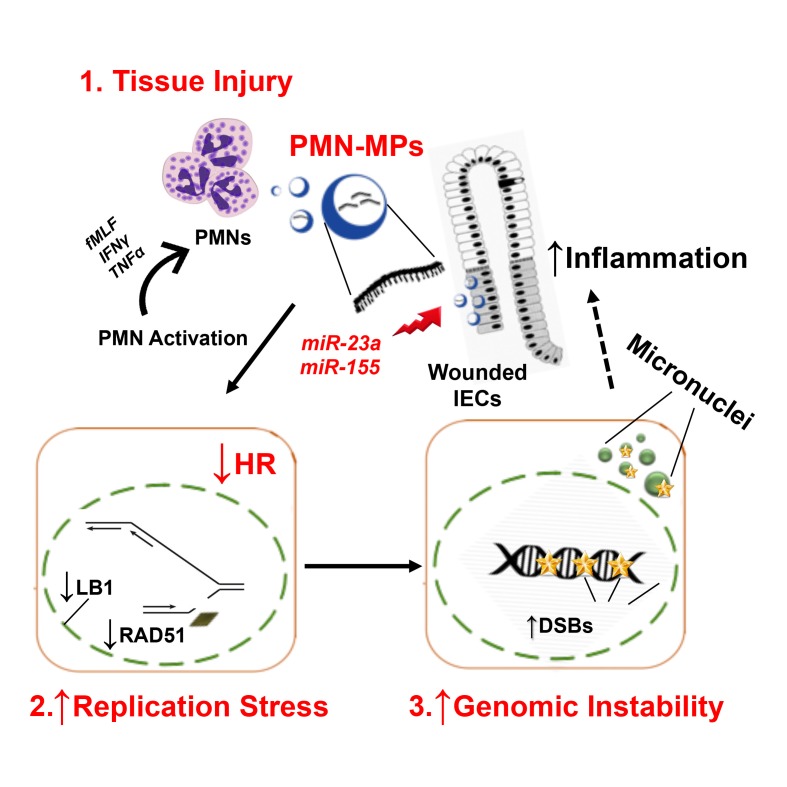
Abstract
Neutrophil (PMN) infiltration of the intestinal mucosa is a hallmark of tissue injury associated with inflammatory bowel diseases (IBDs). The pathological effects of PMNs are largely attributed to the release of soluble mediators and reactive oxygen species (ROS). We identified what we believe is a new, ROS-independent mechanism whereby activated tissue-infiltrating PMNs release microparticles armed with proinflammatory microRNAs (miR-23a and miR-155). Using IBD clinical samples, and in vitro and in vivo injury models, we show that PMN-derived miR-23a and miR-155 promote accumulation of double-strand breaks (DSBs) by inducing lamin B1–dependent replication fork collapse and inhibition of homologous recombination (HR) by targeting HR-regulator RAD51. DSB accumulation in injured epithelium led to impaired colonic healing and genomic instability. Targeted inhibition of miR-23a and miR-155 in cultured intestinal epithelial cells and in acutely injured mucosa decreased the detrimental effects of PMNs and enhanced tissue healing responses, suggesting that this approach can be used in therapies aimed at resolution of inflammation, in wound healing, and potentially to prevent neoplasia.
Keywords: Gastroenterology, Inflammation
Keywords: DNA repair, Inflammatory bowel disease, Neutrophils
Introduction
Epithelial injury resulting from an exacerbated inflammatory response is a common pathological feature of gastrointestinal disorders, including inflammatory bowel diseases (IBDs) (1, 2). Another important feature of IBD is en masse neutrophil (PMN) infiltration of the intestinal epithelium and their accumulation in the intestinal lumen (3). Although PMNs play a vital role in host defense, dysregulated PMN recruitment can lead to tissue injury. Thus, in IBD, the number of PMNs in the intestinal mucosa has been correlated with disease severity (4).
Pathological effects of PMNs are primarily attributed to the release of soluble mediators, including matrix metalloproteinases (MMPs), neutrophil elastase, and myeloperoxidase (MPO) (5). The latter mediates bacterial killing through the generation of reactive oxygen species (ROS) (6). While redox signaling is an important component of cellular renewal, migration, and proliferation (7), excessive levels of ROS can disrupt tissue homeostasis (8). As such, ROS can induce genotoxic stress by mediating DNA alkylation, generating abasic sites (9), and oxidizing DNA to promote formation of 8-oxoguanine (8oxoG), one of the most prevalent DNA lesions (10) removed by base excision repair (BER) (10). Furthermore, ROS attack on the DNA sugar-phosphate backbone, or deficient BER, induces formation of single- and/or double-strand breaks (DSBs) (11).
DSBs are the most genotoxic DNA lesions induced by irradiation, DNA damaging agents, excessive buildup of ROS, or endogenously due to the collapse of replication forks (12). Without timely repair, DSB accumulation can induce apoptosis or senescence or lead to genomic instability, a hallmark of carcinogenesis (13).
In replicating cells, DSBs are predominately repaired by high-fidelity homologous recombination (HR) in S-phase (14), or alternatively by less active, error-prone nonhomologous end joining (NHEJ) (13), promoting genomic instability (15). Furthermore, under the conditions where DSBs persist, continued activation of the DNA damage response (DDR) can exacerbate inflammation by increasing expression and release of proinflammatory cytokines (e.g., IL-6 and IFN-γ) that promote immune cell recruitment and potentiate tissue injury (16).
Efficient epithelial wound healing is critical for the restoration of tissue homeostasis (17). We have recently shown that immune cells, particularly PMNs, contribute to the regulation of key processes involved in epithelial healing responses, including migration and proliferation (18). We further defined what we believe is a new mechanism for this regulation, whereby tissue-infiltrating PMNs release extracellular vesicles or microparticles (PMN-MPs), delivering a variety of biologically functional molecules such as MMPs or peroxidases to actively modulate epithelial barrier function and wound healing (19, 20).
The release of MPs by PMNs and other cells, including platelets, endothelial cells, and epithelial cells, has recently emerged as new means for cell-to-cell communication during homeostatic and pathological conditions (21). PMN-MPs, ranging from 0.05 μm to 1 μm in diameter, include both exosomes (smaller, cytosol-derived vesicles) and larger membrane-derived particles (22). Although MPs contain and transport a variety of biological mediators, they are known to deliver regulatory micro-RNAs (miRNAs) to neighboring cells (23). miRNAs are short (20–25 bp), noncoding mature RNA sequences that function in the posttranscriptional regulation of gene expression by targeting mRNA for degradation (24). A number of miRNAs were shown to regulate DNA damage repair, activation of DDR, and inflammatory responses (25).
Given the important role PMNs play in the pathology of IBDs, in the present work we used human IBD samples, an acute in vivo injury model, and in vitro coculture approaches to define the contribution of tissue-infiltrating PMNs and PMN-derived MPs in DSB accumulation and consequent functional effects on mucosal injury and resolution of inflammation. We found that MPs released by activated PMNs contain proinflammatory miR-23a and miR-155. Through the activity of these 2 miRNAs, PMNs were found to (a) induce replicative arrest and DSBs, due to miR-23a–mediated downregulation of nuclear envelope protein lamin B1 (LB1), and (b) inhibit HR-mediated DSB repair due to downregulation of a key HR regulator, RAD51. We further showed that persistent, PMN-induced accumulation of DSBs in the mucosa leads to impaired wound healing and can lead to genomic instability. Importantly, our data show that the pathologic effects of PMNs were diminished with specific inhibition of miR-23a and miR-155, suggesting that this approach may be incorporated in the future in therapies aimed at the resolution of inflammation, improvement of tissue healing, and prevention of genomic instability.
Results
Tissue-infiltrating PMNs promote accumulation of DSBs.
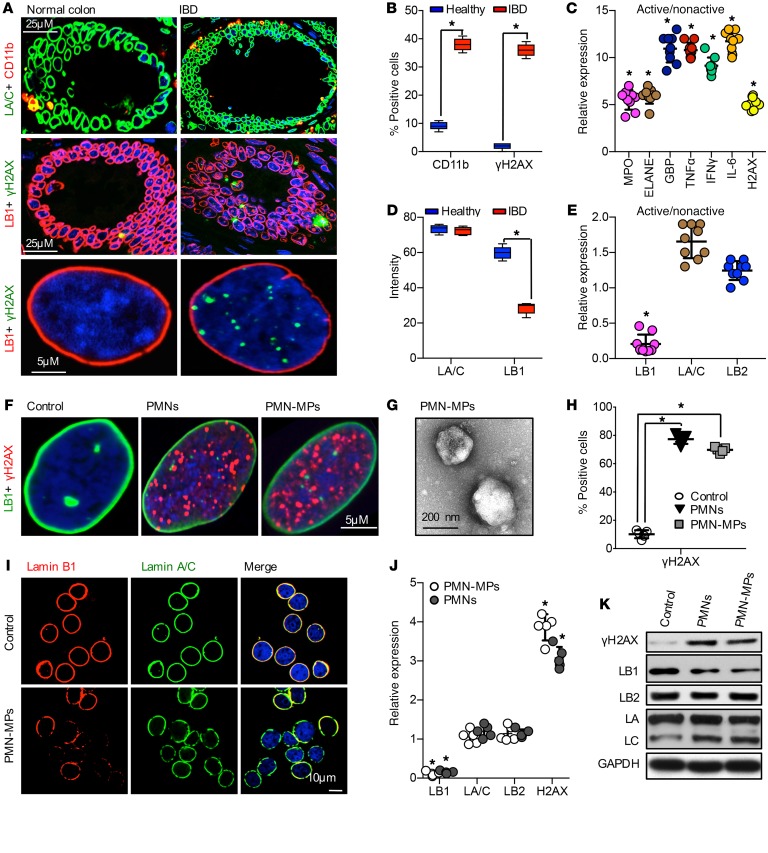
Chronic inflammation is often associated with an increase in DNA damage, and in particular, accumulation of DSBs. Since PMNs can exacerbate inflammation, we asked whether PMN tissue infiltration, which underlies active IBD, promotes DSB accumulation. Immunostaining of clinical samples from patients with active IBD revealed immune cell infiltrates and increased numbers of IECs with γH2AX-positive foci, indicating accumulation of DSBs (Figure 1, A and B; and low magnification images, Supplemental Figure 1A; supplemental material available online with this article; https://doi.org/10.1172/JCI122085DS1). To further confirm the association of PMN-mediated inflammation with DSBs, gene expression analysis of colon biopsies from 9 patients with IBD was performed. High levels of MPO and elastase, indicating PMN infiltration, were accompanied by elevated expression of inflammatory markers, guanylate-binding protein 1 (GBP-1), IFN-γ, TNF-α, and IL-6, and importantly, induction of γH2AX expression (Figure 1C). We further examined the IBD tissue for markers of nuclear integrity, the nuclear envelope proteins: Lamins B1, B2 and Lamin A/C (LB1, LB2, LA/C). A robust downregulation of LB1, but not LA/C or LB2 (Figure 1, A, D, and E), was detected. Since the loss of LB1 has been previously linked to DSB accumulation and genomic instability (26), these data suggest that PMN-mediated inflammation can lead to the accumulation of DSBs, potentially due to LB1 downregulation.
Figure 1. Tissue-infiltrating PMNs promote accumulation of DSBs.
(A) Healthy and IBD colon tissue sections were stained by immunofluorescence for LA/C (green) and CD11b (red) (upper panels), or LB1 (red) and γH2AX (green) (middle and lower panels). Images in lower panels depict representative normal and IBD cell nuclei. (B) Quantitation of immune cell infiltrate (CD11b-positive cells) and nuclear DNA damage (cells positive for γH2AX foci). A total of more than 400 nuclei were examined (n = 3; *P < 0.05). (C) Relative mRNA expression of the indicated genes was determined by qRT-PCR analysis of freshly obtained tissue biopsies from healthy controls and patients with IBD. Data shown as active IBD relative to nonactive controls using GAPDH as a reference gene (n = 9 patients, *P < 0.05). (D) Relative fluorescence intensity in confocal images (as shown in A) was quantified as an index of LA/C and LB1 expression. For each condition, 12 random fields were analyzed (n = 3; *P < 0.05). (E) Relative mRNA expression analysis of human clinical samples. Data are shown as active/nonactive controls using GAPDH as a reference gene (n = 9 patients, *P < 0.05). (F–K) IECs were cocultured for 24 hours with either freshly isolated PMNs or PMN-MPs (ratio of 2 PMNs, or MPs derived from 4 PMNs, to 1 IEC). (F) Representative immunofluorescence images show γH2AX foci (red) and LB1 (green) following treatment for 24 hours with human PMN/PMN-MP. (G) Representative transmission electron microscopy image depicts isolated human PMN-MPs. (H) Quantitation of IECs positive for nuclear γH2AX foci. Total of more than 800 nuclei were analyzed (n = 5; *P < 0.05). (I) IECs were stained for LB1 (red) and LA/C (green) following 24 hours of PMN-MP treatment. (J) mRNA and (K) protein analysis of IECs following 24 hours of coculture with human PMN or PMN-MPs (n = 5, *P < 0.05). One-way ANOVA was used for statistical analyses (P values). Data are mean ± SD from at least 3 independent experiments.
We next used a PMN-IEC coculture setup to determine whether PMNs can directly promote DSBs. Supporting clinical observations, PMN treatment resulted in DSB accumulation in more than 70% of IECs (Figure 1, F and H). Furthermore, during transepithelial migration (TEM), activated PMNs can release microparticles (PMN-MPs) (Figure 1G) containing effector molecules that can potently alter cellular function (19). IEC treatment with PMN-MPs alone resulted in the accumulation of DSBs, similar to the observed PMN effect (Figure 1, F and H). PMN cultures alone (24 hours without IECs) resulted in only a slight elevation in γH2AX, whereas no γH2AX was detected in PMN-MPs (Supplemental Figure 1B), ruling out significant exogenous contribution of PMNs to the observed accumulation of DSBs in IECs. Finally, both PMN and PMN-MP treatment decreased LB1, but not LA/C or LB2, expression at the protein and mRNA levels (Figure 1, I–K), supporting the idea of LB1-dependent DSB accumulation.
PMN-MPs promote ROS-independent generation of DSBs.
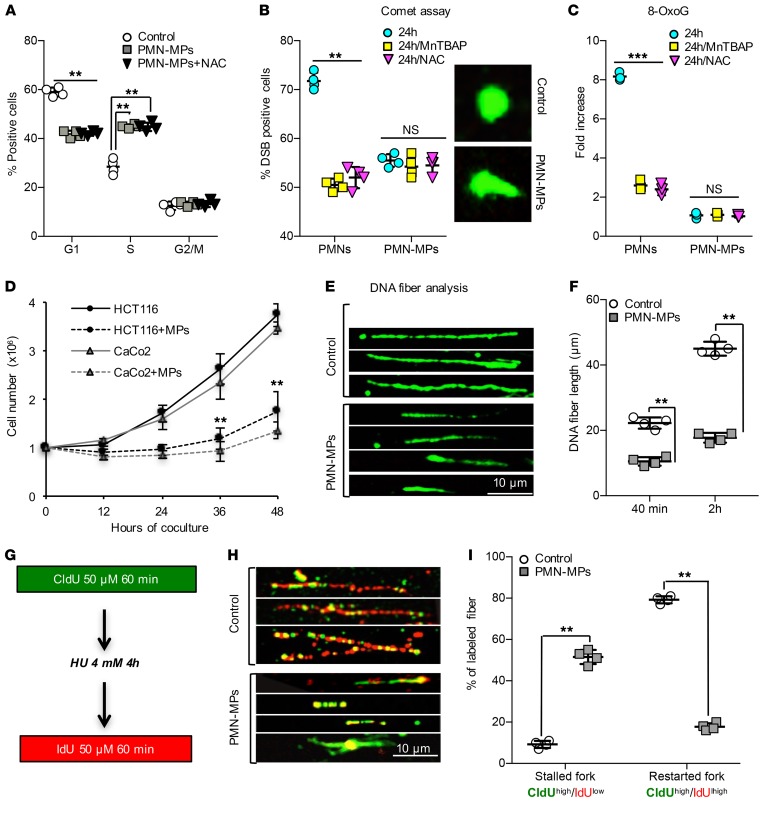
ROS can promote senescence (27) associated with the loss of LB1 (28). To test whether PMN/PMN-MP–mediated loss of LB1 was an outcome of ROS-induced senescence, we examined the effect of PMN and PMN-MP treatment on IEC cycle progression. Both PMNs and PMN-MPs induced S-phase arrest in greater than 45% of IECs (Supplemental Figure 1C and Figure 2A), but did not induce apoptosis (Supplemental Figure 1D). Consistent with ROS generation by PMNs, the addition of the ROS scavenger NAC (N-acetyl cysteine) partly recovered the PMN effect (Supplemental Figure 1C); however, it had no effect on PMN-MP–induced S-phase arrest (Figure 2A). Notably, PMN- and PMN-MP–induced S-phase arrest in IECs was also accompanied by an induction of TP53 and its target gene CDKN1A (p21) (both are known cell-cycle regulators) (Supplemental Figure 1, E and F). However, in contrast to PMNs, PMN-MP treatment (24 hours) did not increase CDKN2A (p16) expression in IECs, a marker of senescence (Supplemental Figure 1, E and F). Consistently, an increased number of β-galactosidase–positive cells (indicating senescent cells) was observed following PMN but not PMN-MP treatment (Supplemental Figure 1G). This suggests that in addition to ROS, PMNs can promote replicative arrest via the release of PMN-MPs, and downregulation of LB1 independently of ROS-induced senescence.
Figure 2. PMN-MPs promote ROS-independent generation of DSBs.
IECs were cocultured with either freshly isolated PMNs or PMN-MPs (ratio of 2 PMNs, or MPs derived from 4 PMNs, to 1 IEC). (A) FACS analysis of IEC cell cycle with or without PMN-MP treatment and the addition of ROS scavenger NAC (2 mM, 24 hours, n = 4, **P < 0.01). (B) Detection of DSBs in PMN- or PMN-MP–treated IECs with or without NAC (2 mM, 24 hours) or MnTBAP (100 μM, 24 hours) by COMET assay. Representative images of normal and DSB-positive nuclei (right panel, n = 3, **P < 0.01). (C) ELISA detecting 8-oxoG in genomic DNA isolated from IECs treated with PMNs, PMN-MPs with or without NAC (2 mM), or MnTBAP (100 μM) (24 hours, n = 4, ***P < 0.001). (D) Proliferation rate of PMN-MP–treated HCT116 and CaCo2 was monitored over indicated times (n = 4, **P < 0.01). (E and F) PMN-MP effect on IEC replication was assessed by single-labeled DNA fiber analysis. Following 24 hours of treatment, BrdU was added to IECs for 40 minutes or 2 hours, and its incorporation was quantified by immunofluorescence labeling from images as shown in E. The length of more than 400 fibers per condition was analyzed for each cell type (n = 4, **P < 0.01). (G–I) For analysis of IEC replication fork stability, DNA fibers were extracted (as described in Supplemental Methods) and double labeled according to schematics depicted in G, where CldU incorporation (H, green) indicates replication before fork stalling and IdU (H, red) indicates recovered replication forks. (H) Representative images of double-labeled fibers of control and PMN-MP–treated cells. (I) The lengths of separated tracks for each cell type were analyzed in over 400 fibers/condition (n = 4, **P < 0.01). Two-tailed Student’s t test was used for statistical analyses (P values). Data are mean ± SD from at least 3 independent experiments.
ROS-independent induction of DSBs by PMN-MPs was further confirmed by the comet assay (Figure 2B). ROS inhibition by the addition of NAC or MnTBAP (manganese (III) tetrakis (4-benzoic acid) porphyri) partially reduced DSBs in PMN-treated IECs, but had no effect on the PMN-MP–induced DSB accumulation (Figure 2B). Importantly, PMN but not PMN-MP treatment induced 8-oxoG accumulation (ROS-induced DNA damage lesions, Figure 2C and Supplemental Figure 1H), which was diminished with ROS inhibition by NAC or MnTBAP (Figure 2C). Similarly, upregulation of BER factors OGG1, MUTYH, and APE1 (activated by ROS) was detected in PMN- but not PMN-MP–treated IECs (Supplemental Figure 1I). Together these data confirm ROS-independent generation of DSBs by PMN-MPs.
PMN-MP–induced DSBs result from collapsed replication forks.
Since PMN-MP treatment induced S-phase arrest without inducing senescence or apoptosis, we asked whether PMN-MPs impair the IEC proliferation rate. PMN-MP treatment decreased the population doubling (PD) time of IECs (~2 fold for HCT116 cells and ~3 fold for CaCo2 cells, Figure 2D). Consistently, decreases in Ki67 (a proliferation factor, Supplemental Figure 1F) and BrdU incorporation (Supplemental Figure 1J) were observed with PMN-MP treatment. Further DNA fiber analysis (DNA combing assay) detected significantly shorter newly synthesized DNA fibers in the presence of PMN-MPs, both with short and long BrdU incubation times (Figure 2, E and F), suggesting that the decreased proliferation rate was due to impaired DNA elongation.
Impaired DNA synthesis may result from decreased stability of replication forks (29). To test this, we examined the ability of DNA replication forks to restart elongation with PMN-MP treatment. Dual labeling of DNA fibers with or without hydroxyurea (HU) to deplete the nucleotide pool was performed (Figure 2G). Upon removal of HU, less than 20% of all analyzed replication forks was able to restart elongation (positive for secondary IdU labeling), resulting in approximately 4.5 times shorter DNA fibers (Figure 2, H and I). This suggests that PMN-MPs induce DSBs by promoting replication fork collapse.
PMN-MPs inhibit HR-mediated DSB repair.
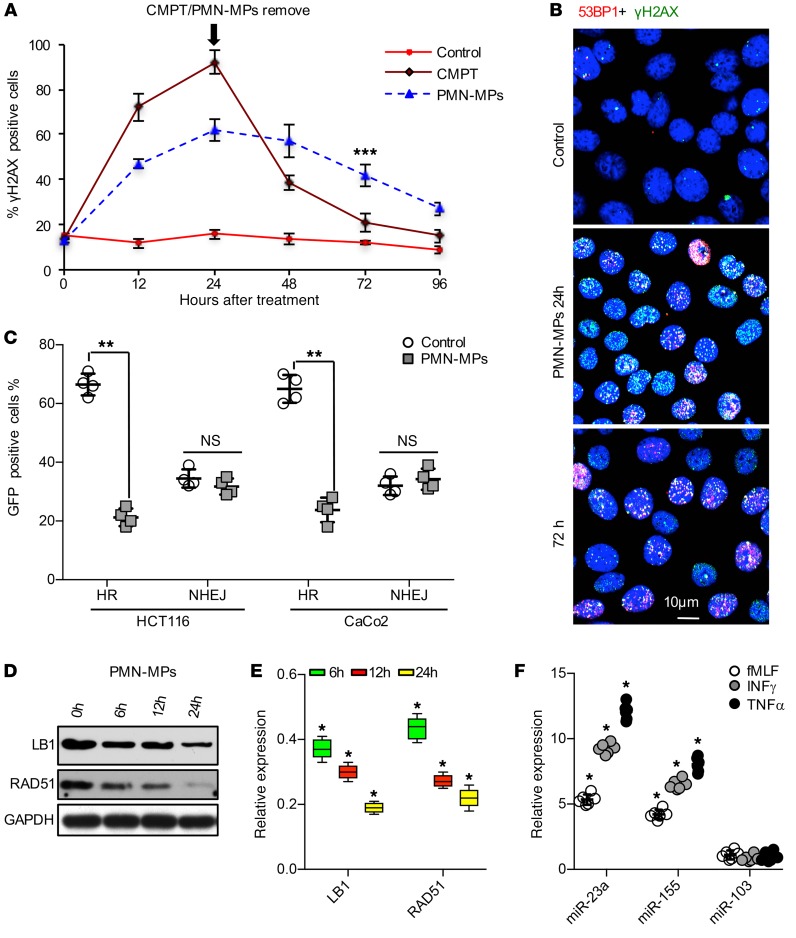
We next asked whether PMN-MP–induced accumulation of DSBs and decreased elongation resulted from compromised DNA damage repair. IECs were treated with either PMN-MPs or a low concentration of camptothecin (CMPT, causes DSBs by mediating replication fork collapse, 2 μM, 24 hours), and the kinetics of DSB repair by evaluating resolution of γH2AX/53BP1 foci was quantified over 72 hours. While CMPT-induced DSBs were efficiently repaired by 72 hours after their removal (remaining <8% of DSB-positive IECs), persistent accumulation of γH2AX/53BP1 foci was observed with PMN-MP treatment (>35%, Figure 3, A and B), suggesting deficient DSB repair. Impaired DSB repair following PMN-MP treatment was further corroborated by comet assay (Supplemental Figure 2A).
Figure 3. PMN-MPs inhibit DSB repair by homologous recombination.
IECs were cocultured with PMN-MPs. (A and B) At the indicated time points IECs were stained for 53BP1 (red) and γH2AX (green), and DSB foci were visualized by confocal microscopy. (A) Kinetics of DSB repair in PMN-MP– or CMPT-treated IECs was quantified from images as shown in (B), for which more than 600 nuclei per time point for each condition were quantified (n = 4, ***P < 0.001). (C) Analysis of IECs for EGFP fluorescence, induced by activation of HR or NHEJ (HCT116 cell line, n = 4; CaCo2 cell line, n = 3; **P < 0.01). (D) Immunoblotting and (E) gene expression analysis of LB1 and RAD51 in PMN-MP treated IECs (n = 5, *P < 0.05). (F) MPs were isolated from PMNs that were stimulated with fMLF (1 μM), TNF-α (10 nM), or IFN-γ (50 nM), and the relative expression of miR-23a, miR-155, and miR-103 was analyzed. Data are shown as fold increase over PMN-MPs that were isolated from unstimulated PMNs. U-6 was used as a reference gene (n = 6, *P < 0.05). One-way ANOVA was used for statistical analyses (P values). Data are mean ± SD from at least 3 independent experiments.
In mammalian cells, DSB repair is mediated by either high-fidelity HR during the S/G2 phase or by error-prone NHEJ, which is active throughout the entire cell cycle (30). Thus, we analyzed HR- versus NHEJ-mediated DSB repair following PMN-MP treatment, using an EGFP reconstitution assay as previously described (31). PMN-MP treatment of IECs (HCT116 and CaCo2, 48 hours) significantly suppressed HR activity without affecting NHEJ (Figure 3C, and representative image Supplemental Figure 2B).
Since DSB repair by either HR or NHEJ requires upstream activation of the DDR (29), we examined the possibility that PMN-MPs impede DDR activation to inhibit HR activity and prevent compensatory NHEJ activation. Immunoblotting analysis revealed activation of key DDR proteins 53BP1, ATM, ATR, and DNA-PKcs in response to PMN-MP–induced accumulation of γH2AX, as well as activation of MRN complex (MRE11, RAD50, NBS1, Supplemental Figure 2C), ruling out this possibility. We further ruled out the possibility that PMN-MPs impede mismatch repair (MMR), which is induced by replication fork stalling (32), to inhibit HR. The expression of MMR genes (MLH1, MSH2, MSH6) and HR genes (FANCD2, BRCA1/2) involved in MMR-HR crosstalk (33, 34) was not altered by PMN or PMN-MP treatment (Supplemental Figure 2D). Together these findings suggest that PMN-MPs specifically suppress HR, leading to a decrease in the DNA elongation rate during replication and accumulation of DSBs.
PMN-MPs mediate downregulation of RAD51 and LB1.
Given the previously suggested role of LB1 in regulating expression of RAD51 (26), a key HR regulator, we examined whether HR inhibition by PMN-MPs was due to its downregulation. As was seen for LB1, RAD51 protein levels were found to be rapidly downregulated (< 12 hours) in IECs treated with PMN-MPs (Figure 3D). We confirmed that PMN-MP–induced downregulation of both proteins was not due to previously suggested mechanisms, including apoptosis-mediated cleavage (35, 36), senescence (28), and autophagy (37) (Supplemental Figure 1, D, F, and G). Importantly, we ruled out the possibility of LB1-mediated downregulation of RAD51 at this early time point, as has been previously suggested (26). Although the transcript levels of both RAD51 and LB1 were robustly reduced as early as 6 hours after PMN-MP treatment (Figure 3E), ChIP analyses detected decreases in LB1 interactions with RAD51 promoter only after 24 hours of PMN-MP treatment (Supplemental Figure 2E). This rapid and simultaneous downregulation of both LB1 and RAD51 may indicate posttranscriptional regulation of transcripts mediated by regulatory miRNAs delivered by PMN-MPs.
PMN-derived miR-23a and miR-155 downregulate LB1 and RAD51.
miRNAs play important roles in regulating cellular processes, including inflammation and DNA repair (38, 39). To test the idea that PMN-MPs can deliver miRNAs to regulate expression of LB1 and RAD51, we performed a targeted screen of activated PMNs and isolated PMN-MPs for the expression of miRNAs with previously established roles in the regulation of DNA repair and inflammation. Analyses of over 20 miRNAs (summarized in Supplemental Table 1) revealed miR-23a and miR-155, which were previously suggested to downregulate LB1 (40) and RAD 51 (41), respectively, to be most abundantly expressed by PMNs (under several activating conditions, including fMLF, IFN-γ, and TNF-α) and by PMN-MPs (Figure 3F). In contrast, miR-103, which also targets RAD51 (42), although upregulated in activated PMNs (Supplemental Table 1) was not found in PMN-MPs (Figure 3F). The clinical relevance of our observations is indicated by the fact that both miR-23a and miR-155 were highly enriched in active IBD biopsies (Supplemental Table 1). De novo miRNA synthesis was indicated by the upregulation of primary miR-23a and miR-155 (pri-miRNA, precursors to mature miRNAs) (24, 43), which was observed in stimulated PMNs but not in PMN-MPs (Supplemental Table 1).
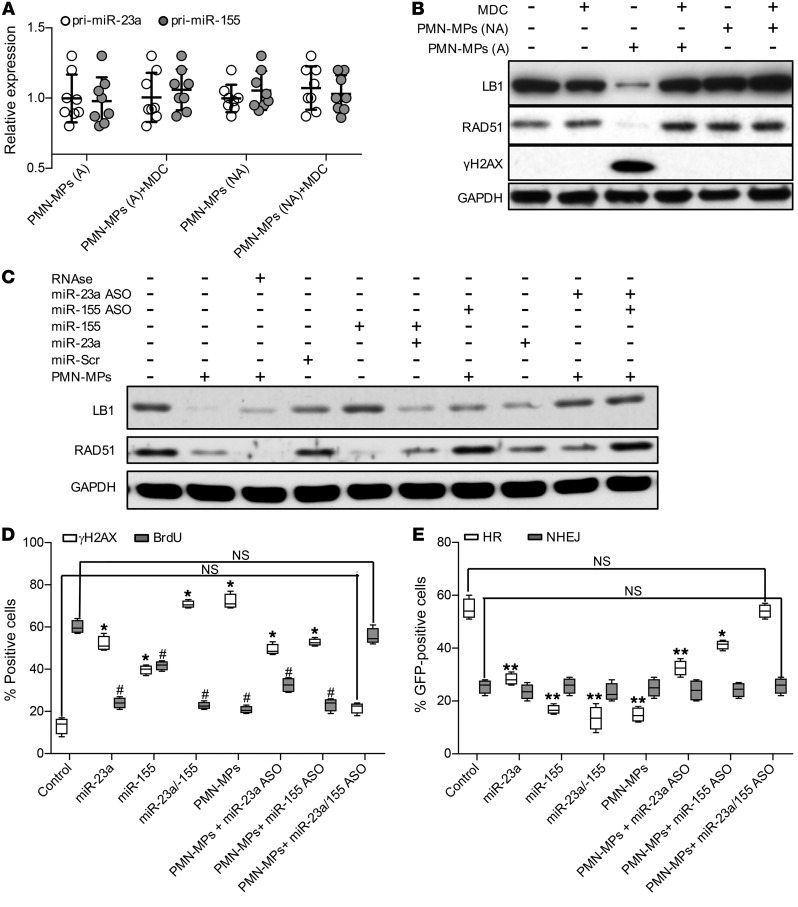
To rule out the possibility that the observed PMN-MP effects on IECs were due to potential DNA transfer, PMN-MPs were treated with DNAse I (with and without NP40-mediated permeabilization) prior to coincubation with IECs. DNAse I treatment did not alter PMN-MP miRNA content, and had no effect on PMN-MP–mediated downregulation of LB1/RAD51 and accumulation of DNA damage (Supplemental Figure 2, F–I). We further confirmed that miR-23a and miR-155 were transferred to IECs by PMN-MP uptake and not due to induction of endogenous miRNA expression in IECs. No upregulation of pri-miR-23a or miR-155 was seen in PMN-MP–treated IECs (Figure 4A). An uptake of control MPs by IECs did not increase miR-23a or miR-155 expression, and had no effect on LB1, RAD51, and DNA damage response (Figure 4B; Supplemental Figure 3A). Similarly, blocking PMN-MP uptake by IECs using monodansylcadaverine (MDC), an inhibitor of endocytosis (44), prevented miRNA upregulation and effector functions (Supplemental Figure 3A; Figure 4B).
Figure 4. PMN-MP–derived miR-23a and miR-155 induce downregulation of RAD51 and LB1.
(A) Expression analysis of pri-miR-23a and miR-155 in IECs treated with MPs derived from either fMLF-activated PMNs (1 μM, activated [A], nonactivated control [NA], or nonactivated PMNs; n = 8). (B) Immunoblotting analysis of IECs treated with active versus nonactive PMN-MPs (24 hours) and with or without IEC pretreatment with the endocytosis inhibitor MDC (50 μM, 45 minutes; n = 3). (C) Immunoblotting of IECs treated with PMN-MPs (24 hours) with or without pretreatment with RNase (10 μg/ml, 45 minutes) and with or without the addition of miR-23a and miR-155 mimics or ASOs (1 nM each; n = 3). (D) PMN-MP–treated IECs (24 hours) were immunostained for γH2AX (a DSB marker) and BrdU incorporation (a replication marker) as described in Methods. At least 700 cells per marker per treatment were analyzed (n = 4; γH2AX, *P < 0.01, different from control; BrdU, #P < 0.01, different from control). (E) Analysis of EGFP reconstitution by HR or NHEJ following the indicated treatment (n = 4, *P < 0.05; **P < 0.01). One-way ANOVA was used for statistical analyses (P values). Data are mean ± SD from at least 3 independent experiments.
After establishing miR-23a and miR-155 delivery by PMN-MPs, we used miR-23a– and miR-155–specific mimics and antisense oligonucleotides (ASOs) to confirm the role of PMN-MP–derived miR-23a and miR-155 in downregulating LB1 and RAD51 expression. Consistent with the suggested respective roles of miR-23a and miR-155 in targeting LB1 and RAD51, administration of each miRNA mimic reduced the protein level of their targets. However, combined treatment, similar to PMN-MPs, resulted in potent downregulation of both proteins. Similarly, combined treatment with both ASOs prevented PMN-MP–mediated downregulation of both LB1 and RAD51 (Figure 4C, Supplemental Figure 2J). Pretreatment of PMN-MPs with physiological levels of RNAse (without detergent-induced PMN-MP permeabilization) did not impair the ability of miR-23a or miR-155 to downregulate LB1 and RAD51 (Figure 4C), suggesting that miRNAs are encapsulated and protected within PMN-MPs. Furthermore, we confirmed that PMN-MP–induced inhibition of HR-mediated DSB repair was due to miR-23a and miR-155 activity. Treatment with miR-23a or miR-155 mimics recapitulated the effect of PMN-MPs, and inhibition of PMN-MP–derived miR-23a or miR-155 with specific ASOs abrogated DSB accumulation (decrease in γH2AX foci; Figure 4D), recovered replication (increased BrdU incorporation; Figure 4D), and recuperated HR efficiency (EGFP reconstitution; Figure 4E). These experiments confirmed that miR-23a and miR-155 delivered by PMN-MPs downregulate LB1 and RAD51 to inhibit HR and promote accumulation of DSBs.
Wound-infiltrating PMNs deliver miR-23a and miR-155 to injured IECs to promote inflammation and downregulate LB1 and RAD51.
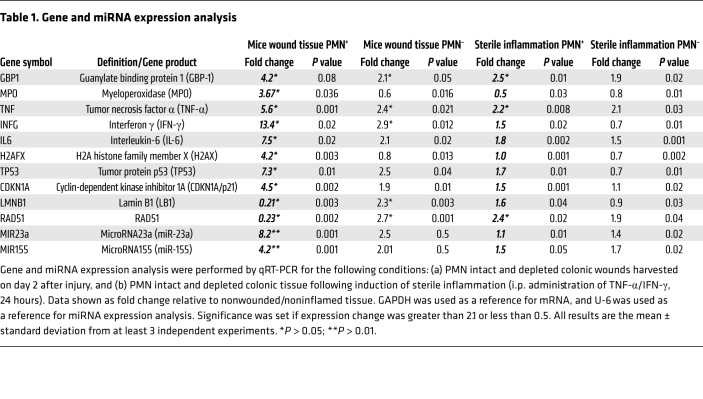
To determine whether wound-infiltrating PMNs promote accumulation of DSBs in vivo, an acute mucosal injury model was used. Superficial wounds (~800 μm in diameter) were inflicted to the dorsal surface of the colon with or without prior Ab-mediated PMN depletion (anti-Ly6G) as described in Methods. Expression analysis by qRT-PCR of extracted PMN-intact wound tissue (day 2 after injury) revealed an induction of inflammation (increased expression of GBP-1 and proinflammatory cytokines TNF-α, IFN-γ, and IL-6), upregulation of H2AX (indicating DNA damage), activation of p53 (increased expression of TP53 and its transcriptional target, CDKN1A, indicating cell-cycle arrest), and robust downregulation of LB1 and RAD51 (Table 1). These changes were significantly attenuated with PMN depletion, suggesting PMN contribution. Importantly, expression of miR-23a and miR-155 was dramatically increased in the presence of infiltrating PMNs and was diminished with PMN depletion (confirmed by MPO expression, Table 1 and flow cytometry, Supplemental Figure 3B). To confirm that PMNs act as a major source of miR-23a and miR-155 in vivo, we induced sterile inflammation (TNF-α/IFN-γ treatment, 24 hours), which does not elicit a robust PMN recruitment into the colonic mucosa. Although increased expression of GBP-1 and IL-6 was observed in extracted colonic IECs, indicating tissue inflammation, miR-23a or miR-155 expression was not induced in the absence of PMN infiltration (no MPO induction, Table 1). Consistent with the role of miR-23a and miR-155 in downregulating LB1 and RAD51, no changes in expression of these genes were seen. Furthermore, confirming physiological relevance, we isolated MPs from colonic wounds (day 2 after injury) and confirmed abundance of miR-23a– and miR-155–positive MPs, which were near-completely diminished with PMN depletion (Supplemental Figure 3, B and C).
Table 1. Gene and miRNA expression analysis.
miR-23a and miR-155 promote accumulation of DSBs in inured colonic epithelium.
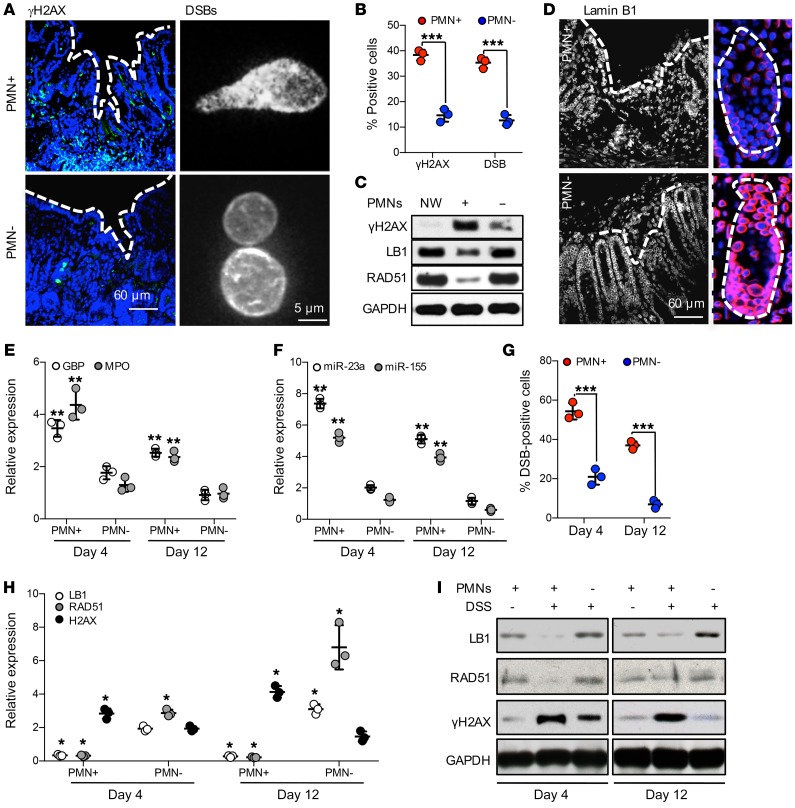
Consistent with the gene expression analyses (Table 1), DSBs (γH2AX-positive foci) were observed in greater than 35% of IECs in PMN-intact colonic wounds (day 4 after injury, representative images, Figure 5A, left panels, and Figure 5B). The presence of DSBs in wound IECs was further confirmed by the comet assay (Figure 5A, right panels, and Figure 5B). PMN depletion significantly reduced DSBs in wound tissue and restored the expression of LB1 and RAD51 proteins in wound IECs (representative immunoblots Figure 5C and confocal images Figure 5D).
Figure 5. Wound-infiltrating PMNs promote inflammation and accumulation of DSBs in injured IECs.
(A–D) Superficial wounds were introduced to the colonic mucosa with or without Ab-mediated PMN depletion, as described in Methods. (A) At day 4 after injury, colonic wounds were extracted and either OCT-fixed, sectioned, and analyzed for DSBs (γH2AX, green, left panel), or the nuclei of wound IECs were analyzed for DSBs by COMET assay (right panel). (B) Quantification of DSBs in tissue and isolated IECs, as shown by representative images in A. More than 400 and 1000 nuclei were analyzed for γH2AX and COMET assay, respectively (n = 3, ***P < 0.001). (C) Colonic wound tissue was analyzed by immunoblotting for γH2AX, LB1, and RAD51 protein expression at day 4 after wounding in PMN intact and depleted mice. (D) OCT-fixed colonic wounds were sectioned and immunostained for LB1 (red). (E–I) Epithelial injury was induced by introduction of DSS 3% (wt/vol) to drinking water. (E) Relative expression analysis for GBP (a marker of inflamed IECs), MPO (a tissue PMN marker), and (F) miRNAs in distal colon tissue 4 and 12 days following initiation of DSS. Data were normalized to nontreated control tissue, using GAPDH and U-6 as reference genes for protein and miRNA expression, respectively (n = 3, **P < 0.01). (G) DSB formation in nuclei of epithelial cells isolated from DSS-injured distal colons (days 4 and 12 following DSS initiation) were analyzed by the COMET assay (n = 3, ***P < 0.001). (H) Gene expression and (I) immunoblotting analyses of LB1, RAD51, and DSB marker H2AX (n = 3, *P < 0.05). Two-tailed Student’s t test and 1-way ANOVA were used for statistical analyses (P values). Data are mean ± SD from at least 3 independent experiments.
The PMN effects in injured epithelium were further confirmed in a dextran sulfate sodium–induced (DSS-induced) injury model simulating human colitis (45). Consistent with the biopsy model, PMN influx in the DSS-colitis model (Figure 5E) was accompanied by significant increases in miR-23a and miR-155 expression (Figure 5F), downregulation of LB1 and RAD51 (Figure 5, H and I), and accumulation of DSBs (as early as day 4 after treatment initiation (Figure 5G). With PMN depletion, no significant increase in miR-23a or miR-155 expression was seen and DSB accumulation was significantly reduced. Intriguingly, during the resolution phase (day 12), levels of LB1 and RAD51 were recovered and DSBs were mostly resolved in PMN-depleted animals. In contrast, in PMN-intact animals, despite decreases in tissue PMNs, DNA damage and DSBs remained elevated (Figure 5, G–I), as did GBP, a marker of IEC inflammatory activation (Figure 5E).
Inhibition of PMN-derived miR-23a and miR-155 prevents accumulation of DSBs and improves mucosal healing.
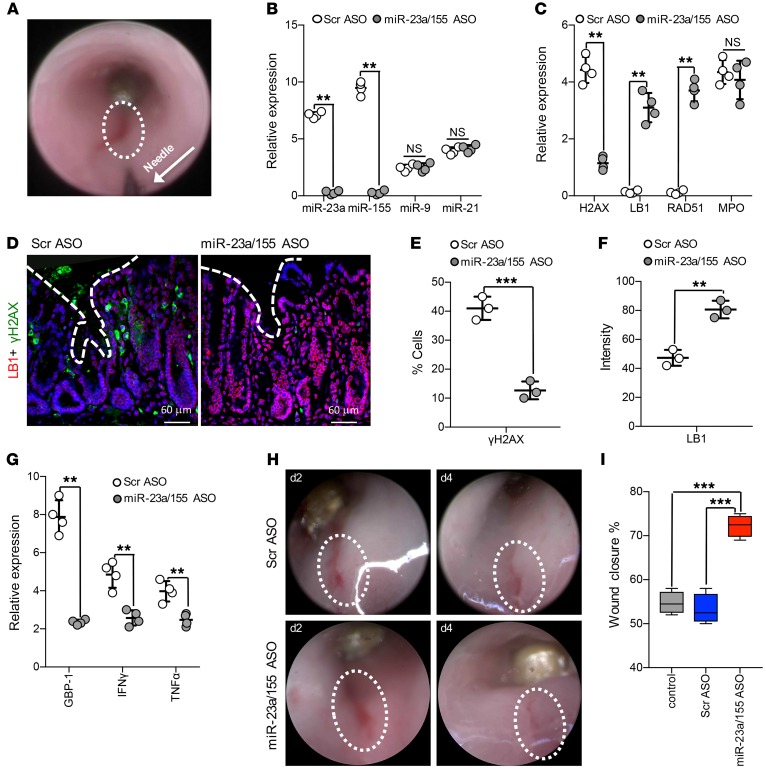
miRNA specificity in mediating the observed responses in injured IECs and the therapeutic potential of miRNA inhibition were confirmed using a biopsy injury model. In this model, synthetically modified miR-23a or miR-155 or scrambled sequence (Scr) ASOs were administered directly into colonic wounds (day 2 after wounding, 1.5 nM in 100 μl/injection) using an endoscopy-guided microinjection system (Figure 6A) (46). ASO treatment of colonic wounds specifically decreased miR-23a and miR-155 expression, but had no effect on several other miRNAs, including miR-9 and miR-21, which are also found on PMN-MPs (Figure 6B and Supplemental Table 1). Importantly, as was seen in cultured IECs, miR-23a and miR-155 ASO treatment of colonic wounds decreased H2AX expression (Figure 6C), the number of γH2AX-positive cells (Figure 6, D and E), and prevented downregulation of RAD51 and LB1 without affecting the expression of other lamins (Figure 6, C, D, and F and Supplemental Figure 3D). Of note, although attenuating the PMN effects, ASO treatment did not affect PMN presence in the tissue (confirmed by MPO expression, Figure 6C), implying specificity for miRNA activity. DSB accumulation in injured tissue can impede healing (47) and promote inflammation by persistent activation of DDR (48). Thus we analyzed colonic wound healing with and without Scr and miR-23a or miR-155 ASO treatment, using high-resolution endoscopic imaging. Treatment with miR-23a or miR-155 ASOs reduced tissue inflammation (decreased expression of GBP1 and inflammatory cytokines IFN-γ and TNF-α, Figure 6G) and significantly improved wound closure (>20% over control ASO, Figure 6, H and I), confirming the therapeutic potential of miR-23a and miR-155 inhibition.
Figure 6. Inhibition of miR-23a and miR-155 promotes DSB repair and improves mucosal wound healing.
Colonic wounds induced by endoscopic biopsies were treated with either Scr (2.5 nM) or with a combination of miR-23a and miR-155 ASOs (1.5 nM each, 24 hours after wounding). ASOs were administered directly into the mucosal wound regions by endoscopy-guided microinjection. (A) Colonic wound and the injection needle tip are outlined by the white circle and the arrow, respectively. (B) Relative expression analysis of the indicated miRNAs and (C) genes of interest (day 4 after injury and 72 hours after ASO administration). Data shown relative to nonwounded and nontreated colonic tissue, using U-6 as reference gene for miRNAs and GAPDH for gene expression (n = 4, **P < 0.01). (D–F) Immunofluorescence analysis of colonic wounds for γH2AX (green) and LB1 (red). Representative images (D) and quantification (E, F) day 4 after injury and 72 hours after ASO administration. For both γH2AX and LB1, more than 400 nuclei were analyzed (n = 3; ***P < 0.001; **P < 0.01). (G) Relative gene expression analysis of colonic wounds (day 4 after injury and 72 hours after ASO administration) for markers of inflammation. Data are shown as fold change relative to nonwounded and nontreated tissue. GAPDH was used as a reference gene (n = 4, **P < 0.01). (H, I) The closure of colonic wounds (day 4 after injury relative to day 1) with or without ASO treatment was quantified from high-resolution images acquired by endoscopy using ImageJ (NIH). (H) Representative images that were used for wound measurements and (I) quantification of wound closure (n = 4; ***P < 0.001). Two-tailed Student’s t test and 1-way ANOVA were used for statistical analyses (P values). Data are mean ± SD from at least 3 independent experiments.
PMN-derived miR-23a and miR-155 promote genomic instability.
Deficient repair and the persistent presence of DSBs in tissue can lead to genomic instability (13). Given the PMN-induced inflammation and recurring tissue injury, as seen in IBD, and our current finding that tissue-infiltrating PMNs promote DSB accumulation, we tested the idea that in the long-term, PMN-derived miRNAs can promote genomic instability.
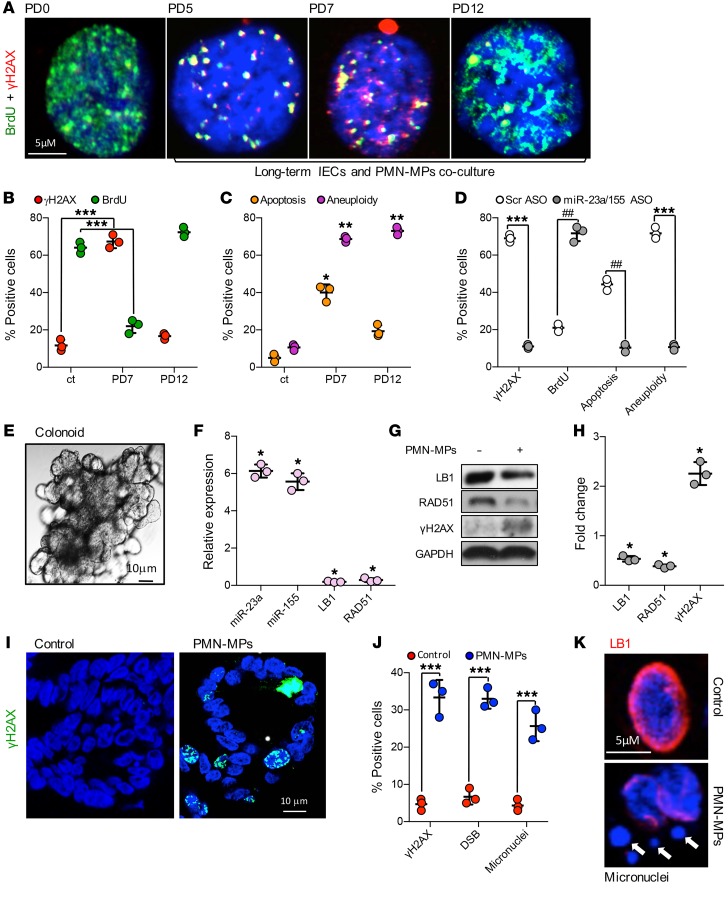
To simulate PMN-induced recurring or chronic inflammation, HCT116 IECs were cocultured with PMN-MPs for 15 population doublings. In these experiments, PMN-MPs were replaced every 48 hours and replication (by BrdU incorporation), DSBs (γH2AX-positive foci), apoptosis, and aneuploidy (as genomic instability marker) in IECs were analyzed every 2 PDs. At PD7, more than 70% of all IECs were positive for DSB foci and the lowest BrdU incorporation rate was observed (Figure 7, A and B). Moreover, as seen in representative images (Figure 7A), frequent colocalization of γH2AX and BrdU was detected, indicating an association of replication fork and DSBs. Importantly, while a persistent deficiency in DSB repair induced by PMN-MPs resulted in increased cell apoptosis (~40%, Figure 7C), an induction of aneuploidy (indicating genomic instability) (49) was observed in more than 70% of surviving IECs (Figure 7C). This confirmed that recurring tissue injury and persistent inhibition of DSB repair by PMNs can lead to genomic instability. Consistent with the ability of miR-23a and miR-155 ASOs to reduce inflammation and prevent accumulation of DSBs, administration of ASOs to long-term PMN-MP and IEC cultures significantly reduced both cell death and aneuploidy (Figure 7D).
Figure 7. Induction of genomic instability by PMN-MP–derived miR-23a and miR-155.
(A–D) Long-term coculture of IECs and PMN-MPs was performed as described in Methods. (A and B) Immunofluorescence detection of BrdU (replication marker; green) and γH2AX (DSB marker; red). Approximately 600 cells were analyzed for each PD per marker, representative images are shown in (A) and quantification is shown in (B) (n = 3; ***P < 0.001). (C) Induction of apoptosis and aneuploidy during long-term IEC and PMN-MP cocultures was determined. At the defined PD time, cells were harvested, stained for Annexin V and propidium iodide (PI), and analyzed by FACS (n = 3, *P < 0.05; **P < 0.01). (D) The effect of miR-23a and miR-155 inhibition by ASOs (1 nM each, replaced every 48 hours) during long-term IEC and PMN-MP coculture on the indicated processes. Approximately 500 cells per PD per marker were analyzed (n = 3, ***P < 0.001, ##P < 0.01). (E–K) Patient-derived colonoids grown in 3D cultures were treated with PMN-MPs (24 hours, derived from fMLF-stimulated PMNs). (E) Representative phase contrast image of a mature colonoid (~10 days in culture) used in experiments. (F) Relative expression analysis of miRNAs and genes of interest following PMN-MP treatment. Data shown relative to nontreated, control colonoids, using U-6 and GAPDH as reference genes, respectively (n = 3, *P < 0.05). (G) Representative immunoblots and (H) quantification of lysed colonoids following PMN-MP treatment. (I–K) 3D colonoids were serially sectioned and processed for immunofluorescence analyses. (I) Representative images of γH2AX (a DSB marker, green) induction, quantified in panel J (left bars). For DSB analysis (J, middle bars), COMET assay was performed on dissociated colonoid epithelial cell nuclei. (K) LB1 downregulation (red) and formation of micronuclei was determined by staining for LB1 (red) and DNA (blue). Micronuclei are indicated by white arrows and quantified in panel J (right bars). For quantification, at least 20 colonoids were analyzed per condition (n = 3, ***P < 0.001). Two-tailed Student’s t test and 1-way ANOVA were used for statistical analyses (P values). Data are mean ± SD from at least 3 independent experiments.
To examine whether a single, inflammatory episode was sufficient to promote genomic instability and confirm our findings in primary human cells, patient-derived colonoids were used (Figure 7E). Patient biopsy-derived crypts were cultured in 3D matrigel until multi-lobed colonoids were formed, and were treated with PMN-MPs (24 hours) as was done with cultured IECs. Acute PMN-MP treatment led to upregulation of mature miR-23a and miR-155, but not pri-miR-23a or miR-155 (Figure 7F, Supplemental Figure 4A), the downregulation of their targets LB1 and Rad51 at mRNA and protein levels, and an induction of DSB marker γH2AX (Figure 7, F–H). Accumulation of DSBs was confirmed by quantification of an increased number of γH2AX-positive nuclear foci and by comet assay (Figure 7, I and J). Importantly, in this experimental model, an increased number of micronuclei (Figure 7J, Supplemental Figure 4B) that were frequently positive for γH2AX (Supplemental Figure 4C) was detected, indicating deficient DSB repair and possible induction of genomic instability (50). Intriguingly, in parallel to PMN-MP–induced DSBs, upregulation of senescence marker CDKN2A/p16 and significant decrease in replication factors Ki67 and PCNA were observed. This suggests that PMN-MPs may also acutely induce replicative senescence due to genomic instability (Supplemental Figure 4D).
Discussion
Timely and efficient resolution of mucosal injury is crucial for reestablishing tissue homeostasis. Thus, identifying new targets that potentiate injury or delay healing are priorities for potential therapeutic development for patients with IBD (51) and other inflammatory disorders of the gastrointestinal tract. Furthermore, given the predisposition of IBD patients suffering from exacerbated inflammation and recurring mucosal injury to developing colitis-associated colorectal cancer, specific inhibition of such targets can help prevent carcinogenesis (52).
In the present work, we defined a new mechanism that contributes to IBD pathogenesis and that can potentially promote carcinogenesis. We found that activated PMNs infiltrating the intestinal mucosa cause accumulation of DSBs by both promoting formation of DSBs and by inhibiting HR during IEC replication, thus impeding tissue recovery. Importantly, although thus far PMN-induced tissue injury, and particularly DNA damage, was mainly considered to be due to excessive generation of ROS, our studies identified an alternative ROS-independent mechanism.
We report for the first time that activated PMNs that infiltrate the intestinal mucosa downregulate LB1 expression in IECs. LB1 is a nuclear envelope protein that serves to preserve the nuclear integrity (53), genome organization, and function, including replication fork stability (54). It has been previously suggested that LB1 directly regulates RAD51 transcription, such that the loss of LB1 in epithelial cells led to deficient DNA repair and genomic instability (26, 55). However, we found that in acute mucosal injury, while causing LB1-dependent DSBs, PMNs also directly downregulate RAD51 to inhibit DSB repair. Although the loss of LB1 has been previously observed in association with cellular senescence (28) and autophagy (37), the precise mechanism of LB1 downregulation is not clear.
We found that PMNs downregulate LB1 as well as RAD51 via the release of miRNAs that are effectively transported by MPs. We confirmed that miRNAs were delivered to IECs through PMN-MP uptake and not due to endogenous induction by IECs. We further determined that treatment with PMN-MPs alone was sufficient to impair DNA synthesis, inhibit HR, and promote DSB accumulation. PMN-MP–induced DSBs resulted from replication fork stalling and collapse due to LB1 loss, and thus were predominately localized to proliferating IECs. Importantly, while PMN effects were partially reversed with the inhibition of ROS (consistent with the ability of PMNs to both generate ROS and release MPs), the effects of PMN-MPs were ROS independent.
We have screened PMNs and PMN-MPs for more than 20 miRNAs that have been previously implicated in regulating DNA damage repair and LB1/RAD51 expression. We found that of all the analyzed miRNAs, miR-23a and miR-155 were among the most highly expressed by activated PMNs and most efficiently packaged into PMN-MPs. Since we observed variable miRNA expression in activated PMNs and PMN-MPs, it is possible that the expression differences will impact the miRNA effector function. Intriguingly, we found that not all miRNAs that were upregulated by activated PMNs were packaged into PMN-MPs. For example, miR-103, which also targets RAD51, although increased in activated PMNs, was not found in PMN-MPs. This implies that not only expression levels but also miRNA packaging into MPs may be specifically regulated by an as-yet-unknown mechanism.
Given the emerging roles of miRNAs in regulating cellular responses, and the fact that each miRNA has numerous targets, identifying such mechanisms would be extremely valuable in designing specific neutralizing strategies in the future. Indeed, target-specificity of various miRNAs may depend on environmental and cellular cues that are yet to be defined. As such, miR-155 in addition to RAD51 can target MMR genes such as MLH1 and MSH6 (56), which when downregulated can contribute to DSB accumulation. Interestingly, in the model of acute colonic injury or in clinical IBD samples, the transcript levels of these genes were not significantly altered despite abundant presence of miR-155 and a robust downregulation of RAD51.
Given the complexity of immune cell function in diseases such as IBD, it is important to note that other resident and infiltrating immune cells, including eosinophils, monocytes, and macrophages, can release MPs to impact the responses of neighboring cells (57); however, whether miRNA upregulation and packaging into MPs is cell-type specific remains to be determined. Furthermore, MP content can also be stimulus dependent (46, 58). Thus we confirmed enrichment of miR-23a and miR-155 in PMNs using several inflammatory conditions, including stimulation with IFN-γ, TNF-α, or bacterial fMLF, all of which are commonly found in inflamed mucosa. Increased levels of miR-23a and miR-155 were similarly observed in active IBD alongside infiltrating PMNs, confirming the physiological relevance of our observations.
Accumulation of DSBs can further lead to persistent activation of DDR, which can in turn trigger increased secretion of proinflammatory cytokines and chemokines (59). The resulting inflammatory milieu can promote continued leukocyte recruitment and activation, perpetuating chronic inflammation. Although both chronic inflammation and genomic instability are tightly linked to carcinogenesis (60, 61), how inflammation is linked to genomic instability is not well defined. Using a long-term coculture system modeling chronic inflammation that is associated with en masse PMN infiltration, we introduce what we believe is a new mechanism whereby PMNs can promote genomic instability (as indicated by increased aneuploidy) via miRNA-dependent DSB accumulation. Intriguingly, despite the robust accumulation of DSBs during long-term coculture, which is expected to lead to cell death (62), the majority of IECs were able to overcome DSBs and survive (~40% apoptosis was detected). Increased survival and significantly elevated aneuploidy in PMN-MP–treated IECs implies an induction of genomic instability, with potential consequent alterations in cell-cycle regulatory/tumor suppressor genes (such as APC and TP53) and carcinogenesis.
We used patient-derived organoids/colonoids to examine whether a single inflammatory episode was sufficient to promote genomic instability. Acute PMN-MP treatment (24 hours) was sufficient to promote accumulation of DSBs (similar to observations in immortalized cell lines) and formation of micronuclei, indicating an induction of genomic instability (50). However, in this setup we also detected upregulation of senescence marker CDKN2A/p16 and a significant decrease in replication factors Ki67 and PCNA, indicating replicative senescence. As replicative senescence induced by genomic instability serves to prevent genomically instable cells to reenter replicative cycle and become transformed (63, 64), in an acute setting this is likely to limit carcinogenesis. These findings imply that recurring PMN-induced tissue injury as seen in IBD and not a single inflammatory episode promotes tumorigenesis.
Finally, we used specific miR-23a and miR-155 mimics and ASOs in cultured IECs and in acute colonic injury model to confirm the synergistic role of miR-23a and miR-155 in mediating accumulation of DSBs. Since miRNAs have multiple downstream targets (65), to avoid systemic effects, miRNA antagonists were successfully administered by localized endoscopy-based technique to the colonic mucosa, producing robust and specific effects. miR-23a and miR-155 ASOs significantly attenuated PMN-induced accumulation of DSBs and enhanced tissue healing. Specific inhibition of miR-23a and miR-155 further prevented long-term PMN-MP–induced genomic instability. To our knowledge, it has not yet been established whether immunosuppressive therapy (primarily used to treat IBD) reduces DSBs in IBD or reduces chances of neoplasia. However, our data clearly indicate that specific inhibition of PMN-miRNA attenuated inflammation and decreased DSB accumulation to promote mucosal healing and prevent the induction of genomic instability. Thus, our work defines PMN miR-23a and miR-155 as new prognostic and therapeutic targets to promote mucosal healing and prevent progression to cancer.
Methods
Generation of PMN-MPs.
Human PMN-MPs were prepared from the supernatants of stimulated PMNs as follows. Freshly isolated PMNs (2 × 107 cells) were stimulated with either fMLF (1 μM), TNF-α (10 nM), or IFN-γ (50 nM) in 200 μl HBSS+ (20 minutes, 37°C). After stimulation, supernatants were cleared of cells and cellular debris by 2-stage centrifugation (400g followed by 3000g spins, 10 minutes) and subjected to ultracentrifugation at 100,000g. Pelleted MPs were washed and prepared for miRNA analysis, transmission electron microscopy, and functional assays. Murine PMN-MPs were isolated from bone marrow–derived PMNs that were enriched to 85%–90% purity using histopaque gradients (1077 and 1119; Sigma-Aldrich) and the centrifugation methods as described above. As determined by a particle analyzer (LSR Fortessa Special Order Research Product), the isolated PMN-MPs consisted of a mixed population of exosomes (~50%) and other larger particles.
Human specimens.
Colonic biopsies were obtained with informed consent from healthy patients with IBD during routine screening procedures with approval by and in accordance with Northwestern University and Mayo Clinic IRB protocols. Generation of human 3D colonoids and processing of human tissues is detailed in the Supplemental Materials.
Animals.
C57BL6J mice (Jackson Laboratories) aged 8–14 weeks were used in all experimental procedures. All mice were maintained under specific pathogen-free conditions at the Northwestern University, Feinberg School of Medicine animal facilities. At the end of all experimental procedures, animals were killed via rapid cervical dislocation.
Biopsy-based in vivo injury model.
For in vivo assays, a biopsy-based acute colonic injury model was used as previously described (46). Briefly, 2–3 superficial wounds along the dorsal surface of the colon were generated by biopsy forceps in anesthetized (ketamine 100 mg/kg and xylazine 5 mg/kg) mice. Wound healing was monitored by a high-resolution endoscope (COLOVIEW Veterinary Endoscope, Karl Storz) and quantified on days 1 and 4 after wounding. When indicated, wound tissue was extracted for molecular and histological analysis. To assess PMN contribution to tissue injury, circulating and tissue PMNs were depleted using Ly6G antibody (200 μg i.p., 24 hours prior to and 24 hours after injury). PMN depletion (~98%) was confirmed by flow cytometric analysis of the extracted tissue (Supplemental Figure 3B). To assess miRNA contribution to mucosal healing, scrambled-sequence and specific miR-23a and miR-155 ASOs (1.5 nM) were injected directly into the wound area 24 hours after wounding using an endoscopy-guided microinjection system.
Dextran sulfate sodium colitis model.
Mice were allowed free access to food and drinking water containing 3% (wt/vol) DSS for 7 days. Daily clinical assessment and histological analysis were performed as previously described (45, 66). For recovery experiments, DSS was withdrawn at day 5 and replaced with regular water. Mice were allowed to recover for an additional 7 days.
Statistical analysis.
Two-tailed Student’s t test and 1-way ANOVA were used for statistical analyses. All results are the mean ± standard deviation from at least 3 separate experiments. Significance was set at a P value of less than 0.05.
Study approval.
All human studies were reviewed and approved by the Northwestern University and Mayo Clinic IRB. All animal studies were reviewed and approved by the Institutional Animal Care and Use Committee at Northwestern University (PHS assurance number A328301).
Author contributions
VBI and RS conceived and designed the experiments. VBI, HLW, TMB, LM, LCM, KRK, JJL, and RS performed the experiments. VBI, RS, TMB, and LM analyzed the data. VBI, LW, SBH, AB, SAA, RDG, and RS contributed reagents, materials, or analysis tools. VBI and RS wrote the paper.
Supplementary Material
Acknowledgments
This work was supported by grants from the National Institutes of Health (DK101675), Digestive Health Foundation, Chicago, a Research Scholar Grant by the American Cancer Society (RSG-17-235-01-CSM), and by the Robert H. Lurie Comprehensive Cancer Center (Eisenberg Scholar grant). We thank the Northwestern University Center for Advanced Microscopy core (supported by a National Cancer Institute Cancer Center Support Grant P30 CA060553, awarded to the Robert H. Lurie Comprehensive Cancer Center) for help with the imaging experiments. The colonoids/organoid work was supported by the Mayo Clinic Center for Cell Signaling in Gastroenterology (National Institute of Diabetes and Digestive and Kidney Diseases, P30DK084567) and Lisa Boardman (clinical core).
Version 1. 01/14/2019
Electronic publication
Version 2. 02/01/2019
Print issue publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
License: Copyright 2019, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2019;129(2):712–726.https://doi.org/10.1172/JCI122085.
See the related Commentary at Neutrophil-derived microRNAs put the (DNA) breaks on intestinal mucosal healing.
Contributor Information
Veronika Butin-Israeli, Email: v-butin-israeli@northwestern.edu.
Lorraine Mascarenhas, Email: Lmasca25@gmail.com.
Joseph J. Lee, Email: JosephLee2018@u.northwestern.edu.
Lindsey C. Mehl, Email: lmehl94@gmail.com.
Kaitlyn R. Knutson, Email: Hawkins.Kaitlyn@mayo.edu.
Stephen A. Adam, Email: s-adam@northwestern.edu.
Arthur Beyder, Email: beyder.arthur@mayo.edu.
Ronen Sumagin, Email: ronen.sumagin@northwestern.edu.
References
- 1.Ma TY. Intestinal epithelial barrier dysfunction in Crohn’s disease. Proc Soc Exp Biol Med. 1997;214(4):318–327. doi: 10.3181/00379727-214-44099. [DOI] [PubMed] [Google Scholar]
- 2.Maloy KJ, Powrie F. Intestinal homeostasis and its breakdown in inflammatory bowel disease. Nature. 2011;474(7351):298–306. doi: 10.1038/nature10208. [DOI] [PubMed] [Google Scholar]
- 3.Xavier RJ, Podolsky DK. Unravelling the pathogenesis of inflammatory bowel disease. Nature. 2007;448(7152):427–434. doi: 10.1038/nature06005. [DOI] [PubMed] [Google Scholar]
- 4.Brazil JC, Louis NA, Parkos CA. The role of polymorphonuclear leukocyte trafficking in the perpetuation of inflammation during inflammatory bowel disease. Inflamm Bowel Dis. 2013;19(7):1556–1565. doi: 10.1097/MIB.0b013e318281f54e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kolaczkowska E, Kubes P. Neutrophil recruitment and function in health and inflammation. Nat Rev Immunol. 2013;13(3):159–175. doi: 10.1038/nri3399. [DOI] [PubMed] [Google Scholar]
- 6.Knaapen AM, Güngör N, Schins RP, Borm PJ, Van Schooten FJ. Neutrophils and respiratory tract DNA damage and mutagenesis: a review. Mutagenesis. 2006;21(4):225–236. doi: 10.1093/mutage/gel032. [DOI] [PubMed] [Google Scholar]
- 7.Holmström KM, Finkel T. Cellular mechanisms and physiological consequences of redox-dependent signalling. Nat Rev Mol Cell Biol. 2014;15(6):411–421. doi: 10.1038/nrm3801. [DOI] [PubMed] [Google Scholar]
- 8.Bani D, Bencini A. Developing ROS scavenging agents for pharmacological purposes: recent advances in design of manganese-based complexes with anti-inflammatory and anti-nociceptive activity. Curr Med Chem. 2012;19(26):4431–4444. doi: 10.2174/092986712803251476. [DOI] [PubMed] [Google Scholar]
- 9.Cadet J, Wagner JR. DNA base damage by reactive oxygen species, oxidizing agents, and UV radiation. Cold Spring Harb Perspect Biol. 2013;5(2):a012559. doi: 10.1101/cshperspect.a012559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.David SS, O’Shea VL, Kundu S. Base-excision repair of oxidative DNA damage. Nature. 2007;447(7147):941–950. doi: 10.1038/nature05978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maynard S, Schurman SH, Harboe C, de Souza-Pinto NC, Bohr VA. Base excision repair of oxidative DNA damage and association with cancer and aging. Carcinogenesis. 2009;30(1):2–10. doi: 10.1093/carcin/bgn250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Neelsen KJ, Lopes M. Replication fork reversal in eukaryotes: from dead end to dynamic response. Nat Rev Mol Cell Biol. 2015;16(4):207–220. doi: 10.1038/nrm3935. [DOI] [PubMed] [Google Scholar]
- 13.Ceccaldi R, Rondinelli B, D’Andrea AD. Repair pathway choices and consequences at the double-strand break. Trends Cell Biol. 2016;26(1):52–64. doi: 10.1016/j.tcb.2015.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Petermann E, Helleday T. Pathways of mammalian replication fork restart. Nat Rev Mol Cell Biol. 2010;11(10):683–687. doi: 10.1038/nrm2974. [DOI] [PubMed] [Google Scholar]
- 15.Nagaria P, Robert C, Rassool FV. DNA double-strand break response in stem cells: mechanisms to maintain genomic integrity. Biochim Biophys Acta. 2013;1830(2):2345–2353. doi: 10.1016/j.bbagen.2012.09.001. [DOI] [PubMed] [Google Scholar]
- 16.Lasry A, Zinger A, Ben-Neriah Y. Inflammatory networks underlying colorectal cancer. Nat Immunol. 2016;17(3):230–240. doi: 10.1038/ni.3384. [DOI] [PubMed] [Google Scholar]
- 17.Sturm A, Dignass AU. Epithelial restitution and wound healing in inflammatory bowel disease. World J Gastroenterol. 2008;14(3):348–353. doi: 10.3748/wjg.14.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sumagin R, Robin AZ, Nusrat A, Parkos CA. Activation of PKCβII by PMA facilitates enhanced epithelial wound repair through increased cell spreading and migration. PLoS ONE. 2013;8(2):e55775. doi: 10.1371/journal.pone.0055775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Butin-Israeli V, et al. Deposition of microparticles by neutrophils onto inflamed epithelium: a new mechanism to disrupt epithelial intercellular adhesions and promote transepithelial migration. FASEB J. 2016;30(12):4007–4020. doi: 10.1096/fj.201600734R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dalli J, et al. Heterogeneity in neutrophil microparticles reveals distinct proteome and functional properties. Mol Cell Proteomics. 2013;12(8):2205–2219. doi: 10.1074/mcp.M113.028589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Buzas EI, György B, Nagy G, Falus A, Gay S. Emerging role of extracellular vesicles in inflammatory diseases. Nat Rev Rheumatol. 2014;10(6):356–364. doi: 10.1038/nrrheum.2014.19. [DOI] [PubMed] [Google Scholar]
- 22.Finkielsztein A, Mascarenhas L, Butin-Israeli V, Sumagin R. Isolation and characterization of neutrophil-derived microparticles for functional studies. J Vis Exp. 2018;(133):e56949. doi: 10.3791/56949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Robbins PD, Morelli AE. Regulation of immune responses by extracellular vesicles. Nat Rev Immunol. 2014;14(3):195–208. doi: 10.1038/nri3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol. 2014;15(8):509–524. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- 25.Chowdhury D, Choi YE, Brault ME. Charity begins at home: non-coding RNA functions in DNA repair. Nat Rev Mol Cell Biol. 2013;14(3):181–189. doi: 10.1038/nrm3523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Butin-Israeli V, et al. Role of lamin b1 in chromatin instability. Mol Cell Biol. 2015;35(5):884–898. doi: 10.1128/MCB.01145-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Muñoz-Espín D, Serrano M. Cellular senescence: from physiology to pathology. Nat Rev Mol Cell Biol. 2014;15(7):482–496. doi: 10.1038/nrm3823. [DOI] [PubMed] [Google Scholar]
- 28.Shimi T, et al. The role of nuclear lamin B1 in cell proliferation and senescence. Genes Dev. 2011;25(24):2579–2593. doi: 10.1101/gad.179515.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Branzei D, Foiani M. The DNA damage response during DNA replication. Curr Opin Cell Biol. 2005;17(6):568–575. doi: 10.1016/j.ceb.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 30.Branzei D, Foiani M. Regulation of DNA repair throughout the cell cycle. Nat Rev Mol Cell Biol. 2008;9(4):297–308. doi: 10.1038/nrm2351. [DOI] [PubMed] [Google Scholar]
- 31.Akyüz N, et al. DNA substrate dependence of p53-mediated regulation of double-strand break repair. Mol Cell Biol. 2002;22(17):6306–6317. doi: 10.1128/MCB.22.17.6306-6317.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kunkel TA, Erie DA. Eukaryotic mismatch repair in relation to DNA replication. Annu Rev Genet. 2015;49:291–313. doi: 10.1146/annurev-genet-112414-054722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mojas N, Lopes M, Jiricny J. Mismatch repair-dependent processing of methylation damage gives rise to persistent single-stranded gaps in newly replicated DNA. Genes Dev. 2007;21(24):3342–3355. doi: 10.1101/gad.455407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peng M, Xie J, Ucher A, Stavnezer J, Cantor SB. Crosstalk between BRCA-Fanconi anemia and mismatch repair pathways prevents MSH2-dependent aberrant DNA damage responses. EMBO J. 2014;33(15):1698–1712. doi: 10.15252/embj.201387530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rao L, Perez D, White E. Lamin proteolysis facilitates nuclear events during apoptosis. J Cell Biol. 1996;135(6 Pt 1):1441–1455. doi: 10.1083/jcb.135.6.1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang Y, et al. Role for caspase-mediated cleavage of Rad51 in induction of apoptosis by DNA damage. Mol Cell Biol. 1999;19(4):2986–2997. doi: 10.1128/MCB.19.4.2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dou Z, et al. Autophagy mediates degradation of nuclear lamina. Nature. 2015;527(7576):105–109. doi: 10.1038/nature15548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wan G, Mathur R, Hu X, Zhang X, Lu X. miRNA response to DNA damage. Trends Biochem Sci. 2011;36(9):478–484. doi: 10.1016/j.tibs.2011.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.O’Connell RM, Rao DS, Chaudhuri AA, Baltimore D. Physiological and pathological roles for microRNAs in the immune system. Nat Rev Immunol. 2010;10(2):111–122. doi: 10.1038/nri2708. [DOI] [PubMed] [Google Scholar]
- 40.Dreesen O, et al. Lamin B1 fluctuations have differential effects on cellular proliferation and senescence. J Cell Biol. 2013;200(5):605–617. doi: 10.1083/jcb.201206121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gasparini P, et al. Protective role of miR-155 in breast cancer through RAD51 targeting impairs homologous recombination after irradiation. Proc Natl Acad Sci U S A. 2014;111(12):4536–4541. doi: 10.1073/pnas.1402604111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang JW, et al. Systematic screen identifies miRNAs that target RAD51 and RAD51D to enhance chemosensitivity. Mol Cancer Res. 2013;11(12):1564–1573. doi: 10.1158/1541-7786.MCR-13-0292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lin S, Gregory RI. MicroRNA biogenesis pathways in cancer. Nat Rev Cancer. 2015;15(6):321–333. doi: 10.1038/nrc3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Neudecker V, et al. Neutrophil transfer of miR-223 to lung epithelial cells dampens acute lung injury in mice. Sci Transl Med. 2017;9(408):eaah5360. doi: 10.1126/scitranslmed.aah5360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chassaing B, Aitken JD, Malleshappa M, Vijay-Kumar M. Dextran sulfate sodium (DSS)-induced colitis in mice. Curr Protoc Immunol. 2014;104:Unit 15.25. doi: 10.1002/0471142735.im1525s104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Slater TW, Finkielsztein A, Mascarenhas LA, Mehl LC, Butin-Israeli V, Sumagin R. Neutrophil microparticles deliver active myeloperoxidase to injured mucosa to inhibit epithelial wound healing. J Immunol. 2017;198(7):2886–2897. doi: 10.4049/jimmunol.1601810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.White RR, et al. Controlled induction of DNA double-strand breaks in the mouse liver induces features of tissue ageing. Nat Commun. 2015;6:6790. doi: 10.1038/ncomms7790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nakad R, Schumacher B. DNA damage response and immune defense: links and mechanisms. Front Genet. 2016;7:147. doi: 10.3389/fgene.2016.00147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gordon DJ, Resio B, Pellman D. Causes and consequences of aneuploidy in cancer. Nat Rev Genet. 2012;13(3):189–203. doi: 10.1038/nrg3123. [DOI] [PubMed] [Google Scholar]
- 50.Pfau SJ, Amon A. Chromosomal instability and aneuploidy in cancer: from yeast to man. EMBO Rep. 2012;13(6):515–527. doi: 10.1038/embor.2012.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.López-Posadas R, Neurath MF, Atreya I. Molecular pathways driving disease-specific alterations of intestinal epithelial cells. Cell Mol Life Sci. 2017;74(5):803–826. doi: 10.1007/s00018-016-2363-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Romano M, et al. From inflammation to cancer in inflammatory bowel disease: molecular perspectives. Anticancer Res. 2016;36(4):1447–1460. [PubMed] [Google Scholar]
- 53.Stewart C, Burke B. Teratocarcinoma stem cells and early mouse embryos contain only a single major lamin polypeptide closely resembling lamin B. Cell. 1987;51(3):383–392. doi: 10.1016/0092-8674(87)90634-9. [DOI] [PubMed] [Google Scholar]
- 54.Butin-Israeli V, Adam SA, Goldman AE, Goldman RD. Nuclear lamin functions and disease. Trends Genet. 2012;28(9):464–471. doi: 10.1016/j.tig.2012.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Butin-Israeli V, Adam SA, Goldman RD. Regulation of nucleotide excision repair by nuclear lamin b1. PLoS ONE. 2013;8(7):e69169. doi: 10.1371/journal.pone.0069169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yamamoto H, et al. Interrelationship between microsatellite instability and microRNA in gastrointestinal cancer. World J Gastroenterol. 2012;18(22):2745–2755. doi: 10.3748/wjg.v18.i22.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bui TM, Mascarenhas LA, Sumagin R. Extracellular vesicles regulate immune responses and cellular function in intestinal inflammation and repair. Tissue Barriers. 2018;6(2):e1431038. doi: 10.1080/21688370.2018.1431038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Johnson BL, Kuethe JW, Caldwell CC. Neutrophil derived microvesicles: emerging role of a key mediator to the immune response. Endocr Metab Immune Disord Drug Targets. 2014;14(3):210–217. doi: 10.2174/1871530314666140722083717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pálmai-Pallag T, Bachrati CZ. Inflammation-induced DNA damage and damage-induced inflammation: a vicious cycle. Microbes Infect. 2014;16(10):822–832. doi: 10.1016/j.micinf.2014.10.001. [DOI] [PubMed] [Google Scholar]
- 60.Negrini S, Gorgoulis VG, Halazonetis TD. Genomic instability--an evolving hallmark of cancer. Nat Rev Mol Cell Biol. 2010;11(3):220–228. doi: 10.1038/nrm2858. [DOI] [PubMed] [Google Scholar]
- 61.Crusz SM, Balkwill FR. Inflammation and cancer: advances and new agents. Nat Rev Clin Oncol. 2015;12(10):584–596. doi: 10.1038/nrclinonc.2015.105. [DOI] [PubMed] [Google Scholar]
- 62.Roos WP, Thomas AD, Kaina B. DNA damage and the balance between survival and death in cancer biology. Nat Rev Cancer. 2016;16(1):20–33. doi: 10.1038/nrc.2015.2. [DOI] [PubMed] [Google Scholar]
- 63.Childs BG, Durik M, Baker DJ, van Deursen JM. Cellular senescence in aging and age-related disease: from mechanisms to therapy. Nat Med. 2015;21(12):1424–1435. doi: 10.1038/nm.4000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lecot P, Alimirah F, Desprez PY, Campisi J, Wiley C. Context-dependent effects of cellular senescence in cancer development. Br J Cancer. 2016;114(11):1180–1184. doi: 10.1038/bjc.2016.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bracken CP, Scott HS, Goodall GJ. A network-biology perspective of microRNA function and dysfunction in cancer. Nat Rev Genet. 2016;17(12):719–732. doi: 10.1038/nrg.2016.134. [DOI] [PubMed] [Google Scholar]
- 66.Leoni G, et al. Annexin A1-containing extracellular vesicles and polymeric nanoparticles promote epithelial wound repair. J Clin Invest. 2015;125(3):1215–1227. doi: 10.1172/JCI76693. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.