Abstract
Ubiquitinylation drives many cellular processes by targeting proteins for proteasomal degradation. Ubiquitin conjugation enzymes promote ubiquitinylation and, thus, degradation of protein substrates. Ubiquitinylation is a well-known posttranslational modification controlling cell-cycle transitions and levels or/and activation levels of ubiquitin-conjugating enzymes change during development and cell cycle. Progression through the cell cycle is tightly controlled by CDK inhibitors such as p27Kip1. Here we show that, in contrast to promoting its degradation, the ubiquitin-conjugating enzyme UBCH7/UBE2L3 specifically protects p27Kip1 from degradation. Overexpression of UBCH7/UBE2L3 stabilizes p27Kip1 and delays the G1-to-S transition, while depletion of UBCH7/UBE2L3 increases turnover of p27Kip1. Levels of p21Cip1/Waf1, p57Kip2, cyclin A and cyclin E, all of which are also involved in regulating the G1/S transition are not affected by UBCH7/UBE2L3 depletion. The effect of UBCH7/UBE2L3 on p27Kip1 is not due to alteration of the levels of any of the ubiquitin ligases known to ubiquitinylate p27Kip1. Rather, UBCH7/UBE2L3 catalyzes the conjugation of heterotypic ubiquitin chains on p27Kip1 that are proteolytically incompetent. These data reveal new controls and concepts about the ubiquitin proteasome system in which a ubiquitin-conjugating enzyme selectively inhibits and may even protect, rather than promote degradation of a crucial cell-cycle regulatory molecule.—Whitcomb, E. A., Tsai, Y. C., Basappa, J., Liu, K., Le Feuvre, A. K., Weissman, A. M., Taylor, A. Stabilization of p27Kip1/CDKN1B by UBCH7/UBE2L3 catalyzed ubiquitinylation: a new paradigm in cell-cycle control.
Keywords: cell division, proteasome, ubiquitin-conjugating enzyme, CDKI
Ubiquitin is an 8.9 kDa protein that is typically attached to lysines on protein substrates (1). Ubiquitinylation is crucial for regulation of cell proliferation (2–9), responses to DNA damage (10), subcellular localization of substrates (11), maintaining protein quality (12–14), and protein–protein interactions (15). A primary function of ubiquitinylation is targeting proteins for degradation via the ubiquitin proteasome system (UPS) (16). Highlighting the requirement for a functional UPS in cell-cycle control are observations that mutations and dysregulation of UPS components, including ubiquitin itself, predispose to malignancies (17–19), hyperproliferative diseases (20), and abnormal differentiation and development (21–23).
The attachment of ubiquitin to substrates is accomplished through the sequential activities of 3 groups of enzymes: ubiquitin-activating enzymes (E1s), ubiquitin-conjugation enzymes (Ubcs or E2s), and ubiquitin ligases (E3s). For many E2s, there is a remarkable level of evolutionary conservation. Each E2 generally works with a limited number of E3s to transfer ubiquitin to substrates, and the combined activities of multiple E2s and E3s confer exquisite substrate specificity to the UPS (24). Raising the levels of Ubc enzymes generally accelerates substrate degradation (25, 26).
Ubiquitinylation can involve the attachment of a single ubiquitin, multiple monoubiquitins, or polymers of ubiquitin, which are built using 1 of the 7 internal lysines or α-NH2 within ubiquitin (27). The assembly into a chain of several ubiquitins linked through lysine 48 (K48) of 1 ubiquitin and the C terminus of another constitutes a signal for UPS degradation (24, 28, 29). Reports indicate that the attachment of mixed ubiquitin chains can lead to impaired proteolysis of substrates (30, 31), whereas others show increased proteolysis with mixed ubiquitin chains (32, 33).
UBCH7/UBE2L3 is unusual among E2s in that it has no direct homolog in yeast, and information regarding its biologic functions is limited. Like many other E2s, UBCH7/UBE2L3 is a core domain or class I E2, with a conserved active-site cysteine but without N- or C-terminal extensions or acidic loops (26). A critical mammalian function of UBCH7/UBE2L3 is indicated because decreased expression of this enzyme in mice results in pre- or perinatal death due to retarded growth and placental vascular defects (22). UBCH7/UBE2L3 also appears to play an important role in inflammatory disease (34–37). Levels and activation states of UBCH7/UBE2L3 change during cell cycle and differentiation. Elevation of UBCH7/UBE2L3 increases the proportion of cells in G1 phase relative to S phase, while its depletion extends S phase (6, 38). Levels of UBCH7/UBE2L3 that are charged with ubiquitin are higher in differentiated tissue. The charged state is associated with enhanced activity of the E2, and such activity usually presages degradation of the target substrate (21, 39–41).
The cell cycle is controlled by the timed UPS-directed degradation of cyclins and cyclin-dependent kinase inhibitors (CDKI). In addition to being a major effector of the G1-to-S transition of the cell cycle, the CDKI p27Kip1 plays critical roles in regulating development in postmitotic cells and tissues (5, 21, 42). Prior studies indicate that p27Kip1 is a primary substrate for degradation by the UPS, particularly on ubiquitinylation by Cdc34/Ubc3/UBE2R1 (9, 43). However, relations between UBCH7/UBE2L3 and p27Kip1 have not been extensively explored. In contrast with prior literature regarding Cdc34/Ubc3/UBE2R1, we find that active UBCH7/UBE2L3 protects p27Kip1 from UPS-dependent degradation.
Here we observe that UBCH7/UBE2L3 catalyzes formation of linkages using different lysines on ubiquitin besides K48 on p27Kip1. We find that instead of promoting the degradation of this well-established UPS substrate, active UBCH7/UBE2L3 protects p27Kip1 from UPS-dependent degradation by promoting the attachment of various proteolytically incompetent ubiquitin chains. We did not detect changes in levels in E3 ligases that have been reported to participate in ubiquitinylation of p27Kip1 and target it for degradation by the proteasome (8, 44–48). The results indicate a new dimension in ubiquitin conjugation biology.
MATERIALS AND METHODS
Cells
Human lens epithelial cells (HLEC SRA 01/04), HeLa, HEK-293, and ARPE-19 cells were grown in DMEM (Thermo Fisher Scientific, Waltham, MA, USA) supplemented with 10% fetal bovine serum, 50 U/ml penicillin and 50 µg/ml streptomycin. Medium for HeLa and HEK-293 cells was supplemented with glucose for a final concentration of 20 mM. For Western blot analysis, cells were lysed in 10 mM Tris–HCl (pH 7.6), 50 mM EDTA, 1% NP-40, 0.1% SDS, 20 mM N-ethylmaleimide, and 2 mM 4-(2-aminoethyl)-benzene-sulfonylfluoride.
Plasmids and small interfering RNA
pAD-Track-UBCH7/UBE2L3 was previously described (6). The mutant UBCH7/UBE2L3 expressing Ser instead of Cys at position 89 was generated from the wild-type (WT) vector using the QuickChange XL Mutagenesis Kit (Stratagene, La Jolla, CA, USA) using the primer 5′-GGCAGGTCTCTCTGCCAGTAATTAGTGCCG-3′. HA-ubiquitin was generated by PCR and subcloned into pcDNA3.1 (+) between HindIII and XhoI sites. Single K to R mutants were generated by site-directed mutagenesis (Quickchange XL; Thermo Fisher Scientific). pFLAG-CMV-p27Kip1 was a kind gift from N. Fujita (Japanese Foundation for Cancer Research, Tokyo, Japan). For transfection, 1 × 105 cells were plated in a 24-well plate 24 h before transfection. On the day of transfection, 1 μg plasmid DNA and 4 μl polyfect transfection reagent (Qiagen, Hilden, Germany) were added to each well. Small interfering RNA (siRNA) for UBCH7/UBE2L3 and nonspecific siRNA were purchased from Qiagen. Sequences of siRNA are as previously described (6).
Cell-cycle analysis
Cells overexpressing UBCH7/UBE2L3 or empty plasmids were treated with 1 mM hydroxyurea (HU) for 18 h. Cells were washed with PBS and returned to culture in complete medium. At time points indicated, cells were collected by trypsin digestion and treated with PBS with 4% paraformaldehyde for 30 min. After treatment, cells were washed, resuspended in 70% ethanol, and stored at −20°C until analysis. Cells were stained with propidium iodide and analyzed for DNA content as described (6).
Antibodies and reagents
Anti-p27Kip1 was purchased from BD Technologies (La Jolla, CA, USA). Anti-Skp2, anti-DDB1, and anti-KPC1 (RNF123) were purchased from Santa Cruz Biotechnology (Dallas, TX, USA). Anti-FLAG and anti-p21Cip1/Waf1 were purchased from MilliporeSigma (Burlington, MA, USA). Anti-Pirh2 was purchased from Bethyl Laboratories (Montgomery, TX, USA). Anti- UBCH7/UBE2L3 and E2 proteins UBCH7/UBE2L3, Cdc34/Ubc3/UBE2R1, and UbcH5C/UBE2D3 were purchased from Boston Biochem (Boston, MA, USA). Anti-E1 (49) and anti-ubiquitin were produced in this laboratory. Anti K63 ubiquitin was purchased from Enzo Life Sciences (Farmingdale, NY, USA).
Real-time quantitative PCR
Primers for UBCH7/UBE2L3, p27Kip1 and GAPDH were purchased from Qiagen. Cells treated with nonsilencing siRNA or UBCH7/UBE2L3-specific siRNA for 72 h and mRNA were prepared with the RNeasy kit (Qiagen) according to the manufacturer’s instructions. Real-time quantitative PCR was performed with the Mx4000 Multiplex PCR machine (Stratagene, San Diego, CA, USA). Signals for UBCH7/UBE2L3 and p27Kip1 were compared to GAPDH for normalization.
p27Kip1 subcloning and production
The pCMV5 vector harboring human p27Kip1 was obtained from J. Massague (Memorial Sloan-Kettering Cancer Center, New York, NY, USA) through Addgene (Cambridge, MA, USA). We subcloned p27Kip1 into the pET-15b vector using the following primers: p27Kip1T_F 5′-ACTCATATGTCAAACGTGCGAG-3′ (histidine tagged); p27Kip1nt_F 5′-ATACCATGGGGTCAAACGTGCGAG-3′ (nontagged) p27Kip1_R 5′-ACCGGATCCTTACGTTTGACGTCTTCTG-3′ (reverse). p27Kip1 was amplified, digested with BamHI and NdeI using the restriction sites engineered in the primers, inserted into pET-15b, and transformed to BL21 (DE3) strain.
Purification [35S]-labeled p27Kip1 protein
In vitro translation reactions were performed using the TNT Coupled Reticulocyte Lysate System (Promega, Madison, WI, USA). In brief, 1 µg of His-tagged p27Kip1 plasmid DNA was translated according to the manufacturer’s instructions. Alternatively, [35S]-labeled p27Kip1 protein was expressed and purified from Escherichia coli. In brief, BL21 (DE3) strain transformed with His-tagged p27Kip1 plasmid was inoculated into Luria-Bertani medium. When OD600 reached 1.0–1.5, bacterial cells were transferred to M9 medium with 60 µCi/ml Expre [35S][35S] label mix (PerkinElmer, Waltham, MA, USA) and 2 mM isopropyl β-D-1-thiogalactopyranoside, and incubated at 37°C for 30 min followed by additional 3.5 h incubation after adding 400 µg/ml rifampicin. Affinity purification of [35S]-His–tagged p27Kip1 was performed using Ni-NTA agarose (Qiagen) according to the manufacturer’s instructions.
Cycloheximide chase
Cells were transfected with plasmids or siRNA for 72 h. Cycloheximide (50 μg/ml) or MG132 (40 μM) was added to inhibit protein synthesis or proteasomal degradation, respectively. At the specified times, cells were lysed in modified RIPA buffer (50 mM Tris, pH 7.4, 150 mM NaCl, 1% Triton X-100, 0.5% sodium deoxycholate, 10 mM iodoacetamide, 50 μM MG132) and complete protease inhibitors or separated into cytosolic and nuclear fractions using the NE-PER Extraction Kit (Thermo Fisher Scientific) and processed for immunoblotting.
In vitro degradation
Experiments performed as in (6, 12). For ATP-dependent degradation, purified [35S]-p27Kip1 was incubated at 37°C for 90 min in a final reaction volume of 25 μl containing 50 mM Tris-HCl, pH 7.6, 5 mM MgCl2, 1 mM DTT, 2 mM ATP, 15 mM creatine phosphate, and 5 U creatine phosphokinase, 10 μg ubiquitin, and 1 μM of E2s with rabbit reticulocyte lysate (Promega or Green Hectares, Oregon, WI, USA), or G0 synchronized HLEC cell extract. Degradation was terminated, and trichloroacetic acid (TCA)-soluble counts per minute (CPM) were quantified using a Cobra II γ counter. Percentage degradation was calculated as [(experimental soluble CPM) − (buffer control soluble CPM)]/(total CPM) × 100. Efforts to obtain UBCH7/UBE2L3 catalyzed ubiquitinylated p27Kip1 for direct degradation assays were frustrated by our inability to isolate and purify sufficient quantities.
In vitro ubiquitinylation of p27Kip1
For ubiquitin conjugation assays, His-tagged p27Kip1 was incubated at 37°C for 90 min in a final reaction volume of 50 μl containing 40 mM Tris-HCl, pH 7.6, 5 mM MgCl2, 40 μM MG132, 2 μM ubiquitin aldehyde, 5 mM ATP, 40–80 μM ubiquitin, 15 mM creatine phosphate, 5 U creatine phosphokinase, and 2 mM DTT in the presence of HLEC G0 lysate, and E2s where indicated. After ubiquitinylation, His-tagged p27Kip1 was affinity purified using Ni-NTA agarose (Qiagen) according to the manufacturer’s instructions. Ni-NTA agarose beads were washed 5 times with 20 mM imidazole before His-tagged p27Kip1 was eluted using 200 mM imidazole. Eluates were mixed with 2 times Laemmli buffer and incubated at 37°C for 60 min. Proteins were resolved by 12% SDS-PAGE and blotted for total ubiquitin. Duplicate samples were set up without the addition of His-tagged p27Kip1 to assess nonspecific binding to the Ni-NTA agarose.
In cell ubiquitinylation of p27Kip1
FLAG p27Kip1 was expressed in HeLa cells together with depletion of UBCH7/UBE2L3. After 48 h, cells were lysed in RIPA buffer (Thermo Fisher Scientific). FLAG-p27Kip1 was immunoprecipitated with anti-FLAG (MilliporeSigma) followed by protein A/G agarose (Santa Cruz Biotechnology). Immunoprecipiates were subject to SDS-PAGE and immunoblotting with anti-ubiquitin or anti K48 ubiquitin (MilliporeSigma).
RESULTS
Expression of UBCH7/UBE2L3 stabilizes p27Kip1
We previously showed that UBCH7/UBE2L3 had an unanticipated relationship with the cell cycle and proliferation: overexpression of UBCH7/UBE2L3 increased the percentage of cells in G1 phase at the expense of S phase (6). We thus hypothesized that overexpression of UBCH7/UBE2L3 would delay the G1-to-S transition. This was tested by overexpressing UBCH7/UBE2L3 or an empty vector and synchronizing with HU. HU inhibits DNA replication and as such synchronizes cells in late G1/early S phase (50). In both cases, >80% of the cells of both transfectants were synchronized at the G1-to-S transition immediately after removal of HU, (t = 0) (Fig. 1A). However, cells overexpressing WT UBCH7/UBE2L3 show about 25% higher proportion of cells with 2 N DNA content, designated here as G1 phase. By 4 h after drug release, cells in which UBCH7/UBE2L3 was overexpressed had a 225% higher level of the G1 cells (P < 0.05, medium blue in Fig. 1A), and 19% fewer cells begin DNA replication compared to cells transfected with the empty vector (P < 0.01, light blue in Fig. 1A). There were no differences in the percentage of cells in G1/M between the transfectants (dark blue in Fig. 1A).
Figure 1.
Expression of UBCH7/UBE2L3 impairs G1-to-S transition via posttranslational stabilization of p27Kip1. A) HeLa cells were transfected with empty vector or vector expressing UBCH7/UBE2L3 (GFP-UBCH7) for 48 h. Cells were synchronized with 2 mM HU for 18 h. Drug was washed out, and cells were allowed to progress through cell cycle and were collected at times indicated. DNA content was measured using propidium iodide and fluorescence-activated cell sorting analysis gated on transfected cells. Data are average of 3 independent experiments. Significance values were calculated by paired Student’s t test. B) HEK-293 cells were transfected with vector expressing FLAG-p27Kip1 and vectors expressing WT UBCH7/UBE2L3 (UBCH7) or active site mutant of UBCH7/UBE2L3 (C89S UBCH7). After 48 h of expression, cells were collected and probed for anti-E1 (loading control) anti FLAG-p27 or anti UBCH7/UBE2L3, as indicated. C) HeLa cells were treated with siRNA for UBCH7/UBE2L3 or control siRNA for 72 h. mRNA was collected and subject to real-time quantitative PCR. Signals for UBCH7/UBE2L3 and p27Kip1 were compared to GAPDH levels and then normalized to NS siRNA signal. Data are presented as means ± sem, n = 3. D) WT, but not mutant UBCH7/UBE2L3, stabilizes p27Kip1 in cell-free assays. All reactions contain ATP-generating system, cell lysate, ubiquitin, and [35S]-p27Kip1. Degradation of p27Kip1 without addition of E2 was set as 100%. Average of 10 independent experiments ± sem.
The G1-to-S transition is controlled by CDK activators, cyclins A and E, and the CDKIs p27Kip1, p21Cip1/Waf1, and p57Kip2. No changes in cyclin A levels were found on UBCH7/UBE2L3 depletion, as previously reported (6) (Supplemental Fig. S1A). Comparable findings, particularly the invariant levels of cyclin E on alteration of UBCH7/UBE2L3, were corroborated in diverse cells including HeLa, HEK-293, and human lens epithelial cells (HLEC) (Supplemental Fig. S1B). Thus, we hypothesized that UBCH7/UBE2L3 might be affecting the G1-to-S transition by affecting the levels of the CDKIs. Unexpectedly, but consistent with UBCH7/UBE2L3 inhibiting the G1-to-S transition, overexpression of WT UBCH7/UBE2L3 increased the level of p27Kip1 by more than 6-fold (Fig. 1B, lane 2). In contrast, there was no stabilization of p27Kip1 on overexpression of mutant UBCH7/UBE2L3 in which the active-site cysteine had been mutated to serine, C89S UBCH7/UBE2L3, indicating that UBCH7/UBE2L3 activity is critical for the stabilization of p27Kip1 (Fig. 1B).
Next we asked if UBCH7/UBE2L3 is actively involved in the transcriptional regulation of p27Kip1. UBCH7/UBE2L3 siRNA treatment decreased UBCH7/UBE2L3 mRNA, as expected. However, p27Kip1 mRNA levels were not decreased (Fig. 1C) on UBCH7/UBE2L3 depletion. Together, these data suggest that the activity of UBCH7/UBE2L3 is necessary and sufficient to stabilize p27Kip1, and that the increase in p27Kip1 on overexpression of UBCH7/UBE2L3 is due to a posttranscriptional, and likely a posttranslational, process.
Our finding that UBCH7/UBE2L3 increases the level of p27Kip1 at a posttranscriptional level was counterintuitive because Ubc enzymes usually participate in ubiquitinylation and subsequent degradation of substrates, rather than increasing their levels. Therefore, we asked whether UBCH7/UBE2L3 directly affects p27Kip1 degradation using an in vitro degradation assay. Consistent with the observed increase in p27Kip1, addition of WT UBCH7/UBE2L3 inhibited the ATP-dependent degradation of radiolabeled p27Kip1 by 50% relative to unsupplemented lysate or ∼35% relative to when inactive enzyme was added (Fig. 1D). The inhibition of the degradation of p27Kip1 was not significant in the presence of the active site mutant of UBCH7/UBE2L3 (C89S). Together, these data suggest that rather than catalyze degradation, active UBCH7/UBE2L3 inhibits the degradation of p27Kip1. In contrast with UBCH7/UBE2L3, addition of the cognate E2 for the SKP1/Cullin/F-box containing (SCF) complex, Cdc34/Ubc3/UBE2R1 (3, 9), increased the degradation of p27Kip1, as expected (Supplemental Fig. S1C). The UbcH5/UBE2D family of E2s can function with many E3s to support the ubiquitinylation and degradation of many substrates in vitro, including when the physiologic E2 is unknown because they cooperate functionally with many E3s. Specifically relevant to this work, the UbcH5/UBE2D family has been shown to work with both the SCF and the Pirh2 E3 ligases that are known to ubiquitinylate p27Kip1 (46, 51). UbcH5C/UBE2D3 (Supplemental Fig. S1C) had no effect on ATP-dependent p27Kip1 degradation, consistent with the idea that the inhibition of p27Kip1 degradation we observe with UBCH7/UBE2L3 is specific and not an activity shared by all class I E2s.
In order to determine if the direct relationship between levels of p27Kip1 and UBCH7/UBE2L3 is generalizable, we decreased the steady-state level of UBCH7/UBE2L3 in HeLa, HLEC, HEK-293, and retinal pigmented epithelial cells (Fig. 2A, B). p27Kip1 was destabilized in each cell type. This was corroborated by the observation of a decrease in p27Kip1 with 3 different siRNA sequences targeting UBCH7/UBE2L3 (Supplemental Fig. S1D), indicating that the stabilization of p27Kip1 is related to UBCH7/UBE2L3 activity.
Figure 2.
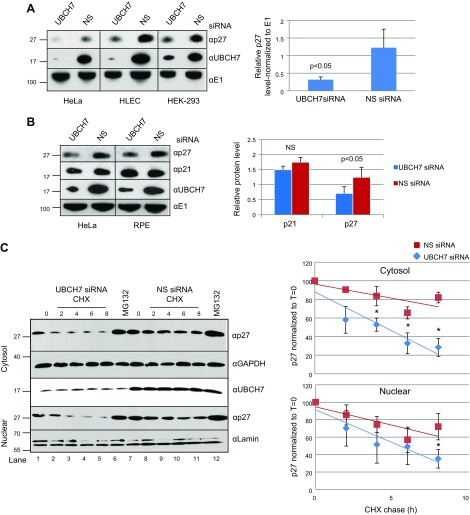
UBCH7/UBE2L3 depletion increases proteasomal turnover of p27Kip1 but not p21cip1/waf1. Cells were depleted of UBCH7/UBE2L3 for 72 h using siRNA. Lysates from treated cell lines were blotted for E1, UBCH7/UBE2L3 (UBCH7), p21, or p27 as indicated. A) UBCH7/UBE2L3 depletion results are evident decreased p27 levels. Right: quantification of 11 experiments from 3 cell lines ± sem. Significance value calculated by paired Student’s t test. B) p27, but not p21, is diminished after UBCH7/UBE2L3 depletion. Right: quantification from 5 independent experiments ± sem. Significance value was calculated by paired Student’s t test. C) UBCH7/UBE2L3 depletion increases proteasomal turnover of p27Kip1. After 72 h depletion, HeLa cells were treated with cycloheximide (CHX) or MG132, as indicated, for up to 8 h. Cytosolic and nuclear fractions were separated and subjected to immunoblot as indicated. Quantification from 4 independent experiments. *P < 0.05.
The CDKIs p21Cip1/Waf1 and p57Kip2 also control the G1-to-S transition in many types of cells. When UBCH7/UBE2L3 was depleted, no significant changes were found in p21Cip1/Waf1 (Fig. 2B) or p57Kip2 (Supplemental Fig. S1E). These CDKIs are usually degraded at the G1-to-S transition by the SCFSkp2 functioning with Cdc34/Ubc3/UBE2R1. This is the E2–E3 pair most frequently associated with degradation of p27Kip1. Additionally, we did not see an effect of UBCH7/UBE2L3 depletion on another SCF substrate, β-catenin (Supplemental Fig. S1F). Thus, the UBCH7/UBE2L3 effect appears to be specific to p27Kip1, and the change in cell cycle that we observed on UBCH7/UBE2L3 overexpression is likely mediated by p27Kip1, not p21Cip1/Waf1, p57Kip2, cyclins A or E, or other SCF substrates.
To explicitly assess whether the decrease in p27Kip1 observed on depletion of UBCH7/UBE2L3 in cells is a consequence of proteasomal degradation, we depleted cells of UBCH7/UBE2L3 for 72 h and then treated with either cycloheximide or with an inhibitor of the proteasome MG132. Because the nuclear and cytosolic pools of p27Kip1 have been reported to be differently regulated (52), cells were fractionated (Supplemental Fig. S3), and the stability of p27Kip1 was examined. Depletion of UBCH7/UBE2L3 significantly increased degradation of p27Kip1 in the cytosolic fraction (Fig. 2C, lanes 1–5 to 7–11; P < 0.05 for trend) as well as, more modestly, in the nuclear fraction. As expected for a UPS substrate, MG132 stabilized p27Kip1 in both fractions to the level observed in nondepleted cells (Fig. 2C, lane 6 vs. 7). Combined, the depletion and overexpression data indicate that endogenous levels of catalytically active UBCH7/UBE2L3 controls the stability of p27Kip1 in a counterintuitive manner by preventing the proteasomal degradation of p27Kip1.
These results suggest that increased expression of UBCH7/UBE2L3 can overcome the degradation signal directed by the ubiquitinylation of p27Kip1 by Cdc34/Ubc3/UBE2R1 and associated E3s including SCFSkp2. Indeed, this was found to be the case in in vitro degradation assays (Supplemental Fig. S4).
UBCH7/UBE2L3 does not affect levels of E3s reported to target p27Kip1 for degradation
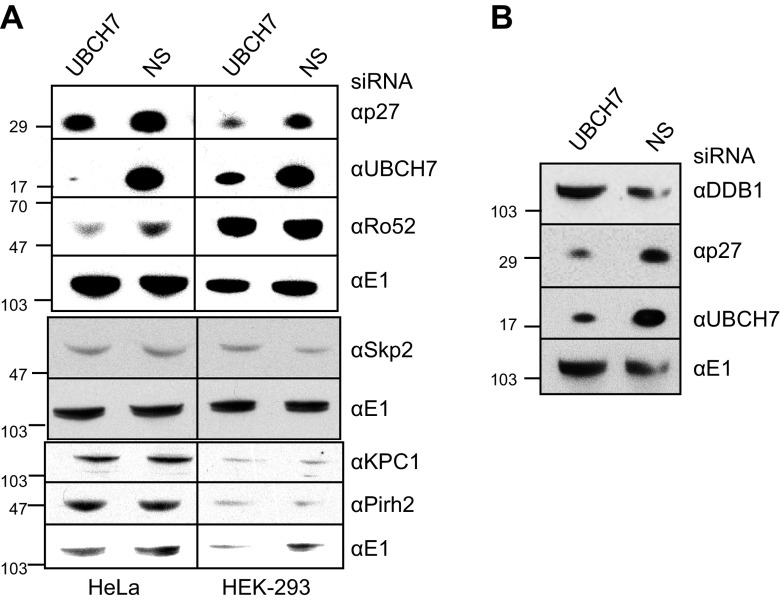
One means by which UBCH7/UBE2L3 could stabilize p27Kip1 is by targeting for UPS degradation an E3 that normally ubiquitinylates p27Kip1. Seven E3s or subunits have been shown to be involved in ubiquitinylation and proteasomal degradation of p27Kip1. These are the SCFSkp2 complex, the KPC1/2 complex, Pirh2, Ro52, DDB1, and possibly E6AP and HHARI/ARIH1 (8, 44–48, 53). Levels of Skp2 are known to be controlled by the UPS, and alterations in Skp2 concentration have been shown to affect the levels of p27Kip1 (54). The KPC1 ubiquitin ligase is also known to be regulated by ubiquitin-dependent degradation (55). Thus, we knocked down UBCH7/UBE2L3 and assessed whether E3 levels were increased. As expected, a decrease of p27Kip1 on UBCH7/UBE2L3 depletion was observed in both HeLa and HEK-293 cells (Fig. 3A, top). No increases in Skp2, KPC1, Pirh2, or Ro52 were observed on UBCH7/UBE2L3 depletion (Fig. 3A). The modest decrease in Ro52 levels on UBCH7/UBE2L3 depletion observed in HeLa cells is unlikely to contribute to the decrease in p27Kip1 because a decrease in the concentration of this E3 would lead to an increase in p27Kip1. The E3 subunit DDB1 is also not affected by UBCH7/UBE2L3 depletion (Fig. 3B). Additionally, levels of the UBCH7/UBE2L3 partner E3 HHARI/ARIH1 is not altered on UBCH7/UBE2L3 knockdown (Supplemental Fig. S2). Thus, UBCH7/UBE2L3 depletion does not stabilize any of the E3s known to ubiquitinylate p27Kip1. One report indicates that E6AP may affect the ubiquitinylation of p27Kip1 and target it for degradation (48), but we previously observed that E6AP levels were unchanged on UBCH7/UBE2L3 depletion (6). Thus, it seems unlikely that the increase of p27Kip1 levels that are associated with increased UBCH7/UBE2L3 is due to an E6AP-dependent process.
Figure 3.
UBCH7/UBE2L3 depletion does not alter levels of E3s known to target p27Kip1 for proteasomal degradation. HeLa and HEK-293 cells were treated with siRNA specific for UBCH7/UBE2L3 or control siRNA for 72 h. Cells were lysed and probed for p27Kip1, UBCH7/UBE2L3, and E1 as loading control. A) Lysates from HeLa and HEK-293 cells were probed in 3 separate Western blots for E3s. B) HeLa lysates were probed for DDB1, p27Kip1, UBCH7/UBE2L3, and E1 as noted.
We also asked whether levels of any of the E3s known to ubiquitinylate other substrates in a UBCH7/UBE2L3-dependent manner are affected by UBCH7/UBE2L3 depletion. We did not see changes in the levels of the UBCH7/UBE2L3 E3 partners NEDD4 or TRAF6 (Supplemental Fig. S2). Although Parkin is a known UBCH7/UBE2L3 partner, because of the very low expression level in HeLa cells (56), it is unlikely that Parkin is involved in the regulation of p27Kip1.
UBCH7/UBE2L3 forms ubiquitin conjugates of p27Kip1 that resist degradation
The finding that UBCH7/UBE2L3 stabilizes p27Kip1 elicited the hypothesis that UBCH7/UBE2L3 inhibits the ubiquitinylation of p27Kip1. Using a cell-free assay, we compared the ubiquitinylation of p27Kip1 in the presence of UBCH7/UBE2L3 to ubiquitinylation catalyzed by Cdc34/Ubc3/UBE2R1, the canonical E2 that ubiquitinylates p27Kip1 in conjunction with the SCFskp2 complex to target p27Kip1 for proteasomal degradation (57). When p27Kip1 was added to the lysates, there was no significant increase in overall ubiquitin conjugate levels (Fig. 4A, left, lanes 1–4 vs. 5–8), indicating that such an addition did not cause a major perturbation in overall ubiquitinylation capabilities under various conditions. Addition of Cdc34/Ubc3/UBE2R1 (lane 3) and UBCH7/UBE2L3 (Fig. 4A, lane 4) catalyzed the formation of high-mass conjugates of many proteins, as expected (Fig. 4A, left, lanes 3, 4 vs. 2). These are found at the top of the resolving gel and as very high mass conjugates that remain in the stacking gel (Fig. 4A, left).
Figure 4.
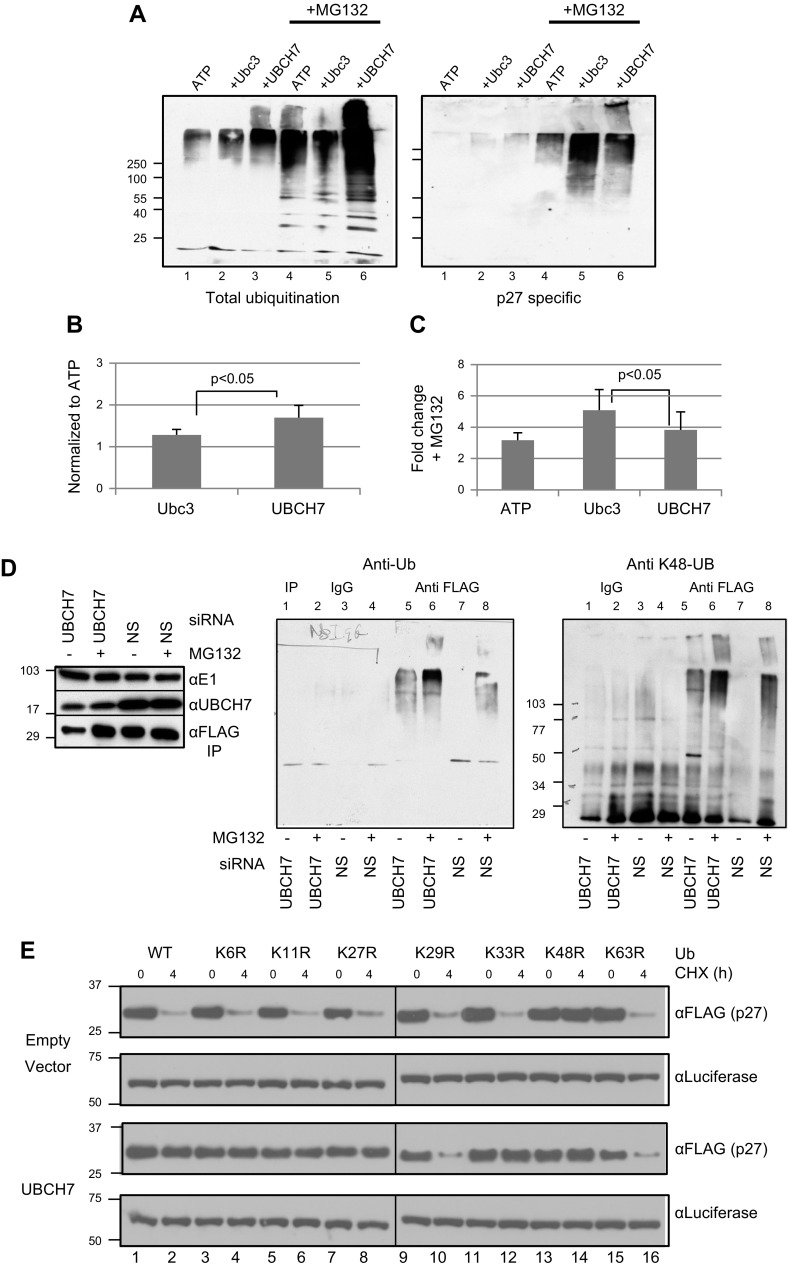
UBCH7/UBE2L3 catalyzes ubiquitinylation of p27Kip1. In vitro ubiquitinylation assay using HELC G0 lysate and His6-p27Kip1. Ubiquitinylation was carried out in presence of 500 nM Cdc34/Ubc3/UBE2R1 (Ubc3) or UBCH7/UBE2L3 (UBCH7) as indicated; ATP and ubiquitin were added (+) in lanes as noted. Immunoblots are for total ubiquitin. Brackets identify high mass conjugates in stacking gel. A) Ubiquitinylation with WT ubiquitin. Left: total ubiquitinylation; right, after Ni-NTA agarose purification. First 4 lanes in each blot contained His6-p27Kip1; lanes 5–8 had no His6-p27Kip1. B) p27Kip1 was isolated as in (A), and ubiquitinylation was performed with K-only mutant ubiquitins: K11O, K29O, K63O, K48R or K48O. C) Quantification from 3 (K48R) or 4 (K48O) independent experiments.
Unexpectedly, however, on p27Kip1 isolation by Ni-NTA agarose purification, the addition of UBCH7/UBE2L3 increased p27Kip1-ubiquitin conjugates to a similar or even greater extent than Cdc34/Ubc3/UBE2R1 (Fig. 4A, right, lane 4 vs. 3). The ubiquitin conjugates formed on p27Kip1 by UBCH7/UBE2L3 were consistently of higher mass compared to those catalyzed by Cdc34/Ubc3/UBE2R1 (percentage of conjugates in the stacking gel UBCH7/UBE2L3 = 28.3% vs. Cdc34/Ubc3/UBE2R1 = 17.1%, P < 0.003; percentage of conjugates <100 kDa UBCH7/UBE2L3 = 27.7% vs. Cdc34/Ubc3/UBE2R1 = 39.5%, P < 0.001, n = 13. Supplemental Fig. S5A, lane 2, left vs. right panels). No ubiquitin conjugates were observed in the isolated Ni-NTA agarose purification samples to which His6-p27Kip1 was not added (Fig. 4A, right, lanes 5–8).
Given the surprising finding that UBCH7/UBE2L3 catalyzes formation and stabilization of high-mass ubiquitin p27Kip1 conjugates, we hypothesized that the conjugates catalyzed by UBCH7/UBE2L3 on p27Kip1 might be resistant to proteasomal degradation, as has been demonstrated for a limited number of ubiquitinylated substrates (28–31). We tested the ability of UBCH7/UBE2L3 to catalyze the attachment of noncanonical ubiquitin chains on p27Kip1 by using WT and mutant ubiquitins in our conjugation assays. Using ubiquitin mutants that have only one lysine available for chain formation dictates the type of ubiquitin chain formed. UBCH7/UBE2L3 clearly catalyzes formation of ubiquitin adducts on p27Kip1 using K11-only, K29-only, K48-only, and K63-only ubiquitins (Fig. 4B; note that K48-only data are at far right; Supplemental Fig. S5A, top left, lanes 4, 6, 8, 9). By comparison, Cdc34/Ubc3/UBE2R1 is only able to enhance conjugate formation of p27Kip1 with WT and K48-only ubiquitin (Fig. 4B and Supplemental Fig. S5A, top right, lanes 2, 8). These data suggest that p27Kip1 ubiquitin conjugates, catalyzed by UBCH7/UBE2L3 with WT ubiquitin, are a mixture of different ubiquitin chain linkages. Our observation of enhanced ubiquitinylation when specific ubiquitins and E2s are added indicates that the lysates have only limited levels of endogenous ubiquitin and E2s (58, 59). Thus, the ubiquitinylation activity of our lysates is dictated by the type of ubiquitin added, and the ubiquitinylation patterns reflect the activity of the E2s.
Next, we did reciprocal ubiquitinylations by interrogating conjugation catalyzed by UBCH7/UBE2L3 and Cdc34/Ubc3/UBE2R1 activity using ubiquitin mutants in which only a single lysine was mutated to arginine. These K-to-R mutant ubiquitins preclude formation of ubiquitin linkages through the mutated lysine, but they allow ubiquitylation on all remaining lysines. Consistent with the observation that Cdc34/Ubc3/UBE2R1 makes primarily K48-linked ubiquitin conjugates (60), we observe that Cdc34/Ubc3/UBE2R1 is significantly impaired in its ability to catalyze ubiquitinylation of total cellular proteins with the K48R mutant ubiquitin or to enhance levels of conjugates above endogenous levels (Fig. 4B, lane 3 vs. 2 in each panel; Supplemental Fig. S5D, right, lane 8 vs. 2). Ubiquitin mutants in lysines other than K48 have K48 available and consequently do enter into polyubiquitinylated conjugates in the presence of Cdc34/Ubc3/UBE2R1. The ability of the K-to-R mutant ubiquitins that have K48 available to enter into conjugates is even more obvious when only p27Kip1 conjugates are examined (Fig. 4B, right panels). However, no conjugates of p27Kip1 are formed with K48R-Ub with Cdc34/Ubc3/UBE2R1 (Fig. 4B and Supplemental Fig. S5C). In contrast, UBCH7/UBE2L3 is able to catalyze ubiquitinylation of total endogenous substrates as well as of p27Kip1 specifically with all the K-to-R ubiquitin mutants (Fig. 4B and Supplemental Fig. S5C, D, left). These results confirm that UBCH7/UBE2L3 catalyzes the formation of a variety of ubiquitin chains on p27Kip1, while Cdc34/Ubc3/UBE2R1 primarily catalyzes the formation of primarily K48-linked chains on p27Kip1.
We next tested the hypothesis that conjugates of p27Kip1 that are synthesized in the presence of UBCH7/UBE2L3 are not targeted to the proteasome for degradation, whereas Cdc34/Ubc3/UBE2R1-catalyzed p27Kip1 conjugates would be degraded. To do this, we performed ubiquitinylation assays in the presence or absence of MG132. Consistently, in the absence of MG132, more conjugates of total proteins, and of p27Kip1 specifically, accumulated in the presence of UBCH7/UBE2L3 than when Cdc34/Ubc3/UBE2R1 was added (Fig. 5A, B, lane 3 vs. 2), in keeping with the notion that UBCH7/UBE2L3 catalyzes ubiquitin conjugates that are degradation incompetent, whereas conjugates formed by Cdc34/Ubc3/UBE2R1 are recognized and degraded by the proteasome. The addition of proteasome inhibitor results in a marked accumulation of total ubiquitin conjugates of endogenous substrates when either E2 is added, consistent with the paradigm that many ubiquitin conjugates are destined for proteasomal degradation (Fig. 5A, left, lanes 4–6). However, the extent of the accumulation of total ubiquitin conjugates is greater for the UBCH7/UBE2L3 (Fig. 5A, left; lanes 6, 5). The same patterns are observed for conjugates of p27Kip1, and the effects of UBCH7/UBE2L3 are even more obvious for the highest-mass conjugates of p27Kip1 (Fig. 5A, right, lane 6 vs. 5, B). However, the extent of increase in all conjugates of p27Kip1 on addition of MG132 is consistently greater for Cdc34/Ubc3/UBE2R1 vs. the accumulation of conjugates in the presence of UBCH7/UBE2L3, consistent with Cdc34/Ubc3/UBE2R1 forming degradation competent conjugates with K48-linked ubiquitin chains on p27Kip1 (Fig. 5A, C) that are indeed destined for proteasomal degradation (Supplemental Fig. S5A, right). As noted earlier, we also observed an increase in the amount of conjugates formed on p27Kip1 with UBCH7/UBE2L3 in the presence of MG132, suggesting that some of these p27Kip1 conjugates are in fact targeted for proteasomal degradation. Taken together, our data indicate that UBCH7/UBE2L3 catalyzes the attachment of ubiquitin chains on p27Kip1, some of which are not degraded by the proteasome, whereas other UBCH7/UBE2L3 catalyzed ubiquitinylations on other substrates are proteolytically competent.
Figure 5.
UBCH7/UBE2L3 catalyzes attachment of nonproteasome targeting chains on p27Kip1. In vitro ubiquitinylation assays were performed in presence and absence of MG132. A) Left: total ubiquitination. Right: p27Kip1 specific ubiquitinylation. B) Quantification of conjugates on p27Kip1 in absence of MG132, normalized to amount of conjugation (6, 12), formed in absence of additional E2. C) Fold change of ubiquitin conjugates on p27Kip1 in presence compared to absence of MG132. B, C) Average of 8 to 10 independent experiments. Significance values calculated by paired Student’s t test; data presented as means ± sem. D) Ubiquitinylation of p27Kip1 in cells depleted of UBCH7/UBE2L3. Left: lysate or anti FLAG immunoprecipitates probed for E1 (loading control) UBCH7/UBE2L3 (UBCH7) or FLAG (p27Kip1), as indicated. Middle, immunoprecipitates probed with anti-ubiquitin. Right, immunoprecipitates probed with anti-K48 ubiquitin. Representative of 4 independent experiments. E) K29 and K63 ubiquitin chains protect p27Kip1 from degradation in UBCH7/UBE2L3-dependent manner. HEK-293 cells were transfected with UBCH7/UBE2L3 or empty vector (EV) and FLAG-p27Kip1 with WT or K-to-R mutant ubiquitin. Cells were treated for 4 h with cycloheximide (CHX) and probed with anti-FLAG. Luciferase was cotransfected and used as loading and transfection control. Representative of 3 independent experiments.
Next we asked whether the absence of UBCH7/UBE2L3 affects the ubiquitinylation and proteasomal degradation of p27Kip1 in live cells. Thus, we expressed FLAG-p27, knocked down UBCH7/UBE2L3 using siRNA, and did or did not treat cells with MG132 to stabilize substrates targeted for degradation. In the absence of MG132, knockdown of UBCH7/UBE2L3 decreased p27Kip1 levels (Fig. 5D, left, lane 1 vs. 3, bottom row), whereas it was stabilized when the proteasome was inhibited (Fig. 5D, left, lane 2 vs. 1, bottom row). There was no effect of MG132 on p27Kip1 if UBCH7/UBE2L3 was not diminished (Fig. 5D, lanes 3, 4 vs. 2). To examine p27Kip1-specific ubiquitinylation, immunoprecipitates were probed with anti-ubiquitin (Fig. 5D, middle, lanes 5–8). Control immunoprecipitates with isotype-matched IgG did not show any ubiquitin signal (Fig. 5D, middle, lanes 1–4). Depletion of UBCH7/UBE2L3 increased ubiquitin conjugates of p27Kip1 (Fig. 5D, middle, lane 5 vs. 7). This suggests that in the absence of UBCH7/UBE2L3, Cdc34/Ubc3/UBE2R1 has more access and catalyzes formation of more, presumably K48 linked, ubiquitinylated p27Kip1. Consistent with this hypothesis, the amount of ubiquitinylation is further increased on MG132 treatment (Fig. 5D, middle, lane 6 vs. 5). This was further corroborated by observation of increased K48-linked chains on p27Kip1, a hallmark of Cdc34/Ubc3/UBE2R1-directed ubiquitinylation (60), on depletion of UBCH7/UBE2L3 (Fig. 5D, right, lanes 5, 6 vs. 7, 8). As expected, for K48-linked, proteasome-competent substrate ubiquitin conjugates, inhibition of the proteasome resulted in stabilization of K48-linked ubiquitin conjugates of p27Kip1 (Fig. 5D, right, lanes 6, 8 vs. 5, 7), and much of the K48-linked ubiquitinylated p27Kip1 is degraded in the absence of proteasome inhibition (lane 7).
We then tested the hypothesis that UBCH7/UBE2L3 catalyzes formation of non-K48 linked chains to protect p27kip1 from proteasomal degradation in cells. New protein synthesis was limited by treatment with cycloheximide for 4 h. In the absence of added UBCH7/UBE2L3, p27kip1 is degraded in the presence of WT ubiquitin (Fig. 5E, top; lane 2 vs. 1). When K-to-R ubiquitins were expressed in the absence of UBCH7/UBE2L3, degradation of p27Kip1 was observed, suggesting that these mutant ubiquitins do not significantly interfere with endogenous ubiquitin for formation of conjugates of p27kip1 that are recognized for degradation. As might be anticipated, the only exception to this is K48R-Ub, which precludes formation of canonical K48 proteasome targeting chains. However, when UBCH7/UBE2L3 is overexpressed, p27kip1 is stabilized, with expression of WT and most K-to-R mutants suggesting that in live cells and in cell-free conditions, UBCH7/UBE2L3 catalyzes formation of a preponderance of proteolytically incompetent ubiquitin conjugates on p27Kip1 (Fig. 5E, third panel). In the presence of UBCH7/UBE2L3, all the K-to-R mutant ubiquitins are integrated into conjugates (Supplemental Fig. S5C, D). Strikingly however, expression of either K29R or K63R does not result in stabilization of p27Kip1 in the presence of UBCH7/UBE2L3. Taken together, these data suggest that it is likely that both K29 and K63 of ubiquitin are involved in generation of proteasome-resistant ubiquitinylated p27Kip1.
We then asked whether UBCH7/UBE2L3 could catalyze K63-linked ubiquitinylation of p27Kip1 in cells. Consistent with our in vitro ubiquitinylation data (Fig. 4B), overexpression of UBCH7/UBE2L3 catalyzed K63 ubiquitinylation of p27Kip1 (Supplemental Fig. S6).
DISCUSSION
UBCH7/UBE2L3 is critical for survival, and its levels and/or the extent to which it is activated vary during cell cycle (6, 38, 39). Here we document a novel, noncanonical role for UBCH7/UBE2L3 in catalyzing the attachment of degradation-resistant ubiquitin chains to, and stabilizing, p27Kip1, resulting in delayed cell-cycle entry and progression in HeLa (Fig. 1), retinal pigmented epithelial cells, HLEC, and HECT 293 but purportedly not in A529 cells (61), a result we were unable to replicate (data not shown). The stabilization of substrates on elevation of levels of a Ubc enzyme in live cells contrasts with prior indications that E2s promote the degradation of proteolytic substrates, including the specific promotion of degradation of p27Kip1 by the major cell cycle regulating E2 and Cdc34/Ubc3/UBE2R1, and the degradation of mitotic regulators by E2s, UbcH10, and Ube2S (32).
The functional ramifications of this noncanonical ubiquitin conjugation activity are profound because p27Kip1 is involved in regulating proliferation (45, 62), development, differentiation (21, 63, 64), cell migration (65), senescence (66, 67), disease prognosis (68, 69), cell–cell communication (70), and glucocorticoid responses (71), and is regulated in response to the unfolded protein response (42). Our data may also help rationalize the defect in the placental labyrinth that is seen in UBCH7/UBE2L3 knockdown mutant mice (22). A similar placental defect was also observed in p27Kip1−/−/p57Kip2−/+ mice (72). Both mice have altered placental vasculature, likely resulting in decreased maternal/fetal nutrient exchange. Thus, the placental defect in the UBCH7/UBE2L3 mutant mice might be due to diminished levels of p27Kip1. Unfortunately, these animals are no longer in existence. It is plausible that this UBCH7/UBE2L3-induced dysregulation of p27Kip1 and cell cycle is mechanistically analogous to the stabilization of p27Kip1, the resulting inhibition of Cdk1, and the abnormal proliferation, differentiation and growth phenotypes that are observed when K6W-ubiquitin is expressed (5, 21, 42).
The paradigm for the role of ubiquitinylation in proteasomal degradation is that increased ubiquitinylation enhances proteasomal degradation, as exemplified by K48 linked poly-ubiquitin chains. The results in this report indicate that UBCH7/UBE2L3 catalyzes attachment of non–proteasome-targeting ubiquitin chains to p27Kip1 and possibly other substrates. These appear to include conjugates involving ubiquitin chains with K11, K29, K48, and K63 linkages (Fig. 4 and Supplemental Figs. S5 and S6). Such attachments of non–K48-linked ubiquitin chains can occur even when only WT ubiquitin is present and does not exclude formation of canonical proteolysis-competent ubiquitin chains (Figs. 4 and 5) (73). Support for this noncanonical function for UBCH7/UBE2L3 is indicated because only WT, but not inactive mutant UBCH7/UBE2L3, protects p27Kip1 from degradation in multiple experimental settings, both in live cells and in cell-free situations (Fig. 1). Corroboration is indicated by the observations that on tissue differentiation, when p27Kip1 rises (38), UBCH7/UBE2L3 is in an ubiquitin-charged state, whereas depletion of UBCH7/UBE2L3 increases proteasomal degradation of p27Kip1 in multiple cell types (Fig. 2). These findings are also consistent with precedents showing that UBCH7/UBE2L3 can catalyze formation of linear (74), K11 (75), K48 (76), and K63-based chains (77, 78) on other substrates.
Our data further suggest that K29- and K63-linked ubiquitin chains protect p27Kip1 by precluding the formation of proteolytically competent K48-linked chains. An analog is the ubiquitinylation of Myc by 1 E3 ligase, which inhibits the ubiquitinylation by another E3, with unanticipated protection from degradation (79). Our data are also consistent with cell-free–based findings that E3s, MuRF1, Mdm2, and ChIP can catalyze formation of forked chains on particular substrates with the promiscuous E2 UbcH5/UBE2D (80) as well as with enhanced stabilization of green fluorescent protein (GFP) in assays that used E3/E2 combinations Cul3/UbcH5/UBE2D vs. Rsp5/UBCH7/UBE2L3 (78). RNF4 catalyzed attachment of K11/K33 chains that inhibit degradation of β-catenin (30), and SCFFbl12 catalyzes the attachment of K48/K63 mixed chains to inhibit degradation of p21Cip1/Waf1 (31). The stabilization of p27Kip1 that we observed in the presence of UBCH7/UBE2L3 is in contrast to the mixed ubiquitin chains, involving K63, that are assembled on TXNIP and that allow for degradation. Thus, these findings indicate that the status of E2s is another critical aspect of cellular context that determines ubiquitination and degradation of a substrate (33).
Seven E3s have been shown to directly or indirectly regulate turnover of p27Kip1, and we determined the relations between each of them and the levels of UBCH7/UBE2L3 and p27Kip1. Levels of the F box Skp2 (45), Ro52 (44), DDB1 (47), the KPC1/KPC2 complex (8), Pirh2 (46), and E6AP (48) were not altered on UBCH7/UBE2L3 depletion (Fig. 3) (6). Depletion of UBCH7/UBE2L3 did not alter levels of E3s that ubiquitinylate other substrates with the aid of UBCH7/UBE2L3. These include E6AP (6) NEDD4, TRAF6, and HHARI/ARIH1 (Supplemental Fig. S2). The latter collaborates with UBCH7/UBE2L3 to ubiquitinylate and target for degradation p27Kip1 and a number of cullin ring ligase substrates (53). We did not observe changes in β-catenin and several other cullin ring ligase substrates on UBCH7/UBE2L3 depletion (Supplemental Fig. S1). Our findings distinguish the effects of limiting UBCH7/UBE2L3 from limiting the E3 HHARI/ARIH1, which led to the accumulation rather than degradation of a number of cullin substrates, including p27Kip1 (53), and from substrate-E2 interactions that alter degradation and function of Nrf2 (81).
Although it has been suggested that UBCH7/UBE2L3 can catalyze ubiquitinylation only with HECT and RING/HECT hybrid domain E3s (82), other investigations indicate that UBCH7/UBE2L3, along with the SCF, a RING E3 ligase, catalyzes ubiquitinylation of the E7 oncoprotein to target it for degradation (83), and UBCH7/UBE2L3 can ubiquitinylate Trk1 with the RING domain E3 TRAF6 (77). These observations, together with findings that UBCH7/UBE2L3 cannot add ubiquitins directly to substrates, as is necessary with RING domain E3s, suggest that the ability of UBCH7/UBE2L3 to ubiquitinylate substrates with RING domain E3s such as the SCF may involve other as yet unidentified partners.
We also asked whether other cell-cycle regulatory substrates are subject to the same regulation by UBCH7/UBE2L3, but we did not see comparable protection of other CDKIs such as p21Cip1/Waf1 (Fig. 2), p57Kip2 (Supplemental Fig. S1E), or CDK activators cyclin A or cyclin E (Supplemental Fig. S1A, B).
Here we demonstrate for the first time that increased UBCH7/UBE2L3 results in increased ubiquitinylation but diminished degradation of p27Kip1, thus indicating a previously unappreciated ubiquitinylation-based biology that is required for cell proliferation, differentiation, and development, as well as maintaining protein quality and homeostasis. Specifically, stabilization of cell-cycle regulatory proteins by the attachment of mixed ubiquitin chains appears to be another mechanism to regulate the cell cycle. These data have pharmacologic ramifications, as dysregulation of p27Kip1 is prognostic of a number of cancers and senescence (68, 69), and it is crucial to understand regulation of this CDKI to design new drugs and therapies to control cell proliferation. Our observations are consistent with the possibility that transient ubiquitinylation, like glutathiolation (84), may be exploited to temporarily protect proteins against premature degradation or processing, and they require answers to understand changes in levels of E2s during the cell cycle (39).
Supplementary Material
This article includes supplemental data. Please visit http://www.fasebj.org to obtain this information.
ACKNOWLEDGMENTS
Funding for this work was provided by U.S. National Institutes of Health (NIH) National Eye Institute Grants RO1 EY 13250, RO1 EY21212, and RO1 EY26979, U.S. Department of Agriculture contract 1950-510000-060-03A (to A.T.), and the NIH Center for Cancer Research–National Cancer Institute Intramural Research Program (to Y.C.T. and A.M.W.). The authors thank E. Bejarano-Fernandez (Tufts University) for critical review and W. Yang (Brandeis University, Waltham, MA, USA) for expert technical assistance. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. The authors declare no conflicts of interest.
Glossary
- CDKI
cyclin-dependent kinase inhibitor
- CPM
counts per minute
- E1
ubiquitin-activating enzyme
- E2
ubiquitin-conjugation enzyme
- E3
ubiquitin ligase
- GFP
green fluorescent protein
- HLEC
human lens epithelial cell
- HU
hydroxyurea
- siRNA
small interfering RNA
- SCF
SKP1/Cullin/F-box containing
- TCA
trichloroacetic acid
- Ubc
ubiquitin conjugating
- UPS
ubiquitin proteasome system
- WT
wild type
Footnotes
This article includes supplemental data. Please visit http://www.fasebj.org to obtain this information.
AUTHOR CONTRIBUTIONS
E. A. Whitcomb, J. Basappa, and A. Taylor conceived the study; E. A. Whitcomb, Y. C. Tsai, J. Basappa, K. Liu, and A. K. Le Feuvre designed the experiments; E. A. Whitcomb, Y. C. Tsai, J. Basappa, K. Liu, and A. K. Le Feuvre performed the experiments; E. A. Whitcomb, Y. C. Tsai, J. Basappa, K. Liu, A. K. Le Feuvre, A. M. Weissman, and A. Taylor analyzed the data; and E. A. Whitcomb and A. Taylor wrote the manuscript.
REFERENCES
- 1.Hershko A., Ciechanover A., Rose I. A. (1979) Resolution of the ATP-dependent proteolytic system from reticulocytes: a component that interacts with ATP. Proc. Natl. Acad. Sci. USA 76, 3107–3110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu Q., Shang F., Zhang X., Li W., Taylor A. (2006) Expression of K6W-ubiquitin inhibits proliferation of human lens epithelial cells. Mol. Vis. 12, 931–936 [PubMed] [Google Scholar]
- 3.Topper L. M., Bastians H., Ruderman J. V., Gorbsky G. J. (2001) Elevating the level of Cdc34/Ubc3 ubiquitin-conjugating enzyme in mitosis inhibits association of CENP-E with kinetochores and blocks the metaphase alignment of chromosomes. J. Cell Biol. 154, 707–717; erratum: 158, 371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Williamson A., Wickliffe K. E., Mellone B. G., Song L., Karpen G. H., Rape M. (2009) Identification of a physiological E2 module for the human anaphase-promoting complex. Proc. Natl. Acad. Sci. USA 106, 18213–18218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu K., Lyu L., Chin D., Gao J., Sun X., Shang F., Caceres A., Chang M. L., Rowan S., Peng J., Mathias R., Kasahara H., Jiang S., Taylor A. (2015) Altered ubiquitin causes perturbed calcium homeostasis, hyperactivation of calpain, dysregulated differentiation, and cataract. Proc. Natl. Acad. Sci. USA 112, 1071–1076; erratum: 112, E817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Whitcomb E. A., Dudek E. J., Liu Q., Taylor A. (2009) Novel control of S phase of the cell cycle by ubiquitin-conjugating enzyme H7. Mol. Biol. Cell 20, 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rape M., Kirschner M. W. (2004) Autonomous regulation of the anaphase-promoting complex couples mitosis to S-phase entry. Nature 432, 588–595 [DOI] [PubMed] [Google Scholar]
- 8.Kamura T., Hara T., Matsumoto M., Ishida N., Okumura F., Hatakeyama S., Yoshida M., Nakayama K., Nakayama K. I. (2004) Cytoplasmic ubiquitin ligase KPC regulates proteolysis of p27(Kip1) at G1 phase. Nat. Cell Biol. 6, 1229–1235 [DOI] [PubMed] [Google Scholar]
- 9.Liu Q., Shang F., Whitcomb E., Guo W., Li W., Taylor A. (2006) Ubiquitin-conjugating enzyme 3 delays human lens epithelial cells in metaphase. Invest. Ophthalmol. Vis. Sci. 47, 1302–1309 [DOI] [PubMed] [Google Scholar]
- 10.Al-Hakim A., Escribano-Diaz C., Landry M. C., O’Donnell L., Panier S., Szilard R. K., Durocher D. (2010) The ubiquitous role of ubiquitin in the DNA damage response. DNA Repair (Amst.) 9, 1229–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carter S., Bischof O., Dejean A., Vousden K. H. (2007) C-terminal modifications regulate MDM2 dissociation and nuclear export of p53. Nat. Cell Biol. 9, 428–435 [DOI] [PubMed] [Google Scholar]
- 12.Dudek E. J., Shang F., Valverde P., Liu Q., Hobbs M., Taylor A. (2005) Selectivity of the ubiquitin pathway for oxidatively modified proteins: relevance to protein precipitation diseases. FASEB J. 19, 1707–1709 [DOI] [PubMed] [Google Scholar]
- 13.Marques C., Guo W., Pereira P., Taylor A., Patterson C., Evans P. C., Shang F. (2006) The triage of damaged proteins: degradation by the ubiquitin–proteasome pathway or repair by molecular chaperones. FASEB J. 20, 741–743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shang F., Nowell T. R., Jr., Taylor A. (2001) Removal of oxidatively damaged proteins from lens cells by the ubiquitin–proteasome pathway. Exp. Eye Res. 73, 229–238 [DOI] [PubMed] [Google Scholar]
- 15.Sobhian B., Shao G., Lilli D. R., Culhane A. C., Moreau L. A., Xia B., Livingston D. M., Greenberg R. A. (2007) RAP80 targets BRCA1 to specific ubiquitin structures at DNA damage sites. Science 316, 1198–1202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hershko A., Ciechanover A. (1998) The ubiquitin system. Annu. Rev. Biochem. 67, 425–479 [DOI] [PubMed] [Google Scholar]
- 17.Hashizume R., Fukuda M., Maeda I., Nishikawa H., Oyake D., Yabuki Y., Ogata H., Ohta T. (2001) The RING heterodimer BRCA1-BARD1 is a ubiquitin ligase inactivated by a breast cancer–derived mutation. J. Biol. Chem. 276, 14537–14540 [DOI] [PubMed] [Google Scholar]
- 18.Berlingieri M. T., Pallante P., Sboner A., Barbareschi M., Bianco M., Ferraro A., Mansueto G., Borbone E., Guerriero E., Troncone G., Fusco A. (2007) UbcH10 is overexpressed in malignant breast carcinomas. Eur. J. Cancer 43, 2729–2735 [DOI] [PubMed] [Google Scholar]
- 19.Eliseeva E., Pati D., Diccinanni M. B., Yu A. L., Mohsin S. K., Margolin J. F., Plon S. E. (2001) Expression and localization of the CDC34 ubiquitin-conjugating enzyme in pediatric acute lymphoblastic leukemia. Cell Growth Differ. 12, 427–433 [PubMed] [Google Scholar]
- 20.Lisztwan J., Imbert G., Wirbelauer C., Gstaiger M., Krek W. (1999) The von Hippel-Lindau tumor suppressor protein is a component of an E3 ubiquitin-protein ligase activity. Genes Dev. 13, 1822–1833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Caceres A., Shang F., Wawrousek E., Liu Q., Avidan O., Cvekl A., Yang Y., Haririnia A., Storaska A., Fushman D., Kuszak J., Dudek E., Smith D., Taylor A. (2010) Perturbing the ubiquitin pathway reveals how mitosis is hijacked to denucleate and regulate cell proliferation and differentiation in vivo. PLoS One 5, e13331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harbers K., Müller U., Grams A., Li E., Jaenisch R., Franz T. (1996) Provirus integration into a gene encoding a ubiquitin-conjugating enzyme results in a placental defect and embryonic lethality. Proc. Natl. Acad. Sci. USA 93, 12412–12417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nakayama K., Nagahama H., Minamishima Y. A., Matsumoto M., Nakamichi I., Kitagawa K., Shirane M., Tsunematsu R., Tsukiyama T., Ishida N., Kitagawa M., Nakayama K., Hatakeyama S. (2000) Targeted disruption of Skp2 results in accumulation of cyclin E and p27(Kip1), polyploidy and centrosome overduplication. EMBO J. 19, 2069–2081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pickart C. M., Fushman D. (2004) Polyubiquitin chains: polymeric protein signals. Curr. Opin. Chem. Biol. 8, 610–616 [DOI] [PubMed] [Google Scholar]
- 25.Patel D., McCance D. J. (2010) Compromised spindle assembly checkpoint due to altered expression of Ubch10 and Cdc20 in human papillomavirus type 16 E6- and E7-expressing keratinocytes. J. Virol. 84, 10956–10964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Van Wijk S. J. L., Timmers H. T. M. (2010) The family of ubiquitin-conjugating enzymes (E2s): deciding between life and death of proteins. FASEB J. 24, 981–993 [DOI] [PubMed] [Google Scholar]
- 27.Kirkpatrick D. S., Denison C., Gygi S. P. (2005) Weighing in on ubiquitin: the expanding role of mass-spectrometry-based proteomics. Nat. Cell Biol. 7, 750–757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grice G. L., Lobb I. T., Weekes M. P., Gygi S. P., Antrobus R., Nathan J. A. (2015) The proteasome distinguishes between heterotypic and homotypic lysine-11-linked polyubiquitin chains. Cell Rep. 12, 545–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shang F., Deng G., Liu Q., Guo W., Haas A. L., Crosas B., Finley D., Taylor A. (2005) Lys6-modified ubiquitin inhibits ubiquitin-dependent protein degradation. J. Biol. Chem. 280, 20365–20374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thomas J. J., Abed M., Heuberger J., Novak R., Zohar Y., Beltran Lopez A. P., Trausch-Azar J. S., Ilagan M. X. G., Benhamou D., Dittmar G., Kopan R., Birchmeier W., Schwartz A. L., Orian A. (2016) RNF4-dependent oncogene activation by protein stabilization. Cell Rep. 16, 3388–3400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tsuruta F., Takebe A., Haratake K., Kanemori Y., Kim J., Endo T., Kigoshi Y., Fukuda T., Miyahara H., Ebina M., Baba T., Chiba T. (2016) SCFFbl12 increases p21Waf1/Cip1 expression level through atypical ubiquitin chain synthesis. Mol. Cell. Biol. 36, 2182–2194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meyer H. J., Rape M. (2014) Enhanced protein degradation by branched ubiquitin chains. Cell 157, 910–921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ohtake F., Tsuchiya H., Saeki Y., Tanaka K. (2018) K63 ubiquitylation triggers proteasomal degradation by seeding branched ubiquitin chains. Proc. Natl. Acad. Sci. USA 115, E1401–E1408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chiaroni-Clarke R. C., Munro J. E., Chavez R. A., Pezic A., Allen R. C., Akikusa J. D., Piper S. E., Saffery R., Ponsonby A. L., Ellis J. A. (2014) Independent confirmation of juvenile idiopathic arthritis genetic risk loci previously identified by immunochip array analysis. Pediatr. Rheumatol. Online J. 12, 53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Orozco G., Eyre S., Hinks A., Bowes J., Morgan A. W., Wilson A. G., Wordsworth P., Steer S., Hocking L., Thomson W., Worthington J., Barton A.; UKRAG Consortium (2011) Study of the common genetic background for rheumatoid arthritis and systemic lupus erythematosus. Ann. Rheum. Dis. 70, 463–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang Y., Zhu Y. F., Wang Q., Xu J., Yan N., Xu J., Shi L. F., He S. T., Zhang J. A. (2016) The haplotype of UBE2L3 gene is associated with Hashimoto’s thyroiditis in a Chinese Han population. BMC Endocr. Disord. 16, 18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fransen K., Visschedijk M. C., van Sommeren S., Fu J. Y., Franke L., Festen E. A., Stokkers P. C., van Bodegraven A. A., Crusius J. B., Hommes D. W., Zanen P., de Jong D. J., Wijmenga C., van Diemen C. C., Weersma R. K. (2010) Analysis of SNPs with an effect on gene expression identifies UBE2L3 and BCL3 as potential new risk genes for Crohn’s disease. Hum. Mol. Genet. 19, 3482–3488 [DOI] [PubMed] [Google Scholar]
- 38.Guo W., Shang F., Liu Q., Urim L., West-Mays J., Taylor A. (2004) Differential regulation of components of the ubiquitin–proteasome pathway during lens cell differentiation. Invest. Ophthalmol. Vis. Sci. 45, 1194–1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu Q., Shang F., Guo W., Hobbs M., Valverde P., Reddy V., Taylor A. (2004) Regulation of the ubiquitin proteasome pathway in human lens epithelial cells during the cell cycle. Exp. Eye Res. 78, 197–205 [DOI] [PubMed] [Google Scholar]
- 40.Deffenbaugh A. E., Scaglione K. M., Zhang L., Moore J. M., Buranda T., Sklar L. A., Skowyra D. (2003) Release of ubiquitin-charged Cdc34-S–Ub from the RING domain is essential for ubiquitination of the SCF(Cdc4)-bound substrate Sic1. Cell 114, 611–622 [DOI] [PubMed] [Google Scholar]
- 41.Craney A., Rape M. (2013) Dynamic regulation of ubiquitin-dependent cell cycle control. Curr. Opin. Cell Biol. 25, 704–710 [DOI] [PubMed] [Google Scholar]
- 42.Lyu L., Whitcomb E. A., Jiang S., Chang M. L., Gu Y., Duncan M. K., Cvekl A., Wang W. L., Limi S., Reneker L. W., Shang F., Du L., Taylor A. (2016) Unfolded-protein response–associated stabilization of p27(Cdkn1b) interferes with lens fiber cell denucleation, leading to cataract. FASEB J. 30, 1087–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Block K., Appikonda S., Lin H. R., Bloom J., Pagano M., Yew P. R. (2005) The acidic tail domain of human Cdc34 is required for p27Kip1 ubiquitination and complementation of a cdc34 temperature sensitive yeast strain. Cell Cycle 4, 1421–1427 [DOI] [PubMed] [Google Scholar]
- 44.Sabile A., Meyer A. M., Wirbelauer C., Hess D., Kogel U., Scheffner M., Krek W. (2006) Regulation of p27 degradation and S-phase progression by Ro52 RING finger protein. Mol. Cell. Biol. 26, 5994–6004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nakayama K., Nagahama H., Minamishima Y. A., Miyake S., Ishida N., Hatakeyama S., Kitagawa M., Iemura S., Natsume T., Nakayama K. I. (2004) Skp2-mediated degradation of p27 regulates progression into mitosis. Dev. Cell 6, 661–672 [DOI] [PubMed] [Google Scholar]
- 46.Hattori T., Isobe T., Abe K., Kikuchi H., Kitagawa K., Oda T., Uchida C., Kitagawa M. (2007) Pirh2 promotes ubiquitin-dependent degradation of the cyclin-dependent kinase inhibitor p27Kip1. Cancer Res. 67, 10789–10795 [DOI] [PubMed] [Google Scholar]
- 47.Iovine B., Iannella M. L., Bevilacqua M. A. (2011) Damage-specific DNA binding protein 1 (DDB1) is involved in ubiquitin-mediated proteolysis of p27Kip1 in response to UV irradiation. Biochimie 93, 867–875 [DOI] [PubMed] [Google Scholar]
- 48.Mishra A., Godavarthi S. K., Jana N. R. (2009) UBE3A/E6-AP regulates cell proliferation by promoting proteasomal degradation of p27. Neurobiol. Dis. 36, 26–34 [DOI] [PubMed] [Google Scholar]
- 49.Shang F., Deng G., Obin M., Wu C. C., Gong X., Smith D., Laursen R. A., Andley U. P., Reddan J. R., Taylor A. (2001) Ubiquitin-activating enzyme (E1) isoforms in lens epithelial cells: origin of translation, E2 specificity and cellular localization determined with novel site-specific antibodies. Exp. Eye Res. 73, 827–836 [DOI] [PubMed] [Google Scholar]
- 50.Borel F., Lacroix F. B., Margolis R. L. (2002) Prolonged arrest of mammalian cells at the G1/S boundary results in permanent S phase stasis. J. Cell Sci. 115, 2829–2838 [DOI] [PubMed] [Google Scholar]
- 51.Wu K., Kovacev J., Pan Z.-Q. (2010) Priming and extending: a UbcH5/Cdc34 E2 handoff mechanism for polyubiquitination on a SCF substrate. Mol. Cell 37, 784–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Connor M. K., Kotchetkov R., Cariou S., Resch A., Lupetti R., Beniston R. G., Melchior F., Hengst L., Slingerland J. M. (2003) CRM1/Ran-mediated nuclear export of p27(Kip1) involves a nuclear export signal and links p27 export and proteolysis. Mol. Biol. Cell 14, 201–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Scott D. C., Rhee D. Y., Duda D. M., Kelsall I. R., Olszewski J. L., Paulo J. A., de Jong A., Ovaa H., Alpi A. F., Harper J. W., Schulman B. A. (2016) Two distinct types of E3 ligases work in unison to regulate substrate ubiquitylation. Cell 166, 1198–1214.e1124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wei W., Ayad N. G., Wan Y., Zhang G. J., Kirschner M. W., Kaelin W. G., Jr (2004) Degradation of the SCF component Skp2 in cell-cycle phase G1 by the anaphase-promoting complex. Nature 428, 194–198 [DOI] [PubMed] [Google Scholar]
- 55.Lu Y., Adegoke O. A., Nepveu A., Nakayama K. I., Bedard N., Cheng D., Peng J., Wing S. S. (2009) USP19 deubiquitinating enzyme supports cell proliferation by stabilizing KPC1, a ubiquitin ligase for p27Kip1. Mol. Cell. Biol. 29, 547–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Narendra D., Tanaka A., Suen D.-F., Youle R. J. (2008) Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J. Cell Biol. 183, 795–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.King R. W., Deshaies R. J., Peters J. M., Kirschner M. W. (1996) How proteolysis drives the cell cycle. Science 274, 1652–1659 [DOI] [PubMed] [Google Scholar]
- 58.Shang F., Gong X., Taylor A. (1997) Activity of ubiquitin-dependent pathway in response to oxidative stress. Ubiquitin-activating enzyme is transiently up-regulated. J. Biol. Chem. 272, 23086–23093 [DOI] [PubMed] [Google Scholar]
- 59.Jahngen J. H., Haas A. L., Ciechanover A., Blondin J., Eisenhauer D., Taylor A. (1986) The eye lens has an active ubiquitin–protein conjugation system. J. Biol. Chem. 261, 13760–13767 [PubMed] [Google Scholar]
- 60.Petroski M. D., Deshaies R. J. (2005) Mechanism of lysine 48-linked ubiquitin-chain synthesis by the cullin–RING ubiquitin–ligase complex SCF-Cdc34. Cell 123, 1107–1120 [DOI] [PubMed] [Google Scholar]
- 61.Ma X., Zhao J., Yang F., Liu H., Qi W. (2017) Ubiquitin conjugating enzyme E2 L3 promoted tumor growth of NSCLC through accelerating p27kip1 ubiquitination and degradation. Oncotarget 8, 84193–84203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nakayama K.-I., Hatakeyama S., Nakayama K. (2001) Regulation of the cell cycle at the G1-S transition by proteolysis of cyclin E and p27Kip1. Biochem. Biophys. Res. Commun. 282, 853–860 [DOI] [PubMed] [Google Scholar]
- 63.Deschênes C., Vézina A., Beaulieu J. F., Rivard N. (2001) Role of p27(Kip1) in human intestinal cell differentiation. Gastroenterology 120, 423–438 [DOI] [PubMed] [Google Scholar]
- 64.Muñoz-Alonso M. J., Acosta J. C., Richard C., Delgado M. D., Sedivy J., León J. (2005) p21Cip1 and p27Kip1 induce distinct cell cycle effects and differentiation programs in myeloid leukemia cells. J. Biol. Chem. 280, 18120–18129 [DOI] [PubMed] [Google Scholar]
- 65.Larrea M. D., Wander S. A., Slingerland J. M. (2009) p27 as Jekyll and Hyde: regulation of cell cycle and cell motility. Cell Cycle 8, 3455–3461 [DOI] [PubMed] [Google Scholar]
- 66.Wagner M., Hampel B., Hütter E., Pfister G., Krek W., Zwerschke W., Jansen-Dürr P. (2001) Metabolic stabilization of p27 in senescent fibroblasts correlates with reduced expression of the F-box protein Skp2. Exp. Gerontol. 37, 41–55 [DOI] [PubMed] [Google Scholar]
- 67.Park S. H., Lim J. S., Jang K. L. (2011) All-trans retinoic acid induces cellular senescence via upregulation of p16, p21, and p27. Cancer Lett. 310, 232–239 [DOI] [PubMed] [Google Scholar]
- 68.Chu I. M., Hengst L., Slingerland J. M. (2008) The Cdk inhibitor p27 in human cancer: prognostic potential and relevance to anticancer therapy. Nat. Rev. Cancer 8, 253–267 [DOI] [PubMed] [Google Scholar]
- 69.Guan X., Wang Y., Xie R., Chen L., Bai J., Lu J., Kuo M. T. (2010) p27(Kip1) as a prognostic factor in breast cancer: a systematic review and meta-analysis. J. Cell. Mol. Med. 14, 944–953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shi Q., Gu S., Yu X. S., White T. W., Banks E. A., Jiang J. X. (2015) Connexin controls cell-cycle exit and cell differentiation by directly promoting cytosolic localization and degradation of E3 ligase Skp2. Dev. Cell 35, 483–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kullmann M. K., Grubbauer C., Goetsch K., Jäkel H., Podmirseg S. R., Trockenbacher A., Ploner C., Cato A. C., Weiss C., Kofler R., Hengst L. (2013) The p27-Skp2 axis mediates glucocorticoid-induced cell cycle arrest in T-lymphoma cells. Cell Cycle 12, 2625–2635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang P., Wong C., DePinho R. A., Harper J. W., Elledge S. J. (1998) Cooperation between the Cdk inhibitors p27(KIP1) and p57(KIP2) in the control of tissue growth and development. Genes Dev. 12, 3162–3167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kulathu Y., Komander D. (2012) Atypical ubiquitylation—the unexplored world of polyubiquitin beyond Lys48 and Lys63 linkages. Nat. Rev. Mol. Cell Biol. 13, 508–523 [DOI] [PubMed] [Google Scholar]
- 74.Stieglitz B., Morris-Davies A. C., Koliopoulos M. G., Christodoulou E., Rittinger K. (2012) LUBAC synthesizes linear ubiquitin chains via a thioester intermediate. EMBO Rep. 13, 840–846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.David Y., Ziv T., Admon A., Navon A. (2010) The E2 ubiquitin-conjugating enzymes direct polyubiquitination to preferred lysines. J. Biol. Chem. 285, 8595–8604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kim H. C., Huibregtse J. M. (2009) Polyubiquitination by HECT E3s and the determinants of chain type specificity. Mol. Cell. Biol. 29, 3307–3318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Geetha T., Kenchappa R. S., Wooten M. W., Carter B. D. (2005) TRAF6-mediated ubiquitination regulates nuclear translocation of NRIF, the p75 receptor interactor. EMBO J. 24, 3859–3868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Reichard E. L., Chirico G. G., Dewey W. J., Nassif N. D., Bard K. E., Millas N. E., Kraut D. A. (2016) Substrate ubiquitination controls the unfolding ability of the proteasome. J. Biol. Chem. 291, 18547–18561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Popov N., Schülein C., Jaenicke L. A., Eilers M. (2010) Ubiquitylation of the amino terminus of Myc by SCF(β-TrCP) antagonizes SCF(Fbw7)-mediated turnover. Nat. Cell Biol. 12, 973–981 [DOI] [PubMed] [Google Scholar]
- 80.Kim H. T., Kim K. P., Lledias F., Kisselev A. F., Scaglione K. M., Skowyra D., Gygi S. P., Goldberg A. L. (2007) Certain pairs of ubiquitin-conjugating enzymes (E2s) and ubiquitin-protein ligases (E3s) synthesize nondegradable forked ubiquitin chains containing all possible isopeptide linkages. J. Biol. Chem. 282, 17375–17386 [DOI] [PubMed] [Google Scholar]
- 81.Plafker K. S., Nguyen L., Barneche M., Mirza S., Crawford D., Plafker S. M. (2010) The ubiquitin-conjugating enzyme UbcM2 can regulate the stability and activity of the antioxidant transcription factor Nrf2. J. Biol. Chem. 285, 23064–23074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wenzel D. M., Lissounov A., Brzovic P. S., Klevit R. E. (2011) UBCH7 reactivity profile reveals parkin and HHARI to be RING/HECT hybrids. Nature 474, 105–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Oh K. J., Kalinina A., Wang J., Nakayama K., Nakayama K. I., Bagchi S. (2004) The papillomavirus E7 oncoprotein is ubiquitinated by UbcH7 and Cullin 1- and Skp2-containing E3 ligase. J. Virol. 78, 5338–5346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Jahngen-Hodge J., Obin M. S., Gong X., Shang F., Nowell T. R., Jr., Gong J., Abasi H., Blumberg J., Taylor A. (1997) Regulation of ubiquitin-conjugating enzymes by glutathione following oxidative stress. J. Biol. Chem. 272, 28218–28226 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.