Abstract
Background
MicroRNAs (miRNAs) can regulate the post-transcriptional level of gene expression. It has been documented that downregulation of miR-206 is significant in human gastric cancer (GC), whereas its role in GC cell biological behaviors, including proliferation, migration, and invasion, has not been thoroughly investigated. MiR-206 levels have a negative association with lymph node metastasis and tumor invasion, and patients with higher miR-206 expression have better prognoses. Functional studies demonstrated that miR-206 overexpression significantly suppresses GC cell proliferation, migration, and invasion, and induces apoptosis in vitro.
Materials and methods
MiR-206 and MUC1 were determined by RNA extraction, quantitative real-time polymerase chain reaction, and luciferase reporter gene assays. The viability of GC cells was tested using the Cell Counting Kit 8 assay. Transwell invasion and migration assays detected GC cancer cell proliferation, invasion, and migration. Flow cytometry was applied to analyze apoptotic cells. FACS analysis was applied to detect the mitochondrial membrane potential of cells. Western blotting assay determined protein levels.
Results
The luciferase reporter gene assay demonstrated that miR-206 might directly bind to the 3′UTR of the MUC1 gene and suppress MUC1 expression. Furthermore, MUCI expression was upregulated and inversely associated with miR-206 levels in GC tissues. More importantly, the miR-206-mediated suppression of proliferation, migration, and invasion, and the induction of apoptosis, were abrogated by MUC1 overexpression.
Conclusion
Our data demonstrated that miR-206 may exert antitumor activities through inhibiting the expression of MUC1, which may serve as an effective and potential target for GC treatment.
Keywords: miR-206, MUC1, gastric cancer, metastasis
Introduction
Gastric cancer (GC) is one of the most common malignancies suffered by human beings; however, its pathogenesis remains unknown. Many factors such as lifestyle, Helicobacter pylori (HP) infection, polyps, gastric ulcers, genetic disorders, and residual stomach tissues may be involved in the tumorigenesis of the stomach.1,2 Adenocarcinoma is the primary pathological type of GC; other pathological types include squamous cell, adenosquamous, carcinoid, small-cell, and other carcinoma, of which the latter are relatively rare. Treatment varies in different types of GC. There are no specific symptoms in the early stage of GC. When the symptoms become quite apparent, most patients are usually diagnosed at an advanced stage, sometimes with distant metastasis, leading to higher mortality and a lower 5-year survival rate.3
MicroRNAs (miRNAs) are a class of endogenous, conserved, non-coding, small RNA, which may regulate the expression of genes at post-transcriptional levels either by directly degrading the target mRNA or by indirectly inhibiting the translation of the target mRNA.4 MiRNAs are able to bind to the 3′UTR of the target mRNAs in a complementary base-pairing manner, which has been demonstrated to contribute to cell apoptosis, proliferation, and differentiation.5,6 Increasing evidence suggests that abnormal miRNA expression is frequently observed in various tumors, and that miRNAs can act as tumor suppressor or oncogenic genes, depending on their target gene function.7,8 In general, tumor-suppressive miRNAs are usually downregulated, whereas oncogenic miRNAs are usually upregulated, in cancers.9,10 For instance, miR-122 may inhibit the tumorigenesis and metastasis of hepatocellular carcinoma, which is found to be significantly downregulated in liver cancers.11 MiR-223, found to be significantly upregulated in GC, may promote invasion and metastasis.12 Until now, miRNAs were believed to regulate more than 50% of protein-coding genes. It has been demonstrated that many miRNAs, including miR-206, are abnormally expressed in GC.13,14 Although the downregulation of miR-206 in GC has been documented, the role of miR-206 in GC cells has not been fully understood.
In the present study, we observed that the downregulation of miR-206 was significant in GC. Exogenous miR-206 overexpression may lead to the inhibition of proliferation and metastasis as well as the promotion of apoptosis in GC. It is further confirmed that MUC1, a glycoprotein, is a direct target of miR-206, which is upregulated in GCs and may induce proliferation and metastasis via activation of the WNT signaling pathway.
Materials and methods
GC specimen collection
Tumor tissues and corresponding non-tumorous tissues were harvested from 60 GC patients who underwent radical resection at the First Affiliated Hospital of Bengbu Medical College from June 2011 to June 2012. Immediately following resection, liquid nitrogen was used to preserve all tissues. Table 1 shows the clinicopathologic features of all cases. All patients provided written informed consent and approved the use of the specimens for this study. The ethics committee of the First Affiliated Hospital of Bengbu Medical College approved the study protocol.
Table 1.
Correlation between the clinicopathologic characteristics and expression of miR-206 in GC
| Variables | No of cases | miR-206 expression
|
P-value | |
|---|---|---|---|---|
| Low (n %) | High (n %) | |||
|
| ||||
| Age (years) | P>0.05 | |||
| <60 | 30 | 18 (60.0) | 12 (40.0) | |
| ≥60 | 30 | 17 (56.7) | 13 (43.3) | |
| Gender | P>0.05 | |||
| Male | 32 | 19 (59.4) | 13 (40.6) | |
| Female | 28 | 16 (57.1) | 12 (42.9) | |
| Tumor size | P>0.05 | |||
| <5 cm | 38 | 25 (65.8) | 13 (34.2) | |
| ≥5 cm | 22 | 10 (45.5) | 12 (54.5) | |
| TNM stage | P=0.002 | |||
| I–II | 42 | 19 (45.2) | 23 (54.8) | |
| III–IV | 18 | 16 (88.9) | 2 (11.1) | |
| Lymph node metastasis | P=0.028 | |||
| Absence | 46 | 23 (50.0) | 23 (50.0) | |
| Presence | 14 | 12 (85.7) | 2 (14.3) | |
Abbreviation: GC, gastric cancer.
Cell culture and treatment
NCI-N87, SGC-7901, MKN-45, HGC-27 (human GC cell lines), and GES-1 (a normal gastric epithelial cell line) provided by the American Type Culture Collection (ATCC; Rockville, MD, USA) were maintained in RPMI-1640 medium (Gibco, Logan, UT, USA) containing 1% penicillin/ streptomycin and 10% FBS (HyClone) in a humidified incubator containing 5% CO2 at 37°C.
GenePharma (Shanghai, People’s Republic of China) provided the miR-206 mimic, miR-206 inhibitor, and miR-206 control. The cDNA of human MUC1 amplified by PCR was inserted into the pcDNA 3.0 vector (pcDNA-MUC1; Invitrogen, Grand Island, NY, USA). Small-interfering RNA (siRNA) targeting human MUC1 (GenePharma, People’s Republic of China) and the siRNA-negative control (100 nM) were transfected into GC cells with Lipofectamine 2000 (Invitrogen).
RNA extraction and quantitative real-time polymerase chain reaction (qRT-PCR) for miR-206 and MUC1
Total RNA extracted from cells and tumor tissues using the TRIzol reagent (Invitrogen, USA) were reverse-transcribed into cDNA using PrimeScript™ RT Master Mix (TaKaRa, Japan). For the detection of miR-206 and MUC1 mRNA levels, qRT-PCR was conducted using the SYBR® PrimeScript™ miRNA RT-PCR Kit (Takara, Japan). Endogenous U6 snRNA and GAPDH were applied as the internal control for miRNAs and mRNAs. The primers for MUC1 and GAPDH are shown in Table 2 and Supplementary material 1. The 2−∆∆Ct method was utilized to quantify miR-206 or MUC1 mRNA expression. Sixty patients were classified into the high miR-206 expression group (25 cases) and the low miR-206 expression group (35 cases) according to the mean value in all GC samples.
Table 2.
Primers for qRT-PCR
| Gene | Sequences |
|---|---|
| MUC1 forward | 5′-TCCTTTCTCTGCCCAGTCTG-3′ |
| MUC1 reverse | 5′-GTGTGGTAGGTGGGGTACTC-3′ |
| GAPDH forward | 5′-GTGTGGTAGGTGGGGTACTC-3′ |
| GAPDH reverse | 5′-ACCCTTCCACGATCCCAAAT-3′ |
Abbreviation: qRT-PCR, quantitative real-time polymerase chain reaction.
Luciferase reporter gene assay for miR-206 and MUC1
PCR was utilized to amplify the wild-type 3′UTR fragment of MUC1 mRNA, which contains potential binding sites for miR-206. Subsequently, the amplified wild-type 3′UTR fragment of MUC1 mRNA was cloned into the pGL3 vector (Promega, Madison, WI, USA) upstream of the luciferase reporter gene. A site-directed mutagenesis kit (Takara, Japan) was used to induce mutation to the complementary site of the seed region of miR-206 (CAUUCC to AACCAA) to construct the plasmid-containing mutant-type MUC1 3′UTR (pGL3-MUC1-3′UTR-MUT). For luciferase assays, miR-206 mimics, inhibitors, or miR-NC, as well as pGL3-MUC1-3′UTR-WT or pGL3-MUC1-3′UTR-MUT, were utilized to transfect HEK293-T cells (5×104 cells per well) plated in a 24-well plate using Lipofectamine 2000 (Invitrogen, USA). After a 48-hour transfection period, the collected HEK293-T cells were analyzed using the dual-luciferase reporter assay system (Promega, USA). The Renilla luciferase reporter plasmid (pRL-TK) activity was assayed for normalization. The exact transfection is shown in Table 3.
Table 3.
Luciferase reporter gene assay for miR-206 and MUC1
| pGL3-MUC1-3′UTR-WT + miR-206 mimics + pRL-TK | pGL3-MUC1-3′UTR-MUT + miR-206 mimics + pRL-TK |
| pGL3-MUC1-3′UTR-WT + miR-206 inhibitor + pRL-TK | pGL3-MUC1-3′UTR-MUT + miR-206 inhibitor + pRL-TK |
| pGL3-MUC1-3′UTR-WT + miR-NC + pRL-TK | pGL3-MUC1-3′UTR-MUT + miR-NC + pRL-TK |
Abbreviation: miR-NC, miR-normal control.
Cell Counting Kit-8 (CCK-8) assay
The viability of GC cells after transfection was tested using CCK-8 assays. In brief, 5×103 cells/wells were plated in 96-well plates. 0, 2, 4, 6, 8, 12, 24, and 48 hours after treatment, 10 µL/ well of CCK-8 solution was added 2–4 hours before the end of incubationat 37°C. The OD values with an absorbance of 450 nm were measured by using a microplate reader (BioTek, USA).
Cell migration and invasion assays
The metastasis of GC cells after transfection was tested using 8-µm Transwell chambers (Millipore, USA) coated with/ without a Matrigel matrix (Corning, USA). As the miR-206 expression of NCI-N87 after transfection with the miR-206 mimic was upregulated the most of the four cell lines assayed, the NCI-N87 was chosen for subsequent studies. We injected 3×104 cells in 200 µL RPMI-1640 medium into the upper chamber containing a non-coated/Matrigel-coated membrane. Subsequently, the lower chamber was filled with 800 µL of RPMI-1640 medium with 10% FBS as a chemoattractant. After a 24-hour incubation period, residual cells on the upper surface of the membrane were removed, 4% methanol and crystal violet, respectively, were utilized to fix and stain cells that adhered to the lower surface. We used a light microscope (using the 10× objective lens; Olympus, Japan) to score cells visually in five random fields.
Cell apoptosis assay
The apoptosis of NCI-N87 cells was detected by Annexin V– fluorescein isothiocyanate (FITC)/propidium iodide (PI) double staining. In brief, transfected cells were harvested at a given time; then Annexin V-FITC and PI were utilized to stain cells for 15 minutes before flow cytometry was applied to analyze the percentage of apoptotic cells.
Detection of mitochondrial membrane potential
The alteration of the mitochondrial membrane potential was measured as follows: transfected cells were incubated in six-well plates for 24, 48, and 72 hours. Then, the fluorescent probe, JC-1 (2 mg/mL), was utilized to incubate approximately 1×106 cells for 20 minutes at 37°C. Fluorescence-activated cell sorting analysis (FACS; Becton Dickinson, Franklin Lakes, NJ, USA) was applied to detect the mitochondrial membrane potential of cells.
Western blotting analysis
A RIPA sample buffer (Beyotime, People’s Republic of China) supplemented with protease inhibitors (Complete, EDTA-free; Roche, USA) and phenylmethylsulfonyl fluoride (Beyotime, People’s Republic of China) were utilized to extract total proteins from cells and tissues. Thereafter, 30 µg/well of protein were electrophoresed on SDS-PAGE before transferring it to the polyvinylidende difluoride membrane. The membranes were respectively incubated with primary anti-MUC1 (1:200 dilution; sc-52347, Santa Cruz Biotechnology, USA) and anti-GAPDH (1:200 dilution; sc-47724, Santa Cruz Biotechnology, USA) at 4°C overnight after being blocked with 5% non-fat milk. GAPDH served as an endogenous control. Then, goat anti-mouse immunoglobulin G (IgG)-horseradish peroxidase (HRP; 1:1,000; Abcam, USA) was applied to incubate membranes. The protein bands were visualized by an enhanced chemiluminescent (ECL) detection reagent (Thermo Fisher, USA). Images were obtained and Gel Doc 2000 (BioRad, USA) was applied to quantify the optical densities of the bands.
Statistical analysis
All data were analyzed using SPSS 19.0 (SPSS Inc, Chicago, IL, USA). The difference between groups was analyzed by one-way ANOVA, Kaplan–Meier log-rank, Student’s t-, and chi-squared tests. P<0.05 represented statistical significance.
Results
Downregulation of miR-206 in both GCcell lines and tissues
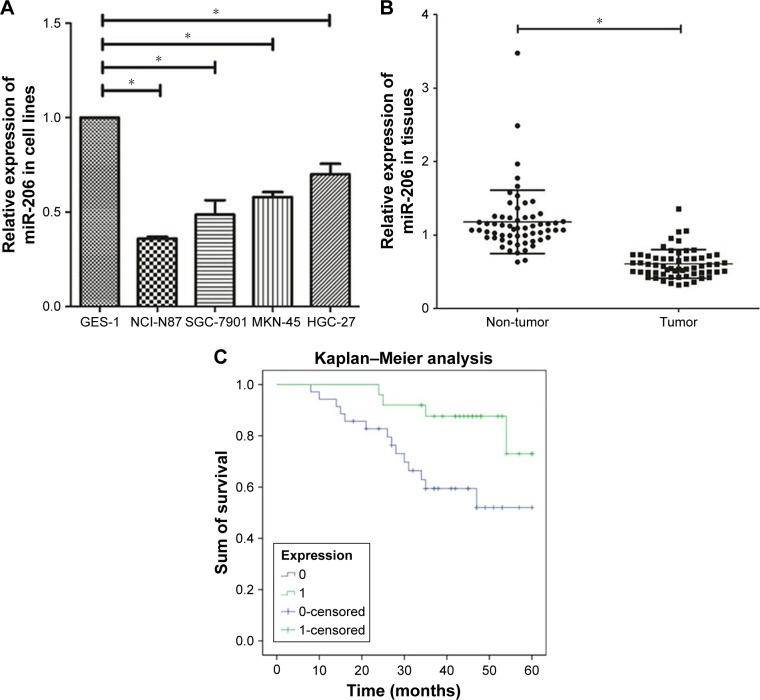
We observed that downregulation of miR-206 was aberrant in GC cells as compared with GES-1 cells (Figure 1A; P<0.001). The level of miR-206 in NCI-N87 decreased the most among the four GC cell lines; therefore, it was selected for further studies. When compared to corresponding adjacent non-tumor tissues, the miR-206 level in GC tissues clearly decreased (P<0.001; Figure 1B). Furthermore, the correlation between miR-206 expression and clinicopathologic features of GC patients was investigated, and the expression of miR-206 had a negative association with the TNM stage and lymph node metastasis (Table 1). Kaplan–Meier analysis showed that the survival of patients was positively correlated with miR-206 expression (P=0.017; Figure 1C).
Figure 1.
Downregulation of miR-206 in GC cell lines and tissues.
Notes: (A) Downregulation of miR-206 was significant in NCI-N87, SGC-7901, MKN-45, and HGC-27 cells compared to GES-1. (B) Downregulation of miR-206 was clear in GC tissues compared with corresponding non-cancerous tissues. (C) The survival of patients has a positive correlation with high expression of miR-206. *P<0.05.
Abbreviation: GC, gastric cancer.
Suppression of miR-206 on GC cell proliferation, migration, and invasion
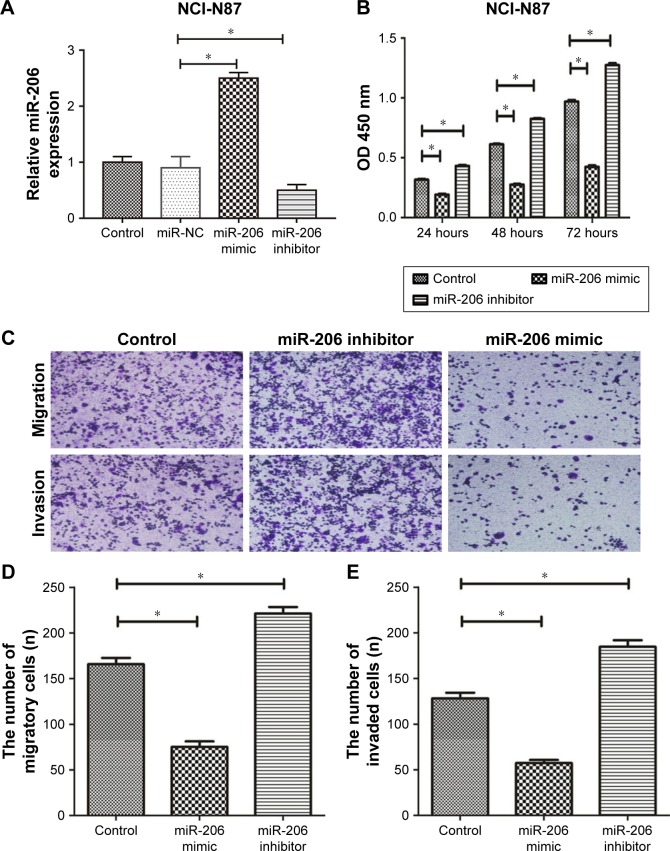
Then, we transfected NCI-N87 cells with a miR-206 mimic and miR-206 inhibitor to regulate miR-206 expression, which was also confirmed by qRT-PCR (Figure 2A). The CCK-8 assay indicated that the proliferation of NCI-N87 cells could be significantly suppressed by miR-206 mimics and enhanced by miR-206 inhibitors in a concentration- and time-dependent manner (Figure 2B). Furthermore, Transwell assays indicated that the migration and invasion capacity of NCI-N87 cells can be significantly inhibited by miR-206 mimics and enhanced by miR-206 inhibitors (Figure 2C–E). Taken together, the data suggested that miR-206 overexpression could significantly suppress the proliferation, migration, and invasion of GC cells.
Figure 2.
The suppression effect of miR-206 on the proliferation, migration, and invasion of gastric cancer cells.
Notes: (A) The expression of miR-206 in NCI-N87 cells transfected with miR-206 mimic, miR-NC and miR-206 inhibitors. (B–E) The proliferation (B), migration (C and D), and invasion (C and E) of NCI-N87 cells were significantly decreased due to the overexpression of miR-206. *P<0.05. Magnification, 10×.
Abbreviation: miR-NC, miR-normal control.
MiR-206 depolarizes transmembrane potential depolarization and induces apoptosis of GC cell
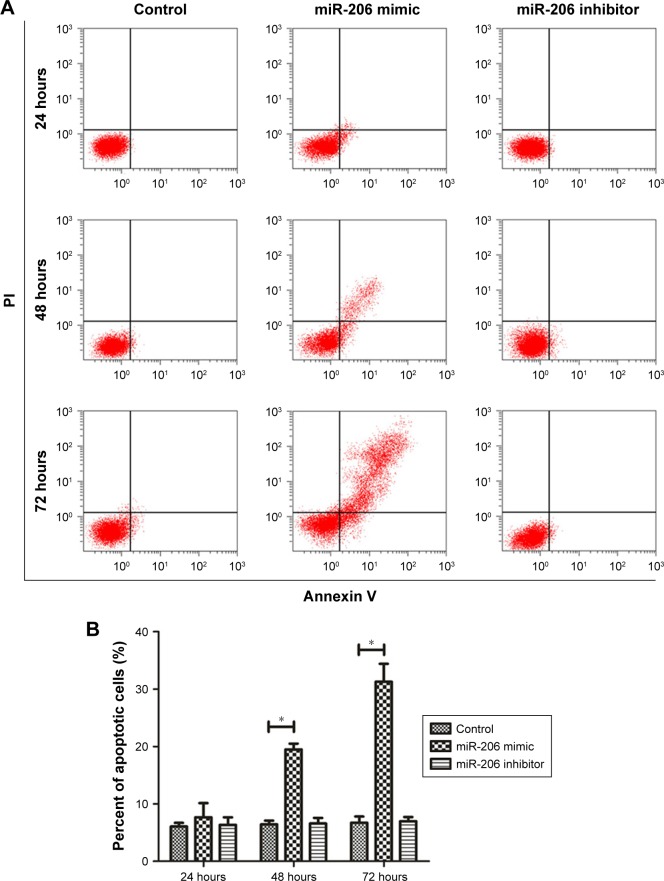
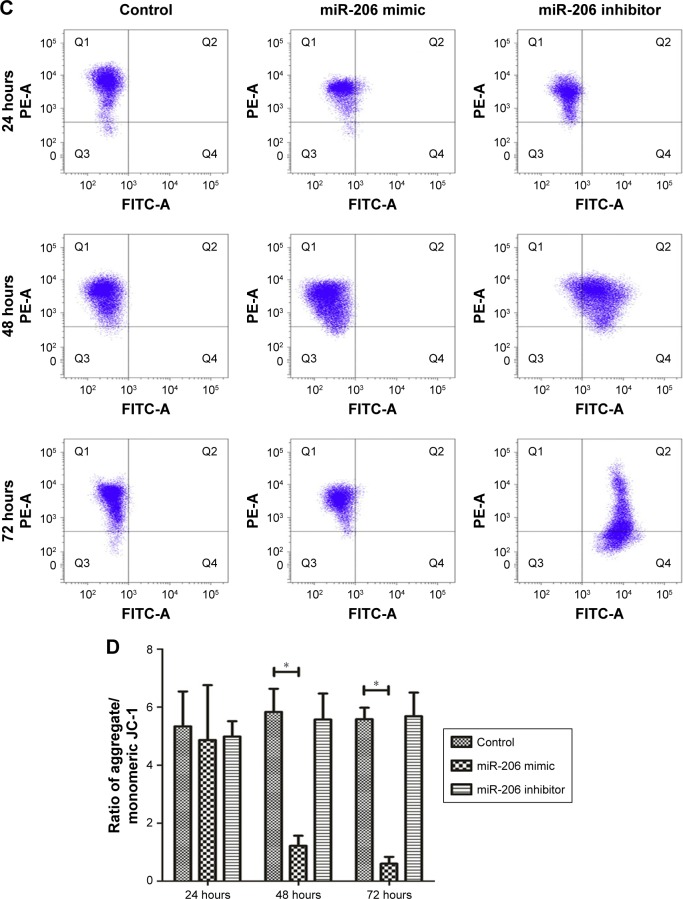
As indicated by the apoptosis assays, miR-206 overexpression promoted NCI-N87 cell apoptosis in vitro. Moreover, miR-206 mimics clearly increased the proportion of apoptotic NCI-N87 cells (*P<0.05 from 48 to 72 hours; Figure 3A and B). The JC-1 assay (Figure 3C and D) revealed that, after transfection with miR-206 mimics, green fluorescence was increased, whereas red fluorescence was reduced, indicating that miR-206 promoted the collapse of the transmembrane potential of NCI-N87 cells.
Figure 3.
MiR-206 overexpression promoted mitochondria-mediated apoptosis.
Notes: (A) Annexin V/PI fluorescence in NCI-N87 cells was detected using flow cytometry. (B) The population of apoptotic cells after transfection at 24, 48, and 72 hours. (C) JC-1 fluorescence in NCI-N87 cells was detected using flow cytometry. (D) The ratio of aggregate and monomeric JC-1 fluorescence (green/red fluorescence) in NCI-N87 cells after transfection at 24, 48, and 72 hours. *P<0.05.
Abbreviations: FITC, fluorescein isothiocyanate; PI, propidium iodide; PE, phycoerythrin.
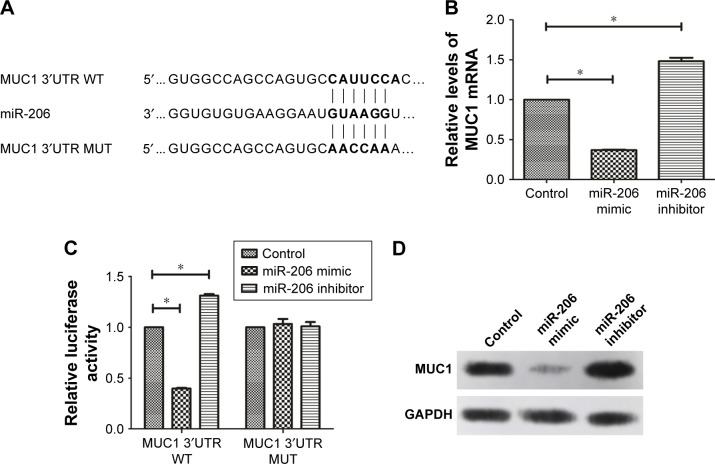
MUC1 is a direct target of miR-206 in GC
The TargetScan, miRDB, and miRanda databases predicted the possible targets of miR-206. Our findings showed that the assumed miR-206-binding sites were located in the 3′UTR of the MUC1 transcript. This was confirmed by constructing the plasmids containing the wild-type or mutant-type MUC1 3′UTR (Figure 4A). The dual luciferase assay indicated that the luciferase activity of the MUC1-3′UTR-WT was clearly reduced when miR-206 was overexpressed, whereas the luciferase reporter activity of the MUC1-3′UTR-WT increased following the reduction of endogenous levels of miR-206 by treating GC cells with the miR-206 inhibitor. In addition, miR-206 mimics or miR-206 inhibitors do not affect the luciferase reporter activity of MUC1-3′UTR-MUT (Figure 4C). Furthermore, the mRNA (Figure 4B) and protein (Figure 4D) levels of MUC1 were significantly decreased by miR-206 mimics and increased by miR-206 inhibitors in NCI-N87 cells. These findings demonstrated that miR-206 can directly targeting MUC1 and suppress its expression in GC cells.
Figure 4.
MUC1 is a direct target of miR-206 in GC cells.
Notes: (A) The binding sites of the MUC1 3′UTR WT and MUT with the miR-206 are shown. (B) NCI-N87 cells after transfection with the miR-206 mimic, miR-206 inhibitor, and MUT-MUC1-3′UTR or WT-MUC1 3′UTR reporter plasmids were utilized for luciferase assays. (C, D) The overexpression of miR-206 suppresses MUC1 expression at the mRNA and protein levels in NCI-N87 cells. *P<0.05.
Abbreviations: GC, gastric cancer; MUT, mutant-type; WT, wild-type.
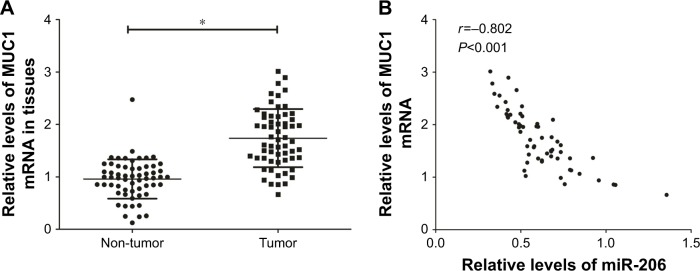
MUC1 expression is upregulated and inversely associated with miR-206 levels in GC tissues
The expression of tissue MUC1 mRNA was examined using qRT-PCR. The upregulation of MUC1 expression was significant in GC specimens compared with corresponding non-tumor tissues (Figure 5A). In addition, the Pearson correlation analysis confirmed that MUC1 mRNA levels had a negative correlation with miR-206 expression in 60 GC tissues (r=−0.802, P<0.001; Figure 5B).
Figure 5.
Upregulation of MUC1 has a negative correlation with miR-206 expression in GC tissues.
Notes: (A) The high mRNA level of MUC1 was significant in GC tissues. (B) The negative correlation between MUC1 mRNA levels and miR-206 in GC tissues (n=60). *P<0.05.
Abbreviation: GC, gastric cancer.
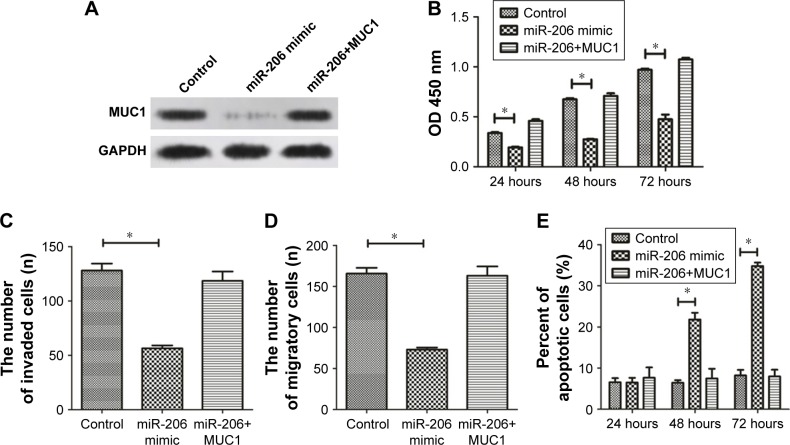
MUC1 overexpression abrogates the antitumor effect of miR-206 in GC
We conducted rescue experiments to validate whether miR-206 exerts tumor-suppressive functions by targeting MUC1. MiR-206 mimics and pcDNA-MUC1 were both transfected into NCI-N87 cells, and the MUC1 protein level was examined by Western blotting analysis (Figure 6A). The study outcomes demonstrated that MUC1 overexpression abrogates the miR-206-mediated suppressive effects on proliferation, migration, and invasion as well as induction of apoptosis of NCI-N87 cells (Figure 6B–E). These findings suggested that the tumor-suppressive effects of miR-206, at least in part, were achieved by targeting MUC1.
Figure 6.
Overexpression of MUC1 abrogates the antitumor effect of miR-206 against GC.
Notes: (A) MUC1 expression in NCI-N87 cells pre-transfected with miR-206 mimic and transfected with or without the MUC1 overexpression plasmid. (B–E) Cell proliferation, migration, invasion, and apoptosis in NCI-N87 cells after transfection with miR-206 with or without MUC1 overexpression were assessed respectively by CCK-8, Transwell migration, Transwell invasion, and Annexin V/PI staining assays. *P<0.05.
Abbreviations: CCK-8, Cell Counting Kit-8; GC, gastric cancer.
Discussion
In recent years, many miRNAs have been demonstrated to contribute to GC tumorigenesis and development, which serve as an effective diagnostic and therapeutic target for GC.10,15 Therefore, it is of great importance to explore potential miRNAs associated with the development of GC as it may provide an opportunity for better diagnosis and treatment as well as to improve patient outcome. We found that the downregulation of miR-206 in GC tissues has a negative association with lymph node metastasis and TNM stage. Moreover, patients with higher miR-206 expression showed a better prognosis. Functional studies showed that miR-206 overexpression may exert inhibitory activities against cell proliferation, migration, and invasion of GC cells. Furthermore, MiR-206 overexpression may promote cell apoptosis. It is well recognized that the mitochondrial apoptosis pathway plays a central role in the anticancer activity of many anticancer drugs or compounds. A study by Zhang et al reported that Euphorbia factor L2 exerts a tumor-suppressive role against A549 cells through activation of the mitochondrial pathway.16 Moreover, other studies demonstrated that a large number of anticancer drug candidates suppress the biological function of cancer cells by inducing the mitochondrial apoptosis pathway.17–19 Consistent with previous studies, in the present study, our findings reveal that miR-206 overexpression could effectively promote NCI-N78 cells apoptotic cell death that is mediated by an intrinsic apoptotic mechanism, supported by the loss of mitochondrial membrane potential following miR-206 mimic transfection. The results from other studies, together with ours, confirm the antitumor effect of miR-206 in many tumors.
It is reported that miR-206 expression is usually deregulated in tumors, including bladder cancer,20 breast cancer,21 cervical cancer,22 and head and neck squamous cell carcinoma.23 In this study, we observed the downregulation of miR-206 in both GC cell lines and tissues. The molecular mechanisms for miR-206 deregulation have not been fully deconstructed yet. Among the mechanisms for miRNA regulation, promoter hypermethylation is a major factor responsible for the downregulation of miRNA expression.24 We found that 5-aza-2′-deoxycytidine (5-aza-dC) increases miR-206 expression in GC cells, which indirectly suggests that promoter hypermethylation is involved in miR-206 deregulation in GC. In addition, 5-aza-dC may inhibit the proliferation of GC cells, which suggests a possible application of DNA methyltransferase inhibitor for use in targeted therapy for GC.
It has been demonstrated that miR-206 may exert an antitumor effect by targeting various oncogenes. MiR-206 may target 14-3-3ζ and inhibit the growth and angiogenesis of non-small cell lung cancer cells;25 miR-206 may inhibit cell proliferation by targeting GAK in renal cancer cells.26 Moreover, we assessed the potential target of miR-206. The bioinformatics methods (TargetScan, miRDB, and miRanda) predicted that some potential genes are downstream target genes of miR-206 including MUC1, Cyclin D1, Cyclin D2, CDK9, and TIMP3 (Table 4; Supplementary material 2). Among the targets of miR-206, we focused on MUC1 because the expression level of MUC1 was the highest in the gastric cells. We detected the effects of miR-206 on mRNA and protein levels of MUC1 by qRT-PCR and Western blotting, and found MUC1 expression was apparently affected by miR-206. Dual luciferase assays indicated that the luciferase activity of MUC1-3′UTR-WT was apparently reduced by miR-206 mimics and increased by miR-206 inhibitors. MUC1, a membrane-bound glycoprotein, is a major component of the ductal cell surface of normal glandular cells. MUC1 acts as an oncogene, which is overexpressed and aberrantly glycosylated in many tumors such as hepatocellular,27 lung,28 breast,29 and colorectal30 cancer. However, the role of the MUC1 gene in tumorigenesis has not been completely elucidated. Loss of polarity and MUC1 overexpression interferes with cell adhesion and promotes cancer cell metastases.31 It is suggested that MUC1 overexpression promotes cell proliferation by activation of the WNT signaling pathway.32 Our previous study found that the MUC1 gene may induce resistance of HER2-positive GC cells to trastuzumab.33 The abovementioned studies suggest that MUC1 may be involved in many biological tumor cell behaviors.
Table 4.
Some potential genes are predicted targets for miR-206 in target scan
| Target gene | Context++ score | Context++ score percentile | Aggregate Pct |
|---|---|---|---|
|
| |||
| MUC1 | −0.03 | −0.03 | <0.1 |
| IGF1 | −0.57 | −0.80 | 0.92 |
| ATG13 | −0.26 | −0.26 | 0.67 |
| SERP1 | −0.77 | −0.80 | 0.97 |
In the present study, according to the results from luciferase activity assays, Western blotting and qRT-PCR, we have identified MUC1 as a direct target of miR-206. Moreover, we found that the MUC1 mRNA level has an inverse correlation with miR-206 expression in GC tissues. Notably, rescue experiments showed that MUC1 overexpression abrogates the suppression of miR-206 on cell proliferation and metastasis, and the induction of miR-206 on apoptosis in GC cells. Taken together, our data suggested that the antitumor activities of miR-206 on GC cells partially is associated with its inactivation of MUC1.
However, this study has been unable to demonstrate that the underlying molecular mechanism that contribute to the anticancer effect of miR-206 or the oncogenic role of MUC1 such as the mechanism by which miR-206 triggers GC cells apoptosis via regulating MUC1, the extrinsic pathway, the mitochondria-mediated intrinsic pathway, or others. Moreover, the network between circRNAs, lncRNAs, and miRNAs has attracted much attention from researches; it has been reported that circRNAs or lncRNA might function as a ceRNAs through sponging target miRNAs and they may indirectly modulate miRNA target genes. Collectively, there is abundant room for further progress in determining the role of miR-206 on GC. Future studies on the current topic are, therefore, recommended.
Conclusion
Our data indicated that downregulation of miR-206 is significant in GC tissues and cells, and that downregulation of miR-206 has an association with lymph node metastasis and TNM stage. Moreover, we found that GC cell proliferation, migration, and invasion were significantly inhibited after restoration of miR-206. Further, xenograft tumor growth in vivo was inhibited.
Supplementary Materials
Acknowledgments
This study was supported by the Key Program of Foundation for Young Talents in Colleges and Universities of Anhui Province (Hefei, China; grant no. 2013SQRL053ZD), Hospital Level Project of Bengbu Medical College (grant no. Byky1340 and Byky1361), Natural Science Research General Project of Anhui Province (grant no. KJ2015B042by).
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Compare D, Rocco A, Nardone G. Risk factors in gastric cancer. Eur Rev Med Pharmacol Sci. 2010;14(4):302–308. [PubMed] [Google Scholar]
- 2.Kelley JR, Duggan JM. Gastric cancer epidemiology and risk factors. J Clin Epidemiol. 2003;56(1):1–9. doi: 10.1016/s0895-4356(02)00534-6. [DOI] [PubMed] [Google Scholar]
- 3.Orditura M, Galizia G, Sforza V, et al. Treatment of gastric cancer. World J Gastroenterol. 2014;20(7):1635–1649. doi: 10.3748/wjg.v20.i7.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu W, Sun M, Zou GM, Chen J. MicroRNA and cancer: Current status and prospective. Int J Cancer. 2007;120(5):953–960. doi: 10.1002/ijc.22454. [DOI] [PubMed] [Google Scholar]
- 5.Wang Y, Lee CG. MicroRNA and cancer – focus on apoptosis. J Cell Mol Med. 2009;13(1):12–23. doi: 10.1111/j.1582-4934.2008.00510.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466(7308):835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shenouda SK, Alahari SK. MicroRNA function in cancer: oncogene or a tumor suppressor? Cancer Metastasis Rev. 2009;28(3–4):369–378. doi: 10.1007/s10555-009-9188-5. [DOI] [PubMed] [Google Scholar]
- 8.Mcmanus MT. MicroRNAs and cancer. Semin Cancer Biol. 2003;13(4):253–258. doi: 10.1016/s1044-579x(03)00038-5. [DOI] [PubMed] [Google Scholar]
- 9.Calin GA, Croce CM. MicroRNA-cancer connection: the beginning of a new tale. Cancer Res. 2006;66(15):7390–7394. doi: 10.1158/0008-5472.CAN-06-0800. [DOI] [PubMed] [Google Scholar]
- 10.Shrestha S, Hsu SD, Huang WY, et al. A systematic review of microRNA expression profiling studies in human gastric cancer. Cancer Med. 2014;3(4):878–888. doi: 10.1002/cam4.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nakao K, Miyaaki H, Ichikawa T. Antitumor function of microRNA-122 against hepatocellular carcinoma. J Gastroenterol. 2014;49(4):589–593. doi: 10.1007/s00535-014-0932-4. [DOI] [PubMed] [Google Scholar]
- 12.Li X, Zhang Y, Zhang H, et al. miRNA-223 promotes gastric cancer invasion and metastasis by targeting tumor suppressor EPB41L3. Mol Cancer Res. 2011;9(7):824–833. doi: 10.1158/1541-7786.MCR-10-0529. [DOI] [PubMed] [Google Scholar]
- 13.Zhang L, Liu X, Jin H, et al. miR-206 inhibits gastric cancer proliferation in part by repressing cyclinD2. Cancer Lett. 2013;332(1):94–101. doi: 10.1016/j.canlet.2013.01.023. [DOI] [PubMed] [Google Scholar]
- 14.Yang Q, Zhang C, Huang B, et al. Downregulation of microRNA-206 is a potent prognostic marker for patients with gastric cancer. Eur J Gastroenterol Hepatol. 2013;25(8):953–957. doi: 10.1097/MEG.0b013e32835ed691. [DOI] [PubMed] [Google Scholar]
- 15.Zhu X, Lv M, Wang H, Guan W. Identification of circulating microRNAs as novel potential biomarkers for gastric cancer detection: a systematic review and meta-analysis. Dig Dis Sci. 2014;59(5):911–919. doi: 10.1007/s10620-013-2970-9. [DOI] [PubMed] [Google Scholar]
- 16.Lin M, Tang S, Zhang C, et al. Euphorbia factor L2 induces apoptosis in A549 cells through the mitochondrial pathway. Acta Pharm Sin B. 2017;7(1):59–64. doi: 10.1016/j.apsb.2016.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang JY, Lin MT, Tung HY, et al. Bruceine D induces apoptosis in human chronic myeloid leukemia K562 cells via mitochondrial pathway. Am J Cancer Res. 2016;6(4):819–826. [PMC free article] [PubMed] [Google Scholar]
- 18.Liu K, Gao H, Wang Q, et al. Hispidulin suppresses cell growth and metastasis by targeting PIM1 through JAK2/STAT3 signaling in colorectal cancer. Cancer Sci. 2018;109(5):1369–1381. doi: 10.1111/cas.13575. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 19.Gao H, Gao MQ, Peng JJ, Han M, Liu KL, Han YT. Hispidulin mediates apoptosis in human renal cell carcinoma by inducing ceramide accumulation. Acta Pharmacol Sin. 2017;38(12):1618–1631. doi: 10.1038/aps.2017.154. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 20.Huang B, Zhai W, Hu G, et al. MicroRNA-206 acts as a tumor suppressor in bladder cancer via targeting YRDC. Am J Transl Res. 2016;8(11):4705–4715. [PMC free article] [PubMed] [Google Scholar]
- 21.Yin K, Yin W, Wang Y, et al. MiR-206 suppresses epithelial mesenchymal transition by targeting TGF-β signaling in estrogen receptor positive breast cancer cells. Oncotarget. 2016;7(17):24537–24548. doi: 10.18632/oncotarget.8233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen AH, Qin YE, Tang WF, Tao J, Song HM, Zuo M. MiR-34a and miR-206 act as novel prognostic and therapy biomarkers in cervical cancer. Cancer Cell Int. 2017;17:63. doi: 10.1186/s12935-017-0431-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koshizuka K, Hanazawa T, Fukumoto I, et al. Dual-receptor (EGFR and c-MET) inhibition by tumor-suppressive miR-1 and miR-206 in head and neck squamous cell carcinoma. J Hum Genet. 2017;62(1):113–121. doi: 10.1038/jhg.2016.47. [DOI] [PubMed] [Google Scholar]
- 24.Gulyaeva LF, Kushlinskiy NE. Regulatory mechanisms of microRNA expression. J Transl Med. 2016;14(1):143. doi: 10.1186/s12967-016-0893-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xue D, Yang Y, Liu Y, et al. MicroRNA-206 attenuates the growth and angiogenesis in non-small cell lung cancer cells by blocking the 14-3-3ζ/STAT3/HIF-1α/VEGF signaling. Oncotarget. 2016;7(48):79805–79813. doi: 10.18632/oncotarget.12972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wei C, Wang S, Ye ZQ, Chen ZQ. miR-206 inhibits renal cell cancer growth by targeting GAK. J Huazhong Univ Sci Technolog Med Sci. 2016;36(6):852–858. doi: 10.1007/s11596-016-1674-8. [DOI] [PubMed] [Google Scholar]
- 27.Wang J, Ni WH, Hu KB, et al. Targeting MUC1 and JNK by RNA interference and inhibitor inhibit the development of hepatocellular carcinoma. Cancer Sci. 2017;108(3):504–511. doi: 10.1111/cas.13144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li J, Hu YM, Du YJ, et al. Expressions of MUC1 and vascular endothelial growth factor mRNA in blood are biomarkers for predicting efficacy of gefitinib treatment in non-small cell lung cancer. BMC Cancer. 2014;14:848. doi: 10.1186/1471-2407-14-848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Patel DS, Khandeparkar SGS, Joshi AR, et al. Immunohistochemical Study of MUC1, MUC2 and MUC5AC Expression in Primary Breast Carcinoma. . J Clin Diagn Res. 2017;11(4):EC30–EC34. doi: 10.7860/JCDR/2017/26533.9707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang HS, Wang LH. The expression and significance of Gal-3 and MUC1 in colorectal cancer and colon cancer. Onco Targets Ther. 2015;8:1893–1898. doi: 10.2147/OTT.S83502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.von Mensdorff-Pouilly S, Snijdewint FG, Verstraeten AA, Verheijen RH, Kenemans P. Human MUC1 mucin: a multifaceted glycoprotein. Int J Biol Markers. 2000;15(4):343–356. doi: 10.1177/172460080001500413. [DOI] [PubMed] [Google Scholar]
- 32.Gnemmi V, Bouillez A, Gaudelot K, et al. MUC1 drives epithelial-mesenchymal transition in renal carcinoma through Wnt/β-catenin pathway and interaction with SNAIL promoter. Cancer Lett. 2014;346(2):225–236. doi: 10.1016/j.canlet.2013.12.029. [DOI] [PubMed] [Google Scholar]
- 33.Deng M, Jing DD, Meng XJ. Effect of MUC1 siRNA on drug resistance of gastric cancer cells to trastuzumab. Asian Pac J Cancer Prev. 2013;14(1):127–131. doi: 10.7314/apjcp.2013.14.1.127. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.