Abstract
X-linked dominant incontinentia pigmenti (IP) and X-linked recessive anhidrotic ectodermal dysplasia with immunodeficiency (EDA-ID) are caused by loss-of-function and hypomorphic IKBKG (also known as NEMO) mutations, respectively. We describe a European mother with mild IP and a Japanese mother without IP, whose 3 boys with EDA-ID died from ID. We identify the same private variant in an intron of IKBKG, IVS4+866 C>T, which was inherited from and occurred de novo in the European mother and Japanese mother, respectively. This mutation creates a new splicing donor site, giving rise to a 44-nucleotide pseudoexon (PE) generating a frameshift. Its leakiness accounts for NF-κB activation being impaired but not abolished in the boys’ cells. However, aberrant splicing rates differ between cell types, with WT NEMO mRNA and protein levels ranging from barely detectable in leukocytes to residual amounts in induced pluripotent stem cell–derived (iPSC-derived) macrophages, and higher levels in fibroblasts and iPSC-derived neuronal precursor cells. Finally, SRSF6 binds to the PE, facilitating its inclusion. Moreover, SRSF6 knockdown or CLK inhibition restores WT NEMO expression and function in mutant cells. A recurrent deep intronic splicing mutation in IKBKG underlies a purely quantitative NEMO defect in males that is most severe in leukocytes and can be rescued by the inhibition of SRSF6 or CLK.
Keywords: Genetics, Infectious disease
Keywords: Molecular diagnosis, NF-kappaB
Introduction
The IKBKG gene, also known as NEMO, encodes the NF-κB essential modulator (NEMO) (1, 2). NEMO encodes the third regulatory subunit of the IκB kinase (IKK) complex and is also called IKKγ (3–5). NEMO was discovered in 1998 as a component required for the activation of NF-κB via the canonical pathway (1–4, 6). In 2000, loss-of-function mutations of IKBKG were shown to abolish canonical NF-κB activation and to underlie X-linked dominant (XD) incontinentia pigmenti (IP) (phenotype MIM #308300), a multisystem disorder affecting the skin and its appendages in all affected women, and the CNS in most but not all women (7, 8). XD-IP has an estimated prevalence of 1 to 10 cases per million births. A defining feature of the IP phenotype and the IP-causing genotype is that they are both lethal in utero in males. The absence of a functional NEMO protein renders cells apoptotic, resulting in skewed X inactivation in most if not all of the cells of affected females and early fetal death in affected males. In 2001, hypomorphic IKBKG mutations were found to impair NF-κB activation and to cause X-linked recessive (XR) anhidrotic ectodermal dysplasia with immunodeficiency (EDA-ID) (phenotype MIM #300291) (9, 10). Affected men display typical signs of EDA, including sparse hair, eyebrows, and eyelashes, hypohidrosis, hypodontia, and conical incisors, together with an ID not seen in patients with mutations of ectodysplasin A or its receptor chains (11). Female carriers in kindreds with EDA-ID are asymptomatic or show mild signs of IP, mostly restricted to the persistence of cutaneous Blaschko lines and sparse or conical incisors (9, 10, 12–14). Indeed, although EDA-ID and IP are clearly different disorders in men, they have a phenotypic overlap in women, in whom very mild forms of IP may be caused by severely hypomorphic IKBKG mutations (8). XD-IP and XR-EDA-ID are allelic, as they are caused by different mutations of the same gene.
Approximately 78% of known cases of XD-IP are due to a recurrent deletion (NEMOΔ4–10), removing exons 4–10 and causing losses of both the production and function of NEMO (7, 15, 16). In two-thirds of these patients, the deletion occurs de novo in the mother’s germline (17). Other typical XD-IP–causing lesions include frameshift indels (n = 32), nonsense mutations (n = 16), and essential splice site mutations that are not leaky and cause frameshift or in-frame deletions of 1 or more exons (n = 6), whereas missense mutations (n = 7) and in-frame indels (n = 1) (8, 18) are much rarer. By contrast and consistent with their hypomorphic nature, most mutations underlying XR-EDA-ID are missense mutations or in-frame indels. In total, 57 IKBKG mutations have been reported to date: 27 are missense, 5 are in-frame indels, and 1 is a stop-loss mutation, whereas only 7 nonsense mutations and 9 frameshift indels have been identified. Fifteen of the 16 nonsense mutations and frameshift indels affect the last 3 exons (3 in exon 8, 2 in exon 9, 10 in exon 10) encoding the ZF domain of NEMO, the truncation of which does not abolish NEMO activity (8–10, 19–21). The remaining mutation is a premature stop codon at position 38, which is hypomorphic because of the reinitiation of translation (13, 22). The other EDA-ID–causing lesions include splicing mutations that are in-frame and/or leaky (n = 7) and an exon duplication (n = 1), resulting in the expression of residual levels of dysfunctional mutant NEMO proteins, with or without residual expression of the full-length protein (23–26). Finally, 2 mutations of the exon 1b splice site (c.‑16+1 G>T, c.-16 G>C) encoding the 5′-UTR affects only 1 of the 4 alternative transcripts. The mechanism underlying the hypomorphism of this mutation is unclear (27, 28). Intriguingly, no mutations outside the IKBKG exons and their flanking intron splice sites, including the promoter and introns, have been reported in families with IP or EDA-ID. Another form of EDA-ID is autosomal dominant (AD) and caused by gain-of-function mutations of the NFKBIA gene (29, 30). However, approximately 10% of sporadic and familial cases of both IP and EDA-ID remain genetically unexplained.
The human IKBKG gene is located on chromosome Xq28, close to the IKBKGP pseudogene, which is located 71 kb away, closer to the telomeric region on the opposite strand. IKBKGP has 8 exons identical to the corresponding IKBKG exons, but it lacks exons 1 (a, b, c, d) and 2 (which contains the IKBKG initiation codon) (Figure 1A). IKBKGP is not transcribed, as it lacks promoter and transcription initiation sites. The 8 introns separating the IKBKGP exons also have nucleotide sequences very similar to those of IKBKG (8,879 of 8,884 nucleotides are identical). The detection of IKBKG mutants has therefore always been hindered by the presence of IKBKGP (7, 31–33). Sanger sequencing of the mRNA products of IKBKG typically overcomes this problem, as the related pseudogene is not transcribed. However, this approach is most suitable for the detection of mutations within exons and their flanking regions, which are likely to alter the mRNA structure. Consequently, noncoding IKBKG mutations have not been reported, neither in families with IP nor in families with EDA-ID. Deep intronic causal mutations of other disease-causing genes were detected by a gene-specific approach in patients without mutations of the exons of a specific gene (e.g., β-thalassemia [refs. 34, 35], Duchenne muscular dystrophy [ref. 36], Usher syndrome [ref. 37], chronic granulomatous disease [ref. 38]). Moreover, the genetic etiology of X-linked recessive reticulate pigmentary disorder (XR-PDR), a syndromic primary ID, was recently determined through the identification of a recurrent deep single-nucleotide mutation within 1 of the introns of the POLA1 gene (39, 40). Disease-causing deep intronic mutations typically create a pseudoexon (PE), which is rarely canonical, often resulting in suboptimal splicing (37). Targeted genetic studies have recently suggested that deep intronic disease-causing mutations may be more common than previously thought, because at least 77 genes have been found to contain disease-causing mutations of this type (41, 42). We studied 2 families, a European family in which the mother had very mild IP and lost 2 sons to EDA-ID at the ages of 1 and 9 months, and a Japanese family in which the mother was healthy but lost a son to EDA-ID at the age of 6 months. In all 3 boys, the ID was much more severe than the manifestations of EDA. No mutations were detected in the exons or flanking intronic regions of IKBKG.
Figure 1. Pedigrees.
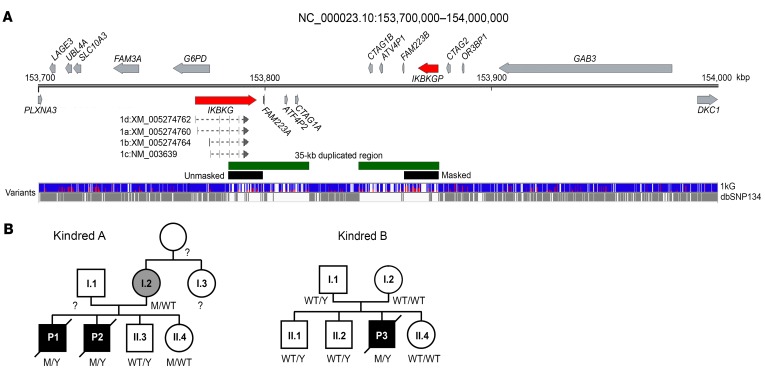
(A) Schematic representation of IKBKG and the surrounding region between positions X: 153,700,000 and X: 154,000,000. IKBKG and IKBKGP are indicated in red, the 35-kb duplication in green, and the region masked for the reanalysis in black. The variants reported in the 1kG and dbSNP 134 databases are shown in blue/red and gray, respectively. (B) Pedigrees of kindreds A and B. Patients with EDA-ID are indicated in black, and the mother with mild IP is indicated in gray.
Results
A family with EDA-ID and IP without mutations in the exons of IKBKG.
We first investigated 2 brothers born to a European mother with very mild IP (case report, Figure 1B, kindred A) at 36 and 37 weeks of gestation. P1 died at the age of 21 days from sepsis and meningitis due to Klebsiella, with no antecedents, and his younger brother P2 died at the age of 9 months from respiratory syncytial virus pneumonitis after a history of recurrent viral, bacterial, and fungal infections in the absence of fever or biological signs of inflammation (case report, Supplemental Table 1; supplemental material available online with this article; https://doi.org/10.1172/JCI124011DS1). P2 had mild signs of EDA, including dry skin and sparse hair, whereas P1 was too young to be evaluated for such features. A younger brother (A.II.3), now aged 15 years, has remained healthy, as has his newborn sister, who is now 6 months of age (A.II.4). Given the mild IP in the mother, signs of EDA in at least 1 boy, and the early death from infection of 2 boys, an IKBKG mutation was suspected. Skewed X-inactivation was observed in the mother’s NK cells, monocytes, granulocytes, and, to a lesser extent, dermal fibroblasts (Supplemental Figure 1). An analysis of X-chromosome microsatellites and copy number variations (CNVs) in the mother, P1, P2, and their healthy brother (A.II.3) confirmed that a haplotype on Xq28 encompassing IKBKG was common to the 2 infants who died, but not to their healthy brother, and was inherited from their mother (Supplemental Figure 2). Sanger sequencing of the IKBKG exons at the genomic level (without discrimination between the IKBKG and IKBKGP exons) (Figure 1A) and of the amplified IKBKG cDNA (including all 4 transcripts encoding the only validated protein isoform) identified no rare or private variants in the patients. Major genomic rearrangements (translocations, large deletions or insertions) were also excluded by Southern blotting (Supplemental Figure 3), high-density arrays for CNV detection, and FISH (data not shown). Finally, whole-exome sequencing (WES) identified no candidate mutations in IKBKG or elsewhere on the X chromosome.
A recurrent deep mutation in an intron of IKBKG.
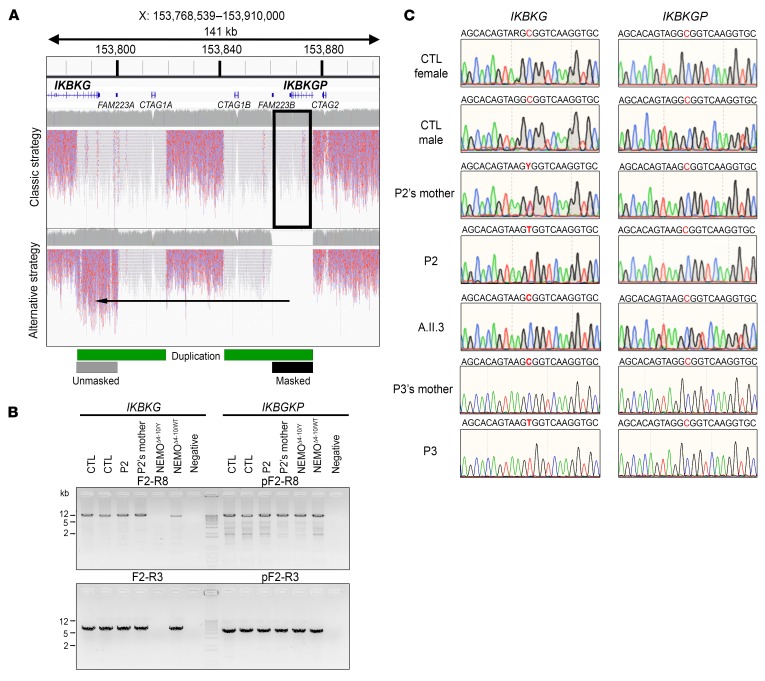
We thus sequenced the whole genome of P2 and his mother. Next-generation sequencing (NGS) software cannot call variants on long stretches of repetitive DNA (43). The region of Xq28 containing the IKBKG locus is characterized by a 35-kb low-copy repeat (LCR) (Figure 1A). Variants at the IKBKG locus are thus partly uncalled. One solution for overcoming the problem posed by this 35-kb duplication (7, 17) is to mask the duplicated region containing IKBKGP (chrX: 153,860,736-153,876,549) artificially, replacing these nucleotides by a stretch of N nucleotides in the reference genome. This results in the raw reads obtained with the modified hg19-b37 reference human genome mapping only to the IKBKG locus, making it easier for Genome Analysis Toolkit (GATK) software to call the variants (Figure 2A; see Methods). The application of this strategy to align whole-genome sequencing (WGS) raw data for IKBKG for P2 and his mother revealed the presence of only 1 private variant at position X: 153,787,731 (IVS4+866 C>T) (Supplemental Figure 4, A and B). We then amplified the IKBKG and IKBKGP loci with specific PCR primers (Figure 2B), which confirmed the hemizygous nature of IVS4+866 C>T in P2, its heterozygous nature in his mother, and its absence from P2’s healthy brother (Figure 2C). In parallel, specific amplification of the IKBKGP locus and sequencing of the position equivalent to IVS4+866 C>T (X: 153,873,196 for IKBKGP) showed the sequence present to be WT in P2, his mother, and all 3 healthy controls tested. No variants at the IVS4+866 C>T position in IKBKG or the equivalent position in IKBKGP were identified in public databases (1000 Genomes [1kG], n = 2,504 individuals, gnomAD, n = 15,496 individuals, Bravo, n = 62,784 individuals). We thus selected the deep intronic variant IVS4+866 C>T as a candidate mutation at the IKBKG locus in this family. We performed Sanger sequencing on intron 4 in 4 sporadic, genetically undiagnosed, unrelated cases involving 3 women with IP and a boy with severe ID and subtle signs of EDA. We found the exact same mutation in the boy (P3). P3 was from Japan and died of recurrent bacterial infections at the age of 6 months (Figure 1B; see clinical report in the Supplemental Data). His mother is healthy and displays no signs of IP. Familial segregation patterns confirmed the presence of the IVS4+866 C>T mutation at the IKBKG locus and its de novo occurrence (Figure 2C). These findings therefore indicate that the IVS4+866 C>T mutation of IKBKG is recurrent as a result of a hotspot rather than a founder effect, providing further support for the hypothesis that it is disease causing.
Figure 2. Genomic strategies to identify IKBKG variants.
(A) Comparison of WGS mapping results between the classical and alternative (masking of the IKBKGP locus) strategies. The red/blue reads are the mapped sequences that can be used for variant calls (mappy quality score [MQ] >20), and the gray/white reads are the mapped sequences for which no variant could be called (MQ = 0). The duplicated region is indicated with a green bar and the masked region with a black bar. (B) Specific amplification by PCR of the full-length (top) or partial (bottom) IKBKG locus. gDNA from 2 controls (CTL), a patient (P2) and his mother, and from 2 NEMOΔ4–10 (from 1 male and 1 female patient) SV40 immortalized fibroblast lines was used as the template. (C) IKBKG and IKBKGP DNA sequence electropherograms for controls, SV40-immortalized fibroblasts from a patient (P2) and his mother and a healthy brother (A.II.3), and for the leukocytes from a patient (P3) and his mother.
Identification of rare and common variants at the IKBKG locus by WGS with 1kG Project data.
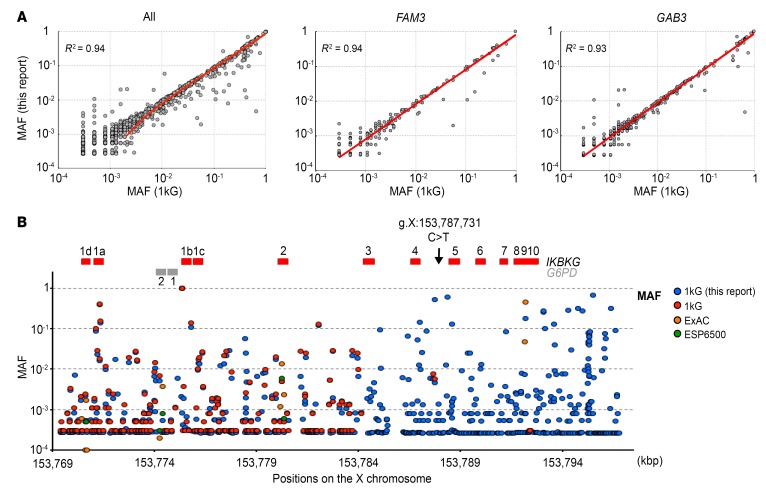
Intriguingly, the specific amplification of IKBKG also revealed the presence of another variant at X: 153,787,729, IVS4+864 A>G, in 2 of the 3 unrelated control genomic DNA (gDNA) samples tested. The lack of reported variants at positions X: 153,787,729 (IVS4+864 A>G) and X: 153,787,731 (IVS4+866 C>T) in the gnomAD and Bravo databases does not necessarily imply that the apparently private mutation IVS4+866 C>T is not really a rare variant or even a polymorphism. Indeed, only 65 variants were reported in gnomAD for the 12.6-kb duplicated region of the genome encompassing IKBKG (from intron 2 to the 3′-UTR), whereas it is generally estimated that the mean frequency of single nucleotide variants is 60/kb in the human genome, based on 1kG Project data (Figure 1A) (44). We therefore assessed allelic diversity at the IKBKG locus. We applied the strategy described above to the 1kG Project data for 2504 healthy individuals, to reanalyze the IKBKG locus (Figure 2A). Using the sex of the individuals, as recorded in the database, we estimated the minor allele frequency (MAF) of all variants. We first validated our set of variants by comparing our calculated MAF with that obtained from the 1kG data. Excluding the duplicated region containing IKBKG and IKBKGP from this analysis, we found a very strong correlation (R2 = 0.94) for all reported variants and for the genes immediately upstream of IKBKG (FAM3; R2 = 0.94) and downstream of IKBKGP (GAB3; R2 = 0.93) (Figure 3A). Focusing on the IKBKG region (X: 153,784,100–153,799,445), we identified a total of 509 variants (Supplemental Table 2). Only 24 of these variants were reported in the 1kG, ESP6500, and gnomAD databases (Figure 3B). Forty-five variants reported in the 1kG, ESP6500, and gnomAD databases were not identified in our analysis. We classified the 509 variants into groups on the basis of zygosity calls. In total, 126 variants were hemizygous in males or homozygous in females, or both, implying that the variants were carried by both IKBKG and IKBKGP (Supplemental Figure 5). Of the identified variants, 51 were located in introns, 67 were downstream of the last exon, or, more rarely, 6 were in the 3′-UTR, and 1 was within a splicing region. We found only 1 missense (E355K) mutation to be hemizygous, albeit with weak coverage (4 reads). However, we found this variant in the heterozygous state in 2 unrelated females, suggesting that it may be a true rare variant. The remaining 383 variants had been called as heterozygous in females or males or both. In such cases, it was not possible to attribute the variant to either IKBKG or IKBKGP by this approach. Of these variants, 189 were located in introns, 140 were downstream of the last exon, or, more rarely, 20 were in the 3′-UTR, and 1 was within the splicing region. Surprisingly, however, 33 of these variants were located in coding exons, and this group included 3 nonsense or indel frameshift variants, 21 missense variants, and 9 synonymous variants (Supplemental Table 3). Given the clinical impact of deleterious mutations of IKBKG, the 3 most deleterious variants (nonsense and frameshift indels) identified were probably located in IKBKGP. We thus report what to our knowledge is the first comprehensive genetic analysis of the IKBKG locus, with a MAF for each variant and population (with a factor 2 approximation) (Supplemental Figure 5 and Supplemental Table 2). In this analysis, we did not find IVS4+866 C>T, whereas IVS4+864 A>G was clearly shown to be a polymorphism, with a MAF of 0.51 in the general population. This approach also confirmed that the 2 families with EDA-ID carried a private IKBKG variant, consistent with this variant being an IP-causing one in the European mother and EDA-ID causing in her sons and in P3.
Figure 3. Reanalysis of the IKBKG locus with 1kG Project data.
(A) Correlation between the MAF of each variant in this study and the MAF reported in the 1kG Project. Plots show all the variants except for the IKBKG locus and the correlation for variants of FAM3 and GAB3, respectively. (B) Magnification of the IKBKG locus, for a comparison of the variants identified in this study (blue circles) with those reported in the various databases (red, orange, and green circles correspond to 1kG, ExAC, and ESP6500 databases, respectively). IKBKG exons are depicted by red bars and G6PD by gray bars.
The IVS4+866 C>T mutation creates a suboptimal donor site for PE inclusion.
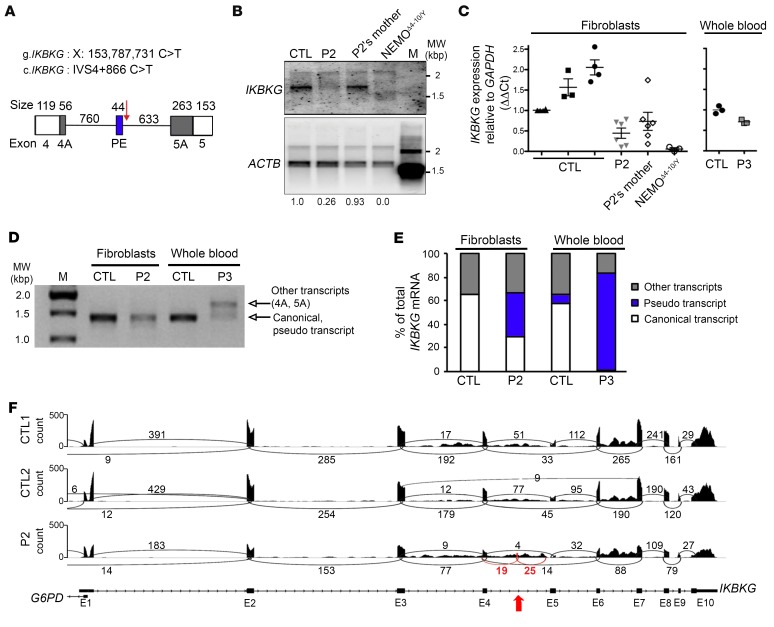
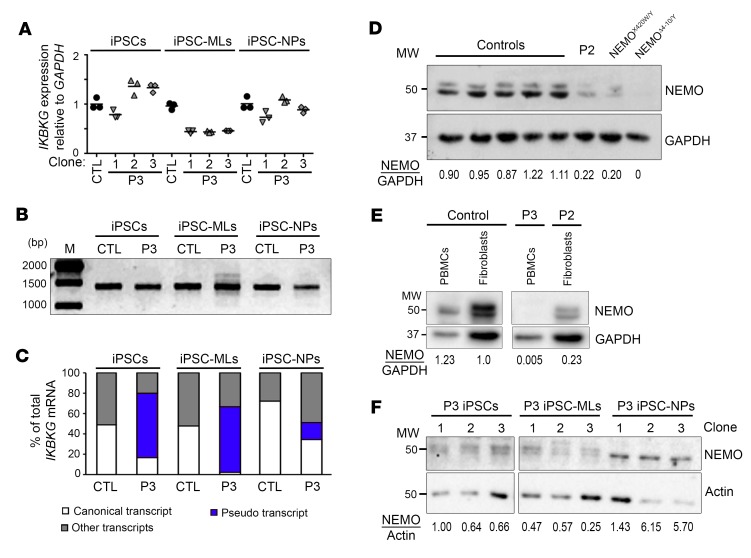
We analyzed the functional consequences of IVS4+866 C>T. In silico analyses with ESE Finder (45) and similar software predicted potential splice donor sites following introduction of the IVS4+866 C>T mutation (Figure 4A and Supplemental Figure 6, A–C). We detected an acceptor site upstream of the mutation (+817) that, in the presence of IVS4+866 C>T, resulted in base pairing between the +861 to +867 stretch of IKBKG intron 4 and the U1snRNA (Supplemental Figure 7), but with no pairing at positions –1, –2, or –3. Exon trapping showed that the IVS4+866 C>T mutation led to the inclusion of a PE within intron 4 of IKBKG. Direct sequencing confirmed the presence of a 44-nt PE by identification of the cryptic acceptor site with a conserved branch point (YUNAY sequence) (46), polypyrimidine tract (PPT) (C/U = 81.8%), and junction site (AG/A) (Supplemental Figures 7 and 8). Conversely, the A/G SNP at the IVS4+864 position had no effect on pairing with the pseudouridine of U1snRNA at the +4 position (Supplemental Figure 7) and thus had no effect on the splicing of IKBKG intron 4 (Supplemental Figure 8). We then analyzed the consequences of IVS4+866 C>T in terms of the mRNA present in the patients’ cells. We first analyzed the amounts of IKBKG transcripts by Northern blotting. Immortalized mRNAs from SV40-transformed fibroblasts from a healthy control, P2, his mother, and a woman with IKBKGΔ4–10 were hybridized with full-length IKBKG cDNA. P2 had 75% less mRNA than did the controls, whereas his mother had only slightly less mRNA than did the controls (Figure 4B). This result was confirmed by reverse transcription quantitative PCR (RT-qPCR), which showed P2 to have approximately 75% as much IKBKG mRNA as the controls (Figure 4C). The whole-blood sample obtained from P3 contained approximately 30% less IKBKG mRNAs than did a sample from a healthy control (Figure 4C). We then analyzed the entire coding sequence (CDS) of IKBKG using cDNA prepared from the fibroblasts of P2 and whole-blood cells from P3 and healthy controls. This analysis confirmed the lower levels of normal transcripts and the presence of aberrant transcripts in the patients’ cells (Figure 4D). We identified and quantified IKBKG mRNA structures by TA cloning and Sanger sequencing of molecular clones of these amplicons from the controls, P2, and P3. In control SV40-immortalized fibroblasts, 65% of the cDNAs obtained were full length and 32% had skipped exon 5 (Supplemental Table 4). In SV40-immortalized fibroblasts from P2, we detected not only the WT transcript but also new aberrant transcripts, accounting for approximately one-third of the total mRNA from this gene. All these new aberrant transcripts contained the 44-nt PE (Figure 4A), either alone or with the 3′ or 5′ end of intron 4 (depicted as exons 4A and 5A in Figure 4A, Figure 4E, Supplemental Figure 7B, Supplemental Figure 9, and Supplemental Table 4). The same approach showed a whole-blood sample from P3 to contain only 1% full-length WT transcript, strongly contrasting with the 58% detected in a sample from a healthy donor (Figure 4E and Supplemental Table 4). RNA-Seq data analysis of the mRNAs extracted from the immortalized fibroblasts of P2 and 2 controls confirmed the lower levels of IKBKG mRNA (approximately 50% lower) in P2, together with the presence of these new transcripts (Figure 4F). The fibroblasts and whole-blood cells from P2 and P3 contained mutant IKBKG transcripts (carrying a 44-nt PE) encoding proteins with a premature stop codon, with moderate and profound decreases, respectively, in WT full-length transcript levels
Figure 4. The IVS4+866 C>T intronic mutation causes aberrant IKBKG splicing.
(A) Description of the impact of the mutation on the IKBKG transcript, focusing on the junction of exons 4 and 5. The PE (blue rectangle) was found in P2 and P3 but not in the controls. White rectangles indicate exons 4 and 5, and black rectangles indicate the alternative donor site (exon 4A) or the alternative acceptor site (exon 5A) found in the patients and controls. The IVS4+866 C>T mutation is indicated by a red arrow. (B) Northern blot analysis of purified mRNA from a control, P2 and his mother, and NEMOΔ4–10/Y SV40-immortalized fibroblasts. The mRNAs were hybridized with a full-length 32P-labeled IKBKG or ACTB cDNA. (C) RT-qPCR analysis of total cDNA from controls, P2 and his mother, and NEMOΔ4–10/Y SV40-immortalized fibroblasts, and of whole-blood cells from a control and P3. Data are expressed as ΔΔCt normalized against GAPDH. The mean ± SD of the data obtained from 8 independent experiments is shown for the fibroblasts. The mean of triplicate experiments is shown for whole-blood cells and is representative of 2 independent experiments. (D) Full-length IKBKG amplification of purified mRNA from control and P2 SV40-immortalized fibroblasts as well as from whole-blood cells from a control and P3. (E) Schematic representation of IKBKG transcripts obtained after TA cloning experiments on controls, P2, and P3 from D. Approximately 100 clones were sequenced, and the results are expressed as percentages. (F) Sashimi plot of the RNA-Seq data from SV40-immortalized control (CTL1 and CTL2) and P2 fibroblasts. The red arrow indicates the position of IVS4+866 C>T and the new exon. Black and red numbers indicates the number of reads overlapping 2 consecutive exons. E1, E2, etc., exon 1, exon 2, etc.; M, molecular weight ladder; MW, molecular weight.
Cell type–dependent effect of IVS4+866 C>T on aberrant IKBKG splicing.
We investigated in greater detail the cell type dependence of WT IKBKG expression in the context of the IVS4+866 C>T mutation. Our previous results were highly consistent with the cell type–dependent pattern of the X-skewing inactivation observed in the affected mother of P1 and P2, as some leukocyte subsets (monocytes, NK cells, and granulocytes) displayed greater skewing than others (B and T cells) and than fibroblasts (Supplemental Figure 1). We hypothesized that the intensity of the alternative splicing generated by IVS4+866 C>T might be dependent on cell type. We tested this hypothesis by using induced pluripotent stem cell (iPSC) lines established from the leukocytes of P3. We measured the levels of WT and alternative IKBKG transcripts in undifferentiated iPSCs, iPSC-derived myeloid cells (iPSC-MLs), and iPSC-derived neuronal precursor cells (iPSC-NPs) (Supplemental Figure 10). RT-qPCR on IKBKG revealed that transcript levels in P3 iPSC-MLs were one-half those in control cells (Figure 5A). The amplification of IKBKG cDNA demonstrated the presence of aberrant products in iPSC-MLs (Figure 5B). TA cloning revealed that the ratio of full-length WT to aberrant IKBKG transcripts differed markedly between undifferentiated iPSCs, iPSC-MLs, and iPSC-NPs (Figure 5C). iPSCs from P3 produced approximately 17% WT transcripts, whereas P3-derived iPSC-NPs produced approximately 35% WT transcripts, a level similar to that obtained with dermal fibroblasts from P2 (Figure 4E). WT transcript levels were as low as 3% in P3-derived iPSC-MLs, a level similar to that reported for whole-blood cells from P3 (Figure 4E). Interestingly, transcripts containing the PE plus exon 5A were abundant in whole-blood cells from P3 and in iPSC-MLs (Figure 4A and Supplemental Table 4), suggesting that the alternative acceptor site for exon 5A was better recognized in whole-blood cells and iPSC-MLs than in the other cell types tested. We also quantified the NEMO protein levels in these cell lines, confirming lower (approximately 25% lower) but detectable levels of WT protein in SV40-immortalized fibroblasts from P2, whereas this protein was almost undetectable in PBMCs from P3 (Figure 5, D and E). Quantification of NEMO protein levels against β-actin showed them to be approximately 40% lower in iPSC-MLs from P3 than in undifferentiated iPSCs and approximately 6.6 times higher in iPSC-NPs (Figure 5F). NEMO protein levels therefore depended on cell type and were correlated with WT mRNA levels (Figure 5C). Collectively, these findings indicate that the IVS4+866 C>T mutation affects NEMO protein levels in a cell type–specific manner, with a more pronounced defect in hematopoietic (leukocytes, iPSC-MLs) than nonhematopoietic (fibroblasts, iPSCs, iPSC-NPs) cells. These findings are consistent with the clinical phenotypes of the 3 boys, the affected mother of P1 and P2, and the documented skewing of X inactivation in the cells of this woman.
Figure 5. The level of aberrant IKBKG splicing caused by IVS4+866 C>T mutation is cell type dependent.
(A) RT-qPCR analysis of total cDNA from iPSCs, iPSC-MLs, and iPSC-NPs obtained from a control and P3. Data were obtained in triplicate and are expressed as ΔΔCt normalized against GAPDH. Representative results of 2 independent experiments are shown. (B) RT-qPCR amplification of full-length IKBKG from RNA extracted from control and P3-derived iPSCs, iPSC-MLs, and iPSC-NPs. (C) Percentage of IKBKG transcripts obtained after TA cloning experiments on P3-derived iPSCs, iPSC-MLs, and iPSC-NPs from B. (D–F) NEMO protein levels in total cell extracts from (D) SV40-immortalized fibroblasts established from controls, P2, a patient with a hypomorphic IKBKG mutation (NEMOX420W/Y), and a patient with complete NEMO deficiency (NEMOΔ4–10/Y), (E) frozen PBMCs from a control and P3, and (F) P3-derived iPSCs, iPSC-MLs, or iPSC-NPs with 3 independent clones. Numbers indicate the expression levels of NEMO relative to GAPDH (D and E) or β-actin (F).
Impaired NF-κB activation in mutant fibroblasts and iPSC-derived macrophages.
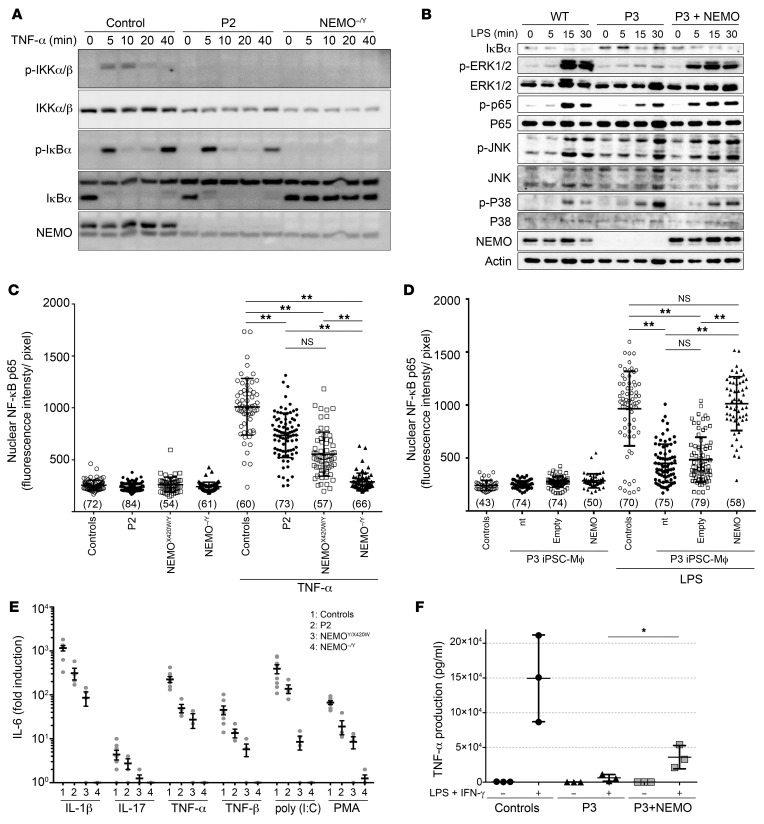
Cells from P2 and P3 had low to undetectable levels of WT NEMO protein and no detectable mutant NEMO protein. We assessed the functional consequences of this profound and purely quantitative defect by analyzing NF-κB activation in SV40-immortalized fibroblasts from P2 upon TNF receptor (TNFR) and IL-1 and TLR stimulation (Figure 6A and Supplemental Figure 11A). We found that IκBα degradation was normal, despite the very low levels of IKKα/β phosphorylation in response to stimulation with either TNF-α or IL-1β (Figure 6A). In P3 iPSC-derived macrophages (iPSC-Mϕ), we observed that NF-κB activation upon stimulation with LPS was impaired (Figure 6B). The JNK and p38 MAPK pathways, which are TAK1 dependent and NEMO independent, were normally activated, whereas the ERK/MAPK pathway, which is NEMO dependent, was strongly affected (Figure 6B). We then measured p65 translocation to the nucleus by analyzing immunofluorescence in both cell types. Fibroblasts from P2 displayed abnormally low levels of p65 nuclear translocation (Figure 6C and Supplemental Figure 11B), and these levels were even lower in iPSC-Mϕ from P3 (Figure 6D and Supplemental Figure 11C). We then used fibroblasts from P2 and performed an EMSA to determine the amounts of nuclear and DNA-binding NF-κB dimers in response to TNF-α or IL-1β. The cells from P2 showed abnormally low levels of DNA-binding dimers in response to high concentrations of either agonist (Supplemental Figure 11D). We then performed ELISA to measure IL-6 and IL-8 production in cells treated with IL-1β, TNF-α, TNF-β, IL-17, poly (I:C), and PMA (Figure 6E, Supplemental Figure 11E). The production of IL-6 in response to all these stimuli was weaker in cells from P2, and this decrease was most marked in response to IL-1β. The impairment we saw in P2’s cells was similar to that observed in X420W-hemizygous cells from a patient with a severe form of EDA-ID (9). Similarly, in P3-derived iPSC-Mϕ, we found that TLR4 stimulation was impaired, as demonstrated by the quantification of TNF-α production (Figure 6F). Complementation of iPSCs from P3 with WT NEMO, but not empty plasmid, followed by differentiation into Mϕ (Supplemental Figure 11F), fully restored NF-κB pathway activation in terms of the phosphorylation of ERK (p-ERK) and p65 (p-p65) (Figure 6B) and p65 nuclear translocation (Figure 6D, and Supplemental Figure 11G), while it partially restored TNF-α production (Figure 6F) upon LPS stimulation. The activation of P3-derived PBMCs by LPS confirmed the inability of leukocytes to produce TNF-α in response to LPS stimulation (Supplemental Figure 11H). Flow cytometric analysis of the leukocytes from P3 revealed that monocytes (CD14+) responded poorly to LPS in terms of TNF-α production (Supplemental Figure 11I). Overall, NF-κB responses to various stimuli were impaired in fibroblasts from P2, which had 25% of the normal level of WT NEMO protein, a phenotype similar to that of another EDA-ID patient with expression of only the X420W-mutated NEMO protein (9). We observed a similar but more pronounced defect in P3-derived iPSC-Mϕ, which had very low levels of WT NEMO expression. Collectively, these findings suggest that the presence of 25% less of the normal levels of WT NEMO severely impairs but does not abolish NF-κB activation in fibroblasts, leukocytes, and iPSC-Mϕ. The mutation found in the 2 kindreds studied here is therefore severely hypomorphic but not loss of function, even in hematopoietic cells, in which the protein was barely, if at all, detectable, accounting for both the severe form of EDA-ID seen early in the lives of the 3 boys who died and the mild IP seen in the European mother.
Figure 6. NEMO insufficiency impairs NF‑κB activation in patients’ fibroblasts and iPSC-derived Mϕ.
(A) Impaired NF-κB activation in response to TNF-α. Time course for TNF-α–stimulated SV40‑immortalized fibroblasts, showing impaired IKKα/β phosphorylation (p-IKKα/β) but no impairment of IκBα degradation in P2 relative to the control. (B) Impaired NF-κB and ERK activation in response to LPS in P3-derived iPSC-Mϕ. Time course for LPS-stimulated iPSC-Mϕ, showing impaired phosphorylation of p65 and ERK1/2, but not of p38 or JNK, which are 2 proteins activated independently of NEMO. NF‑κB and ERK activation was rescued by reexpression of WT NEMO (P3 + NEMO). (C and D) Impaired NF-κB p65 nuclear translocation (C) in P2-derived SV40-immortalized fibroblasts and (D) P3-derived iPSC-Mϕ. NF-κB p65 nuclear translocation was rescued by reexpression of WT NEMO (P3 + NEMO). Data are shown as the mean ± SD of cells from more than 4 random fields taken from 1 well per condition, and the number of cells analyzed in each condition is shown in parentheses. Representative results of 3 independent experiments are shown. **P < 0.001, by 1-way ANOVA followed by Turkey’s test for multiple comparisons. (E) Impaired IL-6 production in response to TNF-α, TNF-β, IL-1β, poly (I:C), IL-17, and PMA in SV40-immortalized fibroblasts from P2 and 2 reported NEMO-deficient patients (NEMOX420W/Y and NEMOΔ4–10/Y). n = 3 or 4. (F) Impaired TNF-α production in P3-derived iPSC-Mϕ. Control and P3-derived iPSC-Mϕ were stimulated with LPS and IFN-γ for 4 hours before TNF-α determination by ELISA. TNF-α production of P3-derived iPSC-Mϕ was restored by reexpression of WT NEMO. Data are shown as the mean ± SD of 3 independent clones and are representative of 2 independent experiments. *P < 0.05, by unpaired, 2-tailed Student’s t test.
CDC-like kinase inhibitor restores the normal splicing of IKBKG.
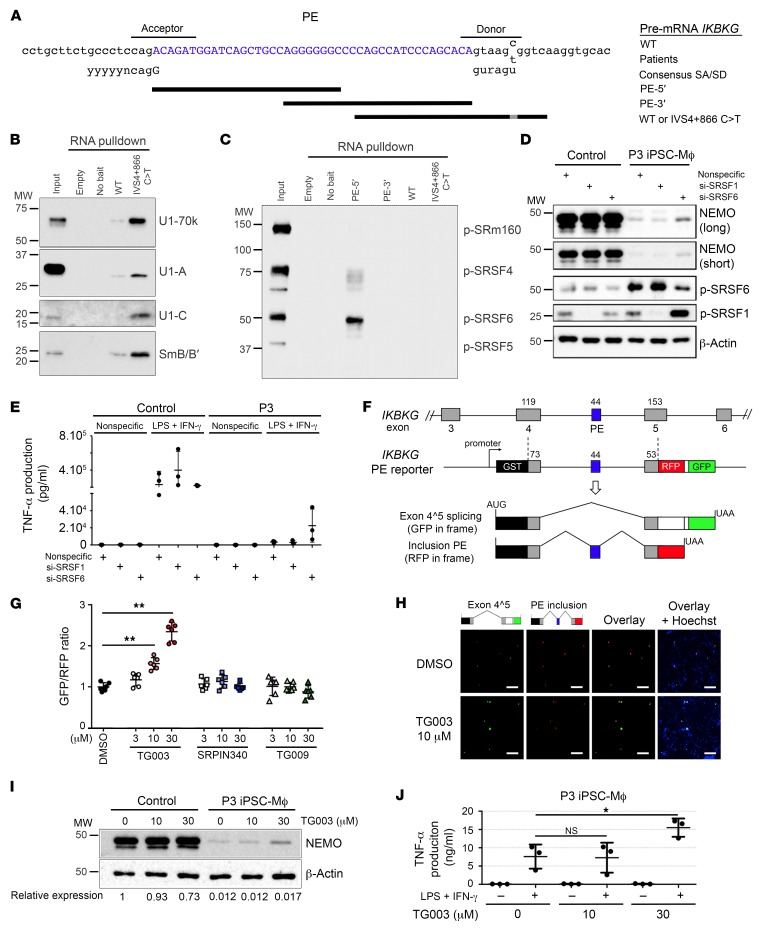
We analyzed the mechanisms underlying the PE inclusion caused by the mutation IVS4+866 C>T. RNA-pulldown assays showed the PE donor to have a greater affinity for U1snRNP, with the detection of U1-70k, U1-A, U1-C, and SmB/B′ subunits following the introduction of IVS4+866 C>T (Figure 7, A and B). However, the absence of base pairing to U1snRNA at positions –1, –2, and –3 renders this donor suboptimal (Supplemental Figure 7A) and suggests that its recognition is assisted by a splicing enhancer. Serine/arginine-rich splicing factor (SRSF) belongs to a major family of trans-acting splicing factors that act as exonic splicing enhancers (ESEs), facilitating spliceosomal complex formation at suboptimal cassette exons (47, 48). RNA-binding assays for the identification of the SRSF-binding site revealed a high affinity of SRSF6 for the PE sequence (Figure 7C), suggesting the presence of an SRSF6-dependent cis regulatory element. We therefore hypothesized that the inhibition of SRSF6 activity might repress PE inclusion and restore the expression of functional IKBKG. We observed a partial restoration of NEMO expression in iPSC-Mϕ from P3 following the siRNA-mediated depletion of SRSF6 (Figure 7D), but we observed no such effect with depletion of SRSF1 protein, which was used as a control because it does not associate with the PE (Figure 7D). Accordingly, the impaired ability of P3-derived iPSC-Mϕ to produce TNF-α was partially rescued by the knockdown of SRSF6, but not SRSF1 (Figure 7E and Supplemental Figure 12). We then assessed the therapeutic potential of small-molecule inhibitors of the CDC-like kinases (CLKs) and SR protein kinases (SRPKs) responsible for the phosphorylation and activation of SRSFs (49). We used the SPREADD (splicing reporter assay for disease genes with dual color) splicing reporter system (50) for quantitative monitoring of the inclusion of the IKBKG PE. The GFP signal indicated exon 4^5 splicing, whereas the red fluorescent protein (RFP) signal identified the PE inclusion product (Figure 7F). By comparing splice-regulating compounds with the SPREADD reporter system, we were able to confirm the dose-dependent suppression of the IKBKG PE by the CLK inhibitor TG003 (51, 52), but not by its inactive analog, TG009 (51) (Figure 7, G and H, and Supplemental Figure 13). Conversely, we found that the SRPK inhibitor SRPIN340 (53) had no effect on IKBKG PE inclusion or skipping (Figure 7G), indicating a dominant role for CLKs in the regulation of the IKBKG PE. Finally, we investigated the consequences of CLK inhibition in iPSC-Mϕ from P3 and showed that TG003 treatment partially restored NEMO expression (Figure 7I) and TNF-α production in response to stimulation with LPS and IFN-γ (Figure 7J). Collectively, these results indicate that SRSF6-dependent ESEs facilitate recognition of the suboptimal PE donor created by the IVS4+866 C>T mutation and that the CLK inhibitor TG003 exerts its therapeutic effects by restoring the normal splicing of IKBKG.
Figure 7. The CLK inhibitor TG003 suppresses IVS4+866 C>T–induced PE inclusion to restore the production of a functional NEMO protein.
(A) Genomic sequence surrounding the 44-bp PE of IKBKG. Black bars indicate the RNA sequences used for the pulldown assays (PE-5′ and PE-3′, WT, and IVS4+866 C>T). (B) Western blot of RNA-pulldown products (WT and IVS4+866 C>T) for U1snRNP subunits (U1-70k, U1-A, U1-C, and SmB/B′). (C) Western blot of RNA-pulldown products (PE-5′, PE-3′, WT, and IVS4+866 C>T) for phosphorylated SR proteins. (D) Western blot of P3-derived iPSC-Mϕ transfected with nonspecific siRNA or SRSF1-specific or SRSF6-specific siRNA. β-Actin served as the internal control. (E) TNF-α production by P3-derived iPSC-Mϕ transfected with nonspecific, SRSF1-specific, or SRSF6-specific siRNA for 72 hours was evaluated 4 hours after stimulation with LPS and IFN-γ (n = 3). (F) Diagram of the SPREADD reporter for IKBKG exons 4–5 with the IVS4+866 C>T mutation. The GFP signal indicates IKBKG exon 4^5 splicing, whereas the RFP signal indicates inclusion of the 44-nt PE. (G) The intensities of green and red fluorescence were quantified in HeLa cells transfected with the IKBKG (IVS4+866 C>T) SPREADD vector and treated with small-molecule compounds (TG003, TG009, and SRPIN340) or not (0.1% DMSO) for 24 hours. Dot plots represent the GFP/RFP ratio of 6 random fields (80–100 fluorescence-positive cells/field) from a single experiment. **P < 0.001. (H) Representative fluorescence micrographs of cells treated with 10 μM TG003 or 0.1% DMSO following transfection with the IKBKG (IVS4+866 C>T) SPREADD vector. Nuclei were counterstained with Hoechst 33342. Scale bars: 200 μm. (I) Western blot of NEMO and β-actin for protein extracts from P3-derived or control iPSC-Mϕ. (J) TNF-α production by P3-derived iPSC-Mϕ stimulated with LPS and IFN-γ for 4 hours, with or without TG003 pretreatment. Data are presented as the mean ± SD of 3 independent experiments using a representative clone. *P < 0.05, by 1-way ANOVA followed by Dunnett’s test.
Discussion
We report what to our knowledge is the first deep intronic variant of IKBKG as a genetic etiology of mild XD-IP in a mother and severe XR-EDA-ID in 3 boys from 2 unrelated families. IKBKG mutations were first reported in families with IP or EDA-ID in 2000, but no genetic etiology has yet been identified for a small but significant proportion of these individuals (10%) (7, 18). IVS4+866 C>T is located deep enough in the intron to be missed by classical approaches, including WES and the Sanger sequencing of exons and flanking intronic regions. We believe this is the first disease-causing mutation of IKBKG to be identified outside the exons and flanking intronic regions. This mutation is also located outside the intronic regions covered by WES. Moreover, the newly created mRNA was not detectable by Sanger sequencing of the cDNA obtained from patients’ fibroblasts, given the nonsense-mediated decay of the mutant transcripts, which were present at much lower levels than were WT transcripts. These findings contrast with those of all previous reports concerning IKBKG mutations or their impact on mRNA structure based on Sanger sequencing after cDNA-PCR (8–10, 13, 19–22, 24–27, 54–59). IVS4+866 C>T underlies a purely quantitative defect of the NEMO protein, given both the suboptimal nature of the new splicing donor site and the nonsense-mediated decay of the mutant transcripts, resulting in a detectable expression of the WT protein and a total absence of the mutant protein. We provide what we believe to be the first description of EDA-ID due to a purely quantitative NEMO defect (IP often being caused by a complete lack of NEMO protein). These findings provide insight into the levels of human NEMO proteins required for correct canonical NF-κB activation. The most closely related known mutation is located in the 5′-UTR encoding exon 1b, the expression of which is selectively impaired (transcript 1b) (27). Interestingly, patients with this 5′-UTR mutation develop ID without EDA (27, 28), as previously reported for a few other mutations of the IKBKG coding sequence (12, 21–23, 60). Little is known about how expression of the 4 main transcripts of IKBKG generated by alternative splicing of the first exon is regulated (61).
The detection and validation of disease-causing mutations located deep within introns is challenging (43). WGS is the best approach for identifying such mutations, particularly given the size and number of introns in the human genome (62). WES is not a good alternative, because it is designed to capture exons and flanking regions and to filter out duplicated regions and tandem repeats (63, 64). RNA-Seq is a possible alternative but is subject to several limitations: (a) mutations creating a new splice site are typically absent from the encoded mRNAs; (b) mutant mRNAs can be difficult to detect because of their instability (42); and (c) WT mRNA levels may be only slightly lower than usual in the tested cells, in which the splicing mutation may be more leaky than in untested cell types underlying the disease phenotype. Careful quantification of WT mRNA levels by RNA-Seq and RT-qPCR may suggest a deep regulatory mutation, provided that the most appropriate cell type is studied, as shown here. Overall, WGS has the advantage of an unbiased capture of the entire genome, provided that the region studied is analyzed thoroughly (63, 65), as illustrated here for IKBKG. Moreover, WGS can be used to compare any candidate variant with a revised list of existing variants (66–68). Indeed, we used the raw WGS data from the 1kG Project (66) to define an initial list of rare and common variants of IKBKG, with estimated MAFs, population by population. This work provides a simple platform for estimating the potential impact of any variant of IKBKG. For example, we found that 3 variants previously reported to be disease causing (E57K, D113N, and Q183H) (16, 69–71) are actually present in the general population and are therefore unlikely to be responsible for IP or EDA-ID. Two of these variants have not been found in the homozygous state in females or in the hemizygous state in males. However, E57K is frequent (MAF = 1.1 × 10–3) and has been found in 67 hemizygous males, strongly suggesting that this variant does not cause EDA-ID, despite having been described as hypomorphic (69, 70, 72).
This and other reports (e.g., for Usher syndrome; ref. 37) highlight the importance of searching for deep intronic mutations by WGS before considering alternative genetic etiologies, particularly for conditions in which most patients are found to have mutations of a single gene, as is the case for IKBKG (42). AD EDA-ID due to IκBα GOF mutations is very rare (29, 30). The discovery of a novel genetic etiology may itself require searches for deep intronic mutations, as illustrated by XR-PDR (39, 40). This approach may also be of interest for other known disease-causing loci, which, like IKBKG, are part of a single, long duplicated region (e.g., neurofibromatosis 1 [NF1] [ref. 73] and Charcot-Marie-Tooth disease [CMT1A] [refs. 74 and 75]). The number of variants in such duplications is typically underestimated. As much as 5.4% of the human genome is duplicated (≥1 kb and ≥90% identity) (75, 76 ). At least 1 gene associated with a primary ID (NCF1, encoding P47-phox) (76) and 29 genes associated with other diseases are completely or partially duplicated in the human genome, resulting in a scarcity of reliable information concerning these loci (76). In the present work, we provide proof of principle that the paucity of information for duplicated regions can be overcome by reanalyzing NGS data, not only to facilitate the detection and selection of disease-causing mutations, but also to identify common and rare variants in specific populations. Pre-mRNAs contain 10 times as many noncoding nucleotides, mostly in introns, than do coding nucleotides, but the potential impact of intronic mutations is probably underestimated. Damage other than interference with the splicing process may also occur. In this respect, one of the difficulties in the identification of deep intronic variants is the paucity of software capable of reliably predicting the impact of such mutations. For example, the IVS4+866 C>T mutation is considered benign by MutationTaster (77) and has a combined annotation-dependent depletion (CADD) score of only 1.02, well below the mutation significance cutoff (MSC) of IKBKG and the CADD score of all other proven disease-causing IKBKG mutations (78–80). Moreover, none of the splicing software used to annotate the new splice site created by IVS4+866 C>T can be used to scan WGS data. Overall, our findings argue for the development of computational and experimental approaches for the detection and validation of deep intronic mutations in patients with unexplained genetic disorders.
This study is also interesting in terms of the cell type–dependent variability in aberrant splicing levels observed, hinting at potential therapeutic approaches. Indeed, residual WT IKBKG mRNA levels ranged from barely detectable in leukocytes, to 3% in iPSC-Mϕ, and up to 35% in iPSC-NPs. We found that a NEMO protein level of approximately 25% was insufficient for correct NF-κB activation in fibroblasts. However, the clinical consequences, in terms of IP and EDA, were mild. The WT NEMO levels in leukocytes and iPSC-Mϕ were lower and more detrimental. This unequal distribution probably explains the severity of the ID in the 3 boys who died from infection in their first year of life. This observation has interesting implications in terms of human genetics. Coding nonsynonymous mutations or mutations that impair canonical splice sites are more likely to have similar consequences across cell types, in terms of protein levels, than mutations that create noncanonical or suboptimal splice sites in the middle of introns, as shown in this work. Deep intronic mutations that create splice sites probably generate a considerable diversity of cellular and clinical phenotypes. In this case, it is possible to draw conclusions, thanks to a unique feature of the IVS4+866 C>T mutation, which is recurrent because of a hotspot, in 2 families of different ancestries (British and Japanese). Both the cellular and clinical phenotypes can be attributed, with confidence, to the mutation itself. It has a robust impact, as it was detected in the context of 2 different genetic backgrounds. The rest of the patients’ genetic makeup is unlikely to be the main driver of this particular phenotype.
We have also determined the mechanisms underlying the creation of the IKBKG PE. The IVS4+866 C>T mutation creates a suboptimal donor site by improving base pairing to U1snRNA, and CLK-regulated SRSF6 provides the ESE activity required for PE donor recognition. However, the differences in aberrant IKBKG splicing between cell types (Figure 4E and Figure 5C) cannot be explained solely by the level of SRSF6 expression, as no clear inverse correlation was found between the expression levels of WT IKBKG and SRSF6 in iPSCs, iPSC-Mϕ, or iPSC-NPs (data not shown). Instead, this observation suggests that variations in the expression levels and/or activity of various CLK isoforms mediate the cell type–dependent pattern of IKBKG splicing. Indeed, we found that the levels of IKBKG transcripts generated varied considerably between cell types in the healthy controls (Figure 4E, Figure 5C, and Supplemental Table 4). Furthermore, the aberrant splicing caused by IVS4+886 C>T was more common in hematopoietic cells, which produce smaller numbers of WT full-length IKBKG transcripts. This variation in IKBKG splicing highlights the importance of using appropriate cell types for mRNA analysis, whether for diagnostic purposes or for the evaluation of therapeutic compounds. This observation also suggests that deep intronic mutations that create suboptimal splice sites probably underlie a considerable diversity of cellular and clinical phenotypes, including some involving known disease-causing loci. For example, the IVS4+866 C>T mutation underlies a very severe ID with very mild developmental phenotypes, whereas other deep intronic mutations in IKBKG might underlie severe developmental phenotypes in the absence of overt ID.
These findings led us to develop a therapeutic strategy that involves the use of a CLK inhibitor to overcome the PE inclusion caused by IVS4+886 C>T. The restoration of NEMO protein production and function, with normal responses to LPS in patients’ iPSC-Mϕ, as shown by assessments of cytokine production, provided a first demonstration of proof of concept for this approach. In addition to the IVS4+866 C>T mutation of IKBKG, recent WGS studies have suggested that many disease-causing deep intronic mutations have gone undiscovered in WES studies (41, 42, 81, 82). As highlighted by the IVS4+866 C>T mutation of IKBKG, those PEs often result from weak or suboptimal splice sites (donor and/or acceptor), as the sites used are not authentic or canonical splice sites in most cases (42). PE recognition also depends on splicing enhancers in cis and their corresponding trans-acting factors, such as SRSF6, for the IVS4+866 C>T–dependent PE. These factors facilitate the splicing reaction by promoting the recruitment of spliceosomal components. In this respect, other genetic disorders attributable to similar PEs would also benefit from the CLK-targeting strategy described here. Indeed, we have confirmed similar therapeutic effects for small-molecule compounds targeting CLK and other factors by preventing the recognition of pathogenic PEs in other genetic disorders (our unpublished observations). Moreover, the target selectivity of CLK inhibitors, each of which affects only a subset of SR protein–dependent exons, with minimal effects on transcriptome profiles (ref. 83 and our unpublished observations), is also a considerable advantage over conventional spliceosomal inhibitors, which target constitutive splicing factors (e.g., spliceostatin A and E7107 for SF3B) (84–86), resulting in a general toxicity due to a global arrest of splicing reactions (87–89). Collectively, our findings pave the way for novel treatments of PE-associated human genetic diseases through the selective manipulation of splicing with rationally designed small-molecule compounds.
Methods
A complete description of the methods is provided in the Supplemental Methods.
Statistics.
A 1-way ANOVA followed by Turkey’s post test was used for comparisons between multiple groups. Dunnett’s test was used for comparisons between a reference and multiple treatment groups. For comparisons of 2 groups, an unpaired, 2-tailed Student’s t test was used. For all analyses, a P value of less than 0.05 was considered significant.
Study approval.
All experiments involving human subjects were conducted in accordance with local, national, and international regulations and were approved by the French ethics committee ANSM (the French National Agency for the Safety of Medicines and Health Products) and the French Ministry of Research (protocol C10-16), by the ethics commitee of Rockefeller University (protocol JCA-0698), and by the ethics committee of Kyoto University Hospital (protocols R0091, G0259, and G0457). Informed consent was obtained from all participants included in this study.
Author contributions
B. Boisson performed experiments, analyzed and interpreted data, designed the study, and wrote the manuscript. TY attended the patient, analyzed and interpreted data, designed and organized the study, and prepared the manuscript. JLC analyzed and interpreted data, designed the study, and wrote the manuscript. RN and TH analyzed and interpreted data, designed the study, and critically reviewed the manuscript. YH and MA performed experiments, analyzed and interpreted data, and prepared the manuscript. ARG, HN, and TS attended the patients and analyzed and interpreted data. MH organized the splicing study, analyzed and interpreted data, and prepared the manuscript. MKS organized the iPSC study, analyzed and interpreted data, and critically reviewed the manuscript. KI attended the patients, analyzed and interpreted data, and critically reviewed the manuscript. SO analyzed and interpreted data and critically reviewed the manuscript. LA and AP critically reviewed the manuscript. JB, TT, and OO performed experiments and analyzed and interpreted data. MB, YK, JI, MO, and MC performed experiments. B. Bigio, HH, YI, and YL analyzed and interpreted WGS and RNA-Seq data.
Supplementary Material
Acknowledgments
We thank the members of the Human Genetics of Infectious Diseases Laboratory their for helpful discussions, especially Stéphanie Boisson-Dupuis and Capucine Picard. We thank Caroline Deswarte (Laboratory of Human Genetics of Infectious Diseases, Necker Branch, INSERM U1163, Necker Hospital for Sick Children, Paris, France ) for her technical help. We also thank Yelena Nemiroskaya, Dominick Papandrea, Mark Woollett (1St. Giles Laboratory of Human Genetics of Infectious Diseases, Rockefeller Branch, The Rockefeller University, New York, New York, USA), Cécile Patissier, Céline Desvallées (Laboratory of Human Genetics of Infectious Diseases, Necker Branch, INSERM U1163, Necker Hospital for Sick Children, Paris, France), Shinobu Yoshida (Department of Pediatrics, Omihachiman Community Medical Center), Hirofumi Shibata (Department of Pediatrics, Kyoto University Graduate School of Medicine), Saiko Shibata (Department of Drug Discovery Medicine, Kyoto University Graduate School of Medicine), and Miki Tsutsui(Department of Anatomy and Developmental Biology, Kyoto University Graduate School of Medicine) for their assistance. This work was supported by the St. Giles Foundation; the Rockefeller University; INSERM, Paris Descartes University; the HHMI; the National Institute of Allergy and Infectious Diseases (NIAID) of the NIH (P01AI061093); the French National Research Agency, under the “Investments for the Future” program (ANR-10-IAHU-01) and the PNEUMOPID project (ANR 14-CE15-0009-01); Grants-in-Aid for Scientific Research of the Japan Society for the Promotion of Science (JSPS) (15H05721); the Platform Program for the Promotion of Genome Medicine (JP18kk0305003h0003); the Basis for Supporting Innovative Drug Discovery and Life Science Research (BINDS) (JP18am0101092) program of the Japan Agency for Medical Research and Development (AMED); a grant to the Core Center for iPS Cell Research of the Research Center Network for Realization of Regenerative Medicine (AMED); the AMED Program for Intractable Diseases Research Utilizing Disease-specific iPS Cells (17935423 and 17935244); the AMED Practical Research Project for Rare/Intractable Diseases (17930095); and a “Research on Measures for Intractable Diseases” grant from the Japanese Ministry of Health, Labor, and Welfare. ARG was partly supported by the National Institute for Health Research (NIHR) Newcastle Biomedical Research Centre based at the Newcastle Hospitals National Health Service (NHS) Foundation Trust and Newcastle University (the views expressed are those of the author and not necessarily those of the NHS, the NIHR, or the Department of Health).
Version 1. 11/13/2018
In-Press Preview
Version 2. 12/18/2018
Electronic publication
Version 3. 02/01/2019
Print issue publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
License: Copyright 2019, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2019;129(2):583–597.https://doi.org/10.1172/JCI124011.
Contributor Information
Bertrand Boisson, Email: bebo283@rockefeller.edu.
Yoshitaka Honda, Email: yohonda@kuhp.kyoto-u.ac.jp.
Masahiko Ajiro, Email: ajiro.masahiko.6e@kyoto-u.ac.jp.
Jacinta Bustamante, Email: jacinta.bustamante@inserm.fr.
Matthieu Bendavid, Email: matthieu.bendavid@aphp.fr.
Andrew R. Gennery, Email: andrew.gennery@newcastle.ac.uk.
Yuri Kawasaki, Email: yuri.kawasaki@cira.kyoto-u.ac.jp.
Jose Ichishima, Email: ichisima.jose@cira.kyoto-u.ac.jp.
Mitsujiro Osawa, Email: mitsujiro.osawa@cira.kyoto-u.ac.jp.
Hiroshi Nihira, Email: nihira@kuhp.kyoto-u.ac.jp.
Takeshi Shiba, Email: shivago@kuhp.kyoto-u.ac.jp.
Takayuki Tanaka, Email: tatanaka@kuhp.kyoto-u.ac.jp.
Maya Chrabieh, Email: maya.chrabieh@inserm.fr.
Benedetta Bigio, Email: bbigio@rockefeller.edu.
Hong Hur, Email: hhur@mail.rockefeller.edu.
Yuval Itan, Email: yitan@rockefeller.edu.
Yupu Liang, Email: liangy@rockefeller.edu.
Satoshi Okada, Email: saok969@gmail.com.
Kazushi Izawa, Email: kizawa@kuhp.kyoto-u.ac.jp.
Ryuta Nishikomori, Email: rnishiko@kuhp.kyoto-u.ac.jp.
Osamu Ohara, Email: ohara@kazusa.or.jp.
Toshio Heike, Email: heike317@kuhp.kyoto-u.ac.jp.
Laurent Abel, Email: laurent.abel@inserm.fr.
Anne Puel, Email: anne.puel@inserm.fr.
Megumu K. Saito, Email: msaito@cira.kyoto-u.ac.jp.
Jean-Laurent Casanova, Email: Jean-Laurent.Casanova@rockefeller.edu.
Masatoshi Hagiwara, Email: hagiwara.masatoshi.8c@kyoto-u.ac.jp.
Takahiro Yasumi, Email: yasumi@kuhp.kyoto-u.ac.jp.
References
- 1.Yamaoka S, et al. Complementation cloning of NEMO, a component of the IkappaB kinase complex essential for NF-kappaB activation. Cell. 1998;93(7):1231–1240. doi: 10.1016/S0092-8674(00)81466-X. [DOI] [PubMed] [Google Scholar]
- 2.Rothwarf DM, Zandi E, Natoli G, Karin M. IKK-gamma is an essential regulatory subunit of the IkappaB kinase complex. Nature. 1998;395(6699):297–300. doi: 10.1038/26261. [DOI] [PubMed] [Google Scholar]
- 3.Ghosh S, Hayden MS. New regulators of NF-kappaB in inflammation. Nat Rev Immunol. 2008;8(11):837–848. doi: 10.1038/nri2423. [DOI] [PubMed] [Google Scholar]
- 4.Oeckinghaus A, Ghosh S. The NF-kappaB family of transcription factors and its regulation. Cold Spring Harb Perspect Biol. 2009;1(4):a000034. doi: 10.1101/cshperspect.a000034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang Q, Lenardo MJ, Baltimore D. 30 Years of NF-κB: a blossoming of relevance to human pathobiology. Cell. 2017;168(1–2):37–57. doi: 10.1016/j.cell.2016.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hayden MS, Ghosh S. NF-κB, the first quarter-century: remarkable progress and outstanding questions. Genes Dev. 2012;26(3):203–234. doi: 10.1101/gad.183434.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smahi A, et al. Genomic rearrangement in NEMO impairs NF-kappaB activation and is a cause of incontinentia pigmenti. The International Incontinentia Pigmenti (IP) Consortium. Nature. 2000;405(6785):466–472. doi: 10.1038/35013114. [DOI] [PubMed] [Google Scholar]
- 8.Fusco F, et al. Alterations of the IKBKG locus and diseases: an update and a report of 13 novel mutations. Hum Mutat. 2008;29(5):595–604. doi: 10.1002/humu.20739. [DOI] [PubMed] [Google Scholar]
- 9.Döffinger R, et al. X-linked anhidrotic ectodermal dysplasia with immunodeficiency is caused by impaired NF-kappaB signaling. Nat Genet. 2001;27(3):277–285. doi: 10.1038/85837. [DOI] [PubMed] [Google Scholar]
- 10.Zonana J, et al. A novel X-linked disorder of immune deficiency and hypohidrotic ectodermal dysplasia is allelic to incontinentia pigmenti and due to mutations in IKK-gamma (NEMO) Am J Hum Genet. 2000;67(6):1555–1562. doi: 10.1086/316914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Visinoni AF, Lisboa-Costa T, Pagnan NA, Chautard-Freire-Maia EA. Ectodermal dysplasias: clinical and molecular review. Am J Med Genet A. 2009;149A(9):1980–2002. doi: 10.1002/ajmg.a.32864. [DOI] [PubMed] [Google Scholar]
- 12.Hanson EP, et al. Hypomorphic nuclear factor-kappaB essential modulator mutation database and reconstitution system identifies phenotypic and immunologic diversity. J Allergy Clin Immunol. 2008;122(6):1169–1177.e16. doi: 10.1016/j.jaci.2008.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Puel A, et al. The NEMO mutation creating the most-upstream premature stop codon is hypomorphic because of a reinitiation of translation. Am J Hum Genet. 2006;78(4):691–701. doi: 10.1086/501532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fusco F, et al. EDA-ID and IP, two faces of the same coin: how the same IKBKG/NEMO mutation affecting the NF-κB pathway can cause immunodeficiency and/or inflammation. Int Rev Immunol. 2015;34(6):445–459. doi: 10.3109/08830185.2015.1055331. [DOI] [PubMed] [Google Scholar]
- 15.Aradhya S, et al. A recurrent deletion in the ubiquitously expressed NEMO (IKK-gamma) gene accounts for the vast majority of incontinentia pigmenti mutations. Hum Mol Genet. 2001;10(19):2171–2179. doi: 10.1093/hmg/10.19.2171. [DOI] [PubMed] [Google Scholar]
- 16.Fusco F, et al. Molecular analysis of the genetic defect in a large cohort of IP patients and identification of novel NEMO mutations interfering with NF-kappaB activation. Hum Mol Genet. 2004;13(16):1763–1773. doi: 10.1093/hmg/ddh192. [DOI] [PubMed] [Google Scholar]
- 17.Fusco F, et al. Microdeletion/duplication at the Xq28 IP locus causes a de novo IKBKG/NEMO/IKKgamma exon4_10 deletion in families with Incontinentia Pigmenti. Hum Mutat. 2009;30(9):1284–1291. doi: 10.1002/humu.21069. [DOI] [PubMed] [Google Scholar]
- 18.Conte MI, et al. Insight into IKBKG/NEMO locus: report of new mutations and complex genomic rearrangements leading to incontinentia pigmenti disease. Hum Mutat. 2014;35(2):165–177. doi: 10.1002/humu.22483. [DOI] [PubMed] [Google Scholar]
- 19.Orange JS, et al. Deficient natural killer cell cytotoxicity in patients with IKK-gamma/NEMO mutations. J Clin Invest. 2002;109(11):1501–1509. doi: 10.1172/JCI14858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nishikomori R, et al. X-linked ectodermal dysplasia and immunodeficiency caused by reversion mosaicism of NEMO reveals a critical role for NEMO in human T-cell development and/or survival. Blood. 2004;103(12):4565–4572. doi: 10.1182/blood-2003-10-3655. [DOI] [PubMed] [Google Scholar]
- 21.Filipe-Santos O, et al. X-linked susceptibility to mycobacteria is caused by mutations in NEMO impairing CD40-dependent IL-12 production. J Exp Med. 2006;203(7):1745–1759. doi: 10.1084/jem.20060085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Niehues T, et al. Nuclear factor kappaB essential modulator-deficient child with immunodeficiency yet without anhidrotic ectodermal dysplasia. J Allergy Clin Immunol. 2004;114(6):1456–1462. doi: 10.1016/j.jaci.2004.08.047. [DOI] [PubMed] [Google Scholar]
- 23.Orange JS, et al. Human nuclear factor kappa B essential modulator mutation can result in immunodeficiency without ectodermal dysplasia. J Allergy Clin Immunol. 2004;114(3):650–656. doi: 10.1016/j.jaci.2004.06.052. [DOI] [PubMed] [Google Scholar]
- 24.Ørstavik KH, et al. Novel splicing mutation in the NEMO (IKK-gamma) gene with severe immunodeficiency and heterogeneity of X-chromosome inactivation. Am J Med Genet A. 2006;140(1):31–39. doi: 10.1002/ajmg.a.31026. [DOI] [PubMed] [Google Scholar]
- 25.Karakawa S, et al. Decreased expression in nuclear factor-sB essential modulator due to a novel splice-site mutation causes X-linked ectodermal dysplasia with immunodeficiency. J Clin Immunol. 2011;31(5):762–772. doi: 10.1007/s10875-011-9560-4. [DOI] [PubMed] [Google Scholar]
- 26.Jørgensen SE, et al. Ectodermal dysplasia with immunodeficiency caused by a branch-point mutation in IKBKG/NEMO. J Allergy Clin Immunol. 2016;138(6):1706–1709.e4. doi: 10.1016/j.jaci.2016.05.030. [DOI] [PubMed] [Google Scholar]
- 27.Mooster JL, et al. Immune deficiency caused by impaired expression of nuclear factor-kappaB essential modifier (NEMO) because of a mutation in the 5′ untranslated region of the NEMO gene. J Allergy Clin Immunol. 2010;126(1):127–32.e7. doi: 10.1016/j.jaci.2010.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hsu AP, et al. IKBKG (NEMO) 5′ untranslated splice mutations lead to severe, chronic disseminated mycobacterial infections. Clin Infect Dis. 2018;67(3):456–459. doi: 10.1093/cid/ciy186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Courtois G, et al. A hypermorphic IkappaBalpha mutation is associated with autosomal dominant anhidrotic ectodermal dysplasia and T cell immunodeficiency. J Clin Invest. 2003;112(7):1108–1115. doi: 10.1172/JCI18714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boisson B, Puel A, Picard C, Casanova JL. Human IκBα gain of function: a severe and syndromic immunodeficiency. J Clin Immunol. 2017;37(5):397–412. doi: 10.1007/s10875-017-0400-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bardaro T, et al. Two cases of misinterpretation of molecular results in incontinentia pigmenti, and a PCR-based method to discriminate NEMO/IKKgamma dene deletion. Hum Mutat. 2003;21(1):8–11. doi: 10.1002/humu.10150. [DOI] [PubMed] [Google Scholar]
- 32.Steffann J, et al. A novel PCR approach for prenatal detection of the common NEMO rearrangement in incontinentia pigmenti. Prenat Diagn. 2004;24(5):384–388. doi: 10.1002/pd.889. [DOI] [PubMed] [Google Scholar]
- 33.Frans G, et al. Conventional and single-molecule targeted sequencing method for specific variant detection in IKBKG while bypassing the IKBKGP1 pseudogene. J Mol Diagn. 2018;20(2):195–202. doi: 10.1016/j.jmoldx.2017.10.005. [DOI] [PubMed] [Google Scholar]
- 34.Dobkin C, Bank A. A nucleotide change in IVS 2 of a beta-thalassemia gene leads to a cryptic splice not at the site of the mutation. Prog Clin Biol Res. 1983;134:127–128. [PubMed] [Google Scholar]
- 35.Treisman R, Orkin SH, Maniatis T. Specific transcription and RNA splicing defects in five cloned beta-thalassaemia genes. Nature. 1983;302(5909):591–596. doi: 10.1038/302591a0. [DOI] [PubMed] [Google Scholar]
- 36.Gonorazky H, et al. RNAseq analysis for the diagnosis of muscular dystrophy. Ann Clin Transl Neurol. 2016;3(1):55–60. doi: 10.1002/acn3.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Khan AO, et al. A deep intronic CLRN1 (USH3A) founder mutation generates an aberrant exon and underlies severe Usher syndrome on the Arabian Peninsula. Sci Rep. 2017;7(1):1411. doi: 10.1038/s41598-017-01577-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bustamante J, et al. BCG-osis and tuberculosis in a child with chronic granulomatous disease. J Allergy Clin Immunol. 2007;120(1):32–38. doi: 10.1016/j.jaci.2007.04.034. [DOI] [PubMed] [Google Scholar]
- 39.Starokadomskyy P, et al. DNA polymerase-α regulates the activation of type I interferons through cytosolic RNA:DNA synthesis. Nat Immunol. 2016;17(5):495–504. doi: 10.1038/ni.3409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Meyts I, Casanova JL. A human inborn error connects the α’s. Nat Immunol. 2016;17(5):472–474. doi: 10.1038/ni.3420. [DOI] [PubMed] [Google Scholar]
- 41.Xiong HY, et al. RNA splicing. The human splicing code reveals new insights into the genetic determinants of disease. Science. 2015;347(6218):1254806. doi: 10.1126/science.1254806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vaz-Drago R, Custódio N, Carmo-Fonseca M. Deep intronic mutations and human disease. Hum Genet. 2017;136(9):1093–1111. doi: 10.1007/s00439-017-1809-4. [DOI] [PubMed] [Google Scholar]
- 43.Treangen TJ, Salzberg SL. Repetitive DNA and next-generation sequencing: computational challenges and solutions. Nat Rev Genet. 2011;13(1):36–46. doi: 10.1038/nrg3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Telenti A, et al. Deep sequencing of 10,000 human genomes. Proc Natl Acad Sci U S A. 2016;113(42):11901–11906. doi: 10.1073/pnas.1613365113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cartegni L, Wang J, Zhu Z, Zhang MQ, Krainer AR. ESEfinder: A web resource to identify exonic splicing enhancers. Nucleic Acids Res. 2003;31(13):3568–3571. doi: 10.1093/nar/gkg616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gao K, Masuda A, Matsuura T, Ohno K. Human branch point consensus sequence is yUnAy. Nucleic Acids Res. 2008;36(7):2257–2267. doi: 10.1093/nar/gkn073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fu XD, Ares M. Context-dependent control of alternative splicing by RNA-binding proteins. Nat Rev Genet. 2014;15(10):689–701. doi: 10.1038/nrg3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cartegni L, Chew SL, Krainer AR. Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nat Rev Genet. 2002;3(4):285–298. doi: 10.1038/nrg775. [DOI] [PubMed] [Google Scholar]
- 49.Ohe K, Hagiwara M. Modulation of alternative splicing with chemical compounds in new therapeutics for human diseases. ACS Chem Biol. 2015;10(4):914–924. doi: 10.1021/cb500697f. [DOI] [PubMed] [Google Scholar]
- 50.Yoshida M, et al. Rectifier of aberrant mRNA splicing recovers tRNA modification in familial dysautonomia. Proc Natl Acad Sci U S A. 2015;112(9):2764–2769. doi: 10.1073/pnas.1415525112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Muraki M, et al. Manipulation of alternative splicing by a newly developed inhibitor of Clks. J Biol Chem. 2004;279(23):24246–24254. doi: 10.1074/jbc.M314298200. [DOI] [PubMed] [Google Scholar]
- 52.Nishida A, et al. Chemical treatment enhances skipping of a mutated exon in the dystrophin gene. Nat Commun. 2011;2:308. doi: 10.1038/ncomms1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fukuhara T, et al. Utilization of host SR protein kinases and RNA-splicing machinery during viral replication. Proc Natl Acad Sci U S A. 2006;103(30):11329–11333. doi: 10.1073/pnas.0604616103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jain A, Ma CA, Liu S, Brown M, Cohen J, Strober W. Specific missense mutations in NEMO result in hyper-IgM syndrome with hypohydrotic ectodermal dysplasia. Nat Immunol. 2001;2(3):223–228. doi: 10.1038/85277. [DOI] [PubMed] [Google Scholar]
- 55.Orange JS, Jain A, Ballas ZK, Schneider LC, Geha RS, Bonilla FA. The presentation and natural history of immunodeficiency caused by nuclear factor kappaB essential modulator mutation. J Allergy Clin Immunol. 2004;113(4):725–733. doi: 10.1016/j.jaci.2004.01.762. [DOI] [PubMed] [Google Scholar]
- 56.Ku CL, et al. NEMO mutations in 2 unrelated boys with severe infections and conical teeth. Pediatrics. 2005;115(5):e615–e619. doi: 10.1542/peds.2004-1754. [DOI] [PubMed] [Google Scholar]
- 57.Ku CL, et al. Selective predisposition to bacterial infections in IRAK-4-deficient children: IRAK-4-dependent TLRs are otherwise redundant in protective immunity. J Exp Med. 2007;204(10):2407–2422. doi: 10.1084/jem.20070628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hubeau M, et al. New mechanism of X-linked anhidrotic ectodermal dysplasia with immunodeficiency: impairment of ubiquitin binding despite normal folding of NEMO protein. Blood. 2011;118(4):926–935. doi: 10.1182/blood-2010-10-315234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mizukami T, et al. Successful treatment with infliximab for inflammatory colitis in a patient with X-linked anhidrotic ectodermal dysplasia with immunodeficiency. J Clin Immunol. 2012;32(1):39–49. doi: 10.1007/s10875-011-9600-0. [DOI] [PubMed] [Google Scholar]
- 60.Haverkamp MH, Arend SM, Lindeboom JA, Hartwig NG, van Dissel JT. Nontuberculous mycobacterial infection in children: a 2-year prospective surveillance study in the Netherlands. Clin Infect Dis. 2004;39(4):450–456. doi: 10.1086/422319. [DOI] [PubMed] [Google Scholar]
- 61.Fusco F, Mercadante V, Miano MG, Ursini MV. Multiple regulatory regions and tissue-specific transcription initiation mediate the expression of NEMO/IKKgamma gene. Gene. 2006;383:99–107. doi: 10.1016/j.gene.2006.07.022. [DOI] [PubMed] [Google Scholar]
- 62.International Human Genome Sequencing Consortium. Finishing the euchromatic sequence of the human genome. Nature. 2004;431(7011):931–945. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- 63.Belkadi A, et al. Whole-genome sequencing is more powerful than whole-exome sequencing for detecting exome variants. Proc Natl Acad Sci U S A. 2015;112(17):5473–5478. doi: 10.1073/pnas.1418631112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Meyts I, et al. Exome and genome sequencing for inborn errors of immunity. J Allergy Clin Immunol. 2016;138(4):957–969. doi: 10.1016/j.jaci.2016.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Goodwin S, McPherson JD, McCombie WR. Coming of age: ten years of next-generation sequencing technologies. Nat Rev Genet. 2016;17(6):333–351. doi: 10.1038/nrg.2016.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sudmant PH, et al. An integrated map of structural variation in 2,504 human genomes. Nature. 2015;526(7571):75–81. doi: 10.1038/nature15394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.1000 Genomes Project Consortium. et al. A global reference for human genetic variation. Nature. 2015;526(7571):68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lek M, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536(7616):285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Keller MD, et al. Hypohidrotic ectodermal dysplasia and immunodeficiency with coincident NEMO and EDA mutations. Front Immunol. 2011;2:61. doi: 10.3389/fimmu.2011.00061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Frans G, et al. Functional evaluation of an IKBKG variant suspected to cause immunodeficiency without ectodermal dysplasia. J Clin Immunol. 2017;37(8):801–810. doi: 10.1007/s10875-017-0448-9. [DOI] [PubMed] [Google Scholar]
- 71.Hsiao PF, Lin SP, Chiang SS, Wu YH, Chen HC, Lin YC. NEMO gene mutations in Chinese patients with incontinentia pigmenti. J Formos Med Assoc. 2010;109(3):192–200. doi: 10.1016/S0929-6646(10)60042-3. [DOI] [PubMed] [Google Scholar]
- 72.Gautheron J, et al. Identification of a new NEMO/TRAF6 interface affected in incontinentia pigmenti pathology. Hum Mol Genet. 2010;19(16):3138–3149. doi: 10.1093/hmg/ddq222. [DOI] [PubMed] [Google Scholar]
- 73.Jenne DE, et al. Molecular characterization and gene content of breakpoint boundaries in patients with neurofibromatosis type 1 with 17q11.2 microdeletions. Am J Hum Genet. 2001;69(3):516–527. doi: 10.1086/323043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pentao L, Wise CA, Chinault AC, Patel PI, Lupski JR. Charcot-Marie-Tooth type 1A duplication appears to arise from recombination at repeat sequences flanking the 1.5 Mb monomer unit. Nat Genet. 1992;2(4):292–300. doi: 10.1038/ng1292-292. [DOI] [PubMed] [Google Scholar]
- 75.Carvalho CM, Zhang F, Lupski JR. Evolution in health and medicine Sackler colloquium: Genomic disorders: a window into human gene and genome evolution. Proc Natl Acad Sci U S A. 2010;107(Suppl 1):1765–1771. doi: 10.1073/pnas.0906222107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dittwald P, et al. Inverted low-copy repeats and genome instability — a genome-wide analysis. Hum Mutat. 2013;34(1):210–220. doi: 10.1002/humu.22217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Schwarz JM, Cooper DN, Schuelke M, Seelow D. MutationTaster2: mutation prediction for the deep-sequencing age. Nat Methods. 2014;11(4):361–362. doi: 10.1038/nmeth.2890. [DOI] [PubMed] [Google Scholar]
- 78.Kircher M, Witten DM, Jain P, O’Roak BJ, Cooper GM, Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46(3):310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Itan Y, et al. The human gene damage index as a gene-level approach to prioritizing exome variants. Proc Natl Acad Sci U S A. 2015;112(44):13615–13620. doi: 10.1073/pnas.1518646112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Itan Y, et al. The mutation significance cutoff: gene-level thresholds for variant predictions. Nat Methods. 2016;13(2):109–110. doi: 10.1038/nmeth.3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Li YI, et al. RNA splicing is a primary link between genetic variation and disease. Science. 2016;352(6285):600–604. doi: 10.1126/science.aad9417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hsiao YH, Bahn JH, Lin X, Chan TM, Wang R, Xiao X. Alternative splicing modulated by genetic variants demonstrates accelerated evolution regulated by highly conserved proteins. Genome Res. 2016;26(4):440–450. doi: 10.1101/gr.193359.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sakuma M, Iida K, Hagiwara M. Deciphering targeting rules of splicing modulator compounds: case of TG003. BMC Mol Biol. 2015;16:16. doi: 10.1186/s12867-015-0044-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Folco EG, Coil KE, Reed R. The anti-tumor drug E7107 reveals an essential role for SF3b in remodeling U2 snRNP to expose the branch point-binding region. Genes Dev. 2011;25(5):440–444. doi: 10.1101/gad.2009411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kotake Y, et al. Splicing factor SF3b as a target of the antitumor natural product pladienolide. Nat Chem Biol. 2007;3(9):570–575. doi: 10.1038/nchembio.2007.16. [DOI] [PubMed] [Google Scholar]
- 86.Kaida D, et al. Spliceostatin A targets SF3b and inhibits both splicing and nuclear retention of pre-mRNA. Nat Chem Biol. 2007;3(9):576–583. doi: 10.1038/nchembio.2007.18. [DOI] [PubMed] [Google Scholar]
- 87.Eskens FA, et al. Phase I pharmacokinetic and pharmacodynamic study of the first-in-class spliceosome inhibitor E7107 in patients with advanced solid tumors. Clin Cancer Res. 2013;19(22):6296–6304. doi: 10.1158/1078-0432.CCR-13-0485. [DOI] [PubMed] [Google Scholar]
- 88.Hong DS, et al. A phase I, open-label, single-arm, dose-escalation study of E7107, a precursor messenger ribonucleic acid (pre-mRNA) splicesome inhibitor administered intravenously on days 1 and 8 every 21 days to patients with solid tumors. Invest New Drugs. 2014;32(3):436–444. doi: 10.1007/s10637-013-0046-5. [DOI] [PubMed] [Google Scholar]
- 89.Lee AJ, et al. Severe mycobacterial diseases in a patient with GOF IκBα mutation without EDA. J Clin Immunol. 2016;36(1):12–15. doi: 10.1007/s10875-015-0223-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.