like growth factor binding protein 2 in bipolar disorder: an expression study in peripheral tissues. [E-pub ahead of print] World J. Biol. Psychiatry [DOI] [PubMed] [Google Scholar]
- 33.Mendes K. N., Wang G. K., Fuller G. N., Zhang W. (2010) JNK mediates insulin-like growth factor binding protein 2/integrin α5-dependent glioma cell migration. Int. J. Oncol. 37, 143–153 [DOI] [PubMed] [Google Scholar]
- 34.Deng Y. J., Wang L., Ge L., Duan D., Zhuo Y., Yuan T., Yan W. P., Huang P. Q., Teng X. H., Lu M. (2017) Effects of IGFBP-2 on proliferation and differentiation in neural stem cell line C17.2. J Neurorestoratology 5, 143–153 10.2147/JN.S134363 [DOI] [Google Scholar]
- 35.Levy-Strumpf N., Kimchi A. (1998) Death associated proteins (DAPs): from gene identification to the analysis of their apoptotic and tumor suppressive functions. Oncogene 17, 3331–3340 10.1038/sj.onc.1202588 [DOI] [PubMed] [Google Scholar]
- 36.Koren I., Reem E., Kimchi A. (2010) DAP1, a novel substrate of mTOR, negatively regulates autophagy. Curr. Biol. 20, 1093–1098 10.1016/j.cub.2010.04.041 [DOI] [PubMed] [Google Scholar]
- 37.Neumann H., Kotter M. R., Franklin R. J. (2009) Debris clearance by microglia: an essential link between degeneration and regeneration. Brain 132, 288–295 10.1093/brain/awn109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ziegenfuss J. S., Doherty J., Freeman M. R. (2012) Distinct molecular pathways mediate glial activation and engulfment of axonal debris after axotomy. Nat. Neurosci. 15, 979–987 10.1038/nn.3135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Deiss L. P., Feinstein E., Berissi H., Cohen O., Kimchi A. (1995) Identification of a novel serine/threonine kinase and a novel 15-kD protein as potential mediators of the γ-interferon-induced cell death. Genes Dev. 9, 15–30 10.1101/gad.9.1.15 [DOI] [PubMed] [Google Scholar]
- 40.Feinstein E., Druck T., Kastury K., Berissi H., Goodart S. A., Overhauser J., Kimchi A., Huebner K. (1995) Assignment of DAP1 and DAPK—genes that positively mediate programmed cell death triggered by IFN-γ—to chromosome regions 5p12.2 and 9q34.1, respectively. Genomics 29, 305–307 10.1006/geno.1995.1255 [DOI] [PubMed] [Google Scholar]
- 41.Kumar S., Tomooka Y., Noda M. (1992) Identification of a set of genes with developmentally down-regulated expression in the mouse brain. Biochem. Biophys. Res. Commun. 185, 1155–1161 10.1016/0006-291X(92)91747-E [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.