Abstract
Neoantigen-targeted therapies have typically been based upon personalized neoantigen-specific vaccines; however, in this issue of JCI, van der Lee et al. describe the development of a potential cellular immunotherapy targeting a “public” neoantigen derived from nucleophosmin 1 (NPM1), which is mutated in approximately 30% of acute myeloid leukemias (AMLs). The authors use reverse immunology to predict, and biochemically confirm, NPM1-derived neoepitopes (ΔNPM1) and then generate high-avidity T cell clones and retrovirally transduced T cell populations that kill NPM1-mutated AML. This study provides a general approach to adoptive cellular therapy that can be applied to targeting other tumors with public neoantigens.
Neoantigen targeting for cancer immunotherapy
The advent of high-density sequencing platforms has allowed the mutational landscape of human tumor types to be mapped and annotated in great detail. Studies from The Cancer Genome Atlas (TCGA) have shown that different tumor types harbor vastly different numbers of somatic mutations, and these studies have now provided for us a comprehensive list of targetable recurring mutations (1).
Somatic mutations can clearly lead to aberrant protein activity with downstream cellular consequences; however, they can also generate new immunologically active targets that can be recognized by the host’s adaptive immune system (2). These “neoantigens” are derived from the proteasomally digested peptide fragments from mutated proteins, which are then presented by class I human leukocyte antigens (HLA) on the cancer cell surface (3). Because the host’s T cell repertoire does not encounter the mutated peptides through maturation in the thymus, there exist, in some cases, cytotoxic CD8+ T cells that can bind with high avidity to the mutated peptide/HLA complex and induce cell death through the release of perforin and granzyme (3).
Several different neoantigen-targeting approaches have been undertaken in clinical contexts. One of the most successful strategies that likely involves neoantigen targeting is immune checkpoint inhibition, primarily against PD-1, to allow chronically antigen-exposed, immunologically exhausted, neoantigen-specific T cells to regain effector function (4). While not specific neoantigen-targeting strategies, anti–PD-1 therapies have changed treatment strategies in many cancers, with their greatest efficacy observed in tumors with high mutational burdens, which suggests their efficacy involves enhancing T cell responses to multiple neoantigens (5–7). For tumors with low mutational burdens, other strategies, primarily neoantigen-specific ones, are needed.
Neoantigen-specific immunotherapies have also been developed as single- or multi-epitope vaccines with many formulations that provide truly personalized targeted immunotherapies (8–10). While immune responses to neoantigen vaccines are well described, there are still significant gaps in our understanding of which mutations lead to neoantigens that can elicit robust immune responses following vaccination and lead to clinically relevant outcomes. At this time, it is not clear whether there will be a readily available general solution for determining the hierarchy of neoantigen-specific T cell responses in individual patients, as responses are likely to derive from a combination of tumor-intrinsic features, such as HLA type, neoantigen gene expression, and intratumor heterogeneity as well as the patient’s underlying T cell repertoire.
Neoantigen targeting adoptive cellular therapy (ACT), an alternative to neoantigen vaccines, offers a narrower, but in many ways, more controlled series of immunotherapy parameters that can be manipulated to create high-potency therapeutics. Because ACT requires the complex and expensive genetic modification of T cells, neoantigen selection is crucial, so that a therapy can be provided to the maximum number of patients. Optimal neoantigens for ACT should be developed from genes that are highly expressed in tumors and have common, recurrent (i.e., “public”) driver mutations, which should ensure that the neoantigen will be present in multiple patients and in all intratumor subclones. An additional feature for selection of neoantigens for ACT targeting is that they be presented on the cell surface by HLA molecules that are common in the human population.
Development of a neoantigen-specific cellular therapy
In this issue, van der Lee et al. report the development of a T cell therapy against a neoantigen that has all of the properties that make it an ideal immunotherapy target (11). Roughly one-third of acute myeloid leukemia (AML) cases contain a driver mutation in the nucleophosmin 1 (NPM1) gene (12, 13). The most common mutation is a 4 base-pair insertion into exon 12 of the gene, which leads to a frame shift resulting in a mutant protein that is 4 amino acids longer than the wild-type and whose last 11 amino acids are translated in an alternate reading frame. The C terminus of wild-type NPM1 contains a nuclear localization signal, which is lost in mutated NPM1, leading to increased cytosolic concentration, and presumably increased proteasomal degradation and antigen presentation (12, 13).
van der Lee and colleagues elegantly demonstrate the power of a reverse immunology approach for the discovery of a public neoantigen and the subsequent development of a potential T cell immunotherapeutic (Figure 1). Reverse immunology is predicated on the initial prediction and identification of a target antigen, with the subsequent generation of an antigen-specific T cell clone (as opposed to classical antigen discovery approaches, which start with a T cell clone and work to identify the antigen) (14, 15). Using known genomics data for NPM1 mutations and publicly available computational modeling software, van der Lee et al. predicted candidate peptide epitopes downstream of the NPM1 mutation that could bind to common class I HLA types. Because HLA-A*02:01 is the most common HLA type in the Dutch population, investigations were subsequently focused on epitopes that were predicted to bind this HLA type.
Figure 1. Developing neoantigen-specific T cells by reverse immunology.
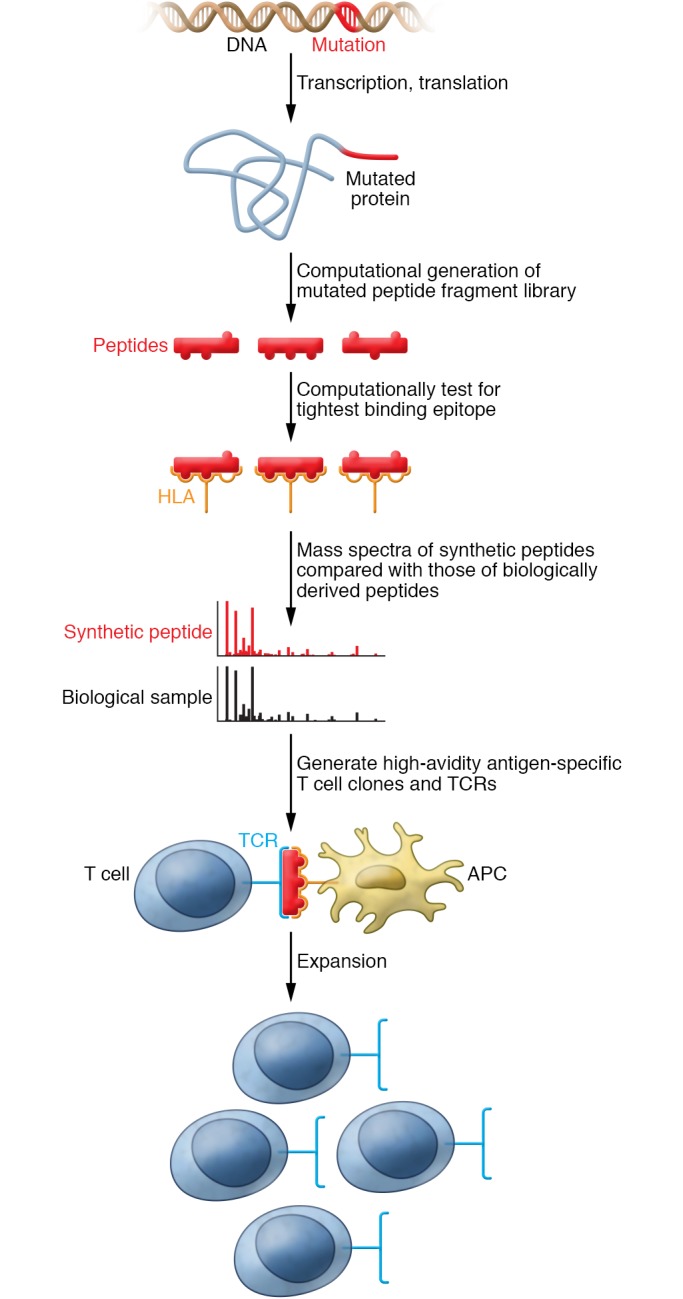
For a “public” neoantigen, a common mutation is identified. In the case of NPM1, this is a 4 base pair insertion. The translational result of the mutation is predicted, which for NPM1 is an out of frame amino acid sequence at the C terminus. The new peptide sequences are tested for predicted binding to an HLA type of interest to determine the most likely neoepitopes. Targeted mass spectrometry is performed to compare peptide epitope sequences from a biological sample (e.g., cell line or primary tumor) to a synthetic peptide standard. High-avidity antigen-specific T cell clones are generated, and their T cell receptors are sequenced. The T cell receptor is incorporated into a viral vector into subsequent generation of T cell populations for use in ACT.
Many neoantigen-based studies have relied solely on computational predictions for “discovery”; however, biochemical confirmation of peptide epitope presentation is necessary to provide confidence for the development of a T cell–based immunotherapeutic. van der Lee and colleagues immunoprecipitated peptide/HLA complexes from primary AML samples and used targeted mass spectrometry to directly compare the fragmentation spectra from the AML samples to those of the synthetic peptides that were predicted to be potential neoantigens.
van der Lee et al. generated T cell clones against the two mass spectrometry–validated epitopes (ΔNPM1) predicted to bind HLA-A*02:01. Interestingly, they were unable to generate any ΔNPM1 neoantigen–specific T cell clones from AML patients using their standard approach of peptide/HLA magnetic-activated cell sorting (MACS) preenrichment, followed by single-cell FACS-based cloning (16). To generate a T cell clone with high avidity toward either neoantigen, they used the same approach on HLA-*A02:01-expressing, healthy donor buffy coats. The T cell receptor (TCR) sequences from these clones were inserted into retroviral vectors, and T cells containing the transgenic neoantigen TCR were produced. These cells had high affinity to the neoantigen/HLA complex and demonstrated cytotoxicity against NPM1-mutated AML both in vitro and in vivo.
Concluding remarks
This study demonstrates a clean method for identifying public neoantigens and generating immunotherapeutics. The general approach of (a) computational prediction of peptide epitopes, (b) targeted mass spectrometric confirmation of epitope presentation, and (c) generation of high-avidity, high-specificity T cell clones could be applied to any public somatic mutations, such as the G12D mutation in KRAS commonly seen in pancreatic cancer (17), the L265P mutation in MYD88 seen in Waldenstrom’s macroglobulinemia (18), and the BRAF V600E mutation seen in melanoma (19), to mention a few.
The work by van der Lee and colleagues lays a clear blueprint for the development of neoantigen-specific T cell–based immunotherapies, and their work also raises several issues that should be investigated in future studies. Conceptually, neoantigens can be derived from multiple genetic perturbations. Single nucleotide variants (SNVs) are the most common and most extensively studied source of neoantigens; however, other genetic events, such as insertions (as evidenced by NPM1), gene fusions (e.g., BCR-ABL), and aberrant mRNA splicing could lead to novel proteins and peptide epitopes. While it is tempting to focus on the more common and easier to characterize SNV-derived neoantigens, van der Lee and colleagues’ work suggests that non–SNV-derived neoantigens are valuable targets as well. It is in fact conceivable, that non–SNV-derived neoantigens may be able to elicit stronger immune responses due to their greater divergence from wild-type sequences. Further investigation needs to be performed to fully elicit what properties, e.g., mutant/wild-type sequence disparity and T cell repertoire, are associated with highly immunogenic neoantigens.
Interestingly, van der Lee et al. were unable to generate ΔNPM1 neoantigen–specific T cells from NPM1-mutated, HLA-A*02:01-expressing AML patients, while neoantigen-specific T cells have been generated from patients with other cancers (20). It is possible that AML can induce deletion of high-avidity ΔNPM1-specific T cells through processes such as activation-induced cell death. Studies investigating mechanisms of tumor immune evasion through both T cell exhaustion/senescence and T cell deletion would greatly improve our understanding of the different mechanisms of tumor immune evasion and their association with different tumor types or microenvironments.
Acknowledgments
I would like to thank members of the Armistead and Ben Vincent labs for helpful discussions regarding this commentary.
Version 1. 01/14/2019
Electronic publication
Version 2. 02/01/2019
Print issue publication
Footnotes
Conflict of interest: The author has declared that no conflict of interest exists.
Reference information: J Clin Invest. 2019;129(2):506–508. https://doi.org/10.1172/JCI126116.
See the related article at Mutated nucleophosmin 1 as immunotherapy target in acute myeloid leukemia.
References
- 1.Hoadley KA, et al. Cell-of-origin patterns dominate the molecular classification of 10,000 tumors from 33 types of cancer. Cell. 2018;173(2):291–304.e6. doi: 10.1016/j.cell.2018.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thorsson V, et al. The immune landscape of cancer. Immunity. 2018;48(4):812–830.e14. doi: 10.1016/j.immuni.2018.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schumacher TN, Schreiber RD. Neoantigens in cancer immunotherapy. Science. 2015;348(6230):69–74. doi: 10.1126/science.aaa4971. [DOI] [PubMed] [Google Scholar]
- 4.Sharpe AH, Pauken KE. The diverse functions of the PD1 inhibitory pathway. Nat Rev Immunol. 2018;18(3):153–167. doi: 10.1038/nri.2017.108. [DOI] [PubMed] [Google Scholar]
- 5.Eggermont AMM, et al. Adjuvant pembrolizumab versus placebo in resected stage III melanoma. N Engl J Med. 2018;378(19):1789–1801. doi: 10.1056/NEJMoa1802357. [DOI] [PubMed] [Google Scholar]
- 6.Hellmann MD, et al. Genomic features of response to combination immunotherapy in patients with advanced non-small-cell lung cancer. Cancer Cell. 2018;33(5):843–852.e4. doi: 10.1016/j.ccell.2018.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rizvi NA, et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015;348(6230):124–128. doi: 10.1126/science.aaa1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carreno BM, et al. Cancer immunotherapy. A dendritic cell vaccine increases the breadth and diversity of melanoma neoantigen-specific T cells. Science. 2015;348(6236):803–808. doi: 10.1126/science.aaa3828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ott PA, et al. An immunogenic personal neoantigen vaccine for patients with melanoma. Nature. 2017;547(7662):217–221. doi: 10.1038/nature22991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sahin U, et al. Personalized RNA mutanome vaccines mobilize poly-specific therapeutic immunity against cancer. Nature. 2017;547(7662):222–226. doi: 10.1038/nature23003. [DOI] [PubMed] [Google Scholar]
- 11.van der Lee DI, et al. Mutated nucleophosmin 1 as immunotherapy target in acute myeloid leukemia. J Clin Invest. 2019;129(2):774–785. doi: 10.1172/JCI97482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Falini B, et al. Cytoplasmic nucleophosmin in acute myelogenous leukemia with a normal karyotype. N Engl J Med. 2005;352(3):254–266. doi: 10.1056/NEJMoa041974. [DOI] [PubMed] [Google Scholar]
- 13.Heath EM, Chan SM, Minden MD, Murphy T, Shlush LI, Schimmer AD. Biological and clinical consequences of NPM1 mutations in AML. Leukemia. 2017;31(4):798–807. doi: 10.1038/leu.2017.30. [DOI] [PubMed] [Google Scholar]
- 14.Gubin MM, Artyomov MN, Mardis ER, Schreiber RD. Tumor neoantigens: building a framework for personalized cancer immunotherapy. J Clin Invest. 2015;125(9):3413–3421. doi: 10.1172/JCI80008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hombrink P, et al. Discovery of T cell epitopes implementing HLA-peptidomics into a reverse immunology approach. J Immunol. 2013;190(8):3869–3877. doi: 10.4049/jimmunol.1202351. [DOI] [PubMed] [Google Scholar]
- 16.Jahn L, et al. TCR-based therapy for multiple myeloma and other B-cell malignancies targeting intracellular transcription factor BOB1. Blood. 2017;129(10):1284–1295. doi: 10.1182/blood-2016-09-737536. [DOI] [PubMed] [Google Scholar]
- 17.Biankin AV, et al. Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature. 2012;491(7424):399–405. doi: 10.1038/nature11547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Poulain S, et al. MYD88 L265P mutation in Waldenstrom macroglobulinemia. Blood. 2013;121(22):4504–4511. doi: 10.1182/blood-2012-06-436329. [DOI] [PubMed] [Google Scholar]
- 19.Long GV, et al. Prognostic and clinicopathologic associations of oncogenic BRAF in metastatic melanoma. J Clin Oncol. 2011;29(10):1239–1246. doi: 10.1200/JCO.2010.32.4327. [DOI] [PubMed] [Google Scholar]
- 20.Tran E, et al. Immunogenicity of somatic mutations in human gastrointestinal cancers. Science. 2015;350(6266):1387–1390. doi: 10.1126/science.aad1253. [DOI] [PMC free article] [PubMed] [Google Scholar]


