Abstract
Background:
Although hepatocellular regeneration is the cornerstone of liver homeostasis, current techniques for assessing such regeneration are limited. A method for visualizing the regeneration process would provide a means for advanced studies. Therefore, we examined the possibility of using Fucci mice for direct visualization of hepatocellular regeneration.
Materials and Methods:
We performed a two-thirds partial hepatectomy in conventional and Fucci (fluorescence ubiquination-based cell cycle indicator) mice. Fucci animals have orange Cdt1 expressed in the G1 phase; green Geminin, S/G2/M phases. Regenerating livers were procured daily for 7 days. Immunohistochemical staining was performed for proliferative Ki67 and mitotic pHH3 serine 10 (pHH3) markers on formalin-fixed, paraffin-embedded tissue sections from conventional mice. The orange Cdt1 and green Geminin fluorescence indicative of the G1 and S/G2/M phases, respectively, were assessed in liver tissues, in vivo and ex vivo, with two-photon laser scanning microscopy.
Results:
Immunostaining with Ki67 and pHH3 revealed a typical profile of hepatocellular regeneration after hepatectomy in conventional mice, although immunostaining required more than a week to process. In contrast, hepatocellular regeneration could be visualized with two-photon microscopy within a few hours in regenerating livers of the Fucci mice. Only orange G1 hepatocytes were seen in the baseline liver specimens; however, multiple bright green and yellow hepatocytes were seen 48 hours after hepatectomy, indicating active hepatocytes in the S/G2/M phases of the cell cycle.
Conclusions:
Hepatocellular regeneration is readily visualized in regenerating livers of Fucci mice. The Fucci model is an exciting tool for advanced studies of hepatocellular and liver regeneration.
Keywords: Fucci mice, hepatocellular regeneration, liver regeneration, partial hepatectomy
Graphical Abstract
Introduction
Hepatocytes have a remarkable ability to regenerate.1 Within a week after a two-thirds partial hepatectomy (2/3-PH) in rodents, the liver is restored to nearly its original volume.2 After a partial hepatectomy (PH), hepatocytes at inactive G0 or resting G1 states enter the cell cycle and progress through S, G2, and M phases, resulting in mitosis and regeneration.3,4 A 2/3-PH provides strong mitogenic stimuli for hepatocellular regeneration and is an optimal experimental model of liver regeneration.5,6 However, recent evidence has shown that not all hepatocytes in proliferative phases undergo cell division.7 How to improve hepatocellular regeneration in acute liver failure and small-for-size liver remains an enigma because the mechanisms of hepatocellular regeneration are inadequately understood.5,8,9
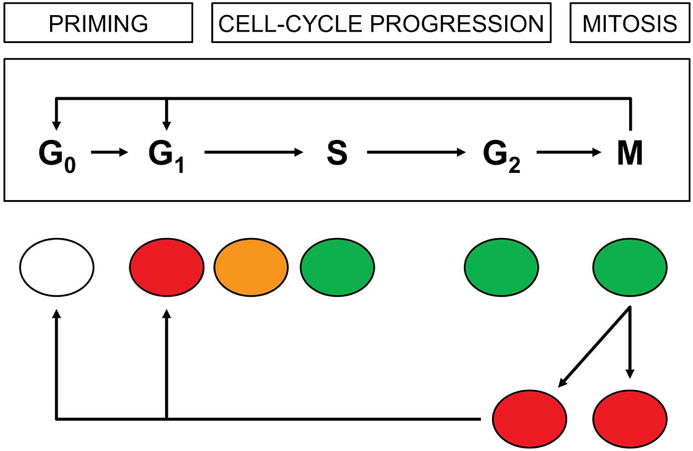
Characterizing hepatocellular regeneration after 2/3-PH typically is done with traditional immunohistochemical techniques, including 5-bromo-2′-deoxyuridine (BrdU) and 5-ethynyl-2′-deoxyuridine (EdU) for nucleotide synthesis, monoclonal antibodies for proliferative proteins of proliferation-related K-67 antigen (Ki67) and proliferating cell nuclear antigen (PCNA), and for cell cycle proteins including cyclin D1 of the G1 phase, cyclin A1 of the S phase, cyclin B1 of the G2 phase, and phosphohistone H3 serine 10 (pHH3) for the M phase.7 However, these techniques require the tissue sample to be fixed and are not applicable for live imaging. Recent advances with fluorescence ubiquitin-based cell cycle indicator (Fucci) technology have allowed direct visualization of cell cycle phases in tissue slices, in vitro cells, and mice.10,11 This technology takes advantage of red fluorescence-tagged chromatin licensing and DNA replication factor 1 (Cdt1) that peaks in expression during the G1 phase and is ubiquinated for degradation by SKP1 (S-phase kinase-associated protein 2)-cullin F-box (SCFskp2) at the onset of the S phase until mid M phase. In contrast, green Geminin accumulates in nuclei during the proliferative S, G2, and M phases until it is degraded by anaphase-promoting complex/cyclosome E3 ubiquitin ligase (APC/C) at the onset of mitosis. The inactive G0 cells are colorless. The overlapped expression of both Cdt1 and Geminin during the transition of G1 into the S phase results in yellow/orange-appearing nuclei (Figure 1).11 Because current techniques for assessing hepatocellular regeneration are limited, we aimed to determine whether a Fucci mouse model could be used to visualize the proliferation and regeneration of hepatocytes after PH.
Figure 1.
Variation in Fucci Fluorescence Colors by Cell-Cycle Phases. Nuclei of resting G1 hepatocytes predominantly express the red Cdt1 gene and thus appear red. As G1 cells transition into the S phase, their nuclei contain both red Cdt1 and green Geminin, giving the nuclei a yellow/orange appearance. When hepatocytes fully enter the S, G2, and M phases, their nuclei express only green Geminin. The cell cycle is repeated as the green hepatocytes divide into 2 daughter red G1 cells. Some G1 cells may enter in and out of the G0 phase of the cell cycle.
Materials and Methods
Animals
Young inbred C57BL/6 mice (male, 2-4 months) (Harlan Laboratories, Indianapolis, Indiana, USA) were housed under specific pathogen-free conditions with 12-hour light/dark cycles, food, and water. Fucci mice (B6;129-Gt[ROSA]26Sor<tm1.1[Fucci2aR]Jkn> [RBRC09613]) were obtained from Riken BRC (Experimental Animal Division, Tsukuba, Japan) and, after quarantine, were housed in the same manner as the C57BL/6 mice. All experimental protocols were approved by the Institutional Animal Care and Use Committee at Mayo Clinic (Protocol No. A68412), and procedures were performed according to institutional guidelines.
Antibodies and Reagents
The primary antibodies used in this study were cyclin D1 (sc-753; Santa Cruz Biotechnology, Inc, Santa Cruz, California, USA); cyclin A (sc-596; Santa Cruz Biotechnology, Inc); cyclin B1 (sc-245; Santa Cruz Biotechnology, Inc); pHH3 (9701A, Cell Signaling Technology, Inc, Danvers, Massachusetts, USA); Ki67 (ab15580; Abcam, Cambridge, Massachusetts, USA); GAPDH (IMG-5019A-2; Imgenex, San Diego, California, USA). Horseradish peroxidase-conjugated secondary antibodies of goat antirabbit (4010-05) and goat antimouse (1012-05) were purchased from Southern Biotech (Birmingham, Alabama, USA).
Two-Thirds Partial Hepatectomy
The mice were anesthetized with 2% to 3% isoflurane and underwent a 2/3-PH, as previously described.6,12 Briefly, following a midabdominal skin incision, surrounding ligaments and membranes were divided. The left-lateral lobe was resected with ligation at its base, and the median lobe was resected with ligation at the level between the gall bladder and the suprahepatic inferior vena cava. After the abdomen was closed, the mice received 1.0 mL of normal saline subcutaneously and were placed on a warming pad until they recovered from anesthesia. The antibiotic cefamandole nafate (30 mg/kg) was administered subcutaneously before and 24 hours after operation. Buprenorphine (0.05 mg/kg) was injected subcutaneously for pain before the procedure and every 12 hours thereafter for 2 days, according to the institutional guideline. After surgery, the mice were maintained with free access to water and food in temperature-controlled conditions.
Immunohistochemistry
Paraffin sections of each liver sample were cut at 5 μm and stained for various cell cycle proteins by using the specific antibodies. All proteins were visualized using the avidin-biotin-peroxidase complex technique (Vectastain ABC Kit and DAB Peroxidase Substrate Kit, Vector Laboratories, Burlingame, California, USA). The tissue was counterstained with hematoxylin. The staining was visualized using an Axiovert 40 CFL inverted microscope (Carl Zeiss MicroImaging, Thornwood, New York, USA) and scanned into a Leica-Aperio system (Aperio Technologies, Vista, California, USA) for digital image analysis.
Digital Image Analysis of Immunohistochemical Stains
Digital images were captured using the Aperio ScanScope XT Slide Scanner (Aperio Technologies) under objective magnification (×20). All slides were evaluated visually on the monitor to determine a semiquantitative estimate of the percentage of positive cells for quality assurance of the Aperio digital image results. The digital Aperio analysis was performed on the same images as those evaluated visually. Aperio Spectrum software was trained to recognize hepatocytes, other cells, and background (glass) through modifications of appropriate algorithms. The modified Aperio Nuclear v9, Aperio color deconvolution v9, or Aperio colocalization algorithms, or a combination, were run as needed for each antibody used. Hepatocytes were defined as positive for regeneration by medium or high intensity in the cytoplasm or the threshold of weak (1+) or higher nuclear staining. Data from the digital analyses were expressed as mean and standard error of the mean.
Two-Photon Laser Scanning Microscopic Imaging
The upright laser scanning microscope (BX61WI, Olympus, Center Valley, Pennsylvania, USA) attached to a Ti:sapphire pulsed laser system (80 MHz repetition rate, <100 fs pulse width) (Spectra Physics, Santa Clara, California, USA) using Prairie View versiib 5.4 software (Bruker, Billerica, Massachusetts, USA) was used for two-photon fluorescence imaging. For fluorescence imaging in vivo, we selectively chose ×20 (numerical aperture [NA], 1.00; working distance [WD], 2 mm; Olympus), and ×40 water-immersion objectives (NA, 0.80; WD, 3.3 mm; Olympus). An 890-nm irradiation wavelength was used to excite Fucci protein, and the emission light was differentiated and detected with 525/50 and 615/50 filters, respectively. The average laser power for imaging was less than 50 mW.
Time-lapse imaging (20-30 image planes with 1.5-2.0 μm axial spacing) was performed for at least 60 minutes to track the mitosis process in the liver; the interval between stack sequences was 5 minutes. Photomultiplier (PMT) settings (including gain and offset) and laser excitation power were kept constant during time-lapse imaging. Three-dimensional (3D) stack images (step size: 1μm) were captured to track and visualize the 3D morphology of liver tissue.
Data Analysis of Two-Photon Laser Scanning Microscopic Imaging
Images were processed using open-source software (Fiji [National Institutes of Health]) and commercial Matlab software (Version 8.5.0 R2015a, MathWorks Inc, Natick, Massachusetts, USA). The intensity-based alignment of images at the different time points (registration) was done by using the imregister function in Matlab and the StackReg plugin in Fiji. First, we performed the maximum intensity project (MIP) from the stack, and the threshold was set to segment and measure cell numbers at the different channels (red: 615/50 filters and green: 525/50 filters) from the MIP image.
Results
Hepatocellular Regeneration in Mice After 2/3-PH
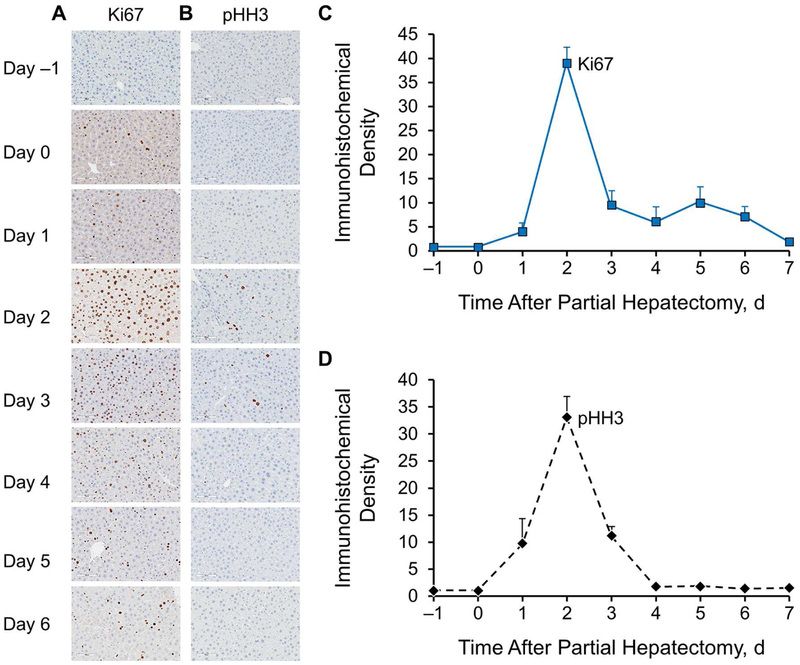
Following 2/3-PH, typically, more than a week was required to allow for visualization of hepatocellular regeneration. We used both proliferative Ki67 and mitotic pHH3 markers to evaluate the regeneration profile. As shown in Figure 2, the peak of hepatocellular regeneration was between 36 and 48 hours. These results are consistent with those of previous reports.7,13 Although the results are not shown, we performed immunostaining for cyclin A1, which is the cell cycle protein for the S phase, and BrdU for nucleotides as an indicator for DNA synthesis. Both showed a similar profile of hepatocellular proliferative activities to that of Ki67.
Figure 2.
Immunohistochemical Profiles of Hepatocellular Regeneration in Regenerating Livers After Partial Hepatectomy in 4-Month-Old Mice. A and B, At baseline (before hepatectomy, day −1) and at day 0 (2 hours) through day 6 (after two-thirds partial hepatectomy), proliferative and mitotic activities were determined by immunostaining for Ki67 and pHH3, respectively (n=6 for each group) (magnification ×400). C and D, The profiles of hepatocellular immunoreactivity of Ki67 and pHH3 show hepatocellular proliferative and mitotic activities, respectively. Ki67 indicates proliferation-related K-67 antigen; pHH3, phosphohistone H3 serine 10.
Visualization of Hepatocellular Regeneration in Fucci Mice
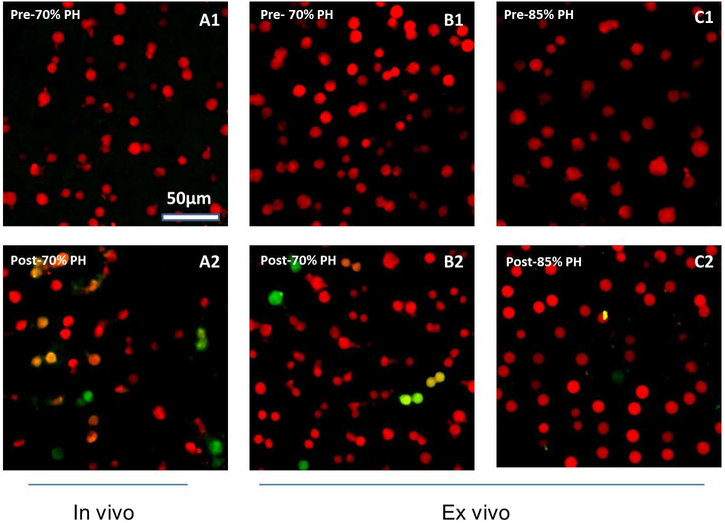
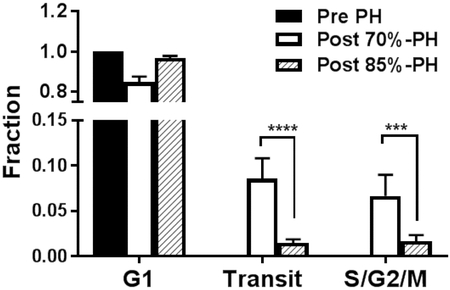
By using Fucci mice, we were able to visualize regeneration in a shorter time. The overall color alteration with cell cycle phases is shown in Figure 1. We consistently observed only red hepatocytes in the pre-PH liver in vivo or ex vivo (Figure 3, A1 and B1), indicating hepatocytes in the resting G1 phase of the cell cycle. At 48 hours after the 2/3-PH, we found green and yellow hepatocytes in the background of red cells (Figure 3, A2 and B2). The quantitation of the cell cycle phase distribution of the hepatocytes is summarized in Figure 3D. We did not observe mitosis in progress; however, we clearly visualized proliferative hepatocytes. The hepatocytes transitioning from G1 to S phases had yellow/red nuclei; and those in S, G2, or M phases, or a combination appeared green.
Figure 3.
Visualization of Hepatocellular Progression Through the Cell Cycle After Partial Hepatectomy. A, In vivo live imaging of the regenerating livers: Hepatocytes in the baseline prehepatectomy liver (A1) were predominantly in the resting G1 phase of the cell cycle; their nuclei appeared red; following 2/3-PH (A2), hepatocytes transitioned into proliferative S, G2, and M phases of the cell cycle. B, Ex vivo live imaging of liver tissues: In the pre 70%-PH liver, hepatocytes were in the G1 phase, ie, red (B1); in the post-70% PH liver (B2), hepatocytes were actively transitioning (yellow/orange) from G1 into S phases and entered S, G2, and M phases (green). C, In both of the livers of pre (C1) and post (C2) 85%-PH, hepatocytes were in mainly the G1 phase, ie, red nuclei; only occasional hepatocytes had a faint green nucleus in the liver remnant following 85%-PH (C2), consistent with a lack of progression through the cell cycle. D, Histogram showing hepatocytes in G1, transit (G1 to S), and S/G2/M phases of the cell cycle. PH indicates partial hepatectomy.
Following 2/3-PH, hepatocytes readily regenerate. In contrast, when more extensive resection of the liver occurs, ie, after 85%-PH, it is established that hepatocytes in the liver remnant are arrested at the G1 phase, leading to failure of liver regeneration.9 Consistent with this, we observed that the hepatocytes in the liver remnant after 85%-PH, a- model of small-for-size liver, were predominantly red (Figure 3, C2), consistent with being in the G1 phase as in the baseline (before) hepatectomy liver (Figure 3, C1). These observations vividly illustrate the primary failure of hepatocellular regeneration in small-for-size liver. Thus, Fucci mice facilitated the visualization of hepatocellular regeneration.
Discussion
In this study, we showed that Fucci technology in mice can allow direct visualization of hepatocellular regeneration in the regenerating liver remnant. The Fucci mouse model thus provides an advanced method for molecular investigations of hepatocellular regeneration, eliminating the need for immunohistochemical staining of formalin-fixed, paraffin-embedded tissues.
The regenerative capacity of the liver is currently assessed by immunostaining for Ki67 or PCNA for proliferative activity, by pHH3 for mitotic activity, and by taking physical measurements, such as liver weight. These conventional techniques are time-consuming and require cutting and fixing the samples. In contrast, transgenic Fucci mice express the red-tagged Cdt1 gene that is present mainly in the G1 phase and green-tagged Geminin that is predominantly expressed during the S, G2, and early M phases.10 This technology allows for clear, live imaging of inactive vs. proliferative cells. Recently, a cell-cycle reporter model was developed with Fucci technology that allows simultaneous reporting of all 4 phases of the cell cycle.14 These novel models are exciting new tools that will allow for more innovative and challenging investigations of hepatocellular regeneration.
By using Fucci mice, the liver can be imaged using two-photon laser scanning microscopy. This technique allows for live imaging of hepatocytes and surrounding nonhepatocellular parenchymal cells in intact animals or in slices of ex situ liver tissue. In future studies, we can use Qtracker 655 vascular labels (ThermoFisher Scientific) to visualize the sinusoids and blood flow.15 In addition, we can use second harmonic generation microscopy to label-free image the extracellular matrix of the sinusoids with infrared laser stimulation.16
After PH, continuous live imaging of the liver in anesthetized mice can be technically demanding; we have done the imaging for up to 4 hours. Image acquisition is sensitive to the slightest positional change, including that from breathing. When only a small liver remnant remains after PH, inhaled isoflurane and fluid supplements must be carefully administered to avoid the animals’ dying from being overanesthetized. Ritsma and colleagues17 have suggested creating an abdominal imaging window for repeated live imaging, which is potentially applicable for longterm assessment of organ function including that of the liver. However, the small liver remnant after PH may be too fragile for such a technique. Alternatively, liver tissues of the small liver remnant can be imaged ex vivo for several hours, thus eliminating concerns of anesthesia, animal movement, and cardiopulmonary support. In addition, the slices of liver tissue can be evaluated with confocal microscopy. Although confocal microscopy is more limited for imaging cellular elements, it is an alternative for ex vivo live imaging.18
A 2/3-PH provides the strongest stimuli for liver regeneration.19 However, why hepatocytes fail to regenerate when the liver remnant is less than 26% in humans is not well understood.20,21 In mice, after 85% or more of the liver was removed in experimental models of small-for-size liver, the hepatocytes became stunted, arrested midway in the cell cycle, and did not complete the cycle.9,22-24 This loss or failure of hepatocellular regenerative ability would lead to death of patients who undergo extended liver resection for cancer treatment and of patients who receive a liver that is small relative to their body size and associated physiologic needs.21 Using Fucci mice, we verified that hepatocytes in the liver remnant after 85%-PH were arrested in the G1 phase.
We aimed to visualize regeneration in progress as a means to demonstrate hepatocellular origin in liver regeneration. Unraveling the mechanisms responsible for regenerative failure or the mechanisms needed to restore or augment hepatocellular regenerative capacity, or both, is clinically important. Fucci mice provide a useful model for further studying how to overcome failure of hepatocellular regeneration in small-for-size livers after massive PH. In addition, combining primary hepatocytes from the Fucci mice with flow cytometry and single-cell sequencing will likely bring about insightful perspectives and an understanding of hepatocellular behavior at a molecular single-cell level.25
This study is limited by its lack of specificity for hepatocytes, because the Fucci markers were present in all mouse cells. However, recent gene targeting of Fucci markers in the mouse Rosa26 locus allowed for specific hepatocellular expression by using adeno-associated viral-thyroxine binding globulin promoter-Cre recombinase (AAV8-TBG-Cre), which is selectively taken up by hepatocytes.26,27 Another limitation is the cost of the Fucci animals. To decrease costs, we used regular mice for initial characterizations and Fucci animals for confirmatory visual and advanced studies with live imaging.
Conclusion
By using Fucci technology, we could directly visualize hepatocellular regeneration in the liver remnant in mice, which avoids the need for immunohistochemical staining. On the basis of our study, we believe that Fucci mice can be used as a model for studying why hepatocytes fail to regenerate in small-for-size livers and after more extensive PH. This failure of hepatocytes to regenerate limits the use of small-for-size livers and split livers in adults requiring liver transplants. Overcoming this limitation would be an important contribution to the field of liver transplantation.
Acknowledgments
The authors would like to thank Laura Lewis-Tuffin of the Cellular Imaging and Flow Cytometry Facility at the Mayo Clinic campus in Jacksonville, Florida, for helpful discussions of imaging options.
This work was supported by a fellowship in the Center of Regenerative Medicine, Liver Regenerative Medicine at Mayo Clinic (T.H.). The research was also supported the National Institutes of Health under award number NIH R21AG052822-01A1 (J.H.N.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Abbreviations
- AAV8-TBG-Cre
adeno-associated viral-thyroxine binding globulin promoter-Cre recombinase
- APC/C
anaphase-promoting complex/cyclosome E3 ubiquitin ligase
- BrdU
5-bromo-2′-deoxyuridine
- Cdt1
chromatin licensing and DNA replication factor 1
- EdU
5-ethynyl-2′-deoxyuridine
- Fucci
fluorescence ubiquination-based cell cycle indicator
- Ki67
proliferation-related K-67 antigen
- MIP
maximum intensity project
- NA
numerical aperture
- PCNA
proliferating cell nuclear antigen
- PH
partial hepatectomy
- pHH3
phosphohistone H3 serine 10
- PMT
photomultiplier
- SCFskp2
SKP1 (S-phase kinase-associated protein 2)-cullin 1 F-box
- 3D
three-dimensional
- 2/3-PH
two-thirds partial hepatectomy
- WD
working distance
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosure
The authors report no proprietary or commercial interest in any product mentioned or concept discussed in this article. The sponsor did not have any involvement in study design; in the collection, analysis and interpretation of the data; in the writing of the report; and in the decision to submit the article for publication.
Conflict of interest: None.
Contributor Information
Dr Yuanxin Chen, Department of Cancer Biology, Mayo Clinic, Jacksonville, Florida.
Dr Toshiyuki Hata, Department of Hepato-Biliary-Pancreatic and Transplant Surgery, Kyoto University Graduate School of Medicine, Kyoto, Japan.
Dr Fatima Rehman, Department of Biology, University of North Florida, Jacksonville, Florida..
Dr Lu Kang, Department of Neuroscience, Mayo Clinic, Jacksonville, Florida.
Dr Liu Yang, Department of Transplantation, Mayo Clinic, Jacksonville, Florida.
Dr Betty Y.S. Kim, Department of Cancer Biology, Department of Neurologic Surgery, Mayo Clinic, Jacksonville, Florida.
Dr Justin H. Nguyen, Department of Transplantation, Mayo Clinic, Jacksonville, Florida.
References
- 1.Overturf K, al-Dhalimy M, Ou CN, Finegold M, Grompe M. Serial transplantation reveals the stem-cell-like regenerative potential of adult mouse hepatocytes. Am J Pathol. 1997;151(5): 1273–1280. [PMC free article] [PubMed] [Google Scholar]
- 2.Higgins GM, Anderson RM. Experimental pathology of the liver. I. Restoration of the liver of the white rat following partial surgical removal. Arch Pathol. 1931;12:186–202. [Google Scholar]
- 3.Fausto N Liver regeneration. J Hepatol. 2000;32(1 Suppl):19–31. [DOI] [PubMed] [Google Scholar]
- 4.Michalopoulos GK, DeFrances MC. Liver regeneration. Science. 1997;276(5309):60–66. [DOI] [PubMed] [Google Scholar]
- 5.Michalopoulos GK. Liver regeneration after partial hepatectomy: critical analysis of mechanistic dilemmas. Am J Pathol. 2010;176(1):2–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mitchell C, Willenbring H. A reproducible and well-tolerated method for 2/3 partial hepatectomy in mice. Nat Protoc. 2008;3(7): 1167–1170. [DOI] [PubMed] [Google Scholar]
- 7.Miyaoka Y, Ebato K, Kato H, Arakawa S, Shimizu S, Miyajima A. Hypertrophy and unconventional cell division of hepatocytes underlie liver regeneration. Curr Biol. 2012;22(13): 1166–1175. [DOI] [PubMed] [Google Scholar]
- 8.Clavien PA, Eshmuminov D. Small for size: Laboratory perspective. Liver Transpl. 2015;21 Suppl 1:S13–14. [DOI] [PubMed] [Google Scholar]
- 9.Lehmann K, Tschuor C, Rickenbacher A, et al. Liver failure after extended hepatectomy in mice is mediated by a p21-dependent barrier to liver regeneration. Gastroenterology. 2012;143(6):1609–1619 e1604. [DOI] [PubMed] [Google Scholar]
- 10.Sakaue-Sawano A, Kurokawa H, Morimura T, et al. Visualizing spatiotemporal dynamics of multicellular cell-cycle progression. Cell. 2008;132(3):487–498. [DOI] [PubMed] [Google Scholar]
- 11.Zielke N, Edgar BA. FUCCI sensors: powerful new tools for analysis of cell proliferation. Wiley Interdiscip Rev Dev Biol. 2015;4(5):469–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hori T, Ohashi N, Chen F, et al. Simple and sure methodology for massive hepatectomy in the mouse. Ann Gastroenterol. 2011;24(4):307–318. [PMC free article] [PubMed] [Google Scholar]
- 13.Satyanarayana A, Geffers R, Manns MP, Buer J, Rudolph KL. Gene expression profile at the G1/S transition of liver regeneration after partial hepatectomy in mice. Cell Cycle. 2004;3(11):1405–1417. [DOI] [PubMed] [Google Scholar]
- 14.Bajar BT, Lam AJ, Badiee RK, et al. Fluorescent indicators for simultaneous reporting of all four cell cycle phases. Nat Methods. 2016;13(12):993–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Porat-Shliom N, Tietgens AJ, Van Itallie CM, et al. Liver kinase B1 regulates hepatocellular tight junction distribution and function in vivo. Hepatology. 2016;64(4):1317–1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chang PE, Goh GBB, Leow WQ, Shen L, Lim KH, Tan CK. Second harmonic generation microscopy provides accurate automated staging of liver fibrosis in patients with non-alcoholic fatty liver disease. PLoS One. 2018;13(6):e0199166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ritsma L, Steller EJ, Ellenbroek SI, Kranenburg O, Borel Rinkes IH, van Rheenen J. Surgical implantation of an abdominal imaging window for intravital microscopy. Nat Protoc. 2013;8(3):583–594. [DOI] [PubMed] [Google Scholar]
- 18.Boisset JC, van Cappellen W, Andrieu-Soler C, Galjart N, Dzierzak E, Robin C. In vivo imaging of haematopoietic cells emerging from the mouse aortic endothelium. Nature. 2010;464(7285):116–120. [DOI] [PubMed] [Google Scholar]
- 19.Jirtle RL, Michalopoulos G. Effects of partial hepatectomy on transplanted hepatocytes. Cancer Res. 1982;42(8):3000–3004. [PubMed] [Google Scholar]
- 20.Schindl MJ, Redhead DN, Fearon KC, et al. The value of residual liver volume as a predictor of hepatic dysfunction and infection after major liver resection. Gut. 2005;54(2):289–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dahm F, Georgiev P, Clavien PA. Small-for-size syndrome after partial liver transplantation: definition, mechanisms of disease and clinical implications. Am J Transplant. 2005;5(11):2605–2610. [DOI] [PubMed] [Google Scholar]
- 22.Panis Y, McMullan DM, Emond JC. Progressive necrosis after hepatectomy and the pathophysiology of liver failure after massive resection. Surgery. 1997;121(2):142–149. [DOI] [PubMed] [Google Scholar]
- 23.Morita T, Togo S, Kubota T, et al. Mechanism of postoperative liver failure after excessive hepatectomy investigated using a cDNA microarray. J Hepatobiliary Pancreat Surg. 2002;9(3):352–359. [DOI] [PubMed] [Google Scholar]
- 24.Zhong Z, Schwabe RF, Kai Y, et al. Liver regeneration is suppressed in small-for-size liver grafts after transplantation: involvement of c-Jun N-terminal kinase, cyclin D1, and defective energy supply. Transplantation. 2006;82(2):241–250. [DOI] [PubMed] [Google Scholar]
- 25.Dulken BW, Leeman DS, Boutet SC, Hebestreit K, Brunet A. Single-Cell Transcriptomic Analysis Defines Heterogeneity and Transcriptional Dynamics in the Adult Neural Stem Cell Lineage. Cell Rep. 2017;18(3):777–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Abe T, Sakaue-Sawano A, Kiyonari H, et al. Visualization of cell cycle in mouse embryos with Fucci2 reporter directed by Rosa26 promoter. Development. 2013;140(1):237–246. [DOI] [PubMed] [Google Scholar]
- 27.Chandler RJ, Tarasenko TN, Cusmano-Ozog K, et al. Liver-directed adeno-associated virus serotype 8 gene transfer rescues a lethal murine model of citrullinemia type 1. Gene Ther. 2013;20(12): 1188–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]